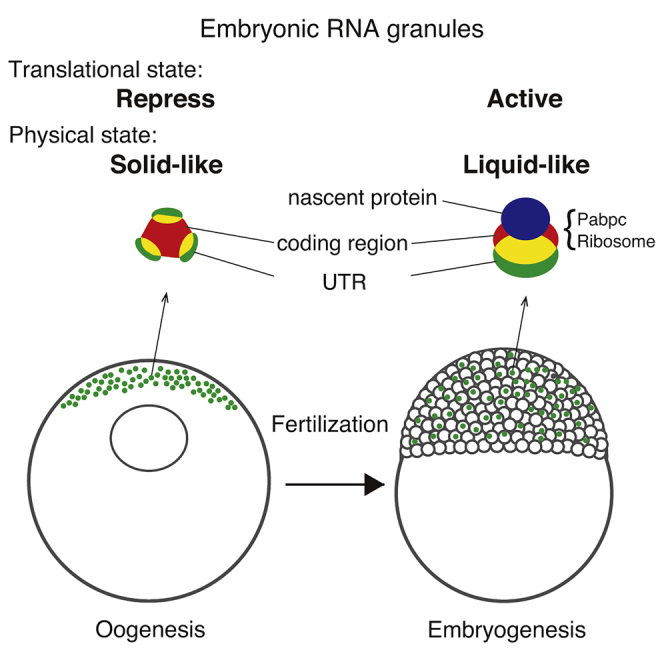
Summary
Fertilized eggs begin to translate mRNAs at appropriate times and placements to control development, but how the translation is regulated remains unclear. Here, we found that pou5f3 mRNA encoding a transcriptional factor essential for development formed granules in a dormant state in zebrafish oocytes. Although the number of pou5f3 granules remained constant, Pou5f3 protein accumulated after fertilization. Intriguingly, signals of newly synthesized peptides and a ribosomal protein became colocalized with pou5f3 granules after fertilization and, moreover, nascent Pou5f3 was shown to be synthesized in the granules. This functional change was accompanied by changes in the state and internal structure of granules. Dissolution of the granules reduced the rate of protein synthesis. Similarly, nanog and sox19b mRNAs in zebrafish and Pou5f1/Oct4 mRNA in mouse assembled into granules. Our results reveal that subcellular compartments, termed embryonic RNA granules, function as activation sites of translation after changing physical properties for directing vertebrate development.
Subject areas: cell biology, organizational aspects of cell biology, functional aspects of cell biology, developmental biology
Graphical abstract
Highlights
-
•
Oocytes accumulate translationally repressed pou5f3 mRNA as granular structures
-
•
These RNA granules exhibit solid-like properties
-
•
After fertilization, pou5f3 RNA granules become to be liquid-like droplets
-
•
Changes in their physical properties promote translation and embryonic development
Cell biology, Organizational aspects of cell biology, Functional aspects of cell biology, Developmental biology
Introduction
Protein syntheses at appropriate times and placements direct various biological processes in almost all organisms including development of animals. Because transcription is silent shortly before fertilization until zygotic genome activation at around the 1000-cell stage in zebrafish and the 2-cell stage in mouse (Hamatani et al., 2004; Harvey et al., 2013; Kane and Kimmel, 1993), timely and spatially controlled protein synthesis after fertilization is achieved by translational activation of the dormant mRNAs stored in eggs. Comprehensive analyses of gene expression demonstrated that zebrafish and mouse eggs accumulate more than eight thousand mRNAs during oogenesis (Aanes et al., 2011; Chen et al., 2011; Harvey et al., 2013). Of these mRNAs, more than two thousand mRNAs are stored as a translationally repressed form (Chen et al., 2011; Luong et al., 2020; Winata and Korzh, 2018). Although when and where distinct mRNAs are translated remain largely unknown, the temporal and spatial control of global translation has been shown to be crucial for the proper promotion of developmental processes including zygotic genome activation, formation of embryonic axes, and cell differentiation (Aoki et al., 2003; Kumari et al., 2013; Sun et al., 2018; Wang and Latham, 1997; Winata and Korzh, 2018; Winata et al., 2018; Zaucker et al., 2020). However, the mechanisms by which translation of the dormant mRNAs is temporally and spatially controlled after fertilization remain largely unresolved.
pou5f3 mRNA encodes the Pou-domain transcription factor Pou5f3 (also known as Pou2/Pou5f1), a homolog of mammalian Oct4/Pou5f1 (Takeda et al., 1994), and was identified as one of the mRNAs extensively translated in polysomes of zebrafish embryos at the cleavage stage (Lee et al., 2013). Identification of pou5f3 mutants in zebrafish that possess the maternally expressed pou5f3 transcript but are deficient in zygotically expressed pou5f3 demonstrated that Pou5f3 is essential for brain morphogenesis (Belting et al., 2001). Moreover, generation and analyses of maternal and zygotic pou5f3 mutants demonstrated that Pou5f3 is essential in early embryogenesis for differentiation of endoderm cells and specification of dorsal-ventral regions (Belting et al., 2011; Lunde et al., 2004; Reim and Brand, 2006; Reim et al., 2004). In addition, Pou5f3 has been shown to trigger zygotic genome activation cooperatively with Nanog and Sox19b, which are also extensively translated in cleavage-stage embryos (Lee et al., 2013; Leichsenring et al., 2013). These studies demonstrated the importance of Pou5f3 synthesis from stored mRNAs after fertilization for directing diverse developmental processes. However, how the translation of pou5f3 mRNA is controlled during embryogenesis remains unknown.
mRNA localization at particular regions within cells has been found in various types of cells including oocytes, neurons, and cultured fibroblasts (Buxbaum et al., 2015; Kloc et al., 2002; Martin and Ephrussi, 2009). Furthermore, studies in which gene expression patterns in Drosophila embryos, larvae, and ovaries were comprehensively analyzed revealed that thousands of mRNAs exhibit subcellular localizations within cells (Jambor et al., 2015; Lecuyer et al., 2007; Wilk et al., 2016). These findings suggest that widespread mRNAs are post-transcriptionally regulated in spatially controlled manners, although the biological significance of most of the mRNA localizations remains unknown. Cytoplasmic RNA granules such as stress granules and processing bodies (P-bodies) are assembled in many types of cells in organisms ranging from yeast to mammals and are thought to function as sites of storage and/or degradation of translationally repressed mRNAs (Buchan and Parker, 2009; Ivanov et al., 2019). Neuronal granules, another type of RNA granules found in neurons, transport mRNAs in a translationally repressed form to dendritic and/or axonal regions (Kiebler and Bassell, 2006; Thomas et al., 2014). We previously showed that zebrafish and mouse oocytes possess cytoplasmic RNA granules consisting of translationally repressed cyclin B1, mos, mad2, or emi2 mRNA (Horie and Kotani, 2016; Kotani et al., 2013, 2017; Takei et al., 2020, 2021). These granules disassembled at different timings after initiation of meiosis, coinciding with the translational activation of assembled mRNA. Collectively, the results of most studies have suggested that cytoplasmic RNA granules function as sites of storage and/or cargo of dormant mRNAs within cells.
In this study, we found that pou5f3 mRNA assembled into cytoplasmic RNA granules in a translationally repressed form in zebrafish oocytes. The granular structure of pou5f3 mRNA persisted after fertilization until, at least, the gastrulation stage, despite an increase in Pou5f3 protein. Staining of translating proteins and a ribosome protein indicated that newly synthesized peptides and the ribosomal protein were not colocalized with pou5f3 RNA granules in fertilized eggs but became colocalized in later stages. Moreover, nascent Pou5f3 polypeptides translated in polysomes were detected within pou5f3 RNA granules. The state and internal structure of granules were found to be changed after fertilization, and their change into liquid droplets was shown to be important for effective translation. These findings provide a model in which RNA granules, termed embryonic RNA granules, function as both repression and activation sites of translation for directing developmental processes.
Results
Zebrafish oocytes store pou5f3 mRNA as RNA granules
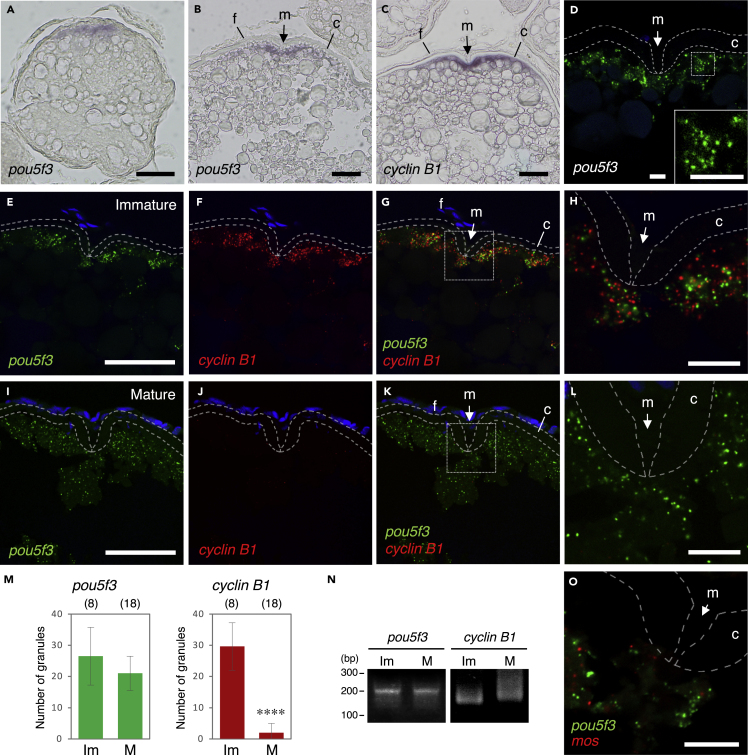
In zebrafish oocytes, pou5f3 mRNA was shown to be localized at the animal polar cytoplasm (Howley and Ho, 2000). We first confirmed the localization of pou5f3 mRNA in growing and fully grown oocytes by in situ hybridization of zebrafish ovaries (Figures 1A and 1B). The micropyle is a structure located at the top of the animal pole, through which a sperm enters into the egg cytoplasm (Figures 1B–1L). The region of localization of pou5f3 mRNA was similar to that of cyclin B1 mRNA (Figure 1C) (Kondo et al., 2001), which encodes Cyclin B1, a regulatory subunit of maturation-promoting factor (MPF). To examine the distribution pattern of pou5f3 mRNA, we performed fluorescence in situ hybridization (FISH) of zebrafish ovaries. pou5f3 mRNA was found to be distributed as granular structures in the animal polar cytoplasm (Figure 1D). No signal was detected with the pou5f3 sense probe (Figures S1A and S1B), confirming the specificity of signals. Interestingly, pou5f3 mRNA and cyclin B1 mRNA appeared to be assembled into different granules in the same region (Figures 1E–1H). The assembly of pou5f3 and cyclin B1 mRNAs into distinct granules in the same region was also observed in growing oocytes (Figures S1C and S1D).
Figure 1.
Zebrafish oocytes store pou5f3 mRNA as RNA granules that are different from those of cyclin B1 mRNA
(A–C) Distributions of pou5f3 mRNA (A and B) and cyclin B1 mRNA (C) in growing (A) and fully grown (B and C) oocytes. m, micropyle; f, follicle cells; c, chorion.
(D) FISH analysis of pou5f3 mRNA (green) in fully grown oocytes. An inset shows an enlarged view of the boxed region. The chorion is outlined by broken lines. DNA is shown in blue.
(E–L) FISH analyses of pou5f3 (green) and cyclin B1 (red) mRNAs in fully grown immature oocytes (E–H) and mature oocytes (I–L). DNA is shown in blue. H and L show enlarged views of the boxed regions in G and K, respectively.
(M) Numbers of pou5f3 (left) and cyclin B1 (right) RNA granules per 100 μm2 in individual immature (Im) and mature (M) oocytes were counted (means ± standard deviations). ∗∗∗∗p < 0.0001 (Student’s t test). The numbers in parentheses indicate the total numbers of oocytes analyzed. Similar results were obtained from three independent experiments.
(N) PAT assay for pou5f3 and cyclin B1 mRNAs in immature (Im) and mature (M) oocytes. Similar results were obtained from three independent experiments.
(O) FISH analysis of pou5f3 (green) and mos (red) mRNAs in fully grown immature oocytes. Bars, (A–C, E–G, and I–K) 50 μm and (D, H, L, and O) 10 μm.
In our previous study, mos and cyclin B1 mRNAs were shown to be assembled into different granules at the animal polar cytoplasm of fully grown oocytes (Horie and Kotani, 2016). pou5f3 RNA granules were also different from mos RNA granules (Figure 1O). In contrast to these mRNAs assembling into granules, α-tubulin and β-actin mRNAs were diffusely distributed in the oocyte cytoplasm (Figures S1E–S1G).
The meiosis of fully grown oocytes is arrested at the prophase of meiosis I. These immature oocytes resume meiosis in response to hormonal stimuli and are arrested again at the metaphase of meiosis II. This process is called oocyte maturation, through which oocytes become matured and acquire fertility. We have shown that translationally repressed mRNAs such as cyclin B1, mos, mad2, and emi2 mRNAs form RNA granules in immature oocytes and that these granules disassemble during oocyte maturation at the times of translational activation (Horie and Kotani, 2016; Kotani et al., 2013; Takei et al., 2020, 2021). Consistent with the results of previous studies, cyclin B1 RNA granules had almost completely disassembled in mature oocytes (Figures 1J and 1M). In contrast, pou5f3 RNA granules remained in mature oocytes (Figures 1I–1M). We then analyzed polyadenylation of pou5f3 mRNA by using a poly(A) test (PAT) assay. Polyadenylation of dormant mRNAs has been shown to direct the translational activation of the mRNAs (Richter and Sonenberg, 2005), and previous studies showed that the poly(A) tails of cyclin B1, mos, mad2, and emi2 are elongated at timings consistent with those of granule disassembly (Horie and Kotani, 2016; Kotani et al., 2013; Takei et al., 2020, 2021). We confirmed that cyclin B1 mRNA was polyadenylated in mature oocytes (Figure 1N). In contrast, the poly(A) tails of pou5f3 mRNA were not elongated (Figure 1N). Taken together, these results demonstrate that pou5f3 RNA granules are assembled and regulated differently from RNA granules consisting of dormant mRNAs that are translated during meiosis.
