Abstract
Computational docking methods can provide structural models of protein–protein complexes, but protein backbone flexibility upon association often thwarts accurate predictions. In recent blind challenges, medium or high accuracy models were submitted in less than 20% of the ‘difficult’ targets (with significant backbone change or uncertainty). Here, we describe recent developments in protein–protein docking and highlight advances that tackle backbone flexibility. In molecular dynamics and Monte Carlo approaches, enhanced sampling techniques have reduced time-scale limitations. Internal coordinate formulations can now capture realistic motions of monomers and complexes using harmonic dynamics. And machine learning approaches adaptively guide docking trajectories or generate novel binding site predictions from deep neural networks trained on protein interfaces. These tools poise the field to break through the longstanding challenge of correctly predicting complex structures with significant conformational change.
Introduction
Protein–protein interactions are involved in nearly all of the biological processes in human health and disease. Understanding the dynamics of binding and the structure of protein complexes at the molecular level can be instrumental in delineating biological mechanisms and developing intervention strategies. Computational protein–protein docking provides a route to predict the three-dimensional structures of protein assemblies or complexes from known structures of individual monomeric proteins.
Docking methods are tested in the blind prediction challenge known as the Critical Assessment of PRediction of Interactions (CAPRI) [1], which in recent rounds pushed the field by including a wide array of target types such as transport proteins, higher order assemblies and host–virus interactions [2,3]. Out of the 28 protein–protein targets evaluated in CAPRI over the past four years [3,2], predictors achieved high quality structures for 11 ‘easy’ targets, defined as those with little backbone motion (unbound to bound Cα root mean square deviation (RMSDBU) of less than 1.5 Å [4]; Figure 1). The remaining 17 targets were categorized as ‘difficult’ (RMSDBU over 2.2 Å and/or poor monomer template availability). For these targets, predictors only achieved acceptable quality in 8 of 17 targets (47%) and high quality in only 2 (12%) [3,2]. Thus, the intrinsic flexibility of biomolecules still confounds the protein docking community at large.
Figure 1.
Performance of protein docking approaches on blind targets in CAPRI Rounds 38–46 [3,2]. Distribution of DockQ scores for the best model submitted by each predictor group (points) for each individual target (x-axis). DockQ measures a combination of intermolecular residue-residue contacts, interface RMSD, and ligand RMSD on a scale of 0 (incorrect) to 1 (matching the experimental structure) [2]. Targets are labelled by their CAPRI target number and, when needed, interface number (after the decimal). The targets are classified into rigid (easy) targets (high-homology monomer templates and under 1.5 Å unbound–bound backbone motion, and flexible targets (poor template availability and/or over 1.5 Å RMSDBU). DockQ scores are color-coded by CAPRI model quality ranking: blue, high; green, medium; yellow, acceptable; gray, incorrect. Data graciously provided by Lensink et al. [3,2].
In this review, we focus on the central docking challenge of capturing larger binding-induced conformational changes. We summarize progress by recent algorithms and frameworks, additionally augmented by growth in databases and computational power (CPU-based and GPU-based). These new methods have achieved greater accuracy on more challenging targets and additionally yielded insight into binding mechanisms. We first present progress in binding site identification and then docking methods including molecular dynamics (MD) and Monte Carlo (MC) approaches, normal modes, and machine learning. Together, these techniques have helped better explore broader regions of conformational space and more thoroughly evaluate the energy landscape to improve protein–protein docking.
Identifying putative binding sites: a global search
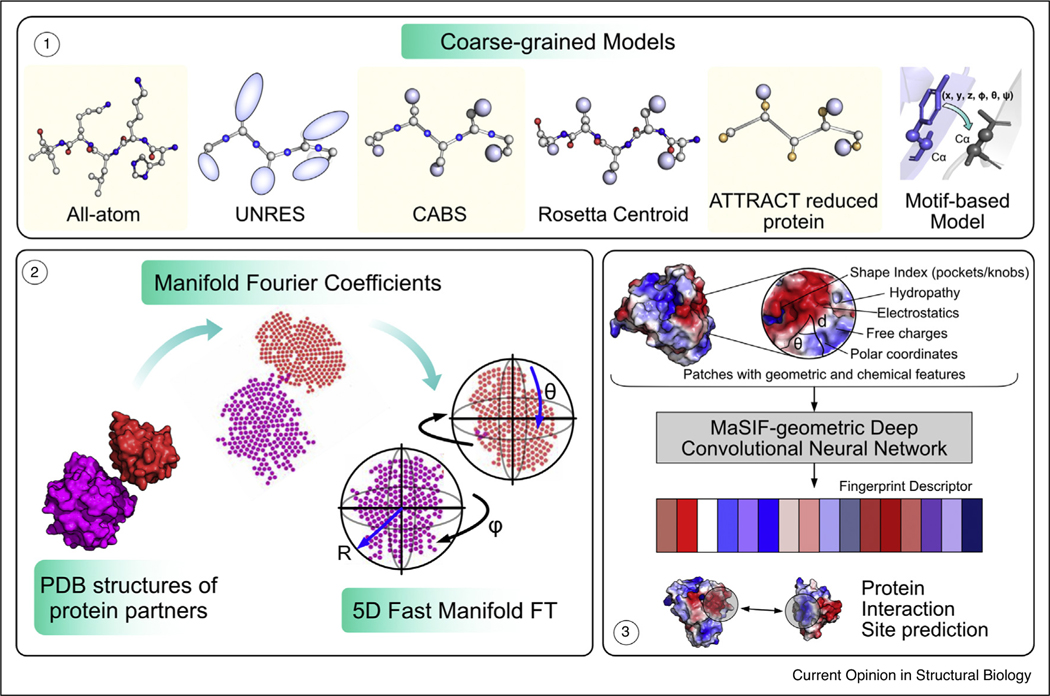
