Abstract
Introduction
Activation of skeletal progenitors upon tissue injury and the subsequent cell fate specification are tightly coordinated in the bone repair process. Although known osteoimmunological signaling networks play important roles in the microenvironment of the bone defect sites, the molecular mechanism underlying the bone repair process has not been fully understood.
Methods
To better understand the behavior of the skeletal progenitors and the heterogeneity of the cells during bone repair at the microenvironmental level, we performed a combinatorial analysis consisting of lineage tracing for skeletal progenitors using the Sox9-CreERT2;R26RtdTomato mouse line followed by single-cell RNA sequencing (scRNA-seq) analysis using a mouse model of calvarial bone repair. To identify a therapeutic target for bone regeneration, further computational analysis was performed focusing on the identification of the cell–cell interactions, followed by pharmacological assessments with a critical-size calvarial bone defect mouse model.
Results
Lineage tracing analysis showed that skeletal progenitors marked by Sox9 were activated upon bone injury and contributed to bone repair by differentiating into osteoblasts. The scRNA-seq analysis characterized heterogeneous cell populations at the bone defect sites; the computational analysis predicted a bifurcated lineage from skeletal progenitors toward osteogenic and adipogenic lineages. Chemokine C–C motif ligand 9 (Ccl9) was identified as a signaling molecule that regulates bone regeneration in the mouse model, possibly through the regulation of adipogenic differentiation at the bone defect site.
Conclusion
Multipotential skeletal progenitors and the direction of the cell differentiation were characterized at single cell resolution in a mouse bone repair model. The Ccl9 signaling pathway may be a key factor directing osteogenesis from the progenitors in the model and may be a therapeutic target for bone regeneration.
Keywords: Bone repair, Lineage tracing, Calvaria, Sox9, Single-cell analysis, Ccl9
Highlights
-
•
Sox9-positive skeletal progenitors contributed to the calvaria bone repair process.
-
•
scRNA-seq analysis revealed a heterogeneous cell population at bone defect sites.
-
•
Skeletal progenitors had a bifurcated lineages of osteogenesis and adipogenesis.
-
•
Ccl9 was identified as an important signaling molecule regulating bone regeneration.
1. Introduction
Bone functions as a framework for the mammalian body and is required for the storage of minerals via a process that tightly coordinates bone-forming osteoblasts and bone-resorbing osteoclasts. Given the dynamics of bone metabolism, bone has relatively high potential for tissue repair, although its potency in this capacity depends on multiple parameters. Meta analyses have clearly shown that age and health conditions, including smoking and diabetes, are related to osteoporosis and bone fracture [1,2]. Diabetes and low bone mineral density have both been shown to contribute to poor bone quality [3]. These issues should be addressed, as osteoporotic fracture is increasing in our super-aging society and is becoming a social burden [4].
Non-union is a problematic complication of fracture, causing functional disabilities including load and movement limitations. For instance, 20% of cases of femoral neck fracture fixation lead to secondary operation due to non-union or avascular necrosis [5]. Surgery for non-union is a particularly challenging problem in orthopaedic traumatology [6].
Treatment for non-union is difficult particularly in critical-sized bone defects such that postoperative bone healing takes a long time. Traditional methods of treatment for critical-sized bone defects are vascularized bone grafts [7], osteoinductive bone lengthening [8], chipping [9], and the Masquelet technique [10]. Even with these methods, non-union sometimes requires reoperation. Vascularized bone grafts and the Masquelet technique require donor sites, and sometimes cause comorbidity including surgical site infection and chronic pain [11]. To avoid such complications, tissue engineering featuring signaling molecules as osteoinductive factors is a promising approach to enhancement of bone healing.
To establish bone regenerative strategies, better understanding of the mechanisms underlying the bone repair process is essential. The bone repair process consists of hematoma formation, inflammation, bone formation and bone remodeling [12]. Both the hematopoietic lineage and mesenchymal lineage are activated upon bone injury. Multiple immune cell types regulate distinct stages of the differentiation of skeletal cell types through the secretion of signaling molecules [[12], [13], [14]]. Although osteoimmunological processes are key to the process of bone healing [15], osteoimmunological targets to promote bone healing remain unknown, because the signaling networks underlying cell–cell interactions in the bone repair process have not been fully understood.
Lineage tracing analysis can be used to characterize the skeletal progenitors in both bone development and bone repair. Multiple progenitors have been defined by distinct markers; these progenitors have distinct properties and potencies for differentiating into skeletal cell types [16]. Among them, Sox9 is expressed in skeletal progenitors that give rise to chondrocytes, osteoblasts and osteocytes during bone development [17]. In adult mice, Sox9-positive cells reside in the periosteum, where the cells are activated upon injury and contribute to bone repair [[17], [18], [19]]. However, the behavior of the skeletal progenitors upon injury has not been well elucidated. In particular, the mechanism by which the multicellular interactions direct the cell fate of the progenitors toward bone-forming osteoblasts is the key to understanding the bone repair process.
In this study, we investigated the mechanism of bone repair by combinatorial analyses of lineage tracing and single-cell RNA-sequencing (scRNA-seq) in a mouse calvaria defect model. We genetically labelled the Sox9-positive skeletal progenitors and observed the behavior of the cells at bone defect sites. We further investigated the heterogeneity of the cells and signaling networks at the microenvironmental level. Cell–cell interaction analysis, followed by pharmacological assessment in a critical-sized bone defect model, revealed a potential therapeutic target for bone regeneration.
2. Methods
2.1. Animal preparation for lineage tracing
Animal experiments were conducted in compliance with the Animal Care and Use Committee of the University of Tokyo (approval number P17-038). The Sox9-CreERT2 knock-in mice were obtained from Riken BioResource Research Center (stock no. RBRC05522, [20]). The R26RtdTomato reporter mice were obtained from the Jackson Laboratory (stock no. 007905, [21]). To generate the bone defect models, mice were anesthetized with 20% ketamine and 8% xylazine in sterilized water. For the bone repair model, 1-mm diameter bone defects were made on the right and left parietal bone with a biopsy punch (BP–10F, Kai Medical) in 8 to 10 -week-old Sox9-CreERT2;R26RtdTomato male mice as previously reported [22]. For Cre recombination, 100 μL of a 20 mg/ml stock of tamoxifen (T5648-1G, Sigma–Aldrich) was used per injection. Two injection schedules were used: 1) tamoxifen injections for 3 consecutive days starting 14 days before surgery (‘Pre’ regimen), 2) tamoxifen injections for 4 consecutive injections starting 1 day prior to surgery (‘Post’ regimen).
2.2. Histological analyses
The Sox9-CreERT2;R26RtdTomato mice were euthanized at either Day 3 or Day 10 after surgery and then the 1-mm calvaria defect bones were exposed. The mice without surgery (Sham) were also euthanized and the calvaria defect bones were exposed at the same timing as Day 10 sample. The tissues were fixed with 4% paraformaldehyde (PFA)/phosphate buffered saline (PBS) for 1 h (h) on ice, washed with PBS, and then decalcified by 20% Ethylenediaminetetraacetic acid (EDTA) (Sigma Aldrich)/PBS at pH 7.5–8.0 for 5 days. Tissues were flash-frozen in O.C.T. compound (4583, SAKURA) and cut into 12-μm sections. For immunohistochemistry, sections were blocked with 3% bovine serum albumin (A7960; Sigma–Aldrich) and 1% heat-inactivated sheep serum (S2263, Sigma–Aldrich) in 0.1% TritonX-100/PBS. Sections were incubated with primary antibodies overnight at 4 °C, followed by incubation with secondary antibodies for 1 h at room temperature. The following antibodies were used: anti-RUNX2 (1:1000, ab76956, Abcam), anti-SP7 (1:1000, ab22552, Abcam) and Alexa Fluor 488 (1:500, secondary anti-rabbit, Life Technologies). Slides were mounted with VECTASHIELD (H-1000, Vector Laboratories). The immunohistochemical (IHC) images were obtained with a Zeiss LSM880 confocal microscope.