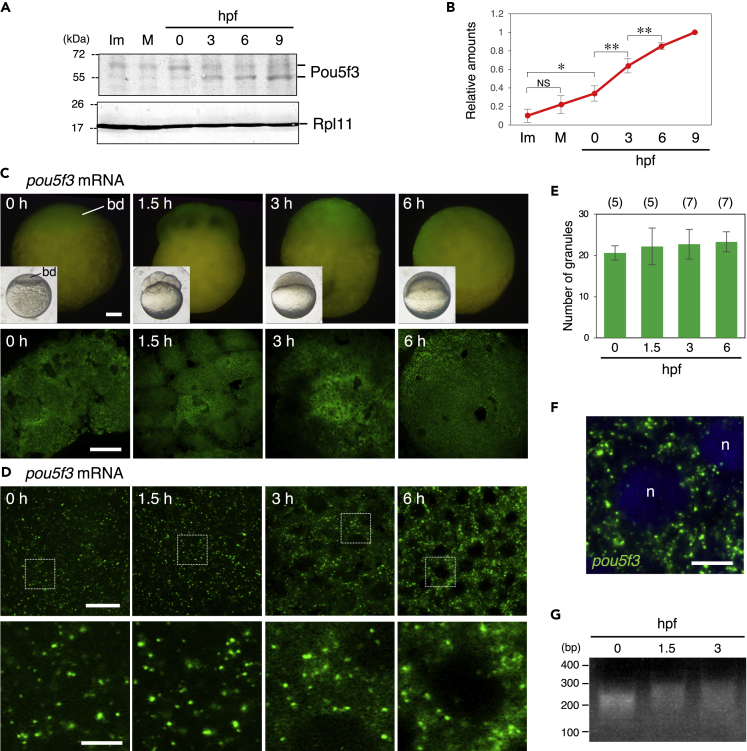
To confirm the synthesis of Pou5f3 protein during zebrafish oogenesis and embryogenesis, we produced antibodies against the N-terminus region of zebrafish Pou5f3 (amino acids 1–122). All three affinity-purified antibodies recognized mainly a protein with a molecular mass of ∼55 kDa and also slightly recognized a protein with a molecular mass of ∼62 kDa in immunoblots of embryos at 3 and 6 h post fertilization (hpf) (Figure S2A, see also Figure 2A). The intensities of both signals were significantly reduced when the antibody was preincubated with recombinant Pou5f3 (Figure S2B) and when the translation of pou5f3 mRNA was inhibited with the antisense MO (Figures S2C and S2D), confirming that the antibodies specifically recognize Pou5f3. The protein with a higher molecular mass may be a phosphorylated form of Pou5f3 as shown in a previous study (Lippok et al., 2014). Time course analysis of oocytes and embryos showed that the amount of Pou5f3 was significantly increased from 3 hpf until 9 hpf (Figures 2A and 2B). No statistical significance was shown between immature and mature oocytes (Figure 2B), although the amount of Pou5f3 was slightly increased between immature and fertilized eggs (0 hpf). Given that poly(A) tails of pou5f3 mRNA were not changed (Figure 1N) and Pou5f3 protein was not accumulated in mature oocytes (Figure 2B), pou5f3 RNA granules observed in immature and mature oocytes exhibit the assembly of mRNAs in a translationally repressed form.
Figure 2.
Zebrafish embryos accumulate Pou5f3 protein during the early stages of development through maintaining the granular structures of pou5f3 mRNA
(A) Time courses of Pou5f3 accumulation in immature (Im) and mature (M) oocytes and in embryos at hours post fertilization (hpf). Ribosomal protein l11 (Rpl11) is a loading control.
(B) The intensities of both Pou5f3 bands were quantified (means ± standard deviations; n = 5). NS, not significant; ∗p < 0.05; ∗∗p < 0.01 (Tukey-Kramer test).
(C and D) Whole mount FISH analyses of pou5f3 mRNA (green) in embryos at 0, 1.5, 3, and 6 h post fertilization.
(C) Upper, lateral views of embryos by fluorescent dissection microscopy. Lower, animal pole views of embryos by confocal microscopy. Insets show bright-field views of live embryos. bd, blastodisc.
(D) Upper, high-resolution confocal images. Lower, enlarged views of the boxed regions in the upper images.
(E) Numbers of granules per 100 μm2 in embryos at 0, 1.5, 3, and 6 hpf were counted (means ± standard deviations). The numbers in parentheses indicate the total numbers of oocytes analyzed. Similar results were obtained from two independent experiments.
(F) Distribution of pou5f3 RNA granules in the cell cytoplasm. DNA is shown in blue. n, nucleus. Bars, (C) 100 μm; (D) upper, 20 μm; lower, 5 μm; (F) 10 μm.
(G) PAT assay for pou5f3 mRNA in embryos at 0, 1.5, and 3 hpf. Similar results were obtained from three independent experiments.
Zebrafish embryos maintain pou5f3 RNA granules during early development
We then analyzed the distribution patterns of pou5f3 mRNA after fertilization by whole mount in situ hybridization. The cortical cytoplasm of zebrafish eggs accumulates at the animal pole, resulting in blastodisc formation and progression of mitotic cleavages in this region (Figure 2C, insets). As reported previously (Takeda et al., 1994), pou5f3 mRNA was ubiquitously detected in the blastodisc and whole cells during early development (Figure 2C). High-resolution images showed that pou5f3 mRNA was present as granular structures in fertilized eggs (0 hpf) (Figure 2D). Intriguingly, the granular structures were maintained in embryos at 1.5, 3, and 6 hpf (Figures 2D and 2E), at which times Pou5f3 protein was significantly accumulated (Figures 2A and 2B). pou5f3 RNA granules were dominantly distributed in the cytoplasm of cells in cleavage-stage embryos (Figure 2F). A PAT assay showed that the poly(A) tails of pou5f3 mRNA were significantly elongated in embryos at 1.5 and 3 hpf (Figure 2G). These results suggest that pou5f3 mRNA starts to be translated after fertilization and that the translational activation is not coupled with apparent granule disassembly.
Signals of newly synthesized peptides become colocalized with pou5f3 RNA granules during early development
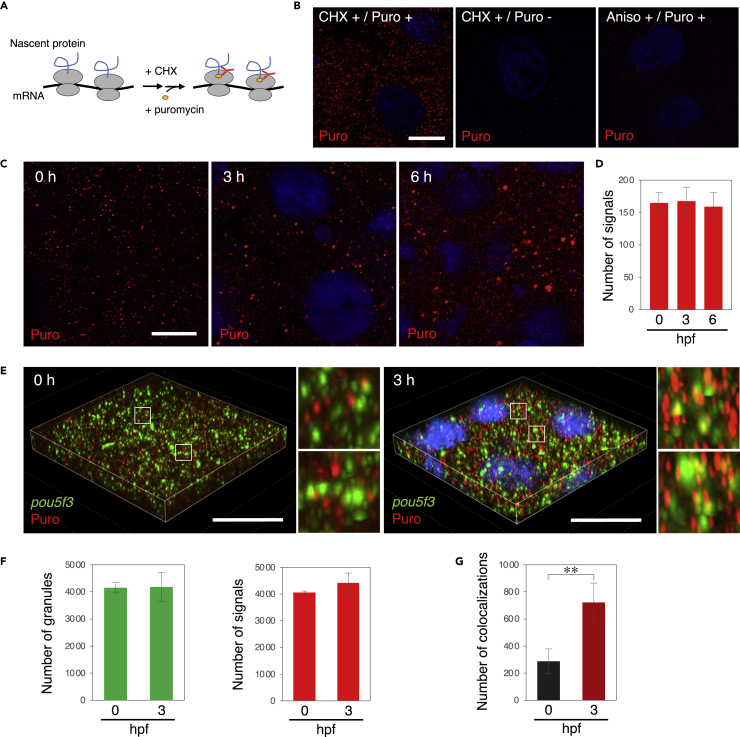
To assess whether the pou5f3 mRNA is translated within granular structures, we used the ribopuromycylation method (David et al., 2012) in embryos. This method is based on properties of the translational inhibitor puromycin, which enters into the ribosome A-site and is subsequently incorporated into the nascent chain C terminus by the peptidyl transferase activity of ribosomes (Figure 3A) (David et al., 2012). Incubation with cycloheximide (CHX) before and during puromycin treatment stabilizes translating ribosomes on mRNAs and detection of puromycin with an anti-puromycin antibody visualizes the newly synthesized peptides (Figure 3A) (David et al., 2012; Moissoglu et al., 2019). This method has been developed using cultured cells but has not been applied to vertebrate embryos.
Figure 3.
Embryos start to translate pou5f3 mRNAs at the sites of granules during the mitotic cleavage stage
(A) A schematic view for visualization of newly synthesized peptides by ribopuromycylation. After pretreatment with CHX, embryos were treated with CHX and puromycin, which was detected by anti-puromycin antibody after washing out free puromycin. Note that this method also enables detection of puromycylated peptides released from polysomes.
(B) Detection of newly synthesized peptides (red) with anti-puromycin antibody in cleavage-stage embryos stimulated with (+) or without (−) CHX and puromycin (Puro). As a control, embryos were pretreated with anisomycin (Aniso) instead of CHX. DNA is shown in blue.
(C) Detection of newly synthesized peptides (red) in embryos at 0, 3, and 6 h post fertilization. DNA is shown in blue.
(D) Numbers of signals of newly synthesized peptides per 3,600 μm2 in embryos at 0, 3, and 6 hpf were counted (means ± standard deviations; n = 3). Similar results were obtained from three independent experiments.
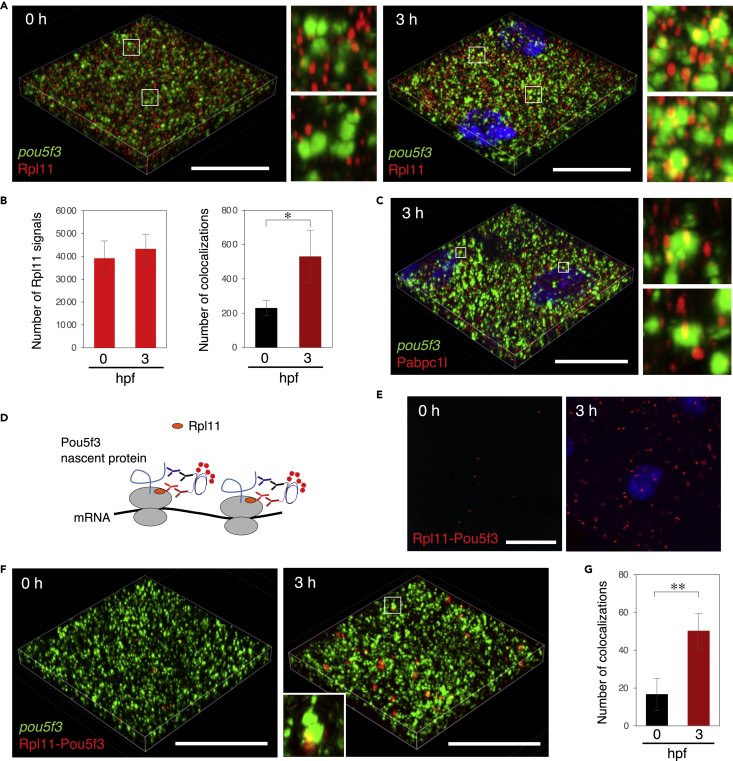
(E) 3D images of SIM for pou5f3 mRNA (green) and newly synthesized peptides (red) in embryos at 0 and 3 h post fertilization. DNA is shown in blue. Insets show enlarged views of the boxed regions. Bars, (B and C) 10 μm, (E) 20 μm.
(F) Numbers of pou5f3 RNA granules (left) and newly synthesized peptides (right) per 28,800 μm3 in embryos at 0 and 3 hpf were counted (means ± standard deviations; n = 3).
(G) Numbers of colocalizations of pou5f3 RNA granules and newly synthesized peptides per 28,800 μm3 in embryos at 0 and 3 hpf were counted (means ± standard deviations; n = 3). Similar results were obtained from two independent experiments. ∗∗p < 0.01 (Student’s t test).
Zebrafish embryos at the cleavage stage were treated with CHX and puromycin after preincubation with CHX and the embryos were then washed with PBS-containing CHX to remove free puromycin. Many spots in the cytoplasm of embryonic cells treated with CHX and puromycin were detected by immunofluorescence with the anti-puromycin antibody, whereas no signal was detected in cells treated with CHX but not puromycin (Figure 3B). In addition, the spots of puromycin were significantly reduced when embryos were treated with puromycin after preincubation with anisomycin, which also stabilizes polysomes but prevents puromycylation by entering into the ribosome A-site and interfering with the peptidyl transferase activity (David et al., 2012), confirming that the signals indeed show the polypeptides that are newly synthesized in ribosomes (Figure 3B). Time course analysis of ribopuromycylation showed that the numbers of signals of newly synthesized peptides were similar in embryos at 0, 3, and 6 hpf (Figures 3C and 3D). Intriguingly, however, the sizes of signals were increased in embryos at 3 and 6 hpf (Figures 3C and S3A). Although these results are basically consistent with the results obtained using Xenopus embryonic extracts, in which the percentage of ribosomes in polysome fractions increased after fertilization (Woodland, 1974), our observations show an increase in the level of translational activity during early development at the subcellular level.
