Abstract
In this review, we summarize and discuss recent advances in understanding the characteristics of tissue-resident memory T cells (TRMs) in the context of solid organ transplantation (SOT). We first introduce the traditionally understood noncirculating features of TRMs and the key phenotypic markers that define this population, then provide a detailed discussion of emerging concepts on the re-circulation and plasticity of TRM in mice and humans. We comment on the potential heterogeneity of transient, temporary resident and permanent resident T cells and potential interchangeable phenotypes between TRM and effector T cells in nonlymphoid tissues. We review the literature on the distribution of TRM in human nonlymphoid organs and association of clinical outcomes in different types of SOT, including intestine, lung, liver, kidney and heart. We focus on both tissue-specific and organ-shared features of donor- and recipient-derived TRMs after transplantation whenever applicable. Studies with comprehensive sample collection, including longitudinal and cross-sectional controls, and applied advanced techniques such as multicolor flow cytometry to distinguish donor and recipient TRMs, bulk and/or single-cell T cell receptor sequencing to track clonotypes and define transcriptome profiles, and functional readouts to define alloreactivity and pro-/anti-inflammatory activities are emphasized. We also discuss important findings on the tissue-resident features of regulatory αβ T cells and unconventional γδ T cells after transplantation. Understanding of TRM in SOT is a rapidly growing field that urges future studies to address unresolved questions regarding their heterogeneity, plasticity, longevity, alloreactivity and roles in rejection and tolerance.
Summary
SOT provides an opportunity to study the dynamics of human tissue-resident T cells, given the abundance of T cells at multiple organ sites and the ability to track donor- and recipient-derived cells based on specific HLA-markers. Many questions regarding bidirectional alloimmune responses between graft and host and the anatomical and environmental diversities of each organ type remain to be addressed. While animal studies provide an opportunity to more precisely manipulate designated factors to investigate relevant mechanisms, not all of the findings can appropriately translate to humans. Important questions include the heterogeneity, plasticity, alloreactivity and persistence of human TRMs in solid organs. Emerging concepts (Figures 1, 2) include the developmental plasticity of TRMs with interchangeable Teff phenotypes and their recirculating features to secondary lymphoid tissues and peripheral blood; the relocation of ex-TRMs to original NLTs and re-differentiation in situ; and the association of rejection and dynamic turnover of intragraft T cells, not only αβ conventional T cells, but also Tregs and γδ T cells. Multiomics provide promising platforms to address the above questions by identifying the phenotype, clonotype, alloreactivity and functional gene profiles of tissue TRMs and their spatial interaction with other cell types and their milieu.
1. Overview of tissue‐resident memory T cells (TRMs) in solid organ transplantation (SOT)
Tissue-resident T cells in nonlymphoid tissues (NLTs) include conventional CD4 and CD8 αβ T cells, regulatory T cells (Tregs), innate lymphoid cells, and several types of unconventional T cells, such as γδ T cells, invariant NKT cells and mucosal-associated invariant T cells.1,2 In this review, we focus on T cells with memory features in such tissues, which are termed TRMs. TRMs differ from their circulating counterparts in phenotype, transcriptional regulation, survival requirements, and function.3,4 TRMs provide a frontline defense against reinfections with pathogens at body surfaces. However, their role in SOT is largely unexplored.5,6 The success of SOT is limited by rejection and risks of infection and cancer, reflecting challenges with immunosuppression.7,8 Given that TRMs are largely excluded from the circulation9 and have lower reliance on costimulation,10,11 they may be shielded from the effects of immunosuppressive drugs,12,13 thereby protecting the organ against infection or promoting tissue homeostasis.14 TRMs carried within the allograft and graft-infiltrating recipient T cells that gradually acquire TRM phenotypes may contribute to graft-versus-host disease (GVHD) and transplant rejection, respectively.5,6,15
Studies from our group and others have demonstrated the presence of both donor- and recipient-derived TRMs in human small intestine,16–19 lung,20,21 liver,22,23 and kidney24 allografts. Remarkably, donor graft-derived tissue lymphocytes can remain within their tissue of origin for months to years after transplantation. In organs highly enriched for TRMs, such as intestines and lungs, the dynamics of donor T cell replacement by the recipient in the graft mucosa are closely associated with clinical outcomes, where slower replacement associates with less rejection and better graft survival.16,17,20 By integrating T cell clonotypes, mixed lymphocyte reaction (MLR)-determined alloreactive clonotype analysis25,26 and single-cell RNA (scRNA) profiling in human intestinal transplantation (ITx), our previous16,17 and ongoing studies27–29 have highlighted the role of bidirectional alloresponses in TRM-enriched grafts in determining clinical outcomes (Section 4.1). Moreover, scRNA-seq studies27–29 provide preliminary evidence for interconversion between TRMs and effector T cells (Teffs) among intragraft T cells after ITx, consistent with the evolving concepts of heterogeneity and recirculating features of TRMs (Section 3). TRMs adapt to local environments that vary in cytokines, metabolites, cell interactions, and matrix proteins. In Section 4, we discuss studies of human TRMs located in gut, lung, liver, kidney and heart and their contribution to clinical outcomes after transplantation. The balance between donor- and recipient-derived T cells in the allograft may not be limited to conventional CD4 and CD8 αβ T cells, but can be extended to Tregs (Section 5) and unconventional γδ T cells (Section 6). This review highlights recent advances and emerging concepts around TRMs in transplant medicine and urges further studies to gain deeper understanding of the impact of TRM on transplant outcomes and develop therapeutic interventions.
2. Definition of TRMs: traditionally nonrecirculating features and key phenotypic markers
Historically, the defining feature of TRMs in both animals and humans is their commitment to peripheral tissue sites and lack of recirculation.2,3,9 Strategies such as parabiosis surgery,30–32 transplantation,33–35 in vivo intravascular antibody staining,36–38 in situ labeling,39,40 T cell depletion,41 and blockade of lymphocyte trafficking32,35,36 have been used in animal studies to assess migration patterns of TRMs. In humans, persistence of donor TRM after different types of SOT, including small intestine,16–19 lung,20,21 liver,22,23 and kidney,24 suggests a similar propensity for TRMs to be retained within tissues. Tissue residency of TRMs is regulated by the induction of a series of retention signals and the repression of tissue egress pathways, consistent with their low migratory and proliferative potential.3,42 Therefore, TRMs are transcriptionally, phenotypically, and functionally distinct from recirculating central memory (TCM) and effector memory (TEM) T cells.3,43
TRMs lack expression of several transcription factors (TFs) and receptors associated with lymph node (LN) homing and recirculation, such as KLF2, KLF3, L-selectin, S1PR1 and CCR7.42 TRMs express surface markers that include C-type lectin CD69 and integrins CD103 (αE) and CD49a (α1). CD69 prevents surface expression of S1PR1, preventing tissue egress.44 CD103 binds to the epithelial cell marker E-cadherin,45 thereby favoring the retention of TRMs in tissues enriched with epithelial cells, such as intestines, lungs and skin. These NLTs with TGF-β-rich environments drive the expression and maintenance of CD103 on TRMs.46 Co-expression of CD69 and CD103 is more frequently seen in CD8 compared to CD4 TRMs.16,20 CD49a, the α subunit of VLA-1, is expressed on skin, lung and intestinal TRMs, likely promoting tissue retention via binding to collagen and laminin.47 Expression of CD49a has been associated with cytotoxic function of CD8 TRMs in human skin.48 Cytotoxic features of TRMs are reflected by their high expression of GZMB, perforin, IFNγ and TNFα.49 Phenotypes of TRMs are controlled by their TF profiles that generally include Runx3, Notch, Blimp1, Hobit, BATF and AHR, although this appears to be subset (CD4 vs CD8) and species (mouse vs human) dependent.49 Despite these common characteristics, identification of TRM is complicated by the fact a single set of phenotypic markers does not appear to be exclusive to this subset. Recent advances in multi-omics technologies will potentially overcome this limitation by measuring a list of TRM signature genes/proteins in combination with clonal tracking50 and even evaluating the environmental milieu of TRM residence in a particular tissue through spatial immune profiling.51,52
Representative flow cytometry gating of TRMs (CD69+CD103+/−) in human NLTs in steady state42,53 and transplantation settings16,17 and schematic representative lists of TRM signature genes42 have been previously presented by us and others. The densities of TRMs in normal human NLTs have been summarized in a review,9 reflected by the percentages of CD69+CD103+/− T cells among total T cells: human skin (70–90%), lungs (60–80%), intestine (80–95%) and liver (60–80%) are highly enriched for TRM. Donor age is also a contributing factor for the composition of TRMs in human NLTs, as younger donors (0–2 years old) have significantly lower proportions of CD8 TRMs in mucosal sites compared with young adults (15–25 years old).53
3. Emerging re-circulating features of TRM in mice and humans
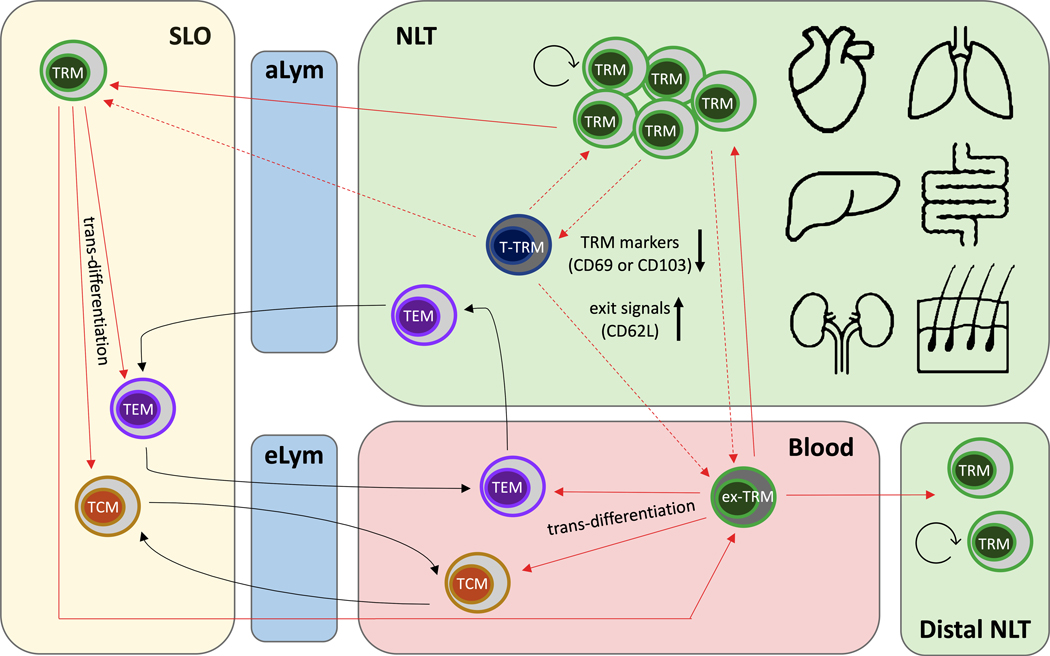
Recent identification of recirculating features of TRMs in mice and humans challenges the previous paradigm that TRMs retain long-term residency in NLTs without participating in systemic recall responses.54 In fact, TRMs exhibit a significant level of developmental plasticity, being capable of tissue egress and re-entry into the circulation in both steady state and inflammatory conditions. Changes in gene expression, such as downregulation of tissue-resident markers (CD69 or CD103) and transient upregulation of exit signals (CD62L), allow “ex-TRMs’ to exit from tissues and re-differentiate to TCM and TEM in the circulating T cell pool.55,56 Interestingly, circulating ex-TRMs retain a propensity to return to their tissue of origin and even populate other NLT sites. In this section, we summarize and discuss the evolving re-circulating features of TRM in murine and human studies (Figure 1).