2.3. Single-cell RNA sequencing preparation
Three Sox9-CreERT2;R26RtdTomato male mice were used in this analysis. As described above, we created 1 mm calvaria bone defects on the parietal bones and injected tamoxifen. On day 10 after surgery, the defect areas were trimmed, and tissues were digested in 0.1% collagenase D, 0.2% dispase and 1 ug/mL DNase I in alpha-Minimum Essential Medium (A10490-01, Life Technologies) at 37 °C under constant agitation at 100 rpm for 10 min. Digestions were subsequently quenched with 20% fetal bovine serum (FBS). This cell digestion process was repeated 6 times. The extracted cells were washed in 2% FBS in PBS and filtered through Flowmi tip strainers (70-μm porosity, SP Bel-Art). For RNA library construction, 10,000 cells were selected. Library construction was performed using the Chromium GEM Single Cell 3′ Reagents kit v3.1 (10x Genomics) in accordance with the manufacturer's protocol. Sequencing was performed on a Novaseq 6000 (Illumina) The read length configuration was 28 × 8 × 91 cycles for Read 1, Index, and Read 2, respectively. Cell Ranger Single Cell Software Suite 3.1.0 was used to perform sample de-multiplexing, barcode processing, and single cell gene counting (Alignment, Barcoding and UMI Count).
2.4. Bioinformatics analysis of scRNA-seq data
R [23] and Seurat 3.2.2 [24] were used for downstream analysis. Cells with total expressed genes in the range of [200, 5000] and with a fraction of mitochondrial gene unique molecular identifiers (UMIs) more than 0.05 were discarded. Seurat's FindVariableFeatures function was used with default parameters to determine highly variable genes, considering 2000 genes. Counts were normalized, and variable genes were calculated using variance stabilizing transformation (VST). The first 30 dimensions were considered for the data object. The data object was then scaled, regressing out the fraction of the mitochondrial gene UMIs. Unsupervised clustering was performed using the Louvain algorithm with multilevel refinement [25]. Resolution in the clustree software [26] was set to 1.1, leading to 20 clusters. Markers for each cluster were calculated with the function “FindAllMarkers” with the arguments logfc.threshold 0.25 and min.pct 0.10. Clusters were annotated according to differentially expressed genes, referring to scCATCH [27]. scRNA-seq data in this study is available at DDBJ BioProject database (https://www.ddbj.nig.ac.jp/index-e.html) with the accession number PRJDB12846.
2.5. Cell trajectories
Cell trajectories were calculated with Monocle 3 0.2.3.0 [28], based on the top 2000 overall variable genes, ranked by scaled dispersion, obtained from Seurat. Velocyto analysis was also performed considering spliced and unspliced reads using velocyto.R by velocyto team [29]. Clusters, cell features, and embedding information were extracted from the Seurat object in R and integrated with the count matrix. RNA velocity was estimated using a gene-relative model with k-nearest neighbor cell pooling (k = 10).
2.6. Inference of regulons and activities, and ligand-receptor analyses
Gene regulatory analysis was performed with SCENIC (v 1.2.0.) implemented in R [30]. Individual regulons were constructed from the data extracted from the Seurat object. Mouse TF ranking database version 9 (motifs-v9-nr.mgi-m0.001-o0.0.tbl) and mouse motif to TF annotation downloaded from (v9) as outlined in the package manual [31] were used as the bases. To run GENIE3, genes were filtered with at least 6 UMI counts across all samples; those that were detected in at least 1% of the cells were filtered. Cell–cell interactions (CCIs) and ligand-receptor (LR) pairs were extracted with scTensor [32]. A variance-stabilizing transformation was performed upon the data matrix converted from the Seurat object. The cell type level average matrix and the LR database were combined, and the calculation of a matrix for each LR pair with the CCI tensor was performed manually [33].
2.7. Bone regeneration in mouse critical sized defects
For the critical-sized bone defect model, a 3-mm diameter bone defect was created with a biopsy punch (BP–30F; Kai Medical) in the parietal bone from 8-week-old wild-type C57BL/6J male mice (Charles River Laboratories Japan) as previously reported [34]. Defects were loaded with an absorbable collagen sponge (3 mm diameter; INFUSE™ Bone Graft, Medtronic) impregnated with 1 ng or 10 ng of a CCL9 antibody (R&D, MAB463-100). Micro-computed tomography (CT) scanning of the bone defect area was performed (Rigaku, Tokyo) using mice at 1-, 7- and 10-weeks post-surgery (n = 3/group). The X-ray source was set at 90 kV and 100 μA, a 1 mm filter, and a tomographic rotation of 360° (rotation step of 0.6°). To assess bone volume (BV, mm3) and bone density (g/cm2), micro-CT scanning of the harvested tissue was performed using SMX-90CT-SV (Shimadzu). The three-dimensional construction software package TRI/3D-BON (Ratoc System Engineering) was used for the quantitative analysis.
2.8. Statistical analysis
Differences between groups were analyzed using Dunnett's test. Values of p < 0.05 were considered significant.
3. Results
3.1. Sox9-positive cells contribute to bone repair through differentiation into osteoblasts
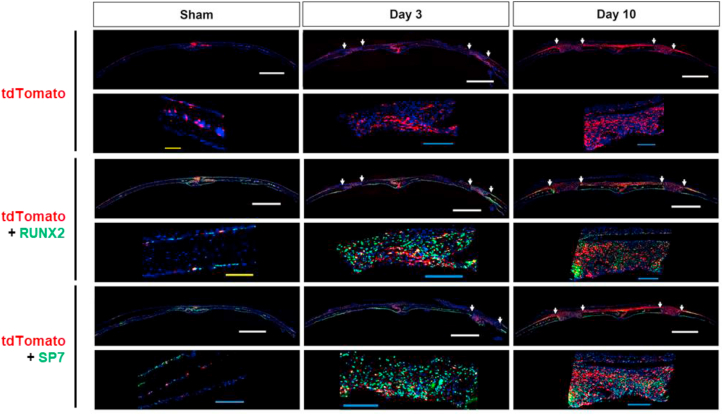
To investigate the behavior of skeletal progenitors upon injury, we performed lineage-tracing analysis. Because Sox9 is a marker of skeletal progenitors during endochondral ossification in mouse embryos and also the periosteum in mouse adults, we hypothesized that Sox9 is a marker of skeletal progenitors in bone repair processes regardless of the mode of ossification. To test this, we used an optimized mouse model of bone repair: we generated Sox9-CreERT2; R26RtdTomato mice and made a 1-mm bone defect in the calvaria [22]. To genetically mark post-existing Sox9-positive cells and their descendants with tdTomato, we injected tamoxifen for 4 consecutive days, starting on the day before surgery and including the day of surgery and the first two post-surgical days. Histological analysis reveled that tdTomato-positive cells were present at the sagittal suture site and in the cranial bone marrow with little expression on the periosteal and dura sides in the sham sample (Fig. 1). At day 3 after the surgery, tdTomato-positive cells were located not only at the sagittal suture but also at the bone defect site. At day 10 after the surgery, tdTomato-positive cells were strongly enriched at the bone defect sites; the tdTomato-positive cells were also widely distributed on the periosteal side throughout the suture (Fig. 1). These results suggest that post-existing Sox9-positive skeletal progenitors were activated upon bone injury; these cells may have either emerged de novo from the bone defect sites and/or migrated into bone defect sites from the suture. Immunohistochemistry revealed that cells in the bone defect sites expressed Runx2 and Sp7, markers of osteoblast precursors; most of the post-existing tdTomato-positive cells expressed with Runx2 or Sp7 (Fig. 1). These results suggest that post-existing Sox9-positive cells contribute to bone repair through differentiation into osteoblast precursors.
