Abstract
Petrochemicals are important hydrocarbons, which are one of the major concerns when accidently escaped into the environment. On one hand, these cause soil and fresh water pollution on land due to their seepage and leakage from automobile and petrochemical industries. On the other hand, oil spills occur during the transport of crude oil or refined petroleum products in the oceans around the world. These hydrocarbon and petrochemical spills have not only posed a hazard to the environment and marine life, but also linked to numerous ailments like cancers and neural disorders. Therefore, it is very important to remove or degrade these pollutants before their hazardous effects deteriorate the environment. There are varieties of mechanical and chemical methods for removing hydrocarbons from polluted areas, but they are all ineffective and expensive. Bioremediation techniques provide an economical and eco-friendly mechanism for removing petrochemical and hydrocarbon residues from the affected sites. Bioremediation refers to the complete mineralization or transformation of complex organic pollutants into the simplest compounds by biological agents such as bacteria, fungi, etc. Many indigenous microbes present in nature are capable of detoxification of various hydrocarbons and their contaminants. This review presents an updated overview of recent advancements in various technologies used in the degradation and bioremediation of petroleum hydrocarbons, providing useful insights to manage such problems in an eco-friendly manner.
Keywords: Bio-surfactants, Bioremediation, GMO, Hydrocarbons, Nanotechnology, Petrochemicals
Introduction
Slapdash exploitation of hydrocarbon fuels to meet the energy demand of the world is continuously harming our environment. Significant increase in the petroleum industry and exploitation of petroleum resources have had a negative impact on the ecosystem. Oil spillage is one such problem caused by the unintentional release of liquid hydrocarbons during oil exploration and transportation to different parts of the world (Abatenh et al. 2017; Gurav et al. 2021). Oil spillage occurs mostly due to natural disasters and anthropogenic activities such as attacks by militant organisations or accidents of ships during their transportation from one Gulf country to another. Incidents of oil spillage are of major concern because they result in consequential contamination of the sea and coastline area (Singh et al. 2015; Wang et al. 2018). Hydrocarbons present in crude oil have many adverse environmental as well as health impacts (Sayed et al. 2021). When crude oil is burned accidentally or as a spill control measure, a number of toxic gases and chemicals such as carbon dioxide, carbon monoxide, nitrogen oxides, sulphur dioxide, particulate matter, lead, polycyclic aromatic hydrocarbons, and volatile organic compounds are released, which have known health impacts (Shigenaka et al. 2015; Jaligamaet al. 2015). Because of the oil spillage, hydrocarbons, including known carcinogens like benzopyrene and polycyclic aromatic hydrocarbons pollute surface water, ground water, ambient air and crops (Moore 2012). Oil spills potentially affect household food security by 60% and also lower the content of ascorbic acid in vegetables and the content of cassava crude protein by 36% and 40%, respectively (Ordinioha and Brisibe 2013). The study also reported the hemotoxic and hepatotoxic nature of crude oil that leads to infertility and cancer.
Conventional physical–chemical methods are not completely effective and have adverse environmental effects. Hence, it is necessary to develop advanced and environmentally friendly technologies to treat these toxic hydrocarbons. Bioremediation is an effective method of cleaning soil and water from petroleum hydrocarbon contaminants (Bhatia et al. 2020; Bhola et al. 2021; Dange et al. 2021; Gurav et al. 2021). Microorganisms play a critical role in the biological disposal of pollutants from soils (Hamad et al. 2021). Special attention is given to the initial oxidation of hydrocarbons and enzymes involved in biodegradation and the identification of intermediates. The complete metabolic pathways and genetic control of biodegradation processes are still unknown.
Polluted sites with hydrocarbons are rehabilitated by microorganisms, which is an appealing solution. Bioremediation technology using indigenous microbial populations is an effective method to clean up contaminated sites. Hydrocarbon-degrading microorganisms include bacteria, fungi, and algae. A number of bacterial species isolated from different environments are known to have high tolerance and hydrocarbon degradation potential (Hassanshahian et al. 2012; Yuniati 2018; Bhatia 2021). Degradation of hydrocarbons can be done through lignolytic, dehydrogenase, and oxygenase enzymes (Hong et al. 2015). The microbes isolated from hydrocarbon-contaminated sites have reported high tolerance to hydrocarbons along with high catabolic activity and greater tolerance to heavy toxic metals, better degradation of pollutants, and high biodegradation ability (Thavamani et al. 2011). This review represents the current bioremediation strategies with cost-effective measures to restore the oil spill-contaminated sites.
Hydrocarbon and their types
Crude oil is a heterogeneous mixture of different classes of volatile liquid hydrocarbons such as alkanes, alkenes, cycloalkanes, and aromatic hydrocarbons (benzene and its derivatives). As hydrocarbons are the main constituents of crude oil, other compounds present are nitrogen, sulphur, and oxygen, along with a minute amount of phosphorus and heavy metals like vanadium and nickel (Yuniati 2018). Depending on the composition, each component of oil has altered physico-chemical properties, as most of these are persistent and hydrophobic in nature and vary in viscosity, solubility, and capacity to absorb. The difference in bioavailability and toxicity also decreases the ease of biodegradation in the following sequence: n-alkanes > methyl alkanes and alkylcyclopentanes > alkylcyclohexanes > cyclic and acyclic isoprenoids (Gros et al. 2014).
Spillage of hydrocarbon
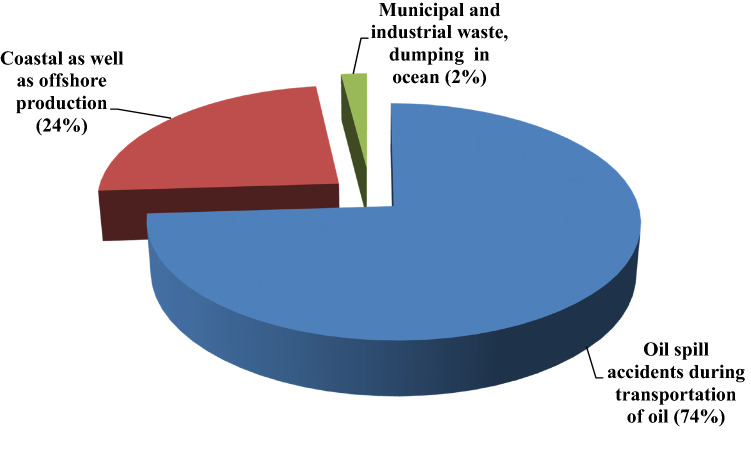
According to the International Tanker Owners Pollution Federation (ITOPF 2017), in the last 10 years from 2007 to 2016, a total of 59,000 tonnes of persistent and non-persistent oil were lost to the environment in 79 tanker accidents worldwide, with an average of 8 oil spills per year. ITOPF is a non-profit organisation that offers information on oil spills from tankers and barges transporting oil. It is estimated that about 2.37 × 106 tonnes of petroleum enter the environment per year, either through anthropogenic or natural activities (Kennisha 1997). A significant portion of this (approximately 65.2%) comes from municipal and industrial waste or dumping in oceans, as well as atmospheric fallout, while the remaining 26.2% comes from oil spill accidents during transportation and dry docking (Fig. 1).
Fig. 1.
Activities contributing to oil spillage
Various activities linked with oil exploration, installation, single point mooring (SPM) and transportation in the coastal area have the potential to create oil spills and pose a hazard to coastal ecosystems, notably those around the Indian Ocean (Kankara and Subramanian 2007). It is estimated that about 8.6% of the petroleum entering the environment comes from coastal as well as offshore production facilities and marine terminals (GESAMP 1993). As soon as the petroleum hydrocarbons enter the environment, the light fraction evaporates, while the higher molecular mass hydrocarbons with complex branched and aromatic structures pose a threat to soil habitat as they are persistent and resistant to biodegradation.
Environmental and economic impacts of oil spillage
Oil spillage is a quantitatively major source of aquatic forms of accidental pollution caused by the maritime sector's transportation. Oil spillage is associated with various environmental, social, and health impacts. In 2015, an oil tanker carrying 1.1 million barrels of oil collapsed near Yemen's coast, which caused huge damage to the environment (Huynh et al. 2021). Oil spills have severe acute and chronic effects on aquatic ecosystems, as oil-affected birds and mammals lose their thermal insulation and buoyancy (Lee et al. 2015). Crude oil components and dispersants used during oil spill clean-up modify the structure and dynamics of planktonic communities in the sea by disrupting the energy transmission between the trophic levels (Almeda et al. 2014). Oil pollution also damages the reproductive system of coral reefs and decreases colony viability, lowering the number of ovarian per polyp on reef corals and resulting in premature planulae shedding (Loya and Rinkevich 1980). Mangrove vegetation is also adversely affected by oil pollution. Duke has reported that about 5.5 million tonnes of oil have killed at least 126,000 ha of mangrove vegetation by entering mangrove-lined coastal waters since 1958 (Duke 2016). Because the accumulation of pollutants in animal and plant tissues can result in offspring mortality or mutation, hydrocarbon-contaminated soil causes significant harm to local ecosystems (Alvarez et al. 1991). These compounds also penetrate and bind to macro and micro components of soil and limit the transport of water and air, which are crucial for remediation by microorganisms (Caravaca and Roldan 2003).
Many oil spills result in the contamination of coastal areas with high amenity values. In addition to the cost of clean-up, enterprises and individuals that rely on coastal resources may suffer significant financial losses. Typically, the tourism and fishing industries endure the most of the consequences. Fisheries and mariculture resources can be severely harmed by oil spills. By fouling gear or obstructing access to fishing grounds, physical contamination can have an impact on stocks and impede commercial operations. Recreational activities such as swimming, boating, angling, and diving are normally disrupted very briefly by oil-contaminated coastlines. Normal trade and such-like activities are likely to resume only after the shorelines are cleaned. However, when the public perception of long-term and widespread contamination persists long after the oil is gone, greater long-term and harmful economic effects can ensue. On the other hand, many other companies' operations and industries could be disrupted, resulting in a loss of earnings (ITOPF 2017).
Remediation of oil spillage
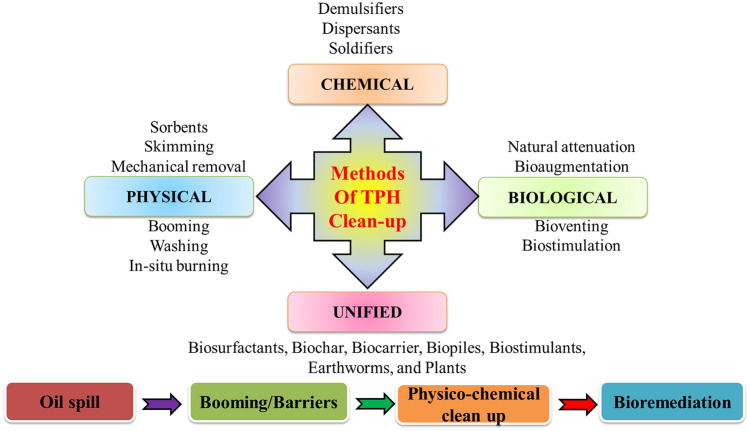
The various clean-up techniques, including in-situ burning, mechanical and chemical remediation, are used (Figs. 2, 3). The use of various cleaning agents, such as organo-clays synthesised by ion exchange, dispersants, chemical absorbents, synthetic surfactants, and additives, has several drawbacks, including higher cost, non-biodegradability, non-recyclability, as well as acute and chronic toxicity to aquatic life (Carmody et al. 2007; Nyman et al. 2007; Lee et al. 2015). The conventional techniques do not lead to the degradation of environmental pollutants and focus unduly on the separation of contaminants. Bioremediation technology has an edge over conventional physico-chemical methods as bioremediation using microbial populations is eco-friendly, cost-effective, and results in total contaminant degradation (Tyagi et al. 2011; Haritash 2020; Sadhana et al. 2021). Although, the bioremediation process also has disadvantages, it has many more benefits with high sustainability and economic feasibility (Kumari et al. 2019; Dutta et al. 2021). The major disadvantages associated with bioremediation are the longer treatment time as that of other remediation technologies, the range of contaminants (which should be biodegradable), and some time it leaves high residual levels which are persistent and toxic. However, the evaluation of performance is difficult because bioremediation has no defined level of a "clean" site, which makes the performance criteria regulations uncertain.