Figure 1. Emerging re-circulating features of TRM in mice and humans.
Recent studies demonstrated that TRMs exhibit a significant level of developmental plasticity, being capable of tissue egress and re-entry into the circulation and SLOs in steady state and/or inflammatory conditions. Some TRMs in NLTs may enter into a transitioning stage (T-TRM) with Teff phenotypes before they egress the tissue to become circulating ex-TRMs or TRMs in SLOs. T-TRMs and ex-TRMs undergo changes in gene expression, such as downregulation of TRM markers (CD69 or CD103) and transient upregulation of exit signals (CD62L), compared to TRMs in NLTs. TRMs in circulation and SLOs can trans-differentiate to TCM and TEM. Circulating ex-TRMs retain a propensity to return to their tissue of origin and even populate distal NLT sites. aLym: afferent lymph. eLym: efferent lymph.
3.1. Murine studies of re-circulating TRMs
Epithelial barrier tissues contain a mixture of resident and recirculating T cells in mice. In herpes simplex virus (HSV)-infected skin, Gebhardt et al57 identified 2 distinct HSV-specific memory subsets: a CD8 T cell population sequestered in the infected epidermis and a dynamic CD4 T cell population that trafficked rapidly through the dermis. Kaede transgenic mouse skin58 carrying photo-convertible protein upon exposure to violet light and parabiotic pairs between CD45.1 or CD45.2 congenic mice were used to track the fate of cutaneous CD4 T cells in secondary lymphoid organs (SLOs) and circulation.59,60 Bromley et al59 demonstrated that a subset of CD4 memory T cells exits from the skin and reenters draining LNs, circulation, distal LNs, and sites of nonspecific cutaneous inflammation. These migrating CD4 T cells expressed a transitional phenotype (CD69−CD103+/−CCR7+/intCD62LintESL+). Collins et al60 demonstrated that a vast majority of skin CD4 T cells equilibrate with the circulation rather than lodge in the tissue at steady state. Almost half of skin-infiltrated CD4 T cells in parabiosis experiments expressed CD69 and CD103, similar to their host counterparts. Photo-converted Kaede CD4+ T cells migrating from the skin to the draining LNs partially expressed CD103 but not CD69, indicating a modulated phenotype of translocating CD4 TRMs. Using the lymphocytic choriomeningitis virus (LCMV) infection model, Masopust et al61 found that CD4 TRMs share overlapping transcriptional signatures and location-specific features with CD8 TRMs, including high CD69 and GZMB expression in the small intestine. A population of bona fide CD4 TRMs specific to LCMV infection was identified in SLO that share transcriptional characteristics with CD4 TRMs from NLTs. CD69+ CD8 TRMs were detected in the red pulp of spleen and medullary area of LNs.62 Utilizing OT-I-Kaede immune chimeras with Vesicular Stomatitis Virus (VSV)-ovalbumin infection, local reactivation in skin and female reproductive tract was shown to induce migration of antigen-specific CD8 TRMs from NLTs to the draining LNs.63
TRMs in NLTs can also give rise to circulating effector and memory T cells and further relocate to the local environment upon reactivation.55,64,65 Restimulated CD8 TRMs in murine intestines undergo retrograde migration to rejoin the circulating pool and exhibit developmental plasticity to differentiate into TCM, TEM and TRM.55 Ex-TRMs downregulated CD69 and CD103, upregulated CD62L and maintained CCR9 expression after infection. They also maintained a heritable capacity to relocate to their tissue of origin during recall responses and re-differentiated into local TRMs, leading to an “outside-in” differentiation model.55 To investigate TRM progeny in secondary responses, Behr et al64 developed a lineage tracer mouse model exploiting the TRM-defining TF Hobit. Reinfection with Listeria monocytogenes-expressing ovalbumin (Lm-OVA) induced local expansion of OT-I TRMs, accumulation of secondary TRMs in draining LNs and a sizeable fraction of circulating secondary memory T cells that developed downstream of TRMs. These secondary TRM responses were substantially impaired by specific ablation of primary local TRMs. OT-1 TRMs reactivated by Lm-OVA lost some TRM markers and their retention profiles (Hobit, CD69, RGS1) and upregulated genes related to egress (S1PR1, KLF2). These ex-Hobit+ secondary memory T cells (ex-TRMs) largely consisted of TEM cells coexpressing KLRG1 and CX3CR1. The same group65 also performed adoptive transfer and LCMV reinfection models to assess secondary responses of TCM and TEM at mucosal sites. Both TCM and TEM appeared compromised in their ability to form CD103+ TRMs in the gut. However, activated intestinal TRM, but not liver TRM, efficiently reformed CD103+ TRMs.
3.2. Human studies of re-circulating TRMs
Studies by Klicznik et al66 challenged the concept of strict tissue compartmentalization of CD4 TRMs in humans. The authors identified a population of circulating CD4 T cells in blood and thoracic duct lymph of healthy individuals with phenotypic, transcriptional, and clonal signatures that suggested that they were ex-TRMs originating from human skin. Using explant cultures from human skin and mass cytometric profiling of circulating CD4 T cells from healthy subjects, a fraction of human circulating CD4 T cells was shown to downregulate CD69, but still express CD103 and cutaneous lymphocyte antigen (CLA), a glycan promoting skin entry. A cluster of these circulating CD4 T cells expressed the skin-tropic chemokine receptors CCR4, CCR6 and CCR10. Clonal analysis demonstrated a greater overlap between CD4+CLA+CD103+ T cells in the blood and skin than other matched subsets. By generating human engineered skin on immunodeficient NSG mice followed by xenografting human skin from healthy donors onto the same NSG mice, the authors confirmed that human cutaneous CD4 TRMs can reenter the circulation and relocate to secondary human skin sites and reassume a TRM phenotype. Recirculating CD4 TRMs (CLA+CD103+) represent a rare population in blood of healthy humans, which, on average, accounts for <2% of circulating CD4+CLA+CD45RA− memory T cells and <0.2% of total CD4+ T cells. Estimated number of CD4+ CLA+CD103+ cells in the blood is between 2×106 and 2×107, which is approximately 250-fold lower than in the skin.66
The concept of recirculating ex-TRMs was also supported by findings in human disease settings.67–72 Diani et al67 found that CCR6+ or CXCR3+ CD8 memory T cells co-expressing CD69 are increased in the blood of psoriatic arthritis (PsA) patients, which was associated with increased systemic inflammation. In vitro transwell migration assays demonstrated preferential migration of CD8 TEMs, with a higher percentage of CXCR3+ cells and a lower percentage of CCR6+ cells, towards synovial fluid of PsA patients. In fact, an accumulation of CXCR3+ CD8 T cells was observed in synovial fluid of PsA patients. A previous study from the same group68 correlated circulating CCR4+ CD8 memory T cells co-expressing CD103 with both systemic inflammation and disease severity in psoriasis patients. These data support the hypothesis that recruitment of specific chemokine receptor-bearing CD8 T cells to inflamed joints is an important downstream event in systemic inflammation, and that a fraction of such cells may constitute recirculating ex-TRMs.