Fig. 1.
Lineage tracing analysis of tdTomato-positive Sox9-lineage with Post regimen and immunohistochemistry of osteoblast markers at the 1-mm defect site in mouse calvaria bone defect model. Confocal microscopy images of coronal section in uninjured calvaria (left; sham), 3 days after 1-mm injured (middle; Day 3) and 10 days after 1-mm injured (right; Day 10) with tile scan and zoom images (n = 2 mice). The region between white arrow shows 1-mm defect. Sections were immunostained with antibodies for RUNX2 (middle) or SP7 (bottom). Scale bar, 1000 μm (white), 200 μm (yellow) and 100 μm (blue).
We next investigated the contribution of pre-existing Sox9-positive cells to bone repair. To do this, we injected tamoxifen 3 consecutive days starting from 14 days before surgery and performed the histological analysis at 10 days after surgery. The analysis revealed that, although certain number of tdTomato-positive cells were observed at day 10, few number of tdTomato-positive cells expressed either Runx2 or Sp7 (Supplementary Fig. 1). This result suggests that the contribution of the pre-existing Sox9-positive cells to bone repair is limited; the injury-induced de novo Sox9-positive cells mostly contributes to the bone repair process.
3.2. Singe-cell profiling of cell population heterogeneity during bone repair
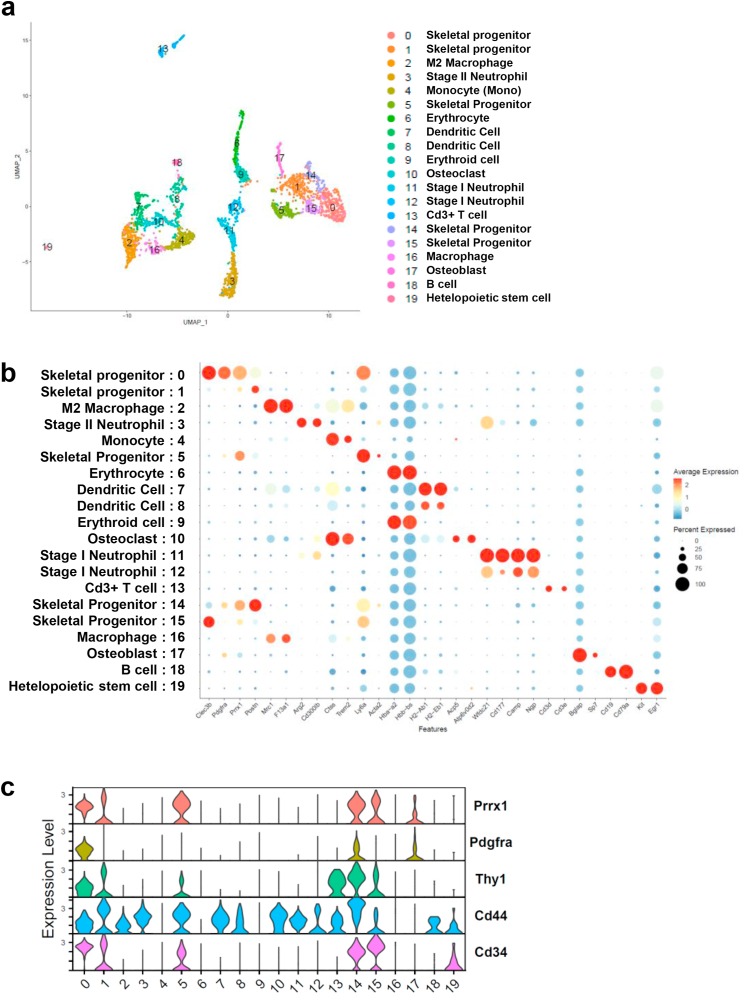
To study the transcriptomic diversity of cell populations during the bone repair process, scRNA-seq was performed with the cells isolated from the area of the 1 mm bone defect at day 10 after bone surgery. We profiled 2977 cells after preprocessing of 3874 cells with the following threshold: the median depth of unique molecular identifiers (UMIs) per cell was 4027; the median number of genes per cell was 1156 (16,274 genes in total). UMAP analysis revealed 20 distinct clusters which were characterized by the expression of marker genes (Fig. 2a and b, Supplemental Table); the clusters included neutrophils (13%, clusters 3 and 11), macrophages (12%, clusters 2 and 16), dendritic cells (10%, clusters 7 and 8), monocytes (8%, cluster 4), osteoclasts (5%, cluster 10), skeletal progenitor cells (32%, clusters 0, 1, 5, 14 and 15) and osteoblasts (3%, cluster 17). Importantly, the skeletal cell-related populations including clusters 0, 1,5, 14, 15 and 17 were clustered together, and possibly represented distinct stages of cell fate specification and differentiation of skeletal cell-types during bone repair. Among them, cells in the clusters 0, 5, 14 and 15 highly expressed Prrx1, Pdgfra, Thy1, Cd44 and Cd34, which are known as mesenchymal stem/progenitor cell markers [35,36], suggesting that the skeletal progenitors were a heterogeneous population (Fig. 2c).
Fig. 2.
scRNA-seq analysis showing the heterogeneity of cells during bone repair at Day 10 of 1-mm calvaria defect. a. Uniform Manifold approximation and Projection (UMAP) plot of cell-type clusters detected by unsupervised graph clustering of 2977 cells after preprocessing, and cell-type definition, color coded with cell clusters. b. Average expression of selected gene markers for definition of clusters with Dot plots. The cluster number and assigned cell-types were shown. c. Violin plots showing expressions of marker genes for skeletal progenitors in all clusters.
