Abstract
A specific fragment of the genome of Tuc2009, a temperate lactococcal bacteriophage, was shown to contain several open reading frames, whose deduced protein products exhibited similarities to proteins known to be involved in DNA replication and modification. In this way, a putative single-stranded binding protein, replisome organizer protein, topoisomerase I, and a methylase were identified. When the genetic information coding for the putative replisome organizer protein of Tuc2009, Rep2009, was supplied on a high-copy-number plasmid vector, it was shown to confer a phage-encoded resistance (Per) phenotype on its lactococcal host UC509.9. The presence of this recombinant plasmid was shown to cause a marked reduction in Tuc2009 DNA replication, suggesting that the observed phage resistance was due to titration of a factor, or factors, required for Tuc2009 DNA replication. Further experiments delineated the phage resistance-conferring region to a 160-bp fragment rich in direct repeats. Gel retardation experiments, which indicated a protein-DNA interaction between this 160-bp fragment and the Rep2009 protein, were performed. UC509.9 strains harboring plasmids with randomly mutated versions of this fragment were shown to display a variable phage resistance phenotype, depending on the position of the mutations.
Lactococcus lactis strains are widely used as starter cultures for many dairy fermentations. Since lactococcal bacteriophages were first identified as a major cause of fermentation failure (46), much research effort has been directed at the development of bacteriophage-resistant starter strains for industrial use (15, 24). Spontaneous bacteriophage-insensitive mutants can be isolated following infection of a bacterial population with a specific phage at a high titer. However, these mutants are only of limited value as starter cultures since their spectrum of resistance tends to be very narrow, while their growth characteristics may also be altered (15, 19).
Naturally occurring phage resistance mechanisms have been identified in wild-type lactococcal strains, and these are often encoded on native conjugative plasmids, which has facilitated the generation of novel resistant starter strains through food grade techniques (5, 39). These resistance mechanisms have been divided into four main groups on the basis of their mode of action (5, 10, 13, 15, 23): adsorption interference, which prevents the adsorption of phage particles to the cell surface; DNA injection blocking, which prevents phages that have successfully attached to the cell surface from injecting their DNA in the cell cytoplasm; restriction-modification, causing intracellular degradation of incoming DNA molecules; and abortive infection, which encompasses a range of mechanisms, any of which may interfere with phage development at any time after the injection of intact phage DNA into the cell up to release of progeny phage from an infected cell.
The advancement of molecular techniques and the increased knowledge of lactococcal phage biology have enabled the development of artificial or “intelligent” phage resistance mechanisms, and a number of these have been described to date.
One such mechanism involves the use of antisense mRNA (21, 22). It was found that plasmid vectors harboring certain conserved phage genes that had been cloned in the reverse orientation behind a strong promoter conferred an abortive infection-type resistance on host cells against a specific phage. It is thought that this resistance system acts through the formation of a nontranslatable double-stranded mRNA hybrid between a sense and its antisense mRNA which is subject to rapid degradation. Another more recently developed resistance mechanism involves the use of a phage-inducible promoter in combination with the LlaI restriction-modification system from the lactococcal plasmid pTR2030 (12). The middle phage-inducible promoter (φ31p) from bacteriophage φ31 was cloned upstream of the LlaIR+ restriction gene so that, upon φ31 infection of a cell harboring this plasmid, the lethal gene product of LlaIR+ is produced, resulting in death of the host cell before the infecting phage has a chance to reproduce itself.
The first of these intelligent systems to be described was termed Per 50, for phage-encoded resistance, and conferred resistance against the small isometric-headed phage, φ50, on L. lactis NCK203 (16). The Per 50 system consists of a piece of φ50 DNA containing the presumptive origin of replication (ori50) cloned into a high-copy-number Streptococcus-Escherichia coli shuttle vector. Following φ50 infection, this ori50-containing plasmid was shown to interfere with intracellular phage DNA replication in NCK203, while the concentration of intracellular plasmid DNA was seen to increase concurrently. This suggested that ori50 supplied on the plasmid was responsible for the depletion of an essential DNA replication function, or functions, and was capable of enhancing plasmid replication. In a later study, it was found that the Per phenotype was directly dependent on copy number of the plasmid carrier, while it was also shown that the Per principle can be applied to other phage-host systems (35).
In this report, we have identified a putative replisome organizer protein from the temperate lactococcal phage Tuc2009, designated Rep2009. A region rich in direct repeats which is contained with the rep2009 gene, designated ori2009, was shown to confer a Tuc2009-resistant phenotype on its lactococcal host, UC509.9. The possible role of ori2009 in Tuc2009 replication was further investigated by DNA mutagenesis and protein-DNA interactions between Rep2009 and ori2009.
MATERIALS AND METHODS
Bacteria, bacteriophages, and plasmids.
Bacterial strains, bacteriophages, and plasmids used in this study are listed in Table 1.
TABLE 1.
List of bacteriophages, bacterial strains and plasmids used in this study
| Phage, bacterial strain, or plasmid | Relevant feature(s) | Source and/or reference |
|---|---|---|
| Phage | ||
| Tuc2009 | Isolated following induction of L. lactis subsp. cremoris UC509 | 2 |
| E. coli | ||
| XL1-Blue | Transformation host | Stratagene |
| L. lactis | ||
| UC509.9 | Prophage-cured derivative of L. lactis subsp. cremoris UC509; host for Tuc2009 | 4 |
| NZ9800 | ΔnisA | 9 |
| Plasmids | ||
| pBluescript | Cloning vector Apr | Stratagene |
| pNZ8048 | L. lactis expression vector; PnisA Cmr (Per−) | 9 |
| pSK220 | pBluescript + 6.9-kb Tuc2009 genomic insert; Apr | 43 |
| pNZRep-1 | pNZ8048 + 783-bp Tuc2009 ORF16; Cmr (Per+) | This study |
| pNZRep-3 | pNZ8048 + 783-bp Tuc2009 ORF16, in reverse orientation to pNZRep-1; Cmr (Per+) | This study |
| pNZ101 | pNZ8048 + 1.5-kb randomly selected Tuc2009 genomic insert; Cmr (Per−) | This study |
| pNZRep-A | pNZ8048 + 283-bp Tuc2009 ORF16 subclone; Cmr (Per−) | This study |
| pNZRep-B | pNZ8048 + 526-bp Tuc2009 ORF16 subclone; Cmr (Per+) | This study |
| pNZRep-C | pNZ8048 + 295-bp Tuc2009 ORF16 subclone; Cmr (Per+) | This study |
| pNZRep-Ci | pNZ8048 + 112-bp Tuc2009 ORF16 subclone; Cmr (Per−) | This study |
| pNZRep-Cii | pNZ8048 + 159-bp Tuc2009 ORF16 subclone; Cmr (Per+) | This study |
| pNZRep-Ciii | pNZ8048 + 219-bp Tuc2009 ORF16 subclone; Cmr (Per+) | This study |
| pNZRep-Civ | pNZ8048 + 187-bp Tuc2009 ORF16 subclone; Cmr (Per+)a | This study |
| pNZRep-Cv | pNZ8048 + 158-bp Tuc2009 ORF16 subclone; Cmr (Per+)a | This study |
| pNZRep-D | pNZ8048 + 553-bp Tuc2009 ORF16 subclone; Cmr (Per+) | This study |
| pNZRep-E | pNZ8048 + 323-bp Tuc2009 ORF16 subclone; Cmr (Per−) | This study |
Slightly reduced Per effect.
