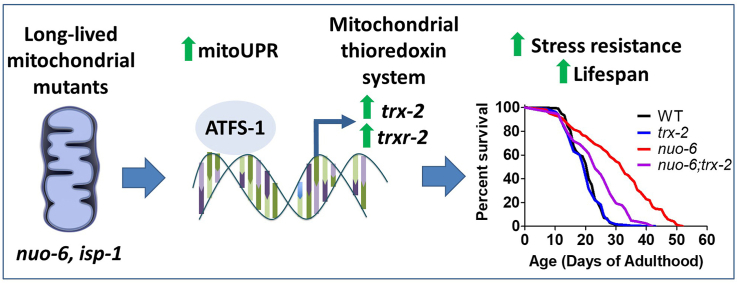
Abstract
Mild impairment of mitochondrial function has been shown to increase lifespan in genetic model organisms including worms, flies and mice. To better understand the mechanisms involved, we analyzed RNA sequencing data and found that genes involved in the mitochondrial thioredoxin system, trx-2 and trxr-2, are specifically upregulated in long-lived mitochondrial mutants but not other non-mitochondrial, long-lived mutants. Upregulation of trx-2 and trxr-2 is mediated by activation of the mitochondrial unfolded protein response (mitoUPR). While we decided to focus on the genes of the mitochondrial thioredoxin system for this paper, we identified multiple other antioxidant genes that are upregulated by the mitoUPR in the long-lived mitochondrial mutants including sod-3, prdx-3, gpx-6, gpx-7, gpx-8 and glrx-5. In exploring the role of the mitochondrial thioredoxin system in the long-lived mitochondrial mutants, nuo-6 and isp-1, we found that disruption of either trx-2 or trxr-2 significantly decreases their long lifespan, but has no effect on wild-type lifespan, indicating that the mitochondrial thioredoxin system is specifically required for their longevity. In contrast, disruption of the cytoplasmic thioredoxin gene trx-1 decreases lifespan in nuo-6, isp-1 and wild-type worms, indicating a non-specific detrimental effect on longevity. Disruption of trx-2 or trxr-2 also decreases the enhanced resistance to stress in nuo-6 and isp-1 worms, indicating a role for the mitochondrial thioredoxin system in protecting against exogenous stressors. Overall, this work demonstrates an important role for the mitochondrial thioredoxin system in both stress resistance and lifespan resulting from mild impairment of mitochondrial function.
Keywords: Aging, Mitochondria, Reactive oxygen species, C. elegans, Antioxidant, Thioredoxin
Graphical abstract
Highlights
-
•
Mitochondrial thioredoxin system genes are specifically upregulated in long-lived mitochondrial mutants.
-
•
Upregulation of mitochondrial thioredoxin genes is mediated by mitochondrial unfolded protein response.
-
•
Mitochondria thioredoxin genes contribute to lifespan extension in long-lived mitochondrial mutants.
-
•
Mitochondrial thioredoxin genes are required for enhanced resistance to oxidative and other stresses.
-
•
Disruption of the cytoplasmic thioredoxin gene has a general detrimental effect on lifespan and stress resistance.
1. Introduction
Aging may be described as a progressive decline in function over time that increases the probability of death. While the discovery of genetic mutations that can extend lifespan in model organisms [[1], [2], [3]] and the identification of genes associated with longevity in humans [4] has advanced our understanding of the aging process, the molecular mechanisms involved remain incompletely understood. The Free Radical Theory of Aging (FRTA) proposes that aging is caused by the accumulation of oxidative damage caused by reactive oxygen species (ROS) that are generated by normal metabolism [5]. While genetic mutations and interventions that increase ROS levels have been shown to decrease lifespan, there are also multiple examples in which increasing ROS levels extends longevity [6].
Mild impairment of mitochondrial function has been shown to increase lifespan in C. elegans [7,8] as well as other model organisms including flies [9] and mice [10,11]. The genetic mutants nuo-6 and isp-1 have point mutations that impair subunits of Complex I and Complex III of the mitochondrial electron transport chain, respectively, leading to increased lifespan [12,13]. These mutants have increased levels of ROS, which are required for their longevity as treatment with antioxidants partially or fully reverts their lifespan to wild-type [14]. While incompletely understood, the mechanism of lifespan extension in these long-lived mitochondrial mutants appears to involve the upregulation of stress response pathways including the DAF-16-mediated stress response [15], the mitochondrial unfolded protein response (mitoUPR) [16], innate immune signaling [17] and the HIF-1-mediated hypoxia response [18], as well as apoptotic signaling [19].
Among the genes that are upregulated by mild mitochondrial impairment in isp-1 worms are antioxidant genes, including genes encoding antioxidant enzymes in the thioredoxin system [20]. Thioredoxin is an oxidoreductase that protects against oxidative stress by reducing disulfide bonds that have formed by ROS, thereby restoring protein function [21,22]. In addition to reducing oxidized proteins, thioredoxin also acts indirectly to decrease the levels of ROS by reducing peroxiredoxin in order to return it to its active state after peroxiredoxin has detoxified hydrogen peroxide or organic peroxides. Once thioredoxin has reduced its target protein, thioredoxin reductase reduces thioredoxin back to its active state using electrons from NADPH. Interestingly, thioredoxins have also been shown to have redox-independent functions. For example, modifying TRX-1 to disrupt its redox activity does not prevent TRX-1 from acting as a chaperone protein [23,24], promoting degradation of the apoptosis-regulating kinase ASK1 [25], facilitating dauer formation in daf-28 mutants [26], mediating avoidance of bacterial pathogens [27], or allowing for the nuclear localization of the oxidative-stress responsive transcription factor SKN-1 and subsequent upregulation of its target gene lips-6 [26].
C. elegans has at least five known thioredoxins (TRX-1, TRX-2, TRX-3, TRX-4 and TRX-5) and two thioredoxin reductases (TRXR-1 and TRXR-2). TRX-1 and TRXR-1 form the cytoplasmic thioredoxin system [28,29], while TRX-2 and TRXR-2 form the mitochondrial thioredoxin system [30]. Expression of TRX-3 is limited to the intestine [31], while the roles of TRX-4 and TRX-5 are not well studied.
In this work we characterize the role of the mitochondrial thioredoxin system in long-lived mitochondrial mutants. We find that trx-2 and trxr-2 are specifically upregulated in nuo-6 and isp-1 mutants, but not other non-mitochondrial, long-lived mutants, through activation of the mitoUPR. Disruption of either trx-2 or trxr-2 significantly decreases the long lifespan of these mitochondrial mutants but has little effect on wild-type worms. In contrast, disrupting the cytoplasmic thioredoxin system decreases lifespan in all strains tested suggesting a non-specific detrimental effect on longevity. Finally, we find that the mitochondrial thioredoxin system is required not only for enhanced resistance to oxidative stress in nuo-6 and isp-1 worms but also for resistance to multiple other external stressors. Overall, this work demonstrates the importance of the mitochondrial thioredoxin system for the long lifespan and enhanced stress resistance resulting from mild mitochondrial impairment.
2. Materials and methods
2.1. Strains
The following strains were used in this research: N2 (WT), trx-1 (ok1449), trxr-1 (sv47), trx-2 (tm2720), trxr-2 (ok2267), isp-1 (qm150), nuo-6 (qm200), isp-1;trx-1, isp-1;trx-2, isp-1;trxr-2, nuo-6;trx-1, nuo-6;trxr-1, nuo-6;trx-2, nuo-6;trxr-2. All strains were kept at 20 °C on nematode growth medium (NGM) plates seeded with OP50 bacteria. Double mutants were generated by standard crossing techniques and confirmed by PCR genotyping for deletion mutations and DNA sequencing for point mutations. We were unable to generate isp-1 trxr-1 worms due to close proximity on the same chromosome.
2.2. Lifespan assay
Pre-fertile young adult worms of each strain were transferred to NGM plates containing 25 μM FUdR (5-fluoro-2′-deoxyuridine) to limit the development of progeny [32]. Plates were incubated at 20 °C and survival was monitored every two to three days. Worms were transferred to new plates after 5 days and once per week thereafter. Worms exhibiting internal hatching or externalization of internal organs were removed from the assay. A minimum of three biological replicates of 50 worms per strains were completed.
2.3. Chronic oxidative stress assay
Resistance to chronic oxidative stress was assessed by exposing worms to 4 mM paraquat (Methyl viologen dichloride hydrate) (see Ref. [33] for full protocol). 100 μM FUdR was included in the plates to prevent internal hatching that is caused by exposure to paraquat. 4 mM paraquat plates were seeded with 10X concentrated OP50 bacteria. Young adult worms were transferred to the plates and survival was monitored every two to three days. The worms were transferred to fresh paraquat plates every week. Three biological replicates of 25 worms per strain were completed.
2.4. Heat stress assay
NGM plates were freshly seeded with OP50 bacteria. Once dried, 25 young adult worms per strain were transferred to the plates and incubated at 37 °C. Survival was monitored every 2 h for a total of 8 h. Three biological replicates of 25 worms per strain were completed.
2.5. Osmotic stress assay
NGM plates were prepared with either 450 mM or 500 mM NaCl. The next day plates were seeded with OP50 bacteria. The following day plates were used for the assay or transferred to 15 °C degrees in a sealed container to minimize evaporation and then utilized the next day. The osmotic stress assay was initiated by transferring at least 21 young adult worms to the plates containing 450 mM or 500 mM NaCl and incubated at 20 °C for 48 h before scoring survival. Worms with internal hatching were censored and excluded from the total number of deaths. A total of five biological replicates were completed.
2.6. Bacterial pathogen stress assay
The Pseudomonas aeruginosa strain PA14 was used to assess bacterial pathogen stress resistance of worms as described previously [17,34]. At least 40 L4 worms of each tested genotype were placed on 35 mm NGM plates containing 100 mg/L (406 μM) FUdR and seeded with 150 μL of OP50 bacteria. Worms were allowed to grow at 20 °C for 72 h. OP50 was grown at 37 °C, 20% culture volume at 200 RPM shaker speed for 12 h. PA14 was streaked to LB plates from a −80 °C storage vial. After growing for 24 h at 37 °C, a single colony of PA14 was placed into LB broth. Next, the culture was grown at 37 °C, 10% culture volume and 200 RPM shaker speed for 16 h 5 μL of PA14 culture was pipetted onto 35 mm NGM plates containing 20 mg/L (81 μM) FUdR. The culture was spread using a glass L-shaped spreader to cover 20–40% of the plate surface area. Plates were allowed to dry near a Bunsen burner flame for 20 min, and were then incubated at 37 °C for 24 h. After 24 h, plates were transferred to 20 °C for 24 h. Adult day 3 worms from the 100 mg/L FUdR plates were transferred to PA14 plates and kept at 20 °C for the duration of the assay. Survival was monitored every other day and dead worms were scored as non-responsive to gentle tap to the worm pick. Dried worms on Petri dish walls and worms with internal hatching and excreted vulvas were censored and excluded from the study. A total of six biological replicates were completed.