To reduce the complexity of the immense conformation space of flexible proteins, coarse-grained models are frequently used to reduce the degrees of freedom (Figure 2). In the extreme, global docking approaches typically first treat protein partners as rigid bodies by restricting to six degrees of freedom (three rotational and three translational). A prime method to exhaustively sample the global 6D space is enumerating and scoring different rigid-body orientations on a dense grid. Approaches such as ClusPro [12] and ZDOCK [13] rely on the fast Fourier transform (FFT) correlation, which projects protein binding partners on a discretized three-dimensional grid. Conventional FFT approaches accelerate sampling only in the translational space and require new FFTs for every rotation. In 2015, Kazennov et al. developed fast manifold Fourier transforms (FMFT) to search arrangements of two rigid bodies in a 5D manifold (Figure 2) [14]. Relative to traditional FFT-based docking, FMFT accelerates calculations 10-fold [10••]. Another shape-based approach is geometric hashing, which indexes point sets or curves to match geometric features under arbitrary transformations like translations, rotations or even scaling [15]. Local 3D Zernike descriptor-based docking (LZerD), one of the top methods in CAPRI, projects 3D surfaces onto spheres to efficiently capture complementarity of protein surfaces [16]. Some rigid-body approaches exploit data from chemical cross-linking experiments [17] or small-angle X-ray scattering (SAXS) [18] to further improve discrimination of generated structures. These approaches provide fast, global exploration of the energy landscape, and in recent CAPRI rounds [3,2], many predictors incorporated these approaches as the first step to identify putative binding patches, and they supplement with other refinement tools to capture backbone flexibility.
Figure 2.
Reducing the degrees of freedom in protein docking. 1. Coarse-grained models: from left to right: some approaches use all-atom representations (except solvent). The UNRES (united residue) model [5] represents the side chains as variable size ellipsoids attached to the Cα atom by peptide linkages and backbone N, C and O atoms are accounted with peptide-bond centers. CABS (Cα, Cβ and side chains) model adds a Cβ atom and approximates rest of the side chain by a single sphere. The Rosetta centroid model [6] uses a CEN atom to represent the side chain while the backbone stays intact. The ATTRACT reduced protein model comprises of 2–3 atoms per residue with only Cα in the backbone and 1–2 atoms in the side chain [7]. Knowledge-based model derived from residue pair transforms of protein motifs from bound complexes in the PDB [8,9••]. 2. Fast manifold Fourier transforms (FMFT): the 5D FMFT method implicitly matches protein shapes over three translations and two rotations in Fourier space (adapted from Padhorny et al. [10••]). 3. MaSIF identifies binding sites using interface ‘fingerprints’ in a geometric deep learning model [11••].
Methods accounting backbone flexibility
Molecular dynamics
Molecular dynamics (MD) is one strategy that is often used after grid-search or template-based approaches for refinement (Figure 3) [19]. Unbiased, all-atom MD simulations can provide a high-resolution, time-resolved microscopic model of protein–protein interactions. MD calculates Newtonian trajectories using physics-based energy functions to simulate protein association and dissociation events. MD use for protein docking has been limited because non-native local minima trap proteins, and dissociation is too slow [20]. Over the past decade, two new modifications to capture conformational changes are steered molecular dynamics (SMD) [21], which utilizes external force constraints, and Markov sampling, which breaks a long MD simulation into multiple short trajectories [22]. To accelerate dissociation of protein partners at suboptimal binding regions, Ostermeir et al. developed a Hamiltonian replica exchange MD protocol (H-REMD) for protein docking [23•]. In H-REMD, biasing potentials are based on the shortest distance between protein partner atoms (defined as ‘ambiguity restraints’). As the biasing potential and associated ambiguity restraints vary across replicas, associated protein partners in one replica are forced to dissociate in another. Pan et al. simulated long timescales in a global search space for a benchmark set of five targets on the special purpose machine Anton [24,25••].Their ‘tempered binding’ protocol updates energy function parameters throughout the simulation: a soft-core van der Waals intermolecular potential is scaled so that long-lived states are dissociated more frequently, improving the sampling efficiency [25••]. Further, Pan et al. found that proteins often follow a repeated dissociation and association pattern rather than probing continually along the surface for the native binding site. Siebenmorgen et al. similarly scaled atomic repulsions with the vdW radii [26••]. They varied the vdW attraction energy across replicas relative to the Lennard-Jones and electrostatic interactions (owing to increased ligand-receptor atom distance). Compared to conventional MD methods, their simulations sampled native-like states 30% more often; resulting in blind docking predictions within 5 Å of native for moderately flexible targets. MD-based docking on proteins that move more than 2.2 Å RMSD upon binding has not yet been reported.
