Abstract
Bacillus thuringiensis accumulates, primarily during sporulation, large quantities of insecticidal protoxins which are deposited as crystalline, intracellular inclusions. Most subspecies contain several plasmid-encoded cry genes, each of which has a unique specificity. The overall toxicity profile of a subspecies depends not only on the array of cry genes present but also on the relative expression of the genes. In general, transcription depends on sporulation-specific sigma factors, but little is known about regulation of expression of the individual genes. In order to determine whether expression of a particular cry gene varies in different subspecies, lacZ fusions to the cry promoters of two protoxin genes (cry1 class) were constructed. Protoxin accumulation and mRNA contents were also measured by performing immunoblotting and Northern analyses, respectively. The expression of a cry1Ab-lacZ fusion, but not the expression of a cry1C-lacZ fusion, was three to four times lower in B. thuringiensis subsp. aizawai strains than in B. thuringiensis subsp. kurstaki or B. thuringiensis subsp. tolworthi. Also, the Cry1Ab antigen and steady-state mRNA contents of B. thuringiensis subsp. aizawai were lower. The regulation of the genes must involve regions upstream of the promoters which are unique to each cry gene since (i) mutations in the upstream region of the cry1Ab gene resulted in enhanced expression in B. thuringiensis subsp. aizawai and (ii) no differences were found when the lacZ fusions contained the cry1Ab promoters but no upstream sequences. The capacity to regulate each of the protoxin genes must be a factor in the overall protoxin composition of a subspecies and thus its toxicity profile.
In most subspecies the crystalline inclusions produced by sporulating cells of Bacillus thuringiensis consist of a mixture of closely related protoxins, each of which is active against a subset of insect larvae (6, 15). The plasmid-encoded cry genes are transcribed throughout much of sporulation by forms of RNA polymerase which function in the mother cells, but there are variations in the types of promoters, as well as in the times of transcription of certain classes of these genes (1, 9).
Each of the many subspecies produces its own array of protoxins, which very often is a mixture of Cry1 types (6, 15). There is evidence that the cry1 genes are transcribed differentially (2) and that the relative amounts of the protoxins in inclusions differ (20, 21). There were also medium-dependent differences in the protoxin yields obtained by Dulmage (13), but since complex media were used, the specific factors involved could not be defined.
The previous reports suggest that regulation of expression of the individual cry genes is probably important for determining the overall toxicity profile of an isolate. In addition to the relative amounts of the various protoxins, inclusion solubility (2, 16) and synergism between certain toxins (19, 31) are also factors to consider. In order to analyze this regulation in more detail, plasmids containing fusions of the cry1 regulatory regions to lacZ were introduced into various B. thuringiensis subspecies. Subspecies-dependent differences in expression were found, and these differences were confirmed by measuring protoxin antigen and mRNA contents. Regions upstream of the promoters were found to be important for this regulation.
MATERIALS AND METHODS
Strains and growth.
The strains used and their origins are listed in Table 1. The presence of the cry1Ab gene in B. thuringiensis subsp. kurstaki HD1, B. thuringiensis subsp. aizawai HD133, B. thuringiensis subsp. aizawai HD112, and B. thuringiensis subsp. tolworthi HD124 had been established previously either by Southern hybridization (26) or by PCR analysis (11, 17, 18). Slot blotting with a specific cry1Ab oligonucleotide (17) was used to demonstrate that the relative amounts of cry1Ab DNA were the same in these subspecies (2, 3).
TABLE 1.
B. thuringiensis strains used
| Straina | Protoxin gene composition | Origin | Reference(s) |
|---|---|---|---|
| B. thuringiensis subsp. kurstaki HD1 | cry1Aa, cry1Ab, cry1Ac, cryIIA, cryIIB | H. Dulmage, lab strainb | 20 |
| B. thuringiensis subsp. kurstaki 80-21 | cry1Aa, cry1Ac, cryIIA, cryIIB | Plasmid-cured HD1 | 2, 7 |
| B. thuringiensis subsp. aizawai HD133 | cry1Aa, cry1Ab, cry1C, cry1D | NRL, lab strainbc | 5, 17, 21 |
| B. thuringiensis subsp. aizawai 5 | cry1Aa, cry1C, cry1D | Plasmid-cured HD133 | 2, 5, 7 |
| B. thuringiensis subsp. aizawai HD112 | cry1Ab, cry1C, cry1Dd | NRL | |
| B. thuringiensis subsp. tolworthi HD124 | cry1Ab, cry1C, cry1Ed | NRL | |
| B. thuringiensis subsp. tolworthi 124-12 | cry1C, cry1E | Plasmid-cured derivative | 2 |
HD, original Howard Dulmage collection of strains.
