Abstract
Gastroenteritis is inflammation of the lining of stomach and intestines and causes significant morbidity and mortality worldwide. Many viruses, especially RNA viruses are the most common cause of enteritis. Innate immunity is the first line of host defense against enteric RNA viruses and virus-induced intestinal inflammation. The first layer of defense against enteric RNA viruses in the intestinal tract is intestinal epithelial cells (IECs), dendritic cells and macrophages under the intestinal epithelium. These innate immune cells express pathogen-recognition receptors (PRRs) for recognizing enteric RNA viruses through sensing viral pathogen-associated molecular patterns (PAMPs). As a result of this recognition type I interferon (IFN), type III IFN and inflammasome activation occurs, which function cooperatively to clear infection and reduce viral-induced intestinal inflammation. In this review, we summarize recent findings about mechanisms involved in enteric RNA virus-induced intestinal inflammation. We will provide an overview of the enteric RNA viruses, their RNA sensing mechanisms by host PRRs, and signaling pathways triggered by host PRRs, which shape the intestinal immune response to maintain intestinal homeostasis.
Keywords: SARS-CoV-2, Diarrhea, Enterocyte, RNA helicase, Cytokine, Pyroptosis, PARP9, Caspase
Introduction
Gastroenteritis is an inflammatory condition of the stomach and intestines that affects people of all ages, with severe complications in young children and elderly people most vulnerable to dehydration [1]. Gastroenteritis causes significant morbidity and mortality worldwide and represents one of major socioeconomic burdens. In children, diarrheal diseases induced by gastroenteritis accounts for 9% of child deaths, making diarrhea the second leading cause of death globally [2]. Since identification in 1972 [3], enteric viruses have been recognized as a leading cause of gastroenteritis worldwide [1]. Enteric viruses are primarily transmitted through the fecal–oral route by contact with the feces. Fecal shedding of enteric viruses can last for months after resolution of gastroenteritis symptoms in human [4–6]. Symptoms during viral gastroenteritis include watery diarrhea, abnormal cramps, nausea or vomiting, and sometimes fever [1]. Viral gastroenteritis pathologies normally are self-limiting and generally resolve within 14 days, but they can turn life-threatening for immunocompromised patients and for infants in developing countries [7].
Many enteric viruses cause gastroenteritis in humans which are mainly RNA viruses [1]. The most common enteric RNA viruses include rotavirus, reovirus, norovirus, enterovirus, astrovirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and torovirus. Both rotavirus and reovirus are double-stranded (ds) RNA virus and are classified in Reoviridae family. Rotavirus infection occurs in small intestinal epithelial cells (IECs) causing intestinal disease mainly in children and has symptoms including vomiting, diarrhea and low-grade fever [8]. Reovirus infection normally does not provoke pathology in humans, whereas reovirus infection can trigger inflammatory responses to cause intestinal inflammation and pathology in mice [9, 10], which makes reovirus a good model virus for investigating pathogenic mechanisms underlying host antiviral immune responses to enteric RNA viruses. Norovirus is single-stranded (ss) RNA virus of the Caliciviridae family and is the most common cause of food-borne gastroenteritis with symptoms of severe vomiting and diarrhea in adults [11]. Globally, norovirus infection causes about 18% of acute gastroenteritis cases and appropriately 200,000 deaths annually [12]. Enteroviruses are ss RNA viruses in Picornaviridae family and include poliovirus, coxsackieviruses, echoviruses and enterovirus 71. Enteroviruses are spread primarily through the fecal–oral route and target the intestinal epithelium to cause gastrointestinal illness such as severe vomiting or diarrhea [13]. Astrovirus is ss RNA virus in Astroviridae family and is recognized as second most common instigator of viral gastroenteritis in children [14]. SARS-CoV-2 and torovirus are ss RNA viruses of the Coronaviridae family and SARS-CoV-2 is the causative agent of the ongoing coronavirus disease 2019 (COVID-19) pandemic, which has infected more than 4 million people, spreading to over 200 countries or regions worldwide [6]. Human torovirus was firstly detected in human gastroenteritis in 1984 [15] and has been associated with viral gastroenteritis in human [16]. A notable proportion of patients with COVID-19 develop gastrointestinal symptoms and nearly half of confirmed COVID-19 patients have detectable SARS-CoV-2 RNA in their fecal samples in human [6, 17, 18]. Moreover, multiple studies have provided direct evidence of intestinal infection by SARS-CoV-2 both in vitro [19, 20] and in vivo in mice and other animals [21–23]. Although these lines of evidence highlight the nature of SARS-CoV-2 gastrointestinal infection, the mechanism of diarrhea in patients with COVID-19 is still largely unknown and needs further studies. There are several available vaccines for preventing SARS-CoV-2 infection, however, COVID-19 induced by SARS-CoV-2 remains a severe global public health problem due to emergence of more mutant and virulent SARS-CoV-2 virus strains. The length of effectiveness of the current vaccines remains unknown.
In this review, we focus on how intestinal innate immunity controls enteric RNA virus-induced intestinal inflammation. We summarize the detailed molecular mechanisms involved in sensing of enteric RNA viruses, intestinal innate immunity triggered by enteric RNA viruses, and the effect on regulation of intestinal inflammation.
Antiviral innate immunity in intestine
Innate immune cells in intestine
The first layer of defense against enteric RNA viruses in the intestinal tract is intestinal epithelial cells (IECs). These cells form a layer facing the luminal surface of the intestinal epithelium. IECs contain six cell lineages from a common stem cell progenitor: enterocytes, goblet cells, Paneth cells, enteroendocrine cells, tuft cells and microfold (M) cells [24] (Fig. 1). Enterocytes are the major cell type of IECs with importance in host defense and nutrient absorption [25]. Goblet cells comprise around 10% of all IECs and secrete mucus for promoting intestinal barrier function [25]. Paneth cells are only found in the small intestine. These cells secrete antimicrobial peptides called defensins, which contribute to host defense against microbes including enteric RNA viruses [26]. Enteroendocrine cells produce and release intestinal hormones into the bloodstream after stimulation that generate systemic effects and stimulate nervous responses [27]. Tuft cells are rare chemosensory cells and are targets of norovirus infection [28]. M cells are specialized cells for antigen sampling [29]. Both tuft and M cells play important role in immune surveillance and response to enteric viruses [28, 29]. Although both small intestine and colon have same intestinal layers, they have some differences. Colon has no plicae circularis, villi and Paneth cells, but has more goblet cells. Underneath the intestinal epithelium, the lamina propria contains immunocompetent cells. Macrophages and dendritic cells (DCs) are major innate immune cells responsible for detecting enteric RNA virus infection as well as present viral antigens to T cells in the lamina propria [30]. T cells and B cells form the adaptive immune system in the lamina propria. Here, we focus only on innate immune cells including IECs, macrophages and DCs and discuss their roles in promoting antiviral innate immunity and controlling intestinal inflammation induced by enteric RNA viruses.
Fig. 1.
