Abstract
Purpose
Tulp2 (tubby‐like protein 2) is a member of the tubby protein family and expressed predominantly in mouse testis. Recently, it was reported that Tulp2 knockout (KO) mice exhibited disrupted sperm tail morphology; however, it remains to be determined how TULP2 deletion causes abnormal tail formation.
Methods
The authors analyzed male fertility, sperm morphology, and motility of two Tulp2 KO mouse lines that were generated using the conventional method that utilizes homologous recombination in embryonic stem (ES) cells as well as the clustered regularly interspaced short palindromic repeats/CRISPR‐associated protein 9 (CRISPR/Cas9) system. Furthermore, the authors observed the spermatogenesis of Tulp2 KO mice in more detail using scanning and transmission electron microscopy (SEM and TEM).
Results
Both mouse lines of Tulp2 KO exhibited male infertility, abnormal tail morphology, and impaired sperm motility. No overt abnormalities were found in the formation of the mitochondrial sheath in Tulp2 KO mice using the freeze‐fracture method with SEM. In contrast, abnormal outer dense fiber (ODF) structure was observed in Tulp2 KO testis with TEM.
Conclusions
TULP2 may play roles in the correct formation and/or maintenance of ODF, which may lead to abnormal tail morphology, impaired sperm motility, and male infertility.
Keywords: flagellum, outer dense fiber, sperm motility, spermiogenesis, tubby‐like protein 2
Observation of sperm morphology in the testis with scanning and transmission electron microscopy (SEM and TEM).
1. INTRODUCTION
Spermatozoa are highly specialized cells and consist of a head containing genetic information and a tail that moves vigorously to reach eggs. The sperm tail can be divided into three parts, midpiece, principal piece, and endpiece. 1 , 2 An axoneme, a motility apparatus that is composed of “9+2” microtubule arrangement, is found throughout the tail and each tail segment is classified according to the accessory structures surrounding the axoneme. The midpiece contains spirally arranged mitochondria called the mitochondrial sheath and outer dense fibers (ODFs) while the principal piece possesses the fibrous sheath and ODFs. The endpiece contains no accessory structures. Abnormal formation of these structures during spermatogenesis could lead to impaired sperm motility and male infertility.
Tubby family proteins are characterized by the tubby domain in the C‐terminus and are evolutionarily conserved in animals and plants. 3 , 4 In mammals, there are five tubby family proteins, TUB, TULP1, TULP2, TULP3, and TULP4. TUB was identified first among tubby family proteins, whose mutation results in obesity, retinal degeneration, and hearing loss in mice. 5 , 6 Tulp1 and Tulp3 knockout (KO) mice have been generated as well with Tulp1 KO mice exhibiting retinal degeneration 7 and Tulp3 KO mice being embryonic lethal. 8 Further analyses indicate that TULP3 functions as an adapter protein for ciliary trafficking of membrane‐associated proteins such as G protein‐coupled receptors 9 , 10 and nephron‐specific Tulp3 conditional KO mice exhibited cystic kidneys. 11 TULP1 is also suggested to be involved in intraflagellar transport in photoreceptor cells. 12
Unlike other TULP family proteins, Tulp2 is expressed predominantly in the mouse and human testes. 13 , 14 Tulp2 KO mice were generated recently and exhibited male infertility due to abnormal sperm tail formation. 14 In this study, we generated Tulp2 KO mice using not only the conventional method that utilizes homologous recombination in embryonic stem (ES) cells but also the clustered regularly interspaced short palindromic repeats/CRISPR‐associated protein 9 (CRISPR/Cas9) system and observed their defective tail formation in the testes using scanning and transmission electron microscopy (SEM and TEM).
2. MATERIALS AND METHOD
2.1. Animals
Mice were purchased from CLEA Japan, Inc. (Tokyo, Japan) or Japan SLC, Inc. Tulp2 KO mice generated in this study are being processed for deposition to the RIKEN BioResource Research Center, Ibaraki, Japan, or the Center for Animal Resources and Development (CARD), Kumamoto University, Kumamoto, Japan. All animal experiments were approved by the Institutional Animal Care and Use Committee of Osaka University (Osaka, Japan) (#Biken‐AP‐H30‐01).