The strain has been maintained in the laboratory for a number of years.
NRL, Howard Dulmage collection kept by L. K. Nakamura, Northern Regional Laboratory, Peoria, Ill.
The gene composition may be incomplete (7).
In order to establish that the regions upstream of the cry1Ab coding sequences were essentially the same in these subspecies, 1,025 bp of each sequence was amplified by PCR (27) by using oligonucleotides 5′GAATGGTTGGCATGCCGAAGACGG and 5′GGTTACTTAAACAATTATAAGG. The approximately 1-kb sequences upstream of the promoters of the three cry1A genes (cry1Aa, cry1Ab, and cry1Ac) in B. thuringiensis subsp. kurstaki HD1 were found to differ at only one base (14), and the sequence of the cry1Ab gene was confirmed (Fig. 1). Based on this sequence, restriction enzymes HpaI, NsiI, NdeI, and NsiI plus NdeI were used to demonstrate that the PCR digestion products of B. thuringiensis subsp. kurstaki HD1, B. thuringiensis subsp. aizawai HD133, and B. thuringiensis subsp. tolworthi HD124 were the same size.
FIG. 1.
Sequence of the region upstream of the cry1A protoxin gene. The BtI and BtII promoters are underlined with one line and two lines, respectively. The start sites of transcription for BtI and BtII are indicated by I and II, respectively. The regions in boldface type are the potential bend and IR (arrows) sites of binding of the pyruvate dehydrogenase E2 protein (32, 33). The GenBank accession no. of this sequence is AF039908.
The plasmid-cured strains 80-21, 5, and HD124-12 and the uncured strain HD112 served as hosts for the lacZ fusion plasmids. A cloned cry1Ab gene (8) was introduced by electroporation (28) into strains 80-21 and 5. This clone and all of the lacZ fusion plasmids were stable in the various strains, as judged by the constant level of resistance to chloramphenicol, the inability to find plasmid deletions after reisolation, and (for the cry1Ab clone) the extent of hybridization with the specific oligonucleotide probe in slot blots (2). Cells were grown in Luria broth (27) for preparation of DNA and in G-Tris medium (4) for all other experiments.
Preparation of nucleic acids.
DNA and RNA were prepared (7, 8) from B. thuringiensis subsp. kurstaki HD1, strain 80-21, B. thuringiensis subsp. aizawai HD112 and HD133, and B. thuringiensis subsp. tolworthi HD124. RNA was isolated from cells 1 h after clumping (early sporulation with no visible endospores), about 1 h later when 40 to 50% of the cells contained phase-dull endospores, and after an additional 90 min when >80% of the cells contained phase-white to phase-bright endospores.
RNA was fractionated in an agarose gel for Northern blotting (27). A nitrocellulose filter was incubated with 35 pmol of 32P-labeled 5′CGGATGCTCATAGAGGAAGAA, an oligonucleotide unique to the cry1Ab gene (17) which had been labeled by using [γ-32P]ATP and polynucleotide kinase (27). The X-ray films were scanned with a PhosphoImager in order to determine relative amounts.
lacZ fusions and β-galactosidase assays.
A plasmid containing the promoter region of the cry1A gene with or without 280 bp upstream of the promoters fused to lacZ has been described previously (29). A 780-bp fragment upstream of the promoters of the cry1C gene (30) was introduced in the same way. Both of these genes contain overlapping promoters, designated BtI and BtII, with the following sequences (in which the boldface segments indicate the sequences of −35 ςK, −35 ςE, −10 ςK, and −10 ςE in that order): TTAGTTGCACTTTGTGCATTTTTTCATAAGATGAGTCATATGTT in cry1A and TTTGTTACGTTTTTTGTATTTTTTCATAAGATGTGTCATATGTT in cry1C. The BtI promoter is recognized by ςE RNA polymerase, and BtII is recognized by ςK RNA polymerase (9). The locations and sequences of the −10 regions are identical in these two genes. The −35 regions differ at one base for the ςE promoter and at two bases for the ςK promoter. Neither of these −35 regions is highly conserved in Bacillus subtilis (25). However, the sequences differ substantially for at least 1 kb upstream of the promoters (Fig. 1) (30, 33).