Figure 3.
Enhanced sampling approaches in protein docking. 1. Temperature replica exchange: MD/MC approaches utilize temperature as the variable parameter across replicas [27,28]. The smoothening of the relatively rugged energy landscape enables sampling of distinct energy basins. 2. Umbrella Sampling methods [29] split the reaction coordinate between an unbound and bound state into multiple windows. This enables biasing molecular dynamics trajectories along the reaction coordinate driving the system from one thermodynamic state to another. 3. Hamiltonian replica change approaches introduce a biasing potential which can be either time-dependent, contact-dependent [23•] or geometry dependent [26••]. Scaling methods: Top: use of contact-dependent ambiguity constraints between protein partners. The weighted distance of the closest contacts of the partners defines bias potentials; Bottom: Bias based on increase in the effective pairwise vdW radii (an illustration to indicate the variable vdW radii across replicas for hamiltonian-based tempering); Hamiltonian REMD: the exchange trajectories with the biasing harmonic potential (red) and the range of potentials used across all the replicas in the system. 4. Conformational flooding/Metadynamics utilize an exhaustive search within a local scope by introducing a funnel-shaped constraint potential [30]. Short metadynamics simulations have been equipped to obtain backbone conformations for ensemble-docking [31].
Monte Carlo methods
In contrast to MD approaches that target flexibility with Newtonian dynamics; Monte Carlo (MC) methods sample by random moves often followed by minimization (MCM) [6]. MC allows a wide variety of conformational move types to sample diverse conformations. MC algorithms have emulated the kinetic binding models, namely key-lock, conformer selection (CS) and induced-fit (IF) mechanisms [6,32,33]. The CS model chooses protein backbones from a pre-generated ensemble, thus this approach has the advantage of docking one partner’s conformations at a time. However, CS docking can fail if the ensemble is devoid of native-like backbone conformations [34]. For targets with RMSDBU up to 2.5 Å, Zhang et al. generated ensembles of 40 structures for MC-based docking [33]. This ensemble docking approach incorporates the ATTRACT coarse-grained protein model (Figure 2) [7] in conjunction with replica-exchange (RE) to sample in backbone as well as rigid body space. Although the ensemble does not always include bound-like conformations of the proteins, their REMC-ensemble docking method obtains higher quality structures than MCM and REMC approaches. RosettaDock4.0 [9••], a conformer selection based MCM approach, modulates backbone swaps with a strategy that modulates rates of sampling of each conformer to handle ensembles of 100 structures for each protein partner (RosettaDock3.0 [32] docked from an ensemble of 10 structures). To diversify backbone conformations, the protocol generates monomer structures by three methods: Firstly, normal modes, secondly, backrub motions [35], and finally, all-atom backbone refinement [36]. Further, to discriminate between near-native and non-native structures, they developed a more accurate coarse-grained energy function with 6-dimensional residue-pair data obtained from protein–protein interfaces in the Protein Data Bank (Figure 2) [8]. Marze et al. report success on 49% of moderately flexible and 31% of flexible targets, the highest local-docking success rates yet reported [9••].
Sampling backbone conformations with normal modes
Since intrinsic fluctuations in proteins contribute to conformational change, some docking approaches utilize harmonic dynamics to capture protein backbone motions [37,38]. Normal modes of vibration represent internal motions of a protein based on a Hookean potential between close residues. Normal mode analysis (NMA) is incorporated in several docking approaches, and there have been recent innovations in the past few years. To mimic induced-fit, Schindler et al. developed iATTRACT [39] by moving interface residues in Cartesian coordinate space subject to NMA-generated harmonic potentials. iATTRACT served as a refinement stage and improved the fraction of native contacts predicted by 70%. For targets with unbound to bound interface RMSD over 4 Å, iATTRACT can achieve acceptable quality models [39]. Population-based methods such as particle swarm optimization (PSO) have also employed NMA. PSO is a heuristic approach that optimizes the multiple degrees of freedom using a set of multiple systems. The SwarmDock algorithm recently incorporated dynamic cross-docking [40•] of multiple backbone conformations within its PSO routine. It obtains an ensemble of conformational states of individual protein partners by using elastic network normal mode calculations and samples with the five lowest frequency non-trivial modes. SwarmDock achieved medium or high quality structures even for difficult targets with i-RMSD between 2.2 and 6 Å along with a challenging prior CAPRI target (T136) [40•,3]. Extending the swarm intelligence methods, the LightDock algorithm uses a ‘glowworm’ swarm optimization to sample different backbone conformations in local regions of the protein surface with an anisotropic network model [41]. LightDock additionally uses multi-scale modeling to combine all-atom and coarse grained scoring functions.