2.7. Use of FUdR
FUdR was utilized in three assays: lifespan, chronic oxidative stress (paraquat) and bacterial pathogen stress (PA14). For each of these assays, the concentration of FUdR used was established in previously published work. The same FUdR concentrations were used in these experiments so that the results in this paper could be accurately compared to previously published findings. For the lifespan assay, we used 25 μM FUdR. This concentration has no effect on the lifespan of wild-type worms and most mutants. For those mutants whose lifespan is affected by FUdR, 25 μM FUdR has only a minimal effect on longevity [32]. Excluding FUdR from lifespan studies results in an increased incidence of internal hatching, and that the percentage of worms undergoing internal hatching is increased in specific genetic mutants. We have found that using 25 μM FUdR provides the optimal balance of minimizing the effect of FUdR on lifespan, while also preventing internal hatching. For the chronic oxidative stress assay involving exposure to 4 mM paraquat we used 100 μM FUdR. Performing this assay without FUdR is not feasible because the majority of worms undergo internal hatching. Finally, for the bacterial pathogenesis assay, we used 20 mg/L FUdR according to a protocol developed in Dr. Keith Blackwell's laboratory [17,34]. Since different concentrations of FUdR were used for different assays, it is possible that the FUdR differentially affected stress resistance and longevity across the different mutants that we examined. Based on our previous experience, the concentration of FUdR used for the lifespan experiment is very unlikely to have had an effect, while the concentrations of FUdR used for the paraquat assay and bacterial pathogen assay are necessary to prevent internal hatching.
2.8. Quantification of mRNA levels by RNA sequencing
RNA sequencing was performed previously [15,16,35]. Raw RNA-seq data is available on NCBI GEO: GSE179825 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE179825.
GSE110984https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE110984.
GSE93724https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE93724.
Raw RNA-seq data was reanalyzed for these experiments with help from the Harvard Chan Bioinformatics as previously described [17,35].
2.9. Quantification of ROS levels
ROS levels were measured using dihydroethidium (DHE; ThermoFisher Scientific, D1168), as previously described [20]. A 30 mM solution of DHE in DMSO was aliquoted and stored at −80 °C. When needed, 5 μl of DHE stock was diluted in 5 mL of PBS to create a 30 μM DHE solution. Age matched day 1 adult worms (approximately 50) were collected in PBS, transferred to a 1.5 mL centrifuge tube and washed 3 times in 1 mL PBS. To immerse worms in a final concentration of 15 μM DHE, 100 μL of PBS was left in the centrifuge tube after the final wash and 100 μL of 30 μM DHE was added to the tube. Centrifuge tubes were wrapped in tinfoil to protect from light and worms were incubated at room temperature, on a shaker for 1 h, then washed 3 times in PBS. For imaging, worms were mounted on a 2% agarose pad and immobilized with 10 mM levamisole. Worms were imaged with the 20 × objective using a Zeiss LSM 780 confocal microscope. A total of 60 worms were imaged over 3 biological replicates, per genotype. Image J was used to quantify the fluorescence intensity of ethidium-labelled ROS in the whole body of the worm. For each image (one worm per image), mean fluorescence intensity of the whole image and background intensity were measured. For each strain, autofluorescence was sampled from approximately 5-10 untreated worms and mean autofluorescence for the strain was determined. Fluorescence intensity of each worm was calculated by subtracting the image's background fluorescence and the strain's mean autofluorescence from the mean fluorescence of the image. As DHE can be oxidized by one electron oxidants, including cytrochromes, it is possible that altered cytochrome content in the mitochondrial mutants affects DHE fluorescence in these worms. Although fluorescence was quantified throughout the entire whole worm, DHE staining resulted in fluorescence primarily in the intestine, which is known to exhibit high levels of dye uptake. The intestinal fluorescence was punctate in appearance, which has been proposed to result from staining of lysosome-related gut granules [36].
2.10. Statistical analysis
All assays were performed with a minimum of three biological replicates. Experimenters were blinded with respect to the genotype of the strains. Within each strain, worms were randomly chosen for inclusion in each assay. Statistical significance was assessed using log-rank test, one-way ANOVA or two-way ANOVA as indicated in the figure legends. Error bars indicate standard error of the mean (SEM).
3. Results
3.1. Mitochondrial thioredoxin system is specifically upregulated in long-lived mitochondrial mutants
Although isp-1 and nuo-6 worms have increased levels of ROS [14], we have previously shown that both of these long-lived mitochondrial mutants have increased resistance to oxidative stress [16,20,37]. Based on the hypothesis that the enhanced resistance to oxidative stress resulted from a stress-induced upregulation of antioxidant genes, we used RNA sequencing (RNA-seq) to examine gene expression in isp-1 mutants. We previously found that a pooled sample containing six biological replicates exhibited higher levels of mRNA for multiple antioxidant genes, including genes from the mitochondrial thioredoxin system [20].
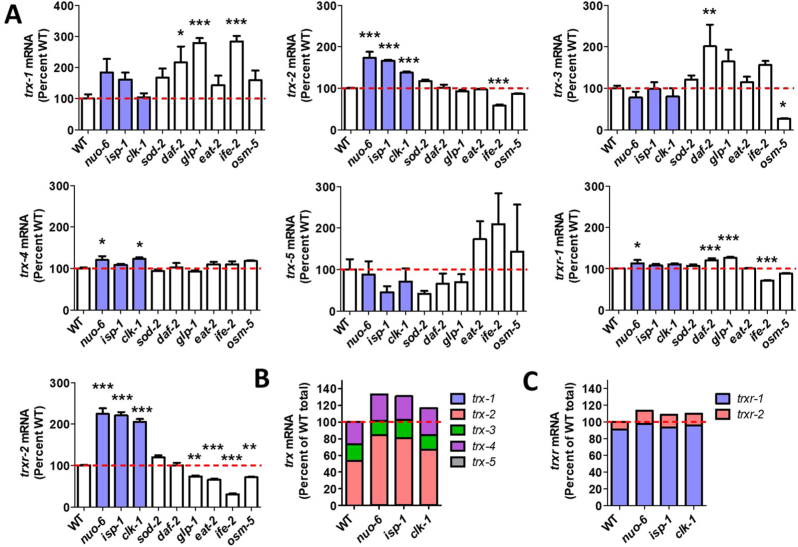
To determine if the increase in expression of mitochondrial thioredoxin genes trx-2 and trxr-2 in isp-1 worms is significant, we analyzed data from a separate RNA-seq experiment containing six biological replicates that were sequenced individually. To determine if the increase is specific to the mitochondrial thioredoxin system, we also examined the expression of non-mitochondrial thioredoxin (trx) and thioredoxin reductase (trxr) genes. We found that trx-2 and trxr-2 have significantly increased expression in isp-1 worms, while all other trx and trxr genes have expression that is equivalent to wild-type (Fig. 1A).
Fig. 1.
Mitochondrial thioredoxin and thioredoxin reductase genes are specifically upregulated in long-lived mitochondrial mutants. To examine the role of thioredoxin and thioredoxin reductase in the longevity of long-lived mutants, we quantified gene expression from RNA sequencing data of six biological replicates per strain. Expression of the mitochondrial thioredoxin system was specifically upregulated in the long-lived mitochondrial mutants nuo-6, isp-1 and clk-1, but not in other non-mitochondrial, long-lived strains (A). Blue bars indicate long-lived mitochondrial mutants. Thioredoxin mRNA expression in long-lived mitochondrial mutants compared to wild-type (B). Thioredoxin reductase mRNA expression in long-lived mitochondrial mutants compared to wild-type (C). Statistical significance was assessed using a one-way ANOVA with Dunnett's multiple comparison test. Error bars indicate SEM. *p < 0.05, **p < 0.01, ***p < 0.001. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Next, we sought to determine if upregulation of the mitochondrial thioredoxin system is specific to isp-1 worms or whether these genes show increased expression in other long-lived mutants. For this purpose, we examined trx and trxr expression in two additional long-lived mitochondrial mutants, clk-1 [7] and nuo-6 [13], as well as six other non-mitochondrial, long-lived mutants from different pathways of lifespan extension, including sod-2 mutants [38]; daf-2 mutants [39]; glp-1 mutants [40]; ife-2 mutants [41,42]; osm-5 mutants [43]; and eat-2 mutants [44]. We found that only the long-lived mitochondrial mutants, clk-1, nuo-6 and isp-1 have significant upregulation of trx-2 and trxr-2 (Fig. 1A). sod-2 mutants, in which the mitochondrial superoxide dismutase gene is disrupted, also exhibit a trend towards increased expression of both trx-2 (17% increase) and trxr-2 (20% increase), which failed to meet significance using a one-way ANOVA and Dunnett's multiple comparison test to compare wild-type to all nine long-lived mutants but is significant using a t-test. The increased expression of trx-2 and trxr-2 in the long-lived mitochondrial mutants results in an overall increase in trx and trxr mRNAs in the long-lived mitochondrial mutants (Fig. 1B and C).
Having shown that genes from the mitochondrial thioredoxin system are specifically upregulated in long-lived mitochondrial mutants, we wondered whether other antioxidant genes exhibit this same pattern, especially those that are localized to the mitochondria. Examining the expression of superoxide dismutase (sod), catalase (ctl), peroxiredoxin (prdx), glutathione peroxidase (gpx) and glutaredoxin (glrx) genes, we found that prdx-3 is also specifically upregulated in clk-1, nuo-6 and isp-1 worms but not in other non-mitochondrial, long-lived mutants (Fig. S1). Like trx-2 and trxr-2, prdx-3 is localized to the mitochondria [45]. gpx-6 was also significantly upregulated in all three mitochondrial mutants but was also upregulated in daf-2 worms (Fig. S1). In addition, four other mitochondrial genes, sod-2, sod-3, gpx-8 and glrx-5 are all significantly upregulated in nuo-6 and isp-1 worms with a trend towards increase in clk-1 worms, which fails to meet significance when correcting for multiple comparisons (Fig. S1).