We then examined the relationship between pou5f3 RNA granules and newly synthesized peptides in embryos by super-resolution microscopy (structured illumination microscopy, SIM) with z stack imaging, which allows a better resolution (<120 nm) than that of conventional confocal microscopy. In this experiment, we focused on the differences between 0 and 3 hpf, since Pou5f3 appeared to be significantly accumulated from 3 hpf (Figure 2B). Three-dimensional (3D) images of SIM showed that pou5f3 RNA granules rarely colocalized with newly synthesized peptides in embryos at 0 hpf, whereas they became colocalized with the newly synthesized peptides in embryos at 3 hpf (Figure 3E). Quantitative analyses of 3D images showed that the number of pou5f3 RNA granules and the number of signals of newly synthesized peptides were not changed in embryos at 0 and 3 hpf (Figure 3F), consistent with the observations in two-dimensional images (Figures 2E and 3D). In contrast, the number of pou5f3 RNA granules colocalizing with newly synthesized peptides became increased in embryos at 3hpf (722 ± 143 granules) compared with the number in embryos at 0 hpf (288 ± 91.6 granules) (Figures 3G and S3B). The percentages of colocalizing granules in total granules were 6.9% at 0 hpf and 18.0% at 3 hpf. These results demonstrate that pou5f3 mRNA is translationally dormant in fertilized eggs and becomes selectively translated during the cleavage stage within granules.
pou5f3 mRNA is translated within embryonic granules
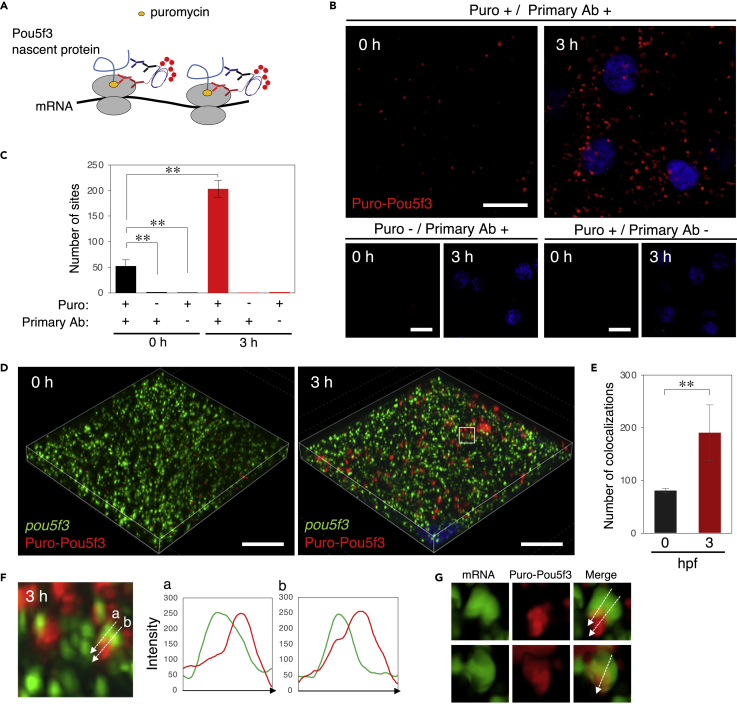
To more directly assess the translational activation of pou5f3 mRNA within granules, we performed puromycylation followed by a proximity ligation assay (Puro-PLA) (tom Dieck et al., 2015) in embryos. Nascent Pou5f3 protein synthesized in polysomes can be visualized by Puro-PLA, since, only when the anti-puromycin and anti-Pou5f3 antibodies are in proximity (<40 nm), linker oligonucleotides fused with secondary antibodies make a circle by a ligase, which is amplified and is finally detected with fluorescently labeled probes (Figure 4A). Antibodies recognizing the N-terminal, but not C-terminal, region of proteins allow detection of translating sites with high sensitivity (tom Dieck et al., 2015).
Figure 4.
Embryos start to translate pou5f3 mRNAs at the sites of granules, termed embryonic RNA granules, during the mitotic cleavage stage
(A) A schematic view for visualization of nascent Pou5f3 protein by Puro-Pou5f3 PLA. Note that this method also enables detection of nascent Pou5f3 peptides released from polysomes.
(B) Detection of Puro-Pou5f3 PLA sites in embryos at 0 and 3 h post fertilization treated with (+) or without (−) puromycin (Puro). After fixation, embryos were incubated with (+) and without (−) anti-puromycin antibody (Primary Ab). DNA is shown in blue.
(C) Numbers of Puro-Pou5f3 PLA sites per 10,000 μm2 in embryos at 0 and 3 hpf were counted (means ± standard deviations; n = 6). Similar results were obtained from two independent experiments. ∗∗p < 0.01 (Tukey-Kramer test).
(D) 3D images of SIM for pou5f3 mRNA (green) and Puro-Pou5f3 PLA sites (red) in embryos at 0 and 3 h post fertilization. DNA is shown in blue.
(E) Numbers of colocalizations of pou5f3 RNA granules and Puro-Pou5f3 PLA sites per 14,400 μm3 in embryos at 0 and 3 hpf were counted (means ± standard deviations; n = 4). Similar results were obtained from two independent experiments. ∗∗p < 0.01 (Student’s t test).
(F) Enlarged views of pou5f3 RNA granules and Puro-Pou5f3 PLA sites in the boxed region in (D). Intensity profiles along the dashed lines are shown in the graphs.
(G) Surface visualization of 3D images of pou5f3 mRNA (green) and Puro-Pou5f3 PLA (red). Merged images are shown (Merge). Bars, (B) 20 μm and (D) 10 μm.
We treated zebrafish embryos with puromycin in a way similar to ribopuromycylation. After fixation, the embryos were incubated with anti-puromycin mouse antibody and anti-Pou5f3 N-terminus rabbit antibody followed by incubation with secondary antibodies. The sites of proximity ligation were labeled with fluorescence probes. Confocal images of Puro-Pou5f3 PLA showed many spots in the cytoplasm of embryonic cells at 3 hpf and a few spots in the blastodiscs of fertilized eggs (Figures 4B and 4C). No spot was detected when the embryos were treated with CHX but not puromycin or when the puromycin-treated embryos were incubated with the secondary antibodies but not with the primary antibodies (Figures 4B and 4C). In addition, reaction with the anti-Pou5f3 antibody followed by detection with a fluorescently labeled secondary antibody showed accumulation of Pou5f3 in the nuclei (Figure S4), which is different from the Puro-Pou5f3 PLA signals (Figure 4B) and consistent with immunostaining of Pou5f3 in the previous study (Lippok et al., 2014). These results confirm that the Puro-Pou5f3 PLA signals indeed show the nascent Pou5f3 peptides. The significant increase in the number of Puro-Pou5f3 PLA sites from 0 to 3 hpf (Figures 4B and 4C) is consistent with the results of immunoblots (Figures 2A and 2B) and supports our findings that pou5f3 mRNA is translationally dormant in oocytes and fertilized eggs and becomes translated in the cleavage stage (Figures 1, 2 and 3).
The Puro-Pou5f3 PLA sites and pou5f3 RNA granules were simultaneously detected by Puro-PLA followed by FISH. Three-dimensional images of SIM showed that only a few pou5f3 RNA granules were overlapped with Puro-Pou5f3 PLA sites in fertilized eggs (Figures 4D and 4E), whereas the number of pou5f3 RNA granules overlapped with Puro-Pou5f3 PLA sites was increased in embryos at 3 hpf (Figures 4D and 4E). Conformation analysis between the pou5f3-RNA granules, which were detected with the full-length RNA probe, and Puro-Pou5f3 PLA sites showed that both signals were largely, but not completely, overlapped and exhibited a direction from RNA granules to Puro-Pou5f3 PLA sites (Figures 4F and 4G).
We further assessed the translational activation of pou5f3 mRNA within granules by staining one of the ribosomal proteins, Rpl11. Rpl11 was distributed as many spots in embryonic cells (Figures S5A and S5B). Simultaneous staining of Rpl11 with pou5f3 mRNA showed that Rpl11 spots were not colocalized with pou5f3 RNA granules in embryos at 0 hpf, whereas Rpl11 spots became colocalized at 3 hpf (Figures 5A and 5B). In addition, a translation factor, Poly(A)-binding protein cytoplasmic 1-like (Pabpc1l), was detected by newly produced antibodies and was found to be distributed as many spots in embryonic cells (Figures S5C and S5D). These Pabpc1l spots were colocalized with pou5f3 RNA granules at 3 hpf (Figure 5C). To detect nascent Pou5f3 peptides synthesized in ribosomes, we performed PLA using anti-Rpl11 rabbit antibody and anti-Pou5f3 mouse antibody (Figure 5D). Rpl11-Pou5f3 PLA detected few spots in embryos at 0 hpf and many spots at 3 hpf (Figure 5E), whereas no spot was detected when the anti-Rpl11 or anti-Pou5f3 antibody was absent (Figures S5E and S5F). Simultaneous detection of Rpl11-Pou5f3 PLA with pou5f3 mRNA demonstrated that nascent Pou5f3 peptides synthesized in ribosomes were found within pou5f3 RNA granules with a direction from RNA granules to Rpl11-Pou5f3 PLA sites (Figure 5F). The number of Rpl11-Pou5f3 PLA sites colocalized with pou5f3 RNA granules was significantly increased in embryos at 3 hpf (Figure 5G).
Figure 5.
Embryos start to translate pou5f3 mRNAs within embryonic RNA granules during the mitotic cleavage stage
(A) 3D images of SIM for pou5f3 mRNA (green) and Rpl11 (red) in embryos at 0 and 3 h post fertilization. DNA is shown in blue. Insets show enlarged views of the boxed regions.
(B) Numbers of Rpl11 spots (left) and numbers of colocalizations of pou5f3 RNA granules and Rpl11 spots (right) per 21,600 μm3 in embryos at 0 and 3 hpf were counted (means ± standard deviations; n = 4). Similar results were obtained from two independent experiments. ∗p < 0.05 (Student’s t test).
(C) 3D images of SIM for pou5f3 mRNA (green) and Pabpc1l (red) in embryos at 3 h post fertilization. DNA is shown in blue. Insets show enlarged views of the boxed regions. Similar results were obtained from two independent experiments.
(D) A schematic view for visualization of pou5f3-translating sites by Rpl11-Pou5f3 PLA. RPL11 is located at the surface of large subunits of ribosomes.
(E) Detection of Rpl11-Pou5f3 PLA in embryos at 0 and 3 h post fertilization. DNA is shown in blue. See Figures S5E and S5F for quantitative analysis.
(F) 3D images of SIM for pou5f3 mRNA (green) and Rpl11-Pou5f3 PLA sites (red) in embryos at 0 and 3 h post fertilization.
(G) Numbers of colocalizations of pou5f3 RNA granules and Rpl11-Pou5f3 PLA sites per 14,400 μm3 in embryos at 0 and 3 hpf were counted (means ± standard deviations; n = 6). Similar results were obtained from two independent experiments. ∗∗p < 0.01 (Student’s t test). Bars, 20 μm.
Altogether, our results demonstrate that pou5f3 mRNA assembles into granular structures that are translationally dormant during oogenesis and in fertilized eggs and are translationally activated through persisting granular structures from the cleavage stage, promoting the progression of development. We termed this type of granule “embryonic RNA granule”.
The state of pou5f3 RNA granules is changed during early development
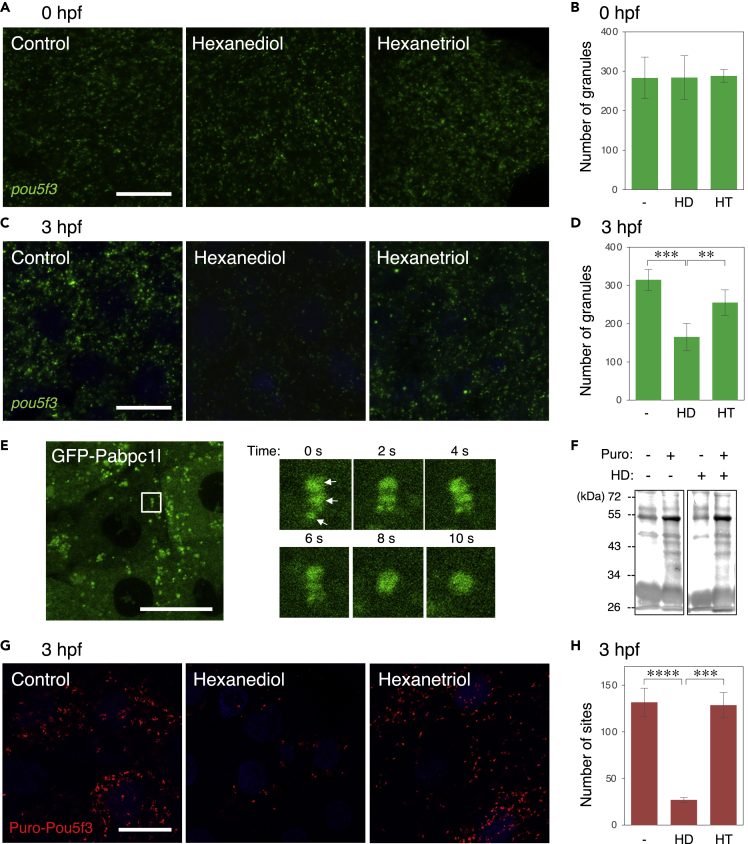
To address the molecular nature of temporal control of mRNA translation in embryonic RNA granules, we assessed whether the phase of granules was changed from 0 to 3 hpf, since the RNA-binding protein Pumilio1 changes its phase from solid-like to liquid-like prior to translational activation of target mRNAs (Takei et al., 2020). Cytoplasmic granules such as P granules in C. elegance embryos and stress granules in many types of cells have been shown to form liquid droplets, which are generated by liquid-liquid phase separation (Shin and Brangwynne, 2017). Treatment of cells with hexanediol dissociates these liquid droplets but does not affect assemblies in a solid-like state (Kroschwald et al., 2015; Updike et al., 2011). We treated zebrafish embryos with hexanediol at 0 and 3 hpf for 20 min and fixed them. As a control, embryos were treated with hexanetriol, a less effective chemical (Updike et al., 2011). FISH analysis showed that neither hexanediol nor hexanetriol affected the assembly of pou5f3 RNA granules in embryos at 0 hpf (Figures 6A and 6B), whereas approximately half of the pou5f3 RNA granules were dissociated by treatment with hexanediol but not by treatment with hexanetriol at 3 hpf (Figures 6C and 6D). The amount of pou5f3 mRNA was not changed by treatment with hexanediol (Figure S6). These results suggest that pou5f3 RNA granules exhibit a solid-like property in fertilized eggs, whereas they become liquid droplets during the mitotic cleavage stage.