Fig. 2.
Proposed methods of clean up spill
Fig. 3.
Improvement of microbes for hydrocarbon bioremediation
Hydrocarbon bioremediation
Microorganisms in both the terrestrial and aquatic environments serve as primary biodegradation agents for environmental significance, majorly against petroleum hydrocarbons. Bacteria, algae, yeast, and fungus that degrade hydrocarbons are found in abundance in marine, freshwater, and soil ecosystems (Fig. 3). In aquatic ecosystems, yeast and bacteria are dominating degraders, whereas in soil, fungi and bacteria appear to be the primary degraders (Hossain et al. 2022). Algae-based bioremediation has a significant capacity for producing biomass and has a greater ability for collection, detoxification, and degradation of xenobiotics and contaminants. Many algal enzymes have been discovered to be involved in the biodegradation of a variety of organic contaminants (Baghour 2019). The intermediate length, n-alkanes (C10–C20), is the favourite substrate, which can be easily degradable to the various petroleum fractions, but shorter chain molecules are more hazardous to bacteria. However, n-alkanes of short length have great aqueous solubility, but they are toxic to microbes or cells, which directly inhibit microbial growth. The toxicity mechanisms are associated with their uptake and dissolution in the cell membrane and have shown more bioavailability of short-chain n-alkanes than midsized or long-chain n-alkanes. Waxes (C20–C40) are hydrophobic solids with long-chain alkanes that are difficult to decompose due to their low solubility and bioavailability in water; alkanes with branched chains also decay more slowly than conventional alkanes (Balba et al. 1998). For creating an artificial microbial-bacterial consortium, a microalgae, i.e., Scenedesmus obliquus GH2, is utilised for crude-oil degradation (Tang et al. 2010b). According to a study on the Rhodococcus genus, it is involved in the degradation of n-alkanes (up to C36) and heavy alkanes in Prestige fuel (Jimenez et al. 2007). Microorganisms’ ability to degrade polyaromatic hydrocarbons (PAH) is largely determined by their molecular weights (Sakhuja et al. 2021).
Various microorganisms have the ability to degrade aromatic compounds, mainly low-molecular weight PAH compounds, i.e., phenanthrene, anthracene, and naphthalene. The degradation rate of cycloalkanes is variable, but it is as slow as that of alkanes, which also involves several microbial species. Bitumen, tars, and asphaltic materials having high-condensed aromatic and cycloparaffinic structures with a high boiling point greatly resist biodegradation (Balba et al. 1998). Hong et al. (2008) investigated the accumulation and biodegradation of two common PAHs, phenanthrene (PHE) and fluoranthene (FLA) by diatoms utilising two algal species, Skeletonema costatum and Nitzschia sp. The scientists discovered that Nitzschia sp. had better accumulation and degradation abilities than S. costatum. FLA degradation was slower in both algal species, showing that FLA was a more resistant PAH molecule. Some organic xenobiotics, such as poly-aromatic hydrocarbons (PAHs) and some fractions of crude oil and coal, are resistant to degradation. Microorganisms, on the other hand, are thought to be capable of decomposing practically all the complex and resistant xenobiotics found on the planet (Hossain et al. 2022). Phytoplankton is high in lipid content and serves as the foundation for benthic and pelagic food chains, which have a role in determining persistent organic pollutants (POPs), i.e., polychlorinated biphenyls (PCBs) in the water column (Lynn et al. 2007).
The most common genera known to be responsible for oil degradation comprise mainly Nocardia, Pseudomonas, Acinetobacter, Flavobacterium, Micrococcus, Arthrobacter, Corynebacterium, Achromobacter, Scenedesmus, Nitzschia, Rhodococcus, Alcaligenes, Mycobacterium, Bacillus, Aspergillus, Mucor, Fusarium, Penicillium, Rhodotorula, and Sporobolomyces (Atlas 1981; Bossert and Bartha 1984; Hassanshahian et al. 2012). The different microorganisms and their hydrocarbon degradation potential are given in Table 1.
Table 1.
Hydrocarbon degradation potential of indigenous microbial population
| Hydrocarbon substrate | Microorganism | Degradation (% age) | Isolated from | References |
|---|---|---|---|---|
| Crude oil | Acinetobacter calcoaceticus | 98 |
Persian Gulf and Caspian Sea |
Hassanshahian et al. (2012) |
| Alcanivorax dieselolei | 98 | |||
| Alcanivorax dieselolei strain PM07 | 71 | |||
| Yarrowia lipolytica PG-20 | 68 | |||
| Yarrowia lipolytica PG-32 | 58 | |||
| C2 strain (99% Citrobacter sedlakii) | 69 | Petroleum polluted soils, Iran | Ghoreishi et al. (2017) | |
| C4 and SI1 (99% Enterobacter hormeachei) | 48 | |||
| SI2 (99% Entrobacter cloacae) | 42 | |||
| TPH | B. subtilis DM-04 | 53.57 | North east India | Das and Mukherjee (2007) |
| P. aeruginosa M | 75 | |||
| Mixture of petroleum hydrocarbons | Bacillus sp. SV9 | 59 | Ankleshwar, India | Verma et al. (2006) |
| Acinetobacter sp. SV4 | 37 | |||
| Pseudomonas sp. SV17 | 35 | |||
| Alkanes |
Pseudomonas strain PS-1 Gordonia sihwensis |
70.69 | Lingala oil field project India | Mittal and Singh (2010), Brown et al. (2016), Imperato et al. (2019) |
| Aromatic hydrocarbons | 45.37 | |||
| Anthracene | Mycobacterium sp. strain PYR-1 | 92 | Contaminated estuarine sediment | Moody et al. (2001) |
| Phenanthrene | 90 | |||
| Phenanthrene | Staphylococcus sp. strain PN/Y | Unknown | Noonmati refinery site, India | Mallick et al. (2007) |
| Acinetobacter baumannii S30 pJES | Hydrocarbon degradation | Unknown | Petro contaminate site | Mishra et al. (2004) |
| Streptomyces coelicolor M145 | n-hexadecane degradation | Unknown | Crude oil contaminate site | Gallo et al. (2012) |
| Acinetobacter sp. BS3 | Aromatic hydrocarbons | Unknown | Crude oil contaminate site | Xie et al. (2014) |
Hydrocarbon-contaminated soils suffer from salinity due to industrial activities (Gao et al. 2015). Soil salinity suppresses microbial growth and reduces the ability of microbes to degrade petroleum pollutants (Hua et al. 2010). Therefore, in such conditions, it is necessary for the microbes to be resistant to salinity. Bacterial communities ("consortia") are more flexible and are efficient degraders of a wider range of pollutants than individual microorganisms (Jobson et al. 1972; Shankar et al. 2014; Li et al. 2016). Recently, Ghoreishi and his co-workers isolated the bacteria from petroleum and kerosene-contaminated soils for the treatment of kerosene and investigated their degradation potential (Ghoreishi et al. 2017). These bacteria were capable of thriving in a medium containing 20% v/v kerosene and decomposing sulphur and carbon compounds in 7 days to 5% v/v kerosene. According to the results of GC analysis, the isolate C2 (97 percent comparable to C. sedlakii) had carbon degradation capacity in kerosene. SI2 (99 percent linked to E. cloacae) also demonstrated a significant proclivity for sulphur degradation in kerosene. These two strains in a microbial consortium could aid in the biodegradation of petroleum-contaminated soil more effectively (Ghoreishi et al. 2017). This finding paves the way for more focused and advanced research in the field of biodegradation, mainly in petroleum biodegradation.
Enzymes involved in hydrocarbon biodegradation
Pollutants in the environment have acute and chronic effects on biotic components, degrading the ecosystem (Bhandari et al. 2021). Oxidase, dehydrogenase, and lignolytic enzymes are involved in the breakdown of hydrocarbons. Aromatic ring hydroxylation is carried out by monooxygenases and dioxygenases; monooxygenases insert one oxygen atom while dioxygenases insert two oxygen atoms into their substrates. Through a series of processes, the ring is then cleaved and transformed into 2-ketoadipate or another chemical that the organism can use (Fan and Krishnamurthy 1995; Kora 2018).
Lignin peroxidase, manganese peroxidase, and laccase are fungal lignolytic extra-cellular enzymes that catalyse the formation of radicals through oxidation for bond disruption in a molecule (Punnapayak et al. 2009; Yadav et al. 2011; Lombard et al. 2014). Temperature, pH, salinity, oxygen availability, nutrition availability, and light conditions all have an impact on these enzymes. The majority of enzymes are most active at mesophilic temperatures, but others have been found to be active even at high temperatures. According to a study by Farnet and colleagues, the laccase activity of the fungus Marasmius quercophilus is best around 80 °C (Farnet et al. 2000). Although enzymes have substrate specificity, lignolytic enzymes are non-specific, creating cation radicals after one electron oxidation on both phenolic and non-phenolic organic compounds (Juhasz et al. 1997; Sadhana et al. 2021). Pleurotus laccase generates hydroxyl radicals, whereas Nematoloma forwardii and Aspergillus oryzae Mn-dependent peroxidase degrades a wide range of polyaromatic and aliphatic compounds to carbon dioxide and polar fission products (Asemoloye et al. 2020).
In a cell free reaction mixture enriched with manganese (II) ions (Mn2+) at a concentration of 200 M, Stropharia coronilla, a litter-decomposing basidiomycete, has also been found to metabolise and mineralize benzo(a) pyrene (BaP) (Steffen et al. 2003; Sharma et al. 2018). This degrading action could be attributed to the lignolytic enzyme manganese peroxidase (MnP), which is activated by the presence of Mn2+ and accelerated by the addition of Tween 80. Tween 80 is a surfactant that stimulates or enhances the release of enzymes into the culture medium. In a cell-free reaction mixture, crude and purified MnP from S. coronilla efficiently oxidized BaP.
Cytochrome P450 is also identified as being used in the microbial decomposition of petroleum hydrocarbons. An alkane hydroxylase enzyme belongs to the ubiquitous heme-thiolate Monooxygenases superfamily (Van-Beilen and Funhoff 2007; Punetha et al. 2022). Eukaryotes have a number of P450 families, each of which contains a huge number of unique P450 forms that can help in the metabolic transformation of a specific substrate. Zimmer et al. (1996) found that only a few kinds of microorganisms possess P450 multiplicity. Candida maltosa, Candida tropicalis, and Candida apicola have all been found to have numerous microsomal Cytochrome P450 types of enzyme (Scheller et al. 1998; Punetha et al. 2022). They are capable of obtaining carbon and energy solely from n-alkanes and other aliphatic hydrocarbons. Some of the enzymes used in hydrocarbon bioremediation are given in Table 2.