3.2. Upregulation of mitochondrial thioredoxin system is mediated by activation of the mitochondrial unfolded protein response
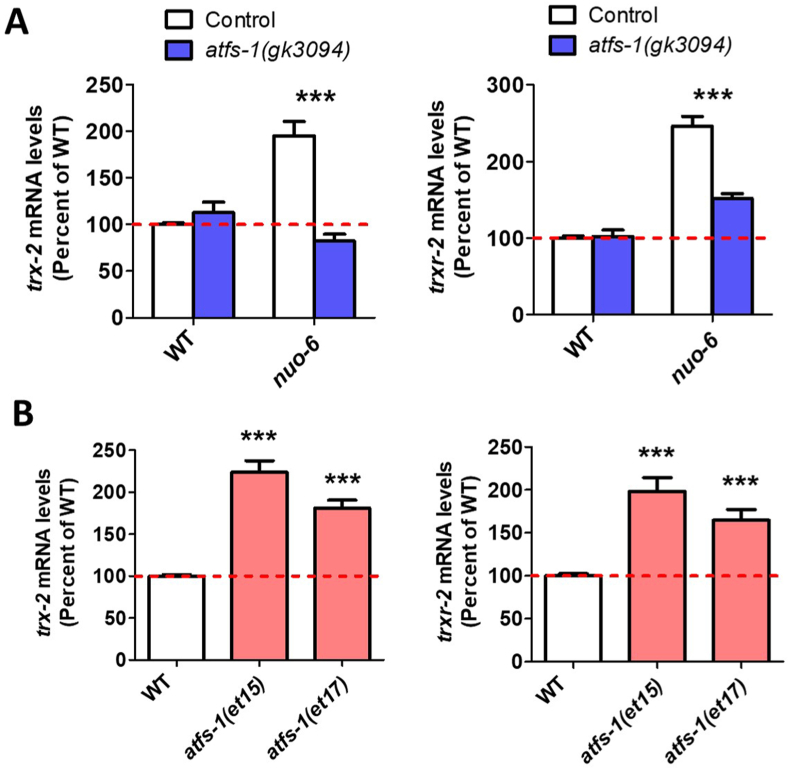
As we have shown that the mitochondrial unfolded protein response (mitoUPR) is activated in the long-lived mitochondrial mutants, clk-1, nuo-6 and isp-1 [16] and that activation of the mitoUPR is sufficient to upregulate target genes from multiple stress response pathways [35], we wondered whether activation of the mitoUPR is responsible for the upregulation of trx-2 and trxr-2 in these mutants. Accordingly, we examined the effect of disrupting the mitoUPR transcription factor ATFS-1 in wild-type and nuo-6 worms. Note that we were not able to test the effects of the atfs-1 mutation on trx-2 and trxr-2 expression in isp-1 mutants because the atfs-1 mutation is highly detrimental in isp-1 mutants. isp-1;atfs-1 double mutants and isp-1 worms treated with atfs-1 RNAi fail to develop to adulthood [16].
While atfs-1(gk3094) deletion mutants have wild-type expression levels of trx-2 and trxr-2, disrupting atfs-1 prevents the upregulation of these genes in nuo-6 mutants (Fig. 2A). We also found that constitutive activation of ATFS-1 is sufficient to increase the expression of trx-2 and trxr-2, independently of mitochondrial impairment, as two constitutively active atfs-1 mutants, et15 and et17 [46], both exhibit upregulation of trx-2 and trxr-2 (Fig. 2B). In addition to genes of the mitochondrial thioredoxin system, we found that six other antioxidant genes (sod-3, prdx-3, gpx-6, gpx-7, gpx-8, glrx-5) exhibit a mitoUPR-dependent upregulation in nuo-6 mutants combined with significant upregulation in at least one constitutively active atfs-1 mutant (Figs. S2 and S3). sod-3 encodes an inducible superoxide dismutase that is localized to the mitochondrial matrix [47]. Although gpx-6 and gpx-7 are predicted to localize to the extracellular matrix and cytosol, respectively, gpx-6, gpx-7 and gpx-8 are all homologs of the mammalian GPX4, the long form of which is targeted to the mitochondria [48]. glrx-5 encodes a mitochondrial matrix-targeted glutaredoxin [49]. Combined, this demonstrates that, in addition to trx-2 and trxr-2, multiple other mitochondrially-targeted antioxidant genes are upregulated by activation of the mitoUPR.
Fig. 2.
Upregulation of mitochondrial thioredoxin and thioredoxin reductase genes is dependent on mitoUPR transcription factor ATFS-1. To examine the role of the mitochondrial unfolded protein response (mitoUPR) in the upregulation of the mitochondrial thioredoxin system, we disrupted the mitoUPR transcription factor ATFS-1 in nuo-6 mutants. Deletion of atfs-1 significantly decreased the expression of both trx-2 and trxr-2 in nuo-6 worms but not wild-type worms (A). Conversely, the levels of trx-2 and trxr-2 are significantly increased in two constitutively active atfs-1 mutants (B). Combined, this indicates that expression of mitochondrial thioredoxin system genes can be modulated by the mitoUPR. Gene expression was measured using RNA sequencing with 3–6 biological replicates per strain. atfs-1(gk3094) is a deletion mutant that results in loss of function. atfs-1(et15) and atfs-1(et17) are constitutively active gain of function mutants. Statistical significance was assessed using a two-way ANOVA with Bonferroni posttest in panel A and a one-way ANOVA with Dunnett's multiple comparison test in panel B. Error bars indicate SEM. ***p < 0.001.
3.3. Disruption of mitochondrial or cytoplasmic thioredoxin systems increases reactive oxygen species
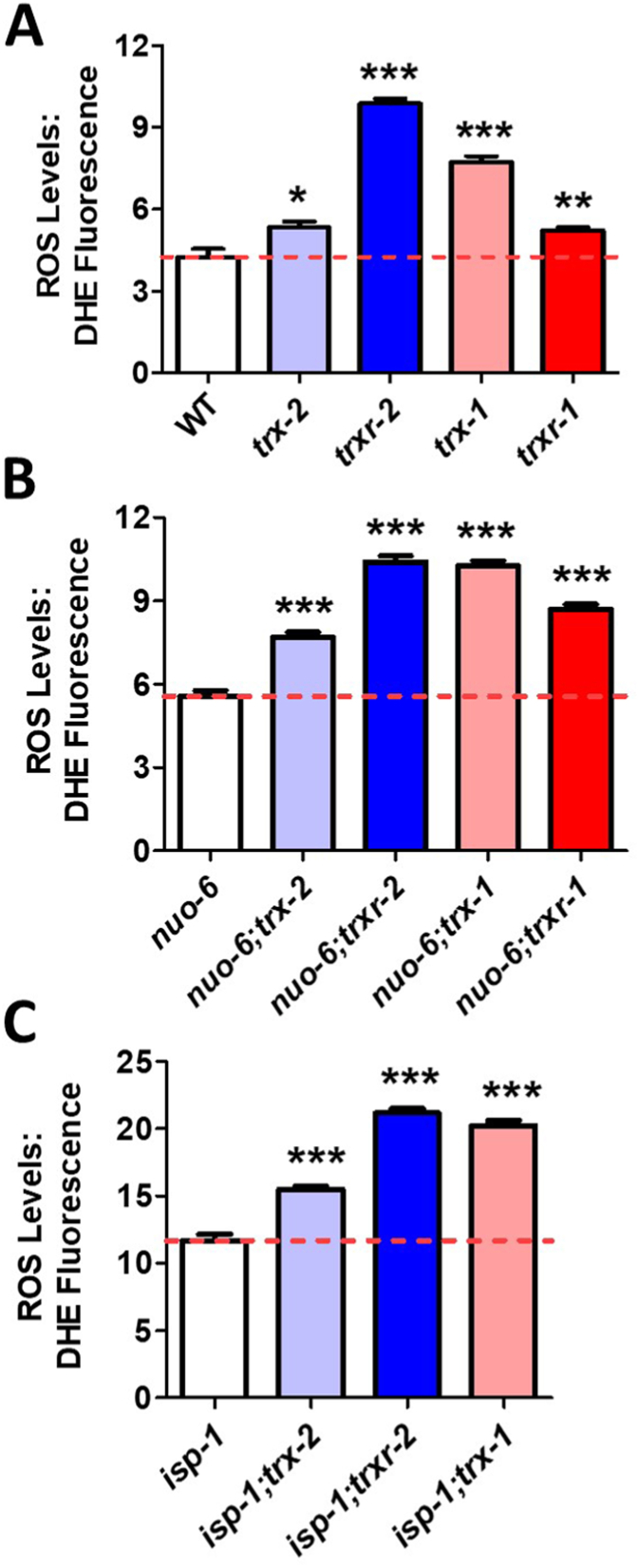
As one of the roles of thioredoxin is to protect against ROS and ROS-induced damage, we wondered whether disruption of thioredoxin genes increases ROS levels in nuo-6 and isp-1 worms. Accordingly, we stained day 1 adult worms with dihydroethidium (DHE), which has been used as an in vivo sensor of ROS because it emits red fluorescence when oxidized [18,36]. Wild-type worms exhibit little or no fluorescence after staining with DHE (Fig. 3A, Fig. S4). Disruption of genes involved in the mitochondrial or cytoplasmic thioredoxin system all significantly increased the levels of ROS, with loss of trxr-2 and trx-1 having the largest effect (Fig. 3A, Fig. S4). Similarly, disruption of trx-2, trxr-2, trx-1 or trxr-1 all further increased the already elevated levels of ROS in nuo-6 (Fig. 3B, Fig. S4) and isp-1 worms (Fig. 3C, Fig. S4), again with loss of trxr-2 and trx-1 resulting in the greatest increase in ROS levels. Combined, these results indicate the disruption of genes involved in the mitochondrial or cytoplasmic thioredoxin system result in increased levels of ROS.
Fig. 3.
Disruption of mitochondrial or cytoplasmic thioredoxin systems increases levels of reactive oxygen species. ROS levels were measured by staining whole worms with dihydroethidium (DHE) at day 1 of adulthood and quantifying the resulting fluorescence. In wild-type worms (A), nuo-6 mutants (B) and isp-1 mutants (C), disruption of mitochondrial or cytoplasmic thioredoxin system genes, including trx-2, trxr-2, trx-1 and trxr-1, all resulted in significantly increased levels of ROS. Disruption of trxr-2 and trx-1 resulted in the greatest increase in ROS levels in all three genotypes. Representative images can be found in Fig. S4. Statistical significance was assessed using a one-way ANOVA with Dunnett's multiple comparison test. Error bars indicate SEM. *p < 0.05, **p < 0.01, ***p < 0.001.