Figure 6.
The state of embryonic RNA granules is changed from solid-like to liquid-like during early stages of development
(A–D) FISH analysis of pou5f3 mRNA (green) in embryos at 0 hpf (A) and 3hpf (C) without (Control) and with hexanediol (Hexanediol) or hexanetriol (Hexanetriol). DNA is shown in blue. Numbers of pou5f3 RNA granules per 3,600 μm2 in embryos at 0 hpf (B) and 3 hpf (D) treated without (−) and with hexanediol (HD) or hexanetriol (HT) were counted (means ± standard deviations; n = 4). Similar results were obtained from two independent experiments. ∗∗p < 0.01, ∗∗∗p < 0.001 (Tukey-Kramer test).
(E) Left, distribution of GFP-Pabc1l in embryos at 3 hpf. Right, time-lapse images of the boxed region in the left image showing fusion of three GFP-Pabp1l spots (arrows) into a larger spherical drop.
(F) Immunoblotting analysis of puromycylated peptides in embryos at 3 hpf treated with (+) or without (−) puromycin (Puro) and hexanediol (HD).
(G) Puro-Pou5f3 PLA sites (red) in embryos at 3 hpf treated without (Control) and with hexanediol (Hexanediol) or hexanetriol (Hexanetriol). DNA is shown in blue. Bars, 20 μm.
(H) Numbers of Puro-Pou5f3 PLA sites per 3,600 μm2 in the embryos treated without (−) and with hexanediol (HD) or hexanetriol (HT) were counted (means ± standard deviations; n = 6). Similar results were obtained from two independent experiments. ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 (Tukey-Kramer test).
The property of pou5f3-translating sites was further analyzed by expressing GFP-Pabpc1l in embryos. Similar to endogenous Pabpc1l, GFP-Pabpc1l was distributed in the cytoplasm as many spots (Figure 6E). Time-lapse observation of these spots demonstrated that several spots often fused into one larger spherical spot (Figure 6E), consistent with the behavior of liquid droplets (Shin and Brangwynne, 2017). The results support the notion that pou5f3-translating sites have properties like those of liquid droplets.
To assess whether the assembly of pou5f3 mRNAs into liquid droplets affects the translational efficiency of mRNAs, we examined newly synthesized Pou5f3 by Puro-Pou5f3 PLA. Hexanediol treatment did not globally affect mRNA translation, since puromycylated peptides detected by immunoblotting were equivalent in both control and hexanediol-treated embryos (Figure 6F). Interestingly, the number of Puro-Pou5f3 PLA sites was significantly reduced in embryos treated with hexanediol but not in embryos treated with hexanetriol (Figures 6G and 6H). The percentage of reduction in Puro-Pou5f3 PLA sites (79.5%) was higher than that of reduction in pou5f3 RNA granules (47.5%, Figure 6D), suggesting that pou5f3 mRNA is effectively translated within liquid-state granules. Because treatment with hexanediol for more than 30 min causes reassembly of liquid droplets due to unknown side effects (Kroschwald et al., 2017), we could not examine the changes in the total amount of Pou5f3 in embryos by treatment with hexanediol. However, the results of Puro-Pou5f3 PLA strongly suggest that assembly of liquid droplet RNA granules promotes translational efficiency of assembled mRNAs.
The internal structure of pou5f3 RNA granules is changed during early development
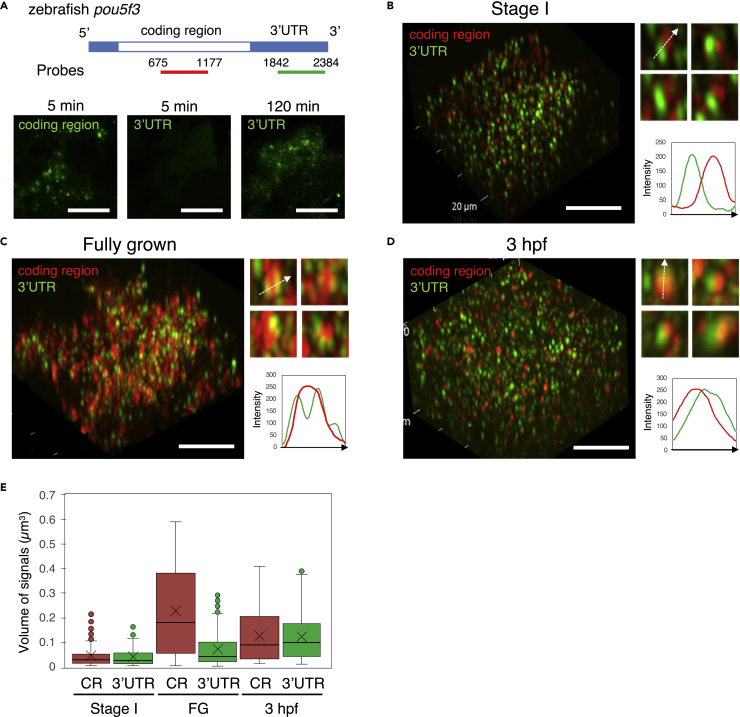
To obtain more insights into the changes in embryonic RNA granules, we next analyzed the internal structure of pou5f3 RNA granules. For this analysis, antisense RNA probes for the coding region and 3′UTR of pou5f3 mRNA in similar lengths (Figure 7A) were synthesized and firstly hybridized with sections of zebrafish ovaries. FISH signals with the antisense RNA probe for the 3′UTR were weak, but the extension of proteinase K treatment increased the intensity of signals (Figure 7A). Three-dimensional images of SIM of double FISH analysis showed that similar sizes of signals for the coding region and 3′UTR were aligned in close proximity in growing oocytes (Figures 7B and 7E). The size of coding region signals became larger in fully grown oocytes and the 3′UTR was concentrated at several sides of the granules (Figures 7C and 7E). These findings suggest that the granules found in growing oocytes are primary units of pou5f3 RNA granules and that these units assemble into larger granules, in which the 3′UTR tails of pou5f3 mRNAs were bundled at the periphery of granules.
Figure 7.
The internal structure of embryonic granules is changed during oogenesis and embryogenesis
(A) Upper, Schematic diagrams of the full length of pou5f3 mRNA and location of antisense RNA probes for the coding region and 3′UTR. Lower, Detection of the coding region and 3′UTR of pou5f3 mRNA (green) with the distinct RNA probes. Before hybridization, ovary sections were treated with proteinase K for 5 min and 120 min.
(B–D) 3D images of SIM for the coding region (red) and 3′UTR (green) in growing (B) and fully grown (C) oocytes and embryos at 3 hpf (D). Insets show representative pou5f3 RNA granules. Intensity profiles along the dashed lines are shown in the graphs. Bars, 10 μm.
(E) Quantitative analysis of the volumes of coding region (CR) and 3′UTR signals in growing oocytes at stage I (Stage I), fully grown oocytes (FG) and embryos at 3 hpf (n ≥ 130). The mean is indicated with an X in a box. Outliers are indicated as circles. Similar results were obtained from two independent experiments.
We then performed whole mount double FISH analysis of embryos in the cleavage stage. In contrast to pou5f3 RNA granules in fully grown oocytes, the sizes of signals of the coding region and 3′UTR were almost the same and both signals were localized at the opposite sites of the granules (Figures 7D and 7E). In addition, the signals of the coding region and 3′UTR were broadly overlapped within granules (Figure 7D). Taken together, these results showed that the internal structure of pou5f3 RNA granules was dynamically changed during the course of oogenesis and embryogenesis.
Assembly of embryonic RNA granules of nanog and sox19b mRNAs in zebrafish and Pou5f1/Oct4 mRNA in mouse
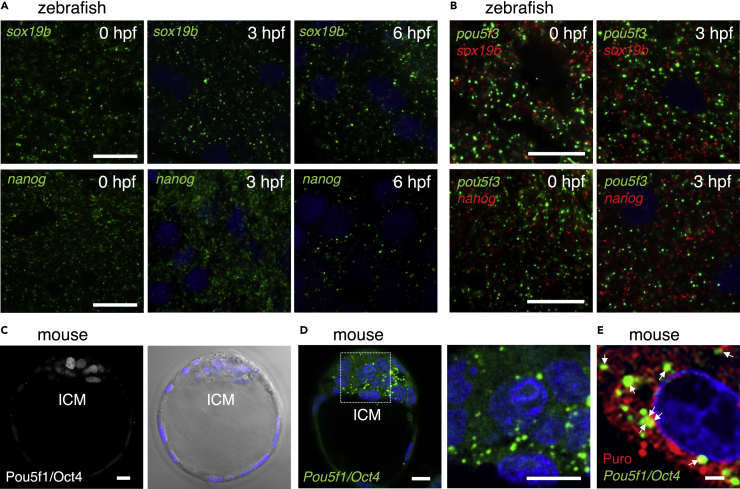
We finally investigated whether translational regulation within embryonic RNA granules is universal or specific to pou5f3 mRNA. In addition to pou5f3 mRNA, sox19b and nanog mRNAs were shown to be extensively translated in polysomes of zebrafish embryos at the cleavage stage (Lee et al., 2013). We found that both sox19b and nanog mRNAs were assembled into granular structures in the cytoplasm of cells at 0, 3, and 6 hpf (Figure 8A). We then investigated whether these mRNAs are translated in the same granules. Double FISH staining showed that pou5f3 and sox19b mRNAs assembled into different granules both in fertilized eggs and embryos at 3 hpf (Figure 8B). Similarly, pou5f3 and nanog mRNAs formed different granules (Figure 8B). These results suggest that the translation of distinct mRNAs is regulated by assembly of different embryonic RNA granules.
Figure 8.
Zebrafish embryos assemble embryonic RNA granules consisting of sox19b or nanog mRNA and mouse embryos assemble those consisting of Pou5f1/Oct4 mRNA
(A) FISH analyses of sox19b (upper) or nanog (lower) mRNA (green) in zebrafish embryos at 0, 3, and 6 hpf. DNA is shown in blue. Similar results were obtained from two independent experiments.
(B) Double FISH analysis of pou5f3 (green) and sox19b (red) (upper) or nanog (red) (lower) mRNAs in zebrafish embryos at 0 and 3 hpf. DNA is shown in blue. Similar results were obtained from two independent experiments.
(C) Immunostaining of Pou5f1/Oct4 (left) in mouse blastocysts. A bright-field image with DNA staining in blue is shown on the right. ICM, inner cell mass.
(D) FISH analysis of Pou5f1/Oct4 mRNA (green) in mouse blastocysts. An enlarged image of the boxed region is shown on the right. DNA is shown in blue. Similar results were obtained from five independent experiments.
(E) Simultaneous detection of Pou5f1/Oct4 mRNA (green) and newly synthesized peptides (red) in mouse blastocysts. DNA is shown in blue. Arrows indicate the regions where both signals overlapped. Bars, (A and B) 20 μm, (C and D) 10 μm, (E) 2 μm.
Mouse Pou5f1/Oct4, a homolog of zebrafish Pou5f3, has been shown to be expressed in blastocysts and to be essential for mammalian embryogenesis (Nichols et al., 1998; Scholer et al., 1989; Takada et al., 2020). We confirmed the expression of Pou5f1/Oct4 in the inner cell mass (ICM) of mouse blastocysts (Figure 8C). FISH analysis showed that Pou5f1/Oct4 mRNA formed granular structures in the cytoplasm of the ICM (Figure 8D). The ribopuromycylation method showed that newly synthesized peptides colocalized with Pou5f1/Oct4 RNA granules (Figure 8E), suggesting the translation of mRNA within the granules. Taken together, our results suggest that translational activation of mRNAs in embryonic RNA granules may be universal in widespread mRNAs and in vertebrates.
Discussion
Translational control of dormant mRNAs during early stages of development
Timings of translational activation and the mechanisms of temporally regulated translation of dormant mRNAs have been extensively analyzed in a period of oocyte maturation using Xenopus oocytes (MacNicol et al., 2015; Mendez and Richter, 2001; Richter, 2007). Compared to the regulation in oocyte maturation, little is known about the timings and mechanisms of translational control of mRNAs after fertilization. Mitotic cell cycles after fertilization were shown to be promoted by temporally controlled translation of cyclin B1 mRNA through timely regulated polyadenylation in Xenopus embryos (Groisman et al., 2000, 2002). A recent study showed the global changes in mRNAs that were incorporated into polysomes during zebrafish embryogenesis and the involvement of polyadenylation in promoting translation of widespread mRNAs (Winata et al., 2018). Our finding that pou5f3 mRNA was polyadenylated after fertilization (Figure 2G) suggests the importance of poly(A) tail elongation in the translational regulation of pou5f3 mRNA. However, our observations also suggest that not all mRNAs carrying long poly(A) tails are translated in embryos at 3 hpf, at which time almost all poly(A) tails of pou5f3 mRNA were elongated (Figure 2G), whereas approximately 80% of pou5f3 RNA granules remained silent (Figures 3E–3G). These results suggest that recruitment of ribosomes to embryonic granules is spatially regulated. It will be interesting to address the relationship between polyadenylation of pou5f3 mRNA and recruitment of ribosomes in temporal and spatial control of mRNA translation.
Translational control within subcellular compartments, embryonic RNA granules
Extensive microscopic studies have suggested that membrane-less compartments such as stress granules, P-bodies, and neuronal granules are involved in translational control of assembled mRNAs. Stress granules are assembled in response to cellular stresses in many types of cells and are thought to be sites of storage of mRNAs (Ivanov et al., 2019). A recent study using live imaging of single mRNA translation demonstrated that mRNAs assembled into stress granules are indeed translationally repressed and that these mRNAs are translated after disassembly of the granules (Moon et al., 2019). In neurons, many translationally repressed mRNAs are packaged into neuronal granules and are transported to their final destinations. These granules disassemble in response to neuronal activity, resulting in translational activation of the mRNAs (Thomas et al., 2014). In Drosophila oocytes, translationally repressed bcd mRNA was shown to be assembled into P-bodies, and the mRNA became translated after disassembly of P-bodies upon fertilization (Weil et al., 2012). In zebrafish and mouse oocytes, translationally repressed mRNAs such as cyclin B1, mos, mad2, and emi2 mRNAs were shown to be assembled into granular structures. These oocyte RNA granules disassembled at the time of translational activation of distinct mRNAs during oocyte maturation (Horie and Kotani, 2016; Kotani et al., 2013; Takei et al., 2020, 2021). Collectively, many types of cytoplasmic RNA granules have been shown to act as repression sites of translation. The assembled mRNAs are translated after dissolution from the granules.