In human celiac disease (CeD), an intestinal autoimmune disease driven by dietary gluten and gluten-specific CD4 T cell responses, Han et al69 identified a large increase in circulating CD38+, αE (CD103)/β7 integrin-expressing CD8+ αβ and γδ T cells after gluten challenge. These T cells had a restricted TCR repertoire. Single-cell analysis70 of γδ and CD8+ αβ TCR sequences from both blood and gut of CeD patients before and during gluten challenge revealed extensive clonotype sharing across tissue and time, even prior to gluten challenge. More expanded clonotypes and clonal sharing between blood and gut were seen in subjects with a challenge-induced surge. However, γδ and CD8+ αβ TCR repertoires between individual patients were rather diverse, suggesting they may not be specific for the gluten antigen. These may be NKG2D+ T cells that exhibit TCR-independent cytolytic activity against epithelial cells expressing stress signals, as described in the intestinal intraepithelium of CeD patients.73 Gluten-specific CD4 TCR repertoires exhibit predominant public features, as 10% of TCRα, TCRβ, or paired TCRαβ amino acid sequences of total 1813 TCRs generated from 17 CeD patients were observed in 2 or more patients, and are shared between blood and gut tissue in CeD patients over decades.71 It is possible that recirculating ex-TRMs from both CD4 and CD8 compartments contribute to these phenomena, consistent with the “outside-in” model proposed in mice.55
In patients undergoing allogeneic hematopoietic stem cell transplantation who developed skin and gastrointestinal GVHD, Strobl et al. demonstrated that there was a population of circulating recipient skin-derived T cells with a TRM phenotype (cTRMs: CD103+CLA+CD69−/lowCD45RO+) that can produce Th2 and Th17 cytokines. Single cell RNA sequencing showed a trend toward increased TRM gene expression among these recipient-derived cTRMs and they were demonstrated to be able to exert damage to keratinocytes in skin and home to distant tissue sites during GVHD, including the gastrointestinal tract, as they expressed the gut-homing marker integrin α4β7.72
3.3. Transient, temporary resident and permanent resident T cells and potential interchangeable phenotypes between TRM and Teff in NLTs
Long-term residency of TRMs in NLTs is overlayed with elements of migration and developmental flexibility, although it is not clear whether these are features of all TRMs or only a subset of them. Systemic distribution of TRMs originating from different NLTs might contribute to broad protection against pathogens that escape local defense.56 It has been proposed that T cells within peripheral tissues may consist of cells at 3 different stages, namely transient, temporary resident, and permanent resident, characterized by rapid, slow and no recirculation between tissue and blood/lymphatics, respectively.74 Given that circulating ex-TRMs undergo changes in gene expression compared to TRMs in NLTs, it is possible that such modulation might occur within tissues before translocation to the circulation. Some TRMs in NLTs may enter into a transitioning stage (T-TRM)75 with Teff phenotypes before they egress the tissue to become circulating ex-TRMs (Figure 1). Our ongoing scRNA-seq studies27–29 provide preliminary evidence that clonally-defined alloreactive and nonalloreactive T cells in intestinal allografts can distribute in different clusters that cover both TRM and Teff phenotypes, supporting the above notion (Figure 2). Epigenetic analysis could help to further describe the identity and plasticity of organ-specific TRMs and their crosstalk with the local environment. The presence of recirculating TRMs might provide diagnostic biomarkers and targets for development of novel therapies for systemic inflammation, autoimmune diseases and allograft rejection.
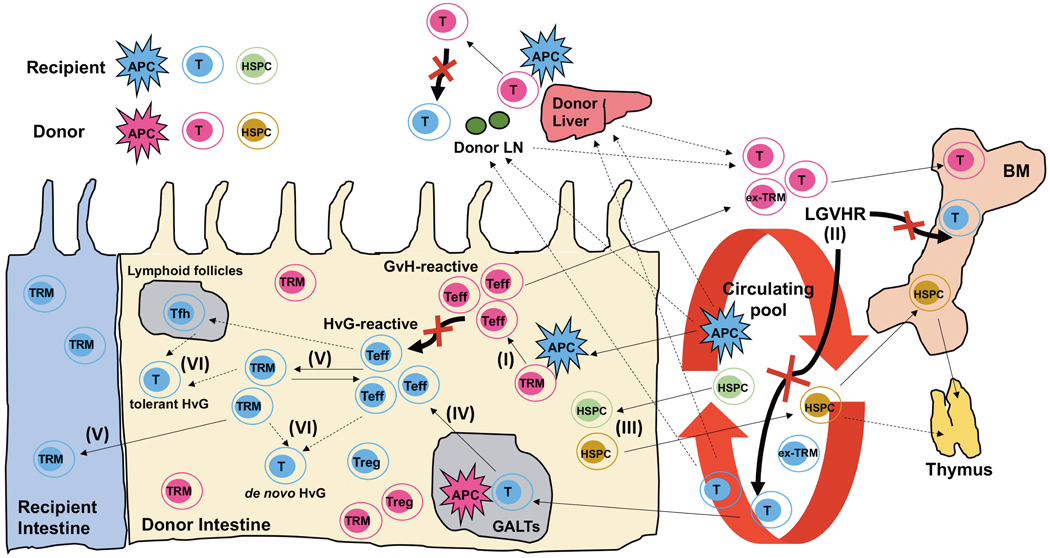
Figure 2. Role of donor- and recipient-derived tissue-resident memory T cell (TRM)-mediated bidirectional alloresponses after human intestinal transplantation (ITx), as determined by integration of T-cell clonotypes, mixed lymphocyte reaction (MLR)-determined alloreactivity, and scRNA profiling.
Major local and systemic immunological events described in our previous and ongoing studies are summarized. (I) Intestinal donor T cells are dominated by a TRM phenotype (CD69+CD103+/−CD28low). Donor GvH-reactive T cells expand within the graft (intestine/liver/LNs), likely acquiring Teff phenotypes (CD69low/−CD103low/−CD28+/high) in response to recipient APCs that enter the graft early and control recipient HvG-reactive T cells locally. Expanded GvH-reactive Teff then migrate into the recipient circulation and BM, where they attack host hematopoietic cells and counteract HvG responses (bold black arrows with red “X” to show inhibition effect). This LGVHR (II) makes hematopoietic “space” for donor cell engraftment early post-Tx and (III) allows the survival and expansion of donor HSPCs from the graft that enter the circulation, BM, and thymus, resulting in de novo donor T cell generation and promoting persistent multilineage chimerism and potentially promoting immune tolerance. Intestinal HSPCs undergo replacement by the recipient from a circulating pool. (IV) Circulating recipient T cells (CD69−CD103−CD28+/high) infiltrating donor intestinal allografts are primed by donor APCs, likely in gut-associated lymphoid tissues (GALTs), and become HvG-reactive T cells with Teff phenotypes that repopulate the graft mucosa early post-Tx. (V) These recipient Teff cells gradually acquire a TRM phenotype during quiescence and seed the entire GI tract, including the residual recipient intestine. They can regain features of circulating Teff during late rejection (eg: upregulation of CD28 and NKG2D). Some of these recipient Teff cells may enter into lymphoid follicles in graft mucosa and become follicular helper T cells (Tfh). (VI) Persistent alloreactive recipient T cells as TRM and/or Tfh may pose a constant risk of recurring rejection or become tolerized (tolerant HvG). De novo-generated HvG-reactive T cells post-Tx and donor- and recipient-derived Tregs may also influence the bidirectional alloresponses after ITx.
4. Distribution of TRM in human NLTs and association with clinical outcomes in different types of SOT
TRMs residing in different organs must accommodate to unique local environments, given that each anatomic site differs in cytokines, nutrients, and composition of epithelial and connective tissues. Murine studies have indicated that tissue topography, which considers tissue spatial structure and cell-cell interaction in a particular microenvironment, such as epidermis (epithelial tissue), exocrine glands (epithelial-connective tissue) and lymphoid organs (connective tissue), may influence CD8 TRM retention and surveillance strategies.76 TRMs rely on chemokine- and integrin-related mechanisms to be retained in epithelial barrier sites.77 However, TRMs in exocrine glands display autonomous motility that is supported by tissue macrophages and independent of chemoattractants and adhesive molecules.78
In considering the role of TRM and graft-versus-host (GvH) alloreactivity in transplant outcomes, it should be remembered that intestine, lung and liver are rich lymphoid organs, whereas kidney and heart are not, which may affect the balance of bidirectional alloresponses after each type of SOT (Figure 2). Blood contamination should also be considered in T cell phenotype and clonotype analysis of vascular organs like liver and kidney. In this section, we discuss human TRMs in different types of SOT and associated graft outcomes.
4.1. TRMs in ITx
As the only long-term option for patients who suffer intestinal failure, ITx is complicated by high rejection rates and consequences of high levels of immunosuppression such as infection, renal dysfunction and de novo malignancy.79,80 Our previous studies showed that peripheral blood macrochimerism, defined as the presence of ≥4% of donor T cells, developed frequently after multivisceral transplantation, usually without causing GVHD. Blood macrochimerism is associated with significantly reduced graft rejection or donor-specific antibody development and slower replacement of donor T cells in the graft by the recipient.16,17,81,82 These observations link local and systemic immunological events. A faster rate of recipient T cell predominance over donor T cells in the graft mucosa correlated with early rejection, which was associated with a preponderance of host-versus-graft (HvG) T cell clones.16,17 Donor T cells persisted in the mucosa for several years in patients lacking rejection, only gradually being replaced by recipient T cells16–19. While intragraft donor T cells were dominated by a TRM phenotype (CD69+CD103+/−CD28low), recipient Teffs, including HvG-reactive T cells, infiltrating the intestinal mucosa slowly acquired a TRM phenotype during quiescence and regained features of circulating Teff during late rejections (eg: upregulation of CD28 and NKG2D).16 The persistence of alloreactive recipient HvG T cells as TRM may pose a constant risk of rejection, perhaps contributing to high intestinal allograft rejection rates (Figure 2).
Studies of intestinal TRMs (CD69+CD161+CD103+/−) after human ITx have also been performed by Jahnsen and colleagues18,19 in patients without rejection. Donor TRMs from duodenal grafts (transplanted with pancreas) were shown to persist for at least 52 weeks. In normal donors,18,19 lamina propria CD8 TRMs demonstrated a polyfunctional profile (IFN-γ+IL-2+TNF-α+) and were potently cytotoxic following stimulation.19 Similarly, the vast majority of lamina propria CD4 TRMs were polyfunctional Th1 cells (IFN-γ+IL-2+TNF-α+), and a fraction produced GZMB and perforin after activation.18
Given that rejection episodes are closely associated with accelerated replacement kinetics of graft T cells after ITx,16 and the potential involvement of recirculating ex-TRMs with re-differentiation plasticity, our group is actively pursuing multiomic studies to integrate T cell clonotypes, alloreactivity and gene expression profiles.16,17,27–29 Clonal and phenotypic tracking of donor and recipient T cells in serial intestinal allograft biopsies, peripheral blood and bone marrow (BM) post-Tx provide a deeper understanding of their tissue origin, migration pattern and phenotypic maturation. We also use a unique platform17 that integrates bulk TCRβ-seq and scRNA-seq that combines 5’ transcriptional analysis with TCRαβ-seq. T cells are further annotated as CD4 or CD8 alloreactive or nonalloreactive or as nonmappable by interrogation of the sequence set defined from pre-Tx MLRs,16,25 allowing functional characterization of known alloreactive T cells within the allograft tissue.