While normal modes have typically been used on individual protein partners before docking, Oliwa and Shen introduced the complex NMA in docking to also sample molecular complex fluctuations [43]. By calculating modes of an encounter complex, this approach focuses on the binding region as it reduces the dimensionality of the search space [44]. One of the problems of NMA is that higher frequency modes often distort protein bonds. To overcome this limitation, Frezza and Lavery developed the internal coordinate NMA (iNMA) approach to move in the torsion angle space, that is, with fixed bond lengths and angles (Figure 4) [45]. With a reduced protein model in an internal coordinate space, they captured larger conformational changes from eigenvectors of low-frequency modes [42•]. iNMA can generate structures within 3 Å of the bound state when starting from the unbound for 39% of single-domain and 45% of multi-domain proteins in their benchmark.
Figure 4.
Internal coordinates NMA captures larger conformational change. (a) Schematic of the bound homodimer (PDB ID: 2EIA) and unbound monomer (1EIA) forms of equine infectious anemia virus (EIAV) capsid protein p26 (RMSDBU of the binding domain is 5.2 Å). (b) Model generated by internal coordinate NMA at maximum amplitude (yellow) retains realistic bond lengths and angles. (c) iNMA with the optimal mode magnitudes yields a structure within 3 Å RMSD of the bound form. Panels (b) and (c) adapted from Frezza and Lavery [42•].
Machine learning methods
Although protein folding has been one prime focus of deep learning methods in biology (e.g. AlphaFold [46] and RaptorX [47]), in recent years, a few studies have explicitly addressed challenges relevant to protein docking [48]. Protein binding sites can be thought of as an information-rich molecular space that can be mined for elucidating protein interactions [49,50].
One approach is to use this information to create score functions for use with traditional docking approaches. For example, Geng et al. used graph representations to train a support vector machine (SVM) on native and non-native protein complex structures to develop a scoring potential (GraphRank) to rank docked poses [51]. And iScore, composed of the GraphRank and HADDOCK [52] scores, achieved top performance in CAPRI scoring rounds (medium or high quality structures for nine out of 13 targets).
Other teams have used deep learning techniques to identify protein interfaces by extrapolating image recognition tools to protein structures. RaptorX-ComplexContact [50] uses a deep residual neural network trained on single-chain proteins to predict contacts between binding partners, achieving the top contact prediction scores in CASP [53]. Another approach is to characterize interaction environments. Townshend et al. created ‘voxels,’ that is, volumetric pixels with local atomic information for every protein surface residue, and with this 3D representation, they trained a deep 3D convolutional neural network (SASNet) on a curated database of bound protein complex structures [54]. Pittala et al. employed graph convolutions with the nodes representing the amino acid residues and edges connecting residues with a Cβ − Cβ distance under 10 Å [55]. They placed geometric and chemical features on both nodes and edges and used a graph neural network to predict epitopes and paratopes in antigen-antibody interfaces. In a unique approach by Gainza et al., a geometric deep learning model (MaSIF) used molecular interaction ‘fingerprints’ calculated using geometric and chemical features of protein surfaces [11••] (Figure 2). Their deep network was composed from geodesic convolutional layers, and they used it to predict binding sites, evaluate alternate docked interfaces, and assess likelihood of a given protein–protein interaction. Relative to conventional rigid docking methods on protein targets, MaSIF-search can perform ultra-fast scanning to identify true ‘binder’ with similar accuracy but significantly faster (4 CPU-minutes versus 45 hours for PatchDock and 93 days for ZDOCK to evaluate a benchmark of 100 bound protein complexes).
In a study to explore how neural networks might be used to generate structures with considerable backbone motion, Degiacomi trained an autoencoder with conformations from MD simulations, compressing the protein motion into a low-dimensional latent space [56•]. By training with simulations of both closed (bound) and apo conformations of a target protein, the autoencoder generated an intermediate closed-apo conformation at 0.8 Å RMSD [56•] from the native state. However, when the autoencoder was trained only with open conformations, the generator could only create structures far from the closed state (over 4.2 Å), limiting the utility of this approach for blind docking. In an approach suitable for blind cases, Cao and Shen developed a Bayesian active learning (BAL) model to quantify uncertainty in protein structure quality, and then they extended their model to flexible protein docking [57•]. The Bayesian framework determines the posterior probability as it samples backbone conformations [43]. Flexibility is captured with low-frequency complex-NMA modes, and in principle it can be extended to higher frequencies that capture loop and hinge motions. Compared to ZDOCK [13] and PSO, BAL improves the interface RMSD of the near-native predictions by 0.5 Å.
Conclusions
In conjunction with experimental data, docking has advanced a range of biological and health applications (e.g. Alzheimer’s disease [58], celiac disease [59], SARS-CoV-2 [60], to name just a few). Over the past few years, docking success rates have improved on ‘difficult’ blind prediction targets, but rates need to be higher for docking to be a reliable stand-alone tool in all cases. Clearly, a diverse and impressive array of tools has steadily advanced toward reliably capturing large conformational changes in protein docking. Docking will be even more impactful when the field finally overcomes this challenge.
Acknowledgements
This work was supported by the National Institutes of Health through grant R01-GM078221. We thank Marc Lensink for generously providing us with data from CAPRI and Sai Pooja Mahajan and Sudhanshu Shanker for helpful comments on the manuscript.