Table 2.
Enzymes involved in hydrocarbon degradation
| Enzyme | Substrate | Strain | References |
|---|---|---|---|
|
Mono-oxygenases Methane, propane and butane mono-oxygenases, CYP153 mono-oxygenases, AlkB non-heme iron mono-oxigenase, flavin-mono-oxigenase AlmA, flavin-dependent mono-oxygenase LadA |
Alkanes, Alkenes, cycloalkenes |
Methylococcus, Methylosinus, Methylocystis, Methylomonas, Methylocella, Pseudomonas putida GPo1 Burkholderia cepacia G4, Ralstonia pickettii PKO1, Pseudomonas mendocina KR1, Pseudomonas stutzeri OX1 |
McDonald et al. (2006), Rojo (2010), Imperato et al. (2019) |
| Di-oxygenases (copper flavin-dependent Di-oxygenase) | Alkanes |
Bacillus sp. Pseudomonas sp. |
Kora (2018) |
| Cytochrome P450 enzymes (Eukaryotic and bacterial) |
C10–C16 alkanes, fatty acids, cycloalkanes |
Candida maltosa, Candida tropicalis, Candida apicola, Acinetobacter, Caulobacter, Mycobacterium |
Scheller et al. (1998), van Beilen and Funhoff (2007), Punetha et al. (2022) |
| Dehydrogenases | Petroleum hydrocarbon | Pseudomonas aeruginosa | Ebadi et al. (2017) |
| Laccase | Polycyclic Aromatic Hydrocarbon |
Marasmius quercophilus, Coriolopsis gallica UAMH 8260 |
Lombard et al. (2014), Sharma et al. (2018), Farnet et al. (2000) |
| Manganese peroxidase | Bezo(a)pyrenes (BaP) |
Nematoloma forwardii, Stropharia coronilla Aspergillus oryzae |
Steffen et al. (2003), Asemoloye et al. (2020) |
| Aldehyde dehydrogenase | Total Petroleum Hydrocarbon | Acinetobacter baumannii S30 pJES | Mishra et al. (2004) |
| Alkane monoxygenase | n-hexadacane | Streptomyces coelicolor M145 | Gallo et al. (2012) |
| Catechol 2,3-dioxygenas | Aromatic Hydrocarbon | Acinetobacter sp. BS3, Pseudomonas putida | Xie et al. (2014), Haritash (2020) |
A group tried the degradation of petrol in an aqueous system with immobilized cells and cell free suspension of Pseudomonas sp. The increase in rhamnolipid production was also observed in this experiment (Sarkar et al. 2005). The amphipathic characteristics of rhamnolipids diminish oil–water interfacial tension systems, allowing better diffusion of water-insoluble n-alkanes in the aqueous phase. The study found that immobilizing cells boosted cell-to-cell interaction with solubilized hydrocarbon droplets and resulted in fast hydrocarbon uptake. A study by Diaz and colleagues found that immobilizing cells of bacteria enhance crude oil biodegradation rate as that of free-living cells (Dıaz et al. 2002; Shahzad et al. 2020).
Immobilized enzymes have higher efficiency, thermostability, half-life and resistance to protease degradation (Sarkar et al. 1989; Solanki et al. 2021). The activity of the immobilised enzyme lasts substantially longer than that of the free enzyme. Furthermore, the immobilised enzyme can be extracted from the reaction fluid and used to convert substrate with minimal activity loss (Guleria et al. 2016; Ruggiero et al. 1989). For the reasons described above, using cell-free enzymes (ideally immobilized) to speed the clean-up of petroleum-contaminated soils holds a lot of potential.
Advances in petroleum hydrocarbon bioremediation
Conventional methods used for oil spill treatment are now replaced by advanced and sustainable techniques such as genetically engineered microorganisms, nanotechnology-based remediation materials, and metabolic engineering to treat toxic bioremediation intermediates (Naeem and Qazi 2020; Dutta et al. 2021; El-Sheshtawy et al. 2022). Use of indigenous microbial communities for effective bioremediation is a current approach as they are resistant to toxic pollutants and can grow in harsh conditions (Karunanithi et al. 2017; Shakya et al. 2021).
For the remediation of highly hydrocarbon-contaminated sites, the application of xenobiotic-resistant genetically modified microorganisms is more suitable as the microbial population is not able to dissipate them completely (Fernandez-Luqueno et al. 2011). Sarkar et al. (2005) and Taccari et al. (2012) describe the efficient techniques involved in hydrocarbon biodegradation in oil spills. Bioaugmentation can increase the degradation efficiency by enhancing the metabolic activity and survival rate of a microbial population by increasing the expression of the alkB gene. Suja and co-workers have studied the combined effects of bioaugmentation and biostimulation and reported the highest TPH degradation of 79% with the fastest rate constant of 0.0390 day1 (78) (Suja et al. 2014; Sui et al. 2021).
Role of biosurfactants in hydrocarbon biodegradation
The main disadvantage of the soil biodegradation process is the restriction of mass transfer and bioavailability of pollutants, which directly hinders microbe access to petroleum components and is responsible for the decrease in biodegradation rate (Onwurah et al. 2007; Karlapudi et al. 2018). Some microorganisms can produce amphipathic molecules called biosurfactants. These surfactants pseudo-solubilize hydrocarbons and desorbed them from the soil matrix into the aqueous phase, which led to better availability of oil for microbial cell uptake. Such microbes have been considered as a source of crude oil contaminate bioremediation in soil (Cameotra and Makkar 1998; Whang et al. 2008; Ganesh and Lin 2009; Rocha et al. 2011; Hua and Wang 2012). Thavasi and his co-workers have reported that biosurfactants are capable of enhancing biodegradation rates by up to 4–5% without adding fertilizers, which also helps in reducing the biodegradation cost (Thavasi et al. 2011).
The application of two biosurfactants was tested in a series of bench-scale experiments, a lipoprotein type biosurfactant surfactin (SF) and a glycolipid biosurfactant rhamnolipid (RL), for increased soil and water biodegradation for removing diesel-contamination has also been described. Bacillus subtilis ATCC 21332 created the surfactin, while Pseudomonas aeruginosa J4 produced the rhamnolipid; both biosurfactants reduced surface tension to < 30 dynes/cm, down from 72 dynes/cm, and increased diesel solubility with increased biosurfactant (Whang et al. 2008; Rikalovic et al. 2015). A consortium of hydrocarbon degrading bacteria sown in crude oil-contaminated soil and added with different combinations of rhamnolipids and nutrients produced a maximum of 77.6% biodegradation (Tahseen et al. 2016).
According to a recent study by Ebadi and colleagues, inoculating contaminated soil with a combination of four biosurfactant-producing Pseudomonas aeruginosa strains improved the adverse effects of salinity on biodegradation and enhanced the rate of degradation of petroleum hydrocarbons by about 30% when compared to non-treated soil. Inoculation of polluted soil with the consortium considerably increases the dehydrogenase enzyme activity (about two-fold) in saline conditions (Ebadi et al. 2017; Mandalenaki et al. 2021). Table 3 lists some of the biosurfactant-producing strains used in hydrocarbon bioremediation.
Table 3.
Biosurfactant producing strains used for degrading hydrocarbons
| Biosurfactants | Micro-organisms | References |
|---|---|---|
| Surfactin |
Bacillus subtilis ATCC 21332 Bacillus subtilis Bacillus licheniformis |
Whang et al. (2008), Karlapudi et al. (2018) |
| Rhamnolipid |
Pseudomonas aeruginosa J4 Pseudomonas fluorescens Pseudomonas aeruginosa MM1011 |
Whang et al. (2008), Amani et al. (2013), Rikalovic et al. (2015) |
| Glycolipid | Aeromonas sp., Bacillus sp., Pseudomonas aeruginosa | Tabatabaee et al. (2005), Rikalovic et al. (2015) |
| Sophorolipids | Candida tropicalis | Imura et al. (2014), Karlapudi et al. (2018) |
| Lipopeptide biosurfactant | Pseudomonas aeruginosa | Thavasi et al. (2011), Ebadi et al. (2017) |
| NA | Bacillus sp., Paenibacillus sp. | Najafi et al. (2015) |
| di-rhamnolipid | Pseudomonas sp. | Varjani and Upasani (2016) |
| NA | Agrobacterium sp. | Ohadi et al. (2017) |
| Lipopeptide biosurfactants | Aeribacillus sp. | Mehetre et al. (2019) |
| Lipopeptide | Acinetobacter sp. | Sharma et al. (2019) |
Role of nanotechnology in bioremediation
Instead of sustainable and economic methods, research is also focused on the use of advanced and inexpensive bio-based materials for the clean-up of petroleum hydrocarbons (Pete et al. 2021). Various nanomaterials and nanostructures have been successfully deployed by various research groups for the treatment of hydrocarbon spills, as shown in Fig. 4. Wu and colleagues created a new form of carbon nanofiber (CNF) aerogel that has a high absorption capacity, more recyclability, and working temperature versatility (up to 400 °C) (Wu et al. 2014). Generally, macroporous materials are used for the absorption of oil spills. Pan and co-workers synthesised new macro-porous and hydrophobic polyvinyl formaldehyde (PVF-H) sponges with improved reusability and oil recovery of 90% (Pan et al. 2014). Nano-TiO2-Induced photocatalysis is a recent technique used for the treatment of water contamination through oil and gas production, transportation, and storage (Liu et al. 2017). For the stabilisation of oil in water emulsion, naturally occurring halloysite clay nanotubes have been reported by Owoseni and colleagues as effective interfacially active carriers for delivering oil spill treatment agents (Owoseni et al. 2014). Therefore, nanotechnology has the ability to provide immense potential in cleaning up pesticides, metal-contaminated sites, and petroleum hydrocarbons (Benjamin and Lima 2019; Pete et al. 2021; Kapoor et al. 2021).
Fig. 4.
Role of nanotechnology in hydrocarbon remediation
Phytoremediation of hydrocarbon
Phytoremediation is an emerging technology that involves the use of plant life for the clean-up of hydrocarbon-contaminated soil and groundwater. Phytoremediation is a cost-effective technique that provides aesthetic value to the site and has long-term applicability. In addition, phytoremediation reduces disturbance on land and removes the liability and transportation expenses involved with off-site treatment and disposal (Das and Chandran 2011; Nedjimi 2021).
The Alabama Department of Environmental Management has approved a location for field-scale total petroleum hydrocarbon degradation. The study used around 1500 cubic yards of soil, with 70% of baseline samples containing more than 100 parts per million of total petroleum hydrocarbons (TPH). Nedunuri et al. (2000) found that when field sites contaminated with total petroleum hydrocarbons (TPH) at 1700–16,000 mg/kg were bioremediated, around 83 percent of the TPH was eliminated after one year. Three coastal trees, milo (Thespesia populnea), kiawe (Prosopis pallida), and kou (Cordia subcordata), as well as the native shrub Scaevola serica, a beach naupaka, survived under diesel fuel soil conditions for clean-up among diverse plants in tropical conditions (USACE 2003). At some locations, organic pollutants are removed by planting grass in a row in between the trees. The thin roots of grasses at the surface of soil are capable of binding effectively with hydrophobic contaminants (TPH, PAHs, and BTEX) and converting them into non-hazardous form. Following harvest, the grass can be composted or burned. The most commonly used legumes to replace the nitrogen requirements of low soils are alfalfa (Medicago sativa) and peas (Pisum sp.). In numerous areas contaminated by petrochemical wastes, reed canary grass (Phalaris arundinacea), fescue (Vulpia myuros), rye (Elymus sp.) and clover (Trifolium sp.) have successfully been utilised (Ahmad 2021; Nero 2021).