We recently demonstrated that donor GvH-reactive T cells expand within the intestinal allograft in response to recipient antigen-presenting cells (APCs) that enter the graft early. These GvH-reactive T cells appear to control recipient HvG-reactive T cells locally, then migrate into the recipient circulation and BM, where they attack host hematopoietic cells and counteract HvG responses17 (Figure 2). This lymphohematopoietic graft-versus-host response (LGVHR) usually occurs without causing GVHD. Single-cell transcriptional profiling of BM-infiltrating donor T cells reveals dominant clusters of cytotoxic Teffs with GvH allorecognition identifiable by TCR sequences. This LGVHR makes hematopoietic “space” for donor cell engraftment early post-Tx and allows the survival and expansion of donor hematopoietic stem and progenitor cells from the graft that enter the circulation, BM, and thymus, resulting in de novo donor T cell generation and promoting persistent multilineage chimerism and potentially promoting immune tolerance.81,82 Individual GvH clones were detected in either the ileal mucosa or PBMCs before detection in recipient BM,17 consistent with an origin in the intestinal allograft, and consistent with our hypothesis that circulating and BM-infiltrating GvH-reactive T cells originated as microbe-reactive TRMs, carried in the intestinal allograft, that cross-react on recipient alloantigens.28 We are currently utilizing the above multiomic platform to test the hypothesis that alloreactive HvG T cells infiltrating the intestinal allograft join the TRM pool and may become Teffs that either participate in recurring rejection and/or are tolerized27 (Figure 2).
In a cohort of patients who developed GVHD after ITx, Weiner et al. described a putative circulating TRM population (CD69+CD62L−HLA−DR+CD57+PD-1+) that they believed to have been derived from the donor intestinal graft and was associated with GVHD.83 Interestingly, the percentages of CD69+ TRMs not only increased in the blood of patients with GVHD compared to those without, but also in parallel increased in the grafted and native intestines of GVHD patients. These were demonstrated to be donor-derived using multiplex immunostaining in the cases involving HLA-A2 host-donor mismatch. These observations support the scenario whereby TRMs with transitioning phenotypes (T-TRMs) translocate from donor intestines to recipient blood and further home to native NLTs to contribute to GVHD pathophysiology.
4.2. TRMs in lung transplantation (LuTx)
Like the intestine, human lung is also an immunological barrier organ that is highly enriched for TRMs. Therefore, bidirectional alloresponses may also contribute to graft outcomes after LuTx. Blood chimerism after LuTx20,84,85 is detected, but is not as robust or persistent as that observed in ITx.17,81,82 Blood chimerism was detected in 3 of twenty LuTx recipients and disappeared by 2 months after transplant.20 However, donor T cells were found to persist in bronchoalveolar lavage (BAL) samples at high frequencies even after 1 year, at levels up to 55% and 85% for CD4 and CD8 T cells, respectively.20 These donor-derived BAL T cells expressed high levels of TRM markers including CD69, CD103 and CD49a. Consistent with observations in ITx patients,16,17, persistence of donor TRMs after human LuTx was associated with better clinical outcomes, as reflected by less acute cellular rejection and primary graft dysfunction.20 Earlier studies reported a much more rapid replacement of donor-derived lung TRM by recipient T cells within 40 days post-Tx when assessed by serial transbronchial biopsies.86 Again, low numbers of donor lymphocytes in the allografted lung seemed to correlate with a worse clinical course. Taken together, monitoring TRM dynamics in lung allograft and BAL samples may be clinically informative.
Not surprisingly, acquisition of TRM phenotypes within graft-repopulating recipient T cells after LuTx6,20,87 also mirrored the phenomenon observed after ITx.16,18,19 A longitudinal study of LuTx showed that 3 to 9 months after transplant, the numbers of CD4 and CD8 T cells in the lung can reach normal healthy donor levels. However, the number of CD3 and CD8 (but not CD4) T cells in post-Tx lung allografts continued to increase over time, regardless of the development of chronic rejection,87 suggesting that graft-associated CD8 T cells participate in both homeostasis and chronic rejection. CMV− LuTx recipients who received CMV+ allografts demonstrated an influx of de novo CMV-specific CD8 T cells into the airways and allografts. These cells were maintained within the transplanted lung at higher frequencies than within PBMCs and were functionally and phenotypically distinct from circulating CMV-specific CD8 T cells.88 A longitudinal study demonstrated low frequencies of TRM markers among recipient BAL T cells at early times (2 to 4 weeks) after LuTx (20 to 40% CD69+ CD4+ and CD8+ T cells); however, by 6 months post-Tx, >50% of recipient BAL T cells were CD69+, with many recipient CD8+ T cells co-expressing CD103. By 3 to 6 months post-Tx, recipient BAL T cells expressed CD69 and CD103 at frequencies similar to those observed in control BAL fluid. Recipient-derived T cells in the lung BAL maintained multifunctional profiles (IFN-γ+ IL-17+ IL-2+GZMB+) associated with mucosal memory T cells. scRNA-seq revealed both non-TRM (putative circulating TEM lacking CD69 and CD103 expression) and TRM-like (CD69+CD103+/−ITGA1+CXCR6+RUNX3+) subpopulations among recipient BAL T cells.20 As in ITx recipients,16 the gradual acquisition of TRM markers by recipient-derived T cells infiltrating lung allografts may reflect the conversion to this phenotype of HvG T cells that pose a constant risk of rejection or the repopulation of donor lung tissue by circulating TEM counterparts of TRMs that acquire TRM phenotypes or by recirculating ex-TRMs.
4.3. TRMs in liver transplantation (LiTx)
The liver is immunologically unique. On 1 hand, it is exposed to a variety of microbes from the systemic circulation or through the portal vein from the gut. On the other hand, it preferentially induces immune tolerance,89 in the context of both MHC-mismatched LiTx (in rodents) and liver infection. However, intrahepatic immune responses can be robustly induced under certain circumstances such as viral or autoimmune hepatitis. Unlike in gut and lung, CD69+ CD8 TRMs in human liver constitute a large proportion (>90%) of CD103− cells and demonstrate low cytotoxicity.90,91 CD103+ liver CD8 TRMs express liver-homing and retention markers CXCR6 and CXCR3, and robustly produce IL-2 and IFNγ upon antigen stimulation.84 Kim et al92 demonstrated that liver CD69+CD103− CD8 T cells have a terminally differentiated TRM phenotype, and their effector functions depend on hypoxia-inducible factor (HIF)-2α, suggesting that they are predominantly located in hypoxic regions. Furthermore, activation of liver CD69+CD103− CD8 T cells with HIF-2α upregulation is observed during acute (hepatitis A) and chronic (cirrhosis) liver pathology. Swadling et al93 found that an increased rate of basal autophagy is a hallmark of human liver CD8 TRMs. Enhanced autophagy in CD8 TRMs can be imprinted by IL-15 or primary hepatic stellate cells and adapts liver CD8 TRMs to combat mitochondrial depolarization and acquire tissue residence. These findings highlight the importance of tissue-specific adaptations of TRMs.
In the LiTx setting, a small population of donor CD4 and CD8 T cells with TRM phenotypes (CD69+CD103+/−CXCR3hi) was detectable in liver grafts even more than a decade after an HLA-mismatched transplant.23 Recipient CD4 and CD8 T cells are also persistently observed.22,23 These graft-repopulating T cells may have a TRM phenotype, although with a less definitive residency program, such as lower levels of CXCR3 compared to donor-derived CD8 TRMs.23 Despite the requirement of unique environmental conditions for liver TRMs, the persistence of intrahepatic donor TRM and the gradual acquisition of TRM phenotypes by graft-infiltrating recipient T cells after LiTx are reminiscent of findings after human ITx and LuTx, underlining the common features of donor- and recipient-derived TRMs in allograft organs. The authors further showed that TRMs lacking CXCR6 expression were detectable in the local draining LNs but did not egress into the hepatic vasculature. Whether these TRMs migrate from the liver graft or represent an independent population developed in situ will need further investigation, for example using TCR clonal tracking.23 While the association of rejection and the dynamic replacement of intragraft donor T cells by the recipient was not explored, studies of two antiviral responses (HBV, CMV) revealed that donor-derived virus-specific CD8 TRMs persist long-term post-Tx and may be supplemented by recipient responses.23
4.4. TRMs in Kidney transplantation (KTx)
Human kidneys contain small numbers of lymphocytes compared to intestine, lungs and liver. The composition and profile of immune cell subsets in human kidneys is largely unknown. Park et al94 identified a predominant CD3+ T cell (47%±12%) population and a low proportion of CD14+ or CD68+ myeloid cells (<10%) in healthy human kidney sections. Kidney T cells included 44% CD4 and 56% CD8 subsets. An average of close to 50% of T cells displayed a TRM phenotype (CD69+CCR7−CD45RA−), while the rest had a TEM phenotype (CD69−CCR7−CD45RA−). It should be borne in mind that non-TRM populations in the healthy kidney may include circulating T cells present in the rich vasculature of the organ. Among kidney TRMs, CD103−CD49a+/− cells were predominant in CD4 cells and CD103−CD49a+/− and CD103+CD49a+ subsets were predominant in CD8 cells.