3.3. The bone repair process was bifurcated into osteogenesis and adipogenesis
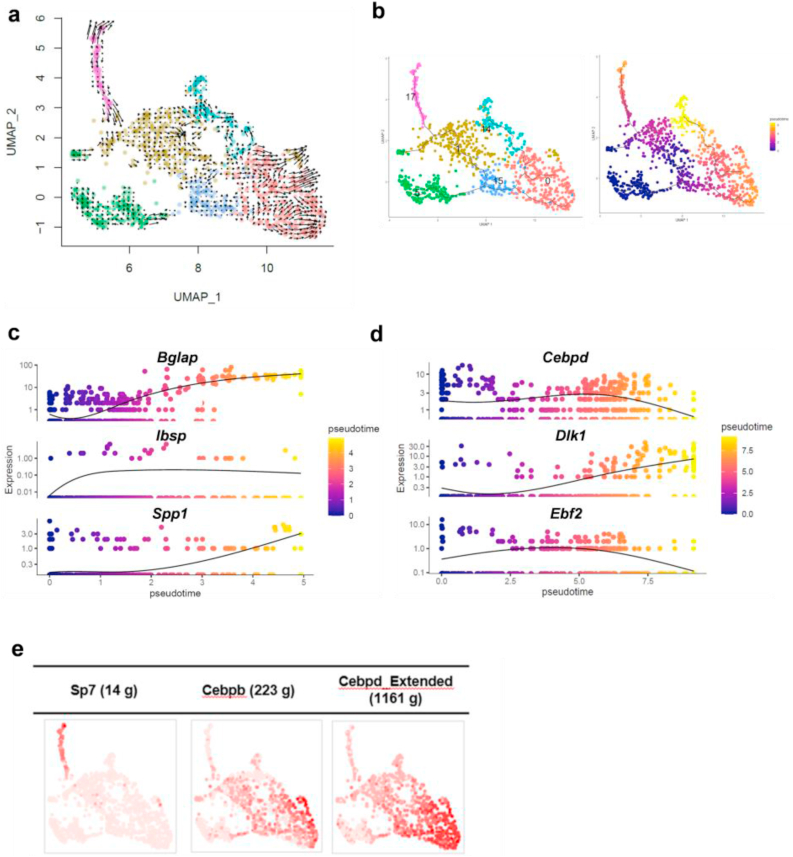
To better understand the dynamics of the skeletal cell fates during the bone repair process, we next performed an analysis of RNA velocity, which is a high-dimensional vector predicting the future state of individual cells, and trajectory analysis with monocle 3 using the aforementioned clusters 0, 1, 5, 14, 15, and 17, which represented skeletal cell-types. The velocity analysis predicted the direction of cell differentiation: it started from cluster 5 and ended in either clusters 0, 14, or 17 (Fig. 3a). For the trajectory analysis, we set cluster 5 as a starting point of the cell lineage based on the marker gene expressions and the results of the velocity analysis. The pseudotime analysis predicted 2 directions from skeletal progenitors of cluster 5 and 15. One was toward cluster 17; the other was toward cluster 14 through cluster 0 (Fig. 3b).
Fig. 3.
Trajectory analysis of skeletal cells at bone defect sites at Day 10 of 1-mm calvaria using subclusters 0, 1, 5, 14, 15 and 17 in scRNA-seq analysis. a. Velocyto analysis in the subclusters. The velocity was shown as arrows which represent RNA velocity kinetics visualizing the direction and acceleration between mature and nascent mRNA. b. Monocle 3 trajectory pseudo-time analysis and pseudotime UMAP latent time plot starting from cluster 5. The line corresponds to the principal graph learned by Monocle 3. c. Pseudotemporal kinetics for osteogenic genes of Bglap, Ibsp and Spp1. The pseudotime was shown as the osteogenic lineage starting from cluster 5. d. Pseudotemporal kinetics for adipogenic genes of Cebpd, Dlk1 and Ebf2. The pseudotime was shown as the adipogenic lineage starting from cluster 5. e. Binary active distribution of regulons of Sp7, an osteogenesis-related transcription factor and Cebpb and Cebpd, adipogenesis-related transcription factors. The intensity of red color in the plot represents the activity of regulons.
We then characterized the 2 lineages by focusing on marker gene expressions in osteogenesis and adipogenesis. The expression profile in one lineage to cluster 17 showed gradual upregulation of osteoblast marker genes including Ibsp, Spp1 and Bglap, suggesting an osteoblast lineage (Fig. 3c). The expression profile in another lineage to clusters 0 and 14 showed that preadipocyte markers Ebf2 and Cebpδ [[37], [38], [39], [40]] were enriched in cluster 0; Dlk1, also known as preadipocyte markers [41,42], was enriched in cluster 14 (Fig. 3d), which suggest that this lineage has an adipogenic signature. Because the mature adipocyte marker Pparγ was mostly undetected and Adiponection was not expressed in any clusters (data not shown), the observed lineage may represent skeletal progenitors directing an adipogenic progenitor or an intermediate stage of adipocyte differentiation [41].
Next, to investigate gene regulatory networks, we used the SCENIC package, in which potent transcription factors and co-factors were predicted by considering their own gene expressions, their downstream target genes and motif enrichment in genomic regions flanking the gene based on public datasets [30]. The putative activities of adipogenesis-related transcription factors Cebpβ and Cebpδ [43] were upregulated in cluster 0, whereas the activity of osteogenesis-related transcription factor Sp7 was upregulated in cluster 17 (Fig. 3e). Taken together, the pseudo-time analyses, velocity analysis and SCENIC results suggest a bifurcation of lineages from skeletal progenitors toward either an osteogenic lineage or an adipogenic lineage during bone repair. This trend was also observed when we used only tdTomato-positive cells in the analysis. tdTomato-positive cells were widely distributed in the subclusters 0, 1, 5, 14, 15, and 17, although cells highly expressing tdTomato gene were enriched in cluster 5. In contrast, Sox9-positive cells were mostly undetected (Supplementary Fig. 2a). These results suggest that Sox9 expression was transient during the repair process and very low at Day 10, whereas the descendants of the Sox9-positive cells contribute both to maintaining the skeletal progenitor states and to the differentiation into osteoblasts and adipogenic cells. The bifurcation of differentiation was also observed in the analysis with only tdTomato-positive cells (Supplementary Figs. 2b–d), suggesting that the Sox9-lineage contributes to both the osteogenic lineage and the adipogenic lineage in the calvaria bone repairing processes.