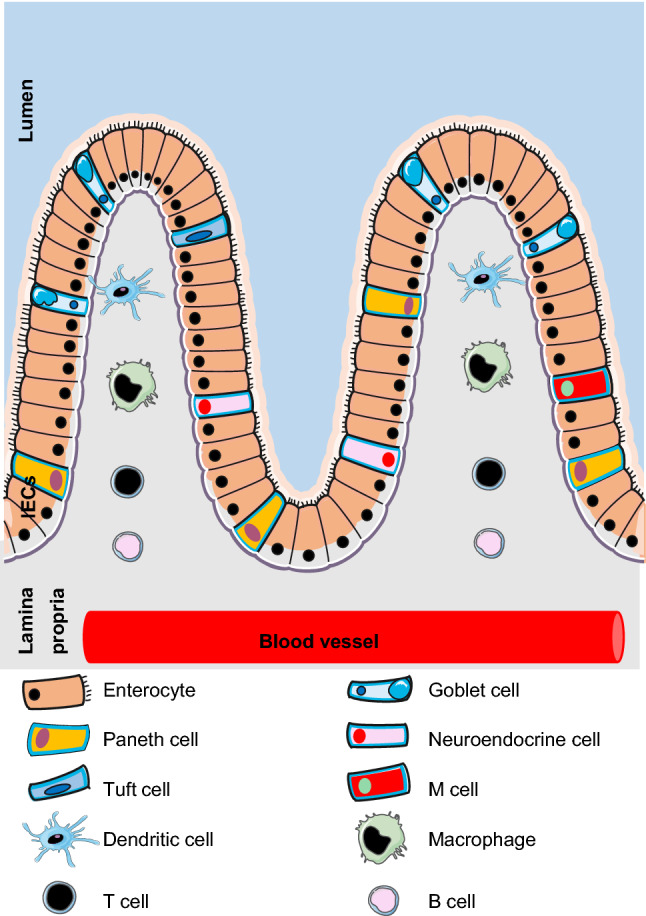
Overview of IECs and immune cells in intestine. The first layer of host defense against enteric RNA viruses in the intestinal tract is intestinal epithelial cells (IECs), which conclude six cell lineages coming from a common stem cell progenitor: enterocytes, goblet cells, Paneth cells, enteroendocrine cells, tuft cells and microfold (M) cells. Underneath the intestinal epithelium, there is the lamina propria containing the immunocompetent cells infiltrated from blood vessel, which include innate immune cells such as dendritic cells (DCs), macrophages and adaptive immune cells such as T cell and B cells
General mechanisms involved in antiviral innate immunity of intestine
Innate immunity is the first line of defense against enteric RNA virus infection in intestine.Intestinal innate immune cells, including IECs, macrophages and DCs, use pathogen-recognition receptors (PRRs) to recognize pathogen-associated molecular patterns (PAMPs) such as viral RNA from enteric RNA viruses [31]. Multiple PRRs have been identified that sense RNA viruses [31], such as some Toll-like receptors (TLRs) recognizing viral RNA in endosome and RNA helicase receptors sensing viral RNA in cytosol. TLR3, TLR7, and TLR8 locate in endosomes and are the major TLRs that detect distinct forms of viral RNA from enteric RNA viruses and initiate antiviral responses [32]. Once sensing viral RNA from enteric RNA viruses, RNA helicase receptors including retinoic acid-inducible gene I (RIG-I) [33, 34], melanoma differentiation-associated gene 5 (MDA5) [35], DEAH-box helicase 9 (DHX9) [36], DHX15 [10, 37, 38], DEAD-box helicase 1 (DDX1)/DDX21/DHX36 complex [39], and DHX33 [40] employ downstream adaptors mitochondrial antiviral-signaling protein (MAVS) [41, 42] or Toll/interleukin-1 (IL-1) receptor (TIR)-domain-containing adaptor-inducing interferon-β (TRIF) to induce type I interferon (IFN) and type III IFN responses [43–45]. Additionally, some RNA helicase receptors recruit downstream adaptors nucleotide-binding oligomerization domain (NOD)-like receptor family pyrin-domain-containing 3 (NLRP3) or NLRP6 to activate the inflammasome response with subsequent release of both IL-1β and IL-18 [10, 40]. Recently, our group identified poly(ADP-ribose) polymerase 9 (PARP9), one of 17 PARP family members, as a noncanonical sensor of RNA viruses resulting in MAVS-independent type I IFN production in mice [46].
The thorough understanding of the mechanisms by which different PRRs recognize enteric RNA viruses for activating the innate immune system will be key to informing the development of putative therapeutic strategies. A classic example of fundamental discoveries translated into an application is when researchers harnessed the ability of TLR agonists (such as TLR9 agonist CpG1018) to initiate the innate immune system to be used as adjuvant candidates to improve the immunogenicity of vaccines [47]. Therefore, agonists or substances that stimulate specific PRRs to recognize enteric RNA viruses and activate strong innate immunity will be developed as promising new vaccine adjuvants or drugs to clinically treat enteric virus-induced diseases.
Type I IFN in antiviral innate immunity
Type I IFN comprises several IFN-α subtypes, IFN-β, IFN-δ, IFN-ε, IFN-κ, IFN-ω, IFN-τ and IFN-ζ [45]. Among the members of type I IFN, IFN-α and IFN-β play crucial roles in limiting viral replication and clearing viral spread. The membrane receptor interferon-α/β receptor (IFNAR) binds secreted IFN-α/β and triggers downstream Janus kinase/signal transducer and activator of transcription (JAK/STAT) signaling cascade resulting in the phosphorylation and heterodimerization of STAT1/2. These STAT heterodimers translocate to the nucleus to drive expression of numerous interferon-stimulated genes (ISGs), which are responsible for clearing enteric RNA viruses by restricting viral replication, inhibiting protein synthesis and priming neighboring cells for a viral attack [48, 49]. Moreover, antiviral type I IFN signaling can induce apoptosis to destroy the replicative niche of enteric RNA viruses [50, 51]. Furthermore, type I IFN induces resistance to virus infections in neighboring cells, mediates killing of enteric RNA virus-infected cells by activating natural killer (NK) cells and cytotoxic T cells (CTLs) and promotes adaptive antiviral immune responses by producing cytokines and chemokines [52, 53]. Although type I IFN plays a critical role in antiviral innate immunity in intestinal macrophages and DCs, it acts in a compartmentalized manner within the small intestine and has a minimal effect on IECs [54].
Type III IFN in antiviral innate immunity
Type III IFN includes IFNλ1, IFNλ2, IFNλ3 and IFNλ4 in humans, whereas only IFNλ2 and IFNλ3 are activated in mice [45]. Type III IFN signal through a heterodimeric receptor complex composed of the IFNλ receptor 1 (IFNλR1) and IL-10Rβ subunit for activating JAK1 and TYK2[55, 56], which lead to the phosphorylation of STAT1/STAT2 and induction of ISGs [57]. Type I IFN receptors are ubiquitously expressed on all nucleated cells, while the expression of type III IFN receptor IFNλR1 subunit is mainly restricted to epithelial cells. Therefore, type III IFN is particularly important in responding to intestinal antiviral responses and to control persistent infections of enteric RNA viruses [10, 58, 59]. In addition to their antiviral activities, both type I IFN and type III IFN protect the intestinal epithelial barrier by inducing the production of tight junction proteins [60]and downregulating intestinal immune responses [61, 62].