Drachenberg and colleagues revealed a correlation of CD8+CD103+ cytolytic T cells (CTLs) that are CD62L−CD11ahiperforin+ with clinical renal allograft rejection by analyses of transplant nephrectomy specimens. These CD103+ CD8 CTLs comprised 40–50% of the graft-infiltrating lymphocyte population during late acute rejection and were also present in biopsies with signs of chronic rejection.95,96 CD103+ CD8 CTLs were biased towards an intratubular localization, while a CD103− subset of graft-infiltrating CD8 T cells that also exhibited a CTL phenotype was restricted to the graft interstitium.96
De Leur et al tracked the turnover dynamics of donor intragraft T cell replacement by the recipient in transplant nephrectomies.24 High proportions (1.7–17.4%) of donor-derived CD4 and CD8 T cells were only observed in early rejecting allografts removed within the first month post-Tx. Grafts that failed greater than 5 months post-Tx mainly contained recipient-derived CD8 TRMs (CD103+/−CCR7−CD45RO+) that produced IFNγ, TNFα and GZMB.
4.5. TRMs in heart transplantation (HTx)
Both protective97 and detrimental98,99 roles of donor T lymphocytes carried in cardiac allografts have been reported in animal models. Meanwhile, studies on the lymphocyte compartment of human hearts are very limited. Hu et al100 created a single-cell atlas of human nondiseased cardiac arteries obtained from HTx patients. T cells were the fourth largest cell population in coronary arteries, at 14.9%. CD4 T cells represented a higher percentage than CD8 T cells in coronary arteries and TRM markers CD69 and CD44 were highly expressed while CD103 and CD49a were poorly expressed. While the authors suggest that these may be specific features of vascular TRM, it is uncertain that these cells are truly TRMs rather than activated T cells. CD8 T cells in human cardiac arteries also highly expressed cytotoxic markers including granzyme family members and perforin, but had low expression of proinflammatory cytokines TNF and IFNγ. Whether these cells play a specific role in the outcome of clinical HTx will require further investigation. Graft-infiltrating T cells after HTx have been associated with rejection in both animals101 and humans.102 However, a lack of longitudinal phenotypic and functional studies makes comparison with other types of human transplants impossible at this time.
5. Tregs with TRM features in transplantation
The presence of Tregs in a variety of NLTs has been documented in both mice and humans, including intestinal mucosa, lung, liver, skin, kidney, adipose tissue and skeletal muscle, where they maintain host homeostasis and self-tolerance.103–107 Emerging data from mouse studies shows that tissue Tregs exhibit unique phenotypic and transcriptional signatures that are controlled by epigenetic reprogramming.105,106,108,109 Tissue Tregs express TRM surface markers such as CD69, CD103 and CCR4 and express the TRM TF Blimp1.110–112 Integrated accessible-chromatin and scRNA-seq109 indicated that adaptation of Tregs to visceral adipose tissue, skeletal muscle and colon reflected a combination of tissue-shared and tissue-specific modulations. Tissue adaptation of human Tregs is still largely undefined. Transcriptional analysis of human skin Tregs demonstrated a TRM phenotype similar to skin-tropic (CLA+) helper CD4 T cells and CD103− CD8 T cells, but these Tregs were distinct from blood-derived CLA+ T cells.113 A comprehensive study to characterize the transcriptome of human mucosal tissue (lung and colon) Tregs from the normal area of cancer resections and their peripheral blood counterparts106 identified TNIP3 as a shared Treg-specific gene that is involved in the regulation of NF-κB signaling. The most prominent genes differentiating lung Treg from gut or blood-derived Treg were Wnt singling genes, suggesting potential crosstalk between lung Tregs with nonmtor immune tissue-specific cells and a role for lung Tregs in epithelial repair and regeneration.
The role of tissue Tregs in mediating tolerance after human SOT was investigated in a limited number of studies. Foxp3+ Tregs in renal allografts with subclinical rejection are associated with significantly better graft function 2 and 3 years post-Tx.114 Urinary Foxp3 mRNA is diagnostic of T cell-mediated rejection (TCMR), and can also predict TCMR reversibility after KTx.115 Direct evidence that tissue Tregs participate in regulating KTx tolerance was reported by our group116,117 in the setting of HLA-haploidentical combined bone marrow and kidney transplants (CKBMT). Enrichment of Tregs measured by Foxp3 expression were found in long-term protocol biopsies of renal allografts from tolerant patients after CKBMT.117 By applying high throughput TCRβ-seq, more than 200 Treg clones identified in sorted circulating Treg populations, were detected in each kidney biopsy from 3 CKBMT subjects.116 Using TCRβ-seq to identify the donor-specific Treg repertoire, we demonstrated expansion of circulating donor-specific Treg clones in tolerant subjects, but not in a patient who failed tolerance, at 6 months post-Tx, implicating donor-specific Tregs in initiating tolerance induction.118 A study in human LuTx patients showed that most Tregs (CD4+CD25+CD127loFoxp3+) in BAL were recipient-derived, even in samples with significant T cell chimerism, suggesting rapid replenishment of Tregs from the circulation after transplant.20 Our ongoing single-cell profiling27–29 of T cells in intestinal allografts has identified a small fraction of tissue Tregs (Foxp3+) with TRM features (RGS1+CXCR6+CCR6+) among both donor- and recipient-derived T cell populations 600–1800 days post-Tx, suggesting long-term residency of donor-derived tissue Tregs and the potential acquisition of TRM features of graft-infiltrating recipient Tregs. Further investigations are needed to understand tissue adaptation of Tregs after transplantation.
6. γδ TRM in transplantation
γδ T cells have both innate and adaptive properties and are implicated in immune surveillance and modulation.119 γδ TCRs can recognize structurally diverse and biologically unrelated antigens mainly through MHC-independent mechanisms,119,120 with only minor, if any, alloreactivity reported in earlier in vitro studies.121,122 Therefore, γδ T cells have garnered interest as mediators of graft-versus-leukemia effects without GVHD in the setting of allogeneic hematopoietic stem cell transplantation.123 Accumulating evidence indicates that innate- and adaptive-like features of human γδ T cells may be driven by differential γδ TCR repertoires, generally defined as Vγ9+δ2+ and non-Vγ9δ2, respectively.124 Immune repertoires can be shaped by tissue compartmentalization, age and history of antigen exposure.124–127 Although γδ T cells only account for <10% of T cells in human peripheral blood, with a dominant semi-invariant TCR Vγ9Vδ2, they are often enriched in human solid organs and barrier sites and have heterogenous TCRs, such as Vγ2/3/4/5/8 and Vδ1/3/5. These can constitute between 10–100% of T cells in gut, lung, liver and skin.124,128 The non-Vγ9δ2 repertoire appears to be shaped by TCR-dependent selection events including CMV infection and cancer.125 This association of V-gene usage with tissue distribution and functional development of human γδ TCR is reminiscent of their mouse counterparts.129,130
The role of γδ T cells in SOT outcomes remains unclear. γδ T cells might contribute to both allograft acceptance and rejection, and could impact infection and post‐Tx malignancy.120 Earlier functional studies were limited to in vitro systems of human peripheral blood-derived γδ T cells,131,132 which are dominated by Vδ2 clonotypes. γδ T cells had been shown to exhibit either direct veto-type suppression of alloreactions,131 or indirect stimulation of alloreactive αβ T cell proliferation by inducing maturation of autologous dendritic cells and B cells into functional APCs.132 A recent study133 demonstrated that the γδ T cell pool in liver explants collected from patients who underwent LiTx for end-stage liver disease included a TRM compartment that is CD69+CXCR3+CXCR6+CD45RAlo and enriched for “private” Vδ1 clonotypes. These liver-resident Vδ1 T cells were found to be polyfunctional and responded to both TCR and cytokine stimuli in vitro. Although a higher ratio of Vδ1/Vδ2 in the peripheral blood of liver allograft recipients has been shown to correlate with stable graft function134 and operational tolerance,135 there are few studies investigating the association of rejection with the turnover dynamics and clonal reconstitution of intragraft γδ T cells after human LiTx.
Our recent published17 and ongoing studies136 have provided further insights into the role of γδ T cells in modulating 2-way alloresponses locally and systemically after human ITx. Single-cell profiling of BM-infiltrating donor γδ T cells revealed a dominant “public” Vδ2+ clonotype with cytotoxic Teff phenotypes similar to their CD8 αβ counterparts. Graft-repopulating recipient γδ T cells show an activated Teff phenotype early post-Tx and gradually develop into TRMs with a dominant “private” Vδ1+ clonotype, likely participating in graft defense and regulating graft rejection. There are still many gaps to explore in this area.
Acknowledgments
Funding
This work was supported in part by National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH) grant #P01 AI106697. J.F. was supported by a Congressionally Directed Medical Research Program (CDMRP) Discovery Award W81XWH-20-1-0159, funded by the Department of Defense (DoD), an R21 grant AI166069 supported by NIH/NIAID and a Nelson Faculty Development Award UR011630-01, funded by the Nelson Family Transplantation Innovation Award Program at Columbia University Irving Medical Center.
Abbreviations
- APCs
antigen-presenting cells
- BAL
bronchoalveolar lavage
- BM
bone marrow
- CeD
celiac disease
- CKBMT
combined bone marrow and kidney transplants
- CLA
cutaneous lymphocyte antigen
- CTLs
cytolytic T cells
- GALTs
gut-associated lymphoid tissues
- GvH
graft-versus-host
- GVHD
graft-versus-host disease
- HIF
hypoxia-inducible factor
- HSPCs
hematopoietic stem and progenitor cells
- HSV
herpes simplex virus
- HTx
heart transplantation
- HvG
host-versus-graft
- ITx
intestinal transplantation
- KTx
Kidney transplantation
- LCMV
lymphocytic choriomeningitis virus
- LGVHR
lymphohematopoietic graft-versus-host responses
- LiTx
liver transplantation
- Lm-OVA
Listeria monocytogenes-expressing ovalbumin
- LNs
lymph nodes
- LuTx
lung transplantation
- MLR
mixed lymphocyte reaction
- NLTs
non-lymphoid tissues
- post-Tx
post-transplant
- pre-Tx
pretransplant
- PsA
psoriatic arthritis
- scRNA-seq
single cell RNA sequencing
- SLOs
secondary lymphoid organs
- SOT
solid organ transplantation
- TCM
central memory T cell
- TCMR
T cell-mediated rejection
- Teffs
effector T cells
- TEM
effector memory T cell
- Tfh
follicular helper T cell
- Tregs
regulatory T cells
- TRM
tissue-resident memory T cell
- T-TRM
transitioning TRM
- Tx
transplantation
Footnotes
Authorship
JF and MS collected data and wrote the review.