Mutations in the −10 region of the BtII promoter (Fig. 1) which resulted in differences from the consensus sequence for both ςK and ςE resulted in inactivation of BtII and a fivefold-greater rate of expression from the BtI promoter (29). One mutant promoter, designated cry1A-272, was used for many of our studies because the β-galactosidase activity with this promoter was much higher than that with the wild-type promoters and thus differences in expression could be readily detected. In all cases in which we observed differences with the cry1A-272 promoter, the differences were confirmed with the wild type.
The upstream sequences were added in the correct orientation as 280- or 780-bp HindIII fragments to the cry1A-272 or cry1A wild-type promoters fused to lacZ (29, 32). The upstream region of the cry1A gene contains an inverted repeat (IR) and a potential bend sequence about 200 to 250 bp from the dual promoters (Fig. 1). These regions were selected previously for mutagenesis on the basis of the footprint of a binding protein (32). The mutations resulted in decreased binding of this protein, as well as altered kinetics of expression of lacZ fusions (32). Each mutant cry1A sequence was introduced as described above for the wild type.
The lacZ fusion plasmids were electroporated into the various B. thuringiensis strains (28) with selection on G-Tris plates containing 7 μg of chloramphenicol per ml. The presence of a functional lacZ gene was established by streaking preparations onto G-Tris plates containing chloramphenicol onto which 0.1 ml of 1% methylumbelliferyl-β-d-galactoside (Sigma) in 50% dimethylformamide had been spread and then examining the plates with a long-wavelength UV lamp.
In order to demonstrate that the lacZ fusion plasmids had not undergone any deletions or rearrangements, they were reisolated from B. thuringiensis transformants by the alkaline lysis procedure (10) and electroporated into the other B. thuringiensis strains. Consequently, β-galactosidase contents were confirmed by using subspecies which contained the same lacZ fusion plasmid. The lacZ fusion plasmids from B. thuringiensis were also transformed into Escherichia coli DH5α. These plasmids were digested with HindIII and BglII (29) in order to establish that no major deletions or rearrangements had occurred.
Duplicate samples of cells grown as described above were removed throughout sporulation at 90- to 120-min intervals until free spores were released. The samples were frozen at −80°C. The optical densities at 600 nm were also determined with a Perkin-Elmer junior model 35 spectrophotometer until the cells became extensively clumped at the end of growth (at least when they were grown on glucose). β-Galactosidase assays were performed with 30- to 50-μl aliquots (29) in duplicate, and the data obtained were converted to Miller units (22). The specific activities were expressed in Miller units per unit of optical density at 600 nm, and the maximum values are reported below. These values are the averages of the values from at least three independent experiments, and the coefficients of variance were less than ±10% in all cases.
Detection of protoxin antigens.
Spores plus inclusions were harvested (usually after 24 h) and washed, and the relative spore concentrations were determined by direct counting in a Petroff-Hauser chamber (in triplicate) and by determining the absorbance at 600 nm. The values agreed well, and equal quantities of spores (plus inclusions) were pelleted and then extracted (5). The inclusions were purified in Renografin gradients, and the protoxins were solubilized as described previously (5, 8).
To prepare cell extracts, B. thuringiensis subsp. kurstaki HD1 and B. thuringiensis subsp. aizawai HD133 were grown at 30°C in 80 ml of G-Tris until about 50 or 80% of the cells contained phase-white to phase-bright endospores. At each time point, 30 ml of cells was harvested, washed once with 10 ml of 1 M KCl–5 mM EDTA (pH 8.0) and twice with 10 ml of deionized water, and resuspended in 80 μl of 6 M urea–1% sodium dodecyl sulfate–5 mM dithiothreitol–2 mM phenylmethylsulfonyl fluoride (pH 9.6) (5). The suspensions were sonicated on ice twice for 40 s each time with a microtip and a Branson model 200 Sonifier. The suspensions were then placed in a boiling water bath for 2 min. The protein contents were determined by using 5-μl portions and the bicinchoninic acid reagent (Pierce Chemical Co.). The samples were first precipitated in 1 ml of 10% trichloroacetic acid, and the pellets were dissolved in 0.2 ml of 0.2 N NaOH. Equal quantities of protein were electrophoresed on sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis gels and transferred to polyvinylidene difluoride membranes for immunoblotting with a Cry1Ab monoclonal antibody plus a rabbit anti-mouse alkaline phosphatase conjugate or a Cry1Ac rabbit polyclonal antibody plus an anti-rabbit alkaline phosphatase conjugate (24).