Media and growth conditions.
L. lactis strains were cultured at 30°C in M17 broth (Difco Laboratories, Detroit, Mich.) (42) containing 0.5% glucose (GM17), or in GSB, a modified version of lactic streptococcus broth (LSB) which contains glucose instead of lactose (2). Medium for plaque assays was prepared as described by Lillehaug (26). Briefly, M17 double-layer agar plates were prepared; these plates contained 5 g of glucose/liter, 5 g of glycine/liter, and 10 mM CaCl2, and 1 and 0.4% agar concentrations were used for the bottom and top layers, respectively. The medium was sterilized by boiling for 5 min in a microwave oven. E. coli strains were grown in Luria-Bertani (LB) medium at 37°C (37). Chloramphenicol at 10 μg/ml and ampicillin at 100 μg/ml were used where appropriate.
Propagation of Tuc2009.
Tuc2009 was propagated on a prophage-free derivative of L. lactis subsp. cremoris UC509, designated UC509.9. UC509.9 was grown to early log phase in GSB, at which point CaCl2 was added to a final concentration of 10 mM prior to the addition of Tuc2009 phage. Incubation was subsequently continued until lysis occurred. Phage particles were collected from this lysate by centrifugation and concentrated by cesium chloride density gradient centrifugation, as described by Sambrook et al. (37).
Plasmid and phage DNA isolation and molecular cloning.
Isolation of E. coli plasmid DNA was accomplished by using the QIAprep spin plasmid miniprep kit (Qiagen, Inc., Chatsworth, Calif.) as described in the manufacturers’ instructions. Phage genomic DNA was isolated from a concentrated phage solution by the method described by Stanley et al. (41). Restriction enzymes and T4 DNA ligase were purchased from Boehringer GmbH (Mannheim, Germany) and used in accordance with the manufacturer’s instructions.
PCR methods.
PCR amplifications were carried out by using Taq DNA polymerase (Boehringer), as described by the manufacturer with a Gene Amp PCR system 2400 thermal cycler (Perkin-Elmer Cetus, Norwalk, Conn.). Suitable restriction sites were incorporated into the 5′ end of synthetic primers when PCR-generated products were used for cloning. Random mutations were introduced into Per-conferring fragments by a method described by Dartois et al. (6), which involves PCR amplification of DNA fragments with limiting amounts of one of the nucleotides.
DNA sequence analysis.
DNA sequence determination of the Tuc2009 genomic subclone in pSK220 was performed with an Applied Biosystems (Foster City, Calif.) 373A automated DNA sequencer using synthetic oligonucleotides (Oligo 1000M; Beckman Instruments) as primers. Assembly of sequences was performed by using the Seqman program of the DNASTAR software package. Database searches were performed by using the BLASTP, BLASTN, and TBLASTN (1) programs with sequences present in the latest releases of the nonredundant databases found at the National Centers for Biotechnology Information website (33a). Sequence alignments were performed by using the Clustal method of the MEGALIGN 3.06 program of the DNASTAR 1996 release software package.
Electroporation procedure.
Electrotransformation of plasmid DNA into E. coli was performed essentially as described by Sambrook et al. (37), whereas electrotransformation of plasmid DNA into L. lactis was performed as described by Wells et al. (45), with the following minor modifications. L. lactis cells were grown in GM17 broth containing 2% (wt/vol) glycine. Plasmid DNA for electrotransformation was prepared by precipitating the DNA from a single miniprep prepared from 2.5 ml of E. coli cells by using the QIAprep spin plasmid miniprep kit (Qiagen), prior to resuspension in 10 μl of storage buffer (10% glycerol, 0.5 M sucrose) and addition to 40 μl of electrocompetent cells.
Bacteriophage assays.
Plaque assays were performed as described by Lillehaug (26). Lysis-in-broth experiments were performed as follows. Strains were grown in GSB (10 ml) to an optical density at 600 nm (OD600) of approximately 0.2, followed by the addition of CaCl2 (final concentration, 10 mM) and 10 μl of a solution containing the desired concentration of Tuc2009 particles, after which the OD600 was monitored over a period of time.
Visualization of intracellular phage DNA replication.
The procedure described by Hill et al. (16) was used to isolate phage DNA at various time points following infection. Phage DNA was run on an agarose gel and transferred to a Hybond N+ nucleic acid transfer membrane (Amersham Corp., Amersham, United Kingdom) by the procedure described by Southern (40). The enhanced chemiluminescence kit (ECL; Amersham) was used to label DNA for use as a probe according to conditions specified by the supplier.
Cloning and overexpression of Rep2009.
A 783-bp fragment exactly encompassing the rep2009 gene was cloned into the NcoI and XbaI sites of the expression plasmid pNZ8048 in order to place it under the control of the nisin-inducible promoter contained on the vector. Restriction sites were incorporated into the rep2009 DNA fragment by using PCR with suitably designed synthetic primers to facilitate cloning, such that the original start codon of the rep2009 gene, GTG, was changed to ATG. This cloning was performed in E. coli, and the integrity of the insert was confirmed by sequencing. The resulting plasmid, called pNZRep-1, was then introduced into L. lactis NZ9800, and Rep2009 was overexpressed as outlined by de Ruyter et al. (9).
Gel retardation assays.
To prepare cell extracts, cells from 50-ml cultures of L. lactis were collected by centrifugation, resuspended in 500 μl of breaking buffer (20 mM Tris [pH 8.0]), 1 mM dithiothreitol), and ruptured by sonication (five consecutive sonication pulses of 30 s; samples placed on ice for 30 s between sonication pulses). Samples were then centrifuged at 15,000 × g for 5 min in a bench top centrifuge to remove cell debris. Protein concentrations of the sample supernatants were determined with a protein assay kit (Bio-Rad Laboratories GmbH, Munich, Germany), with bovine serum albumin as the standard. Gel retardation assays were performed as outlined below. Probes were labelled with polynucleotide kinase by using [γ-32P]ATP. DNA binding was carried out in 20-μl reaction volumes containing 50 mM Tris (pH 8.0), 10% (vol/vol) glycerol, 1 mM EDTA, 5 mM MgCl2, 500 mM KCl, 2 mM dithiothreitol, 50 μg of bovine serum albumin/ml, 75 μg of poly(dI-dC)/ml, probe (approximately 0.3 ng), and 2.5 μg of total protein from cell lysate. Incubation was for 15 min at room temperature, followed by the addition of 5 μl of 50% glycerol, after which the samples were loaded onto a 4% polyacrylamide gel containing 2.5% glycerol. Gels were run in TAE buffer (0.04 M Tris-acetate [pH 7.5], 2 mM EDTA) at 120 V for 3 h, dried, and exposed overnight at −70°C to X-Omat film (Kodak).
RESULTS
Nucleotide sequence determination and identification of ORFs.