Inflammasome in antiviral innate immunity
The inflammasome is a multiprotein complex of the innate immune response that controls caspase-1 activation, which promotes maturation and secretion of the proinflammatory cytokines interleukin 1β (IL-1β) and IL-18, as well as ‘pyroptosis’, a lytic form of cell death [63]. These multiprotein complexes are comprised of three basic components including a sensor such as a NOD-like receptor (NLR), the adaptor protein apoptosis-associated speck-like protein containing a caspase-recruitment domain (ASC) and the intracellular cysteine protease such as caspase-1 [64]. The inflammasomes involved in innate antiviral immunity include those activated through NLRP3 [65, 66], NLRP6 [10, 67] and NLRP9b [68]. The IL-1β, IL-18 and pyroptosis are important for the antiviral functions of inflammasome activation.
Recombinant IL-1β controls viruses in various cell lines [69–72], indicating the crucial role of IL-1β in mediating antiviral functions. Administration of mice with IL-18 before intestinal rotavirus infection diminished subsequent fecal shedding of rotavirus in mice [73], suggesting the critical antiviral role of IL-18 in innate immunity. Pyroptosis is a form of lytic, programmed cell death through cleavage of its substrate Gasdermin D (GSDMD) that forms pores [74–76]. Pyroptosis is an innate immune effector mechanism to facilitate host defense against pathogenic microorganisms, including viruses [77].
Additionally, IL-1β/IL-18 not only functions in the innate immune response containing lymphocyte activation and leukocyte transmigration, but also ensure adaptive immune response by inducing IFN-γ production [78]. Most studies about the antiviral roles of inflammasome activation are investigated in non-enteritic RNA and DNA viruses [79, 80]. There are few reports regarding the antiviral roles of inflammasome in enteric RNA viruses, which need to be further investigated in future.
Therefore, type I IFN, type III IFN and inflammasome responses play multiple roles in antiviral innate immunity against viruses including enteric RNA viruses. Below, we will discuss detailed mechanisms about how several classes of PRRs recognize viral RNA from enteric RNA viruses and induce activation of type I IFN, type III IFN and inflammasome responses in intestine, which normally leads to successful elimination of enteric RNA viruses from intestine.
Role of type I IFN pathway mediated by PRRs sensing enteric RNA viruses
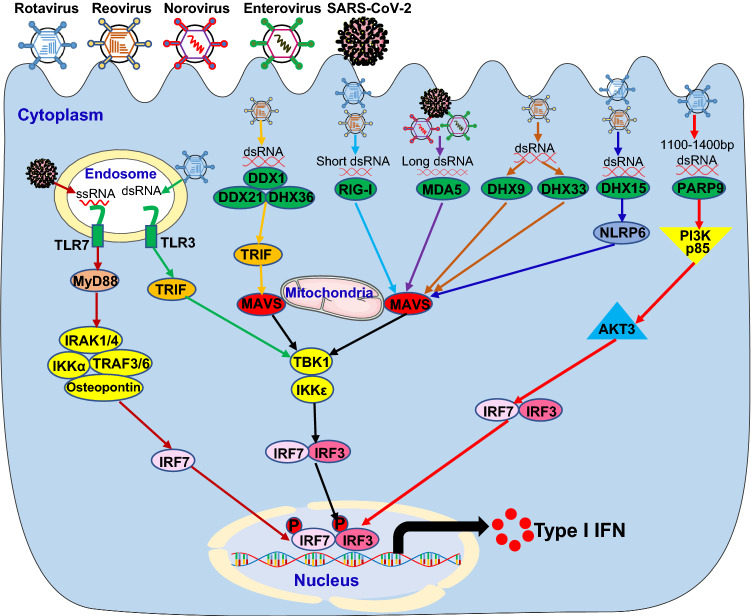
A number of PRRs sense enteric RNA viruses for inducing type I IFN signaling pathway in IECs, DCs and macrophages, introduced below in detail (Fig. 2).
Fig. 2.
Host PRRs sense enteric RNA viruses to induce type I IFN signaling pathway. Invasion by enteric RNA viruses, including rotavirus, reovirus, norovirus, enterovirus and SARS-CoV-2, introduces RNA into the endosome or dsRNA in the cytoplasm. The host pattern recognition receptors (PRRs), including TLR3 and TLR7 in the endosome and DDX1/DDX21/DHX36 complex, RIG-I, MDA5, DHX9, DHX33, DHX15 and PARP9 in the cytoplasm, recognize the RNA molecules and trigger the activation of downstream cascades through their adaptors leading to the induction of type I IFN in innate immune cells of the intestinal tract. MyD88 Myeloid differentiation primary response 88, TRIF TIR-domain-containing adapter-inducing interferon-β, IRAK1/4 Interleukin 1 receptor-associated kinase 1 and 4, IKKα IκB kinase α, TRAF3/6 TNF receptor-associated factor 3 and 6, IRF7 Interferon regulatory factor 7, DDX1 DEAD-Box helicase 1, RIG-I Retinoic acid-inducible gene I, MDA5 Melanoma differentiation-associated protein 5, DHX15 DEAH-Box helicase 15, PARP9 Poly (ADP-ribose) polymerase 9, MAVS Mitochondrial antiviral-signaling protein, TBK1 TANK-binding kinase 1, NLRP6 the NACHT, LRR, and PYD domains-containing protein 6, PI3K p85 Phosphatidylinositol 3-kinase (PI3K) regulatory subunit p85, AKT3 AKT Serine/Threonine kinase 3
Toll-like receptors
Toll-like receptors (TLRs) are a class of transmembrane PRRs containing three structural domains: a leucine-rich repeats (LRRs) motif, a transmembrane domain, and a cytoplasmic Toll/IL-1 receptor (TIR) domain [81]. The LRRs motif is responsible for recognizing pathogens including viruses, while the TIR domain is responsible for initiating downstream signaling[82]. The TLR family contains 10 TLRs (TLR1-10) in humans and 13 TLRs (TLR1-13) in mice. TLR3 recognizes viral double-strand RNA (dsRNA), TLR7 and TLR8 detect viral ssRNA. After binding ligand, TLR3 leads to the recruitment of adaptor protein TRIF, while TLR7 and TLR8 result in the interactions with downstream adaptor MyD88 [82]. TRIF recruits downstream tumor necrosis factor receptor-associated factor 3 (TRAF3), which promotes TANK-binding kinase 1 (TBK1) and IκB kinase ε (IKKε)-mediated activation of IRF3 leading to IFN-β production [77, 78]. However, the activation of TLR7 and TLR8 results in recruitment of downstream adaptors MyD88 and interleukin-1 receptor-associated kinases 1/2/4 (IRAK1/2/4) and subsequent complex formation with TRAF3, TRAF6, IRAK1, IKKα, osteopontin and IRF7, which phosphorylate and activate IRF7 leading to massive production of type I IFN in DCs [83–85].
The first report about the role of TLRs in enteric RNA virus infection is a study regarding host responses to rotavirus infection. Injection of mice with dsRNA from rotavirus induced small intestinal inflammation dependent on activation of the endosomal dsRNA sensor TLR3 [86]. Later, TLR3 knockout mice were found to be highly susceptible to rotavirus infection as adults but not in the neonatal stage in mice [87]. Previous studies using mouse coronavirus showed that TLR7 and MyD88 pathways mediated coronaviral RNA sensing and type I IFN induction in pDCs [88]. TLR7 was subsequently shown to be the primary sensor of MERS-CoV RNA for type I IFN induction in epithelial cells in mice [89].
