Summary
Pseudomonas aeruginosa infections can be difficult to treat and new therapeutics are needed. Bacteriophage therapy is a promising alternative to traditional antibiotics, but large numbers of isolated and characterized phages are lacking. We collected 23 diverse P. aeruginosa isolates from people with cystic fibrosis (CF) and clinical infections, and used them to screen and isolate over a dozen P. aeruginosa-targeting phages from hospital wastewater. Phages were characterized with genome sequencing, comparative genomics, and lytic activity screening against all 23 bacterial host isolates. We evolved bacterial mutants that were resistant to phage infection for four different phages, and used genome sequencing and functional analysis to study them further. We also tested phages for their ability to kill P. aeruginosa grown in biofilms in vitro and ex vivo on CF airway epithelial cells. Overall, this study demonstrates how systematic genomic and phenotypic characterization can be deployed to develop bacteriophages as precision antibiotics.
Subject areas: Microbiology, Virology
Graphical abstract
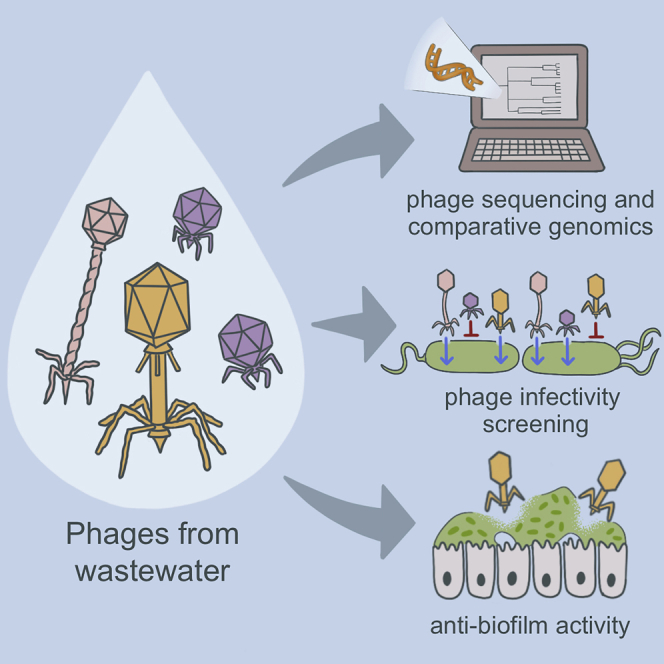
Highlights
-
•
P. aeruginosa-targeting bacteriophages reflect the diversity of their hosts
-
•
Phage resistance drives evolutionary trade-offs in virulence and drug resistance
-
•
Phages can kill bacteria grow in biofilms in vitro and ex vivo
Microbiology; Virology
Introduction
The evolution of multidrug-resistant bacteria continues to outpace the development of new antimicrobials, posing a serious threat to public health. Rates of infection and mortality because of antibiotic-resistant pathogens are continuing to grow in the United States and around the world, despite efforts to curtail their spread ((CDC, 2019); Friedman et al., 2016). Compounding the rise of multidrug-resistant bacterial infections, antibiotic development pipelines at many pharmaceutical companies have slowed or run dry (Singer et al., 2019). To help curtail this growing public health crisis, innovative approaches to antimicrobial therapy are needed.
Pseudomonas aeruginosa is a Gram-negative bacterium that causes a variety of infections, including bacteremia and pneumonia (Moradali et al., 2017). P. aeruginosa chronically colonizes the airways of people with cystic fibrosis (CF), and is associated with increased morbidity and mortality in CF individuals (Hoiby et al., 2010). The P. aeruginosa species encompasses a wide breadth of genomic and phenotypic diversity, and multidrug-resistant strains often evolve during the course of prolonged antibiotic treatment (Winstanley et al., 2016). The success of P. aeruginosa as an opportunistic pathogen, its propensity for developing drug resistance, and the major threat it poses to CF patients, are compelling reasons to develop new and more effective therapies to treat P. aeruginosa infections.