Only a few RNA granules have been demonstrated to act as activation sites of translation. In cultured fibroblast cells, Fus, an RNA-binding protein, directs translation of mRNAs assembled in adenomatous polyposis coli (APC) granules (Yasuda et al., 2013). In yeast, cytoplasmic granules called translation factor mRNA granules actively translate mRNAs that have assembled into granules (Pizzinga et al., 2019). mRNA translation continuously occurs in these granules even while they are transported to their final destination (Moissoglu et al., 2019). Embryonic RNA granules found in this study act as repression sites of translation during oogenesis (Figure 1), whereas they become activation sites during early stages of embryogenesis without disassembling their granular structure (Figures 2, 3, 4, 5, and 6). Thereby, embryonic RNA granules are a novel type of cytoplasmic granules, and our findings provide a novel mode of translational regulation via formation and maintenance of granular structures in temporal and spatial control of mRNA translation.
Recent studies have shown that signals of ribopuromycylation do not indicate the translation sites of mRNAs since puromycylated peptides were rapidly dissociated from polysomes after treatment with puromycin (Enam et al., 2020; Hobson et al., 2020). One possible explanation for the differences in results of those studies and our study is that an in vitro translation system and cultured cells were used in the previous studies, whereas we used zebrafish embryos for this assay. Treatment of rabbit reticulocyte lysate or dish-cultured cells with puromycin rapidly affects the translation of polysomes by spreading quickly throughout the lysate or cytoplasm. In contrast, embryonic cells harbor a large volume of cytoplasm, and thereby it takes a longer time to affect the translation of polysomes after treatment with puromycin. Indeed, a recent study has shown that translation sites of Puro-PLA were overlapped with FISH signals of mRNAs in mouse oocytes, which have a large volume of cytoplasm (Jansova et al., 2021). We have shown that pou5f3 RNA granules were colocalized with the signals of ribopuromycylation and Puro-Pou5f3 PLA (Figures 3 and 4). Moreover, the signals of Rpl11, Pabpc1l, and Rpl11-Pou5f3 PLA were also colocalized with pou5f3 RNA granules (Figure 5). Collectively, these results demonstrate translation of pou5f3 mRNA within RNA granules. Intriguing findings in these experiments were that Rpl11 and Pabpc1l spots were colocalized but not homogenously mixed with pou5f3 RNA granules (Figures 5A and 5C). These observations suggest that Rpl11, Pabpc1l, and pou5f3 mRNA form multiphase droplets as observed in stress granules fused with P-bodies and in nucleoli (Shin and Brangwynne, 2017).
Simultaneous detection of Puro-Pou5f3 PLA sites with pou5f3-RNA granules using the full-length RNA probe showed that both signals exhibited a direction from RNA granules to Puro-Pou5f3 PLA sites (Figures 4F and 4G). These observations suggest the organization of highly ordered structures in the complexes of RNA granules and polysomes, in which coding regions of mRNAs are aligned and translating ribosomes assemble there and synthesize Pou5f3 peptides into the opposite direction. FISH of the coding region and 3′UTR showed the highly ordered internal structure of pou5f3 RNA granules in cleavage-stage embryos (Figure 7D), supporting this notion. Alternatively, the RNA probe was not effectively hybridized with pou5f3 mRNA molecules that are actively translated and thereby surrounded by dense polysomes, because the embryos were not treated with proteinase K for preventing degradation of proteins in this assay to simultaneously detect proteins with mRNA. Future studies such as studies using live imaging of translation are needed to determine the accurate structure of translating sites.
Functional changes in embryonic RNA granules
FISH analysis of the internal structure of pou5f3 RNA granules showed that the coding region is located at the center and the 3′UTR is located at the periphery in fully grown oocytes (Figure 7C). This structure partially resembles that of nuclear granules termed paraspeckles. Within paraspeckles, the 3′ end of a long-noncoding RNA, Neat, is located at the periphery and the central region of Neat is located at the center (West et al., 2016). In the case of pou5f3 RNA granules, location and bundling of the 3′UTR at the periphery seem to be effective in the regulation of mRNA translation, since trans-acting factors tend to bind to the 3′UTR sequences of target mRNAs and modifications in these factors have been shown to direct translational activation. Long-term treatment with proteinase K increased the signals of the 3′UTR (Figure 7A), suggesting the binding of many RNA-binding proteins in this region. An intriguing finding in this analysis is that almost equal sizes of signals of the coding region and the 3′UTR were aligned in close proximity in growing oocytes (Figure 7B). One possible explanation is that this minimum unit shows a single RNA molecule. However, this is unlikely since 1) hexanediol stimulation resulted in disappearance of half of the pou5f3 RNA granules in embryos at 3 hpf despite the fact that the amount of mRNA was not changed (Figures 6C, 6D, and S6), 2) α-tubulin and β-actin mRNAs were diffusely distributed in the oocyte cytoplasm (Figures S1E–S1G), and 3) cyclin B1 RNA granules at the animal polar cytoplasm have been shown to lose granular structures and be dispersed throughout the cytoplasm when the actin filaments were depolymerized (Kotani et al., 2013). Thus, we concluded that our assay is unable to detect single RNA molecules as particles and that the granular signals in growing oocytes indicate assembly of several pou5f3 RNA molecules in a similar structure. Nevertheless, these primary units may assemble into highly ordered structures in fully grown oocytes, also resembling the assembly of paraspeckles (West et al., 2016). These findings suggest the existence of a conserved mechanism by which RNA and binding proteins are assembled into granular structures, although the functional significance of the internal structure remains to be elucidated.
The repression type of embryonic granules exhibited a solid-like property since treatment with hexanediol did not affect the granular structures in embryos at 0 hpf (Figures 6A and 6B). A recent study demonstrated that the solid-like state of RNA-binding proteins promotes translational repression of mRNAs (Shiina, 2019). We previously showed that Pumilio1 in a solid-like state stably represses translation of target mRNAs and that the phase change into a liquid-like state promotes translational activation of them (Takei et al., 2020). Our findings in this study support the notion that RNA granules in a solid-like state effectively repress the translation of assembled mRNAs in the cell cytoplasm. In this state, granular compartments would effectively sequester the assembled mRNAs from translational machinery such as ribosomes. We also showed that pou5f3, cyclin B1, and mos mRNAs formed distinct granules, whereas α-tubulin and β-actin mRNAs were diffusely distributed in the animal polar cytoplasm (Figures 1 and S1). Although the number of mRNAs analyzed by FISH remains small, these observations suggest that translationally repressed mRNAs generally assemble into granular structures to effectively regulate the translational state.
In contrast to oocyte RNA granules such as cyclin B1, mos, mad2, and emi2 RNA granules, embryonic RNA granules did not disassemble and rather the state was changed from solid-like to liquid-like, resulting in granules of liquid droplets (Figure 6). The internal structure of pou5f3 RNA granules was also changed in embryos at 3 hpf, with the coding region and 3′UTR being localized at opposite sites with broad overlapping (Figure 7). The changes in the state and internal structure may be induced by post-translational modifications or changes in trans-acting factors binding to and/or assembling with the mRNAs (Nosella and Forman-Kay, 2021). We also observed changes in the volume of coding region and 3′UTR signals (Figure 7), which may be related to the changes in the state of RNAs and in the number of RNA molecules. These issues need to be addressed in future studies. What then is the role of liquid-state embryonic granules in mRNA regulation? Our results demonstrated that dissolution of liquid-state pou5f3 RNA granules by treatment with hexanediol significantly decreased the rate of translation (Figures 6G and 6H), although we could not rule out a possibility that puromycylated peptides synthesized from diffused mRNAs were not detected in this assay. Nevertheless, one of the functions of liquid-state embryonic granules would be to promote translational efficiency of assembled mRNAs by colocalizing mRNAs, trans-acting factors, and ribosomes in a highly concentrated state. Our time-lapse observations showed that Pabpc1l exhibited properties like a liquid droplet (Figure 6E), whereas immunoblot analysis showed that hexanediol did not affect global translation (Figure 6F). Because other Poly(A)-binding proteins including Pabpc1a were shown to be expressed in zebrafish embryos (Mishima et al., 2012) and only Pabpc1l was isolated as a protein binding to pou5f3 mRNA in our in vitro assay (T.K. unpublished data), global translation would be achieved by other Pabpcs, and Pabpc1l may play a role in the regulation of embryonic RNA granules. Another possible role of embryonic RNA granules is that assembly of mRNAs into liquid droplets allows stable synthesis of the encoded proteins during a long period of development. In this regard, it should be noted that zygotic pou5f3 mutants develop until tail bud stages without obvious defects except a defect in brain morphology (Belting et al., 2001). This implies that maternally deposited pou5f3 mRNA can produce sufficient amounts of Pou5f3 protein to promote diverse developmental processes including progression of gastrulation, specification of endodermal cells, and dorsal-ventral development of embryos, all of which are defective in the maternal and zygotic pou5f3 mutants (Lunde et al., 2004; Reim and Brand, 2006; Reim et al., 2004), until the tail bud stage. Liquid-state pou5f3 RNA granules may function to prevent degradation of maternal mRNAs for long durations to safely promote development. Our findings that certain fractions, but not all, of pou5f3 RNA granules (approximately 20% of total granules) were actively translated in embryos at 3 hpf (Figures 3, 4 and 5) and that the proportion of translated RNA granules increased up to 30% in embryos at 6 hpf (data not shown) support this notion. The remaining granules may be stocks for synthesizing a large amount of Pou5f3 in later stages.
Limitations of the study
Our double FISH analyses suggest that pou5f3, sox19b, and nanog mRNAs form homogeneous RNA granules (Figure 8B). Although these results partially resemble the formation of homogeneous RNA clusters in Drosophila germ granules (Trcek et al., 2015), more comprehensive analyses will be needed to resolve the relationship of these granules. Recent studies using live imaging of single mRNA translation have demonstrated that a small fraction of reporter mRNAs was translated in stress granules, although a large fraction of them was repressed (Moon et al., 2019; Mateju et al., 2020). Because stress granules are large compartments and contain substructures exhibiting a solid- or liquid-like property (Jain et al., 2016; Shiina, 2019), the differential translational state of assembled mRNAs may be related to the differences in the states of substructures. Translation of mRNAs in subcellular compartments was also shown to be coupled with subsequent biological processes such as assembly of co-acting proteins at the same site (Panasenko et al., 2019; Chouaib et al., 2020; Morales-Polanco et al., 2021). It is possible that embryonic RNA granules are regulated in much larger structures and coupled with some biological processes. Future studies are needed to address the regulatory mechanisms including compositions and relationships of the distinct embryonic RNA granules.
Conclusions
We discovered the existence of a novel type of RNA granule, embryonic RNA granule, that may be a universal structure in embryos. Our findings contribute to an understanding of how oocytes and embryos regulate mRNA translation to properly control development and how cells locally and temporally regulate mRNA translation through assembling and changing the properties of membrane-less compartments.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse polyclonal anti-Pou5f3 protein antibody | This paper | N/A |
| Rabbit polyclonal anti-Pou5f3 protein antibody | This paper | N/A |
| Mouse polyclonal anti-PABPC1L protein antibody | This paper | N/A |
| Mouse monoclonal anti-Oct3/4 protein antibody (clone 10) | Santa Cruz Biotechnology | Cat#sc-5279; RRID: AB_628051 |
| Rabbit polyclonal anti-RPL11 protein antibody | Abcam | Cat#ab79352; RRID: AB_2042832 |
| Mouse monoclonal anti-Puromycin antibody (3RH11) | Kerafast | Cat#EQ0001; RRID: AB_2620162 |
| Anti-DIG-alkaline phosphatase antibody | Roche | Cat#11093274910 |
| Anti-DIG-horseradish peroxidase antibody | Roche | Cat#11207733910 |
| Anti-DNP-Alexa 488 antibody | Molecular probes | Cat#A11097; RRID: AB_2314332 |
| Anti-Fluorescein-horseradish peroxidase antibody | Roche | Cat#11426346910 |
| Goat Anti-mouse IgG Alexa 488 antibody | Molecular probes | Cat#A11017 RRID: AB_143160 |
| Goat Anti-mouse IgG Alexa 546 antibody | Molecular probes | Cat#A11003; RRID: AB_141370 |
| Goat Anti-rabbit IgG Alexa 546 antibody | Molecular probes | Cat#A11010; RRID: AB_2534077 |
| Chemicals, peptides, and recombinant proteins | ||
| Trizol reagent | Invitrogen | Cat#15596-018 |
| Tyramide-dinitrophenyl | PerkinElmer | Cat#NEL746A |
| Tyramide-Fluorescein | PerkinElmer | Cat#NEL741 |
| Tyramide-Cy3 | PerkinElmer | Cat#NEL744 |
| Hoechst 33258, Pentahydrate [bis-Benzimide] | Setareh Biotech | Cat#7075 |
| Proteinase K | Sigma | Cat#P2308 |
| Blocking reagent | Roche | Cat#10447200 |
| VECTASHIELD Mounting Medium containing DAPI | Vector Laboratories, Inc | Cat#H1200 |
| T4 RNA ligase | New England Biolabs | Cat#M0204S |
| Puromycin | Invitrogen | Cat#ant-pr-1 |
| 1,6-Hexanediol | Sigma | Cat#240117 |
| 1,2,6-Hexanetriol | Sigma | Cat#T66206 |
| RNasin Plus Rnase Inhibitor | Promega | Cat#N2611 |
| Newborn calf serum | Sigma | Cat#N4762 |
| Critical commercial assays | ||
| Tyramide signal amplification (TSA) Plus DNP system | PerkinElmer | Cat#NEL747A |
| Duolink In situ PLA Probe Anti-Mouse PLUS | Sigma | Cat#DUO92001 |
| Duolink In situ PLA Probe Anti-Rabbit MINUS | Sigma | Cat#DUO92005 |
| Duolink In situ PLA Detection Reagents FarRed | Sigma | Cat#DUO92013 |
| Oligonucleotides | ||
| See Table S1 | See Table S1 | See Table S1 |
| Software and algorithms | ||
| Image J | NIH | imagej.net |
| AIVIA | Leica | aivia-software.com |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Tomoya Kotani (tkotani@sci.hokudai.ac.jp).