2.2. Generation of Tulp2 KO mice using the conventional method
A 4.0‐kb NotI–XhoI fragment as a short arm and a 5.5‐kb AscI–MfeI fragment as a long arm were obtained by polymerase chain reaction (PCR) using genomic DNA derived from C57BL/6N mice as a template. The PCR primers used are shown in Supplementary Table S1. Both arms were inserted into a modified pNT1.1 vector (www.ncbi.nlm.nih.gov/nuccore/JN935771). The linearized targeting vector was electroporated into EGR‐G101 [(CAG/Acr‐EGFP) B6N × (CAG/Acr‐EGFP) B6N] or EGR‐G01 [129S2 × (cag/acr‐EGFP) B6] ES cells 15 and colonies were screened. The homologously recombined ES cells were injected into ICR eight‐cell embryos. The embryos were then incubated in a potassium simplex optimized medium (KSOM) 16 until the next day and transferred into the uterus of pseudopregnant females. The obtained chimeric mice from EGR‐G101 or EGR‐G01 ES cells were mated with C57BL/6N or B6D2F1 females, respectively, for germline transmission. One line was obtained from EGR‐G101 (C57BL/6N‐Tulp2<tm1Osb>/1B) and two lines were obtained from EGR‐G01 (STOCK‐Tulp2<tm1Osb>/3B and STOCK‐Tulp2<tm1Osb>/4A). STOCK‐Tulp2<tm1Osb>/4A line was used for the analyses. Primers used for genotyping are shown in Supplementary Table S1.
2.3. Generation of Tulp2 KO mice using the CRISPR/Cas9 system
Knockout mice were generated using the CRISPR/Cas9 system, as described previously. 17 The guide RNAs (gRNAs) with fewer off‐target sites were designed with the online software CRISPRdirect. 18 The gRNA target sequences are shown in Supplementary Table S1. KO mice were maintained on a B6D2 background. Primers used for genotyping are shown in Supplementary Table S1.
2.4. Fertility analysis of Tulp2 KO lines
Sexually mature wild‐type (WT) or Tulp2 KO male mice were caged individually with three seven‐week‐old female mice for at least five weeks. Male mice were removed after the mating period and females were kept for another three weeks to count the final litters. The numbers of pups and copulation plugs were counted every morning.
2.5. Analysis of morphology and motility of spermatozoa
Spermatozoa from cauda epididymis were suspended in Toyoda, Yokoyama, Hoshi (TYH) medium 19 and placed on MAS‐coated glass slides (Matsunami Glass) for morphology assessment using an Olympus BX53 microscope. For analyzing sperm motility, spermatozoa were incubated in TYH medium for 10 or 120 min and placed in glass chambers (Leja). Sperm motility was analyzed as previously described 20 using the CEROS II sperm analysis system (software version 1.5; Hamilton Thorne Biosciences).
2.6. Histological analysis of testes
Testes were fixed in Bouin's fluid (Polysciences, Inc.) overnight at 4°C, dehydrated by increasing ethanol concentrations, followed by xylene, and embedded in paraffin. Paraffin blocks were sectioned at 5‐μm thickness. The sections were rehydrated and treated with 1% periodic acid for 5 min at room temperature. The sections were then incubated with Schiff's reagent (FUJIFILM Wako Pure Chemical) for 10 min at room temperature, stained with Mayer's hematoxylin solution (FUJIFILM Wako Pure Chemical) for 1 min, and observed with a BX53 microscope (Olympus).
2.7. Ultrastructural analysis using transmission electron microscope (TEM)
Testis and cauda epididymis samples were prepared, as previously described. 21 The ultra‐thin sections were observed using a JEM‐1400plus electron microscope (JEOL) at 80 kV with a CCD Veleta 2K × 2K camera (Olympus).
2.8. Ultrastructural analysis using scanning electron microscope (SEM)
Testis samples were prepared with the freeze‐fracture method, as described previously. 21 , 22 The samples were observed with an S‐4800 field emission scanning electron microscope (FE‐SEM) (Hitachi).
2.9. Statistical analyses
Statistical differences were determined using Welch's t‐test by Microsoft Office Excel (Microsoft Corporation). Differences were considered statistically significant if the p values were <0.05 (*), 0.01 (**), or 0.001 (***). Data represent the mean ± standard deviation (SD).