RIG-I-like receptors
The RIG-I-like receptors (RLRs) are a family of DExD/H box containing RNA helicases and include three family members: RIG-I [33, 34], MDA5 [35] and laboratory of genetics and physiology 2 (LGP2) [90]. RIG-I recognizes short dsRNA or 5’-triphosphate RNA from enteric RNA viruses, whereas MDA5 senses viral dsRNA from enteroviruses in Picornaviridae family and synthetic long dsRNA of more than 2 kb in length [91–94]. Binding of the viral RNA ligands to RIG-I and MDA5 results in engagement of MAVS with large prion-like polymers formation, which is anchored to the outer membrane of mitochondria. MAVS subsequently recruits TBK1 and IκB kinase ε (IKKε) to phosphorylate and activate IRF3 leading to production of type I IFN [42].
In the context of enteric RNA viruses sensed by RLRs, RIG-I and MDA5 are important in restricting rotavirus infection in IECs by inducing IFN-β production [95]. Moreover, knockout of both RIG-I and MDA5 prevented the induction of type I IFN after reovirus infection of conventional DCs [96, 97], suggesting that the type I IFN pathway mediated by RLRs is vital to control reovirus infection. In addition, the production of type I IFN was dependent on MDA5 by DCs since replication of norovirus was controlled by MDA5 in the intestine and lymphoid organs after peroral infection in mice [98]. Furthermore, MDA5 detect viral long dsRNA from enteroviruses, such as coxsackie B virus 3 (CVB3), to induce type I IFN production for controlling viral infection [99–101]. Similarly, MDA5 recognizes viral RNA from coronaviruses critical for host defense against coronavirus infection [102–104].
Other RNA helicases
In addition to RLRs, several other RNA helicases directly detect viral RNA from enteric RNA viruses and initiate antiviral type I IFN response [105]. The RNA helicases DDX1-DDX21- DHX36 complex were shown by our group to recognize reovirus dsRNA using the helicase domain of DDX1, meanwhile DDX21 and DHX36 interact with downstream adaptor TRIF to recruit adaptors TBK1 and IKKε complex for inducing IRF3 activation leading to final type I IFN production in DCs [39]. Our group identified RNA helicases DHX9 and DHX33 as RNA sensors of reovirus to initiate MAVS mediated type I IFN signaling pathway in myeloid DCs [36, 106]. Later, another RNA helicase DHX15 was reported by our group to sense reovirus dsRNA for inducing MAVS-dependent type I IFN production in DCs [37] and IECs and thereby protect mice from intestinal inflammation induced by infection with enteric RNA viruses including reovirus and rotavirus in mice [10].
Noncanonical RNA sensor PARP9
Recently, our group identified poly(ADP-ribose) polymerase 9 (PARP9) as a noncanonical sensor for RNA viruses in DCs and found PARP9 deficient mice had enhanced susceptibility to infections with enteric reovirus because of impaired type I IFN production in mice [46]. Mechanistically, PARP9 recognizes and binds viral dsRNA between 1100 and 1400 bp from reovirus, with resultant recruitment and activation of the downstream phosphoinositide 3-kinase (PI3K) and AKT3 pathway, independent of MAVS. PI3K/AKT3 then activated the IRF3 and IRF7 by phosphorylating IRF3 at Ser385 and IRF7 at Ser437/438 leading to type I IFN production [46].
Role of type III IFN pathway mediated by PRRs sensing enteric RNA viruses
Type III IFN is particularly important in responding to intestinal antiviral responses and controlling virus-induced intestinal inflammation in IECs of the intestinal tract. Although RIG-I and MDA5 have been proposed to induce type III IFN in response to some enteric RNA viruses in DCs, only RNA helicase DHX15 has been identified to sense enteric RNA viruses for inducing type III IFN signaling pathway in IECs both in vitro and in vivo, which is discussed below in detail (Fig. 3).
Fig. 3.
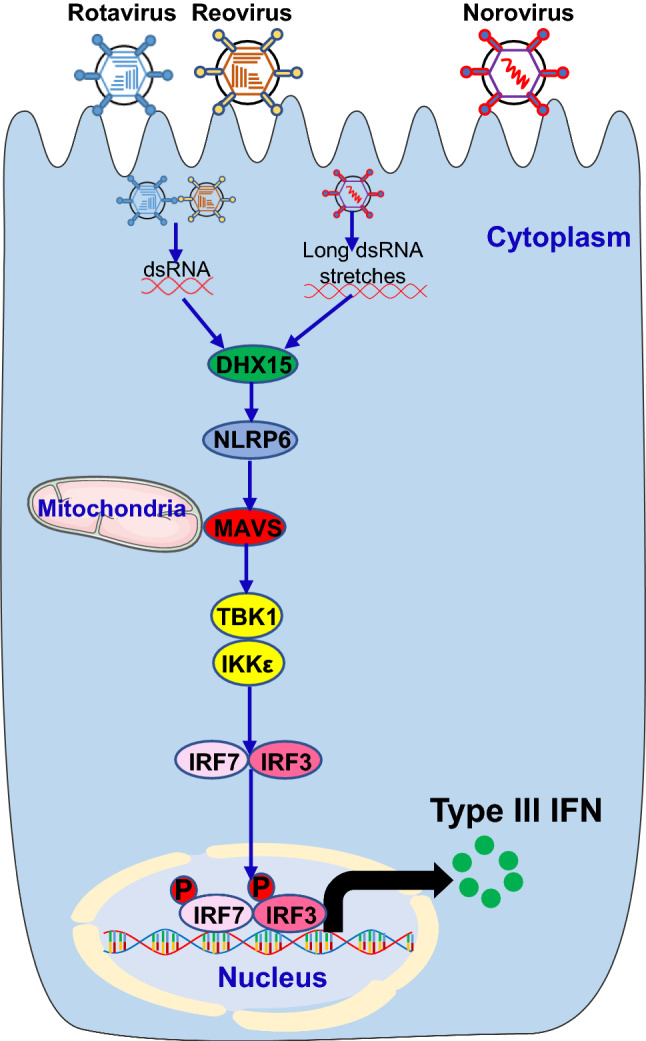
The RNA helicase RNA sensor DHX15 sense enteric RNA viruses to trigger type III IFN signaling pathway. Infection by enteric RNA viruses, including rotavirus, reovirus and norovirus, releases dsRNA in the cytoplasm. The RNA helicase RNA sensor DHX15 in the cytoplasm recognizes the RNA molecules and trigger the activation of downstream cascades through their adaptors leading to the induction of type III IFN in IECs of the intestinal tract
RNA helicase DHX15
IECs are the predominant type III IFN producing and responsive cell type in the intestinal tract [54]. Several groups have reported antiviral effects of type III IFN IFN-λ against rotavirus [54, 107, 108], reovirus [10, 107] and norovirus [109–111] in intestine both in vitro and in vivo. MDA5 is required for type I IFN production by IECs during norovirus infection in mice [98]. The RNA helicase DHX15 induces type III IFN production using a similar pathway as in producing type I IFN [10, 37]. Recently, DHX15 was reported by our group to sense long dsRNA of reovirus for inducing MAVS-dependent type III IFN production in IECs and protect mice from intestinal inflammation induced by infection with enteric RNA viruses including reovirus and rotavirus in mice [10]. DHX15 operates independently of RIG-I and MDA5 to recruit NLRP6 and MAVS complex followed by TBK1/IKKε mediated IRF3/7 activation resulting in type III production as well as ISGs in IECs [10, 67]. In addition, mice lacking NLRP6 failed to control replication of norovirus in the gastrointestinal tract by reducing type III IFN in mice [67].