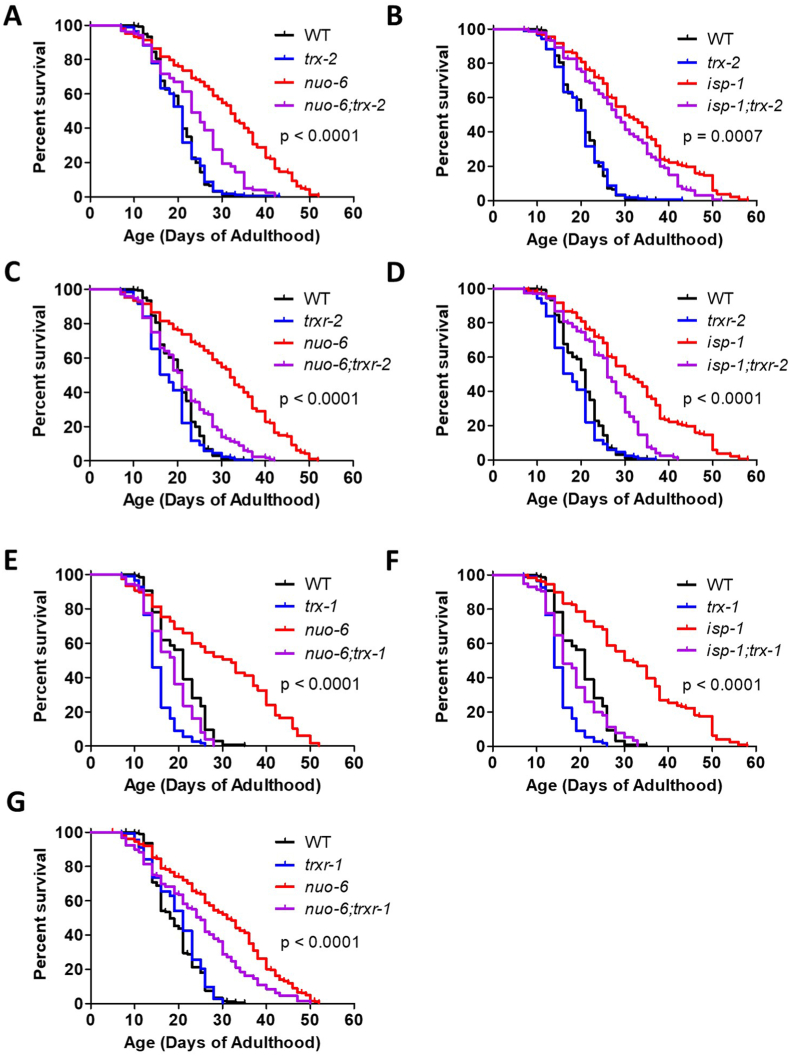
3.4. Mitochondrial thioredoxin system is required for longevity of long-lived mitochondrial mutants
Having shown that trx-2 and trxr-2 are upregulated in nuo-6 and isp-1 mutants, we next sought to determine if the mitochondrial thioredoxin system is contributing to their long lifespans. We found that disruption of trx-2 significantly shortened the lifespan of nuo-6 (Fig. 4A) and isp-1 (Fig. 4B) mutants but did not affect wild-type lifespan. Similarly, deletion of trxr-2 markedly shortened the lifespan of nuo-6 (Fig. 4C) and isp-1 (Fig. 4D) worms but only had a small effect on wild-type lifespan. Combined, this indicates that the mitochondrial thioredoxin system is specifically required for the long lifespan of nuo-6 and isp-1 mutants but largely dispensable for wild-type lifespan.
Fig. 4.
Disruption of mitochondrial thioredoxin system shortens lifespan of long-lived mitochondrial mutants. Deletion of trx-2 (A,B) or trxr-2 (C,D) significantly decreased the lifespan of nuo-6 and isp-1 worms but had little or no effect on the lifespan of wild-type worms. Disruption of trx-1 markedly decreased lifespan in nuo-6, isp-1 and wild-type worms (E,F). Deletion of trxr-1 significantly decreased nuo-6 lifespan but did not decrease the lifespan of wild-type worms (G). isp-1 trxr-1 double mutants could not be generated due to close proximity on the same chromosome. Statistical significance was assessed using a log-rank test. p-values indicate significance of difference between red and purple lines. A minimum of three biological replicates were performed. Raw lifespan data can be found in Table S1. A bar graph of this data can be found in Fig. S5. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
For comparison, we examined the effect of the cytoplasmic thioredoxin system on the lifespan of nuo-6 and isp-1 mutants. Disruption of trx-1 markedly decreased the lifespan of both nuo-6 (Fig. 4E) and isp-1 (Fig. 4F) mutants. In contrast to the mitochondrial thioredoxin genes, trx-1 deletion also shortened wild-type lifespan, as others have observed previously [29,50]. Thus, it appears that loss of trx-1 has a non-specific shortening effect on lifespan. Deletion of trxr-1 decreased the lifespan of nuo-6 mutants but had no effect on wild-type lifespan (Fig. 4G). Note that we were unable to generate isp-1 trxr-1 worms due to the close proximity of the genes on the same chromosome.
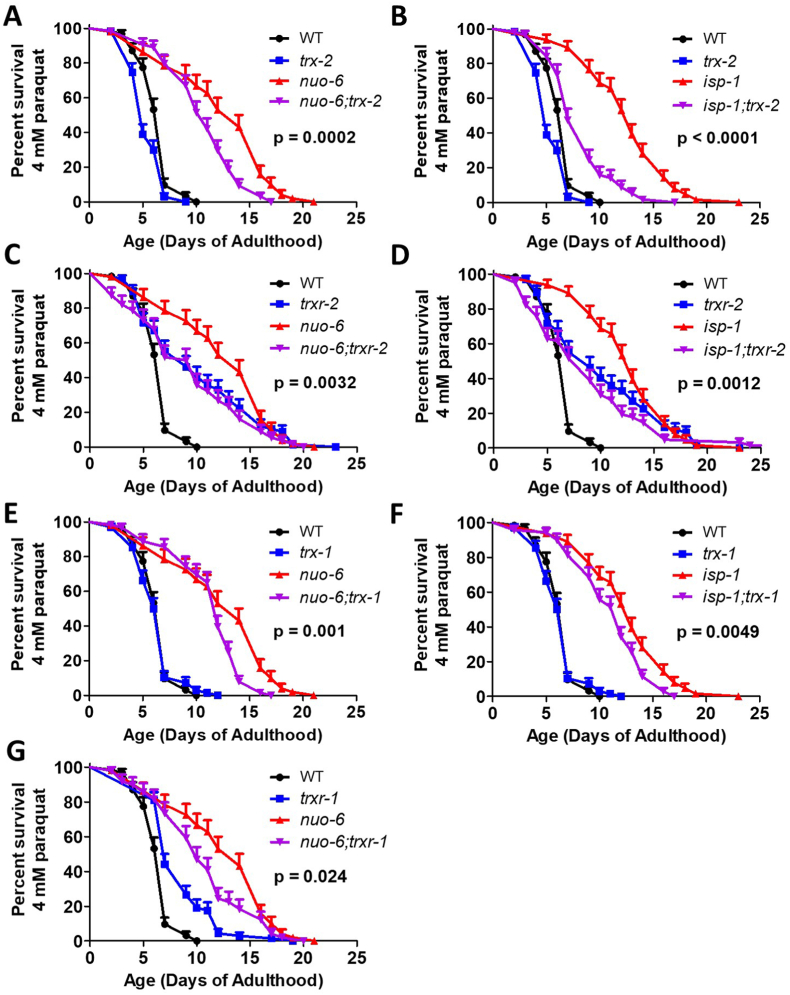
3.5. Mitochondrial thioredoxin system is required for enhanced stress resistance of long-lived mitochondrial mutants
Although trx-2 and trxr-2 are not required for survival of oxidative stress in wild-type worms [30], we wondered whether upregulation of the mitochondrial thioredoxin system contributes to the enhanced resistance to chronic oxidative stress in nuo-6 and isp-1 worms [16,20,37]. We found that deletion of trx-2 significantly reduced the survival of nuo-6 (Fig. 5A) and isp-1 (Fig. 5B) worms exposed to 4 mM paraquat but had only a minor impact on survival in wild-type worms. Disruption of trxr-2 also significantly reduced the survival of nuo-6 (Fig. 5C) and isp-1 (Fig. 5D) worms but increased the survival of wild-type worms. Combined, this indicates that an intact mitochondrial thioredoxin system contributes to the enhanced oxidative stress resistance in isp-1 and nuo-6 worms. Similar to the mitochondrial thioredoxin system, we found that disruption of trx-1 or trxr-1 significantly decreases the survival of the long-lived mitochondrial mutants under oxidative stress (Fig. 5E–G). However, deletion of trx-1 or trxr-1 did not decrease resistance to oxidative stress in wild-type worms.
Fig. 5.
Disruption of mitochondrial thioredoxin system increases sensitivity to oxidative stress in long-lived mitochondrial mutants. Deletion of trx-2 significantly decreases the survival of nuo-6 (A) and isp-1 (B) worms under chronic oxidative stress resulting from exposure to 4 mM paraquat but has little effect in wild-type worms. Disruption of trxr-2 also decreased paraquat survival in nuo-6 (C) and isp-1 (D) worms while increasing survival in wild-type animals. Deletion of trx-1 decreased the paraquat survival of nuo-6 (E) and isp-1 (F) worms but had no effect in wild-type worms. Disruption of trxr-1 decreased the paraquat survival of nuo-6 worms, but increased survival in wild-type animals (G). Statistical significance was assessed using a log-rank test. p-values indicate significance of difference between red and purple lines. Three biological replicates were performed. Error bars indicate SEM. Raw data can be found in Table S1. A bar graph of this data can be found in Fig. S5. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
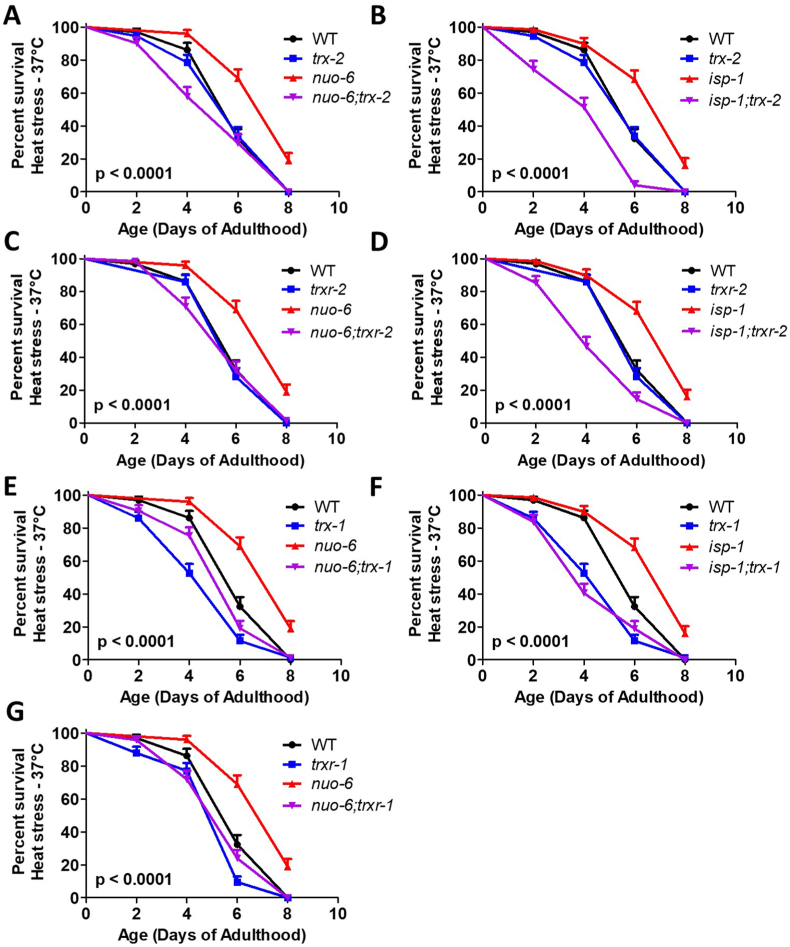
Having shown that the mitochondrial thioredoxin system is important for the enhanced resistance to oxidative stress in nuo-6 and isp-1 worms, we next sought to determine whether these genes also contributed to resistance to other exogenous stressors. For this purpose, we examined resistance to heat stress at 37 °C, resistance to osmotic stress at 450 mM and 500 mM NaCl, and resistance to bacterial pathogens in a slow kill assay where worms are exposed to Pseudomonas aeruginosa strain PA14.