Footnotes
Conflict of interest statement
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Janin J, Henrick K, Moult J, Eyck LT, Sternberg MJ, Vajda S, Vakser I, Wodak SJ: CAPRI: a critical assessment of PRedicted interactions. Proteins: Struct Funct Genet 2003, 52:2–9 10.1002/prot.10381. [DOI] [PubMed] [Google Scholar]
- 2.Lensink MF, Nadzirin N, Velankar S, Wodak SJ: Modeling protein–protein, protein–peptide, and protein-oligosaccharide complexes: CAPRI 7th edition. Proteins: Struct Funct Bioinformatics 2019:1–23 10.1002/prot.25870. [DOI] [PubMed] [Google Scholar]
- 3.Lensink MF, Brysbaert G, Nadzirin N, Velankar S, Chaleil RA, Gerguri T, Bates PA, Laine E, Carbone A, Grudinin S, Kong R, Liu RR, Xu XM, Shi H, Chang S, Eisenstein M, Karczynska A, Czaplewski C, Lubecka E, Lipska A, Krupa P, Mozolewska M, Golon Ł, Samsonov S, Liwo A, Crivelli S, Pagès G, Karasikov M, Kadukova M, Yan Y, Huang SY, Rosell M, Rodríguez-Lumbreras LA, Romero-Durana M, Díaz-Bueno L, Fernandez-Recio J, Christoffer C, Terashi G, Shin WH, Aderinwale T, Maddhuri Venkata Subraman SR, Kihara D, Kozakov D, Vajda S, Porter K, Padhorny D, Desta I, Beglov D, Ignatov M, Kotelnikov S, Moal IH, Ritchie DW, Chauvot de Beauchêne I, Maigret B, Devignes MD, Ruiz Echartea ME, Barradas-Bautista D, Cao Z, Cavallo L, Oliva R, Cao Y, Shen Y, Baek M, Park T, Woo H, Seok C, Braitbard M, Bitton L, Scheidman-Duhovny D, Dapkūnas J, Olechnovič K, Venclovas Č, Kundrotas PJ, Belkin S, Chakravarty D, Badal VD, Vakser IA, Vreven T, Vangaveti S, Borrman T, Weng Z, Guest JD, Gowthaman R, Pierce BG, Xu X, Duan R, Qiu L, Hou J, Ryan Merideth B, Ma Z, Cheng J, Zou X, Koukos PI, Roel-Touris J, Ambrosetti F, Geng C, Schaarschmidt J, Trellet ME, Melquiond AS, Xue L, Jiménez-García B, van Noort CW, Honorato RV, Bonvin AM, Wodak SJ: Blind prediction of homo- and hetero-protein complexes: the CASP13-CAPRI experiment. Proteins: Struct Funct Bioinformatics 2019, 87:12001221 10.1002/prot.25838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kundrotas PJ, Anishchenko I, Dauzhenka T, Kotthoff I, Mnevets D, Copeland MM, Vakser IA: Dockground: a comprehensive data resource for modeling of protein complexes. Protein Sci 2018, 27:172–181 10.1002/pro.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liwo A, Baranowski M, Czaplewski C, Gołaś E, He Y, Jagieła D, Krupa P, Maciejczyk M, Makowski M, Mozolewska MA,Niadzvedtski A, Ołdziej S, Scheraga HA, Sieradzan AK, Slusarz R, Wirecki T, Yin Y, Zaborowski B: A unified coarse-grained model of biological macromolecules based on mean-field multipolemultipole interactions. J Mol Model 2014, 20:2306 10.1007/s00894-014-2306-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang C, Bradley P, Baker D: Protein–protein docking with backbone flexibility. J Mol Biol 2007, 373:503–519 10.1016/j.jmb.2007.07.050. [DOI] [PubMed] [Google Scholar]
- 7.Zacharias M: ATTRACT: protein–protein docking in CAPRI using a reduced protein model. Proteins: Struct Funct Bioinformatics 2005, 60:252–256 10.1002/prot.20566. [DOI] [PubMed] [Google Scholar]
- 8.Fallas JA, Ueda G, Sheffler W, Nguyen V, McNamara DE, Sankaran B, Pereira JH, Parmeggiani F, Brunette TJ, Cascio D, Yeates TR, Zwart P, Baker D: Computational design of selfassembling cyclic protein homo-oligomers. Nat Chem 2017, 9:353–360 10.1038/nchem.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marze NA, Roy Burman SS, Sheffler W, Gray JJ: Efficient flexible backbone protein–protein docking for challenging targets. Bioinformatics 2018, 34:3461–3469 10.1093/bioinformatics/bty355 •• With a novel, six-dimension, coarse-grained score function and adaptive conformer selection, RosettaDock 4.0 succeeds in local docking on 49% of moderately exible and 31% of exible targets, the highest reported todate.
- 10. Padhorny D, Kazennov A, Zerbe BS, Porter KA, Xia B, Mottarella SE, Kholodov Y, Ritchie DW, Vajda S, Kozakov D: Protein–protein docking by fast generalized Fourier transforms on 5D rotational manifolds. Proc Natl Acad Sci U S A 2016, 113:E4286–E4293 10.1073/pnas.1603929113 •• While traditional FFT algorithms transform over three translational degrees of freedom, the fast manifold Fourier transform algorithm encodes an additional two rotational dimensions using spherical functions and radial harmonics. The approach speeds up sampling by an order of magnitude.