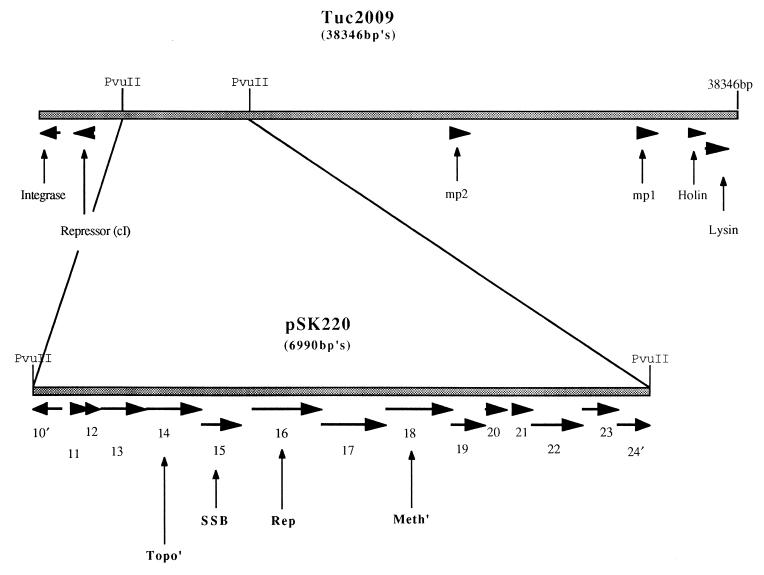
The sequence of the complete genome of Tuc2009 has been determined, and 58 putative open reading frames (ORFs) have been identified (44a). An ORF coding for the putative integrase of Tuc2009 has been designated ORF1, with subsequent ORFs numbered accordingly. Analysis of a PvuII fragment derived from the genome of Tuc2009 and contained within plasmid pSK220 revealed the presence of 13 complete and 2 incomplete putative ORFs (Fig. 1). These were identified on the basis of the adopted criteria that an ORF consists of at least 30 codons preceded by a potential Shine-Dalgarno sequence at an appropriate distance (6 to 15 bp) from one of the commonly used initiation codons (AUG, UUG, GUG) (13, 14, 30). The nucleotide sequences and the deduced amino acid sequences of the putative ORFs were compared to the contents of the available databases. Eleven of the 15 deduced proteins showed significant similarity to sequences in the databanks (see Table 2 for percent similarity). These ORFs are discussed below.
FIG. 1.
Schematic representation of the Tuc2009 genome showing putative ORFs, depicted as arrows, to which functions have been assigned. The direction of the arrow is the same as the direction of the ORF. cI, repressor; mp1 and mp2, major structural proteins. The enlarged section represents all identified ORFs contained within the pSK220 subclone. SSB, putative single-stranded-DNA-binding protein; Rep, putative replisome organizer (Rep2009); Meth′, putative methylase; Topo′, putative topoisomerase I.
TABLE 2.
General features of putative ORFs contained on the pSK220 subclone of Tuc2009 and comparison to sequences from databases
| ORF | Start position (codon) | Stop position (codon) | Size of protein (kDa) | Function or similarity | Pa | Match (%)b | Accession no. (reference) |
|---|---|---|---|---|---|---|---|
| 10 | 4935 (ATG) | 4660 (TAG) | 9.9 | Unknown | |||
| 11 | 5038 (ATG) | 5253 (TAG) | 8.4 | ORF71 from φ31 | 1e−35 | 98 (71) | U51128 (11) |
| 12 | 5255 (ATG) | 5353 (TGA) | 3.7 | Unknown | |||
| 13 | 5369 (ATG) | 5887 (TGA) | 20.1 | Unknown | |||
| 14 | 5896 (ATG) | 6519 (TAA) | 24.0 | ORF11 from TP901-1 | 1e−109 | 92 (207) | Y14232 (27) |
| Gene e12 from bil170 | 2e−60 | 58 (203) | AF009630 (direct submission) | ||||
| ORF35 from sk1 | 3e−62 | 61 (203) | AFO11378 (3) | ||||
| 15 | 6519 (ATG) | 6971 (TAA) | 16.6 | Single-stranded binding protein from: | |||
| B. subtilis | 1e−37 | 67 (110) | P37455 (34) | ||||
| φ PVL | 1e−35 | 51 (157) | AB009866 (20) | ||||
| H. influenzae | 6e−14 | 37 (105) | U04997 (18) | ||||
| E. coli | 7e−12 | 41 (117) | g70816 (38) | ||||
| 16 | 7096 (GTG) | 7878 (TAA) | 29.3 | ORF11 from r1t | 4e−10 | 30 (156) | U38906 (44) |
| 17 | 7878 (ATG) | 8603 (TGA) | 27.0 | Unknown | |||
| 18 | 8600 (ATG) | 9358 (TAA) | 29.4 | Methyltransferase from: | |||
| Bacillus stearothermophilus | 5e−15 | 28 (231) | X97069 (7) | ||||
| Streptococcus pneumoniae | 9e−10 | 25 (259) | P09358 (8) | ||||
| Escherichia coli | 4e−10 | 46 (65) | U48806 (31) | ||||
| 19 | 9348 (TTG) | 9737 (TAA) | 15.8 | ORF14 from r1t | 2e−70 | 97 (128) | U38906 (44) |
| 20 | 9741 (ATG) | 9980 (TAA) | 9.4 | ORF16 from r1t | 2e−37 | 91 (79) | U38906 (44) |
| 21 | 10092 (ATG) | 10271 (TAA) | 6.9 | ORF17 from r1t | 7e−28 | 100 (59) | U38906 (44) |
| 22 | 10261 (ATG) | 10830 (TAG) | 22.4 | ORF18 from r1t | 3e−85 | 83 (189) | U38906 (44) |
| 23 | 10841 (ATG) | 11236 (TAA) | 15.5 | ORF115 from φg1e | 2e−04 | 31 (124) | X98106 (25) |
| 24 | 11229 (ATG) | 11774 (TGA) | 21.0 | ORF1 from L. lactis | 3e−18 | 37 (193) | Y11901 (36) |
| ORF19 from r1t | 7e−06 | 34 (121) | U38906 (44) |
Derived from BLASTP score; P, probability that the observed similarity had occurred by chance. Low P values indicate a high probability of two proteins being structurally related.
The number of amino acids over which the percentage match was determined is shown in parentheses.
(i) ORF11.
The protein specified by ORF11 shows a high degree of similarity to the ORF71 protein product from φ31 (11). This protein has no known function.
(ii) ORF14.
The deduced protein product of ORF14 shows a high degree of similarity to the protein specified by ORF11 of the temperate lactococcal bacteriophage TP901-1, which in turn shows a low degree of similarity to topoisomerase I from two different Mycoplasma species (27). Topoisomerase I enzymes play a role in the relaxing of supercoiled DNA. The deduced ORF14 protein product also shows significant similarity to two other proteins of unknown function from the databases: ORF35 of the lytic lactoccal phage sk1 (3) and ORF14 of another lytic phage, bil170 (accession no AF009630).
(iii) ORF15.