The urgent need for alternative antibacterial strategies has prompted clinicians and scientists to reconsider the use of bacteriophage therapy (Domingo-Calap and Delgado-Martinez, 2018), particularly for treating infections that cannot be resolved with antibiotics alone (Schooley et al., 2017). Recent advances in genomics and genetic engineering have facilitated the development of phage-based therapies that have proven successful in clinical settings (Schooley et al., 2017), including the treatment of P. aeruginosa infections (Aslam et al., 2019, 2020; Khatami et al., 2021; Onsea et al., 2019; Rubalskii et al., 2020; Trend et al., 2017). Here, we used a genetically diverse panel of 23 P. aeruginosa clinical isolates, collected mostly from CF patients, to isolate over a dozen distinct bacteriophages from hospital wastewater. We characterized the genomic and phenotypic diversity of the bacterial isolates and phages, including a subset of evolved phage-resistant bacterial mutants. We also tested the ability of some of the isolated phages to clear bacterial biofilms in vitro and ex vivo. Overall, our findings increase our understanding of the relationship between phages, bacteria, and mammalian hosts in the context of human infection. They can also aid in the rational design of tailored, phage-based therapies for the treatment of P. aeruginosa infections.
Results
P. aeruginosa clinical isolates used for phage screening are genetically and phenotypically diverse
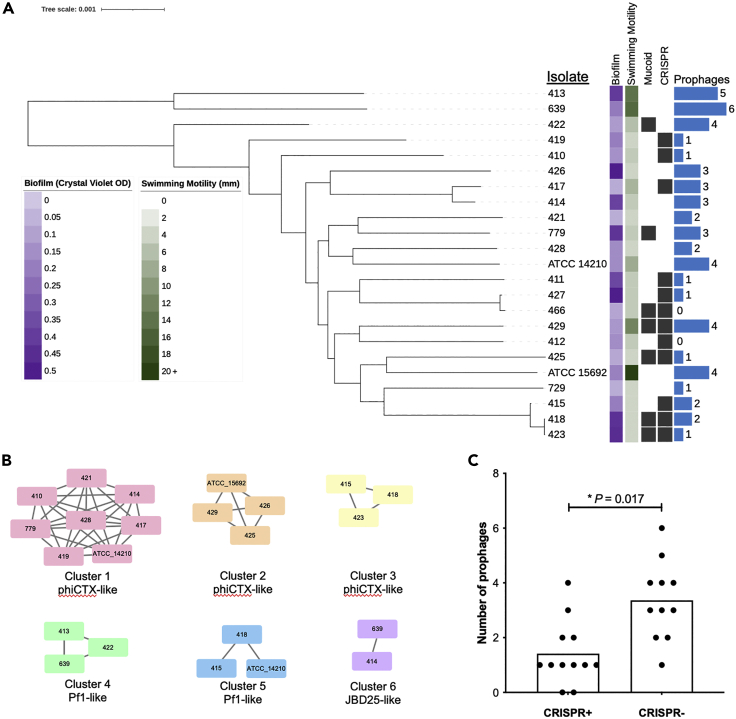
To isolate bacteriophages that could be maximally useful for the treatment of P. aeruginosa infections, we assembled a genetically and phenotypically diverse panel of 23 P. aeruginosa isolates collected from clinical sources (Table S1). Two isolates were purchased from the American Type Culture Collection (ATCC), 20 isolates were collected from adults with cystic fibrosis (CF), and one isolate was collected from the sputum of a hospitalized non-CF patient. All isolates were collected from different patients. The genome of each isolate was sequenced on both the Illumina and Oxford Nanopore MinION platforms, and the resulting sequencing data were hybrid assembled (Wick et al., 2017). Over half (14/23) of the genomes were closed to a single chromosome, and the remaining nine assemblies each contained 10 or fewer contigs (Table S1). Among the 23 isolates, a total of 19 different multi-locus sequence types (STs) were identified (Table S1). A core genome phylogeny of all 23 isolates confirmed that they were highly genetically diverse (Figure 1A). All isolates were tested for their ability to form in vitro biofilms, exhibit swimming motility, and display a mucoid phenotype when grown on LB and Pseudomonas isolation agars. These phenotypes were found to be variable among the collected isolates (Figure 1A), demonstrating that the assembled isolate panel was both genetically and phenotypically diverse.
Figure 1.
Diverse P. aeruginosa clinical isolates used for bacteriophage isolation and screening
(A) Core genome phylogeny of 23 P. aeruginosa isolates used for phage isolation. Isolates were typed for biofilm formation (measured as crystal violet staining intensity, OD550nm), swimming motility (measured as colony diameter, in mm), mucoidy, CRISPR-Cas presence, and prophage abundance. Black squares show the presence of binary features. Scale bar shows number of nucleotide substitutions per site.
(B) Clusters of similar prophages found in the genomes of different P. aeruginosa isolates. Bacterial isolate names are listed inside the nodes of each cluster, and lines connect prophages that share >90% sequence coverage and >90% sequence identity.