Role of inflammasome pathway mediated by PRRs sensing enteric RNA viruses
Three members of the RNA helicase family have been identified to sense enteric RNA viruses and trigger inflammasome activation signaling pathways in IECs and macrophages, which are discussed below in detail (Fig. 4).
Fig. 4.
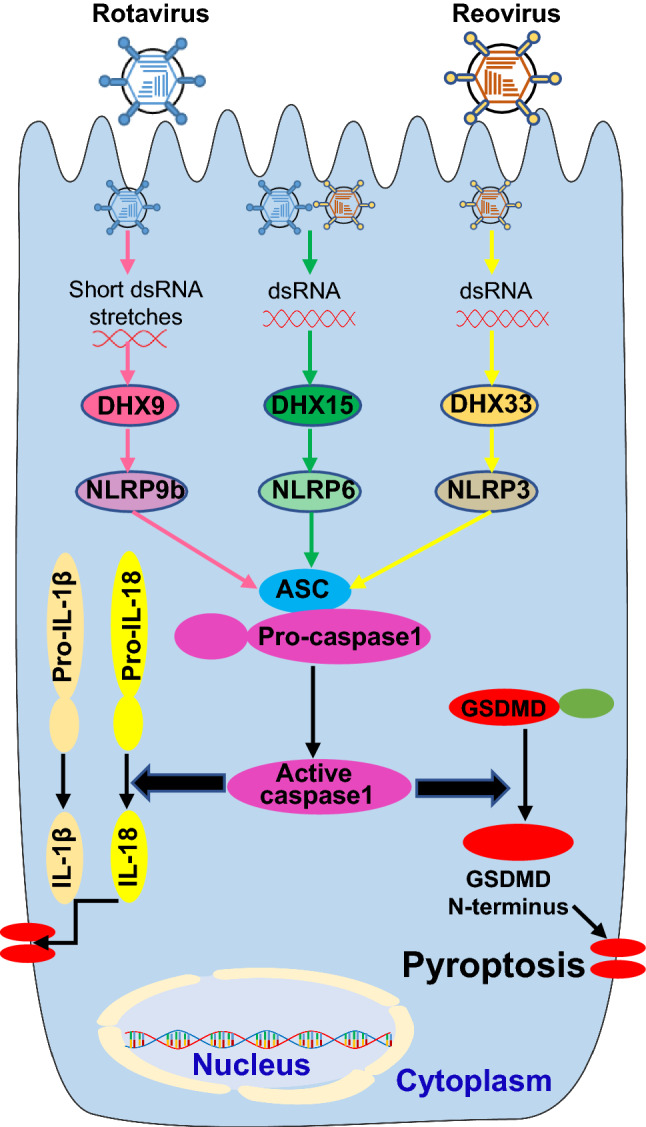
Host PRRs sense enteric RNA viruses to induce inflammasome activation signaling pathway. Entering by enteric RNA viruses, including rotavirus and reovirus, introduces dsRNA in the cytoplasm. The host PRRs in the cytoplasm, including DHX9, DHX15 and DHX33, recognize the RNA molecules and trigger the activation of inflammasome containing ASC (apoptosis-associated speck-like protein containing a caspase recruitment domain), pro-caspase1 and NLRP9b (the NACHT, LRR, and PYD domains-containing protein 9b), NLRP6 or NLRP3, which results in cleavage of pro-caspase1 into active caspase1. The active caspase 1 mediates maturation and secretion of IL-18 and IL-1β cytokines by cleavage of pro-IL-18 and pro-IL-1β. Additionally, active caspase 1 can provoke a lytic form of cell death termed pyroptosis through cleavage of its substrate Gasdermin D (GSDMD) into GSDMD N-terminus that forms pores in the cellular membrane
RNA helicase DHX33
The RNA helicase DHX33 is the first RNA sensor to recognize dsRNA from enteric reovirus followed by recruitment and activation of NLRP3 inflammasome [40]. The NLRP3 recognizes a variety of PAMPs and danger-associated molecular patterns (DAMPs) produced by virus infection to trigger inflammasome-dependent antiviral immune responses for eliminating virus infection [80]. After binding reovirus dsRNA with its helicase domain, DHX33 directly interacts with the NACHT domain of NLRP3 in both human and mouse macrophages. The NLRP3 protein then recruited with ASC (apoptosis-associated speck-like protein containing a caspase recruitment domain) and pro-caspase-1 to form NLRP3–ASC–pro-caspase-1 complex for NLRP3 inflammasome activation and subsequent pro-caspase-1 cleavage, leading to cleavage of pro-IL-1β and pro-IL-18 into mature and secreted IL-1β and IL-18 [40].
RNA helicase DHX15
Recently, DHX15 was reported by our group to sense long dsRNA of reovirus for inducing NLRP6 inflammasome activation in IECs and protect mice from intestinal inflammation induced by infection with enteric RNA viruses including reovirus and rotavirus in mice [10]. NLRP6 is predominantly expressed in IECs [112] and is crucial for maintaining intestinal homeostasis and a healthy intestinal microbiome [113]. However, reovirus was the first enteric RNA virus proposed to activate the NLRP6 inflammasome through its dsRNA. DHX15 binds long dsRNA of reovirus followed by interaction with downstream adaptor NLRP6. The NLRP6 protein then recruits ASC and pro-caspase-1 to form NLRP6–ASC–pro-caspase-1 complex for NLRP6 inflammasome activation and subsequent pro-caspase-1 cleavage, leading to cleavage of pro-IL-18 into mature and secreted IL-18 in IECs [10]. Given that DHX15 induces NLRP6 inflammasome activation after sensing viral RNA, it is likely that DHX15 may also sense viral dsRNA from norovirus and trigger NLRP6 inflammasome activation for controlling norovirus induced intestinal inflammation.
RNA helicase DHX9
Increasing evidence have pointed at critical roles for inflammasomes in preventing infection by enteric RNA viruses. NLRP9b and NLRP6 are the only two NLRPs highly expressed in the ileum of intestine [68]. Similar to NLRP3, NLRP9b could initiate inflammasome formation of NLRP9b, ASC and caspase-1 [114]. Additionally, caspase-1 cleavage was detected in intestine from mice infected with enteric rotavirus [68], suggesting that NLRP9b initiates inflammasome activation upon rotavirus infection. Compared with wild-type mice, NLRP9b, ASC or caspase-1 knockout mice have elevated rotavirus loads and more severe diarrhea [68], further confirming the critical antiviral role of NLRP9b inflammasome. Moreover, mice with NLRP9b deficiency in IECs were more susceptible to rotavirus infection [68], indicating the protective function of NLRP9b inflammasome is important in IECs. Furthermore, rotavirus infection induced activation of NLRP9b inflammasome that led to IL-18 release and GSDMD-dependent pyroptosis, which functions to eliminate rotavirus from intestine in mice [68]. Mechanistically, NLRP9b employs DHX9 to sense short dsRNA stretches from rotavirus for triggering downstream inflammasome activation [68]. DHX9 was previously identified by our group as an RNA sensor for reovirus to induce MAVS-dependent type I IFN production [36].