In the heat stress assay, we found that disruption of either trx-2 or trxr-2 specifically decreased heat stress resistance in the long-lived mitochondrial mutants, nuo-6 and isp-1, but had no effect on resistance to heat stress in wild-type worms (Fig. 6A–D). In contrast, disruption of trx-1 or trxr-1 reduced heat stress resistance in both wild-type worms and long-lived mitochondrial mutants (Fig. 6E–G). Combined these results suggest that the mitochondrial thioredoxin system is specifically required for the enhanced heat stress resistance in nuo-6 and isp-1 worms, while the cytoplasmic thioredoxin system is generally required for survival of heat stress.
Fig. 6.
Disruption of mitochondrial thioredoxin system increases sensitivity to heat stress in long-lived mitochondrial mutants. Disruption of trx-2 markedly reduced heat stress resistance in nuo-6 (A) and isp-1 (B) worms but had no effect on wild-type survival. Similarly, loss of trxr-2 significantly decreased heat stress resistance in both nuo-6 (C) and isp-1 (D) worms but did not affect resistance to heat stress in wild-type worms. In contrast, disruption of trx-1 decreased heat stress resistance in wild-type, nuo-6 (E) and isp-1 (F) worms. The trxr-1 deletion also reduced resistance to heat stress in both wild-type and nuo-6 worms (G). Statistical significance was assessed using a log-rank test. p-values indicate significance of difference between red and purple lines. Three biological replicates were performed. Error bars indicate SEM. Raw data can be found in Table S1. A bar graph of this data can be found in Fig. S5. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
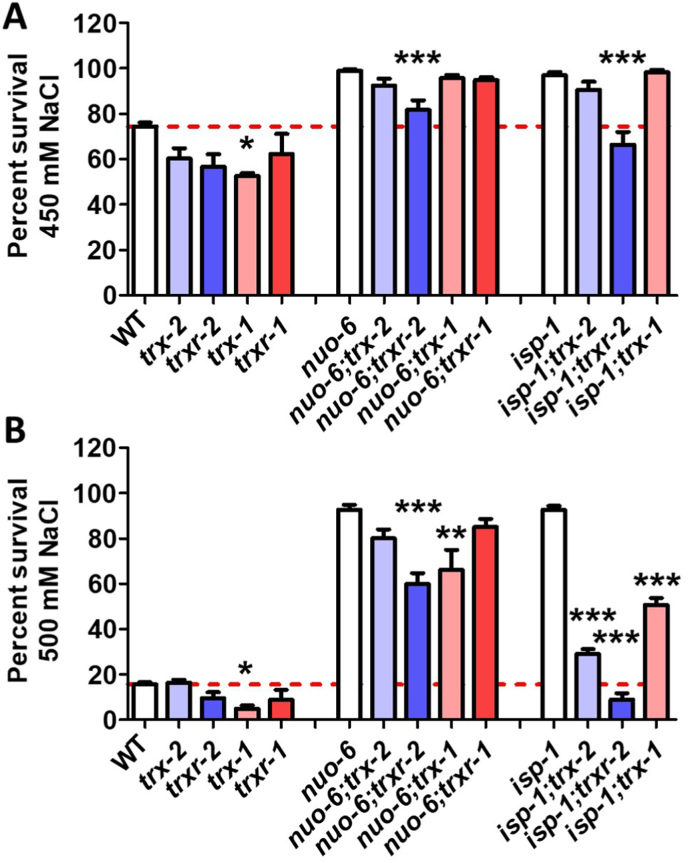
We next examined the role of thioredoxin genes in resistance to osmotic stress. In wild-type worms, disruption of trx-1 resulted in a significant decrease in resistance to osmotic stress (Fig. 7). In both nuo-6 and isp-1 worms, disruption of trxr-2 or trx-1 significantly decreased resistance to osmotic stress (Fig. 7). Deletion of trx-2 also markedly reduced osmotic stress resistance in isp-1 worms but only resulted in a trend towards decrease in nuo-6 worms (Fig. 7). Combined, these results indicate that both the mitochondrial and cytoplasmic thioredoxin systems contribute to the enhanced resistance to osmotic stress in the long-lived mitochondrial mutants, and also appear to have a lesser role in osmotic stress resistance in wild-type worms.
Fig. 7.
Disruption of mitochondrial thioredoxin system increases sensitivity to osmotic stress in long-lived mitochondrial mutants. Resistance to osmotic stress was quantified on plates containing either 450 mM NaCl (A) or 500 mM NaCl (B). In wild-type worms, disruption of trx-1 decreased resistance to osmotic stress. In nuo-6 worms, disruption of trxr-2 or trx-1 significantly decreased resistance to osmotic stress. In isp-1 worms, disruption of trx-2, trxr-2 or trx-1 significantly decreased resistance to osmotic stress. Statistical significance was assessed using a one-way ANOVA with Dunnett's multiple comparison test. Stars indicate significance of difference from the corresponding control strain (white bar; WT, nuo-6 or isp-1). Five biological replicates were performed. Error bars indicate SEM. *p < 0.05, **p < 0.01, ***p < 0.001. Raw data for this figure can be found in Table S1.
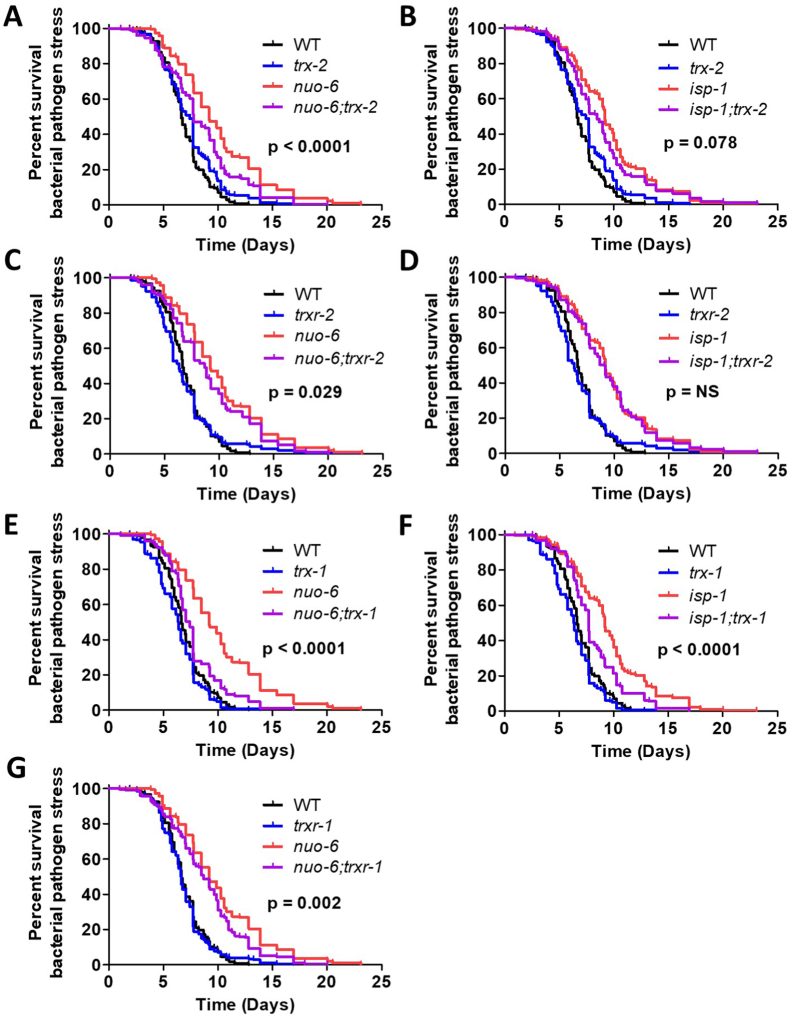
Finally, we measured resistance to bacterial pathogens by exposing worms to P. aeruginosa strain PA14. We found that disruption of genes in the mitochondrial thioredoxin system significantly decreased resistance to bacterial pathogens in nuo-6 worms, but not in wild-type or isp-1 mutants (Fig. 8A-D). In contrast, loss of trx-1 reduced the survival of all three strains when exposed to PA14 (Fig. 8E-F). Deletion of trxr-1 also significantly decreased bacterial pathogen resistance in nuo-6 worms (Fig. 8G). Together, this indicates that disruption of the mitochondrial thioredoxin system specifically decreases resistance to bacterial pathogens in nuo-6 worms, while loss of trx-1 has a general detrimental effect on bacterial pathogen survival.
Fig. 8.
Disruption of mitochondrial thioredoxin system increases sensitivity to bacterial pathogens in long-lived mitochondrial mutants. Survival of bacterial pathogen stress was assessed by exposing worms to P. aeruginosa strain PA14. Disruption of trx-2 significantly reduced the ability of nuo-6 worms to survive bacterial pathogen stress (A), while isp-1;trx-2 worms only exhibited a trend towards decreased survival compared to isp-1 mutants (B). Similarly, loss of trxr-2 reduced bacterial pathogen resistance in nuo-6 worms (C), but not isp-1 worms (D). Deletion of trx-1 decreased the survival of wild-type, nuo-6 (E) and isp-1 (F) worms exposed to P. aeruginosa. Disruption of trxr-1 significantly reduced bacterial pathogen resistance in nuo-6 worms (G). Statistical significance was assessed using a log-rank test. p-values indicate significance of difference between red and purple lines. Six biological replicates were performed. Raw data for this figure can be found in Table S1. A bar graph of this data can be found in Fig. S5. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
4.1. Expression of thioredoxin system genes in long-lived genetic mutants
In this work, we show that trx-2 and trxr-2 are specifically upregulated in the long-lived mitochondrial mutants clk-1, nuo-6 and isp-1 but not in any other examined non-mitochondrial, long-lived mutants. Upregulation of the mitochondrial thioredoxin system is mediated by activation of the mitoUPR, as deletion of atfs-1 reduces expression levels of trx-2 and trxr-2 in mitochondrial mutant worms but not wild-type animals. Moreover, activation of the mitoUPR is sufficient to cause upregulation of the mitochondrial thioredoxin system as two constitutively active atfs-1 gain-of-function mutants exhibit increased expression of trx-2 and trxr-2. Consistent with our results, it was previously shown that spg-7 RNAi, which is known to activate the mitoUPR [51], increases the expression of trx-2 and trxr-2 [30].