3.4. Ccl9 was a novel signaling molecule for bone healing in vivo
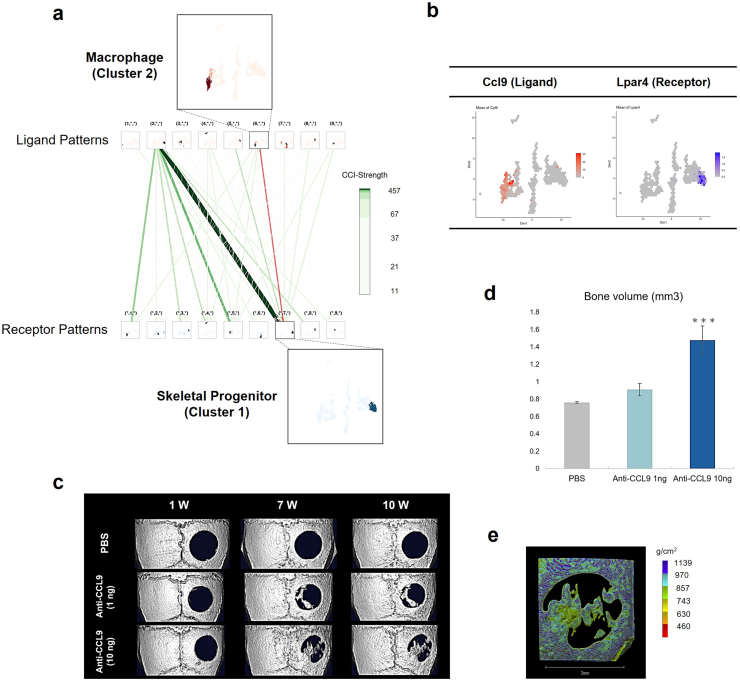
To identify key signaling networks during the bone repair process, we performed ligand-receptor interaction analysis using scTensor. This package is a tensor-based tool that uses tensors to explicitly model ligand–receptor interactions, which is available to extract representative triadic relationships including ligand expression, receptor expression and LR pairs [44]. A total of 20 cell–cell interactions (CCI) and 14,575 LR pairs were identified. To narrow the set of candidate CCIs, we set out the following criteria: (1) LR pair factor value > 0.15%, and (2) P-value < 0.05; this resulted in a total of 458 LR pairs. Given the crucial cell types for both immune responses upon injury and interactions with skeletal cell types in the microenvironment of bone defect sites, we focused on LR pairs between macrophages and skeletal cell types (Fig. 4a and Table 1). Among them, we identified Ccl9 as a candidate signaling for the bone repair process for the following reasons. First, Ccl9 is known to play important roles in adipogenic differentiation [45]. Second, the putative receptor lysophosphatidic acid receptor 4 (Lpar4) was reported to play a role in osteogenesis and bone remodeling in mice [46]. As Lpar4 was expressed in cluster 0, an adipogenic state (Fig. 4b), we hypothesized that inhibition of the Ccl9 signaling pathway inhibits adipogenesis and promotes osteoblast differentiation, contributing to bone regeneration. To examine this hypothesis, a neutralizing antibody for Ccl9 was infused into the collagen sponge [47] implanted in the 3 mm diameter (“critical-sized”) bone defect site in the mouse calvaria [34]. Micro CT analysis showed that the Ccl9 antibody induced bone regeneration in a dose-dependent manner (Fig. 4c and d). Regenerated bone mineral density was comparable to that of intact bone around the defect site (Fig. 4e). These results suggest that manipulation of the Ccl9 signaling pathway may be a therapeutic target for bone regeneration.
Fig. 4.
Identification of CCL9 as a molecule regulating bone repair and investigation of the effects of CCL9 inhibition on bone regeneration with critical bone defect model in mouse calvaria. a. Cell–cell interaction hypergraph. Pink line shows the putative interaction that we selected between the ligand from macrophage as cluster 2 and the receptor in skeletal progenitors as cluster 1. b. Feature plots of the gene expression pair between ligand gene Ccl9 and receptor gene Lpar4. c. μCT analysis in the calvaria 3-mm size defect with indicated treatments at 1 week (1 W), 7 week (7 W) and 10 week (10 W). d. Quantification of bone volume at 10 W determined by μCT analysis (n = 3/sample) Data presented as means ± standard deviation. ∗∗∗p < 0.001 between Anti-CCL9 10 ng and PBS, Dunnett's test. e. Bone density analysis of Anti-CCL9 10 ng sample determined by μCT at 10 W. Scale bar, 3 mm.
Table 1.
LR-pairs in the interaction between macrophages (cluster 2) and skeletal progenitors (cluster 0) at Day 10.
| Ligand Gene | Receptor Gene | LR-pair factor value (%) | P-value | |
|---|---|---|---|---|
| 1 | Pf4 | Mchr1 | 0.499 (0.392%) | 0.036 |
| 2 | Ccl12 | Ackr1 | 0.481 (0.378%) | 0.009 |
| 3 | Fgf17 | Egfr | 0.479 (0.377%) | 0.005 |
| 4 | Amh | Bmpr1a | 0.452 (0.355%) | 0.013 |
| 5 | Fgf9 | Pdgfra | 0.445 (0.349%) | 0.013 |
| 6 | Il10 | Pdgfrb | 0.424 (0.333%) | 0.019 |
| 7 | Gas6 | Axl | 0.369 (0.29%) | 0.029 |
| 8 | Fgf17 | Fgfr1 | 0.368 (0.289%) | 0.001 |
| 9 | Amh | Acvr1 | 0.367 (0.288%) | 0.003 |
| 10 | Cxcl2 | Mchr1 | 0.367 (0.288%) | 0.005 |
| 11 | Cp | Mxra8 | 0.358 (0.281%) | 0.043 |
| 12 | Pf4 | Ackr3 | 0.342 (0.269%) | 0.034 |
| 13 | Ccl6 | Gnrhr | 0.316 (0.249%) | 0.039 |
| 14 | Ccl6 | Mchr1 | 0.316 (0.249%) | 0.025 |
| 15 | Ccl6 | Oxtr | 0.316 (0.249%) | 0.028 |
| 16 | Fgf9 | Flt4 | 0.316 (0.248%) | 0.006 |
| 17 | Fcgr2b | Ly6c1 | 0.292 (0.23%) | 0.045 |
| 18 | Tcn2 | Cnr1 | 0.291 (0.229%) | 0.04 |
| 19 | Npff | Oxtr | 0.289 (0.227%) | 0.048 |
| 20 | Npff | Ptgfr | 0.289 (0.227%) | 0.046 |
| 21 | Hbegf | Egfr | 0.288 (0.226%) | 0.027 |
| 22 | Cxcl2 | Agtr2 | 0.286 (0.225%) | 0.026 |
| 23 | Trf | Gpc3 | 0.286 (0.224%) | 0.014 |
| 24 | Ccl4 | Cnr1 | 0.283 (0.222%) | 0.017 |
| 25 | Ccl9 | Cnr1 | 0.28 (0.22%) | 0.016 |
| 26 | Ccl9 | Gnrhr | 0.28 (0.22%) | 0.018 |
| 27 | Ccl9 | Oxtr | 0.28 (0.22%) | 0.045 |
| 28 | Ccl9 | Ptgfr | 0.28 (0.22%) | 0.024 |
| 29 | Ccl9 | Mchr1 | 0.28 (0.22%) | 0.022 |
| 30 | Pf4 | Lpar1 | 0.274 (0.215%) | 0.006 |
| 31 | Fgf17 | Flt4 | 0.273 (0.214%) | 0.017 |
| 32 | Fgf17 | Fgfrl1 | 0.255 (0.2%) | 0.032 |
| 33 | Igf1 | Pdgfrb | 0.254 (0.2%) | 0.03 |
| 34 | Cxcl2 | Ackr3 | 0.252 (0.198%) | 0.029 |
| 35 | Lama3 | Enpp3 | 0.25 (0.196%) | 0.015 |
| 36 | Lifr | Cntfr | 0.247 (0.194%) | 0.019 |
| 37 | Ccl6 | Agtr2 | 0.247 (0.194%) | 0.028 |
| 38 | Cxcl2 | Gng7 | 0.244 (0.192%) | 0.017 |
| 39 | Trf | Fzd4 | 0.242 (0.19%) | 0.025 |
| 40 | Trf | Egfr | 0.242 (0.19%) | 0.018 |
| 41 | Pf4 | Tas1r1 | 0.242 (0.19%) | 0.025 |
| 42 | Pf4 | Ptger3 | 0.241 (0.19%) | 0.004 |
| 43 | Apoe | Lrp1 | 0.239 (0.188%) | 0.034 |
| 44 | Ccl9 | Lpar4 | 0.234 (0.184%) | 0.001 |
| 45 | Npff | Agtr1a | 0.233 (0.183%) | 0.013 |
| 46 | Bmp2 | Ihh | 0.228 (0.179%) | 0.033 |
| 47 | Cxcl2 | Gng4 | 0.223 (0.175%) | 0.017 |
| 48 | Ccl9 | Agtr2 | 0.218 (0.172%) | 0.032 |
| 49 | Pdgfc | Pdgfra | 0.212 (0.167%) | 0.009 |
| 50 | Tnfsf13 | Tnfrsf11b | 0.211 (0.165%) | 0.011 |
| 51 | Fcgr2b | Cd55 | 0.21 (0.165%) | 0.041 |
| 52 | Ccl2 | Ackr2 | 0.204 (0.16%) | 0.046 |
| 53 | Cxcl2 | Gng13 | 0.203 (0.16%) | 0.025 |
| 54 | Cxcl2 | Lpar1 | 0.202 (0.158%) | 0.017 |
| 55 | Ccl4 | Ackr3 | 0.194 (0.153%) | 0.002 |
| 56 | Bmp2 | Smurf1 | 0.194 (0.152%) | 0.021 |
| 57 | Vwf | Tek | 0.194 (0.152%) | 0.042 |
| 58 | Ccl6 | Gng4 | 0.192 (0.151%) | 0.006 |
LR-pair factor value > 0.15%, and P-value < 0.05.