Concluding remarks
Both type I IFN and especially type III IFN are critical in controlling replication of enteric viruses and viral-induced intestinal inflammation. Despite the incremental progress made in understanding how many PRRs recognize enteric RNA viruses to mediate type I IFN production in IECs, DCs and macrophages, there are few PRRs identified that sense enteric RNA viruses for type III IFN production and inflammasome activation in IECs. Given the immunological importance of IECs in host defense, more efforts should be made to identify more specific PRRs of enteric RNA viruses in IECs and reveal how those intestinal PRRs sense enteric RNA viruses and trigger other signaling pathways that control infection of enteric RNA viruses in IECs in the future. Pyroptosis is well known to control bacterial infection, but mechanism used to control enteric RNA viruses and underlying mechanisms are poorly understood. Given the critical roles of intestinal microbiota in antiviral immunity, the triangle roles of inflammasome, pytoptosis and intestinal microbiota during infection of enteric RNA viruses should be extensively investigated in future work. Furthermore, adaptive immune responses control chronic viral infection in the intestine and innate immunity is required for subsequent initiation and direction of adaptive immune responses. Future efforts should investigate the crosstalk and underlying mechanisms between the innate and adaptive immune responses during infection of enteric RNA viruses. Most important, SARS-CoV-2-induced COVID-19 pandemic is still going on and gastrointestinal symptoms and fecal shedding of SARS-CoV-2 RNA are frequently observed in COVID-19 patients [6, 20]. Gastroenteritis induced by enteric RNA viruses continue to pose a major threat to global public health. Therefore, a thorough understanding of detailed mechanisms of enteric virus sensing, viral-induced intestinal inflammation, and subsequent effects on homeostasis and immune responses in the intestine is critical for designing more effective drugs and vaccines that eliminate enteric RNA virus infection and treat gastroenteritis.
Acknowledgements
We thank Dr. Dorothy E. Lewis (Houston Methodist) for critical reading and editing of this review. We apologize for not including the important contributions from all investigators in the field due to space limitations.
Author contributions
EZ and JX wrote the manuscript with feedback from all authors. MF, CJ and LJM gave their comments and suggestions to the manuscript. JX and ZZ reviewed and modified the manuscript. All authors read and approved the final manuscript.
Funding
Z.Z. was supported by National Institutes of Health grant (R01AI155488). J.X. was supported by American Heart Association Career Development Award (20CDA35260116).
Data availability
Not applicable.
Declarations
Competing interests
The authors declare no conflict of interests.
Ethics approval and consent to participate
Not applicable.
Consent for publication
We all agree to publish this review.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Junji Xing, Email: jxing@houstonmethodist.org.
Zhiqiang Zhang, Email: zzhang@houstonmethodist.org.
References
- 1.Bányai K, et al. Viral gastroenteritis. Lancet. 2018;392(10142):175–186. doi: 10.1016/S0140-6736(18)31128-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black RE, et al. Global, regional, and national causes of child mortality in 2008: a systematic analysis. Lancet. 2010;375(9730):1969–1987. doi: 10.1016/S0140-6736(10)60549-1. [DOI] [PubMed] [Google Scholar]
- 3.Kapikian AZ, et al. Visualization by immune electron microscopy of a 27-nm particle associated with acute infectious nonbacterial gastroenteritis. J Virol. 1972;10(5):1075–1081. doi: 10.1128/jvi.10.5.1075-1081.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kapusinszky B, et al. Nearly constant shedding of diverse enteric viruses by two healthy infants. J Clin Microbiol. 2012;50(11):3427–3434. doi: 10.1128/JCM.01589-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones DL, et al. Shedding of SARS-CoV-2 in feces and urine and its potential role in person-to-person transmission and the environment-based spread of COVID-19. Sci Total Environ. 2020;749:141364. doi: 10.1016/j.scitotenv.2020.141364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo M, et al. Potential intestinal infection and faecal-oral transmission of SARS-CoV-2. Nat Rev Gastroenterol Hepatol. 2021;18(4):269–283. doi: 10.1038/s41575-021-00416-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McClarren RL, et al. Acute infectious diarrhea. Prim Care. 2011;38(3):539–564. doi: 10.1016/j.pop.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 8.Rivero-Calle I, et al. Systemic features of rotavirus infection. J Infect. 2016;72(Suppl):S98–S105. doi: 10.1016/j.jinf.2016.04.029. [DOI] [PubMed] [Google Scholar]
- 9.Bouziat R, et al. Reovirus infection triggers inflammatory responses to dietary antigens and development of celiac disease. Science. 2017;356(6333):44–50. doi: 10.1126/science.aah5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xing J, et al. DHX15 is required to control RNA virus-induced intestinal inflammation. Cell Rep. 2021;35(12):109205. doi: 10.1016/j.celrep.2021.109205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hassan E, Baldridge MT. Norovirus encounters in the gut: multifaceted interactions and disease outcomes. Mucosal Immunol. 2019;12(6):1259–1267. doi: 10.1038/s41385-019-0199-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopman BA, et al. The vast and varied global burden of norovirus: prospects for prevention and control. PLoS Med. 2016;13(4):e1001999. doi: 10.1371/journal.pmed.1001999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wells AI, Coyne CB. Enteroviruses: a gut-wrenching game of entry, detection, and evasion. Viruses. 2019;11(5):460. doi: 10.3390/v11050460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Benedictis P, et al. Astrovirus infections in humans and animals—molecular biology, genetic diversity, and interspecies transmissions. Infect Genet Evol. 2011;11(7):1529–1544. doi: 10.1016/j.meegid.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beards GM, et al. An enveloped virus in stools of children and adults with gastroenteritis that resembles the Breda virus of calves. Lancet. 1984;1(8385):1050–1052. doi: 10.1016/S0140-6736(84)91454-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jamieson FB, et al. Human torovirus: a new nosocomial gastrointestinal pathogen. J Infect Dis. 1998;178(5):1263–1269. doi: 10.1086/314434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheung KS, et al. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and meta-analysis. Gastroenterology. 2020;159(1):81–95. doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Redd WD, et al. Prevalence and characteristics of gastrointestinal symptoms in patients with severe acute respiratory syndrome coronavirus 2 infection in the United States: a multicenter cohort study. Gastroenterology. 2020;159(2):765–767.e2. doi: 10.1053/j.gastro.2020.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zang R, et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci Immunol. 2020 doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamers MM, et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369(6499):50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sia SF, et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583(7818):834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bao L, et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583(7818):830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 23.Shi J, et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368(6494):1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santaolalla R, et al. Innate immunity in the small intestine. Curr Opin Gastroenterol. 2011;27(2):125–131. doi: 10.1097/MOG.0b013e3283438dea. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pelaseyed T, et al. The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol Rev. 2014;260(1):8–20. doi: 10.1111/imr.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clevers HC, Bevins CL. Paneth cells: maestros of the small intestinal crypts. Annu Rev Physiol. 2013;75:289–311. doi: 10.1146/annurev-physiol-030212-183744. [DOI] [PubMed] [Google Scholar]