RESULTS
Subspecies variation in cry gene expression.
Expression of the cry1A-lacZ fusion was three- to fourfold less in either of the two B. thuringiensis subsp. aizawai strains examined, strain 5 (derived from HD133), or HD112 than in B. thuringiensis subsp. kurstaki 80-21 or B. thuringiensis subsp. tolworthi HD124-12 when the organisms were grown in G-Tris supplemented with 0.1% glucose (Table 2). The lacZ fusion plasmid isolated from strain 5 had been used for transformation of strain HD124-12 and was also reintroduced into strain 80-21 in order to confirm the results. Similarly, the lacZ fusion plasmid from the original transformant of strain 80-21 was electroporated into strains 5, 80-21, HD124-12, and HD112, and the results were identical to those shown in Table 2. At the same time, these plasmids were transformed into E. coli DH5α in order to establish that there had been no major changes in the sizes of the plasmids or of the HindIII-BglII restriction fragments (29).
TABLE 2.
Expression of cry1-lacZ fusions in various B. thuringiensis subspecies
| lacZ fusion | Sp act of β-galactosidase ina:
|
|||
|---|---|---|---|---|
| Strain 80-21 | Strain 5 | Strain HD124-12 | Strain HD112 | |
| cry1Ab | 39.0 | 11.0 | 36.0 | 10.0 |
| cry1Cc | 30.0 | 26.0 | NDe | ND |
| Bendd | 31.0 | 32.0 | ND | 19.0 |
| IRd | 30.0 | 29.0 | ND | ND |
| cry1A promoters only | 8.0 | 7.5 | ND | ND |
Average values expressed as the maximum number of Miller units per unit of absorbance at 600 nm at the end of growth; the coefficients of variation were less than ± 10%. The values for strains 80-21 and 5 are the values obtained for the original transformants with plasmids isolated from E. coli. The lacZ fusion plasmid from strain 5 was electroporated into HD124-12. This plasmid and the other plasmids were also reintroduced into strain 80-21, and the plasmids from strain 80-21 were electroporated into strain 5.
A 280-bp region upstream of the promoters was ligated to the cry1A-272–lacZ fusion (29).
A 780-bp region upstream of the cry1C promoters was ligated to the cry1A-272–lacZ fusion.
The 280-bp cry1A upstream region was ligated to cry1A-272–lacZ, but the potential bend or IR region was mutated (Fig. 1) (32). The bend mutation (Fig. 1, boldface region) involved changes in 5′CTCAGTCTGTCTATGTAGAACAGGACAAGTG (changes in boldface type). The IR was mutated to 5′CCTGCAGTTAAGCCTGAATTGTAAATGC.
ND, not determined.
The differences observed with the cry1C-lacZ fusion were marginal (Table 2). This is a hybrid construct containing the cry1A-272 promoters plus the cry1C upstream region. Since the cry1A and cry1C genes have very similar dual overlapping promoter sequences (see above), such a construct should provide a valid assessment of the contribution of the upstream cry1C sequence to transcription in the various subspecies. The subspecies-specific responses of the cry1A and cry1C genes are likely to be due, therefore, to certain unique features of the upstream sequences. These sequences differ substantially in the cry1A (Fig. 1) and cry1C (30) genes.
Further evidence that these upstream sequences have a regulatory function included (i) the fact that the β-galactosidase specific activities for a fusion of only the cry1A promoters to lacZ in the absence of the upstream sequence were the same in strains 80-21 and 5 (Table 2) and (ii) the fact that mutations which substantially changed the potential bend region or the IR in the cry1A upstream sequence (Fig. 1) (33) resulted in somewhat lower specific activities in strain 80-21. However, the specific activities in strain 5 were the same as the specific activities in strain 80-21 and almost threefold higher than the specific activities in the wild type (Table 2). In the other B. thuringiensis subsp. aizawai strain, HD112, there was an approximately twofold increase in the specific activity due to the bend mutation. This strain had a different origin and perhaps a different cry gene composition than the other B. thuringiensis subsp. aizawai strain, HD133, and had not been cured of the plasmid containing the cry1Ab gene.
Measurements of cry1Ab mRNA.