3. RESULTS
3.1. Generation of Tulp2tm1 / tm1 mice and the results of fertility test
To investigate the function of Tulp2 in vivo, we generated KO mice (Tulp1tm1 / tm1 mice) using homologous recombination in ES cells and subsequent generation of chimeric mice. Exon 7 (188 bp) of Tulp2, the longest coding exon among exons whose nucleotide length is not divisible by three, was replaced by a neomycin resistance gene cassette (neo) (Figure 1A). Tulp2tm1 / tm1 mice were obtained by germline transmission and subsequent mating, which was confirmed by genomic PCR (Figure 1B) using primers shown in Figure 1A. Tulp2tm1 / tm1 mice were viable and showed no overt abnormalities.
FIGURE 1.
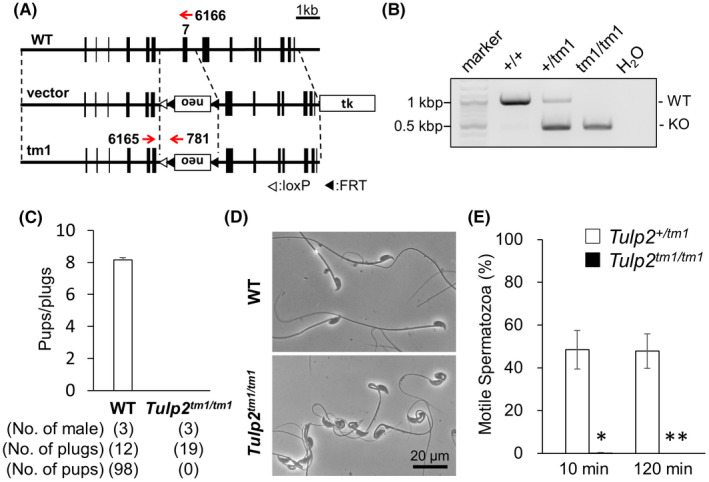
Generation and analyses of Tulp2tm1 / tm1 mice. (A) Targeting scheme for generating Tulp2tm1 / tm1 mice. Exon 7 was replaced with a neomycin resistance cassette (neo). Thymidine kinase (tk) was used for negative selection. (B) Genotyping of Tulp2tm1 / tm1 mice. Primers shown in A were used (#6165 and #6166 for wild type (WT); #6165 and #781 for knockout (KO). Water (H2O) was used as a negative control. (C) Number of pups born per plug detected. (D) Observation of spermatozoa obtained from cauda epididymis. (E) Sperm motility was analyzed 10 and 120 min after incubation in a capacitation medium. Number of males analyzed = 3 each
To analyze the fertility of Tulp2tm1 / tm1 males, the mice were caged with three WT females for more than five weeks. Although vaginal plugs were observed 19 times, no pups were obtained from Tulp2tm1 / tm1 males (Figure 1C), indicating that Tulp2 is essential for male fertility.
3.2. Analyses of sperm morphology and motility in Tulp2tm1 / tm1 mice
To analyze the cause of male infertility in Tulp2tm1 / tm1 males, we observed testis sections with a light microscope, but no apparent differences were found between WT and Tulp2tm1 / tm1 mice (Supplementary Figure S1). We then observed spermatozoa obtained from the cauda epididymis. In Tulp2tm1 / tm1 males, spermatozoa exhibited abnormal bending or coiling in the midpiece (Figure 1D). Furthermore, motile spermatozoa were rarely observed in Tulp2tm1 / tm1 males (Figure 1E) using a computer‐assisted sperm analysis (CASA) system. These results suggest that male infertility in Tulp2tm1 / tm1 mice is caused by abnormal sperm morphology and motility.
3.3. Generation and analyses of Tulp2em1 / em1 mice
It is possible that Tulp2tm1 / tm1 mice still generate functional TULP2 because of alteration in splicing patterns. To confirm that whole deletion of Tulp2 results in the same phenotypes as Tulp2tm1 / tm1 mice, we also generated KO mice (Tulp2em1 / em1 mice) using the CRISPR/Cas9 system that has advanced remarkably in recent years. 23 We designed two gRNAs to delete almost the entire open reading frame of Tulp2 (Figure 2A). Ribonucleoprotein complexes that contain the gRNA and CAS9 were electroporated into 85 pronuclear embryos and 60 two‐cell embryos were transferred into two pseudopregnant females the next day. Four out of nine pups had large deletions, and one male was caged with two WT females to obtain the F1 generation. Subsequent matings resulted in a KO mouse with an 8,420 bp deletion, which was confirmed by PCR (Figure 2B) and Sanger sequencing (Figure 2C). No overt abnormalities were found in the Tulp2em1 / em1 mice.