Use of genetically modified strains in hydrocarbon bioremediation
In situ bioremediation is the combination of traditional microbiology, ecology, genetic engineering, and biochemistry, a promising solution to hydrocarbon bioremediation. A genetically modified microorganism, especially for enzyme specificity, metabolic pathway and its regulation, can be used for bioremediation purposes as well as a biosensor for detection of specific chemical hazardous compounds in the environment (Sheth et al. 2016; Wasilkowski et al. 2012; Mandeep 2020; French et al. 2020).
The first genetically modified strain, Pseudomonas florescens HK44, was utilised for long-term naphthalene contaminated soil bioremediation along with bioluminescence imaging of inoculation cells. The plasmid pUTK21, containing the P. fluorescens HK44 strain, was developed from the P. fluorescens 5R NAH7 plasmid by inserting the Tn4431 transposon element into it. Vibrio fischeri was the source of this transposon, which also carried the luxCDABE gene cassette. As a result, this gene is both responsible for naphthalene breakdown as well as for the production of luminous signals (Sayler and Ripp 2000). Filonov and colleagues created the other P. putida KT2442 (pNF142: TnMod-OTc), a genetically modified strain capable of degrading soil naphthalene (Filonov et al. 2005). Another naphthalene degrading strain Escheriachia coli having bphA2cA1c gene encoding for salicylate oxygenase from S. yanoikuyae B1 (Liu et al. 2019). This strain contains three bacteria:
(1) pTnMod-OTc plasmid conatining E.coli S17-1 with tetracycline resistance gene.
(2) Pseudomonas sp. 142NF (pNF142) capable of depredating naphthalene.
(3) P. putida KT2442 with the green fluorescent protein gene.
These findings demonstrate that genetically modified bacteria are capable of digesting naphthalene and transferring the plasmid pNF142: TnMod-OTc to native microorganisms. The possibility of plasmid pWW0 being transferred into rhizosphere bacteria from the PaW85 strain of Pseudomonas putida is capable of digesting petroleum hydrocarbons (Jussila et al. 2007). In petroleum-contaminated soil, they also confirmed horizontal gene transfer events between PaW85 and Pseudomonas oryzihabitans 29. Lipthay and colleagues looked at how Escherichia coli HB101 and Ralstonia eutropha harbouring the pRO103 plasmid degraded the aromatic hydrocarbon 2,4-dichlorofenoxyacetic acid (2,4-d) (Lipthay et al. 2001). The gene for 2,4-dichlorophenoxyacetic acid/2-oksoglutaric dioxygenase was found on the plasmid. It was proven that the derived transconjugant R. eutropha (pRO103) considerably boosted 2,4-d decomposition in soil. Pseudomonas putidaand Pseudomonas sp. CGMCC2953 having nahH gene cloned in pUC18 and C230 gene cloned in plasmid pK4 responsible for Phenanthrene and Pyrene degradation (Haritash 2020). Mycobacterium sp. strain 6PY1 having pdo gene undertaking the degradation of Pyrene (Sadhana et al. 2021).
Toxic substances in the soil not only impair soil fertility and the number of beneficial microbes, but they also hinder plant growth. As a result, genetically modified microbes can be utilised to both degrade hazardous substances and promote plant development at the same time (Cases and Lorenzo 2005; Pimentel et al. 2011). Yang and his colleagues created genetically modified bacteria that could stimulate maize plant development while also degrading phenol (Yang et al. 2011). The phenol-degrading Pseudomonas aeruginosa SZH16 (unable to boost plant development) and PGPB Pseudomonas fluorescens strains were utilised to create modified bacteria (without the ability to degrade phenol). They discovered horizontal gene transfer, which led to the emergence of recombinant strain P13. This strain was able to increase maize plant growth while also degrading phenol.
Barac et al. (2004) reported the transfer of pTOD, a toluene-degradation plasmid from Burkholderia cepacia G4 (donor) to B. cepacia L.S.2.4 (natural endophyte strain). The findings revealed that genetically modified bacteria had the capacity to degrade toluene and reduce transpiration through the leaves by 50–75% (Taghavi et al. 2005). In a similar study, Taghavi and Barac demonstrated the development of another strain, B. cepacia VM1468, for toluene degradation. This endophyte strain was created by conjugating B. cepacia BU0072 with the pTOM-Bu61 plasmid from B. cepacia BU61. The results revealed five-times lower toluene transpiration than that of control plants. Furthermore, the root and leaf mass were increased by 30%. According to these findings, the pTOMBu61 plasmid might spontaneously transfer to other natural endophytes in plants and accelerate toluene breakdown. Germaine and his colleagues created Pseudomonas putida VM1441, a naphthalene-degrading endophytic bacteria (pNAH7) (Germaine et al. 2009). In comparison to un-inoculated control plants, this strain inoculation in the soil enables a reduction of naphthalene's toxic effects on pea plants. The use of genetic engineering to remove heavy metals has attracted people's interest. Alcaligenes eutrophus AE104 (pEBZ141) was used to remove chromium from industrial wastewater (Srivastava et al. 2010), and Rhodopseudomonas palustris was engineered for Hg2+ removal from heavy metal wastewater (Srivastava et al. 2010; Xu and Pei 2011). Table 4 contains a list of genetically modified bacteria.
Table 4.
Genetically modified microorganisms known for degrading hydrocarbons
| GMMs | Introduced gene | Substrate | References |
|---|---|---|---|
| Alcaligenes eutrophus AE104 | pEBZ141 | Chromium | Srivastava et al. (2010) |
| Rhodopseudomonas palustris | NA | Hg2+ | Xu and Pei (2011) |
| Pseudomonas fluorescens HK44 | transposon Tn4431 insertion into NAH7 plasmid | Naphthalene | Sayler and Ripp (2000) |
| Mycobacterium sp. strain 6PY1 | pdo | Pyrene | Sadhana et al. (2021) |
| Escheriachia coli | bphA2cA1c gene encoding for salicylate oxygenase from S.yanoikuyae B1 | Naphthalene | Liu et al. (2019) |
| P. putida KT2442 (pNF142:TnMod-OTc) | pNF142 plasmid and gfp gene | Naphthalene | Filonov et al. (2005) |
| Pseudomonas putida | nahH gene cloned in pUC18 plasmid | Phenanthrene and Pyrene | Haritash (2020) |
| P. putida PaW85 | plasmid pWW0 | Petroleum | Jussila et al. (2007) |
| Pseudomonas sp. CGMCC2953 | C230 gene cloned in plasmid pK4 | Phenanthrene | Liu et al. (2019) |
| Ralstonia eutropha (pRO103) | pRO103 plasmid | 2,4-D | Lipthay et al. (2001) |
| Recombinant strain P13 | Combination of Pseudomonas aeruginosa SZH16 and PGPB Pseudomonas fluorescens | Phenol | Yang et al. (2011) |
| Bacillus cepacia L.S.2.4 | pTOD plasmid | Toluene | Barac et al. (2004) |
| Pseudomonas sp. | alkB | Alkanes | Imperato et al. (2019) |
| B. cepacia VM1468 | pTOM-Bu61 | Toluene | Taghavi et al. (2005) |
| Sphingomonas sp. GY2B | Pyrene | Zhao et al. (2017) | |
| Streptomyces coelicolor M145 | alkB | n-hexadacane | Gallo et al. (2012) |
| Acinetobacter sp. BS3 | Xyl E gene | Aromatic hydrocarbon | Xie et al. (2014) |
| Pseudomonas putida strain BNF1 | NA | Aromatic hydrocarbon | |
| Pseudoalteromonas haloplanktis TAC125 | NA | Toluene and xylene | Parrilli et al. (2010) |
Unified methods of microbial remediation
The three basic categories of microbial integrated techniques for remediation are physical, chemical, and biological treatments, but due to hydrophobicity and fluidity of petroleum, none of these strategies is effective independently. Hence, integrated approaches are preferred as they are able to remediate hydrocarbon-contaminated land or water systems in a much better way and can boost microbial activity. Various research groups have conducted a number of studies for the breakdown of petroleum-contaminated soil using microbial integrated or combination techniques. To boost the system's degradation rate in petroleum-contaminated areas, several technologies such as chemical oxidation, electric fields, fertilizers, biosurfactants, biochar, bio carrier, bio-piles, biostimulants, earthworms, and plants were used (Mukome et al. 2020; Shahzad et al. 2020). The efficacy of microbial degradation of petroleum pollutants can be increased by combining these physical or chemical approaches with the use of specific microorganisms. The addition of several components to highly polluted petroleum-contaminated soil (10,000 mg/kg) such as biochar, electric field, nutrients, and biosurfactants can increase the clearance rate of petroleum pollutants by more than 60% (Tang et al. 2010a). With a 58% breakdown rate within 162 days (the original oil content was 6.19%), the combination of ryegrass and mixed microbial strains had the best degrading impact (Tang et al. 2010a). Within 60 days, a combination of alfalfa and microorganisms may degrade 63% of petroleum hydrocarbons (the starting oil concentration is 12%) (Shahzad et al. 2020). It has been also found that degradation of PAH from the contaminated sites can also be enhanced by ozone, hydrogen peroxide oxidation which later on helps the microorganisms in rapid degradation of less complex hydrocarbon compounds. These findings proved that the combined approaches are much better than the individual approaches for the treatment of hydrocarbon contaminated soil and water sources.
Challenges and future prospects of hydrocarbon bioremediation
Oil pollution is of great environmental and public concern worldwide due to its long-term effects on ecosystems and health. Despite the fact that bioremediation is a promising approach for cleaning up oil-contaminated sites, a variety of factors influence the degradation process, i.e., type of hydrocarbons, physico-chemical properties of contaminated sites, suitability of the technique used, type of microbes, and other intrinsic as well as extrinsic factors involved. Organic pollutant biodegradation metabolic routes are poorly understood, and inefficient transformation results in the accumulation of increasingly complex intermediate pollutants that resist further degradation by existing pathways (Jones and Voogt 1999; Sadhana et al. 2021). The selection of the inoculum is critical for the complete and effective bioremediation of organic pollutants. Isolation of indigenous microorganisms from polluted sites is resistant to toxic pollutants due to evolved catabolic genes, and they are able to degrade a wide range of pollutants (Korda et al. 1997; Liu et al. 2019). Slower bioremediation rates of microbial populations in contaminated sites are because of harsh environmental conditions where new engineering tools are of prime importance in research to boost the bioremediation process by improving catabolic pathways. Co-contamination with heavy metals is another constraint as heavy metals could adversely affect the microbial population by extending acclimatisation phases and reducing biodegradation rates (Arjoon et al. 2013). The evolution of more heavy metal resistant xenobiotic degrading microbes is underway to eliminate the effect of heavy metal toxicity on bioremediation.