The protein specified by ORF15 shows significant similarity to single-stranded-DNA-binding (SSB) proteins from a wide variety of sources; a number of examples are given in Table 2. SSB proteins, also called helix-destabilizing proteins, play a number of roles in DNA replication, repair, and recombination. One of their main functions is to stabilize the transiently formed single strands of DNA that are generated as the replication fork moves through the DNA during replication (29, 30).
(iv) ORF16.
The deduced protein product of ORF16, designated here as Rep2009, shows similarity to the protein specified by ORF11 of r1t (44) (30% identity), which in turn shows similarity to G38P of the Bacillus subtilis phage SPP1 (47) (26.7% identity). G38P has been demonstrated to function as a replisome organizer in SPP1 DNA replication. Replisome organizer proteins play a key role in the initiation event of DNA replication which occurs at a specific point on the genome, termed the origin of replication (ori), via a protein-DNA interaction (29). It has been demonstrated that a sequence containing several direct repeats (oriL) within gene38 of SPP1 functions as an ori. Sequences containing direct repeats can also be identified within ORF11 of r1t and ORF16 of Tuc2009. It is on this basis that we putatively identify the direct repeat-containing sequence within rep2009 as an ori for Tuc2009, designated ori2009. The deduced protein product of rep2009 shows a higher degree of similarity in the N-terminal region to the protein specified by ORF11 of r1t, whereas a higher degree of similarity is observed in the C-terminal region between this protein and G38P of SPP1. This may represent a modular-type conservation of the genetic material required to initiate DNA replication in the evolution of these phages.
(v) ORF18.
The protein specified by ORF18 shows similarity to type II methyltransferases from a variety of organisms, several examples of which are given in Table 2. This type of methyltransferase methylates exocyclic amino nitrogens on the DNA to form N6-methyladenine or N4-methylcytosine (28). The protein product specified by this ORF may be involved in the methylation of specific sites on newly replicated Tuc2009 DNA, thus protecting it from degradation by host-encoded endonucleases which recognize those sites. A similar methylase enzyme has previously been identified in φ50 (17). It has been suggested that φ50 acquired the structural methylase gene, LlaI, via an in vivo genetic exchange event between the bacteriophage resistance-conferring lactococcal plasmid pTR2030 and the φ50 genome. The acquisition of this functional methylase thus confers a selective advantage on φ50.
(vi) ORF19 to -22.
The deduced protein products of ORF19 to -22 show high degrees of similarity to the proteins specified by ORF14, -16, -17, and -18 of r1t, respectively (44). These proteins have no assigned function.
(vii) ORF23.
The protein specified by ORF23 shows similarity to the deduced protein product of ORF115 of a temperate lactobacillus phage, φg1e. This protein is of no known function.
(viii) ORF24.
The deduced protein product of ORF24 shows similarity to ORF19 of r1t (44) and ORF1 of Lactococcus lactis subsp. cremoris S114 (36). No function has been ascribed to these proteins.
rep2009 confers a Per phenotype.
It has been suggested previously that phage origins of replication, when supplied in trans, confer on the host a phage-resistant phenotype against specific phages (16, 35). To determine whether the putative origin of replication of Tuc2009, ori2009, had the ability to confer a Tuc2009-resistant phenotype on UC509.9, the following experiment was performed. The complete rep2009 gene was cloned on the high-copy-number plasmid pNZ8048. The resulting recombinant plasmid, pNZRep-1, was introduced into UC509.9. Plaque assay and lysis-in-broth experiments were then performed to determine if pNZRep-1 conferred a phage-resistant phenotype.
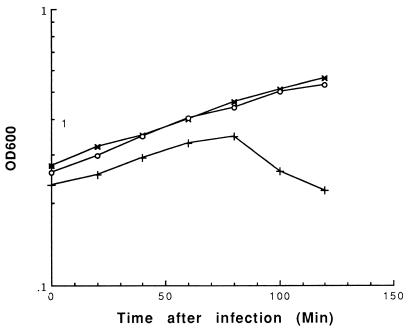
When present in UC509.9, pNZRep-1 strongly inhibited the ability of Tuc2009 to form plaques, the size of which was reduced from 1 to 1.5 mm to pinpoint. The efficiency of plaquing was determined to be 0.1 on UC509.9 harboring pNZRep-1. Lysis-in-broth experiments indicated that UC509.9 carrying pNZRep-1 exhibited an increased resistance to lysis by Tuc2009 at levels of infection that lysed control strains (Fig. 2).
FIG. 2.
Lysis-in-broth experiment. Phage was added at T = 0 at a multiplicity of infection of approximately 0.5, and the OD600 was measured over time. Increasing levels of Tuc2009 infection eventually caused lysis of UC509.9 harboring pNZRep-1 (data not shown). Symbols: ×, UC509.9(pNZRep-1) uninfected; ○, UC509.9(pNZRep-1) plus Tuc2009; +, UC509.9(pNZ8048) plus Tuc2009.
To verify that the observed Per phenotype was specifically due to the presence of the ori2009-containing DNA fragment and not just a general phenomenon displayed by phage fragments distributed throughout the Tuc2009 genome, and also to determine if the orientation of ori2009 had any effect on Per, two additional plasmids were constructed. (i) A 1.5-kb fragment of Tuc2009 DNA was selected randomly and cloned in pNZ8048 to create pNZ101. When UC509.9 harboring pNZ101 was challenged with Tuc2009, no difference in plaque size, efficiency of plaquing, or resistance to lysis in broth was observed when compared with these characteristics in UC509.9 harboring the control plasmid pNZ8048, indicating that the observed Per phenotype is specifically due to the rep2009 gene on pNZRep-1 (data not shown). (ii) The rep2009 gene was cloned in the reverse orientation in pNZ8048, to create pNZRep-3. When UC509.9 containing pNZRep-3 was challenged with Tuc2009, an phenotype identical to that of UC509.9 containing pNZRep-1 was observed, indicating that the phage resistance conferred by ori2009 is orientation independent (data not shown).
The presence of pNZRep-1 affects intracellular Tuc2009 DNA replication.
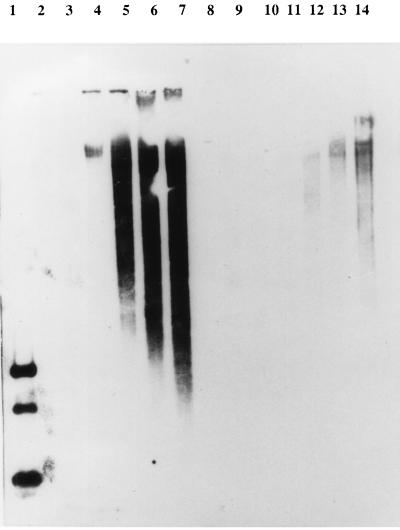
It has been shown for φ31 and φ50 that Per-conferring DNA fragments interfere with intracellular phage DNA replication (16, 35). To determine if this was the case for Tuc2009, total DNA was isolated at 20-min time intervals from two separate phage-infected UC509.9 cultures, one harboring pNZRep-1 and the other harboring the control plasmid pNZ8048. Samples of these DNA preparations were then subjected to agarose gel electrophoresis, followed by Southern blot analysis using a 1.9-kb Tuc2009 DNA PCR-generated fragment, containing ORF11 to -15, as a probe. As can be seen from Fig. 3, following infection of the control strain (time [T] = 0), Tuc2009 DNA replication commences at T = 40 min, followed by a dramatic increase until cell lysis arrests further replication at T = 60 to 80 min (lanes 4 to 7). In contrast, Tuc2009 DNA replication in strain UC509.9 harboring pNZRep-1 is not apparent until T = 60 min and remains at a level that is severely reduced in comparison to that of the control strain with no apparent lysis (lanes 12 to 14).