4. Discussion
Activation of stem cells upon tissue injury followed by the cell fate specification is tightly coordinated in the tissue repair process, with multicellular signaling networks playing important roles at the microenvironmental level. In this study, we investigated the behavior of skeletal progenitor cells upon bone injury, by performing a lineage tracing analysis and single cell transcriptome analysis in a mouse bone repair model. We first confirmed that Sox9-positive skeletal progenitors contributed to the calvaria bone repair process. The Sox9-positive progenitors were activated upon bone injury and contributed to bone repair by differentiating into osteoblasts. Second, scRNA-seq analysis revealed a heterogeneous cell population at the bone defect sites. Computational analysis predicted a bifurcation of lineages from skeletal progenitors toward an osteogenic lineage and an adipogenic lineage. Third, Ccl9 was identified as an important signaling molecule regulating bone regeneration in the mouse model, possibly through the regulating adipogenic differentiation at the bone defect site. The Ccl9 ligand was secreted from macrophages and acted on skeletal cells. Collectively, these findings suggest that Ccl9 signaling could be a therapeutic target in bone regeneration.
This study extends our understanding of the behavior of Sox9-positive progenitors in bone tissue. In addition to the endochondral bones, Sox9 was present in the skeletal progenitors in the calvaria bones, which were generated through intramembranous ossification. Little Sox9 expression was detected in the periosteum before bone injury, whereas the Sox9-positive cells were activated upon injury. However, the origin of the Sox9-positive cells contributing to bone repair at the bone defect sites remains to be clarified. There are three possibilities for their origin as follows: 1) Sox9-positive progenitors emerged de novo upon injury; 2) Sox9 cells that existed in the periosteum proliferated at the site; 3) Sox9 cells migrated from the suture to the bone defect sites. We think 1) is the most probable. This is because tdTomato-positive cells were specifically enriched at the bone defect sites without changing the tdTomato expression in other bone areas at day 3 after bone injury. Over the period of 10 days, Sox9 cells existing in the suture probably migrated to contribute to bone repair after the activation of Sox9 at the defect site. To confirm this, further time-course experiments and testing with different timings of tamoxifen injection will be required. Lineage tracing analysis also showed that not only tdTomato-positive Sox9 lineage, but also tdTomato-negative cells expressed Runx2 or Sp7 in the bone defect regions. To clarify the contribution of the Sox9 lineage to bone repair, further analysis with a mouse line carrying a Cre-inducible diphtheria toxin A will be required in the future. This line enables us to selectively lose the Sox9 lineage when we use the mouse line with Sox9-CreERT2 line.
Our study highlighted the bidirectional differentiation of skeletal progenitors into osteoblast and adipogenic lineages during bone repair. This bidirectional differentiation might be similar to that in bone homeostasis in bone marrow. In bone marrow, osteoblasts and adipocytes are known to be derived from common mesenchymal stem cells (MSCs), and the differentiation of the two lineages is under the modulation of several transcription factors [48]. A previous study also indicated that subcutaneous preadipocytes had the capacity to differentiate into osteoblasts and that osteo-adipocytic transdifferentiation was involved in lipid metabolism [49]. Other research indicated that transdifferentiation of adipogenic-differentiated cells of bone marrow MSCs into osteogenic- or chondrogenic-differentiated cells proceeds via dedifferentiation and correlates with cell cycle arrest genes and deriving genes [50]. These findings suggested that Sox9-positive skeletal progenitors in the calvaria may have similar plasticity to MSCs in the direction of differentiation. However, one limitation in our study was that there was little or no expression of mature adipocyte markers in the scRNA-seq analysis. It remains to be clarified whether the adipogenic cells have the potential to fully differentiate into mature adipocytes, and if so, what the biological functions of adipocytes during bone repair are. Given that recent studies with scRNA-seq analysis revealed heterogeneous cell populations in skeletal tissues in both mice and humans [40,[51], [52], [53], [54], [55]], further integrative analysis will provide better understanding of the identified cell populations in terms of the similarity and specificity of the mesenchymal cells in the calvaria and bone marrow.
This study provides insight into microenvironmental factors that may regulate skeletal cell specification. A signaling interaction between the Ccl9 that was secreted from macrophages and its receptor Lpar4 that was expressed in adipogenic cells highlights the significance of the signaling interaction at the bone defect sites. We hypothesize that the inhibitory action of Ccl9 signaling on the adipogenic cells may enhance osteoblast differentiation; however, the detailed molecular mechanism underlying the bone regeneration has not been revealed yet. Further lineage tracing analysis focusing on the identified adipogenic cells treated with the Ccl9 antibody will be needed to investigate the mechanism in an in vivo setting. In addition, given that the Ccl9 signaling is known to enhance osteoclastogenesis [56,57], investigation of the effects of the Ccl9 antibody on bone remodeling will be required in the future. Further, other chemokines including Ccl2, Ccl4, Ccl7, and Cxcl12 are expressed in macrophages and monocytes [58], suggesting that other complicated signaling interactions may exist during localized bone regeneration.
5. Conclusions
This study revealed Ccl9 as a therapeutic target for bone regeneration. Unraveling the entire networks of the bone repair process and manipulations that optimize the osteoimmunological signaling may result in better outcomes in bone regeneration. Specific cell types and the appropriate timing of the manipulations may improve clinical outcomes of difficult non-union surgery sites in the future.
Acknowledgements
The authors would like to acknowledge Drs. Shoko Onodera and Toshifumi Azuma at Tokyo Dental College, Drs. Shoichiro Tani and Yoshiaki Kitaura at the University of Tokyo for providing technical supports and insightful advice; Kunihiko Kaneko at Fukuyama University for technical support in scRNA-seq analysis; and Asuka Uchida and Nozomi Nagumo for providing technical assistance. This study was supported by Grants-in-Aid for Science Research from the Japan Society for the Promotion of Science (JSPS) (nos. 20H03885, 21K19589, 21H04952, 21K19883) and a Research Grant for Public Health Science. Super-computing resources were provided by the Human Genome Center at the University of Tokyo. This work utilized the core research facility of the Center for Disease Biology and Integrative Medicine at the Graduate School of Medicine, The University of Tokyo.