FIGURE 2.
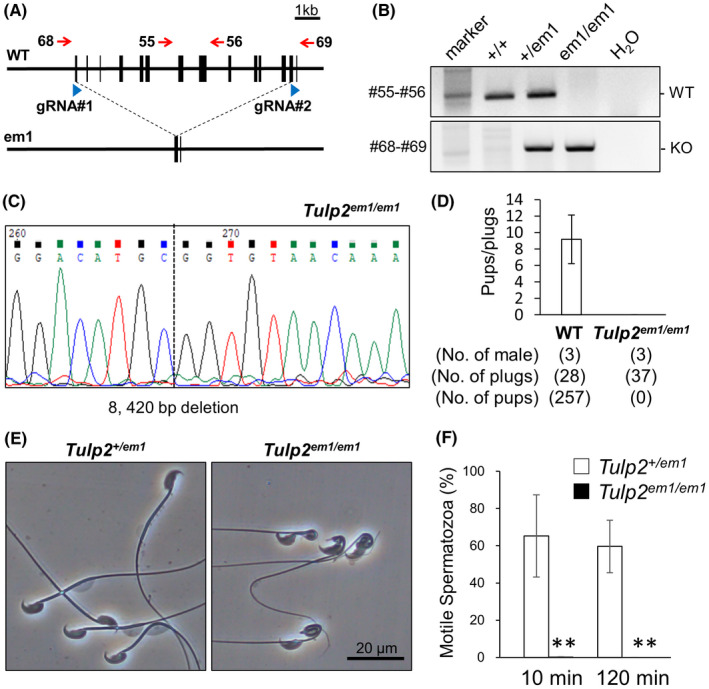
Generation and analyses of Tulp2em1 / em1 mice. (A) Clustered regularly interspaced short palindromic repeats/CRISPR‐associated protein 9 (CRISPR/Cas9) targeting scheme. Two guide RNAs (gRNAs) were designed to delete almost the entire open reading frame of Tulp2. (B) Genotyping of Tulp2em1 / em1 mice. Primers shown in A were used (#55 and #56 for wild type (WT); #68 and #69 for knockout (KO)). Water (H2O) was used as a negative control. (C) Wave pattern sequence of Tulp2em1 / em1 mice. (D) Number of pups born per plug detected. (E) Observation of spermatozoa obtained from cauda epididymis. (F) Sperm motility was analyzed 10 and 120 min after incubation in a capacitation medium. Number of males analyzed = 4 each
Tulp2em1 / em1 males were caged with three females for more than eight weeks. Although vaginal plugs were observed 37 times, no pups were obtained from Tulp2em1 / em1 males (Figure 2D). Furthermore, as consistent with Tulp2tm1 / tm1 males, spermatozoa of Tulp2em1 / em1 mice exhibited abnormal morphology in the midpiece (Figure 2E) and almost no motility with CASA (Figure 2F). These results confirm that Tulp2 is essential for sperm tail formation, motility, and male fertility.
3.4. Analyses of sperm morphology in the epididymis
To further analyze sperm morphological abnormalities in Tulp2tm1 / tm1 mice, we observed spermatozoa in the cauda epididymis using TEM. In the control, mitochondria were helically arranged in the midpiece (Figure 3A). In contrast, the midpiece was disorganized with abnormal mitochondria localization and multiple axonemes enclosed in one plasma membrane in Tulp2tm1 / tm1 mice (Figure 3A), which is consistent with coiling midpieces observed with light microscopy (Figures 1D and 2E).
FIGURE 3.