FIG. 3.
Southern blot analysis showing the effect of pNZRep-1 on intracellular Tuc2009 DNA replication. Lanes: 1, positive control (RsaI digest of Tuc2009 genomic DNA); 2 to 7, total cellular DNA, isolated at 20-min intervals, from Tuc2009-infected UC509.9 harboring pNZ8048 (lane 2, T = 0); 8, blank; 9 to 14, total cellular DNA, isolated at 20-min intervals, from Tuc2009-infected UC509.9 harboring pNZRep-1 (lane 9, T = 0). Tuc2009 was added at a multiplicity of infection of approximately 0.1. The blot was probed with a 1.9-kb PCR-generated Tuc2009 DNA fragment.
Delineation of the Per phenotype-conferring region.
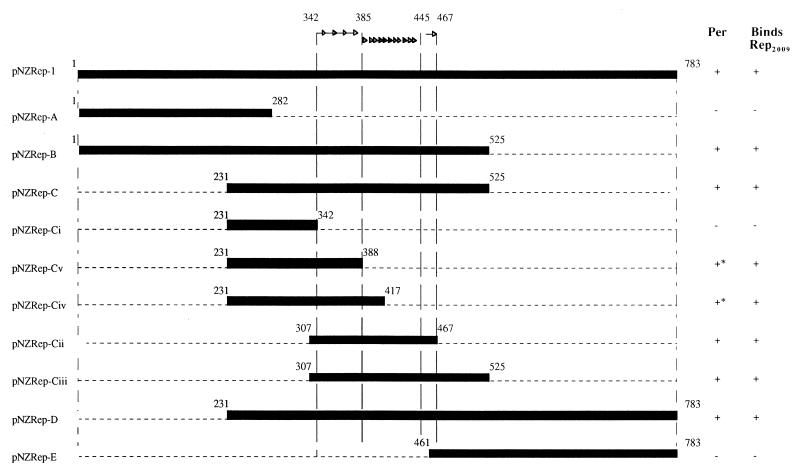
To determine the smallest DNA fragment responsible for conferring the Tuc2009 resistance phenotype, DNA fragments generated by PCR, each spanning a different portion of the rep2009 gene, were cloned separately into pNZ8048 and examined for their Per effect. These experiments indicated that the resistance-conferring region, located within rep2009, could be assigned to an internal 160-bp fragment rich in direct repeats, essentially the region that had been previously designated ori2009 (Fig. 4).
FIG. 4.
Diagram showing the experiment performed to determine the minimum DNA fragment from rep2009 that confers a Per phenotype on UC509.9. Synthetic oligonucleotide primers were designed to generate subfragments spanning the rep2009 gene by PCR. These fragments were then cloned in pNZ8048 and introduced into UC509.9. Plaque assay and lysis-in-broth experiments were performed to determine which fragments conferred a Per phenotype, which is indicated in the column labelled Per. Radioactive probes, made from these rep2009 subfragments, were used in gel retardation experiments to determine if the Rep2009 protein was capable of binding to the fragments (column labelled Binds Rep2009). Arrows indicate the positions of the repeated sequences contained within ori2009. ∗, the Per effect conferred by this DNA fragment was not as strong as that conferred by other Per+ DNA fragments.
Rep2009 specifically interacts with ori2009.
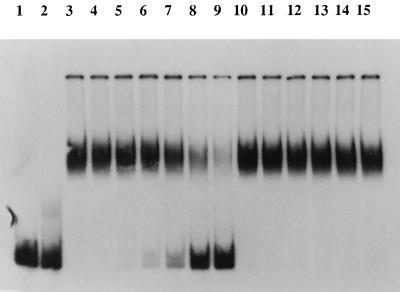
Initiation of DNA replication in SPP1 requires the interaction of G38P with oriL, which is located within gene 38 on the SPP1 genome (32, 47). To determine if a similar DNA-protein interaction between the Rep2009 protein and ori2009 occurs, a series of gel retardation experiments was performed. Crude cell extracts from L. lactis NZ9800 expressing Rep2009 and a γ-32P-labelled 160-bp DNA fragment representing ori2009 were used for these experiments. As can be seen in Fig. 5, the presence of Rep2009 in the cell extract decreases the electrophoretic mobility of the ori2009 DNA fragment in a polyacrylamide gel, indicative of a protein-DNA interaction. To determine if this protein-DNA interaction was specific, increasing amounts of unlabelled ori2009 DNA fragments or of a nonspecific DNA fragment of approximately the same size were incorporated into the binding reaction mixture. As can be seen in Fig. 5 (lanes 4 to 9), increasing amounts of unlabelled ori2009 DNA fragment resulted in the retardation of decreased amounts of the labelled ori2009 DNA fragment to the lower-mobility position, presumably because of competitive binding of the Rep2009 protein to the unlabelled ori2009 DNA fragment. In contrast, addition of comparable amounts of the unlabelled nonspecific DNA fragment did not cause changes in the mobility profile of the labelled ori2009 DNA fragment. These experiments clearly demonstrate that the binding of Rep2009 to ori2009 is the result of a highly specific interaction. Interestingly, the ability of a fragment to confer a Per phenotype always coincided with its ability to interact with the Rep2009 protein. Likewise, fragments that do not confer a Per phenotype were shown to not interact with Rep2009 (Fig. 4).
FIG. 5.
Gel retardation assay showing that the binding between the Rep2009 protein and ori2009 is the result of a specific interaction. Gel retardation was performed as described in Materials and Methods. An 0.3-ng amount of 32P-labeled ori2009 DNA fragment (160-bp Per-conferring region) was used as a probe. Lanes: 1, probe only; 2, probe plus cell extract (2.5 μg of total protein) from L. lactis NZ9800 harboring pNZ8048 (negative control); 3 to 15, probe plus cell extract (2.5 μg of total protein) from L. lactis NZ9800 containing overexpressed Rep2009 protein (approximate Rep2009 protein molecular mass, 29 kDa). Lanes 4 to 9 represent binding assays performed with the labelled ori2009 probe, cell extract, and 1.2, 2.4, 4.8, 9.6, 19.2, and 38.4 ng of unlabelled ori2009 DNA fragment, respectively. Lanes 10 to 15 represent binding assays performed with the labelled ori2009 probe, cell extract, and 1.2, 2.4, 4.8, 9.6, 19.2, and 38.4 ng of an unlabelled nonspecific DNA fragment of approximately the same size as the labelled DNA fragment, respectively.
Random mutations in ori2009 can affect Per but do not eliminate the Rep2009-ori2009 interaction.