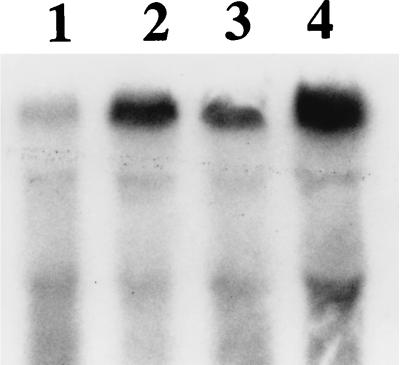
Total RNA from sporulating cells was fractionated in an agarose gel (27), transferred to nitrocellulose, and hybridized to a 32P-labeled oligonucleotide specific for the cry1Ab gene (Fig. 2). The relative contents of cry1Ab mRNA were 0.28:1.00 for HD133 and HD1 (Fig. 2, lanes 1 and 2) and 0.3:1.00 for HD112 and HD124 (lanes 3 and 4). The results obtained with RNA prepared from cells with >80% phase-dull to phase-white endospores are shown in Fig. 2, but the same differences were observed with RNA prepared at two earlier times during sporulation (see above). The differences in steady-state mRNAs were about the same as the differences found with lacZ fusions (Table 2), which confirmed that there were differences in transcription of this cry gene in the subspecies.
FIG. 2.
Northern blot of RNA (5 or 10 μg per lane) prepared from sporulating cells (>80% phase-dull to phase-white endospores) hybridized with 35 pmol of a 32P-labeled oligonucleotide specific for the cry1Ab gene (see Materials and Methods). Lane 1, 5 μg of RNA from B. thuringiensis subsp. aizawai HD133; lane 2, 5 μg of RNA from B. thuringiensis subsp. kurstaki HD1; lane 3, 10 μg of RNA from B. thuringiensis subsp. aizawai HD112; lane 4, 10 μg of RNA from B. thuringiensis subsp. tolworthi HD124.
Measurements of Cry1Ab antigen.
All of the subspecies which we studied contain several cry1 genes, including cry1Ab (Table 1). On the basis of the results obtained with the lacZ fusions, we anticipated that the amount of Cry1Ab antigen in B. thuringiensis subsp. aizawai inclusions should be less than the amount of Cry1Ab antigen in B. thuringiensis subsp. kurstaki or B. thuringiensis subsp. tolworthi inclusions. Protoxins were extracted from purified inclusions and electrophoresed for staining or immunoblotting (Fig. 3). The total amounts of inclusion protein in the three subspecies were about the same (Fig. 3A), but there was considerably less Cry1Ab antigen in B. thuringiensis subsp. aizawai inclusions than in the inclusions of the other organisms (Fig. 3B). A Cry1A polyclonal antibody which also cross-reacted with other Cry1 protoxins was used (Fig. 3C), and no major differences in the total protoxin antigen content were found.
FIG. 3.
Analysis of protoxins extracted from purified inclusions from B. thuringiensis subsp. kurstaki HD1 (lanes 1), B. thuringiensis subsp. aizawai HD133 (lanes 2), and (3) B. thuringiensis subsp. tolworthi HD124 (lanes 3). (A) Stained sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis gel with 2 μg of protein in each lane. The standards used (lane STD) were (from top to bottom) myosin (205 kDa), β-galactosidase (116 kDa), phosphorylase b (97.4 kDa), and bovine serum albumin (66 kDa). (B) Immunoblot obtained with a Cry1Ab monoclonal antibody containing the same amounts of protein. (C) Immunoblot obtained with a Cry1A polyclonal rabbit antibody.
The Cry1Ab antigen content of purified inclusions from B. thuringiensis subsp. aizawai HD133 (Fig. 3B) was considerably less than the content anticipated based on the measurements of β-galactosidase activity (Table 2) or mRNA (Fig. 2). There was some variability in the recovery of Cry1Ab antigen from inclusions, which was attributable to the instability of this protoxin (23). When protoxins were extracted from sporulating cells as well as from spore-inclusion mixtures immediately upon their release, the ratios of Cry1Ab antigen content for B. thuringiensis subsp. kurstaki HD1 and B. thuringiensis subsp. aizawai HD133 were 2.4:1 and 2.8:1 for the cell extracts and 3.0:1.0 (including the 130- and 60-kDa antigens) for the spore-inclusion mixtures (Fig. 4A). These ratios are consistent with the β-galactosidase and mRNA values obtained for these subspecies.