Footnotes
Peer review under responsibility of the Japanese Society for Regenerative Medicine.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.reth.2022.05.001.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Ma L., Oei L., Jiang L., Estrada K., Chen H., Wang Z., et al. Association between bone mineral density and type 2 diabetes mellitus: a meta-analysis of observational studies. Eur J Epidemiol. 2012;27(5):319–332. doi: 10.1007/s10654-012-9674-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ward K.D., Klesges R.C. A meta-analysis of the effects of cigarette smoking on bone mineral density. Calcif Tissue Int. 2001;68(5):259–270. doi: 10.1007/bf02390832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saito M., Kida Y., Kato S., Marumo K. Diabetes, collagen, and bone quality. Curr Osteoporos Rep. 2014;12(2):181–188. doi: 10.1007/s11914-014-0202-7. [DOI] [PubMed] [Google Scholar]
- 4.Hagino H., Sakamoto K., Harada A., Nakamura T., Mutoh Y., Mori S., et al. Nationwide one-decade survey of hip fractures in Japan. J Orthop Sci. 2010;15(6):737–745. doi: 10.1007/s00776-010-1543-4. [DOI] [PubMed] [Google Scholar]
- 5.Slobogean G.P., Sprague S.A., Scott T., Bhandari M. Complications following young femoral neck fractures. Injury. 2015;46(3):484–491. doi: 10.1016/j.injury.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 6.Andrzejowski P., Giannoudis P.V. The 'diamond concept' for long bone non-union management. J Orthop Traumatol. 2019;20(1):21. doi: 10.1186/s10195-019-0528-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Atkins R.M., Madhavan P., Sudhakar J., Whitwell D. Ipsilateral vascularised fibular transport for massive defects of the tibia. J Bone Joint Surg Br. 1999;81(6):1035–1040. doi: 10.1302/0301-620x.81b6.10001. [DOI] [PubMed] [Google Scholar]
- 8.Robert Rozbruch S., Weitzman A.M., Tracey Watson J., Freudigman P., Katz H.V., Ilizarov S. Simultaneous treatment of tibial bone and soft-tissue defects with the Ilizarov method. J Orthop Trauma. 2006;20(3):197–205. doi: 10.1097/00005131-200603000-00006. [DOI] [PubMed] [Google Scholar]
- 9.Matsushita T., Watanabe Y. Chipping and lengthening technique for delayed unions and nonunions with shortening or bone loss. J Orthop Trauma. 2007;21(6):404–406. doi: 10.1097/BOT.0b013e318041f6d1. [DOI] [PubMed] [Google Scholar]
- 10.Masquelet A.C. Induced membrane technique: pearls and pitfalls. J Orthop Trauma. 2017;31(Suppl 5):S36–s38. doi: 10.1097/BOT.0000000000000979. [DOI] [PubMed] [Google Scholar]
- 11.Giladi A.M., Rinkinen J.R., Higgins J.P., Iorio M.L. Donor-site morbidity of vascularized bone flaps from the distal femur: a systematic review. Plast Reconstr Surg. 2018;142(3):363e–372e. doi: 10.1097/PRS.0000000000004691. [DOI] [PubMed] [Google Scholar]
- 12.Ono T., Takayanagi H. Osteoimmunology in bone fracture healing. Curr Osteoporos Rep. 2017;15(4):367–375. doi: 10.1007/s11914-017-0381-0. [DOI] [PubMed] [Google Scholar]
- 13.Deschaseaux F., Sensebe L., Heymann D. Mechanisms of bone repair and regeneration. Trends Mol Med. 2009;15(9):417–429. doi: 10.1016/j.molmed.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 14.Liu H., Li D., Zhang Y., Li M. Inflammation, mesenchymal stem cells and bone regeneration. Histochem Cell Biol. 2018;149(4):393–404. doi: 10.1007/s00418-018-1643-3. [DOI] [PubMed] [Google Scholar]
- 15.Tsukasaki M., Takayanagi H. Osteoimmunology: evolving concepts in bone-immune interactions in health and disease. Nat Rev Immunol. 2019;19(10):626–642. doi: 10.1038/s41577-019-0178-8. [DOI] [PubMed] [Google Scholar]
- 16.Matsushita Y., Ono W., Ono N. Skeletal stem cells for bone development and repair: diversity matters. Curr Osteoporos Rep. 2020;18(3):189–198. doi: 10.1007/s11914-020-00572-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuwahara S.T., Serowoky M.A., Vakhshori V., Tripuraneni N., Hegde N.V., Lieberman J.R., et al. Sox9+ messenger cells orchestrate large-scale skeletal regeneration in the mammalian rib. Elife. 2019;8 doi: 10.7554/eLife.40715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He X., Bougioukli S., Ortega B., Arevalo E., Lieberman J.R., McMahon A.P. Sox9 positive periosteal cells in fracture repair of the adult mammalian long bone. Bone. 2017;103:12–19. doi: 10.1016/j.bone.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murao H., Yamamoto K., Matsuda S., Akiyama H. Periosteal cells are a major source of soft callus in bone fracture. J Bone Miner Metab. 2013;31(4):390–398. doi: 10.1007/s00774-013-0429-x. [DOI] [PubMed] [Google Scholar]
- 20.Soeda T., Deng J.M., de Crombrugghe B., Behringer R.R., Nakamura T., Akiyama H. Sox9-expressing precursors are the cellular origin of the cruciate ligament of the knee joint and the limb tendons. Genesis. 2010;48(11):635–644. doi: 10.1002/dvg.20667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13(1):133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomes P.S., Fernandes M.H. Rodent models in bone-related research: the relevance of calvarial defects in the assessment of bone regeneration strategies. Lab Anim. 2011;45(1):14–24. doi: 10.1258/la.2010.010085. [DOI] [PubMed] [Google Scholar]
- 23.Team R.C.,R. R Foundation for Statistical Computing; 2021. A language and environment for statistical computing. [Google Scholar]
- 24.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902 e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rotta R.a.N., Noack A. Multilevel local search algorithms for modularity clustering. J Exp Algorithmics. 2011;16 [Google Scholar]
- 26.Zappia L., Oshlack A. Clustering trees: a visualization for evaluating clusterings at multiple resolutions. GigaScience. 2018;7(7) doi: 10.1093/gigascience/giy083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shao X., Liao J., Lu X., Xue R., Ai N., Fan X. scCATCH: automatic annotation on cell types of clusters from single-cell RNA sequencing data. iScience. 2020;23(3):100882. doi: 10.1016/j.isci.2020.100882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32(4):381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.La Manno G., Soldatov R., Zeisel A., Braun E., Hochgerner H., Petukhov V., et al. RNA velocity of single cells. Nature. 2018;560(7719):494–498. doi: 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aibar S., Gonzalez-Blas C.B., Moerman T., Huynh-Thu V.A., Imrichova H., Hulselmans G., et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14(11):1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.cisTarget databases. 03 Feb 2021. https://resources.aertslab.org/cistarget/ Available from:
- 32.Tsuyuzaki K., N.K. 2021. scTensor: detection of cell-cell interaction from single-cell RNA-seq dataset by tensor decomposition. R package version 2.4.0. [Google Scholar]
- 33.Koki Tsuyuzaki, K.N. scTensor. Available from: https://www.bioconductor.org/packages/release/bioc/html/scTensor.html.