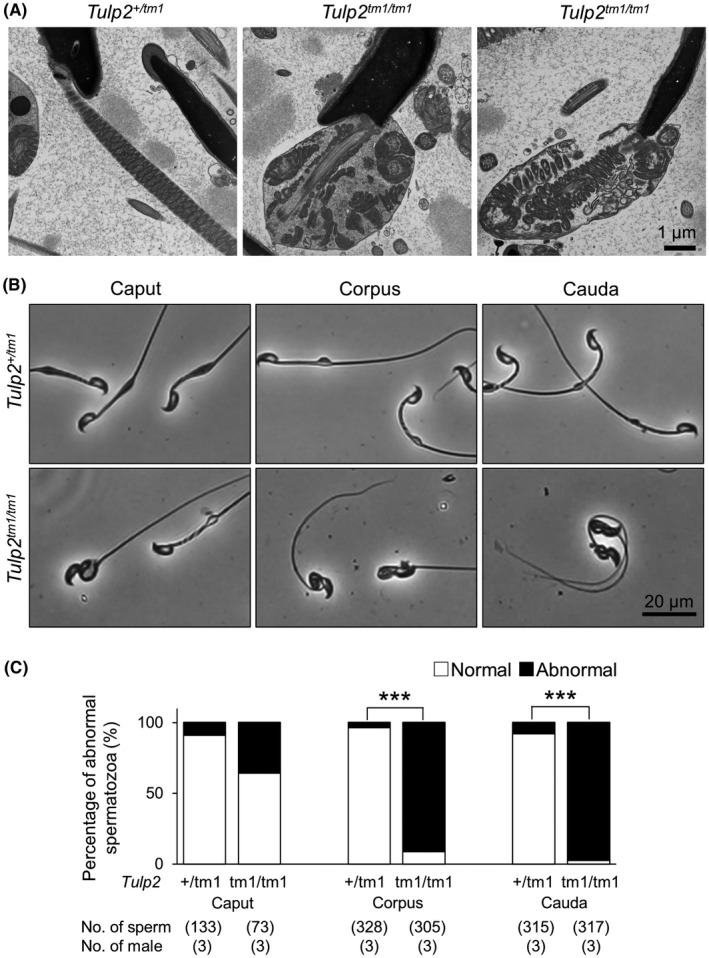
Observation of sperm morphology in the epididymis. (A) Observation of spermatozoa in the cauda epididymis with transmission electron microscopy (TEM). The midpiece was disrupted in Tulp2tm1 / tm1 mice. (B) Representative images of spermatozoa collected from three sections (caput, corpus, and cauda) of epididymis. (C) Percentages of abnormal spermatozoa collected from the three sections of epididymis
To analyze when abnormal midpiece bending or coiling emerged, we observed spermatozoa obtained from caput, corpus, or cauda epididymis. In Tulp2tm1 / tm1 mice, although almost all spermatozoa exhibited abnormal bending or coiling midpieces in both corpus and cauda epididymis (mean ± SD; corpus = 91.2 ± 5.6%, cauda = 97.5 ± 1.8%), fewer abnormal midpieces were observed in caput epididymis (mean ± SD; 35.6 ± 17.9%) (Figure 3B,C). These results indicate that more bending or coiling of the midpieces occurs during sperm transit in the epididymis, although some spermatozoa showed abnormal midpiece bending or coiling already in the caput epididymis.
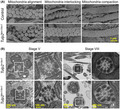
3.5. Analyses of sperm morphology in the testis
Because abnormal midpieces were already observed in the caput epididymis (Figure 3B,C), we analyzed mitochondrial sheath formation in testes using the freeze‐fracture method with SEM. 21 , 22 Unexpectedly, mitochondrial sheath formation (alignment, interlocking, and compaction steps) in Tulp2tm1 / tm1 mice was comparable to that of control mice (Figure 4A). We then observed spermiogenesis in testis using TEM (stage V and stage VIII). Consistent with the freeze‐fracture method, mitochondrial sheath formation is comparable between Tulp2 +/ tm1 and Tulp2tm1 / tm1 mice (Figure 4B). In contrast, we frequently observed an abnormal number of ODFs in Tulp2tm1 / tm1 mice (number of cross‐sections with extra ODFs; Tulp2 +/ tm1 = 0 out of 19 sections, Tulp2tm1 / tm1 = 14 out of 37 sections, all the cross‐sections with extra ODFs showed 10 ODFs, the number of males analyzed = 2 each) (Figure 4B).
FIGURE 4.