- 11. Gainza P, Sverrisson F, Monti F, Rodolà E, Boscaini D, Bronstein MM, Correia BE: Deciphering interaction fingerprints from protein molecular surfaces using geometric deep learning. Nat Methods 2020, 17:184–192 10.1038/s41592-019-0666-6 •• A geometric deep learning model that computes molecular interaction ‘fingerprints’ — geometric and chemical features of protein surface patches — to rapidly identify binding sites (MaSIF-site, MaSIF-ligand) or scan proteininterfaces (MaSIF-search).
- 12.Kozakov D, Hall DR, Xia B, Porter KA, Padhorny D, Yueh C, Beglov D, Vajda S: The ClusPro web server for protein–protein docking. Nat Protoc 2017, 12:255–278 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pierce BG, Wiehe K, Hwang H, Kim BH, Vreven T, Weng Z: ZDOCK server: interactive docking prediction of protein–protein complexes and symmetric multimers. Bioinformatics 2014, 30:1771–1773 10.1093/bioinformatics/btu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kazennov AM, Alekseenko AE, Kozakov D, Padhorny DN, Kholodov YA: Efficient search for the possible mutual arrangements of two rigid bodies with the use of the generalized five-dimensional Fourier transform. Math Models Comput Simul 2015, 7:315–322 10.1134/S2070048215040043. [DOI] [Google Scholar]
- 15.Smith GR, Sternberg MJ: Prediction of protein–protein interactions by docking methods. Curr Opin Struct Biol 2002, 12:28–35 10.1016/S0959-440X(02)00285-3. [DOI] [PubMed] [Google Scholar]
- 16.Venkatraman V, Yang YD, Sael L, Kihara D: Protein–protein docking using region-based 3D Zernike descriptors. BMC Bioinformatics 2009, 10 10.1186/1471-2105-10407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vreven T, Schweppe DK, Chavez JD, Weisbrod CR, Shibata S, Zheng C, Bruce JE, Weng Z: Integrating cross-linking experiments with ab initio protein–protein docking. J Mol Biol 2018, 430:1814–1828 10.1016/j.jmb.2018.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ignatov M, Kazennov A, Kozakov D: ClusPro FMFT-SAXS: ultra-fast filtering using small-angle X-ray scattering data in protein docking. J Mol Biol 2018, 430:2249–2255 10.1016/j.jmb.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 19.Christoffer C, Terashi G, Shin WH, Aderinwale T, Maddhuri Venkata Subramaniya SR, Peterson L, Verburgt J, Kihara D: Performance and enhancement of the LZerD protein assembly pipeline in CAPRI 38–46. Proteins: Struct Funct Bioinformatics 2019:1–14 10.1002/prot.25850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaw DE, Maragakis P, Lindorff-Larsen K, Piana S, Dror RO, Eastwood MP, Bank JA, Jumper JM, Salmon JK, Shan Y, Wriggers W: Atomic-level characterization of the structural dynamics of proteins. Science 2010, 330:341–346 10.1126/science.1187409. [DOI] [PubMed] [Google Scholar]
- 21.Kro l M, Chaleil RAG, Tournier AL, Bates PA: Implicit flexibility in protein docking: cross-docking and local refinement. Proteins 2007, 69:750–757 10.1002/prot.21698. [DOI] [PubMed] [Google Scholar]
- 22.Plattner N, Doerr S, De Fabritiis G, Noé F: Complete protein– protein association kinetics in atomic detail revealed by molecular dynamics simulations and Markov modelling. Nat Chem 2017, 9:1005–1011 10.1038/nchem.2785. [DOI] [PubMed] [Google Scholar]
- 23. Ostermeir K, Zacharias M: Accelerated flexible protein-ligand docking using Hamiltonian replica exchange with a repulsive biasing potential. PLOS ONE 2017, 12 10.1371/journal.pone.0172072 • Hamiltonian replica exchange (H-REMD) modifies parts of the force field across different replicas. In this paper, a repulsive potential between receptor and ligand surface residues promotes transient dissociation on switching replicas, accelerating exploration of the protein surface to identify possible binding sites.
- 24.Shaw DE, Grossman JP, Bank JA, Batson B, Butts JA, Chao JC, Deneroff MM, Dror RO, Even A, Fenton CH, Forte A, Gagliardo J,Gill G, Greskamp B, Ho CR, Ierardi DJ, Iserovich L, Kuskin JS, Larson RH, Layman T, Lee L, Lerer AK, Li C, Killebrew D, Mackenzie KM, Mok SY, Moraes MA, Mueller R, Nociolo LJ, Peticolas JL, Quan T, Ramot D, Salmon JK, Scarpazza DP, Schafer UB, Siddique N, Snyder CW, Spengler J, Tang PTP, Theobald M, Toma H, Towles B, Vitale B, Wang SC, Young C: Anton 2: raising the bar for performance and programmability in a special-purpose molecular dynamics supercomputer. SC’14: Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis 2014:41–53 10.1109/SC.2014.9. [DOI] [Google Scholar]
- 25. Pan AC, Jacobson D, Yatsenko K, Sritharan D, Weinreich TM, Shaw DE: Atomic-level characterization of protein–protein association. Proc Natl Acad Sci U S A 2019, 116:4244–4249 10.1073/pnas.1815431116 •• With long timescale MD simulations using a ‘tempered binding’ protocol that scales a soft-core energy across replicas to promote dissociation of long-lived states, this work found that protein binding occurs through repeated association-dissociation events rather than prolonged in-contact exploration.