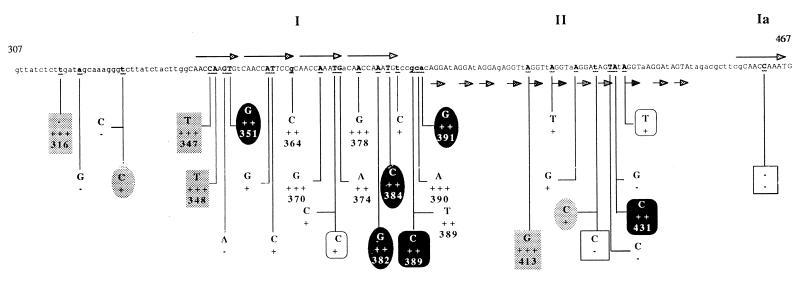
To identify the individual nucleotides within ori2009 which are responsible for conferring the Tuc2009 resistance phenotype, and also to determine if the Per effect is a direct consequence of the Rep2009-ori2009 interaction, random mutations were introduced into ori2009 DNA fragments. A PCR protocol with conditions such that point mutations or deletions occurred with a relatively high frequency was employed (see Materials and Methods). DNA fragments generated in this way were cloned in pNZ8048, introduced into UC509.9, and then tested for their ability to confer a Per phenotype. A number of clones that conferred a weakened level of Tuc2009 resistance on UC509.9 relative to that of the wild-type ori2009 DNA fragment were identified. These clones along with other randomly selected clones that displayed no loss of phage resistance were subjected to DNA sequence analysis. A number of individual clones containing one or more point mutations or deletions that caused a reduction in the efficiency of the Tuc2009-resistant phenotype conferred were identified. In total, 23 clones containing mutations were identified (Fig. 6). Mutations that had the greatest effect on Per were localized around a series of four direct repeats, designated here as region I. The main feature of region I is the presence of two imperfect repeated sequences followed by two perfect repeated sequences. A third copy of the latter sequence is located at the 3′ end of the ori2009 DNA fragment and is designated here as region Ia. One mutation was identified in this region, and this had no effect on Per. Mutations in the series of 11 imperfect direct repeats of 4 nucleotides, designated here as region II, appear to have had little or no effect on the Per phenotype, unless they were accompanied by other mutations in region I. Gel retardation experiments using crude cell lysates from L. lactis NZ9800 expressing Rep2009 and a number of probes representing the randomly mutated versions of the ori2009 DNA fragment were performed. All mutated fragments that completely lost the ability to confer a Per phenotype along with a number of randomly selected mutated fragments that displayed only a slight reduction or no reduction in the Per phenotype conferred were examined in this way. Surprisingly, Rep2009 was found to bind all fragments examined, with an apparent similar affinity. This suggests that the observed Per phenotype of UC509.9 cells harboring pNZRep-1 is not solely a consequence of limitation of Rep2009 binding to ori2009 and that at least one other interaction must take place at ori2009.
FIG. 6.
Diagram showing the DNA sequence of the minimum rep2009 gene subclone, designated ori2009, that confers a Per phenotype on UC509.9. Nucleotides contained in the direct repeats are shown as uppercase letters. Arrows indicate repeated sequences. Underlined nucleotides indicate positions where random nucleotide substitutions or deletions occurred; nucleotides substituted or deleted at that position are indicated as is the effect that the mutations had on Per. Nucleotide substitutions or deletions that are boxed in the same manner indicate positions where more than one mutation has occurred in one fragment, e.g., three nucleotide substitutions (C-T at position 347, A-T at position 348, and A-G at position 413) and one nucleotide deletion (T- − at position 316) are highlighted with a shaded box. This indicates that these four mutations occur on a single DNA fragment. −, mutation had no effect on Per; +, mutation caused slight loss of Per; ++, mutation caused severe loss of Per; +++, mutation caused complete loss of Per.
DISCUSSION
Initiation of DNA replication requires a specific starting point where opening of the double-stranded double helix takes place prior to the recruitment of the replication machinery. The interaction of a sequence-specific duplex DNA-binding protein with a series of direct repeated sequences in DNA represents the primary event for the initiation of replication. These repeated sequences form the origin of replication (ori). The resulting nucleoprotein binding complexes generally consist of 150 to 250 bp of DNA and multimers of the DNA-binding proteins. Subsequently, an A+T-rich region of DNA directly adjacent to the ori becomes denatured and presumably coated with SSB protein. By so marking the origin region and providing available single-stranded DNA, the replication proteins can be recruited to the correct initiation point and nascent strand synthesis can ensue. Examples of this type of initiation of DNA replication include the binding of DnaA to oriC and of λ-O to oriλ in E. coli and λ, respectively (29).
A series of direct repeats was identified within the rep2009 of Tuc2009 as has been shown for oriL, which is contained within gene 38 of SPP1 (32, 47). The Rep2009 protein was putatively identified as a replisome organizer protein through sequence similarities with the deduced protein product of ORF11 of r1t and G38P of SPP1. It has been demonstrated that initiation of DNA replication in SPP1 requires the binding of G38P to oriL. When a piece of DNA encompassing ori2009 is supplied in trans on a high-copy-number plasmid vector, it confers an abortive infection-type resistance on UC509.9 against Tuc2009, similar to that which has been previously reported for φ50 and φ31 (16, 35). We have demonstrated that intracellular Tuc2009 DNA replication is severely inhibited in UC509.9 harboring these plasmids. We have also demonstrated a protein-DNA interaction between the Rep2009 protein and the specific portion of the rep2009 gene that confers Per, ori2009. These data would indicate that the observed phage resistance is due to the titration of Rep2009 proteins away from the true ori on the phage DNA and towards the ori2009 DNA fragments supplied on the plasmid vectors.
However, when DNA fragments representing versions of the ori2009 DNA fragment that no longer conferred a Per phenotype due to the introduction of random mutations in the DNA sequence were used as probes in gel retardation experiments, no decrease in the ability of Rep2009 binding was observed. Data from the mutational analysis of ori2009 lead us to conclude that the DNA region encompassing four direct repeats designated as region I is the most significant contributor to the Per phenotype (Fig. 6). This finding is in good agreement with experimental results which showed that fragments containing deleted versions of other repeated sequences from ori2009 confer a Per phenotype only slightly reduced relative to that of fragments containing all the repeat-containing regions (Fig. 4). Interestingly, gel retardation experiments displayed no discernible difference in the ability of the Rep2009 protein to bind to probes representing ori2009 or these two deleted fragments.
The finding that ori2009 and equivalent sequences in ORF11 of r1t and oriL for SPP1 are located within the genes coding for putative replisome organizer proteins suggests a modular-type evolution of the genetic material necessary for directing initiation of DNA replication in the respective phages. This feature may also represent a means of self-regulating the expression of these proteins, whereby the binding of the protein to the ori contained within the gene could arrest the progression of the RNA polymerase.