(C) Prophage abundance in isolates that are (CRISPR+) or are not (CRISPR-) predicted to encode functional CRISPR-Cas systems. Bars show mean values and p-value is from a two-tailed t-test.
We next assessed the abundance and diversity of prophage sequences in the 23 P. aeruginosa clinical isolates we collected. The genome of each isolate was mined for prophage sequences using the PHASTER online tool (Arndt et al., 2016). Between 0 and six prophages were found in each isolated genome (Figures 1A and Table S2). Prophages varied in length from 5.5 to 74.5 kb and in GC-content from 52.5 to 66.0%. A total of 54 prophage sequences were extracted, and were compared to one another using nucleotide BLAST to assess both the nucleotide identity and coverage across all pairwise comparisons (Figure 1B). A total of six different prophages were found to be present in more than one isolate; these included three distinct phiCTX-like phages and two Pf1-like filamentous phages. Finally, we assessed the number of prophages in isolates that were predicted to have either functional or non-functional Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) loci, as identified by CRISPRCasFinder and DefenseFinder (Table S1). We found that the 11 isolates predicted to have non-functional CRISPR-Cas systems had more prophages compared to isolates with intact CRISPR-Cas loci (Figure 1C, p = 0.017).
P. aeruginosa-targeting bacteriophages isolated from hospital wastewater
We used the 23 P. aeruginosa isolates we collected to screen for lytic bacteriophages in wastewater effluent collected from a Pittsburgh area hospital. A total of 14 phages were isolated on 10 different P. aeruginosa isolates (Table 1). One additional phage, PB1, was purchased from ATCC and was propagated and characterized alongside the newly isolated phages. Because sequencing of the genome of this phage revealed multiple mutations when compared to the PB1 sequence deposited in NCBI, we refer to it here as PSA07/PB1. Phages were picked and repeatedly passaged as single plaques, and were then amplified to generate high-titer stocks. Genomic DNA was extracted from each phage stock, and was sequenced on the Illumina platform. Phage genomes were found to be between 43.7 and 65.9 kb in length and had GC-content ranging from 44.9 to 64.5% (Table 1). Phages were compared to publicly available genomes using PHASTER and NCBI BLAST, and the predicted family and genus of each phage were determined based on similarity to previously described phages. Despite appearing to be lytic on the isolates used to propagate them, three phages (PSA04, PSA20, and PSA21) were predicted to have a lysogenic lifestyle because of the presence of annotated phage integrases. The PSA04 genome was most similar to the JBD44 lysogenic phage (Bondy-Denomy et al., 2016), however the homology between these two phages was not particularly high (Table 1). The PSA20 and PSA21 phage genomes showed moderate sequence similarity to the Yuavirus phages AN14 and LKO4, in which the putative integrase is believed to be a DNA primase (Evseev et al., 2020). The lack of previously described lysogenic activity among Yuavirus phages is consistent with our observations of lytic behavior for phages PSA20 and PSA21.