- 27.Cox HM. Neuroendocrine peptide mechanisms controlling intestinal epithelial function. Curr Opin Pharmacol. 2016;31:50–56. doi: 10.1016/j.coph.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 28.Schneider C, et al. Regulation of immune responses by tuft cells. Nat Rev Immunol. 2019;19(9):584–593. doi: 10.1038/s41577-019-0176-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mabbott NA, et al. Microfold (M) cells: important immunosurveillance posts in the intestinal epithelium. Mucosal Immunol. 2013;6(4):666–677. doi: 10.1038/mi.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cerovic V, et al. Intestinal macrophages and dendritic cells: what's the difference? Trends Immunol. 2014;35(6):270–277. doi: 10.1016/j.it.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 31.Ablasser A, Hur S. Regulation of cGAS- and RLR-mediated immunity to nucleic acids. Nat Immunol. 2020;21(1):17–29. doi: 10.1038/s41590-019-0556-1. [DOI] [PubMed] [Google Scholar]
- 32.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11(5):373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 33.Myong S, et al. Cytosolic viral sensor RIG-I is a 5'-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323(5917):1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5(7):730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 35.Yoneyama M, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175(5):2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Z, et al. DHX9 pairs with IPS-1 to sense double-stranded RNA in myeloid dendritic cells. J Immunol. 2011;187(9):4501–4508. doi: 10.4049/jimmunol.1101307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu H, et al. DHX15 senses double-stranded RNA in myeloid dendritic cells. J Immunol. 2014;193(3):1364–1372. doi: 10.4049/jimmunol.1303322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mosallanejad K, et al. The DEAH-box RNA helicase DHX15 activates NF-kappaB and MAPK signaling downstream of MAVS during antiviral responses. Sci Signal. 2014;7(323):ra40. doi: 10.1126/scisignal.2004841. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Z, et al. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity. 2011;34(6):866–878. doi: 10.1016/j.immuni.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitoma H, et al. The DHX33 RNA helicase senses cytosolic RNA and activates the NLRP3 inflammasome. Immunity. 2013;39(1):123–135. doi: 10.1016/j.immuni.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6(10):981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 42.Seth RB, et al. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122(5):669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 43.Schlee M, Hartmann G. Discriminating self from non-self in nucleic acid sensing. Nat Rev Immunol. 2016;16(9):566–580. doi: 10.1038/nri.2016.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zanoni I, et al. Interferon (IFN)-λ takes the helm: immunomodulatory roles of type III IFNs. Front Immunol. 2017;8:1661. doi: 10.3389/fimmu.2017.01661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lazear HM, et al. Shared and distinct functions of type I and type III interferons. Immunity. 2019;50(4):907–923. doi: 10.1016/j.immuni.2019.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xing J, et al. Identification of poly (ADP-ribose) polymerase 9 (PARP9) as a noncanonical sensor for RNA virus in dendritic cells. Nat Commun. 2021;12(1):2681. doi: 10.1038/s41467-021-23003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ong GH, et al. Exploration of pattern recognition receptor agonists as candidate adjuvants. Front Cell Infect Microbiol. 2021;11:745016. doi: 10.3389/fcimb.2021.745016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kisseleva T, et al. Signaling through the JAK/STAT pathway, recent advances and future challenges. Gene. 2002;285(1–2):1–24. doi: 10.1016/s0378-1119(02)00398-0. [DOI] [PubMed] [Google Scholar]
- 49.Wang W, et al. Transcriptional regulation of antiviral interferon-stimulated genes. Trends Microbiol. 2017;25(7):573–584. doi: 10.1016/j.tim.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Balachandran S, et al. Alpha/beta interferons potentiate virus-induced apoptosis through activation of the FADD/Caspase-8 death signaling pathway. J Virol. 2000;74(3):1513–1523. doi: 10.1128/jvi.74.3.1513-1523.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mattei F, et al. Regulation of immune cell homeostasis by type I interferons. Cytokine Growth Factor Rev. 2010;21(4):227–236. doi: 10.1016/j.cytogfr.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 52.Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25(3):373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 53.Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev. 2009;227(1):75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pott J, et al. IFN-lambda determines the intestinal epithelial antiviral host defense. Proc Natl Acad Sci U S A. 2011;108(19):7944–7949. doi: 10.1073/pnas.1100552108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kotenko SV, et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4(1):69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 56.Sheppard P, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003;4(1):63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 57.Lazear HM, et al. Interferon-λ: immune functions at barrier surfaces and beyond. Immunity. 2015;43(1):15–28. doi: 10.1016/j.immuni.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ingle H, et al. Distinct effects of type I and III interferons on enteric viruses. Viruses. 2018;10(1):46. doi: 10.3390/v10010046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee S, Baldridge MT. Interferon-lambda: a potent regulator of intestinal viral infections. Front Immunol. 2017;8:749. doi: 10.3389/fimmu.2017.00749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.LeMessurier KS, et al. Type I interferon protects against pneumococcal invasive disease by inhibiting bacterial transmigration across the lung. PLoS Pathog. 2013;9(11):e1003727. doi: 10.1371/journal.ppat.1003727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Broggi A, et al. IFN-λ suppresses intestinal inflammation by non-translational regulation of neutrophil function. Nat Immunol. 2017;18(10):1084–1093. doi: 10.1038/ni.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cho H, Kelsall BL. The role of type I interferons in intestinal infection, homeostasis, and inflammation. Immunol Rev. 2014;260(1):145–167. doi: 10.1111/imr.12195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dubois H, et al. General strategies in inflammasome biology. Curr Top Microbiol Immunol. 2016;397:1–22. doi: 10.1007/978-3-319-41171-2_1. [DOI] [PubMed] [Google Scholar]
- 64.Franchi L, et al. The inflammasome: a caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat Immunol. 2009;10(3):241–247. doi: 10.1038/ni.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Allen IC, et al. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity. 2009;30(4):556–565. doi: 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thomas PG, et al. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity. 2009;30(4):566–575. doi: 10.1016/j.immuni.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang P, et al. Nlrp6 regulates intestinal antiviral innate immunity. Science. 2015;350(6262):826–830. doi: 10.1126/science.aab3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu S, et al. Nlrp9b inflammasome restricts rotavirus infection in intestinal epithelial cells. Nature. 2017;546(7660):667–670. doi: 10.1038/nature22967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Iwata M, et al. Interleukin-1 (IL-1) inhibits growth of cytomegalovirus in human marrow stromal cells: inhibition is reversed upon removal of IL-1. Blood. 1999;94(2):572–578. [PubMed] [Google Scholar]
- 70.Randolph-Habecker J, et al. Interleukin-1-mediated inhibition of cytomegalovirus replication is due to increased IFN-beta production. J Interferon Cytokine Res. 2002;22(7):765–772. doi: 10.1089/107999002320271350. [DOI] [PubMed] [Google Scholar]
- 71.Van Damme J, et al. Homogeneous interferon-inducing 22K factor is related to endogenous pyrogen and interleukin-1. Nature. 1985;314(6008):266–268. doi: 10.1038/314266a0. [DOI] [PubMed] [Google Scholar]