Materials availability
Plasmids and antibodies generated in this study are available from the Lead contact with a completed Materials Transfer Agreement, but there are restrictions to the availability of antibodies due to the lack of an external centralized repository for its distribution and our need to maintain the stock.
Experimental model and subject details
Adult zebrafish were maintained at 28°C on a 14 hours light/10 hours dark cycle following standard methods. Embryos were obtained by natural spawning and staged according to timings post fertilization and morphological criteria. Embryos used in this study were less than 9 hpf, and sex cannot be determined at these stages and is unlikely to influence the biological processes under study. All animal experiments in this study were approved by the Committee on Animal Experimentation, Hokkaido University.
Method details
Isolation of ovaries and oocytes
Zebrafish ovaries were dissected from adult females in zebrafish Ringer’s solution (116 mM NaCl, 2.9 mM KCl, 1.8 mM CaCl2, and 5 mM HEPES; pH 7.2). Oocytes were manually isolated from ovaries with forceps under a dissecting microscope. Oocyte maturation was induced by stimulation with 1 μg/mL of 17α, 20β-dihydroxy-4-pregnen-3-one, an MIH in fish. For in situ hybridization analysis, ovaries and embryos were fixed with 4% paraformaldehyde in PBS (4% PFA/PBS) overnight at 4°C. For the poly(A) test (PAT) assay, oocytes and embryos were extracted with Trizol reagent (Invitrogen, 15596-018) and total RNA was used for RNA ligation-coupled RT-PCR. For immnoblotting analysis, oocytes and embryos were homogenized with an equal volume of ice-cold extraction buffer (EB: 100 mM β-glycerophosphate, 20 mM HEPES, 15 mM MgCl2, 5 mM EGTA, 1 mM dithiothreitol, 100 μM (p-amidinophenyl) methanesulfonyl fluoride, and 3 μg/mL leupeptin; pH 7.5). After centrifugation at 15,000 g for 10 min at 4°C, the supernatant was collected and used for SDS-PAGE analysis.
Section in situ hybridization
Section in situ hybridization of zebrafish ovaries with or without the tyramide signal amplification (TSA) Plus DNP system (PerkinElmer, NEL747A) was performed according to the procedure reported previously (Takei et al., 2018). Briefly, fixed ovaries were dehydrated, embedded in paraffin, and cut into 10-μm-thick sections. Digoxigenin (DIG)-labeled antisense RNA probes for the full lengths of pou5f3, cyclin B1, α-tubulin and β-actin were used for detection of pou5f3, cyclin B1, α-tubulin and β-actin gene transcripts, respectively. No signal was detected with sense probes. After hybridization and washing, samples were incubated with anti-DIG-alkaline phosphatase (AP) antibody (Roche, 11093274910) (1:2000 dilution) for 30 min. After washing, the samples were reacted with a mixture of nitro blue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolyl phosphate (BCIP). The samples were then mounted with glycerol and observed under an Axioskop microscope (Carl Zeiss). For FISH with TSA, samples hybridized with RNA probes were incubated with anti-DIG-horseradish peroxidase (HRP) antibody (Roche, 11207733910) (1:500 dilution) for 30 min. The reaction with tyramide-dinitrophenyl (DNP) (PerkinElmer, NEL746A) was performed according to the manufacturer’s instructions. The samples were then incubated overnight with anti-DNP-Alexa 488 antibody (Molecular Probes, A11097) (1:500 dilution). To detect nuclei, the samples were incubated with 10 μg/mL Hoechst 33258 for 10 min. After being mounted with a Prolong Antifade Kit (Molecular probes, P7481), the samples were observed under an LSM 5 LIVE confocal microscope (Carl Zeiss).
Double FISH of pou5f3 and cyclin B1 mRNAs was performed as follows. A fluorescein-labeled antisense RNA probe for cyclin B1 was used for detection of the cyclin B1 gene transcript. Ten-μm-thick sections of ovaries were hybridized with a mixture of pou5f3 and cyclin B1 antisense RNA probes. After detection of the DIG-labeled antisense pou5f3 RNA probe as described above, the samples were incubated with 1% H2O2 in methanol for 30 min for inactivating HRP. After rehydration and washing with PBS, the samples were incubated with anti-Fluorescein-HRP antibody (Roche, 11426346910) (1:200 dillution) for 30 min. The reaction with tyramide-Cy3 (PerkinElmer, NEL744) was performed according to the manufacturer’s instructions. After staining with Hoechst 33258, the samples were mounted and observed under the LSM 5 LIVE confocal microscope. The number of pou5f3 and cyclin B1 RNA granules was quantified using ImageJ software.
Double FISH of the coding region and 3′UTR of pou5f3 was performed as follows. A fluorescein-labeled antisense RNA probe for the coding region and a DIG-labeled antisense RNA probe for the 3′UTR were used for detection of the coding region and 3′UTR of the pou5f3 gene transcript, respectively. Ten-μm-thick sections of ovaries were treated with 1 μg/mL proteinase K (Sigma, P2308) for 2 hours at 37°C before hybridization. The samples were then hybridized with a mixture of coding region and 3′UTR antisense RNA probes. No signal was detected with sense probes. After detection of the DIG- and fluorescein-labeled RNA probe as described above, the samples were mounted and observed under an N-SIM super-resolution microscope (Nikon). The volume of signals was measured by AIVIA software (Leica).
Collection, fixation and section in situ hybridization of mouse embryos were performed according to the procedure reported previously (Takada et al., 2020). The samples were observed under the LSM 5 LIVE confocal microscope.
Whole mount in situ hybridization
Zebrafish embryos at appropriate time points were fixed with 4% PFA/PBS overnight at 4°C. The chorions of fixed embryos were removed with forceps in PBS and the embryos were treated with 100% methanol overnight at −20°C. After rehydration and washing with PBS containing 0.1% Tween 20, the embryos were incubated with hybridization buffer - (HB-) (50% formamide, 5x SSC, 0.1% Tween 20) for 5 min at 55°C. The samples were then prehybridized with HB+ (5 mg/mL torula RNA, 50 μg/mL heparin in HB-) for 1 hour at 55°C and hybridized with the antisense pou5f3 RNA probe in HB+ overnight at 65°C. The embryos were washed once with HB- for 30 min at 65°C, twice with 2x SSCT for 30 min at 65°C, and twice with 0.2x SSCT for 30 min at 65°C. The embryos were then incubated with Maleic acid buffer (150 mM Maleic acid, 100 mM NaCl; pH 7.5) containing 0.1% Tween 20 (MBST) for 5 min and with 2% Blocking reagent (Roche, 10447200) in MBST for 5 hours. The embryos were incubated with anti-DIG-HRP antibody (1:700 dilution) overnight at 4°C. After washing, the embryos were incubated with tyramide-DNP (PerkinElmer, NEL746A) according to the manufacturer’s instructions, followed by incubation with the anti-DNP-Alexa 488 antibody. After being mounted with VECTASHIELD Mounting Medium containing DAPI (Vector Laboratories, Inc, H1200), the samples were observed under the LSM 5 LIVE confocal microscope. The number of pou5f3 granules was quantified using ImageJ software.
Double FISH of pou5f3 and sox19b or nanog mRNAs was performed as follows. After hybridization with the fluorescein-labeled antisense RNA probe for sox19b or nanog and the DIG-labeled antisense RNA probe for pou5f3, the embryos were incubated with anti-Fluorescein-HRP antibody (1:200 dillution) for 3 hours, followed by incubation with tyramide-Cy3 according to the manufacturer’s instructions. The embryos were then incubated with 1% H2O2 in methanol for 30 min for inactivating HRP. After rehydration and washing with PBS, the embryos were incubated with anti-DIG-HRP antibody (1:500 dillution) for 3 hours, followed by incubation with tyramide-Fluorescein (PerkinElmer, NEL741) according to the manufacturer’s instructions. After being mounted, the samples were observed under the LSM 5 LIVE confocal microscope. To perform double FISH of the coding region and 3′UTR of pou5f3, the fixed embryos were treated with 5 μg/mL proteinase K for 5 min at room temperature before hybridization. The embryos were hybridized with the fluorescein-labeled antisense RNA probe for the coding region and a DIG-labeled antisense RNA probe for the 3′UTR, and the signals were detected as described above. After being mounted, the samples were observed under the N-SIM super-resolution microscope. The volume of signals was measured by AIVIA software.
Poly(A) test assay
RNA ligation-coupled RT-PCR was performed according to the procedure reported previously (Charlesworth et al., 2004; Kotani et al., 2013). Two μg of total RNA extracted from pools of 15 zebrafish oocytes or embryos was ligated to 0.4 μg of P1 anchor primer (primer sequences are shown in Table S1) in a 10-μL reaction solution using T4 RNA ligase (New England Biolabs, M0204) for 30 min at 37°C. The ligase was inactivated for 5 min at 92°C. Half of the RNA ligation reaction was used in a 25-μL reverse transcription reaction using the Superscript III First Strand Synthesis System with a P1′ primer. Four μL of the cDNA was used for the 1st PCR with the P1′ primer and a zpou5f3-PAT-f1 primer or a zcyclin B1-PAT-f1 primer for 17 cycles. Two μL of the 1st PCR reaction was used for the 2nd PCR with the P1′ primer and a zpou5f3-PAT-f2 primer or a zcyclin B1-3′UTR-f primer for 35 cycles. The PCR product was resolved on a 2% agarose gel in TAE buffer. We confirmed that the increase in PCR product length was due to elongation of the poly(A) tails by cloning the 2nd PCR products and sequencing them.
Production of antibodies and immunoblotting
The DNA sequence encoding a part of zebrafish Pou5f3 (residues 1-122) was amplified by PCR and ligated into the pET21 vector to produce a histidine (His)-tagged protein. The recombinant protein was expressed in E. coli and purified by SDS-PAGE, followed by electroelution in Tris-glycine buffer without SDS. The purified protein was dialyzed against 1 mM HEPES (pH 7.5), lyophilized, and used for injection into two mice and one rabbit. The obtained antisera were affinity-purified with recombinant Pou5f3-His protein electroblotted onto a membrane (Immobilon; EMD Millipore, IPVH00010). Similarly, a part of Pabpc1l (residues 6-348) was expressed in E. coli, purified, and used for injection into two mice. The obtained antisera were affinity-purified with recombinant Pabpc1l protein electroblotted onto the membrane. The crude extracts from oocytes and embryos at appropriate time points were separated by SDS-PAGE, blotted onto the Immobilon membrane, and probed with the anti-Pou5f3 mouse and rabbit antibodies and with the anti-Pabpc1l mouse antibody. Rpl11 was detected by immunoblotting the crude extracts with anti-Rpl11 rabbit antibody (Abcam; ab79352).
Immunofluorescence
Zebrafish embryos were fixed with 4% PFA/PBS overnight at 4°C. After removing the chorions, the embryos were incubated with blocking buffer (4% skim-milk, 0.1% Tween 20 in TBS) for 1 hour. The embryos were then incubated with affinity-purified anti-Pou5f3 rabbit antibody (1:50 dilution), anti-Rpl11 rabbit antibody (1:200 dilution) or affinity-purified anti-Pabpc1l mouse antibody (1:50 dilution) overnight at room temperature, followed by incubation with anti-rabbit IgG Alexa-546 antibody (1:200 dilution) or anti-mouse IgG Alexa-488 antibody (1:200 dilution) for 1 hour. Fixation and immunofluorescence of mouse embryos were performed according to the procedure reported previously (Takada et al., 2020). After being mounted, the samples were observed under the LSM 5 LIVE confocal microscope.
Ribopuromycylation
The chorions of embryos at 0 and 3 hpf were removed with forceps and the embryos were pretreated with 355 μM cycloheximide or 40 μM anisomycin for 15 min in E3 solution (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4). The embryos were then incubated with 2 μM puromycin (Invitrogen, ant-pr-1) and 355 μM cycloheximide for 5 min in E3 solution. The embryos were incubated for 2 min with permeabilization buffer (50 mM Tris-HCl, 5 mM MgCl2, 25 mM KCl, 0.015% digitonin; pH 6.8) containing 355 μM cycloheximide or 40 μM anisomycin, 1 mM dithiothreitol, 100 μM (p-amidinophenyl) methanesulfonyl fluoride, 3 μg/mL leupeptin, and 8 U/mL RNasin Plus RNase Inhibitor (Promega, N2611). The embryos were fixed with 4% PFA/PBS for 1 hour and then washed with PBS overnight at 4°C. The embryos were incubated for 15 min with blocking buffer (0.05% saponin, 10 mM glycine in PBS) containing 5% newborn calf serum (Sigma, N4762). The embryos were then incubated with anti-Puromycin antibody (Kerafast; 3RH11) (1:300 dilution in blocking buffer) for 1 hour. After washing, the embryos were incubated with anti-mouse IgG-Alexa 546 antibody (Molecular probes) (1:200 dilution in blocking buffer) for 1 hour. After being mounted with VECTASHIELD Mounting Medium containing DAPI, the samples were observed under the LSM 5 LIVE confocal microscope. The number of signals of newly synthesized peptides was quantified using ImageJ software.
Simultaneous detection of newly synthesized peptides and pou5f3 RNA granules was performed as follows. After hybridization with the DIG-labeled antisense pou5f3 RNA probe, the newly synthesized peptides were detected with anti-Puromycin antibody as described above. After washing, the embryos were incubated with anti-DIG-HRP antibody and the signals were detected as described in whole mount in situ hybridization. The samples were mounted and observed under the N-SIM super-resolution microscope. The number and volume of individual signals and the number of colocalizations of both signals were analyzed by AIVIA software.