On the basis of the current information available, a detailed model for Tuc2009 DNA replication remains unclear. However, we can putatively assign functions to some of the ORFs identified. The protein specified by ORF14 of Tuc2009 shows a high degree of similarity to the deduced protein product of ORF11 of TP901-1; this protein in turn shows low similarity to topoisomerase I from two different Mycoplasma species. The protein product of ORF14 of Tuc2009 may represent a highly diverged type of topoisomerase I enzyme. These enzymes are responsible for the unwinding of supercoiled DNA, thereby making the double helix accessible to the replication machinery.
The role of SSB proteins in DNA replication, as in DNA recombination and repair, is well understood. In DNA replication, they serve to stabilize the single strands of DNA that are transiently formed as the replication fork moves through the double helix (29). The deduced protein product of ORF15 shows a high degree of homology to these proteins, and therefore we assume that this protein does play the role of SSB proteins in Tuc2009 DNA replication.
The protein specified by ORF18 of Tuc2009 shows similarity to a range of type II methyltransferases; this protein most likely plays a role in methylating newly replicated DNA, thus protecting it from degradation by host-encoded endonucleases. It is possible, as has been suggested for φ50 (17), that the incorporation of this methylase into the Tuc2009 genome is the result of a recombinogenic event between the host and phage genomic DNA.
We envisage that the Rep2009 protein plays the part of replisome organizer in Tuc2009 DNA replication. We propose that the series of direct repeats contained within the rep2009 gene sequence ori2009 represents a possible ori for Tuc2009 and that DNA replication in Tuc2009 is initiated by the interaction of Rep2009 with ori2009. The results from gel retardation experiments with the mutated ori2009 DNA fragments that no longer conferred a Per phenotype but retained the ability to bind Rep2009 indicate that ori2009 is the target for at least one other phage-encoded protein. The titration of this protein (or proteins) appears to be responsible for the Per phenotype, and future work will be aimed at the identification of this important replication factor.
ACKNOWLEDGMENTS
We thank Aine Healy and Sinead Geary for their contributions to oligonucleotide synthesis and DNA sequence analysis, and we thank Liam Burgess for photographic work. We also thank Maarten van de Guchte for the construction of plasmid pSK220 and Michiel Kleerebezem for supplying plasmid pNZ8048.
This work was supported by a European Community Biotechnology grant (contract BIO4-CT96-0402). Douwe van Sinderen is the recipient of an EMBO fellowship (ALTF 431-1995).
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search program. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arendt E K, Daly C, Fitzgerald G F, van de Guchte M. Molecular characterization of lactococcal bacteriophage Tuc2009 and identification and analysis of genes encoding lysin, a putative holin, and two structural proteins. Appl Environ Microbiol. 1994;60:1875–1883. doi: 10.1128/aem.60.6.1875-1883.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chandry P S, Moore S C, Boyce J D, Davidson B E, Hillier A J. Analysis of the DNA sequence, gene expression, origin of replication and modular structure of the Lactococcus lactis lytic bacteriophage sk1. Mol Microbiol. 1997;26:49–64. doi: 10.1046/j.1365-2958.1997.5491926.x. [DOI] [PubMed] [Google Scholar]
- 4.Costello V A. Characterization of bacteriophage-host interactions in Streptococcus cremoris UC503 and related lactic streptococci. Ph.D. thesis. Cork, Ireland: National University of Ireland, University College; 1988. [Google Scholar]
- 5.Daly C, Fitzgerald G F, Davis R. Biotechnology of lactic acid bacteria with special reference to bacteriophage resistance. Antonie Leeuwenhoek. 1996;70:99–110. doi: 10.1007/BF00395928. [DOI] [PubMed] [Google Scholar]
- 6.Dartois V, Debarbouille M, Kunst F, Rapoport G. Characterization of a novel member of the degS-degU regulon affected by salt stress in Bacillus subtilis. J Bacteriol. 1998;180:1855–1861. doi: 10.1128/jb.180.7.1855-1861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Degtyarev S K, Netesova N A, Abdurashitov M A, Shevchenko A V. Primary structure and strand specificity of BstF5I-1 DNA methyltransferase which recognises 5′-GGATG-3′. Gene. 1997;187:217–219. doi: 10.1016/s0378-1119(96)00752-4. [DOI] [PubMed] [Google Scholar]
- 8.de la Campa A G, Kale P, Springhorn S S, Lacks S A. Proteins encoded by the DpnII restriction gene cassette. Two methylases and an endonuclease. J Mol Biol. 1987;196:457–469. doi: 10.1016/0022-2836(87)90024-6. [DOI] [PubMed] [Google Scholar]
- 9.de Ruyter P G G A, Kuipers O P, de Vos W M. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl Environ Microbiol. 1996;62:3662–3667. doi: 10.1128/aem.62.10.3662-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dinsmore P K, Klaenhammer T R. Bacteriophage resistance in Lactococcus. Mol Biotechnol. 1995;4:297–314. doi: 10.1007/BF02779022. [DOI] [PubMed] [Google Scholar]
- 11.Dinsmore P K, Klaenhammer T R. Molecular characterization of a genomic region in a Lactococcus bacteriophage that is involved in its sensitivity to the phage defense mechanism AbiA. J Bacteriol. 1997;179:2949–2957. doi: 10.1128/jb.179.9.2949-2957.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Djordjevic G M, O’Sullivan D J, Walker S A, Conkling M A, Klaenhammer T R. A triggered-suicide system designed as a defense against bacteriophages. J Bacteriol. 1997;179:6741–6748. doi: 10.1128/jb.179.21.6741-6748.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garvey P, van Sinderen D, Twomey D P, Hill C, Fitzgerald G F. Molecular genetics of bacteriophage and natural phage defense systems in the genus Lactococcus. Int Dairy J. 1995;5:905–947. [Google Scholar]
- 14.Harley C B, Reynolds R P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987;15:2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hill C. Bacteriophage and bacteriophage resistance in lactic acid bacteria. FEMS Microbiol Rev. 1993;12:87–108. [Google Scholar]
- 16.Hill C, Miller L A, Klaenhammer T R. Cloning, expression, and sequence determination of a bacteriophage fragment encoding bacteriophage resistance in Lactococcus lactis. J Bacteriol. 1990;172:6419–6426. doi: 10.1128/jb.172.11.6419-6426.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hill C, Miller L A, Klaenhammer T R. In vivo genetic exchange of a functional domain from a type II A methylase between lactococcal plasmid pTR2030 and a virulent bacteriophage. J Bacteriol. 1991;173:4363–4370. doi: 10.1128/jb.173.14.4363-4370.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jarosik G P, Hansen E J. Cloning and sequencing of the Haemophilus influenzae ssb gene encoding single-strand DNA-binding protein. Gene. 1994;146:101–103. doi: 10.1016/0378-1119(94)90841-9. [DOI] [PubMed] [Google Scholar]