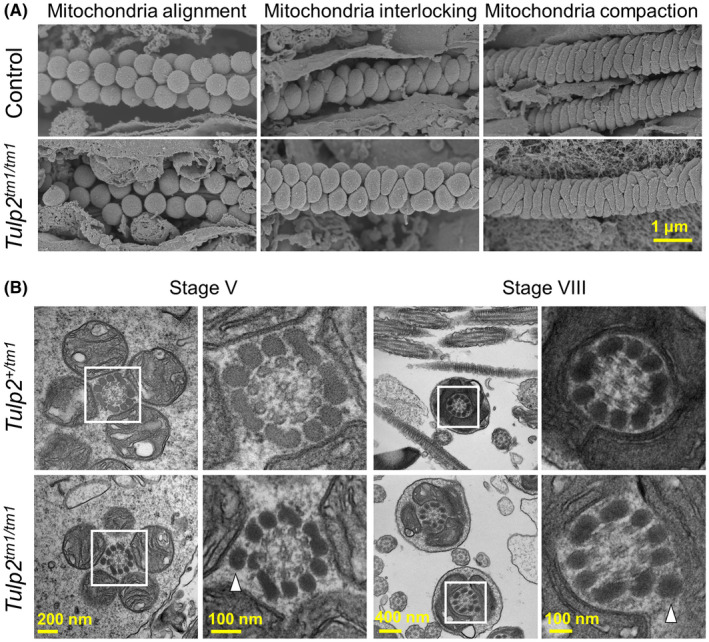
Observation of sperm morphology in the testis. (A) Formation of the mitochondrial sheath during spermatogenesis observed with scanning electron microscopy (SEM). Spherical mitochondria align around the axoneme (left), change their shape in the mitochondrial interlocking step (middle), and form the mitochondrial sheath in the mitochondrial compaction step (right). Wild‐type (WT) and Tulp2 +/ tm1 mice were used as controls. (B) Observation of spermatids with transmission electron microscopy (TEM). Abnormal extra outer dense fibers (ODFs) are highlighted with white arrowheads. Higher magnification images of the boxed areas are shown to the right
4. DISCUSSION
By generating two mouse lines of Tulp2, we confirmed that Tulp2 is essential for male fertility. Further analyses indicate that Tulp2 KO mice exhibited abnormal tail formation and impaired sperm motility. These results are consistent with those of the different mouse line of Tulp2 KO that was reported recently. 14 The tail structures of Tulp2tm1 / tm1 mice were more disorganized during sperm transit in the epididymis (Figure 3B,C). Because Tulp2 is not expressed in the epididymis, 14 it is unlikely that TULP2 deletion in the epididymis causes this abnormality. Rather, impaired tail formation in the testis may lead to this phenotype because some spermatozoa exhibited disrupted midpieces already in the caput epididymis (Figure 3B,C). Further morphological abnormalities of midpieces during the epididymal transit have also been reported in the KO mice of other genes such as Gk2 and Armc12. 21 , 22
Because the mitochondrial sheath was disrupted in the caput spermatozoa of Tulp2tm1 / tm1 mice (Figure 3B,C), we observed the formation of mitochondrial sheath during spermatogenesis using the freeze‐fracture method with SEM; however, no overt abnormalities were found in Tulp2tm1 / tm1 mice (Figure 4A). In contrast, we observed numbers of extra ODFs with TEM (Figure 4B). Because of this abnormal extra ODF, sperm tail structures including mitochondria sheath may be unstable and become disrupted during the later stages of spermatogenesis and sperm transit in the epididymis. Supporting this idea, extra ODFs and abnormal mitochondrial sheath structures were also observed in Cfap44 KO mice 24 and infertile patients with CFAP58 loss‐of‐function variants. 25 Because it is suggested that CFAP44 (cilia‐ and flagella‐associated protein 44) plays a role in the connection between ODFs and the axoneme 24 and CFAP58 (cilia‐ and flagella‐associated protein 58) influences ODF protein transportation, 25 TULP2 may be involved in these processes.
TULP1 and TULP3 are thought to be involved in intraflagellar transport in cilia. 9 , 10 , 11 , 12 Because sperm flagella possess similar axonemal structures to cilia, it is tempting to hypothesize that TULP2 plays roles in intraflagellar transport of molecules that are involved in the correct ODF formation. However, a recent study indicates that TULP2 could not rescue impaired intraflagellar transport completely in Tulp3 KO RPE‐1 cells although TUB could rescue the phenotype, 26 suggesting that TULP2 may not be involved in intraflagellar transport. However, it is still possible that TULP2 functions in intraflagellar transport of sperm flagella in a mechanism different from TULP3 in RPE‐1 cells. It is also possible that TULP2 functions as a transcription factor as suggested by the function of TUB 27 or functions as an RNA‐binding protein as recently suggested with the analyses of Tulp2 KO mice, 14 which impairment may lead to abnormal transcription and/or translation of molecules associated with ODFs. Although Cfap58 or Cfap44 was not listed, a recent study detected 1446 differentially expressed genes between WT and Tulp2 KO testes using RNA sequencing, some of which were related to the cytoskeleton. 14
In summary, we reveal that mitochondrial sheath formation is not disrupted, but ODF structure is abnormal in Tulp2 KO mice, which may lead to impaired sperm motility and male infertility. Further analysis of TULP2 may provide a better understanding of the mechanism that regulates the sperm tail formation and may lead to better treatment for male infertility.