Disclosure
The authors declare no conflicts of interest.
References
- 1.Prosser AC, Kallies A, Lucas M. Tissue-resident lymphocytes in solid organ transplantation: innocent passengers or the key to organ transplant survival? Transplantation. 2018;102(3):378–386. [DOI] [PubMed] [Google Scholar]
- 2.Sun H, Sun C, Xiao W, et al. Tissue-resident lymphocytes: from adaptive to innate immunity. Cellular & Molecular Immunology. 2019;16(3):205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schenkel JM, Masopust D. Tissue-resident memory T cells. Immunity. 2014;41(6):886–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fan X, Rudensky AY. Hallmarks of tissue-resident lymphocytes. Cell. 2016;164(6):1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beura LK, Rosato PC, Masopust D. Implications of Resident Memory T Cells for Transplantation. Am J Transplant. 2017;17(5): 1167–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turner DL, Gordon CL, Farber DL. Tissue-resident T cells, in situ immunity and transplantation. Immunol Rev. 2014;258(1):150–166. [DOI] [PubMed] [Google Scholar]
- 7.Enderby C, Keller CA. An overview of immunosuppression in solid organ transplantation. Am J Manag Care. 2015;21(1 suppl):s12–23. [PubMed] [Google Scholar]
- 8.Pilch NA, Bowman LJ, Taber DJ. Immunosuppression trends in solid organ transplantation: The future of individualization, monitoring, and management. Pharmacotherapy. 2021;41(1):119–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szabo PA, Miron M, Farber DL. Location, location, location: Tissue resident memory T cells in mice and humans. Sci Immunol. 2019;4(34). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhai Y, Meng L, Gao F, et all. Allograft rejection by primed/memory CD8+ T cells is CD154 blockade resistant: therapeutic implications for sensitized transplant recipients. J Immunol. 2002;169(8):4667–4673. [DOI] [PubMed] [Google Scholar]
- 11.Valujskikh A, Pantenburg B, Heeger PS. Primed allospecific T cells prevent the effects of costimulatory blockade on prolonged cardiac allograft survival in mice. Am J Transplant. 2002;2(6): 501–509. [DOI] [PubMed] [Google Scholar]
- 12.Zeevi A, Husain S, Spichty KJ, et al. Recovery of functional memory T cells in lung transplant recipients following induction therapy with alemtuzumab. Am J Transplant. 2007;7(2):471–475. [DOI] [PubMed] [Google Scholar]
- 13.Pearl JP, Parris J, Hale DA, et al. Immunocompetent T-cells with a memory-like phenotype are the dominant cell type following antibody-mediated T-cell depletion. Am J Transplant. 2005;5(3):465–474. [DOI] [PubMed] [Google Scholar]
- 14.Nicosia M, Fairchild RL, Valujskikh A. Memory T Cells in Transplantation: Old Challenges Define New Directions. Transplantation. 2020;104(10):2024–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Ruiz P. Solid organ transplant-associated acute graft-versus-host disease. Arch Pathol Lab Med. 2010;134(8):1220–1224. [DOI] [PubMed] [Google Scholar]
- 16.Zuber J, Shonts B, Lau SP, et al. Bidirectional intragraft alloreactivity drives the repopulation of human intestinal allografts and correlates with clinical outcome. Sci Immunol. 2016;1(4):eaah3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu J, Zuber J, Shonts B, et al. Lymphohematopoietic graft-versus-host responses promote mixed chimerism in patients receiving intestinal transplantation. J Clin Invest. 2021;131(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bartolomé-Casado R, Landsverk OJB, Chauhan SK, et al. CD4(+) T cells persist for years in the human small intestine and display a T(H)1 cytokine profile. Mucosal Immunol. 2021;14(2):402–410. [DOI] [PubMed] [Google Scholar]
- 19.Bartolomé-Casado R, Landsverk OJB, Chauhan SK, et al. Resident memory CD8 T cells persist for years in human small intestine. J Exp Med. 2019;216(10): 2412–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snyder ME, Finlayson MO, Connors TJ, et al. Generation and persistence of human tissue-resident memory T cells in lung transplantation. Sci Immunol. 2019;4(33). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bellmàs Sanz R, Hitz A, Wiegmann B, et al. Donor T and NK cells with a special tissue-resident memory phenotype migrate into the periphery of lung transplant recipients - a potential feature for tolerance development. J Heart Lung Transplant. 2020;39(4, supplement): S198. [Google Scholar]
- 22.Taubert R, Danger R, Londoño MC, et al. Hepatic infiltrates in operational tolerant patients after liver transplantation show enrichment of regulatory T cells before proinflammatory genes are downregulated. Am J Transplant. 2016;16(4):1285–1293. [DOI] [PubMed] [Google Scholar]
- 23.Pallett LJ, Burton AR, Amin OE, et al. Longevity and replenishment of human liver-resident memory T cells and mononuclear phagocytes. J Exp Med. 2020;217(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Leur K, Dieterich M, Hesselink DA, et al. Characterization of donor and recipient CD8+ tissue-resident memory T cells in transplant nephrectomies. Sci Rep. 2019;9(1):5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris H, DeWolf S, Robins H, et al. Tracking donor-reactive T cells: Evidence for clonal deletion in tolerant kidney transplant patients. Sci Transl Med. 2015;7(272): 272ra210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeWolf S, Grinshpun B, Savage T, et al. Quantifying size and diversity of the human T cell alloresponse. JCI Insight. 2018;3(15). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu J, Wang Z, Martinez M, et al. Single cell immune profiling of human intestinal allografts reveals heterogeneity and alloreactivity of resident memory T cells in association with graft outcomes. Transplantation. 2020;104(S3):S72–S73. [Google Scholar]
- 28.Fu J, Zuber J, Shonts B, et al. Clonal and functional analysis reveals the capacity of allograft t cells to join the circulating pool after human intestinal transplantation. Transplantation. 2018;102:S420–S421. [Google Scholar]
- 29.Fu J, Wang Z, Martinez M, et al. O-40: single-cell immune profiling of human intestinal allografts reveals differential contributions of HvG T-cell clones in quiescent vs chronically rejecting allografts. Transplantation. 2021;105(7S). [Google Scholar]
- 30.Teijaro JR, Turner D, Pham Q, et al. Cutting edge: Tissue-retentive lung memory CD4 T cells mediate optimal protection to respiratory virus infection. J Immunol). 2011;187(11):5510–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang X, Clark RA, Liu L, et al. Skin infection generates non-migratory memory CD8+ T(RM) cells providing global skin immunity. Nature. 2012;483(7388):227–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klonowski KD, Williams KJ, Marzo AL, et al. Dynamics of blood-borne CD8 memory T cell migration in vivo. Immunity. 2004;20(5):551–562. [DOI] [PubMed] [Google Scholar]
- 33.Gebhardt T, Wakim LM, Eidsmo L, et al. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat Immunol. 2009;10(5):524–530. [DOI] [PubMed] [Google Scholar]
- 34.Masopust D, Choo D, Vezys V, et al. Dynamic T cell migration program provides resident memory within intestinal epithelium. J Exp Med. 2010;207(3):553–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glennie ND, Yeramilli VA, Beiting DP, et al. Skin-resident memory CD4+ T cells enhance protection against Leishmania major infection. J Exp Med. 2015;212(9):1405–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turner DL, Bickham KL, Thome JJ, et al. Lung niches for the generation and maintenance of tissue-resident memory T cells. Mucosal Immunol. 2014;7(3):501–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson KG, Sung H, Skon CN, et al. Cutting edge: intravascular staining redefines lung CD8 T cell responses. J Immunol. 2012;189(6):2702–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steinert EM, Schenkel JM, Fraser KA, et al. Quantifying memory CD8 T cells reveals regionalization of immunosurveillance. Cell. 2015;161(4):737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wakim LM, Woodward-Davis A, Bevan MJ. Memory T cells persisting within the brain after local infection show functional adaptations to their tissue of residence. Proc Natl Acad Sci USA. 2010;107(42):17872–17879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ely KH, Cookenham T, Roberts AD, et al. Memory T cell populations in the lung airways are maintained by continual recruitment. J Immunol. 2006;176(1):537–543. [DOI] [PubMed] [Google Scholar]
- 41.Schenkel JM, Fraser KA, Vezys V, et al. Sensing and alarm function of resident memory CD8⁺ T cells. Nat Immunol. 2013;14(5):509–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar BV, Ma W, Miron M, et al. Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep. 2017;20(12):2921–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Masopust D, Soerens AG. Tissue-resident T cells and other resident leukocytes. Ann Rev Immunol. 2019;37: 521–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bankovich AJ, Shiow LR, Cyster JG. CD69 suppresses sphingosine 1-phosophate receptor-1 (S1P1) function through interaction with membrane helix 4. J Biol Chem. 2010;285(29): 22328–22337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Erle DJ. Intraepithelial lymphocytes. Scratching the surface. Curr Biol. 1995;5(3):252–254. [DOI] [PubMed] [Google Scholar]