A number of genetic engineering approaches have been introduced in the field of microbial bioremediation in recent years to evolve the desired genetic manipulation of native soil microorganisms and metabolic pathways (Imperato et al. 2019). Rayu et al. (2012) describe metagenomics as an advanced field that enhances the microbial bioremediation potential by obtaining novel pollutant-degrading genes and enzymes involved in the bioremediation process. Although a number of approaches have been developed in the field of microbial remediation of hydrocarbon bioremediation, much is yet to be explored. Better understanding of the metabolic pathways, complete degradation of organic pollutants, use of bio-based treating agents, and flexible legislation for the proper use of genetically engineered microbes can make the bioremediation process more sustainable and economic.
Conclusion
Biodegradation is a method for converting substances to less hazardous forms with minimal chemical and energy input in an environmentally friendly manner. Microorganisms (bacteria, fungus, and algae) can be used singly or in combination to repair oil-contaminated environments. Environmental circumstances, the quantity and kind of microorganisms present, and the chemical structure and nature of the molecule being degraded determine the rate of bioremediation of a contaminant. Exposure to greater levels of pollutants can lead to the acclimatisation of hydrocarbon-degrading microbial species and genetic alterations that can lead to improved degradation and the induction of degradation capacity. An integrated method of physical, chemical, and biological degradation should be applied to rehabilitate contaminated places in an environmentally sustainable manner.
Acknowledgements
Authors acknowledges the support received from Department of Microbiology, CSKHPKV Palampur and Department of Biotechnology, Himachal Pradesh University Summer Hill Shimla to conduct this study.
Declarations
Conflict of interest
There is no conflict of interest between the authors.
References
- Abatenh E, Gizaw B, Tsegaye Z, Wassie M. The role of microorganisms in bioremediation—a review. Open J Environ Biol. 2017;2(1):038-046. doi: 10.17352/ojeb. [DOI] [Google Scholar]
- Ahmad A. Phytoremediation of heavy metals and total petroleum hydrocarbon and nutrients enhancement of Typha latifolia in petroleum secondary effluent for biomass growth. Environ Sci Pollut Res. 2021;29(4):5777–5786. doi: 10.1007/s11356-021-16016-5. [DOI] [PubMed] [Google Scholar]
- Almeda R, Hyatt C, Buskey EJ. Toxicity of dispersant Corexit 9500A and crude oil to marine microzooplankton. Ecotoxicol Environ Saf. 2014;106:76–85. doi: 10.1016/j.ecoenv.2014.04.028. [DOI] [PubMed] [Google Scholar]
- Alvarez PJJ, Anid PJ, Vogel TM. Kinetics of aerobic biodegradation of benzene and toluene in sandy aquifer material. Biodegradation. 1991;2:43–45. doi: 10.1007/BF00122424. [DOI] [PubMed] [Google Scholar]
- Amani H, Müller MM, Syldatk C, Hausmann R. Production of microbial rhamnolipid by Pseudomonas aeruginosa MM1011 for ex situ enhanced oil recovery. Appl Biochem Biotechnol. 2013;170(5):1080–1093. doi: 10.1007/s12010-013-0249-4. [DOI] [PubMed] [Google Scholar]
- Arjoon A, Olaniran AO, Pillay B. Co-contamination of water with chlorinated hydrocarbons and heavy metals: challenges and current bioremediation strategies. Int J Environ Sci Technol. 2013;10(2):395–412. doi: 10.1007/s13762-012-0122-y. [DOI] [Google Scholar]
- Asemoloye MD, Tosi S, Daccò C, Wang X, Xu S, Marchisio MA, Gao W, Jonathan SG, Pecoraro L. Hydrocarbon degradation and enzyme activities of Aspergillus oryzae and Mucor irregularis isolated from nigerian crude oil-polluted sites. Microorganisms. 2020;8(12):1912. doi: 10.3390/microorganisms8121912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlas RM. Microbial degradation of petroleum hydrocarbons: an environmental perspective. Microbial Rev. 1981;45:180–209. doi: 10.1128/mr.45.1.180-209.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baghour M (2019) Algal degradation of organic pollutants. In: Martínez L, Kharissova O, Kharisov B (eds) Handbook of ecomaterials. Springer, Cham. 10.1007/978-3-319-68255-6_86
- Balba MT, Al-Awadhi N, Al-Daher R. Bioremediation of oil-contaminated soil: microbiological methods for feasibility assessment and field evaluation. J Microbiol Methods. 1998;32:155–164. doi: 10.1016/S0167-7012(98)00020-7. [DOI] [Google Scholar]
- Barac T, Taghavi S, Borremans B, Provoost A, Oeyen L, Colpaert JV, Vangronsveld J, van der Lelie D. Engineered endophytic bacteria improve phytoremediation of water-soluble, volatile, organic pollutants. Nat Biotechnol. 2004;22(5):583–588. doi: 10.1038/nbt960. [DOI] [PubMed] [Google Scholar]
- Benjamin SR, Lima FD. Current trends in nanotechnology for bioremediation. Int J Environ Pollut. 2019;66:19–40. doi: 10.1504/IJEP.2019.104526. [DOI] [Google Scholar]
- Bhandari S, Poudel DK, Marahata R, Dawadi S, et al. Microbial enzymes used in bioremediation. J Chem. 2021 doi: 10.1155/2021/8849512. [DOI] [Google Scholar]
- Bhatia SK. Wastewater based microbial biorefinery for bioenergy production. Sustainability. 2021;13:9214. doi: 10.3390/su13169214. [DOI] [PubMed] [Google Scholar]
- Bhatia RK, Sakhuja D, Mundhem S, Walia A. Renewable energy products through bioremediation of wastewater. Sustainability. 2020;12(7501):1–24. doi: 10.3390/su12187501. [DOI] [Google Scholar]
- Bhola S, Arora K, Kulshrestha S, Mehariya S, Bhatia RK, Parneet K, Kumar P. Established and emerging producers of PHA: redefining the possibility. Appl Biochem Biotechnol. 2021;193(11):3812–3854. doi: 10.1007/s12010-021-03626-5. [DOI] [PubMed] [Google Scholar]
- Bossert I, Bartha R. Fate of petroleum in soil ecosystem. In: Atlas RM, editor. Petroleum microbiology. New York: Macmillan Publishing Co; 1984. pp. 441–473. [Google Scholar]
- Brown LM, Gunasekera TS, Striebich RC, Ruiz ON. Draft genome sequence of Gordonia sihwensis strain 9, a branched alkane-degrading bacterium. Genome Announc. 2016;4:e00622–e716. doi: 10.1128/genomeA.00622-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameotra SS, Makkar RS. Synthesis of biosurfactants in extreme conditions. Appl Microbiol Biotechnol. 1998;50:520–529. doi: 10.1007/s002530051329. [DOI] [PubMed] [Google Scholar]
- Caravaca F, Roldan A. Assessing changes in physical and biological properties in a soil contaminated by oil sludges under semiarid Mediterranean condition. Geoderma. 2003;117:53–61. doi: 10.1016/S0016-7061(03)00118-6. [DOI] [Google Scholar]
- Carmody O, Frost R, Xi Y, Kokot S. Adsorption of hydrocarbons on organo-clays-implications for oil spill remediation. J Colloid Interface Sci. 2007;305(1):17–24. doi: 10.1016/j.jcis.2006.09.032. [DOI] [PubMed] [Google Scholar]
- Cases I, de Lorenzo V. Genetically modified organisms for the environment: stories of success and failure and what we have learned from them. Int J Microbiol. 2005;8(3):213–222. [PubMed] [Google Scholar]
- Dange P, Pandit S, Jadhav D, Shanmugam P, Gupta PK, Kumar S, Kumar M, Yang YH, Bhatia SK. Recent developments in microbial electrolysis cell-based biohydrogen production utilizing wastewater as a feedstock. Sustainability. 2021;13(16):8796. doi: 10.3390/su13168796. [DOI] [Google Scholar]
- Das N, Chandran P. Microbial degradation of petroleum hydrocarbon contaminants: an overview. Biotechnol Res Int. 2011;2011:1–13. doi: 10.4061/2011/941810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das K, Mukherjee AK. Crude petroleum-oil biodegradation efficiency of Bacillus subtilis and Pseudomonas aeruginosa strains isolated from a petroleum-oil contaminated soil from North-East India. Bioresour Technol. 2007;98:1339–1345. doi: 10.1016/j.biortech.2006.05.032. [DOI] [PubMed] [Google Scholar]
- Dıaz MP, Boyd KG, Grigson SJW, Burgess JG. Biodegradation of crude oil across a wide range of salinities by an extremely halotolerant bacterial consortium MPD-M, immobilized onto polypropylene fibers. Biotechnol Bioeng. 2002;79(2):145–153. doi: 10.1002/bit.10318. [DOI] [PubMed] [Google Scholar]
- Duke NC. Oil spill impacts on mangroves: Recommendations for operational planning and action based on a global review. Mar Pollut Bull. 2016;109(2):700–715. doi: 10.1016/j.marpolbul.2016.06.082. [DOI] [PubMed] [Google Scholar]
- Dutta K, Shityakov S, Khalifa I. New trends in bioremediation technologies toward environment-friendly society: a mini-review. Front Bioeng Biotechnol. 2021;9:666858. doi: 10.3389/fbioe.2021.666858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebadi A, Azam N, Sima K, Olamaee M, Hashemi M, Nasrabadi RG. Effective bioremediation of a petroleum-polluted saline soil by a surfactant-producing Pseudomonas aeruginosa consortium. J Adv Res. 2017;8(6):627–633. doi: 10.1016/j.jare.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sheshtawy HS, Aman D, Nassar HN. A novel bioremediation technique for petroleum hydrocarbons by bacterial consortium immobilized on goethite-chitosan nanocomposite. Soil Sediment Contam Int J. 2022;31(2):176–199. doi: 10.1080/15320383.2021.1916737. [DOI] [Google Scholar]
- Fan CY, Krishnamurthy S. Enzymes for enhancing bioremediation of petroleum-contaminated soils: a brief review. J Air Waste Manag Assoc. 1995;45(6):453–460. doi: 10.1080/10473289.1995.10467375. [DOI] [PubMed] [Google Scholar]
- Farnet AM, Criquet S, Tagger S, Gil G, Petit JL. Purification, partial characterization, and reactivity with aromatic compounds of two laccases from Marasmius quercophilus strain 17. Can J Microbiol. 2000;46(3):189–194. doi: 10.1139/w99-138. [DOI] [PubMed] [Google Scholar]
- Fernandez-Luqueno F, Valenzuela-Encinas C, Marsch R, Martínez-Suárez C, Vázquez-Núñez E, Dendooven L. Microbial communities to mitigate contamination of PAHs in soil: possibilities and challenges: a review. Environ Sci Pollut Res Int. 2011;18(1):12–30. doi: 10.1007/s11356-010-0371-6. [DOI] [PubMed] [Google Scholar]
- Filonov AE, Akhmetov LI, Puntus IF, Esikova TZ, Gafarov AB, Izmalkova TY, Sokolov SL, Kosheleva IA, Boronin AM. The construction and monitoring of genetically tagged, plasmid-containing, naphthalene-degrading strains in soil. Microbiology. 2005;74(4):526–532. doi: 10.1007/s11021-005-0088-6. [DOI] [PubMed] [Google Scholar]
- French KE, Zhou Z, Terry N. Horizontal ‘gene drives’ harness indigenous bacteria for bioremediation. Sci Rep. 2020;10:15091. doi: 10.1038/s41598-020-72138-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo G, Lo PL, Giovanni R, La RR, Andrea S, Paola Q, Maria PA. Differential proteomic analysis of an engineered Streptomyces coelicolor strain reveals metabolic pathways supporting growth on n-hexadecane. Appl Microbiol Biotechnol. 2012;94(5):1289–1301. doi: 10.1007/s00253-012-4046-8. [DOI] [PubMed] [Google Scholar]