In addition to trx-2 and trxr-2, we also identified multiple other antioxidant genes that are upregulated by mitoUPR activation including sod-3, prdx-3, gpx-6, gpx-7, gpx-8 and glrx-5, many of which are targeted to the mitochondria. We have previously shown that disruption of sod-3 shortens the long lifespan of isp-1 mutants [20]. In future studies, it will be interesting to determine the extent to which the other genes we identified are also required for the longevity of long-lived mitochondrial mutants.
Interestingly, we find that different thioredoxin genes are upregulated in other long-lived mutants. Most notably we observed increased expression of trx-1 in daf-2, glp-1 and ife-2 mutants. As overexpression of trx-1 is sufficient to increase lifespan [28,29], this upregulation of trx-1 may contribute to the extended lifespan in these strains. Previous studies have also shown that dietary deprivation increases trx-1 expression in ASJ neurons [28] and that heat stress induces the expression of trxr-1 and trxr-2 [52]. As both dietary deprivation [53] and a short exposure to heat stress [54] extend lifespan, it is possible that upregulation of components of the thioredoxin system is contributing to longevity.
4.2. Effect of thioredoxin system genes on longevity
Previous studies have shown that disruption of trx-2, trxr-2 or both together do not affect lifespan in a wild-type background [30,52]. Our results confirm that the mitochondrial thioredoxin system is expendable for wild-type longevity. In contrast, deletion of either trx-2 or trxr-2 significantly decreases the lifespan of the long-lived mitochondrial mutants, nuo-6 and isp-1, indicating that the mitochondrial thioredoxin system is specifically needed for lifespan extension when mitochondrial function is impaired. These genes do not appear to be required for all long-lived mutants as disruption of trx-2 or trxr-2 does not decrease the lifespan of long-lived insulin-IGF1 receptor daf-2 mutants [30,39].
Similar to nuo-6 and isp-1 mutants, RNAi against spg-7, the worm homolog of SPG7/paraplegin, increases expression of the mitochondrial thioredoxin system [30] and increases lifespan [55]. In contrast, constitutive activation of ATFS-1 reduces lifespan [35,56] despite also increasing expression of the mitochondrial thioredoxin system. In the case of the constitutively active ATFS-1 mutants, it is possible that other changes in these worms may be masking the beneficial effect of increasing the expression of the mitochondrial thioredoxin system. A role for the mitochondrial thioredoxin system in longevity is also suggested in other organisms, including rodents and primates, where the levels of mitochondrial thioredoxin reductase are correlated with lifespan [57].
Similar to the mitochondrial thioredoxin system, we found that disruption of the cytoplasmic thioredoxin gene trx-1 markedly decreases the lifespan of the long-lived mitochondrial mutants. However, as we and others have observed that trx-1 also decreases wild-type lifespan [29,50], the effect of trx-1 deletion on lifespan appears to be due to a non-specific toxic effect. Consistent with the conclusion of a general toxic effect, trx-1 deletion decreases lifespan in multiple other long-lived mutants including eat-2 dietary restriction mutants, daf-2 insulin-IGF1 receptor mutants, and osm-5 chemosensory mutants [28].
The strong effect of trx-1 on lifespan and stress resistance is surprising given that it accounts for less than 1% of the total thioredoxin mRNA in the worm and its expression is limited to a few neurons (ASI/ASJ) and cells in the posterior intestine [29,50]. Thus, it is likely that disruption of trx-1 is having non-cell autonomous effects on longevity. Unlike trx-1, disruption of trxr-1 does not affect lifespan in wild-type worms [52]. Thus, the decreased lifespan of nuo-6;trxr-1 mutants suggests a specific requirement of trxr-1 for nuo-6 lifespan.
The importance of thioredoxin systems for longevity is conserved across species. In Drosophila, overexpression of mitochondrial thioredoxin reductase (Trxr-2) extends lifespan, while overexpression of the cytoplasmic thioredoxin reductase (Trxr-1) has no effect on longevity [57]. Also in Drosophila, the female-specific thioredoxin deadhead (dhd) is required for meiosis and embryonic development [58], while disruption of Trx-2 results in shortened lifespan [59]. Both mitochondrial and cytoplasmic forms of the thioredoxin reductase Trxr-1 are required for viability in flies, and hemizygosity of Trxr-1 leads to decreased lifespan [60]. Overexpression of TXNIP thioredoxin-interacting protein, which is a negative regulator of thioredoxin, also decreases lifespan in flies [61]. In mice, disruption of cytoplasmic thioredoxin (Txn1/Trx1), mitochondrial thioredoxin (Txn2/Trx2), cytoplasmic thioredoxin reductase (Txnrd1/TrxR1) or mitochondrial thioredoxin reductase (Txnrd2/TrxR2) all lead to embryonic lethality [[62], [63], [64], [65]].
4.3. Effect of mitochondrial thioredoxin system genes on stress resistance
While disruption of the mitochondrial thioredoxin system did not decrease resistance to oxidative stress in wild-type worms, both trx-2 and trxr-2 were found to be required for the enhanced oxidative stress resistance in nuo-6 and isp-1 mutants. Consistent with our results, one previous study found that disruption of trx-2 or trxr-2 did not decrease resistance to oxidative stress in wild-type worms [30]. However, a different study did observe reduced oxidative stress resistance in trxr-2 mutants [52]. It is unclear why differing results were obtained for trxr-2 but could be due to differences in the assay parameters. For example, the concentration of paraquat that we used here (4 mM) is much less than that used in the study that observed decreased oxidative stress resistance (40 mM). The oxidative stress imparted by 4 mM paraquat acts as a chronic low-level stress, while 40 mM paraquat acts as an acute stress. With a chronic stress there is time to upregulate genes involved in antioxidant defense while defense against an acute stress likely relies more on baseline levels of antioxidant defense genes. Other differences between our study and the previous study may also have influenced the outcome including the use of solid media here versus liquid media, and a longitudinal analysis here versus analysis of a single time point.
Similar to the mitochondrial thioredoxin system, disruption of trx-1 or trxr-1 did not decrease resistance to oxidative stress in wild-type worms but did reduce oxidative stress resistance in nuo-6 and isp-1 mutants. Previous studies have shown that deletion of trx-1 decreases resistance to oxidative stress in wild-type worms [50] (in an acute assay involving exposure to 50 mM paraquat in liquid), while trxr-1 mutants exhibit wild-type survival of oxidative stress [52,66]. Combined, our results suggest that the mitochondrial and cytoplasmic thioredoxin systems are dispensable for oxidative stress resistance in wild-type worms but are important for the enhanced resistance to oxidative stress in the long-lived mitochondrial mutants. While it is perhaps surprising that thioredoxin system genes are not required for normal oxidative stress resistance in wild-type animals, this may at least partially be explained by these proteins performing other roles within the organism. For example, it has been shown that TRXR-1 does not protect against oxidative stress but instead acts with GSR-1 to remove old cuticle by modulating the oxidation state of cuticle proteins [66].
It was interesting to observe a paradoxical increase in resistance to oxidative stress in trxr-2 and trxr-1 mutants. As our results show that deletion of either trxr-2 or trxr-1 results in a significant increase in ROS levels, one possibility is that the elevated levels of ROS are activating stress response pathways leading to increased resistance to paraquat, similar to what is observed in nuo-6 and isp-1 mutants. In the trx-2 and trx-1 mutants, which also have elevated ROS levels, the mutations may have detrimental effects that mask the positive effect of ROS-mediated signaling. In the case of trx-1, it is interesting that deletion of trx-1 did not decrease resistance to paraquat, despite decreasing resistance to all of the other stresses we examined. It is possible that the elevation of ROS levels in trx-1 may activate stress response pathways that allow for wild-type survival of paraquat stress but perhaps have less of an impact on other types of stress that aren't directly related to ROS.
An alternative explanation for the enhanced resistance to paraquat in the thioredoxin reductase mutants, trxr-2 and trxr-1, is that the loss of thioredoxin reductase decreases the amount of superoxide generated from paraquat thereby reducing its toxicity. The toxic effects of paraquat are believed to be primarily due to the generation of the ROS superoxide through redox cycling. A similar redox cycling mechanism is involved for juglone, which also induces oxidative stress through generation of superoxide. It has been shown that thioredoxin reductase can participate in redox cycling with juglone to generate superoxide and that disruption of thioredoxin reductase reduces superoxide generation from juglone [67]. If thioredoxin reductase is also involved in the generation of superoxide from paraquat, then disruption of trxr-2 and trxr-1 may be increasing the survival of wild type worms on paraquat by decreasing superoxide generation. In nuo-6 and isp-1 worms, thioredoxin reductase may be needed to defend against internally generated ROS, which is elevated compared to wild-type worms at baseline, such that loss of trxr-2 or trxr-1 is still detrimental overall despite decreasing paraquat-induced ROS.
In wild-type worms, disruption of either component of the cytoplasmic thioredoxin system decreases resistance to heat stress, while deletion of trx-2 or trxr-2 does not decrease heat stress resistance, consistent with an earlier study [30]. In contrast, disruption of either the mitochondrial or the cytoplasmic thioredoxin systems decreases resistance to heat stress in the long-lived mitochondrial mutants. This suggests that the cytoplasmic thioredoxin system is generally required for surviving heat stress, while the mitochondrial thioredoxin system is specifically required for the enhanced heat stress resistance in nuo-6 and isp-1 worms.
Deletion of trx-1 decreased resistance to osmotic stress in wild-type, nuo-6 and isp-1 worms. Since trx-1 is primarily expressed in ASJ neurons [29,50] and key signaling steps in osmotic stress response take place in ASJ neurons [68], TRX-1 may be playing a role in initiating the transcriptional response to osmotic stress. Further studies will be required to determine the exact role of TRX-1 on osmotic stress resistance and whether it is directly involved in signaling or indirectly involved by protecting other signaling proteins from damage. Unlike trx-1, disruption of genes in the mitochondrial thioredoxin system only decreases osmotic stress resistance in the long-lived mitochondrial mutants but not wild-type worms. Thus, similar to heat stress, the mitochondrial thioredoxin system appears to be specifically required for the increased resistance to osmotic stress in the long-lived mitochondrial mutants.