- 46.Qiu Z, Chu TH, Sheridan BS. TGF-β: many paths to CD103(+) CD8 T cell residency. Cells. 2021;10(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haddadi S, Thanthrige-Don N, Afkhami S, et al. Expression and role of VLA-1 in resident memory CD8 T cell responses to respiratory mucosal viral-vectored immunization against tuberculosis. Sci Rep. 2017;7(1): 9525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheuk S, Schlums H, Gallais Sérézal I, et al. CD49a Expression Defines Tissue-Resident CD8(+) T Cells Poised for Cytotoxic Function in Human Skin. Immunity. 2017;46(2): 287–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akbaba H. Resident Memory T Cells. Cells of the Immune System: IntechOpen; 2020. [Google Scholar]
- 50.He S, Wang LH, Liu Y, et al. Single-cell transcriptome profiling of an adult human cell atlas of 15 major organs. Genome Biol. 2020;21(1): 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dubois A, Gopee N, Olabi B, Haniffa M. Defining the Skin Cellular Community Using Single-Cell Genomics to Advance Precision Medicine. J Invest Dermatol. 2021;141(2): 255–264. [DOI] [PubMed] [Google Scholar]
- 52.Kleshchevnikov V, Shmatko A, Dann E, et al. Comprehensive mapping of tissue cell architecture via integrated single cell and spatial transcriptomics. bioRxiv. 2020: 2020.2011.2015.378125. [Google Scholar]
- 53.Thome JJ, Bickham KL, Ohmura Y, et al. Early-life compartmentalization of human T cell differentiation and regulatory function in mucosal and lymphoid tissues. Nature medicine. 2016;22(1): 72–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Samat AAK, van der Geest J, Vastert SJ, van Loosdregt J, van Wijk F. Tissue-Resident Memory T Cells in Chronic Inflammation-Local Cells with Systemic Effects? Cells. 2021;10(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fonseca R, Beura LK, Quarnstrom CF, et al. Developmental plasticity allows outside-in immune responses by resident memory T cells. Nat Immunol. 2020;21(4): 412–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gratz IK, Campbell DJ. Resident memory T cells show that it is never too late to change your ways. Nat Immunol. 2020;21(4): 359–360. [DOI] [PubMed] [Google Scholar]
- 57.Gebhardt T, Whitney PG, Zaid A, et al. Different patterns of peripheral migration by memory CD4+ and CD8+ T cells. Nature. 2011;477(7363): 216–219. [DOI] [PubMed] [Google Scholar]
- 58.Tomura M, Yoshida N, Tanaka J, et al. Monitoring cellular movement in vivo with photoconvertible fluorescence protein “Kaede” transgenic mice. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(31): 10871–10876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bromley SK, Yan S, Tomura M, Kanagawa O, Luster AD. Recirculating memory T cells are a unique subset of CD4+ T cells with a distinct phenotype and migratory pattern. Journal of immunology (Baltimore, Md : 1950). 2013;190(3): 970–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Collins N, Jiang X, Zaid A, et al. Skin CD4(+) memory T cells exhibit combined cluster-mediated retention and equilibration with the circulation. Nat Commun. 2016;7: 11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beura LK, Fares-Frederickson NJ, Steinert EM, et al. CD4(+) resident memory T cells dominate immunosurveillance and orchestrate local recall responses. The Journal of experimental medicine. 2019;216(5): 1214–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schenkel JM, Fraser KA, Masopust D. Cutting edge: resident memory CD8 T cells occupy frontline niches in secondary lymphoid organs. Journal of immunology (Baltimore, Md : 1950). 2014;192(7): 2961–2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beura LK, Wijeyesinghe S, Thompson EA, et al. T Cells in Nonlymphoid Tissues Give Rise to Lymph-Node-Resident Memory T Cells. Immunity. 2018;48(2): 327–338.e325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Behr FM, Parga-Vidal L, Kragten NAM, et al. Tissue-resident memory CD8(+) T cells shape local and systemic secondary T cell responses. Nat Immunol. 2020;21(9): 1070–1081. [DOI] [PubMed] [Google Scholar]
- 65.Behr FM, Beumer-Chuwonpad A, Kragten NAM, Wesselink TH, Stark R, van Gisbergen K. Circulating memory CD8(+) T cells are limited in forming CD103(+) tissue-resident memory T cells at mucosal sites after reinfection. Eur J Immunol. 2021;51(1): 151–166. [DOI] [PubMed] [Google Scholar]
- 66.Klicznik MM, Morawski PA, Hollbacher B, et al. Human CD4(+)CD103(+) cutaneous resident memory T cells are found in the circulation of healthy individuals. Science immunology. 2019;4(37). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Diani M, Casciano F, Marongiu L, et al. Increased frequency of activated CD8(+) T cell effectors in patients with psoriatic arthritis. Sci Rep. 2019;9(1): 10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sgambelluri F, Diani M, Altomare A, et al. A role for CCR5(+)CD4 T cells in cutaneous psoriasis and for CD103(+) CCR4(+) CD8 Teff cells in the associated systemic inflammation. J Autoimmun. 2016;70: 80–90. [DOI] [PubMed] [Google Scholar]
- 69.Han A, Newell EW, Glanville J, et al. Dietary gluten triggers concomitant activation of CD4+ and CD8+ αβ T cells and γδ T cells in celiac disease. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(32): 13073–13078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Risnes LF, Eggesbø LM, Zühlke S, et al. Circulating CD103(+) γδ and CD8(+) T cells are clonally shared with tissue-resident intraepithelial lymphocytes in celiac disease. Mucosal Immunol. 2021. [DOI] [PubMed] [Google Scholar]
- 71.Risnes LF, Christophersen A, Dahal-Koirala S, et al. Disease-driving CD4+ T cell clonotypes persist for decades in celiac disease. J Clin Invest. 2018;128(6): 2642–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Strobl J, Gail LM, Kleissl L, et al. Human resident memory T cells exit the skin and mediate systemic Th2-driven inflammation. The Journal of Experimental Medicine. 2021;218(11): e20210417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jabri B, Sollid LM. T Cells in Celiac Disease. Journal of immunology (Baltimore, Md : 1950). 2017;198(8): 3005–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gebhardt T, Palendira U, Tscharke DC, Bedoui S. Tissue-resident memory T cells in tissue homeostasis, persistent infection, and cancer surveillance. Immunological Reviews. 2018;283(1): 54–76. [DOI] [PubMed] [Google Scholar]
- 75.Clegg J, Soldaini E, Bagnoli F, McLoughlin RM. Targeting Skin-Resident Memory T Cells via Vaccination to Combat Staphylococcus aureus Infections. Trends in immunology. 2021;42(1): 6–17. [DOI] [PubMed] [Google Scholar]
- 76.Stein JV, Ruef N, Wissmann S. Organ-Specific Surveillance and Long-Term Residency Strategies Adapted by Tissue-Resident Memory CD8(+) T Cells. Front Immunol. 2021;12: 626019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McCully ML, Kouzeli A, Moser B. Peripheral Tissue Chemokines: Homeostatic Control of Immune Surveillance T Cells. Trends in immunology. 2018;39(9): 734–747. [DOI] [PubMed] [Google Scholar]
- 78.Stolp B, Thelen F, Ficht X, et al. Salivary gland macrophages and tissue-resident CD8(+) T cells cooperate for homeostatic organ surveillance. Science immunology. 2020;5(46). [DOI] [PubMed] [Google Scholar]
- 79.Sudan D. The current state of intestine transplantation: indications, techniques, outcomes and challenges. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2014;14(9): 1976–1984. [DOI] [PubMed] [Google Scholar]
- 80.Trentadue G, Dijkstra G. Current understanding of alloimmunity of the intestinal graft. Curr Opin Organ Transplant. 2015;20(3): 286–294. [DOI] [PubMed] [Google Scholar]
- 81.Zuber J, Rosen S, Shonts B, et al. Macrochimerism in Intestinal Transplantation: Association With Lower Rejection Rates and Multivisceral Transplants, Without GVHD. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2015;15(10): 2691–2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fu J, Zuber J, Martinez M, et al. Human Intestinal Allografts Contain Functional Hematopoietic Stem and Progenitor Cells that Are Maintained by a Circulating Pool. Cell Stem Cell. 2019;24(2): 227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Weiner J, Svetlicky N, Kang J, et al. CD69+ resident memory T cells are associated with graft-versus-host disease in intestinal transplantation. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2021;21(5): 1878–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rowntree LC, Bayliss J, Nguyen TH, Kotsimbos TC, Mifsud NA. Human leucocyte antigen-defined microchimerism early post-transplant does not predict for stable lung allograft function. Clin Exp Immunol. 2013;172(3): 483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Paantjens AW, van de Graaf EA, Heerkens HD, et al. Chimerism of dendritic cell subsets in peripheral blood after lung transplantation. J Heart Lung Transplant. 2011;30(6): 691–697. [DOI] [PubMed] [Google Scholar]
- 86.Bittmann I, Dose T, Baretton GB, et al. Cellular chimerism of the lung after transplantation. An interphase cytogenetic study. Am J Clin Pathol. 2001;115(4): 525–533. [DOI] [PubMed] [Google Scholar]
- 87.Zheng L, Orsida B, Whitford H, et al. Longitudinal comparisons of lymphocytes and subtypes between airway wall and bronchoalveolar lavage after human lung transplantation. Transplantation. 2005;80(2): 185–192. [DOI] [PubMed] [Google Scholar]
- 88.Pipeling MR, West EE, Osborne CM, et al. Differential CMV-specific CD8+ effector T cell responses in the lung allograft predominate over the blood during human primary infection. Journal of immunology (Baltimore, Md : 1950). 2008;181(1): 546–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Crispe IN. Immune tolerance in liver disease. Hepatology (Baltimore, Md). 2014;60(6): 2109–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pallett LJ, Davies J, Colbeck EJ, et al. IL-2(high) tissue-resident T cells in the human liver: Sentinels for hepatotropic infection. The Journal of experimental medicine. 2017;214(6): 1567–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stelma F, de Niet A, Sinnige MJ, et al. Human intrahepatic CD69 + CD8+ T cells have a tissue resident memory T cell phenotype with reduced cytolytic capacity. Sci Rep. 2017;7(1): 6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim JH, Han JW, Choi YJ, et al. Functions of human liver CD69(+)CD103(-)CD8(+) T cells depend on HIF-2α activity in healthy and pathologic livers. J Hepatol. 2020;72(6): 1170–1181. [DOI] [PubMed] [Google Scholar]