- 26. Siebenmorgen T, Engelhard M, Zacharias M: Prediction of protein–protein complexes using replica exchange with repulsive scaling. J Comput Chem 2020:1436–1447 10.1002/jcc.26187 •• Using a novel replica exchange scheme with variable van der Waals radii for interface residue atoms, the RS-REMD approach promotes dissociation in some replicas, which improves sampling for both global searches and refinement.
- 27.Liu P, Kim B, Friesner RA, Berne BJ: Replica exchange with solute tempering: a method for sampling biological systems in explicit water. Proc Natl Acad Sci 2005, 102:13749–13754 10.1073/pnas.0506346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Z, Lange OF: Replica exchange improves sampling in low-resolution docking stage of RosettaDock. PLOS ONE 2013, 8:e72096 10.1371/journal.pone.0072096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kästner J: Umbrella sampling. Wiley Interdisc Rev: Comput Mol Sci 2011, 1:932–942 10.1002/wcms.66. [DOI] [Google Scholar]
- 30.Limongelli V, Bonomi M, Parrinello M: Funnel metadynamics as accurate binding free-energy method. Proc Natl Acad Sci U S A 2013, 110:6358–6363 10.1073/pnas.1303186110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Basciu A, Malloci G, Pietrucci F, Bonvin AMJJ, Vargiu AV: Hololike and druggable protein conformations from enhanced sampling of binding pocket volume and shape. J Chem Inform Model 2019, 59:1515–1528 10.1021/acs.jcim.8b00730. [DOI] [PubMed] [Google Scholar]
- 32.Chaudhury S, Gray JJ: Conformer selection and induced fit in flexible backbone protein–protein docking using computational and NMR ensembles. J Mol Biol 2008, 381: 10.1016/j.jmb.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Z, Ehmann U, Zacharias M: Monte Carlo replica-exchange based ensemble docking of protein conformations. Proteins: Struct Funct Bioinformatics 2017, 85:924–937 10.1002/prot.25262. [DOI] [PubMed] [Google Scholar]
- 34.Kuroda D, Gray JJ: Pushing the backbone in protein–protein docking. Structure 2016, 24:1821–1829 10.1016/j.str.2016.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith CA, Kortemme T: Backrub-like backbone simulation recapitulates natural protein conformational variability and improves mutant side-chain prediction. J Mol Biol 2008, 380:742–756 10.1016/j.jmb.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tyka MD, Keedy Da, Andre I, Dimaio F, Song Y, Richardson DC, Richardsonb JS, Baker D: Alternate states of proteins revealed by detailed energy landscape mapping. J Mol Biol 2011, 405:607–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zacharias M, Sklenar H: Harmonic modes as variables to approximately account for receptor flexibility in ligand-receptor docking simulations: application to DNA minor groove ligand complex. J Comput Chem 1999, 20:287–300. [Google Scholar]
- 38.Zacharias M: Accounting for conformational changes during protein–protein docking. Curr Opin Struct Biol 2010, 20:180–186 10.1016/j.sbi.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 39.Schindler CEM, de Vries SJ, Zacharias M: iATTRACT: simultaneous global and local interface optimization for protein–protein docking refinement. Proteins: Struct Funct Bioinformatics 2015, 83:248–258 10.1002/prot.24728. [DOI] [PubMed] [Google Scholar]
- 40. Torchala M, Gerguri T, Chaleil RAG, Gordon P, Russell F, Keshani M, Bates PA: Enhanced sampling of protein conformational states for dynamic cross-docking within the protein–protein docking server SwarmDock. Proteins: Struct Funct Bioinformatics 2020, 88:962–972 10.1002/prot.25851 • A hybrid conformational-selection/induced-_t approach for dynamic cross-docking in SwarmDock, a particle swarm optimization algorithm. Ensembles are pre-generated with NMA and undergo cross-docking while sampling alter-nate protein conformations using low frequency normal modes.
- 41.Jimènez-García B, Roel-Touris J, Romero-Durana M, Vidal M, Jimènez-Gonzalez D, Fernandez-Recio J: LightDock: a new multi-scale approach to protein–protein docking. Bioinformatics 2018, 34:49–55 10.1093/bioinformatics/btx555. [DOI] [PubMed] [Google Scholar]
- 42. Frezza E, Lavery R: Internal coordinate normal mode analysis: a strategy to predict protein conformational transitions. J Phys Chem B 2019, 123:1294–1301 10.1021/acs.jpcb.8b11913 • This work employs NMA in the internal coordinate space with a reduced protein model to capture large conformational changes of proteins with a faster compute time and no distortion of protein bonds.