Similar to heat stress and osmotic stress, disruption of trx-1 decreased resistance to bacterial pathogen stress in wild-type, nuo-6 and isp-1 worms, indicating a general effect on bacterial pathogen resistance. In contrast, disruption of trx-2 or trxr-2 specifically reduced bacterial pathogen survival in nuo-6 mutants. Overall, the trx-1 deletion has general detrimental effects on stress resistance and lifespan in both wild-type worms and the long-lived mitochondrial mutants, while disruption of the mitochondrial thioredoxin system has little or no effect in wild-type worms, but specifically reduces the enhanced lifespan and stress resistance in nuo-6 and isp-1 mutants.
In interpreting the results of the different stress resistance assays in mitochondrial mutants and mutants in which the mitochondrial thioredoxin system is disrupted, it is important to consider that at least some of these stresses may exhibit a specific interaction with the mitochondria. For example, some of the toxins produced by P. aeruginosa affect the mitochondria, while a number of the stresses require energy to mount defenses.
4.4. Thioredoxins and thioredoxin reductases can act independently of one another
In this work, we observed several differences between disrupting the mitochondrial versus the cytoplasmic thioredoxin system. These results highlight the need for an active thioredoxin system in both the mitochondria and cytoplasm. This is consistent with thioredoxin's function of reducing disulfide bonds in oxidized proteins in order to restore function. Since these proteins cannot pass through a biological membrane without a specific transporter, it is necessary to be able to restore their function in the cellular compartment that they are in. Thioredoxin also acts indirectly to detoxify hydrogen peroxide through its ability to reactivate peroxiredoxins. While hydrogen peroxide is able to pass through membranes, it may be important to have thioredoxin present in the cellular compartment in which it is generated to allow peroxiredoxin to detoxify it before it has reacted with components of the cell.
Another interesting finding of this work is that we observed different results when we disrupted thioredoxin and thioredoxin reductase genes within the same thioredoxin system (see Table S2 for summary of all results). In the mitochondrial thioredoxin system, deletion of trx-2 was found to decrease resistance to oxidative stress, while deletion of trxr-2 had the opposite effect. In the case of the cytoplasmic thioredoxin system, disruption of trx-1 decreased lifespan, bacterial pathogen resistance and osmotic stress resistance, while trxr-1 deletion did not affect any of these phenotypes. These results suggest that thioredoxins and or thioredoxin reductases have additional roles that are independent of the other. One possible explanation for these differences is the fact that thioredoxins have redox-independent functions [[23], [24], [25], [26], [27]]. Disruption of the thioredoxin gene would affect both redox-dependent and redox-independent functions, while disruption of the thioredoxin reductase genes would be predicted to only affect the redox-dependent functions of its corresponding thioredoxin. An alternative explanation is that thioredoxin reductases are acting on other targets, possibly other thioredoxins (TRX-3, TRX-4, TRX-5) or other proteins such that disruption of thioredoxin reductases has broader effects than disrupting individual thioredoxins.
5. Conclusions
Genes from the mitochondrial thioredoxin system are specifically upregulated in long-lived mitochondrial mutants in C. elegans through activation of the mitoUPR. The upregulation of these genes contributes to the long lifespan of these strains as well as their enhanced resistance to exogenous stressors, as disruption of trx-2 or trxr-2 decreases the lifespan and stress resistance of long-lived mitochondrial mutants nuo-6 and isp-1 but does not have a detrimental effect in wild-type worms. In contrast, disruption of the cytoplasmic thioredoxin system decreases lifespan and stress resistance in both mitochondrial mutants and wild-type worms indicating a non-specific detrimental effect. Overall, this work highlights the importance of the thioredoxin system for lifespan and resistance to stress.
Author contributions
Conceptualization: JVR. Methodology: NHG, AT, SKS, UA, AM, AA, JVR. Investigation: NHG, AT, SKS, UA, AM, AA, JVR. Analysis: NHG, AT, SKS, UA, AM, AA, JVR. Visualization NHG, AT, SKS, UA, AM, AA, JVR. Writing – original draft: NHG, JVR. Writing – review and editing: NHG, AT, SKS, UA, AM, AA, JVR. Supervision: JVR.
Conflicts of interest
The authors declare that no conflicts of interest exist.
Materials & correspondence
Correspondence and material requests should be addressed to Jeremy Van Raamsdonk.
Funding
This work was supported by the National Institute of General Medical Sciences (NIGMS; https://www.nigms.nih.gov/; JVR) by grant number R01 GM121756, the Canadian Institutes of Health Research (CIHR; http://www.cihr-irsc.gc.ca/; JVR) and the Natural Sciences and Engineering Research Council of Canada (NSERC; https://www.nserc-crsng.gc.ca/index_eng.asp; JVR). JVR received a salary award from Fonds de Recherche du Quebec Santé (FRQS) and Parkinson Quebec. AT received scholarships from NSERC and FRQS. SKS received a scholarship from FRQS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgments
Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P30 OD010440). We would also like to acknowledge the C. elegans knockout consortium and the National Bioresource Project of Japan for providing strains used in this research.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2022.102335.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Friedman D.B., Johnson T.E. A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics. 1988;118(1):75–86. doi: 10.1093/genetics/118.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clancy D.J., Gems D., Harshman L.G., et al. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292(5514):104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- 3.Holzenberger M., Dupont J., Ducos B., et al. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421(6919):182–187. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- 4.Willcox B.J., Donlon T.A., He Q., et al. FOXO3A genotype is strongly associated with human longevity. Proc. Natl. Acad. Sci. U. S. A. 2008;105(37):13987–13992. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harman D. Aging: a theory based on free radical and radiation chemistry. J. Gerontol. 1956;11(3):298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 6.Shields H.J., Traa A., Van Raamsdonk J.M. Beneficial and detrimental effects of reactive oxygen species on lifespan: a comprehensive review of comparative and experimental studies. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.628157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lakowski B., Hekimi S. Determination of life-span in Caenorhabditis elegans by four clock genes. Science. 1996;272(5264):1010–1013. doi: 10.1126/science.272.5264.1010. [DOI] [PubMed] [Google Scholar]
- 8.Dillin A., Hsu A.L., Arantes-Oliveira N., et al. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298(5602):2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- 9.Copeland J.M., Cho J., Lo T., Jr., et al. Extension of Drosophila life span by RNAi of the mitochondrial respiratory chain. Curr. Biol. 2009;19(19):1591–1598. doi: 10.1016/j.cub.2009.08.016. [DOI] [PubMed] [Google Scholar]
- 10.Liu X., Jiang N., Hughes B., et al. Evolutionary conservation of the clk-1-dependent mechanism of longevity: loss of mclk1 increases cellular fitness and lifespan in mice. Genes Dev. 2005;19(20):2424–2434. doi: 10.1101/gad.1352905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dell'agnello C., Leo S., Agostino A., et al. Increased longevity and refractoriness to Ca(2+)-dependent neurodegeneration in Surf1 knockout mice. Hum. Mol. Genet. 2007;16(4):431–444. doi: 10.1093/hmg/ddl477. [DOI] [PubMed] [Google Scholar]
- 12.Feng J., Bussiere F., Hekimi S. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev. Cell. 2001;1(5):633–644. doi: 10.1016/s1534-5807(01)00071-5. [DOI] [PubMed] [Google Scholar]
- 13.Yang W., Hekimi S. Two modes of mitochondrial dysfunction lead independently to lifespan extension in Caenorhabditis elegans. Aging Cell. 2010;9(3):433–447. doi: 10.1111/j.1474-9726.2010.00571.x. [DOI] [PubMed] [Google Scholar]
- 14.Yang W., Hekimi S. A mitochondrial superoxide signal triggers increased longevity in Caenorhabditis elegans. PLoS Biol. 2010;8(12):e1000556. doi: 10.1371/journal.pbio.1000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Senchuk M.M., Dues D.J., Schaar C.E., et al. Activation of DAF-16/FOXO by reactive oxygen species contributes to longevity in long-lived mitochondrial mutants in Caenorhabditis elegans. PLoS Genet. 2018;14(3) doi: 10.1371/journal.pgen.1007268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Z., Senchuk M.M., Dues D.J., et al. Mitochondrial unfolded protein response transcription factor ATFS-1 promotes longevity in a long-lived mitochondrial mutant through activation of stress response pathways. BMC Biol. 2018;16(1):147. doi: 10.1186/s12915-018-0615-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campos J.C., Wu Z., Rudich P.D., et al. Mild mitochondrial impairment enhances innate immunity and longevity through ATFS-1 and p38 signaling. EMBO Rep. 2021 doi: 10.15252/embr.202152964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee S.J., Hwang A.B., Kenyon C. Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Curr. Biol. 2010;20(23):2131–2136. doi: 10.1016/j.cub.2010.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yee C., Yang W., Hekimi S. The intrinsic apoptosis pathway mediates the pro-longevity response to mitochondrial ROS in C. elegans. Cell. 2014;157(4):897–909. doi: 10.1016/j.cell.2014.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dues D.J., Schaar C.E., Johnson B.K., et al. Uncoupling of oxidative stress resistance and lifespan in long-lived isp-1 mitochondrial mutants in Caenorhabditis elegans. Free Radic. Biol. Med. 2017;108:362–373. doi: 10.1016/j.freeradbiomed.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lillig C.H., Holmgren A. Thioredoxin and related molecules-from biology to health and disease. Antioxidants Redox Signal. 2007;9(1):25–47. doi: 10.1089/ars.2007.9.25. Epub 2006/11/23. [DOI] [PubMed] [Google Scholar]
- 22.Lee S., Kim S.M., Lee R.T. Thioredoxin and thioredoxin target proteins: from molecular mechanisms to functional significance. Antioxidants Redox Signal. 2013;18(10):1165–1207. doi: 10.1089/ars.2011.4322. Epub 2012/06/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jurado P., de Lorenzo V., Fernandez L.A. Thioredoxin fusions increase folding of single chain Fv antibodies in the cytoplasm of Escherichia coli: evidence that chaperone activity is the prime effect of thioredoxin. J. Mol. Biol. 2006;357(1):49–61. doi: 10.1016/j.jmb.2005.12.058. [DOI] [PubMed] [Google Scholar]