- 93.Swadling L, Pallett LJ, Diniz MO, et al. Human Liver Memory CD8(+) T Cells Use Autophagy for Tissue Residence. Cell Rep. 2020;30(3): 687–698.e686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Park J-G, Na M, Kim M-G, et al. Immune cell composition in normal human kidneys. Scientific Reports. 2020;10(1): 15678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hadley GA, Rostapshova EA, Gomolka DM, et al. Regulation of the epithelial cell-specific integrin, CD103, by human CD8+ cytolytic T lymphocytes. Transplantation. 1999;67(11): 1418–1425. [DOI] [PubMed] [Google Scholar]
- 96.Hadley GA, Charandee C, Weir MR, Wang D, Bartlett ST, Drachenberg CB. CD103+ CTL accumulate within the graft epithelium during clinical renal allograft rejection. Transplantation. 2001;72(9): 1548–1555. [DOI] [PubMed] [Google Scholar]
- 97.Ko S, Deiwick A, Jäger MD, et al. The functional relevance of passenger leukocytes and microchimerism for heart allograft acceptance in the rat. Nature medicine. 1999;5(11): 1292–1297. [DOI] [PubMed] [Google Scholar]
- 98.Harper IG, Ali JM, Harper SJ, et al. Augmentation of Recipient Adaptive Alloimmunity by Donor Passenger Lymphocytes within the Transplant. Cell Rep. 2016;15(6): 1214–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Win TS, Rehakova S, Negus MC, et al. Donor CD4 T cells contribute to cardiac allograft vasculopathy by providing help for autoantibody production. Circ Heart Fail. 2009;2(4): 361–369. [DOI] [PubMed] [Google Scholar]
- 100.Hu Z, Liu W, Hua X, et al. Single-Cell Transcriptomic Atlas of Different Human Cardiac Arteries Identifies Cell Types Associated With Vascular Physiology. Arterioscler Thromb Vasc Biol. 2021;41(4): 1408–1427. [DOI] [PubMed] [Google Scholar]
- 101.Fischbein MP, Yun J, Laks H, et al. Role of CD8+ lymphocytes in chronic rejection of transplanted hearts. J Thorac Cardiovasc Surg. 2002;123(4): 803–809. [DOI] [PubMed] [Google Scholar]
- 102.Ouwehand AJ, Baan CC, Vaessen LM, et al. Characteristics of graft-infiltrating lymphocytes after human heart transplantation. HLA mismatches and the cellular immune response within the transplanted heart. Hum Immunol. 1994;39(4): 233–242. [DOI] [PubMed] [Google Scholar]
- 103.Panduro M, Benoist C, Mathis D. Tissue Tregs. Annual review of immunology. 2016;34: 609–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Burzyn D, Benoist C, Mathis D. Regulatory T cells in nonlymphoid tissues. Nat Immunol. 2013;14(10): 1007–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Delacher M, Imbusch CD, Weichenhan D, et al. Genome-wide DNA-methylation landscape defines specialization of regulatory T cells in tissues. Nat Immunol. 2017;18(10): 1160–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Niedzielska M, Israelsson E, Angermann B, et al. Differential gene expression in human tissue resident regulatory T cells from lung, colon, and blood. Oncotarget. 2018;9(90): 36166–36184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ali N, Rosenblum MD. Regulatory T cells in skin. Immunology. 2017;152(3): 372–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mackay LK, Kallies A. Transcriptional Regulation of Tissue-Resident Lymphocytes. Trends in immunology. 2017;38(2): 94–103. [DOI] [PubMed] [Google Scholar]
- 109.DiSpirito JR, Zemmour D, Ramanan D, et al. Molecular diversification of regulatory T cells in nonlymphoid tissues. Science immunology. 2018;3(27): eaat5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vasanthakumar A, Moro K, Xin A, et al. The transcriptional regulators IRF4, BATF and IL-33 orchestrate development and maintenance of adipose tissue-resident regulatory T cells. Nat Immunol. 2015;16(3): 276–285. [DOI] [PubMed] [Google Scholar]
- 111.Cretney E, Xin A, Shi W, et al. The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nat Immunol. 2011;12(4): 304–311. [DOI] [PubMed] [Google Scholar]
- 112.Ichikawa T, Hirahara K, Kokubo K, et al. CD103(hi) T(reg) cells constrain lung fibrosis induced by CD103(lo) tissue-resident pathogenic CD4 T cells. Nat Immunol. 2019;20(11): 1469–1480. [DOI] [PubMed] [Google Scholar]
- 113.Li J, Olshansky M, Carbone FR, Ma JZ. Transcriptional Analysis of T Cells Resident in Human Skin. PloS one. 2016;11(1): e0148351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bestard O, Cruzado JM, Rama I, et al. Presence of FoxP3+ regulatory T Cells predicts outcome of subclinical rejection of renal allografts. J Am Soc Nephrol. 2008;19(10): 2020–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Luan D, Dadhania DM, Ding R, et al. FOXP3 mRNA Profile Prognostic of Acute T-cell-mediated Rejection and Human Kidney Allograft Survival. Transplantation. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sprangers B, DeWolf S, Savage TM, et al. Origin of Enriched Regulatory T Cells in Patients Receiving Combined Kidney-Bone Marrow Transplantation to Induce Transplantation Tolerance. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2017;17(8): 2020–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kawai T, Cosimi AB, Spitzer TR, et al. HLA-Mismatched Renal Transplantation without Maintenance Immunosuppression. New England Journal of Medicine. 2008;358(4): 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Savage TM, Shonts BA, Obradovic A, et al. Early expansion of donor-specific Tregs in tolerant kidney transplant recipients. JCI Insight. 2018;3(22). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kalyan S, Kabelitz D. Defining the nature of human gammadelta T cells: a biographical sketch of the highly empathetic. Cell Mol Immunol. 2013;10(1): 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sullivan LC, Shaw EM, Stankovic S, Snell GI, Brooks AG, Westall GP. The complex existence of gammadelta T cells following transplantation: the good, the bad and the simply confusing. Clin Transl Immunology. 2019;8(9): e1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lamb LS Jr., Musk P, Ye Z, et al. Human gammadelta(+) T lymphocytes have in vitro graft vs leukemia activity in the absence of an allogeneic response. Bone Marrow Transplant. 2001;27(6): 601–606. [DOI] [PubMed] [Google Scholar]
- 122.Schilbach KE, Geiselhart A, Wessels JT, Niethammer D, Handgretinger R. Human gammadelta T lymphocytes exert natural and IL-2-induced cytotoxicity to neuroblastoma cells. J Immunother. 2000;23(5): 536–548. [DOI] [PubMed] [Google Scholar]
- 123.Minculescu L, Sengeløv H. The role of gamma delta T cells in haematopoietic stem cell transplantation. Scand J Immunol. 2015;81(6): 459–468. [DOI] [PubMed] [Google Scholar]
- 124.Papadopoulou M, Sanchez Sanchez G, Vermijlen D. Innate and adaptive γδ T cells: How, when, and why. Immunol Rev. 2020;298(1): 99–116. [DOI] [PubMed] [Google Scholar]
- 125.Khairallah C, Chu TH, Sheridan BS. Tissue Adaptations of Memory and Tissue-Resident Gamma Delta T Cells. Front Immunol. 2018;9: 2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sant S, Jenkins MR, Dash P, et al. Human gammadelta T-cell receptor repertoire is shaped by influenza viruses, age and tissue compartmentalisation. Clin Transl Immunology. 2019;8(9): e1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Xu W, Monaco G, Wong EH, et al. Mapping of gamma/delta T cells reveals Vdelta2+ T cells resistance to senescence. EBioMedicine. 2019;39: 44–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nielsen MM, Witherden DA, Havran WL. γδ T cells in homeostasis and host defence of epithelial barrier tissues. Nature reviews Immunology. 2017;17(12): 733–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Carding SR, Egan PJ. Gammadelta T cells: functional plasticity and heterogeneity. Nature reviews Immunology. 2002;2(5): 336–345. [DOI] [PubMed] [Google Scholar]
- 130.Prinz I, Silva-Santos B, Pennington DJ. Functional development of γδ T cells. Eur J Immunol. 2013;43(8): 1988–1994. [DOI] [PubMed] [Google Scholar]
- 131.Nagai M, Azuma E, Qi J, et al. Suppression of alloreactivity with gamma delta T-cells: relevance to increased gamma delta T-cells following bone marrow transplantation. Biomed Pharmacother. 1998;52(3): 137–142. [DOI] [PubMed] [Google Scholar]
- 132.Petrasca A, Doherty DG. Human Vδ2(+) γδ T cells differentially induce maturation, cytokine production, and alloreactive t cell stimulation by dendritic cells and B cells. Front Immunol. 2014;5:650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hunter S, Willcox CR, Davey MS, et al. Human liver infiltrating γδ T cells are composed of clonally expanded circulating and tissue-resident populations. J Hepatol. 2018;69(3):654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yu X, Liu Z, Wang Y, et al. Characteristics of Vδ1(+) and Vδ2(+) γδ T cell subsets in acute liver allograft rejection. Transpl Immunol. 2013;29(1–4):118–122. [DOI] [PubMed] [Google Scholar]
- 135.Martínez-Llordella M, Puig-Pey I, Orlando G, et al. Multiparameter immune profiling of operational tolerance in liver transplantation. Am J Transplant. 2007;7(2):309–319. [DOI] [PubMed] [Google Scholar]
- 136.Fu J, Fang Z, Gorur A, et al. O-37: Immune profiling of γδ T cells locally and systemically after human intestinal transplantation reveals unique innate and adaptive features. Transplantation. 2021;105(7S). [Google Scholar]