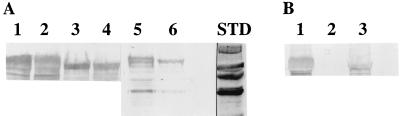
FIG. 4.
Immunoblots with a Cry1Ab monoclonal antibody of extracts from sporulating cells (lanes 1 to 4 in panel A) or spore-inclusion mixtures (lanes 5 and 6 in panel A; lanes 1 to 3 in panel B). (A) Extracts of sporulating cells of B. thuringiensis subsp. kurstaki HD1 (lanes 1 and 2) and B. thuringiensis subsp. aizawai HD133 (lanes 3 and 4) were prepared when either 30% of the cells (lanes 1 and 3) or 60% of the cells (lanes 2 and 4) contained phase-bright endospores. Equal quantities (50 μg) of protein in the extracts were electrophoresed. Lanes 5 and 6 contained extracts of washed spore-inclusion mixtures of each strain. (B) Extracts of spore-inclusion mixtures of strain 80-21 transformed with a clone of the cry1Ab gene (lane 1), strain 80-21 (lane 2), and strain 5 transformed with the same clone (lane 3). For all of the spore-inclusion mixture extractions, the same quantity of spores was used. Lane STD contained (from top to bottom) 200-, 116-, 90-, and 65-kDa standards. See Materials and Methods for details concerning strain construction and extraction procedures. The staining intensities were quantitated with a General Dynamics ImageQuant apparatus.
Differences in the amounts of Cry1Ab antigen may also be due to small differences in the sequences of the Cry1Ab protoxins in the two subspecies and thus in the extent of reactivity with the monoclonal antibody prepared against the B. thuringiensis subsp. kurstaki HD1 Cry1Ab protoxin. In order to examine this possibility, a clone of the cry1Ab gene from B. thuringiensis subsp. kurstaki HD1 (8) was introduced into strains 80-21 and 5. Protoxins were extracted from spore-inclusion mixtures, and the Cry1Ab antigen contents were determined with the monoclonal antibody (Fig. 4B). The ratio of the major reactive bands at 130 kDa was >3.0:1.0, which confirmed that the expression of this cry gene was subspecies dependent.
DISCUSSION
Subspecies-dependent differences in the expression of a cry1 gene were established with lacZ fusions and were confirmed by measuring cry1Ab mRNA and Cry1Ab protoxin antigen contents of sporulating cells and inclusion-spore extracts. In all cases, the cells were grown with glucose as the major carbon source. Similar differences were found when other carbon sources were used, although the absolute β-galactosidase specific activities differed (12).
The cry1Ab gene is present on a 40- to 50-kDa plasmid in all of the subspecies examined (2, 7). The copy numbers appear to be very similar based on hybridization in slot blots of total DNA with a cry1Ab-specific probe (2). In addition, differences in transcription were found with identical lacZ fusion plasmids, as well as with the same clone of the cry1Ab gene (Fig. 4B).
This subspecies-dependent regulation is attributable to the region upstream of the promoters since (i) there were no differences when only the promoters were fused to lacZ, (ii) fusion of the cry1C upstream region did not result in any difference, and (iii) the low level of transcription in strain 5 was enhanced by mutations in the potential bend and IR regions (Fig. 1) in the cry1A upstream sequence (Table 2). The segments used for mutagenesis were selected because a DNA binding protein identified as the E2 subunit of pyruvate dehydrogenase footprinted to these sites (32). There were decreases in the rates of β-galactosidase synthesis (as well as in the maximum specific activities, as shown in Table 2) in strains containing lacZ fusions with either the bend or the IR region mutated (32).
There were similar differences in steady-state cry1Ab mRNAs (Fig. 2), as well as in the relative accumulation of the Cry1Ab protoxin in sporulating cells, between B. thuringiensis subsp. kurstaki and B. thuringiensis subsp. aizawai (Fig. 4A). The amounts of this protoxin in spore-inclusion extracts and especially in purified inclusions were somewhat variable. When Cry1Ab is the only protoxin produced, it is unstable, but it is stabilized by disulfide cross-linking to other protoxins in an inclusion (2, 23). Perhaps this protoxin does not cross-link as well with the Cry1C and Cry1D protoxins in B. thuringiensis subsp. aizawai as it does with the more closely related Cry1Aa and Cry1Ac protoxins in B. thuringiensis subsp. kurstaki and thus is more unstable in the former subspecies.