- 24.Du H., Kim S., Hur Y.S., et al. A cytosolic thioredoxin acts as a molecular chaperone for peroxisome matrix proteins as well as antioxidant in peroxisome. Mol. Cell. 2015;38(2):187–194. doi: 10.14348/molcells.2015.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y., Min W. Thioredoxin promotes ASK1 ubiquitination and degradation to inhibit ASK1-mediated apoptosis in a redox activity-independent manner. Circ. Res. 2002;90(12):1259–1266. doi: 10.1161/01.res.0000022160.64355.62. [DOI] [PubMed] [Google Scholar]
- 26.Sanzo-Machuca Á, Monje Moreno J.M., Casado-Navarro R., et al. Redox-dependent and redox-independent functions of Caenorhabditis elegans thioredoxin 1. Redox Biol. 2019;24 doi: 10.1016/j.redox.2019.101178. Epub 2019/04/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hao Y., Yang W., Ren J., et al. Thioredoxin shapes the C. elegans sensory response to Pseudomonas produced nitric oxide. Elife. 2018;7 doi: 10.7554/eLife.36833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fierro-Gonzalez J.C., Gonzalez-Barrios M., Miranda-Vizuete A., et al. The thioredoxin TRX-1 regulates adult lifespan extension induced by dietary restriction in Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 2011;406(3):478–482. doi: 10.1016/j.bbrc.2011.02.079. [DOI] [PubMed] [Google Scholar]
- 29.Miranda-Vizuete A., Fierro Gonzalez J.C., Gahmon G., et al. Lifespan decrease in a Caenorhabditis elegans mutant lacking TRX-1, a thioredoxin expressed in ASJ sensory neurons. FEBS Lett. 2006;580(2):484–490. doi: 10.1016/j.febslet.2005.12.046. [DOI] [PubMed] [Google Scholar]
- 30.Cacho-Valadez B., Munoz-Lobato F., Pedrajas J.R., et al. The characterization of the Caenorhabditis elegans mitochondrial thioredoxin system uncovers an unexpected protective role of thioredoxin reductase 2 in beta-amyloid peptide toxicity. Antioxidants Redox Signal. 2012;16(12):1384–1400. doi: 10.1089/ars.2011.4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jimenez-Hidalgo M., Kurz C.L., Pedrajas J.R., et al. Functional characterization of thioredoxin 3 (TRX-3), a Caenorhabditis elegans intestine-specific thioredoxin. Free Radic. Biol. Med. 2014;68:205–219. doi: 10.1016/j.freeradbiomed.2013.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Raamsdonk J.M., Hekimi S. FUdR causes a twofold increase in the lifespan of the mitochondrial mutant gas-1. Mech. Ageing Dev. 2011;132(10):519–521. doi: 10.1016/j.mad.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Senchuk M.M., Dues D.J., Van Raamsdonk J.M. Measuring oxidative stress in Caenorhabditis elegans: paraquat and juglone sensitivity assays. Bio-protocol. 2017;7(1) doi: 10.21769/BioProtoc.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Z., Isik M., Moroz N., et al. Dietary restriction extends lifespan through metabolic regulation of innate immunity. Cell Metabol. 2019;29(5):1192–1205. doi: 10.1016/j.cmet.2019.02.013. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soo S.K., Traa A., Rudich P.D., et al. Activation of mitochondrial unfolded protein response protects against multiple exogenous stressors. Life Sci Alliance. 2021;4(12) doi: 10.26508/lsa.202101182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei Y., Kenyon C. Roles for ROS and hydrogen sulfide in the longevity response to germline loss in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 2016;113(20):E2832–E2841. doi: 10.1073/pnas.1524727113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soo S.K., Rudich P.D., Mistry M., et al. Genetic basis of enhanced stress resistance in long-lived mutants highlights key role of innate immunity in determining longevity. bioRxiv. 2021 doi: 10.1101/2021.07.19.452975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Raamsdonk J.M., Hekimi S. Deletion of the mitochondrial superoxide dismutase sod-2 extends lifespan in Caenorhabditis elegans. PLoS Genet. 2009;5(2) doi: 10.1371/journal.pgen.1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kenyon C., Chang J., Gensch E., et al. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 40.Hsin H., Kenyon C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature. 1999;399:362–366. doi: 10.1038/20694. [DOI] [PubMed] [Google Scholar]
- 41.Syntichaki P., Troulinaki K., Tavernarakis N. eIF4E function in somatic cells modulates ageing in Caenorhabditis elegans. Nature. 2007;445(7130):922–926. doi: 10.1038/nature05603. [DOI] [PubMed] [Google Scholar]
- 42.Hansen M., Taubert S., Crawford D., et al. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6(1):95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- 43.Apfeld J., Kenyon C. Regulation of lifespan by sensory perception in Caenorhabditiselegans. Nature. 1999:804–809. doi: 10.1038/45544. [DOI] [PubMed] [Google Scholar]
- 44.Lakowski B., Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 1998;95:13091–13096. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ranjan M., Gruber J., Ng L.F., et al. Repression of the mitochondrial peroxiredoxin antioxidant system does not shorten life span but causes reduced fitness in Caenorhabditis elegans. Free Radic. Biol. Med. 2013 doi: 10.1016/j.freeradbiomed.2013.05.025. [DOI] [PubMed] [Google Scholar]
- 46.Rauthan M., Ranji P., Aguilera Pradenas N., et al. The mitochondrial unfolded protein response activator ATFS-1 protects cells from inhibition of the mevalonate pathway. Proc. Natl. Acad. Sci. U. S. A. 2013;110(15):5981–5986. doi: 10.1073/pnas.1218778110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hunter T., Bannister W.H., Hunter G.J. Cloning, expression, and characterization of two manganese superoxide dismutases from Caenorhabditis elegans. J. Biol. Chem. 1997;272(45):28652–28659. doi: 10.1074/jbc.272.45.28652. [DOI] [PubMed] [Google Scholar]
- 48.Liang H., Ran Q., Jang Y.C., et al. Glutathione peroxidase 4 differentially regulates the release of apoptogenic proteins from mitochondria. Free Radic. Biol. Med. 2009;47(3):312–320. doi: 10.1016/j.freeradbiomed.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jeong D.E., Lee D., Hwang S.Y., et al. Mitochondrial chaperone HSP-60 regulates anti-bacterial immunity via p38 MAP kinase signaling. EMBO J. 2017;36(8):1046–1065. doi: 10.15252/embj.201694781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jee C., Vanoaica L., Lee J., et al. Thioredoxin is related to life span regulation and oxidative stress response in Caenorhabditis elegans. Gene Cell. 2005;10(12):1203–1210. doi: 10.1111/j.1365-2443.2005.00913.x. [DOI] [PubMed] [Google Scholar]
- 51.Yoneda T., Benedetti C., Urano F., et al. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J. Cell Sci. 2004;117(Pt 18):4055–4066. doi: 10.1242/jcs.01275. [DOI] [PubMed] [Google Scholar]
- 52.Yu D., Pendergraff H., Liu J., et al. Single-stranded RNAs use RNAi to potently and allele-selectively inhibit mutant Huntingtin expression. Cell. 2012;150(5):895–908. doi: 10.1016/j.cell.2012.08.002. Epub 2012/09/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaeberlein T.L., Smith E.D., Tsuchiya M., et al. Lifespan extension in Caenorhabditis elegans by complete removal of food. Aging Cell. 2006;5(6):487–494. doi: 10.1111/j.1474-9726.2006.00238.x. [DOI] [PubMed] [Google Scholar]
- 54.Dues D.J., Andrews E.K., Schaar C.E., et al. Aging causes decreased resistance to multiple stresses and a failure to activate specific stress response pathways. Aging (Albany NY) 2016;8(4):777–795. doi: 10.18632/aging.100939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Curran S.P., Ruvkun G. Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 2007;3(4):e56. doi: 10.1371/journal.pgen.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bennett C.F., Vander Wende H., Simko M., et al. Activation of the mitochondrial unfolded protein response does not predict longevity in Caenorhabditis elegans. Nat. Commun. 2014;5:3483. doi: 10.1038/ncomms4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pickering A.M., Lehr M., Gendron C.M., et al. Mitochondrial thioredoxin reductase 2 is elevated in long-lived primate as well as rodent species and extends fly mean lifespan. Aging Cell. 2017;16(4):683–692. doi: 10.1111/acel.12596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Salz H.K., Flickinger T.W., Mittendorf E., et al. The Drosophila maternal effect locus deadhead encodes a thioredoxin homolog required for female meiosis and early embryonic development. Genetics. 1994;136(3):1075–1086. doi: 10.1093/genetics/136.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsuda M., Ootaka R., Ohkura C., et al. Loss of Trx-2 enhances oxidative stress-dependent phenotypes in Drosophila. FEBS Lett. 2010;584(15):3398–3401. doi: 10.1016/j.febslet.2010.06.034. [DOI] [PubMed] [Google Scholar]
- 60.Missirlis F., Phillips J.P., Jackle H. Cooperative action of antioxidant defense systems in Drosophila. Curr. Biol. 2001;11(16):1272–1277. doi: 10.1016/s0960-9822(01)00393-1. [DOI] [PubMed] [Google Scholar]
- 61.Oberacker T., Bajorat J., Ziola S., et al. Enhanced expression of thioredoxin-interacting-protein regulates oxidative DNA damage and aging. FEBS Lett. 2018;592(13):2297–2307. doi: 10.1002/1873-3468.13156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matsui M., Oshima M., Oshima H., et al. Early embryonic lethality caused by targeted disruption of the mouse thioredoxin gene. Dev. Biol. 1996;178(1):179–185. doi: 10.1006/dbio.1996.0208. [DOI] [PubMed] [Google Scholar]
- 63.Nonn L., Williams R.R., Erickson R.P., et al. The absence of mitochondrial thioredoxin 2 causes massive apoptosis, exencephaly, and early embryonic lethality in homozygous mice. Mol. Cell Biol. 2003;23(3):916–922. doi: 10.1128/MCB.23.3.916-922.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jakupoglu C., Przemeck G.K., Schneider M., et al. Cytoplasmic thioredoxin reductase is essential for embryogenesis but dispensable for cardiac development. Mol. Cell Biol. 2005;25(5):1980–1988. doi: 10.1128/MCB.25.5.1980-1988.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Conrad M., Jakupoglu C., Moreno S.G., et al. Essential role for mitochondrial thioredoxin reductase in hematopoiesis, heart development, and heart function. Mol. Cell Biol. 2004;24(21):9414–9423. doi: 10.1128/MCB.24.21.9414-9423.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stenvall J., Fierro-Gonzalez J.C., Swoboda P., et al. Selenoprotein TRXR-1 and GSR-1 are essential for removal of old cuticle during molting in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 2011;108(3):1064–1069. doi: 10.1073/pnas.1006328108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu J., Cheng Q., Arner E.S. Details in the catalytic mechanism of mammalian thioredoxin reductase 1 revealed using point mutations and juglone-coupled enzyme activities. Free Radic. Biol. Med. 2016;94:110–120. doi: 10.1016/j.freeradbiomed.2016.02.013. [DOI] [PubMed] [Google Scholar]
- 68.Burton N.O., Dwivedi V.K., Burkhart K.B., et al. Neurohormonal signaling via a sulfotransferase antagonizes insulin-like signaling to regulate a Caenorhabditis elegans stress response. Nat. Commun. 2018;9(1):5152. doi: 10.1038/s41467-018-07640-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.