- Ganesh A, Lin J (2009) Diesel degradation and biosurfactant production by Gram-positive isolates. Afr J Biotechnol 8(21):5847–5854
- Gao Y, Wang J, Guo S, Hu YL, Li T, Mao R. Effects of salinization and crude oil contamination on soil bacterial community structure in the Yellow River Delta region, China. Appl Soil Ecol. 2015;86:165–173. doi: 10.1016/j.apsoil.2014.10.011. [DOI] [Google Scholar]
- Germaine KJ, Keogh E, Ryan D, Dowling DN. Bacterial endophyte mediated naphthalene phytoprotection and phytoremediation. FEMS Microbiol Lett. 2009;296(2):226–234. doi: 10.1111/j.1574-6968.2009.01637.x. [DOI] [PubMed] [Google Scholar]
- GESAMP Imact of oil and related chemicals and wastes on the marine environment. Rep Stud GESAMP. 1993;59:180. [Google Scholar]
- Ghoreishi G, Alemzadeh A, Mojarrad M, Djavaheri M. Bioremediation capability and characterization of bacteria isolated from petroleum contaminated soils in Iran. Sust Environ Res. 2017;27:195–202. doi: 10.1016/j.serj.2017.05.002. [DOI] [Google Scholar]
- Gros J, Reddy CM, Aeppli C, Nelson RK, Carmichael CA, Arey JS. Resolving biodegradation patterns of persistent saturated hydrocarbons in weathered oil samples from the Deepwater Horizon disaster. Environ Sci Technol. 2014;48(3):1628–1637. doi: 10.1021/es4042836. [DOI] [PubMed] [Google Scholar]
- Guleria S, Walia A, Chauhan A, Shirkot CK. Immobilization of Bacillus amyloliquefaciens SP1 and its alkaline protease in various matrices for effective hydrolysis of casein. 3Biotech. 2016;6:208. doi: 10.1007/s13205-016-0519-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurav R, Bhatia SK, Choi TR, Choi YK, Kim HJ, Song HS, Park SL, Lee HS, Lee SM, Choi KY, Yang YH. Adsorptive removal of crude petroleum oil from water using floating pinewood biochar decorated with coconut oil-derived fatty acids. Sci Total Environ. 2021;10(781):146636. doi: 10.1016/j.scitotenv.2021.146636. [DOI] [PubMed] [Google Scholar]
- Hamad AA, Moubasher HA, Moustafa YM, Mohamed NH, Rhim EHAE. Petroleum hydrocarbon bioremediation using native fungal isolates and consortia. Sci World J. 2021 doi: 10.1155/2021/6641533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haritash AK. A comprehensive review of metabolic and genomic aspects of PAH-degradation. Arch Microbiol. 2020;202:2033–2058. doi: 10.1007/s00203-020-01929-5. [DOI] [PubMed] [Google Scholar]
- Hassanshahian M, Tebyanian H, Cappello S. Isolation and characterization of two crude oil-degrading yeast strains, Yarrowia lipolytica PG-20 and PG-32, from the Persian Gulf. Mar Pollut Bull. 2012;64:1386–1391. doi: 10.1016/j.marpolbul.2012.04.020. [DOI] [PubMed] [Google Scholar]
- Hong YW, Yuan DX, Lin QM, Yang TL. Accumulation and biodegradation of phenanthrene and fluoranthene by the algae enriched from a mangrove aquatic ecosystem. Mar Pollut Bull. 2008;56:1400–1405. doi: 10.1016/j.marpolbul.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Hong Y, Liao D, Chen J, Khan S, Su J, Li H. A comprehensive study of the impact of polycyclic aromatic hydrocarbons (PAHs) contamination on salt marsh plants Spartina alterniflora: implication for plant-microbe interactions in phytoremediation. Environ Sci Pollut Res Int. 2015;22(9):7071–7081. doi: 10.1007/s11356-014-3912-6. [DOI] [PubMed] [Google Scholar]
- Hossain MF, Akter MA, Rahman Sohan MS, Sultana DN, Reza MA, Faisal Hoque KM. Bioremediation potential of hydrocarbon degrading bacteria: isolation, characterization, and assessment. Saudi J Biol Sci. 2022;29:211–216. doi: 10.1016/j.sjbs.2021.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua F, Wang H. Uptake modes of octadecane by Pseudomonas sp. DG17 and synthesis of biosurfactant. J Appl Microbiol. 2012;112(1):25–37. doi: 10.1111/j.1365-2672.2011.05178.x. [DOI] [PubMed] [Google Scholar]
- Hua X, Wang J, Wu Z, Zhang H, Li H, Xing X. A salt tolerant Enterobacter cloacae mutant for bioaugmentation of petroleum and salt-contaminated soil. Biochem Eng J. 2010;49(2):201–206. doi: 10.1016/j.bej.2009.12.014. [DOI] [Google Scholar]
- Huynh BQ, Kwong LH, Kiang MV, Chin ET, Mohareb AM, Jumaan AO, Basu S, Geldsetzer P, Karaki FM, Rehkopf DH. Public health impacts of an imminent red sea oil spill. Nat Sustain. 2021;4(12):1084–1091. doi: 10.1038/s41893-021-00774-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imperato V, Portillo-Estrada M, McAmmond BM, Douwen Y, Van Hamme JD, Gawronski SW, Vangronsveld J, Thijs S. Genomic diversity of two hydrocarbon-degrading and plant growth-promoting Pseudomonas species isolated from the oil field of bóbrka (Poland) Genes. 2019;10(6):443. doi: 10.3390/genes10060443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imura T, Morita T, Fukuoka T, Ryu M, Igarashi K, Hirata Y, Kitamoto D. Spontaneous vesicle formation from sodium salt of acidic sophorolipid and its application as a skin penetration enhancer. J Oleo Sci. 2014;2014:13117. doi: 10.5650/jos.ess13117. [DOI] [PubMed] [Google Scholar]
- ITOPF (2017) International Tanker Owners Pollution Federation Limited. http://www.itopf.com/knowledge-resources/data-statistics/statistics/
- Jaligama S, Chen Z, Saravia J, Yadav N, Lomnicki SM, Dugas TR, Cormier SA. Exposure to deepwater horizon crude Oil burnoff particulate matter induces pulmonary inflammation and alters adaptive immune response. Environ Sci Technol. 2015;49(14):8769–8776. doi: 10.1021/acs.est.5b01439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez N, Viñas M, Bayona JM, Albaiges J, Solanas AM. The Prestige oil spill: bacterial community dynamics during a field biostimulation assay. Appl Microbiol Biotechnol. 2007;77:935–945. doi: 10.1007/s00253-007-1229-9. [DOI] [PubMed] [Google Scholar]
- Jobson A, Cook FD, Westlake DWS. Microbial utilization of crude oil. Appl Microbiol. 1972;23:1082–1089. doi: 10.1128/am.23.6.1082-1089.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KC, de Voogt P. Persistant Organic Pollutants (POPs): state of the science. Environ Pollut. 1999;100:209–222. doi: 10.1016/s0269-7491(99)00098-6. [DOI] [PubMed] [Google Scholar]
- Juhasz AL, Btitz ML, Stanley GA. Degradation of fluoranthrene, pyren, benz(a)anthracene and dibenz(a, h)anthracene by Burkholderia cepacia. J Appl Microbiol. 1997;83:189–198. doi: 10.1046/j.1365-2672.1997.00220.x. [DOI] [Google Scholar]
- Jussila MM, Zhao J, Suominen L, Lindström K. TOL plasmid transfer during bacterial conjugation in vitro and rhizoremediation of oil compounds in vivo. Environ Pollut. 2007;146(2):510–524. doi: 10.1016/j.envpol.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Kankara RS, Subramanian BR. Oil spill sensitivity analysis and risk assessment for Gulf of Kachchh, India, using integrated modeling. J Coast Res. 2007;235:1251–1258. doi: 10.2112/04-0362.1. [DOI] [Google Scholar]
- Kapoor RT, Salvadori MR, Rafatullah M, Siddiqui MR, Khan MA, Alshareef SA. Exploration of microbial factories for synthesis of nanoparticles—a sustainable approach for bioremediation of environmental contaminants. Front Microbiol. 2021;12:658294. doi: 10.3389/fmicb.2021.658294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlapudi AP, Venkateswarulu TC, Tammineedi J, Kanumuri L, Ravuru BK, Ramu Dirisala V, Kodali VP. Role of biosurfactants in bioremediation of oil pollution-a review. Petroleum. 2018;4(3):241–249. doi: 10.1016/j.petlm.2018.03.007. [DOI] [Google Scholar]
- Karunanithi S, Sivaganesh A, Dhailappan AK, Packiasamy R. Biodegradation of hydrocarbon pollutant soil by indigenous microbes. Biodegradation. 2017;4(3):1038–1047. [Google Scholar]
- Kennisha MJ (1997). Practical handbook of estuarine and marine pollution. CRC Press, London, p 524
- Kora AJ. Growth and metabolic characteristics of hydrocarbon degrading bacteria isolated from an oil refinery soil. Indian J Geo Marine Sci. 2018;47(5):1029–1035. [Google Scholar]
- Korda A, Santas P, Tenente A, Santas R. Petroleum hydrocarbon bioremediation: sampling and analytical techniques, in situ treatments and commercial microorganisms currently used. Appl Microbiol Biotechnol. 1997;48(6):677–686. doi: 10.1007/s002530051115. [DOI] [PubMed] [Google Scholar]
- Kumari A, Kaur R, Kaur R (2019) A review on fate and remediation techniques of oil spills. Intern J Res Pharmaceut Sci 10(1):111–116. http://scopeindex.org/handle/sc/1595
- Lee K, Boufadel M, Chen B, Foght J, Hodson P, Swanson S, Venosa A. The behaviour and environmental impacts of crude oil released into aqueous environments. Ottawa: The Royal Society of Canada; 2015. [Google Scholar]
- Li X, Zhao L, Adam M. Biodegradation of marine crude oil pollution using a salt-tolerant bacterial consortium isolated from Bohai Bay, China. Mar Pollut Bull. 2016;105(1):43–50. doi: 10.1016/j.marpolbul.2016.02.073. [DOI] [PubMed] [Google Scholar]
- Lipthay JR, Barkay T, Sørensen SJ. Enhanced degradation of phenoxyacetic acid in soil by horizontal transfer of the tfdA gene encoding a 2,4-dichlorophenoxyacetic acid dioxygenase. FEMS Microbiol Ecol. 2001;35(1):75–84. doi: 10.1111/j.1574-6941.2001.tb00790.x. [DOI] [PubMed] [Google Scholar]
- Liu B, Chen B, Zhang B. Oily wastewater treatment by nano-TiO2-induced photocatalysis: seeking more efficient and feasible solutions. IEEE Nanotechnol Mag. 2017;11(3):4–15. doi: 10.1109/MNANO.2017.2708818. [DOI] [Google Scholar]
- Liu L, Bilal M, Duan X, Iqbal HMN. Mitigation of environmental pollution by genetically engineered bacteria—current challenges and future perspectives. Sci Total Environ. 2019;667:444–454. doi: 10.1016/j.scitotenv.2019.02.390. [DOI] [PubMed] [Google Scholar]
- Lombard V, Golaconda RH, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loya Y, Rinkevich B. Effects of oil pollution on coral reef communities. Mar Ecol Prog Ser. 1980;56:167–180. doi: 10.3354/meps003167. [DOI] [Google Scholar]
- Lynn SG, Price DJ, Birge WJ, Kilham SS. Effect of nutrient availability on the uptake of PCB congener2,2',6,6'-tetrachlorobiphenyl by a diatom (Stephanodiscus minutulus) and transfer to a zooplankton (Daphnia pulicaria) Aquat Toxicol. 2007;83:24–32. doi: 10.1016/j.aquatox.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Mallick S, Chatterjee S, Dutta TK (2007) A novel degradation pathway in the assimilation of phenanthrene by Staphylococcus sp. Strain PN/Y via meta-cleavage of 2-hydroxy-1-naphthoic acid: formation of trans-2, 3-dioxo-5-(2'-hydroxyphenyl)-pent-4-enoicacid. Microbiology 153(7): 2104–2115. Doi: 10.1099/mic.0.2006/004218-0 [DOI] [PubMed]