It is known that media can influence protoxin accumulation (13), so there may be catabolic properties unique to each subspecies which account for the differences in expression of the cry1Ab gene. This regulation may involve the relative amount of soluble E2 present in sporulating cells of each subspecies. This protein binds to cry gene upstream regions, and it could regulate transcription (32). Alternatively, or in addition, the protoxin compositions of the various subspecies may be a factor due to the competition among the cry genes for limiting transcription components (such as ςE and ςK). This possibility is difficult to evaluate without information about the protoxin gene complement of each subspecies, the number of genes transcribed, and the extent of transcription of each gene (especially if the same sigma factors are used). Other unspecified subspecies differences may also influence the relative transcription of the genes. Whatever regulatory mechanism is involved, the gene-specific differences and the abilities of mutations in the cry1A upstream region to overcome low-level expression of the lacZ fusion in B. thuringiensis subsp. aizawai must be accounted for.
There are obvious practical implications for the differential expression of cry genes. Growth and sporulation conditions could affect the overall protoxin composition of inclusions and thus the toxicity profile. This regulation is likely to be significant to B. thuringiensis in its natural environment, a possibility which is worth exploring.
ACKNOWLEDGMENTS
This research was supported by grant MCB-9600584 from the National Science Foundation and by Abbott Laboratories. Ping Cheng was a visiting scientist supported by the Ministry of Agriculture, People’s Republic of China.
REFERENCES
- 1.Agaisse H, Lereclus D. How does Bacillus thuringiensis produce so much insecticidal crystal protein? J Bacteriol. 1995;177:6027–6032. doi: 10.1128/jb.177.21.6027-6032.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aronson A. The protoxin composition of Bacillus thuringiensis insecticidal inclusions affects solubility and toxicity. Appl Environ Microbiol. 1995;61:4057–4060. doi: 10.1128/aem.61.11.4057-4060.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aronson, A. Unpublished results.
- 4.Aronson A I, Angelo W, Holt S C. Regulation of extracellular protease production in Bacillus cereus T: characterization of mutants producing altered amounts of protease. J Bacteriol. 1971;106:1016–1025. doi: 10.1128/jb.106.3.1016-1025.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aronson A I, Han E-S, McGaughey W, Johnson D. The solubility of inclusion proteins from Bacillus thuringiensis is dependent upon protoxin composition and is a factor in toxicity to insects. Appl Environ Microbiol. 1991;57:981–986. doi: 10.1128/aem.57.4.981-986.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aronson A I. The two faces of Bacillus thuringiensis: insecticidal proteins and post-exponential survival. Mol Microbiol. 1993;7:489–496. doi: 10.1111/j.1365-2958.1993.tb01139.x. [DOI] [PubMed] [Google Scholar]
- 7.Aronson A I. Flexibility in the protoxin composition of Bacillus thuringiensis. FEMS Microbiol Lett. 1994;117:21–28. doi: 10.1111/j.1574-6968.1994.tb06737.x. [DOI] [PubMed] [Google Scholar]
- 8.Arvidson H, Dunn P E, Strand S, Aronson A I. Specificity of Bacillus thuringiensis for lepidopteran larvae: factors involved in vivo and in the structure of a purified protoxin. Mol Microbiol. 1989;3:1533–1543. doi: 10.1111/j.1365-2958.1989.tb00139.x. [DOI] [PubMed] [Google Scholar]
- 9.Baum J A, Malvar T. Regulation of insecticidal crystal protein production in Bacillus thuringiensis. Mol Microbiol. 1995;18:1–12. doi: 10.1111/j.1365-2958.1995.mmi_18010001.x. [DOI] [PubMed] [Google Scholar]
- 10.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carozzi N B, Kramer V C, Warren G W, Evola S, Koziel M G. Prediction of insecticidal activity of Bacillus thuringiensis strains by polymerase chain reaction product profiles. Appl Environ Microbiol. 1991;57:3057–3061. doi: 10.1128/aem.57.11.3057-3061.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng, P., and A. Aronson. Unpublished results.
- 13.Dulmage H T. Production of the spore-δ-endotoxin complex by variants of Bacillus thuringiensis in two fermentation media. J Invertebr Pathol. 1970;16:385–389. doi: 10.1016/0022-2011(70)90157-6. [DOI] [PubMed] [Google Scholar]
- 14.Geiser, M. Personal communication.