CONFLICT OF INTEREST
Yuki Oyama, Haruhiko Miyata, Keisuke Shimada, Tamara Larasati, Yoshitaka Fujihara, and Masahito Ikawa declare that they have no conflict of interest.
HUMAN RIGHTS STATEMENTS AND INFORMED CONSENT
This article does not contain any studies with human subjects performed by any of the authors.
ANIMAL RIGHTS
All institutional and national guidelines for the care and use of laboratory animals were followed.
APPROVAL BY ETHICS COMMITTEE
All animal experiments performed in this study were approved by the Institutional Animal Care and Use Committee of Osaka University (Osaka, Japan) (#Biken‐AP‐H30‐01).
Supporting information
Supplementary Information
ACKNOWLEDGEMENTS
We thank the Department of Experimental Genome Research and Animal Resource Center for Infectious Diseases for creating a perfect environment for these experiments. We also thank Dr. Julio M. Castaneda for critical reading of the manuscript and Dr. Ko Miyoshi for constructive comments on the project. This work was supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT)/Japan Society for the Promotion of Science (JSPS) KAKENHI grants (JP21K19569 to H.M., JP20K16107 to K.S., JP20KK0155, JP21H02397, and JP21K19198 to Y.F., JP19H05750 and JP21H05033 to M.I.); Japan Agency for Medical Research and Development (AMED) grant JP21gm5010001 to M.I.; Takeda Science Foundation grants to H.M., K.S., Y.F., and M.I.; Senri Life Science Foundation grants to H.M. and Y.F.; Mochida Memorial Foundation for Medical and Pharmaceutical Research grant to Y.F.; The Sumitomo Foundation Grant for Basic Science Research Projects to Y.F.; Eunice Kennedy Shriver National Institute of Child Health and Human Development (grants R01HD088412 and P01HD087157 to M.I.); and the Bill & Melinda Gates Foundation (INV‐001902 to M.I.).
Oyama Y, Miyata H, Shimada K, Larasati T, Fujihara Y, Ikawa M. TULP2 deletion mice exhibit abnormal outer dense fiber structure and male infertility. Reprod Med Biol. 2022;21:e12467. doi: 10.1002/rmb2.12467
Contributor Information
Haruhiko Miyata, Email: hmiya003@biken.osaka-u.ac.jp.
Masahito Ikawa, Email: ikawa@biken.osaka-u.ac.jp.
REFERENCES
- 1. Eddy EM, Toshimori K, O'Brien DA. Fibrous sheath of mammalian spermatozoa. Microsc Res Tech. 2003;61(1):103‐115. [DOI] [PubMed] [Google Scholar]
- 2. Miyata H, Morohoshi A, Ikawa M. Analysis of the sperm flagellar axoneme using gene‐modified mice. Exp Anim. 2020;69(4):374‐381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mukhopadhyay S, Jackson PK. The tubby family proteins. Genome Biol. 2011;12(6):225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang M, Xu Z, Kong Y. The tubby‐like proteins kingdom in animals and plants. Gene. 2018;642:16‐25. [DOI] [PubMed] [Google Scholar]
- 5. Noben‐Trauth K, Naggert JK, North MA, Nishina PM. A candidate gene for the mouse mutation tubby. Nature. 1996;380(6574):534‐538. [DOI] [PubMed] [Google Scholar]
- 6. Kleyn PW, Fan W, Kovats SG, et al. Identification and characterization of the mouse obesity gene tubby: a member of a novel gene family. Cell. 1996;85(2):281‐290. [DOI] [PubMed] [Google Scholar]
- 7. Ikeda S, Shiva N, Ikeda A, et al. Retinal degeneration but not obesity is observed in null mutants of the tubby‐like protein 1 gene. Hum Mol Genet. 2000;9(2):155‐163. [DOI] [PubMed] [Google Scholar]
- 8. Ikeda A, Ikeda S, Gridley T, Nishina PM, Naggert JK. Neural tube defects and neuroepithelial cell death in Tulp3 knockout mice. Hum Mol Genet. 2001;10(12):1325‐1334. [DOI] [PubMed] [Google Scholar]
- 9. Badgandi HB, Hwang SH, Shimada IS, Loriot E, Mukhopadhyay S. Tubby family proteins are adapters for ciliary trafficking of integral membrane proteins. J Cell Biol. 2017;216(3):743‐760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Han S, Miyoshi K, Shikada S, et al. TULP3 is required for localization of membrane‐associated proteins ARL13B and INPP5E to primary cilia. Biochem Biophys Res Commun. 2019;509(1):227‐234. [DOI] [PubMed] [Google Scholar]