- Mandalenaki A, Kalogerakis N, Antoniou E. Production of high purity biosurfactants using heavy oil residues as carbon source. Energies. 2021 doi: 10.3390/en14123557. [DOI] [Google Scholar]
- Mandeep SP. Microbial nanotechnology for bioremediation of industrial wastewater. Front Microbiol. 2020 doi: 10.3389/fmicb.2020.590631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald IR, Miguez CB, Rogge G. Diversity of soluble methane monooxygenase-containing methanotrophs isolated from polluted environments. FEMS Microbiol Lett. 2006;255(2):225–232. doi: 10.1111/j.1574-6968.2005.00090.x. [DOI] [PubMed] [Google Scholar]
- Mehetre GT, Dastager SG, Dharne MS. Biodegradation of mixed polycyclic aromatic hydrocarbons by pure and mixed cultures of biosurfactant producing thermophilic and thermo-tolerant bacteria. Sci Total Environ. 2019;679:52–60. doi: 10.1016/j.scitotenv.2019.04.376. [DOI] [PubMed] [Google Scholar]
- Mishra S, Sarma PM, Lal B. Crude oil degradation efficiency of a recombinant Acinetobacter baumannii strain and its survival in crude oil-contaminated soil microcosm. FEMS Microbiol Lett. 2004;235(2):323–331. doi: 10.1016/j.femsle.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Mittal A, Singh P. A feasibility study for assessment of in-situ bioremediation potential of a crude oil degrading Pseudomonas Consortium. Int J Sci Res. 2010;2(1):127–137. doi: 10.3329/jsr.v2i1.2601. [DOI] [Google Scholar]
- Moody JD, Freeman JP, Doerge DR, Cerniglia CE. Degradation of Phenanthrene and anthracene by cell suspension of Mycobacterium sp. PYR-1. Appl Environ Microbiol. 2001;67:1476–1483. doi: 10.1128/AEM.67.4.1476-1483.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JW. Inorganic contaminants of surface water: research and monitoring priorities. Berlin: Springer Science & Business Media; 2012. [Google Scholar]
- Mukome FND, Buelowa MC, Shang U, Peng J, Rodriguez M, Mackay DM, Pignatello JJ, Sihota N, Hoelen TP, Parikh SJ. Biochar amendment as a remediation strategy for surface soils impacted by crude oil. Environ Pollut. 2020;265:115006. doi: 10.1016/j.envpol.2020.115006. [DOI] [PubMed] [Google Scholar]
- Naeem U, Qazi MA. Leading edges in bioremediation technologies for removal of petroleum hydrocarbons. Environ Sci Pollut Res Int. 2020;27(22):27370–27382. doi: 10.1007/s11356-019-06124-8. [DOI] [PubMed] [Google Scholar]
- Najafi AR, Roostaazad R, Soleimani M, Arabian D, Moazed MT, Rahimpour MR, Mazinani S. Comparison and modification of models in production of biosurfactant for Paenibacillus alvei and Bacillus mycoides and its effect on MEOR efficiency. J Pet Sci Eng. 2015;128:177–183. doi: 10.1016/J.PETROL.2015.02.019. [DOI] [Google Scholar]
- Nedjimi B. Phytoremediation: a sustainable environmental technology for heavy metals decontamination. SN Applied Sciences. 2021 doi: 10.1007/s42452-021-04301-4. [DOI] [Google Scholar]
- Nedunuri KV, Govindaraju RS, Banks MK, Schwab AP, Chen Z. Evaluation of phytoremediation for field-scale degradation of total petroleum hydrocarbons. J Environ Eng. 2000;126(6):483–490. doi: 10.1061/(ASCE)0733-9372(2000)126:6(483). [DOI] [Google Scholar]
- Nero BF (2021) Phytoremediation of petroleum hydrocarbon-contaminated soils with two plant species: Jatropha curcas and Vetiveria zizanioides at Ghana Manganese Company Ltd. Int J Phytoremed. 10.1080/15226514.2020.1803204 [DOI] [PubMed]
- Nyman JA, Klerks PL, Bhattacharyya S. Effects of chemical additives on hydrocarbon disappearance and biodegradation in freshwater marsh microcosms. Environ Pollut. 2007;149(2):227–238. doi: 10.1016/j.envpol.2006.12.028. [DOI] [PubMed] [Google Scholar]
- Ohadi M, Dehghannoudeh G, Shakibaie M, Banat IM, Pournamdari M, Forootanfar H. Isolation, characterization, and optimization of biosurfactant production by an oil-degrading Acinetobacter junii B6 isolated from an Iranian oil excavation site. Biocatal Agric Biotechnol. 2017;12:1–9. doi: 10.1016/j.bcab.2017.08.007. [DOI] [Google Scholar]
- Onwurah INE, Ogugua VN, Onyike NB, Ochonogor AE, Otitoju OF. Crude oils spills in the environment, effects and some innovative clean-up biotechnologies. Int J Environ Res. 2007;1:307–320. [Google Scholar]
- Ordinioha B, Brisibe S. The human health implications of crude oil spills in the Niger delta, Nigeria: an interpretation of published studies. Niger Med J. 2013;54(1):10–15. doi: 10.4103/0300-1652.108887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owoseni O, Nyankson E, Zhang Y, Adams SJ, He J, McPherson GL, John VT. Release of surfactant cargo from interfacially-active halloysite clay nano tubes for oil spill remediation. Langmuir. 2014;30(45):13533–13541. doi: 10.1021/la503687b. [DOI] [PubMed] [Google Scholar]
- Pan Y, Shi K, Peng C, Wang W, Liu Z, Ji X. Evaluation of hydrophobic polyvinyl-alcohol formaldehyde sponges as absorbents for oil spill. ACS Appl Mater Interfaces. 2014;6(11):8651–8659. doi: 10.1021/am5014634. [DOI] [PubMed] [Google Scholar]
- Parrilli E, Papa R, Tutino ML, Sannia G. Engineering of a psychrophilic bacterium for the bioremediation of aromatic compounds. Bioengineer Bugs. 2010;1(3):213–216. doi: 10.4161/bbug.1.3.11439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pete AJ, Bharti B, Benton MG. Nano-enhanced bioremediation for oil spills: a review. ACS ES&T Engineer. 2021;6:928–946. doi: 10.1021/acsestengg.0c00217. [DOI] [Google Scholar]
- Pimentel MR, Molina G, Dionísio AP, Maróstica MRJ, Pastore GM. The use of endophytes to obtain bioactive compounds and their application in biotransformation process. Biotechnol Res Int. 2011;2011:1–11. doi: 10.4061/2011/576286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punetha A, Saraswat S, Rai JPN. An insight on microbial degradation of benzo[a]pyrene: current status and advances in research. World J Microbiol Biotechnol. 2022;38:61. doi: 10.1007/s11274-022-03250-3. [DOI] [PubMed] [Google Scholar]
- Punnapayak H, Prasongsuk S, Messner K, Danmek K, Lotrakul P. Polycyclic aromatic hydrocarbons (PAHs) degradation by laccase from a tropical white rot fungus Ganoderma lucidum. Afr J Biotechnol. 2009;8:5897–5900. doi: 10.4314/ajb.v8i21.66070. [DOI] [Google Scholar]
- Rayu S, Karpouzas DG, Singh BK. Emerging technologies in bioremediation: constraints and opportunities. Biodegradation. 2012;23(6):917–926. doi: 10.1007/s10532-012-9576-3. [DOI] [PubMed] [Google Scholar]
- Rikalovic MG, Vrvić MM, Karadžić IM. Rhamnolipid biosurfactant from Pseudomonas aeruginosa: from discovery to application in contemporary technology. J Serbian Chem Soc. 2015;80(3):279–304. doi: 10.2298/JSC140627096R. [DOI] [Google Scholar]
- Rocha C, Pedregosa A, Laborda F. Biosurfactant-mediated biodegradation of straight and methyl-branched alkanes by Pseudomonas aeruginosa ATCC 55925. AMB Express. 2011;1(1):9. doi: 10.1186/2191-0855-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo F. Enzymes for aerobic degradation of alkanes. In: Timmis KN, editor. Handbook of hydrocarbon and lipid microbiology. Berlin, Heidelberg: Springer; 2010. pp. 781–793. [Google Scholar]
- Ruggiero P, Sarkar JM, Bollag JM. Detoxification of 2,4-dichlorophenol by a laccase immobilization on soil or clay. Soil Sci. 1989;147(5):361–310. doi: 10.1097/00010694-198905000-00007. [DOI] [Google Scholar]
- Sadhana S, JaiVarshini E, Nikita Reddy S, Shruthi S. Crude oil bioremediation—genetically modified microorganisms for poly-aromatic hydrocarbon degradation. App Ecol Environ Sci. 2021;9(8):769–785. doi: 10.12691/aees-9-8-8. [DOI] [Google Scholar]
- Sakhuja D, Hemant G, Rathour RK, Kumar P, Bhatt AK. Cost-effective production of biocatalysts using inexpensive plant biomass: a review. 3Biotech. 2021;11:280. doi: 10.1007/s13205-021-02847-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar JN, Leunuwitcz A, Bollag JM. Immobilization of enzymes on clay and soil. Soil Biol Biochem. 1989;21(2):222–230. doi: 10.1016/0038-0717(89)90098-9. [DOI] [Google Scholar]
- Sarkar D, Ferguson M, Datta R, Birnbaum S. Bioremediation of petroleum hydrocarbons in contaminated soils: comparison of biosolids addition, carbon supplementation, and monitored natural attenuation. Environ Pollut. 2005;136(1):187–195. doi: 10.1016/j.envpol.2004.09.025. [DOI] [PubMed] [Google Scholar]
- Sayed K, Baloo L, Sharma NK. Bioremediation of total petroleum hydrocarbons (TPH) by bioaugmentation and biostimulation in water with floating oil spill containment booms as bioreactor basin. Int J Environ Res Public Health. 2021;18(5):2226. doi: 10.3390/ijerph18052226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayler GS, Ripp S. Field applications of genetically engineered microorganisms for bioremediation processes. Curr Opin Biotechnol. 2000;11(3):286–289. doi: 10.1016/s0958-1669(00)00097-5. [DOI] [PubMed] [Google Scholar]
- Scheller U, Zimmer T, Becher D, Schauer F, Schunck WH. Oxygenation cascade in conversion of n-alkanes to α, ω-dioic acids catalyzed by cytochrome P450 52A3. J Biol Chem. 1998;273(49):32528–32534. doi: 10.1074/jbc.273.49.32528. [DOI] [PubMed] [Google Scholar]
- Shahzad A, Siddiqui S, Bano A, Sattar S, Hashmi MZ, Qin M, Shakoor A. Hydrocarbon degradation in oily sludge by bacterial consortium assisted with alfalfa (Medicago sativa L.) and maize (Zea mays L.) Arab J Geosci. 2020;13:1–12. doi: 10.1007/s12517-020-05902-w. [DOI] [Google Scholar]
- Shakya M, Kothari M, Kumar S, Sandhu SS. Bioremediation of petroleum hydrocarbon by microorganisms: a review. Res J Life Sci Bioinform Pharmaceut Chem Sci. 2021;7:51–64. doi: 10.26479/2021.0702.04. [DOI] [Google Scholar]
- Shankar S, Kansrajh C, Dinesh MG, Satyan RS, Kiruthika S, Tharanipriya A. Application of indigenous microbial consortia in bioremediation of oil-contaminated soils. Int J Environ Sci Technol. 2014;11(2):367–376. doi: 10.1007/s13762-013-0366-1. [DOI] [Google Scholar]
- Sharma B, Dangi AK, Shukla P. Contemporary enzyme based technologies for bioremediation: a review. J Environ Manag. 2018;210:10–22. doi: 10.1016/j.jenvman.2017.12.075. [DOI] [PubMed] [Google Scholar]
- Sharma S, Verma R, Pandey LM. Crude oil degradation and biosurfactant production abilities of isolated Agrobacterium fabrum SLAJ731. Biocatal Agric Biotechnol. 2019;21:101322. doi: 10.1016/j.bcab.2019.101322. [DOI] [Google Scholar]
- Sheth RU, Cabral V, Chen SP, Wang HH. Manipulating bacterial communities by in situ microbiome engineering. Trends Genet. 2016;32:189–200. doi: 10.1016/j.tig.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigenaka G, Overton E, Meyer B, Gao H, Miles S (2015) Physical and chemical characteristics of in-situ burn residue and other environmental oil samples collected during the deepwater horizon spill response. In: Interspil Conference pp: 1–2.