- 72.Orzalli MH, et al. An antiviral branch of the IL-1 signaling pathway restricts immune-evasive virus replication. Mol Cell. 2018;71(5):825–840.e6. doi: 10.1016/j.molcel.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang B, et al. Viral infection. Prevention and cure of rotavirus infection via TLR5NLRC4-mediated production of IL-22 and IL-18. Science. 2014;346(6211):861–865. doi: 10.1126/science.1256999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Aglietti RA, et al. GsdmD p30 elicited by caspase-11 during pyroptosis forms pores in membranes. Proc Natl Acad Sci U S A. 2016;113(28):7858–7863. doi: 10.1073/pnas.1607769113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ding J, et al. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature. 2016;535(7610):111–116. doi: 10.1038/nature18590. [DOI] [PubMed] [Google Scholar]
- 76.Zhang Y, et al. Plasma membrane changes during programmed cell deaths. Cell Res. 2018;28(1):9–21. doi: 10.1038/cr.2017.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kuriakose T, Kanneganti TD. Current Topics in Microbiology Immunology. Berlin Heidelberg: Springer; 2019. Pyroptosis in Antiviral Immunity; pp. 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dinarello CA. Immunological and inflammatory functions of the interleukin-1 family. Annu Rev Immunol. 2009;27:519–550. doi: 10.1146/annurev.immunol.021908.132612. [DOI] [PubMed] [Google Scholar]
- 79.Chen IY, Ichinohe T. Response of host inflammasomes to viral infection. Trends Microbiol. 2015;23(1):55–63. doi: 10.1016/j.tim.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 80.Spel L, Martinon F. Detection of viruses by inflammasomes. Curr Opin Virol. 2021;46:59–64. doi: 10.1016/j.coviro.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kawasaki T, Kawai T. Toll-like receptor signaling pathways. Front Immunol. 2014;5:461. doi: 10.3389/fimmu.2014.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Takeda K, et al. Toll-like receptors. Annu Rev Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 83.Honda K, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434(7034):772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 84.Kawai T, et al. Interferon-alpha induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat Immunol. 2004;5(10):1061–1068. doi: 10.1038/ni1118. [DOI] [PubMed] [Google Scholar]
- 85.Shinohara ML, et al. Osteopontin expression is essential for interferon-alpha production by plasmacytoid dendritic cells. Nat Immunol. 2006;7(5):498–506. doi: 10.1038/ni1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhou R, et al. Recognition of double-stranded RNA by TLR3 induces severe small intestinal injury in mice. J Immunol. 2007;178(7):4548–4556. doi: 10.4049/jimmunol.178.7.4548. [DOI] [PubMed] [Google Scholar]
- 87.Pott J, et al. Age-dependent TLR3 expression of the intestinal epithelium contributes to rotavirus susceptibility. PLoS Pathog. 2012;8(5):e1002670. doi: 10.1371/journal.ppat.1002670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cervantes-Barragan L, et al. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood. 2007;109(3):1131–1137. doi: 10.1182/blood-2006-05-023770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Channappanavar R, et al. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J Clin Invest. 2019;129(9):3625–3639. doi: 10.1172/JCI126363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Komuro A, Horvath CM. RNA- and virus-independent inhibition of antiviral signaling by RNA helicase LGP2. J Virol. 2006;80(24):12332–12342. doi: 10.1128/JVI.01325-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hornung V, et al. 5'-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314(5801):994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 92.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441(7089):101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 93.Loo YM, et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82(1):335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schlee M, et al. Recognition of 5' triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31(1):25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Broquet AH, et al. RIG-I/MDA5/MAVS are required to signal a protective IFN response in rotavirus-infected intestinal epithelium. J Immunol. 2011;186(3):1618–1626. doi: 10.4049/jimmunol.1002862. [DOI] [PubMed] [Google Scholar]
- 96.Kato H, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205(7):1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Holm GH, et al. Retinoic acid-inducible gene-I and interferon-beta promoter stimulator-1 augment proapoptotic responses following mammalian reovirus infection via interferon regulatory factor-3. J Biol Chem. 2007;282(30):21953–21961. doi: 10.1074/jbc.M702112200. [DOI] [PubMed] [Google Scholar]
- 98.McCartney SA, et al. MDA-5 recognition of a murine norovirus. PLoS Pathog. 2008;4(7):e1000108. doi: 10.1371/journal.ppat.1000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang JP, et al. MDA5 and MAVS mediate type I interferon responses to coxsackie B virus. J Virol. 2010;84(1):254–260. doi: 10.1128/JVI.00631-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Triantafilou K, et al. Visualisation of direct interaction of MDA5 and the dsRNA replicative intermediate form of positive strand RNA viruses. J Cell Sci. 2012;125(Pt 20):4761–4769. doi: 10.1242/jcs.103887. [DOI] [PubMed] [Google Scholar]
- 101.Feng Q, et al. MDA5 detects the double-stranded RNA replicative form in picornavirus-infected cells. Cell Rep. 2012;2(5):1187–1196. doi: 10.1016/j.celrep.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Roth-Cross JK, et al. Murine coronavirus mouse hepatitis virus is recognized by MDA5 and induces type I interferon in brain macrophages/microglia. J Virol. 2008;82(20):9829–9838. doi: 10.1128/JVI.01199-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zalinger ZB, et al. MDA5 is critical to host defense during infection with murine coronavirus. J Virol. 2015;89(24):12330–12340. doi: 10.1128/JVI.01470-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hackbart M, et al. Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proc Natl Acad Sci U S A. 2020;117(14):8094–8103. doi: 10.1073/pnas.1921485117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fullam A, Schröder M. (2013) DExD/H-box RNA helicases as mediators of anti-viral innate immunity and essential host factors for viral replication. Biochim Biophys Acta. 1829;8:854–865. doi: 10.1016/j.bbagrm.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu Y, et al. The interaction between the helicase DHX33 and IPS-1 as a novel pathway to sense double-stranded RNA and RNA viruses in myeloid dendritic cells. Cell Mol Immunol. 2014;11(1):49–57. doi: 10.1038/cmi.2013.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hernández PP, et al. Interferon-λ and interleukin 22 act synergistically for the induction of interferon-stimulated genes and control of rotavirus infection. Nat Immunol. 2015;16(7):698–707. doi: 10.1038/ni.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lin JD, et al. Distinct roles of type I and type III interferons in intestinal immunity to homologous and heterologous rotavirus infections. PLoS Pathog. 2016;12(4):e1005600. doi: 10.1371/journal.ppat.1005600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Baldridge MT, et al. Expression of of Ifnlr1 on intestinal epithelial cells is critical to the antiviral effects of interferon lambda against norovirus and reovirus. J Virol. 2017 doi: 10.1128/JVI.02079-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Baldridge MT, et al. Commensal microbes and interferon-λ determine persistence of enteric murine norovirus infection. Science. 2015;347(6219):266–269. doi: 10.1126/science.1258025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nice TJ, et al. Interferon-λ cures persistent murine norovirus infection in the absence of adaptive immunity. Science. 2015;347(6219):269–273. doi: 10.1126/science.1258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sellin ME, et al. Inflammasomes of the intestinal epithelium. Trends Immunol. 2015;36(8):442–450. doi: 10.1016/j.it.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 113.Elinav E, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145(5):745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mullins B, Chen J. NLRP9 in innate immunity and inflammation. Immunology. 2021;162(3):262–267. doi: 10.1111/imm.13290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.