Simultaneous detection of newly synthesized peptides and Pou5f1/Oct4 RNA granules in mouse blastocyst-stage embryos was performed as follows. Mouse blastocyst-stage embryos were recovered from uteri on day 4 of pregnancy. The embryos were pretreated with 100 μg/mL cycloheximide for 15 min in M2 medium at 37°C. The embryos were then incubated with 100 μg/mL puromycin and 100 μg/mL cycloheximide for 15 min in M2 medium at 37°C, followed by fixation with 4% PFA/PBS for 10 min at 4°C. The embryos were transferred into oviducts and then the oviducts were fixed with 4% PFA/PBS overnight at 4°C. Fixed oviducts were dehydrated, embedded in paraffin, and cut into 10-μm thick sections. After detection of Pou5f1/Oct4 mRNA as reported previously (Takada et al., 2020), the sections were microwaved for 10 min (500 W) with 0.01 M citric acid (pH 6.0) containing 0.05% Tween 20, followed by cooling down for 40 min. The samples were then incubated with anti-Puromycin antibody (1:300 dilution in Blocking buffer) for 1 hour. After washing, the samples were incubated with anti-mouse IgG-Alexa 546 antibody (1:200 dilution in Blocking buffer) for 1 hour. After staining of DNA with 10 μg/mL Hoechst 33258, the samples were mounted with Fluoro-KEEPER Antifade Reagent (Nacalai Tesque) and observed under the LSM 5 LIVE confocal microscope.
Puro-Pou5f3 PLA
The proximity ligation assay (PLA) was performed according to the manufacturer’s instructions (Sigma). Briefly, after ribopuromycylation treatment and fixation with PFA/PBS, the embryos were incubated with Duolink Blocking Solution (Sigma) for 1 hour. The embryos were then incubated with anti-Puromycin mouse antibody (1:300) and affinity-purified anti-Pou5f3 rabbit antibody (1:50) diluted in Duolink Antibody Diluent (Sigma) for 90 min. After washing with Wash Buffer A (10 mM Tris-HCl, 0.15M NaCl, 0.05% Tween 20; pH 6.8), the embryos were incubated with Duolink PLA Probes (mouse PLUS and rabbit MINUS; Sigma, DUO092001 and DUO09002) diluted in Duolink Antibody Diluent (1:5) for 1 hour at 37°C. After washing with Wash Buffer A, the embryos were incubated with a Ligase (Sigma, DUO092013) diluted in Ligation Buffer (1:40) (Sigma, DUO092013) for 30 min at 37°C. Amplification and binding of the FarRed-labeled probe were performed with a Polymerase (Sigma, DUO092013) diluted in Amplification Buffer (1:80) (Sigma, DUO092013) for 100 min at 37°C. Amplification was stopped by washing with Wash Buffer B (0.2 M Tris-HCl, 0.1 M NaCl; pH 6.8), followed by washing with PBS. After being mounted with VECTASHIELD Mounting Medium containing DAPI, the samples were observed under the LSM 5 LIVE confocal microscope. The number of Puro-Pou5f3 PLA sites was quantified using ImageJ software.
Simultaneous detection of Puro-Pou5f3 PLA sites and pou5f3 RNA granules was performed as follows. After hybridization with the DIG-labeled antisense pou5f3 RNA probe, the Puro-Pou5f3 PLA sites were detected as described above. After washing, the embryos were incubated with anti-DIG-HRP antibody and the signals were detected as described in whole mount in situ hybridization. The samples were mounted and observed under the N-SIM super-resolution microscope. The number and volume of individual signals and the number of colocalizations of both signals were analyzed by AIVIA software.
Rpl11-Pou5f3 PLA
After fixation of embryos with 4% PFA/PBS and removal of the chorions, the PLA of Rpl11 and Pou5f3 proteins was performed with the anti-Rpl11 rabbit antibody (1:200 dilution) and anti-Pou5f3 mouse antibody (1:50 dilution) as described above. Simultaneous detection of Rpl11-Pou5f3 PLA sites and pou5f3 RNA granules was performed as follows. After hybridization with the DIG-labeled antisense pou5f3 RNA probe, Rpl11-Pou5f3 sites were detected. After washing, the embryos were incubated with anti-DIG-HRP antibody and the signals were detected as described in whole mount in situ hybridization. The samples were mounted and observed under the N-SIM super-resolution microscope. The number of individual signals and the number of colocalizations of both signals were analyzed by AIVIA software.
Hexanediol treatment
To dissociate lipid droplets, embryos were treated with 1,6-hexanediol (10% w/v in E3 solution) for 20 min at 28°C. As a control, embryos were treated with 1,2,6-hexanetriol (10% w/v in E3 solution) for 20 min at 28°C. Hexanediol and hexanetriol were dissolved in E3 solution as stocks and diluted in E3 solution before use. After fixation with 4% PFA/PBS overnight at 4°C, the embryos were analyzed by whole mount in situ hybridization. Crude extracts of the embryos were analyzed by immunoblotting with the anti-puromycin antibody. Puro-PLA was performed after hexanediol and hexanetriol treatment as described above.
Quantitative RT-PCR
The amount of pou5f3 and β-actin mRNAs was quantified by using a real-time PCR system with PowerUP SYBR Green Master Mix (Applied Biosystems) according to the manufacturer’s instructions. Total RNA extracted from 50 embryos at 3 hpf was used for cDNA synthesis using the FastGene Scriptase Ⅱ (NIPPON Genetics). The pou5f3 and β-actin transcripts were amplified with the cDNA and primer sets specific to pou5f3 (zpou5f3-qPCR-f1 and zpou5f3-qPCR-r1 primers) and β-actin (zβ-actin-qPCR-f1 and zβ-actin-qPCR-r1 primers). Primers can be found in Table S1.
mRNA injection
The full-length Pabpc1l was cloned into pCS2-GFP-N to produce Pabpc1l fused with GFP at the N-terminus of Pabpc1l. mRNA encoding GFP-Pabpc1l was synthesized with an mMESSAGE mMACHINE SP6 kit (Ambion) and dissolved in distilled water. One nl of 50 ng/μL mRNA was injected into fertilized eggs by using Femtojet (Eppendorf). The distribution of GFP-Pabpc1l was observed under an LSM 980 confocal microscope (Carl Zeiss).
Quantification and statistical analysis
Image analyses
Single confocal optical images (Figures 1, 2, 3B, 3C, 4B, 5E, 6, 7A, 8, S1, S4 and S5) were acquired by the confocal microscopes, LSM 5 LIVE and LSM 980, and analyzed by Image J software. After removing intensities lower than 30% level of the top intensity as diffused background signals, the number and area of signals were quantified using the Analyze Particles tool in Image J. Signals larger than 0.2 μm in diameter were counted. Z-stack images (Figures 3E, 4D, 4F, 4G, 5A, 5C, 5F, and 7B–7D) were acquired by the N-SIM super-resolution microscope and were analyzed by AIVIA software after reconstruction of 3D images with intensities in binary 16-bit type. The number and volume of signals of FISH, ribopuromycylation, immunofluorescence and PLA were quantified using the 3D Object Analysis recipe in AIVIA. Detection values were set up as follows. Image Smoothing Filter Size was 9. Min Edge Intensity was 1700 for pou5f3 mRNA, 1800 for ribopuromycylation, 1650 for Puro-Pou5f3 PLA, 2000 for Rpl11, and 1750 for Rpl11-Pou5f3 PLA. Fill Holes Size was 0. Partition values were set up as follows. Object Radius was 0.1-3. Mesh Smoothing Factor was 2. Min Edge to Center Distance was 0.1. The Pixel Colocalization tool in AIVIA enables detection of spatial overlaps, i.e., colocalized spaces in 3D from two input channels of the same image (Figure S3B). Input channels were set up as follows. Input Coloc Channel 1 was z-stack images of pou5f3 mRNA signal, and Input Coloc Channel 2 was z-stack images of ribopuromycylation, Puro-Pou5f3 PLA, Rpl11, or Rpl11-Pou5f3 PLA signal. Detection and partition values were similar to those above.
Statistical analyses
All the results were expressed as mean ± standard deviations. Statistical analysis compering two samples was performed using Student’s t-test, and that compering multiple groups was performed using Tukey-Kramer test as indicated in the figure legends. Significance was defined as p < 0.05 (∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05).
Acknowledgments
We thank Dr. K. Kobayashi at Nikon Imaging Center in Hokkaido University for technical support on super-resolution microscopy. This work was supported by Grant-in-Aid for Scientific Research KAKENHI grant numbers 16K07242, 19K22376, and 21H02398 (to T.K.) from Japan Society for the Promotion of Science (JSPS) and was in part supported by a grant from JSPS grant number JP16H06280.
Autor contributions
Conceptualization, K.S. and T.K.; Investigation, K.S., M.S., A.I., K.M., and Y.T.; Methodology and Resources, K.Y.; Supervision, Funding acquisition and Writing, T.K.
Declaration of interests
The authors declare no competing interests.
Published: June 17, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104344.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Aanes H., Winata C.L., Lin C.H., Chen J.P., Srinivasan K.G., Lee S.G., Lim A.Y., Hajan H.S., Collas P., Bourque G., et al. Zebrafish mRNA sequencing deciphers novelties in transcriptome dynamics during maternal to zygotic transition. Genome Res. 2011;21:1328–1338. doi: 10.1101/gr.116012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki F., Hara K.T., Schultz R.M. Acquisition of transcriptional competence in the 1-cell mouse embryo: requirement for recruitment of maternal mRNAs. Mol. Reprod. Dev. 2003;64:270–274. doi: 10.1002/mrd.10227. [DOI] [PubMed] [Google Scholar]
- Belting H.G., Hauptmann G., Meyer D., Abdelilah-Seyfried S., Chitnis A., Eschbach C., Soll I., Thisse C., Thisse B., Artinger K.B., et al. Spiel ohne grenzen/pou2 is required during establishment of the zebrafish midbrain-hindbrain boundary organizer. Development. 2001;128:4165–4176. doi: 10.1242/dev.128.21.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belting H.G., Wendik B., Lunde K., Leichsenring M., Mossner R., Driever W., Onichtchouk D. Pou5f1 contributes to dorsoventral patterning by positive regulation of vox and modulation of fgf8a expression. Dev. Bio. 2011;356:323–336. doi: 10.1016/j.ydbio.2011.05.660. [DOI] [PubMed] [Google Scholar]
- Buchan J.R., Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxbaum A.R., Haimovich G., Singer R.H. In the right place at the right time: visualizing and understanding mRNA localization. Nat. Rev. Mol. Cell Biol. 2015;16:95–109. doi: 10.1038/nrm3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth A., Cox L.L., MacNicol A.M. Cytoplasmic polyadenylation element (CPE)- and CPE-binding protein (CPEB)-independent mechanisms regulate early class maternal mRNA translational activation in Xenopus oocytes. J. Biol. Chem. 2004;279:17650–17659. doi: 10.1074/jbc.m313837200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Melton C., Suh N., Oh J.S., Horner K., Xie F., Sette C., Blelloch R., Conti M. Genome-wide analysis of translation reveals a critical role for deleted in azoospermia-like (Dazl) at the oocyte-to-zygote transition. Genes Dev. 2011;25:755–766. doi: 10.1101/gad.2028911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouaib R., Safieddine A., Pichon X., Imbert A., Kwon O.S., Samacoits A., Traboulsi A.M., Robert M.C., Tsanov N., Coleno E., et al. A dual protein-mRNA localization screen reveals compartmentalized translation and widespread co-translational RNA targeting. Dev. Cell. 2020;54:773–791.e5. doi: 10.1016/j.devcel.2020.07.010. [DOI] [PubMed] [Google Scholar]
- David A., Dolan B.P., Hickman H.D., Knowlton J.J., Clavarino G., Pierre P., Bennink J.R., Yewdell J.W. Nuclear translation visualized by ribosome-bound nascent chain puromycylation. J. Cell Biol. 2012;197:45–57. doi: 10.1083/jcb.201112145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enam S.U., Zinshteyn B., Goldman D.H., Cassani M., Livingston N.M., Seydoux G., Green R. Puromycin reactivity does not accurately localize translation at the subcellular level. Elife. 2020;9:e60303. doi: 10.7554/elife.60303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman I., Huang Y.S., Mendez R., Cao Q.P., Theurkauf W., Richter J.D. CPEB, Maskin, and cyclin B1 mRNA at the mitotic apparatus. Cell. 2000;103:435–447. doi: 10.1016/s0092-8674(00)00135-5. [DOI] [PubMed] [Google Scholar]
- Groisman I., Jung M.Y., Sarkissian M., Cao Q., Richter J.D. Translational control of the embryonic cell cycle. Cell. 2002;109:473–483. doi: 10.1016/s0092-8674(02)00733-x. [DOI] [PubMed] [Google Scholar]
- Hamatani T., Carter M.G., Sharov A.A., Ko M.S. Dynamics of global gene expression changes during mouse preimplantation development. Dev. Cell. 2004;6:117–131. doi: 10.1016/s1534-5807(03)00373-3. [DOI] [PubMed] [Google Scholar]
- Harvey S.A., Sealy I., Kettleborough R., Fenyes F., White R., Stemple D., Smith J.C. Identification of the zebrafish maternal and paternal transcriptomes. Development. 2013;140:2703–2710. doi: 10.1242/dev.095091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobson B.D., Kong L., Hartwick E.W., Gonzalez R.L., Sims P.A. Elongation inhibitors do not prevent the release of puromycylated nascent polypeptide chains from ribosomes. Elife. 2020;9:e60048. doi: 10.7554/elife.60048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie M., Kotani T. Formation of mos RNA granules in the zebrafish oocyte that differ from cyclin B1 RNA granules in distribution, density and regulation. Eur. J. Cell Biol. 2016;95:563–573. doi: 10.1016/j.ejcb.2016.10.001. [DOI] [PubMed] [Google Scholar]
- Howley C., Ho R.K. mRNA localization patterns in zebrafish oocytes. Mech. Dev. 2000;92:305–309. doi: 10.1016/s0925-4773(00)00247-1. [DOI] [PubMed] [Google Scholar]