- 11. Hwang SH, Somatilaka BN, Badgandi H, et al. Tulp3 regulates renal cystogenesis by trafficking of cystoproteins to Cilia. Curr Biol. 2019;29(5):790‐802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hagstrom SA, Adamian M, Scimeca M, Pawlyk BS, Yue GH, Li TS. A role for the tubby‐like protein 1 in rhodopsin transport. Invest Ophth Vis Sci. 2001;42(9):1955‐1962. [PubMed] [Google Scholar]
- 13. North MA, Naggert JK, Yan YZ, NobenTrauth K, Nishina PM. Molecular characterization of TUB, TULP1, and TULP2, members of the novel tubby gene family and their possible relation to ocular diseases. Proc Natl Acad Sci USA. 1997;94(7):3128‐3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zheng MM, Chen X, Cui YQ, et al. TULP2, a New RNA‐binding protein, is required for mouse spermatid differentiation and male fertility. Front Cell Dev Biol. 2021;9:623738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fujihara Y, Kaseda K, Inoue N, Ikawa M, Okabe M. Production of mouse pups from germline transmission‐failed knockout chimeras. Transgenic Res. 2013;22(1):195‐200. [DOI] [PubMed] [Google Scholar]
- 16. Ho YG, Wigglesworth K, Eppig JJ, Schultz RM. Preimplantation development of mouse embryos in KSOM ‐ augmentation by amino‐acids and analysis of gene‐expression. Mol Reprod Dev. 1995;41(2):232‐238. [DOI] [PubMed] [Google Scholar]
- 17. Abbasi F, Miyata H, Shimada K, et al. RSPH6A is required for sperm flagellum formation and male fertility in mice. J Cell Sci. 2018;131(19):jcs221648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Naito Y, Hino K, Bono H, Ui‐Tei K. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off‐target sites. Bioinformatics. 2015;31(7):1120‐1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Muro Y, Hasuwa H, Isotani A, et al. Behavior of mouse spermatozoa in the female reproductive tract from soon after mating to the beginning of fertilization. Biol Reprod. 2016;94(4):80. [DOI] [PubMed] [Google Scholar]
- 20. Miyata H, Oura S, Morohoshi A, et al. SPATA33 localizes calcineurin to the mitochondria and regulates sperm motility in mice. Proc Natl Acad Sci USA. 2021;118(35):e2106673118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shimada K, Kato H, Miyata H, Ikawa M. Glycerol kinase 2 is essential for proper arrangement of crescent‐like mitochondria to form the mitochondrial sheath during mouse spermatogenesis. J Reprod Dev. 2019;65(2):155‐162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shimada K, Park S, Miyata H, et al. ARMC12 regulates spatiotemporal mitochondrial dynamics during spermiogenesis and is required for male fertility. Proc Natl Acad Sci USA. 2021;118(6):e2018355118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Abbasi F, Miyata H, Ikawa M. Revolutionizing male fertility factor research in mice by using the genome editing tool CRISPR/Cas9. Reprod Med Biol. 2018;17(1):3‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Coutton C, Vargas AS, Amiri‐Yekta A, et al. Mutations in CFAP43 and CFAP44 cause male infertility and flagellum defects in Trypanosoma and human. Nat Commun. 2018;9(1):686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. He XJ, Liu CY, Yang XY, et al. Bi‐allelic loss‐of‐function variants in CFAP58 cause flagellar axoneme and mitochondrial sheath defects and asthenoteratozoospermia in humans and mice. Am J Hum Genet. 2020;107(3):514‐526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hong JJ, Kim KE, Park SY, Bok J, Seo JT, Moon SJ. Differential roles of tubby family proteins in ciliary formation and trafficking. Mol Cells. 2021;44(8):591‐601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Santagata S, Boggon TJ, Baird CL, et al. G‐protein signaling through tubby proteins. Science. 2001;292(5524):2041‐2050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