- 15.Hofte H, Whiteley H R. Insecticidal crystal proteins of Bacillus thuringiensis. Microbiol Rev. 1989;53:242–255. doi: 10.1128/mr.53.2.242-255.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaquet F, Hutter R, Luthy P. Specificity of Bacillus thuringiensis delta-endotoxins. Appl Environ Microbiol. 1987;53:500–504. doi: 10.1128/aem.53.3.500-504.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Juarez-Perez V M, Ferrandis M D, Frutos R. PCR-based approach for the detection of novel Bacillus thuringiensis crystal genes. Appl Environ Microbiol. 1997;63:2997–3002. doi: 10.1128/aem.63.8.2997-3002.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuo W-S, Chak K-F. Identification of novel cry-type genes from Bacillus thuringiensis strains on the basis of restriction fragment length polymorphism of the PCR-amplified DNA. Appl Environ Microbiol. 1996;62:1369–1377. doi: 10.1128/aem.62.4.1369-1377.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee M K, Curtiss A, Alacantra E, Dean D H. Synergistic effects of the Bacillus thuringiensis toxins Cry1Aa and Cry1Ac on the gypsy moth, Lymantria dispar. Appl Environ Microbiol. 1996;62:583–586. doi: 10.1128/aem.62.2.583-586.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masson L, Pre’fontaine G, Pe’loquin L, Lau P C K. Comparative analysis of the individual protoxin constituents in P1 crystals of Bacillus thuringiensis subsp. kurstaki isolates NRD12 and HD1. Biochem J. 1989;269:507–512. doi: 10.1042/bj2690507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masson L, Erlandson M, Puzstai-Carey M, Brousseau R, Juarez-Perez V, Frutos R. A holistic approach for determining the entomopathogenic potential of Bacillus thuringiensis strains. Appl Environ Microbiol. 1998;64:4782–4788. doi: 10.1128/aem.64.12.4782-4788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 23.Minnich S A, Aronson A I. Regulation of protoxin synthesis in Bacillus thuringiensis. J Bacteriol. 1984;158:447–454. doi: 10.1128/jb.158.2.447-454.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohammed S I, Johnson D E, Aronson A I. Altered binding of the Cry1Ac toxin to larval membranes but not to the toxin-binding protein in Plodia interpunctella selected for resistance to different Bacillus thuringiensis isolates. Appl Environ Microbiol. 1996;62:4168–4173. doi: 10.1128/aem.62.11.4168-4173.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moran C P., Jr . RNA polymerase and transcription factors. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C: American Society for Microbiology; 1993. pp. 653–668. [Google Scholar]
- 26.Prefontaine G, Fast P, Lau P C K, Hefford M A, Hanna Z, Brousseau R. Use of oligonucleotide probes to study the relatedness of delta-endotoxin genes among Bacillus thuringiensis subspecies and strains. Appl Environ Microbiol. 1987;53:2808–2814. doi: 10.1128/aem.53.12.2808-2814.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Schurter W, Geiser M, Mathé D. Efficient transformation of Bacillus thuringiensis and B. cereus via electroporation: transformation of acrystalliferous strains with a cloned δ-endotoxin gene. Mol Gen Genet. 1989;218:177–181. doi: 10.1007/BF00330581. [DOI] [PubMed] [Google Scholar]
- 29.Sedlak, M., T. Walter, and A. Aronson. The function of overlapping promoters in the regulation of Bacillus thuringiensis protoxin genes. Submitted for publication.
- 30.Smith G P, Ellar D J. Novel sequence elements associated with the cry1C gene from Bacillus thuringiensis subsp. aizawai. Nucleic Acids Res. 1993;16:6240. [Google Scholar]
- 31.Tabashnik B E. Evaluation of synergism among Bacillus thuringiensis toxins. Appl Environ Microbiol. 1992;58:334–346. doi: 10.1128/aem.58.10.3343-3346.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walter T, Aronson A. Specific binding of the E2 subunit of pyruvate dehydrogenase to the upstream regions of Bacillus thuringiensis protoxin genes. J Biol Chem. 1999;274:7901–7906. doi: 10.1074/jbc.274.12.7901. [DOI] [PubMed] [Google Scholar]
- 33.Walter T M. Regulation of cry gene transcription in Bacillus thuringiensis by a DNA binding protein which recognizes DNA elements upstream of the cry1Ab, cry1C and cry1D genes. PhD. thesis. West Lafayette, Ind: Purdue University; 1995. [Google Scholar]