- 43.Oliwa T, Shen Y: cNMA: a framework of encounter complex-based normal mode analysis to model conformational changes in protein interactions. Bioinformatics 2015, 31:i151i160 10.1093/bioinformatics/btv252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen H, Sun Y, Shen Y: Predicting protein conformational changes for unbound and homology docking: learning from intrinsic and induced flexibility. Proteins: Struct Funct Bioinformatics 2017, 85:544–556 10.1002/prot.25212. [DOI] [PubMed] [Google Scholar]
- 45.Frezza E, Lavery R: Internal normal mode analysis (iNMA) applied to protein conformational flexibility. J Chem Theory Comput 2015, 11:5503–5512 10.1021/acs.jctc.5b00724. [DOI] [PubMed] [Google Scholar]
- 46.Senior AW, Evans R, Jumper J, Kirkpatrick J, Sifre L, Green T, Qin C, Žídek A, Nelson AW, Bridgland A, Penedones H, Petersen S, Simonyan K, Crossan S, Kohli P, Jones DT, Silver D, Kavukcuoglu K, Hassabis D: Improved protein structure prediction using potentials from deep learning. Nature 2020, 577:706–710 10.1038/s41586-019-1923-7. [DOI] [PubMed] [Google Scholar]
- 47.Wang S, Sun S, Li Z, Zhang R, Xu J: Accurate de novo prediction of protein contact map by ultra-deep learning model. PLOS Comput Biol 2017, 13:1–34 10.1371/journal.pcbi.1005324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao W, Mahajan SP, Sulam J, Gray JJ: Deep Learning in Protein Structural Modeling and Design. 2020arXiv:2007.08383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fout A, Byrd J, Shariat B, Ben-Hur A: Protein interface prediction using graph convolutional networks. Advances in Neural Information Processing Systems 2017-December (NIPS) 2017:6531–6540. [Google Scholar]
- 50.Zeng H, Wang S, Zhou T, Zhao F, Li X, Wu Q, Xu J: ComplexContact: a web server for inter-protein contact prediction using deep learning. Nucleic Acids Res 2018, 46: W432–W437 10.1093/nar/gky420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Geng C, Jung Y, Renaud N, Honavar V, Bonvin AMJJ, Xue LC: iScore: a novel graph kernel-based function for scoring protein–protein docking models. Bioinformatics 2019, 36:112121 10.1093/bioinformatics/btz496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dominguez C, Boelens R, Bonvin AM: HADDOCK: a protein–protein docking approach based on biochemical or biophysical information. J Am Chem Soc 2003, 125:1731–1737 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 53.Kryshtafovych A, Schwede T, Topf M, Fidelis K, Moult J: Critical assessment of methods of protein structure prediction (CASP)-Round XIII. Proteins 2019, 87:1011–1020 10.1002/prot.25823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Townshend R, Bedi R, Suriana P, Dror R: End-to-end learning on 3D protein structure for interface prediction. Advances in Neural Information Processing Systems 32 2019:15642–15651. [Google Scholar]
- 55.Pittala S, Bailey-Kellogg C: Learning context-aware structural representations to predict antigen and antibody binding interfaces. Bioinformatics (Oxford, England) 2020, 36:3996–4003 10.1093/bioinformatics/btaa263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Degiacomi MT: Coupling molecular dynamics and deep learning to mine protein conformational space. Structure 2019, 27:1034–1040 10.1016/j.str.2019.03.018 e3 • This paper describes a unique method of generating plausible motions of a protein using a generative neural network (autoencoder). When trained with conformations from an MD simulation, the autoencoder can quickly generate interpolated structures.
- 57. Cao Y, Shen Y: Bayesian active learning for optimization and uncertainty quantification in protein docking. J Chem Theory Comput 2020, 16:5334–5347 10.1021/acs.jctc.0c00476 • With a framework to quantify uncertainty in docked models, the Bayesian approach uses a posterior distribution to guide sampling to likely lowenergy conformations.
- 58.Frost CV, Zacharias M: From monomer to fibril: Abeta-amyloid binding to Aducanumab antibody studied by molecular dynamics simulation. Proteins: Struct Funct Bioinformatics 2020:1–15 10.1002/prot.25978. [DOI] [PubMed] [Google Scholar]
- 59.Høydahl LS, Richter L, Frick R, Snir O, Gunnarsen KS, Landsverk OJB, Iversen R, Jeliazkov JR, Gray JJ, Bergseng E, Foss S, Qiao S-WW, Lundin KEA, Jahnsen J, Jahnsen FL, Sandlie I, Sollid LM, Løset GÅ : Plasma cells are the most abundant gluten peptide mhc-expressing cells in inflamed intestinal tissues from patients with celiac disease. Gastroenterology 2019, 156:1428–1439 10.1053/j.gastro.2018.12.013e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cleri F, Lensink M, Blossey R: DNA aptamers block the receptor binding domain at the spike protein of SARS-CoV-2. chemRxiv 2020. 10.26434/chemrxiv.12696173.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]