- Ivanov P., Kedersha N., Anderson P. Stress granules and processing bodies in translational control. Cold Spring Harb. Perspect. Biol. 2019;11:a032813. doi: 10.1101/cshperspect.a032813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S., Wheeler J.R., Walters R.W., Agrawal A., Barsic A., Parker R. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell. 2016;164:487–498. doi: 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambor H., Surendranath V., Kalinka A.T., Mejstrik P., Saalfeld S., Tomancak P. Systematic imaging reveals features and changing localization of mRNAs in Drosophila development. Elife. 2015;4:e05003. doi: 10.7554/elife.05003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansova D., Aleshkina D., Jindrova A., Iyyappan R., An Q., Fan G., Susor A. Single molecule RNA localization and translation in the mammalian oocyte and embryo. J. Mol. Biol. 2021;433:167166. doi: 10.1016/j.jmb.2021.167166. [DOI] [PubMed] [Google Scholar]
- Kane D.A., Kimmel C.B. The zebrafish midblastula transition. Development. 1993;119:447–456. doi: 10.1242/dev.119.2.447. [DOI] [PubMed] [Google Scholar]
- Kiebler M.A., Bassell G.J. Neuronal RNA granules: movers and makers. Neuron. 2006;51:685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Kloc M., Zearfoss N.R., Etkin L.D. Mechanisms of subcellular mRNA localization. Cell. 2002;108:533–544. doi: 10.1016/s0092-8674(02)00651-7. [DOI] [PubMed] [Google Scholar]
- Kondo T., Kotani T., Yamashita M. Dispersion of cyclin B mRNA aggregation is coupled with translational activation of the mRNA during zebrafish oocyte maturation. Dev. Biol. 2001;229:421–431. doi: 10.1006/dbio.2000.9990. [DOI] [PubMed] [Google Scholar]
- Kotani T., Maehata K., Takei N. Regulation of translationally repressed mRNAs in zebrafish and mouse oocytes. Results Probl. Cell Differ. 2017;63:297–324. doi: 10.1007/978-3-319-60855-6_13. [DOI] [PubMed] [Google Scholar]
- Kotani T., Yasuda K., Ota R., Yamashita M. Cyclin B1 mRNA translation is temporally controlled through formation and disassembly of RNA granules. J. Cell Biol. 2013;202:1041–1055. doi: 10.1083/jcb.201302139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald S., Maharana S., Mateju D., Malinovska L., Nuske E., Poser I., Richter D., Alberti S. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. Elife. 2015;4:e06807. doi: 10.7554/elife.06807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald S., Maharana S., Simon A. Hexanediol: a chemical probe to investigate the material properties of membrane-less compartments. Matters. 2017 doi: 10.19185/matters.201702000010. [DOI] [Google Scholar]
- Kumari P., Gilligan P.C., Lim S., Tran L.D., Winkler S., Philp R., Sampath K. An essential role for maternal control of Nodal signaling. Elife. 2013;2:e00683. doi: 10.7554/elife.00683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuyer E., Yoshida H., Parthasarathy N., Alm C., Babak T., Cerovina T., Hughes T.R., Tomancak P., Krause H.M. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Lee M.T., Bonneau A.R., Takacs C.M., Bazzini A.A., DiVito K.R., Fleming E.S., Giraldez A.J. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature. 2013;503:360–364. doi: 10.1038/nature12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichsenring M., Maes J., Mossner R., Driever W., Onichtchouk D. Pou5f1 transcription factor controls zygotic gene activation in vertebrates. Science. 2013;341:1005–1009. doi: 10.1126/science.1242527. [DOI] [PubMed] [Google Scholar]
- Lippok B., Song S., Driever W. Pou5f1 protein expression and posttranslational modification during early zebrafish development. Dev. Dyn. 2014;243:468–477. doi: 10.1002/dvdy.24079. [DOI] [PubMed] [Google Scholar]
- Lunde K., Belting H.G., Driever W. Zebrafish pou5f1/pou2, homolog of mammalian Oct4, functions in the endoderm specification cascade. Curr. Biol. 2004;14:48–55. doi: 10.1016/j.cub.2003.11.022. [DOI] [PubMed] [Google Scholar]
- Luong X.G., Daldello E.M., Rajkovic G., Yang C.R., Conti M. Genome-wide analysis reveals a switch in the translational program upon oocyte meiotic resumption. Nucleic Acids Res. 2020;48:3257–3276. doi: 10.1093/nar/gkaa010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNicol M.C., Cragle C.E., Arumugam K., Fosso B., Pesole G., MacNicol A.M. Functional integration of mRNA translational control programs. Biomolecules. 2015;5:1580–1599. doi: 10.3390/biom5031580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K.C., Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateju D., Eichenberger B., Voigt F., Eglinger J., Roth G., Chao J.A. Single-molecule imaging reveals translation of mRNAs localized to stress granules. Cell. 2020;183:1801–1812.e13. doi: 10.1016/j.cell.2020.11.010. [DOI] [PubMed] [Google Scholar]
- Mendez R., Richter J.D. Translational control by CPEB: a means to the end. Nat. Rev. Mol. Cell Biol. 2001;2:521–529. doi: 10.1038/35080081. [DOI] [PubMed] [Google Scholar]
- Mishima Y., Fukao A., Kishimoto T., Sakamoto H., Fujiwara T., Inoue K. Translational inhibition by deadenylation-independent mechanisms is central to microRNA-mediated silencing in zebrafish. Proc. Natl. Acad. Sci. USA. 2012;109:1104–1109. doi: 10.1073/pnas.1113350109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moissoglu K., Yasuda K., Wang T., Chrisafis G., Mili S. Translational regulation of protrusion-localized RNAs involves silencing and clustering after transport. Elife. 2019;8:e44752. doi: 10.7554/elife.44752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon S.L., Morisaki T., Khong A., Lyon K., Parker R., Stasevich T.J. Multicolour single-molecule tracking of mRNA interactions with RNP granules. Nat. Cell Biol. 2019;21:162–168. doi: 10.1038/s41556-018-0263-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Polanco F., Bates C., Lui J., Casson J., Solari C.A., Pizzinga M., Forte G., Griffin C., Garner K.E., Burt H.E., et al. Core Fermentation (CoFe) granules focus coordinated glycolytic mRNA localization and translation to fuel glucose fermentation. iScience. 2021;24:102069. doi: 10.1016/j.isci.2021.102069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J., Zevnik B., Anastassiadis K., Niwa H., Klewe-Nebenius D., Chambers I., Scholer H., Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Nosella M.L., Forman-Kay J.D. Phosphorylation-dependent regulation of messenger RNA transcription, processing and translation within biomolecular condensates. Curr. Opin. Cell Biol. 2021;69:30–40. doi: 10.1016/j.ceb.2020.12.007. [DOI] [PubMed] [Google Scholar]
- Panasenko O.O., Somasekharan S.P., Villanyi Z., Zagatti M., Bezrukov F., Rashpa R., Cornut J., Iqbal J., Longis M., Carl S.H., et al. Co-translational assembly of proteasome subunits in NOT1-containing assemblysomes. Nat. Struct. Mol. Biol. 2019;26:110–120. doi: 10.1038/s41594-018-0179-5. [DOI] [PubMed] [Google Scholar]
- Pizzinga M., Bates C., Lui J., Forte G., Morales-Polanco F., Linney E., Knotkova B., Wilson B., Solari C.A., Berchowitz L.E., et al. Translation factor mRNA granules direct protein synthetic capacity to regions of polarized growth. J. Cell Biol. 2019;218:1564–1581. doi: 10.1083/jcb.201704019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reim G., Brand M. Maternal control of vertebrate dorsoventral axis formation and epiboly by the POU domain protein Spg/Pou2/Oct4. Development. 2006;133:2757–2770. doi: 10.1242/dev.02391. [DOI] [PubMed] [Google Scholar]
- Reim G., Mizoguchi T., Stainier D.Y., Kikuchi Y., Brand M. The POU domain protein spg (pou2/Oct4) is essential for endoderm formation in cooperation with the HMG domain protein casanova. Dev. Cell. 2004;6:91–101. doi: 10.1016/s1534-5807(03)00396-4. [DOI] [PubMed] [Google Scholar]
- Richter J.D. CPEB: a life in translation. Trends Biochem. Sci. 2007;32:279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Richter J.D., Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- Scholer H.R., Hatzopoulos A.K., Balling R., Suzuki N., Gruss P. A family of octamer-specific proteins present during mouse embryogenesis: evidence for germline-specific expression of an Oct factor. EMBO J. 1989;8:2543–2550. doi: 10.1002/j.1460-2075.1989.tb08392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina N. Liquid- and solid-like RNA granules form through specific scaffold proteins and combine into biphasic granules. J. Biol. Chem. 2019;294:3532–3548. doi: 10.1074/jbc.ra118.005423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y., Brangwynne C.P. Liquid phase condensation in cell physiology and disease. Science. 2017;357:eaaf4382. doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- Sun J., Yan L., Shen W., Meng A. Maternal Ybx1 safeguards zebrafish oocyte maturation and maternal-to-zygotic transition by repressing global translation. Development. 2018;145:dev166587. doi: 10.1242/dev.166587. [DOI] [PubMed] [Google Scholar]
- Takada Y., Iyyappan R., Susor A., Kotani T. Posttranscriptional regulation of maternal Pou5f1/Oct4 during mouse oogenesis and early embryogenesis. Histochem. Cell Biol. 2020;154:609–620. doi: 10.1007/s00418-020-01915-4. [DOI] [PubMed] [Google Scholar]
- Takeda H., Matsuzaki T., Oki T., Miyagawa T., Amanuma H. A novel POU domain gene, zebrafish pou2: expression and roles of two alternatively spliced twin products in early development. Genes Dev. 1994;8:45–59. doi: 10.1101/gad.8.1.45. [DOI] [PubMed] [Google Scholar]
- Takei N., Nakamura T., Kawamura S., Takada Y., Satoh Y., Kimura A.P., Kotani T. High-sensitivity and high-resolution in situ hybridization of coding and long non-coding RNAs in vertebrate ovaries and testes. Biol. Proced. Online. 2018;20:6. doi: 10.1186/s12575-018-0071-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei N., Sato K., Takada Y., Iyyappan R., Susor A., Yamamoto T., Kotani T. Tdrd3 regulates the progression of meiosis II through translational control of Emi2 mRNA in mouse oocytes. Curr. Res. Cell Biol. 2021;2:100009. doi: 10.1016/j.crcbio.2021.100009. [DOI] [Google Scholar]
- Takei N., Takada Y., Kawamura S., Sato K., Saitoh A., Bormann J., Yuen W.S., Carroll J., Kotani T. Changes in subcellular structures and states of pumilio 1 regulate the translation of target Mad2 and cyclin B1 mRNAs. J. Cell Sci. 2020;133:jcs249128. doi: 10.1242/jcs.249128. [DOI] [PubMed] [Google Scholar]
- Thomas M.G., Pascual M.L., Maschi D., Luchelli L., Boccaccio G.L. Synaptic control of local translation: the plot thickens with new characters. Cell Mol. Life Sci. 2014;71:2219–2239. doi: 10.1007/s00018-013-1506-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- tom Dieck S., Kochen L., Hanus C., Heumuller M., Bartnik I., Nassim-Assir B., Merk K., Mosler T., Garg S., Bunse S., et al. Direct visualization of newly synthesized target proteins in situ. Nat. Methods. 2015;12:411–414. doi: 10.1038/nmeth.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trcek T., Grosch M., York A., Shroff H., Lionnet T., Lehmann R. Drosophila germ granules are structured and contain homotypic mRNA clusters. Nat. Commun. 2015;6:7962. doi: 10.1038/ncomms8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updike D.L., Hachey S.J., Kreher J., Strome S. P granules extend the nuclear pore complex environment in the C. elegans germ line. J. Cell Biol. 2011;192:939–948. doi: 10.1083/jcb.201010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Latham K.E. Requirement for protein synthesis during embryonic genome activation in mice. Mol. Reprod. Dev. 1997;47:265–270. doi: 10.1002/(sici)1098-2795(199707)47:3<265::aid-mrd5>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Weil T.T., Parton R.M., Herpers B., Soetaert J., Veenendaal T., Xanthakis D., Dobbie I.M., Halstead J.M., Hayashi R., Rabouille C., Davis I. Drosophila patterning is established by differential association of mRNAs with P bodies. Nat. Cell Biol. 2012;14:1305–1313. doi: 10.1038/ncb2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West J.A., Mito M., Kurosaka S., Takumi T., Tanegashima C., Chujo T., Yanaka K., Kingston R.E., Hirose T., Bond C., et al. Structural, super-resolution microscopy analysis of paraspeckle nuclear body organization. J. Cell Biol. 2016;214:817–830. doi: 10.1083/jcb.201601071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk R., Hu J., Blotsky D., Krause H.M. Diverse and pervasive subcellular distributions for both coding and long noncoding RNAs. Genes Dev. 2016;30:594–609. doi: 10.1101/gad.276931.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winata C.L., Korzh V. The translational regulation of maternal mRNAs in time and space. FEBS Lett. 2018;592:3007–3023. doi: 10.1002/1873-3468.13183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winata C.L., Lapinski M., Pryszcz L., Vaz C., Bin Ismail M.H., Nama S., Hajan H.S., Lee S.G.P., Korzh V., Sampath P., et al. Cytoplasmic polyadenylation-mediated translational control of maternal mRNAs directs maternal-to-zygotic transition. Development. 2018;145:dev159566. doi: 10.1242/dev.159566. [DOI] [PubMed] [Google Scholar]
- Woodland H.R. Changes in the polysome content of developing Xenopus laevis embryos. Dev. Biol. 1974;40:90–101. doi: 10.1016/0012-1606(74)90111-0. [DOI] [PubMed] [Google Scholar]
- Yasuda K., Zhang H., Loiselle D., Haystead T., Macara I.G., Mili S. The RNA-binding protein Fus directs translation of localized mRNAs in APC-RNP granules. J. Cell Biol. 2013;203:737–746. doi: 10.1083/jcb.201306058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaucker A., Kumari P., Sampath K. Zebrafish embryogenesis - a framework to study regulatory RNA elements in development and disease. Dev. Biol. 2020;457:172–180. doi: 10.1016/j.ydbio.2019.01.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.