- 34.Aalami O.O., Nacamuli R.P., Lenton K.A., Cowan C.M., Fang T.D., Fong K.D., et al. Applications of a mouse model of calvarial healing: differences in regenerative abilities of juveniles and adults. Plast Reconstr Surg. 2004;114(3):713–720. doi: 10.1097/01.prs.0000131016.12754.30. [DOI] [PubMed] [Google Scholar]
- 35.Farahani R.M., Xaymardan M. Platelet-derived growth factor receptor alpha as a marker of mesenchymal stem cells in development and stem cell Biology. Stem Cells Int. 2015:362753. doi: 10.1155/2015/362753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilk K., Yeh S.A., Mortensen L.J., Ghaffarigarakani S., Lombardo C.M., Bassir S.H., et al. Postnatal calvarial skeletal stem cells expressing PRX1 reside exclusively in the calvarial sutures and are required for bone regeneration. Stem Cell Rep. 2017;8(4):933–946. doi: 10.1016/j.stemcr.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moseti D., Regassa A., Kim W.K. Molecular regulation of adipogenesis and potential anti-adipogenic bioactive molecules. Int J Mol Sci. 2016;17(1):124. doi: 10.3390/ijms17010124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajakumari S., Wu J., Ishibashi J., Lim H.W., Giang A.H., Won K.J., et al. EBF2 determines and maintains brown adipocyte identity. Cell Metab. 2013;17(4):562–574. doi: 10.1016/j.cmet.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang W., Kissig M., Rajakumari S., Huang L., Lim H.W., Won K.J., et al. Ebf2 is a selective marker of brown and beige adipogenic precursor cells. Proc Natl Acad Sci U S A. 2014;111(40):14466–14471. doi: 10.1073/pnas.1412685111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwalie P.C., Dong H., Zachara M., Russeil J., Alpern D., Akchiche N., et al. A stromal cell population that inhibits adipogenesis in mammalian fat depots. Nature. 2018;559(7712):103–108. doi: 10.1038/s41586-018-0226-8. [DOI] [PubMed] [Google Scholar]
- 41.Ghaben A.L., Scherer P.E. Adipogenesis and metabolic health. Nat Rev Mol Cell Biol. 2019;20(4):242–258. doi: 10.1038/s41580-018-0093-z. [DOI] [PubMed] [Google Scholar]
- 42.Mitterberger M.C., Lechner S., Mattesich M., Kaiser A., Probst D., Wenger N., et al. DLK1(PREF1) is a negative regulator of adipogenesis in CD105(+)/CD90(+)/CD34(+)/CD31(-)/FABP4(-) adipose-derived stromal cells from subcutaneous abdominal fat pats of adult women. Stem Cell Res. 2012;9(1):35–48. doi: 10.1016/j.scr.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 43.Ambele M.A., Dhanraj P., Giles R., Pepper M.S. Adipogenesis: a complex interplay of multiple molecular determinants and pathways. Int J Mol Sci. 2020;21(12):4283. doi: 10.3390/ijms21124283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsuyuzaki K., Ishii M., Nikaido I. bioRxiv; 2019. Uncovering hypergraphs of cell-cell interaction from single cell RNA-sequencing data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim C.-S., Kawada T., Yoo H., Kwon B.-S., Yu R. Macrophage inflammatory protein-related protein-2, a novel CC chemokine, can regulate preadipocyte migration and adipocyte differentiation. FEBS (Fed Eur Biochem Soc) Lett. 2003;548(1–3):125–130. doi: 10.1016/s0014-5793(03)00728-2. [DOI] [PubMed] [Google Scholar]
- 46.Xie Y., Wang X., Wu X., Tian L., Zhou J., Li X., et al. Lysophosphatidic acid receptor 4 regulates osteogenic and adipogenic differentiation of progenitor cells via inactivation of RhoA/ROCK1/beta-catenin signaling. Stem Cells. 2020;38(3):451–463. doi: 10.1002/stem.3128. [DOI] [PubMed] [Google Scholar]
- 47.Administration, U.S.F.a.D. Summary of safety and effectiveness data for infuse Bone Graft/LT-Cage™ Lumbar Tapered Fusion Device (PMA Number P000058) 2021. https://www.accessdata.fda.gov/cdrh_docs/pdf/p000058b.pdf Available from: [Google Scholar]
- 48.Takahashi T. Overexpression of Runx2 and MKP-1 stimulates transdifferentiation of 3T3-L1 preadipocytes into bone-forming osteoblasts in vitro. Calcif Tissue Int. 2011;88(4):336–347. doi: 10.1007/s00223-011-9461-9. [DOI] [PubMed] [Google Scholar]
- 49.Schilling T., Küffner R., Klein-Hitpass L., Zimmer R., Jakob F., Schütze N. Microarray analyses of transdifferentiated mesenchymal stem cells. J Cell Biochem. 2008;103(2):413–433. doi: 10.1002/jcb.21415. [DOI] [PubMed] [Google Scholar]
- 50.Almalki S.G., Agrawal D.K. Key transcription factors in the differentiation of mesenchymal stem cells. Differentiation. 2016;92(1–2):41–51. doi: 10.1016/j.diff.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qu R., He K., Fan T., Yang Y., Mai L., Lian Z., et al. Single-cell transcriptomic sequencing analyses of cell heterogeneity during osteogenesis of human adipose-derived mesenchymal stem cells. Stem Cells. 2021;39(11):1478–1488. doi: 10.1002/stem.3442. [DOI] [PubMed] [Google Scholar]
- 52.Holmes G., Gonzalez-Reiche A.S., Lu N., Zhou X., Rivera J., Kriti D., et al. Integrated transcriptome and network analysis reveals spatiotemporal dynamics of calvarial suturogenesis. Cell Rep. 2020;32(1):107871. doi: 10.1016/j.celrep.2020.107871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huber A.K., Patel N., Pagani C.A., Marini S., Padmanabhan K.R., Matera D.L., et al. Immobilization after injury alters extracellular matrix and stem cell fate. J Clin Invest. 2020;130(10):5444–5460. doi: 10.1172/JCI136142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pagani C.A., Huber A.K., Hwang C., Marini S., Padmanabhan K., Livingston N., et al. Novel lineage-tracing system to identify site-specific ectopic bone precursor cells. Stem Cell Rep. 2021;16(3):626–640. doi: 10.1016/j.stemcr.2021.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Farmer D.T., Mlcochova H., Zhou Y., Koelling N., Wang G., Ashley N., et al. The developing mouse coronal suture at single-cell resolution. Nat Commun. 2021;12(1):4797. doi: 10.1038/s41467-021-24917-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lean J.M., Murphy C., Fuller K., Chambers T.J. CCL9/MIP-1gamma and its receptor CCR1 are the major chemokine ligand/receptor species expressed by osteoclasts. J Cell Biochem. 2002;87(4):386–393. doi: 10.1002/jcb.10319. [DOI] [PubMed] [Google Scholar]
- 57.Okamatsu Y., Kim D., Battaglino R., Sasaki H., Spate U., Stashenko P. MIP-1 gamma promotes receptor-activator-of-NF-kappa-B-ligand-induced osteoclast formation and survival. J Immunol. 2004;173(3):2084–2090. doi: 10.4049/jimmunol.173.3.2084. [DOI] [PubMed] [Google Scholar]
- 58.Edderkaoui B. Potential role of chemokines in fracture repair. Front Endocrinol (Lausanne), 2017;8:39. doi: 10.3389/fendo.2017.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.