- Singh A, Asmath H, Chee CL, Darsan J. Potential oil spill risk from shipping and the implications for management in the Caribbean Sea. Mar Pollut Bull. 2015;93(1):217–227. doi: 10.1016/j.marpolbul.2015.01.013. [DOI] [PubMed] [Google Scholar]
- Solanki P, Putatunda C, Kumar A, Bhatia R, Walia A. Microbial proteases: ubiquitous enzymes with innumerable uses. 3Biotech. 2021;11(10):428. doi: 10.1007/s13205-021-02928-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava NK, Jha MK, Mall ID, Singh D. Application of genetic engineering for chromium removal from industrial wastewater. Int J Chem Biol Engineer. 2010;3:3. doi: 10.5281/zenodo.1080197. [DOI] [Google Scholar]
- Steffen KT, Hatakka A, Hofrichter M. Degradation of Benzo(a)pyrene by the litter-decomposing basidiomycete Stropharia coronilla: role of manganese peroxidase. Appl Environ Microbiol. 2003;69(7):3957–3964. doi: 10.1128/AEM.69.7.3957-3964.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui X, Wang X, Li Y, Ji H. Remediation of petroleum-contaminated soils with microbial and microbial combined methods: advances, mechanisms, and challenges. Sustainability. 2021;13(16):9267. doi: 10.3390/su13169267. [DOI] [Google Scholar]
- Suja F, Rahim F, Taha MR, Hambali N, Razali MR, Khalid A, Hamzah A. Effects of local microbial bioaugmentation and biostimulation on the bioremediation of total petroleum hydrocarbons (TPH) in crude oil contaminated soil based on laboratory and field observations. Int Biodeterior Biodegradation. 2014;90:115–122. doi: 10.1016/j.ibiod.2014.03.006. [DOI] [Google Scholar]
- Tabatabaee A, Assadi MM, Noohi AA, Sajadian VA. Isolation of biosurfactant producing bacteria from oil reservoirs. Iran J Environ Health Sci Eng. 2005;2(1):6–12. [Google Scholar]
- Taccari M, Milanovic V, Comitini F, Casucci C, Ciani M. Effects of biostimulation and bioaugmentation on diesel removal and bacterial community. Int Biodeterior Biodegradation. 2012;66(1):39–46. doi: 10.1016/j.ibiod.2011.09.012. [DOI] [Google Scholar]
- Taghavi S, Barac T, Greenberg B, Borremans B, Vangronsveld J, van der Lelie D. Horizontal gene transfer to endogenous endophytic bacteria from poplar improves phytoremediation of toluene. Appl Environ Microbiol. 2005;71(12):8500–8505. doi: 10.1128/AEM.71.12.8500-8505.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahseen R, Afzal M, Iqbal S, Shabir G, Khan QM, Khalid ZM, Banat IM. Rhamnolipids and nutrients boost remediation of crude oil-contaminated soil by enhancing bacterial colonization and metabolic activities. Int Biodeterior Biodegrad. 2016;115:192–198. doi: 10.1016/j.ibiod.2016.08.010. [DOI] [Google Scholar]
- Tang J, Wang R, Niu X, Zhou Q. Enhancement of soil petroleum remediation by using a combination of ryegrass (Lolium perenne) and different microorganisms. Soil Tillage Res. 2010;110:87–93. doi: 10.1016/j.still.2010.06.010. [DOI] [Google Scholar]
- Tang X, He LY, Tao XQ, Dang Z, Guo CL, Lu GN, Yi XY. Construction of an artificial microalgal-bacterial consortium that efficiently degrades crude oil. J Hazard Mater. 2010;181:1158–1162. doi: 10.1016/j.jhazmat.2010.05.033. [DOI] [PubMed] [Google Scholar]
- Thavamani P, Megharaj M, Krishnamurti GSR, McFarland R, Naidu R. Finger printing of mixed contaminants from former manufactured gas plant (MGP) site soils: implications to bioremediation. Environ Int. 2011;37(1):184–189. doi: 10.1016/j.envint.2010.08.017. [DOI] [PubMed] [Google Scholar]
- Thavasi R, Jayalakshmi S, Banat IM. Effect of biosurfactant and fertilizer on biodegradation of crude oil by marine isolates of Bacillus megaterium, Corynebacterium kutscheri and Pseudomonas aeruginosa. Bioresource Technol. 2011;102(2):772–778. doi: 10.1016/j.biortech.2010.08.099. [DOI] [PubMed] [Google Scholar]
- Tyagi M, da Fonseca MMR, de Carvalho CC. Bioaugmentation and biostimulation strategies to improve the effectiveness of bioremediation. Proc Biodegrad. 2011;22(2):231–241. doi: 10.1007/s10532-010-9394-4. [DOI] [PubMed] [Google Scholar]
- US Army Corps of Engineers (2003) Agriculturally based bioremediation of petroleum-contaminated soils and shallow groundwater in Pacific Island ecosystems
- van Beilen JB, Funhoff EG. Alkane hydroxylases involved in microbial alkane degradation. Appl Microbiol Biotechnol. 2007;74(1):13–21. doi: 10.1007/s00253-006-0748-0. [DOI] [PubMed] [Google Scholar]
- Varjani SJ, Upasani VN. Carbon spectrum utilization by an indigenous strain of Pseudomonas aeruginosa NCIM 5514: production, characterization and surface active properties of biosurfactant. Bioresour Technol. 2016;221:510–516. doi: 10.1016/j.biortech.2016.09.080. [DOI] [PubMed] [Google Scholar]
- Verma S, Bhargava R, Pruthi V. Oily sludge degradation by bacteria from Ankleshwar, India. Int Biodeterior Biodegrad. 2006;57(4):207–213. doi: 10.1016/J.IBIOD.2006.02.004. [DOI] [Google Scholar]
- Wang Y, Liang J, Wang J, Gao S. Combining stable carbon isotope analysis and petroleum-fingerprinting to evaluate petroleum contamination in the Yanchang oilfield located on loess plateau in China. Environ Sci Pollut Res. 2018;25:2830–2841. doi: 10.1007/s11356-017-0500-6. [DOI] [PubMed] [Google Scholar]
- Wasilkowski D, Swędzioł Z, Mrozik A. The applicability of genetically modified microorganisms in bioremediation of contaminated environments. Chemik. 2012;66(8):817–826. [Google Scholar]
- Whang L, Liu P, Ma C, Cheng S. Application of biosurfactants, rhamnolipid, and surfactin, for enhanced biodegradation of diesel-contaminated water and soil. J Hazard Mater. 2008;151(1):155–163. doi: 10.1016/j.jhazmat.2007.05.063. [DOI] [PubMed] [Google Scholar]
- Wu ZY, Li C, Liang HW, Zhang YN, Wang X, Chen JF, Yu SH. Carbon nanofiber aerogels for emergent clean-up of oil spillage and chemical leakage under harsh conditions. Sci Rep. 2014;4(1):4079. doi: 10.1038/srep04079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Yu F, Wang Q, Gu X, Chen W. Cloning of catechol 2, 3-dioxygenase gene and construction of a stable genetically engineered strain for degrading crude oil. Indian J Microbiol. 2014;54(1):59–64. doi: 10.1007/s12088-013-0411-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Pei J. Construction and characterization of a photosynthetic bacterium genetically engineered for Hg2+ uptake. Bioresour Technol. 2011;102:3083–3088. doi: 10.1016/j.biortech.2010.10.051. [DOI] [PubMed] [Google Scholar]
- Yadav M, Singh S, Sharma J, Deo Singh K. Oxidation of polyaromatic hydrocarbons in systems containing water miscible organic solvents by the lignin peroxidase of Gleophyllum striatum MTCC-1117. Environ Technol. 2011;32:1287–1294. doi: 10.1080/09593330.2010.535177. [DOI] [PubMed] [Google Scholar]
- Yang L, Wang Y, Song J, Zhao W, He X, Chen J, Xiao M. Promotion of plant growth and in situ degradation of phenol by an engineered Pseudomonas fluorescens strain in different contaminated environments. Soil Biol Biochem. 2011;43(5):915–922. doi: 10.1016/j.soilbio.2011.01.001. [DOI] [Google Scholar]
- Yuniati MD. Bioremediation of petroleum-contaminated soil: a review. IOP Conf Series: Earth Environ Sci. 2018;118(2018):012063. doi: 10.1088/1755-1315/118/1/012063. [DOI] [Google Scholar]
- Zhao Q, Yue S, Bilal M, Hu H, Wang W, Zhang X. Comparative genomic analysis of 26 Sphingomonas and Sphingobium strains: dissemination of bioremediation capabilities, biodegradation potential and horizontal gene transfer. Sci Total Environ. 2017;609:1238–1247. doi: 10.1016/j.scitotenv.2017.07.249. [DOI] [PubMed] [Google Scholar]
- Zimmer T, Ohkuma M, Ohta A, Takagi M, Schunck WH. The CYP52 multigene family of Candida maltose encodes functionally diverse n-alkane-inducible cytochromes p450. Biochem Biophys Res Commun. 1996;224(3):784–789. doi: 10.1006/bbrc.1996.1100. [DOI] [PubMed] [Google Scholar]