- 19.Jarvis A W. Bacteriophages of lactic acid bacteria. J Dairy Sci. 1989;72:3406–3428. [Google Scholar]
- 20.Kaneko J, Kimura T, Kawakami Y, Tomita T, Kamio Y. Panton-Valentine leukocidin genes in a phage-like particle isolated from mitomycin C-treated Staphylococcus aureus V8 (ATCC 49775) J Biosci Biotechnol Biochem. 1997;61:1960–1962. doi: 10.1271/bbb.61.1960. [DOI] [PubMed] [Google Scholar]
- 21.Kim S G, Batt C A. Antisense mRNA-mediated bacteriophage resistance in Lactococcus lactis subsp. lactis. Appl Environ Microbiol. 1991;57:1109–1113. doi: 10.1128/aem.57.4.1109-1113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim S G, Bor Y C, Batt C A. Bacteriophage resistance in Lactococcus lactis ssp. lactis using antisense ribonucleic acid. J Dairy Sci. 1992;75:1761–1767. doi: 10.3168/jds.S0022-0302(92)77935-1. [DOI] [PubMed] [Google Scholar]
- 23.Klaenhammer T R. Interactions of bacteriophages with lactic streptococci. Adv Appl Microbiol. 1984;30:1–29. [Google Scholar]
- 24.Klaenhammer T R, Fitzgerald G F. Bacteriophage and bacteriophage resistance. In: Gasson M J, de Vos W M, editors. Genetics and biotechnology of lactic acid bacteria. Glasgow, United Kingdom: Blackie Academic and Professional; 1994. pp. 106–168. [Google Scholar]
- 25.Kodaira K I, Oki M, Kakikawa M, Watanabe N, Hirakawa M, Yamada K, Taketo A. Genome structure of the Lactobacillus temperate phage phi g1e: whole genome sequence and the putative promoter/repressor system. Gene. 1997;187:45–53. doi: 10.1016/s0378-1119(96)00687-7. [DOI] [PubMed] [Google Scholar]
- 26.Lillehaug D. An improved plaque assay for poor plaque-producing temperate lactococcal bacteriophages. J Appl Microbiol. 1997;83:85–90. doi: 10.1046/j.1365-2672.1997.00193.x. [DOI] [PubMed] [Google Scholar]
- 27.Madsen P L, Hammer K. Temporal transcription of the lactococcal temperate phage TP901-1 and DNA sequence of the early promoter region. Microbiology. 1998;144:2203–2215. doi: 10.1099/00221287-144-8-2203. [DOI] [PubMed] [Google Scholar]
- 28.Malone T, Blumenthal R M, Cheng X. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases and suggests a catalytic mechanism for these enzymes. J Mol Biol. 1995;253:618–632. doi: 10.1006/jmbi.1995.0577. [DOI] [PubMed] [Google Scholar]
- 29.Marians K J. Prokaryotic DNA replication. Annu Rev Biochem. 1992;61:673–719. doi: 10.1146/annurev.bi.61.070192.003325. [DOI] [PubMed] [Google Scholar]
- 30.Meyer R R, Laine P S. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990;54:342–380. doi: 10.1128/mr.54.4.342-380.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mise K, Nakajima K. Isolation of restriction enzyme EcoVIII, an isoschizomer of HindIII, produced by Escherichia coli E1585-68. Gene. 1984;30:79–85. doi: 10.1016/0378-1119(84)90107-0. [DOI] [PubMed] [Google Scholar]
- 32.Missich R, Weise F, Chai S, Lurz R, Xiomara P, Alonso J C. The replisome organiser (G38P) of Bacillus subtilis bacteriophage SPP1 forms specialised nucleoprotein complexes with two discrete distant regions of the SPP1 genome. J Mol Biol. 1997;270:50–64. doi: 10.1006/jmbi.1997.1060. [DOI] [PubMed] [Google Scholar]
- 33.Moran C P, Lang N, Le Grice S F J, Lee G, Stephens M, Sonenshein A L, Pero J, Losick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilus. Mol Gen Genet. 1982;186:339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- 33a.National Centers for Biotechnology Information. 24 November 1997, copyright date. [Online.] http://www.ncbi.nlm.nih.gov. [18 February 1999, last date accessed.]
- 34.Ogasawara N, Nakai S, Yoshikawa H. Systematic sequencing of the 180 kilobase region of the Bacillus subtilis chromosome containing the replication origin. DNA Res. 1994;1:1–14. doi: 10.1093/dnares/1.1.1. [DOI] [PubMed] [Google Scholar]
- 35.O’Sullivan D J, Hill C, Klaenhammer T R. Effect of increasing the copy number of bacteriophage origins of replication, in trans, on incoming phage proliferation. Appl Environ Microbiol. 1993;59:2449–2456. doi: 10.1128/aem.59.8.2449-2456.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prevots F, Tolou S, Delpech B, Kaghad M, Daloyau M. Nucleotide sequence and analysis of the new chromosomal abortive infection gene abiN of Lactococcus lactis subsp. cremoris S114. FEMS Microbiol Lett. 1998;159:331–336. doi: 10.1111/j.1574-6968.1998.tb12879.x. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 38.Sancar A, Williams K R, Chase J W, Rupp W D. Sequences of the ssb gene and protein. Proc Natl Acad Sci USA. 1981;78:4274–4278. doi: 10.1073/pnas.78.7.4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanders M E, Leonhard P J, Sing W D, Klaenhammer T R. Conjugal strategy for construction of fast acid-producing, bacteriophage-resistant lactic streptococci for use in dairy fermentations. Appl Environ Microbiol. 1986;52:1001–1007. doi: 10.1128/aem.52.5.1001-1007.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 41.Stanley E, Fitzgerald G F, Le Marrec C, Fayard B, van Sinderen D. Sequence analysis and characterization of φO1205, a temperate bacteriophage infecting Streptococcus thermophilus CNRZ1205. Microbiol. 1997;143:3417–3429. doi: 10.1099/00221287-143-11-3417. [DOI] [PubMed] [Google Scholar]
- 42.Terzaghi B E, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van de Guchte M, Daly C, Fitzgerald G F, Arendt E K. Identification of the putative repressor-encoding gene cI of the temperate lactococcal bacteriophage Tuc2009. Gene. 1994;144:93–95. doi: 10.1016/0378-1119(94)90209-7. [DOI] [PubMed] [Google Scholar]
- 44.van Sinderen D, Karsens H, Kok J, Terpstra P, Ruiters M H J, Venema G, Nauta A. Sequence analysis and molecular characterization of the temperate lactococcal bacteriophage r1t. Mol Microbiol. 1996;19:1343–1355. doi: 10.1111/j.1365-2958.1996.tb02478.x. [DOI] [PubMed] [Google Scholar]
- 44a.van Sinderen, D. Unpublished results.
- 45.Wells J M, Wilson P W, Le Page R W F. Improved cloning vectors and transformation procedure for Lactococcus lactis. J Appl Bacteriol. 1993;74:629–636. doi: 10.1111/j.1365-2672.1993.tb05195.x. [DOI] [PubMed] [Google Scholar]
- 46.Whitehead H R, Cox G A. The occurrence of bacteriophage in lactic streptococci. NZ J Dairy Sci Technol. 1935;16:319–320. [Google Scholar]
- 47.Xiomara P, Weise F, Chai S, Luder G, Alonso J C. Analysis of cis and trans acting elements required for the initiation of DNA replication in the Bacillus subtilis bacteriophage SPP1. J Mol Biol. 1994;236:1324–1340. doi: 10.1016/0022-2836(94)90061-2. [DOI] [PubMed] [Google Scholar]