Table 1.
Genome characteristics of P. aeruginosa-targeting bacteriophages
| Phage | Sourcea | Host | Length (bp) | %GC | Predicted Family | Predicted Genus | NCBI Similar Phage (Accession)b | Coverage/Identity | BioSample |
|---|---|---|---|---|---|---|---|---|---|
| PSA04 | HWW | 418 | 48544 | 59.2 | Siphoviridae | - | JBD44 (NC_030929) | 68%/98% | SAMN18741767 |
| PSA07/PB1 | ATCC | ATCC 15692 | 65891 | 54.9 | Myoviridae | Pbunavirus | PB1 (NC_011810) | 100%/100% | SAMN18741768 |
| PSA09 | HWW | 410 | 62015 | 55.5 | Myoviridae | Pbunavirus | Pa193 (NC_050148) | 99%/93% | SAMN18741769 |
| PSA11 | HWW | ATCC 14210 | 48815 | 44.9 | Podoviridae | - | PA11 (NC_007808) | 97%/99% | SAMN18741770 |
| PSA13 | HWW | 427 | 45731 | 52.6 | Podoviridae | Bruynoghevirus | Pa222 (MK837011) | 99%/93% | SAMN18741771 |
| PSA16 | HWW | 466 | 45623 | 52.5 | Podoviridae | Bruynoghevirus | Pa222 (MK837011) | 98%/98% | SAMN18741772 |
| PSA20 | HWW | 639 | 62296 | 64.4 | Siphoviridae | Yuavirus | AN14 (KX198613) | 95%/98% | SAMN18741773 |
| PSA21 | HWW | 639 | 62243 | 64.5 | Siphoviridae | Yuavirus | LKO4 (NC_041934) | 96%/97% | SAMN18741774 |
| PSA25 | HWW | 426 | 64290 | 55.5 | Myoviridae | Pbunavirus | LBL3 (NC_011165) | 99%/95% | SAMN18741775 |
| PSA28 | HWW | 428 | 48440 | 58.3 | Siphoviridae | - | PMBT28 (MG641885) | 96%/86% | SAMN18741776 |
| PSA31 | HWW | 411 | 45505 | 52.5 | Podoviridae | Bruynoghevirus | Pa222 (MK837011) | 98%/97% | SAMN18741777 |
| PSA34 | HWW | 427 | 43749 | 52.3 | Podoviridae | Bruynoghevirus | Pa222 (MK837011) | 98%/98% | SAMN18741778 |
| PSA37 | HWW | 639 | 45506 | 52.5 | Podoviridae | Bruynoghevirus | Pa222 (MK837011) | 98%/97% | SAMN18741779 |
| PSA39 | HWW | 423 | 47030 | 64.2 | Siphoviridae | Yuavirus | LKO4 (NC_041934) | 95%/98% | SAMN18741780 |
| PSA40 | HWW | 466 | 45506 | 52.5 | Podoviridae | Bruynoghevirus | Pa222 (MK837011) | 98%/97% | SAMN18741781 |
HWW = Hospital wastewater, ATCC = American type culture collection.
Most similar phage based on BLAST to the NCBI nr database.
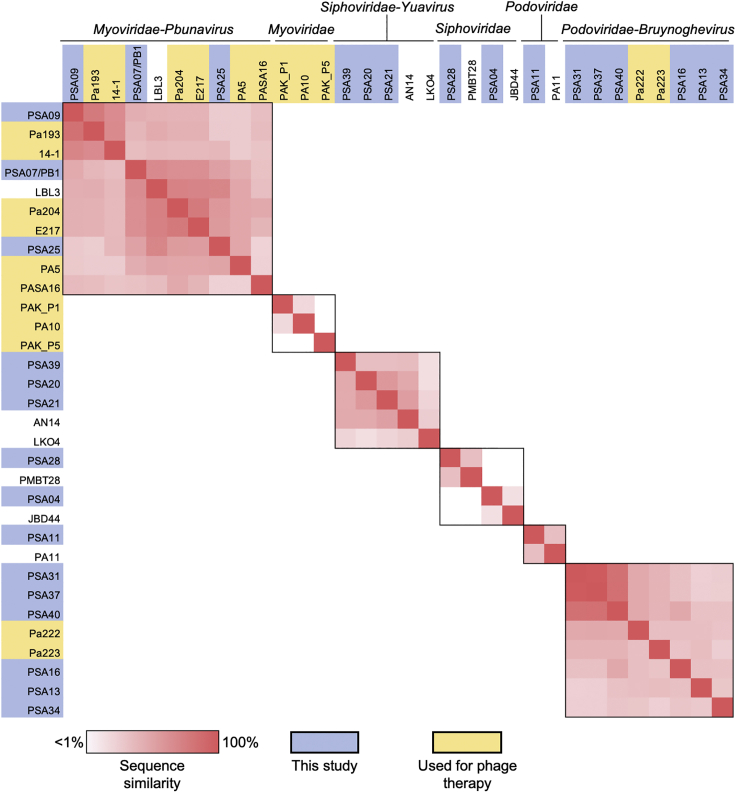
Next, we compared the genomes of the isolated phages to one another using nucleotide BLAST, as well as to the genomes of publicly available phages that were most similar to them and the genomes of phages previously used in phage therapy for P. aeruginosa (Table 1, Figure 2) (Aslam et al., 2019, 2020; Khatami et al., 2021; Onsea et al., 2019; Rubalskii et al., 2020). Phages within the same genus showed varying degrees of genomic similarity with one another, and no similarity was observed across different families or genera. Phages previously used for phage therapy belonged to the Myoviridae and Podoviridae families. The three Myoviridae phages we isolated were predicted to belong to the Pbunavirus genus, and showed modest similarity with phage LBL3 as well as other Pbunavirus phages used for phage therapy. Six of the phages we isolated belonged to the Bruynoghevirus genus within the Podoviridae family; three of these phages (PSA31, PSA37, and PSA40) were highly similar to one another, despite having been isolated on three different P. aeruginosa isolates and from three different wastewater samples (Figure S1). These data suggest that Bruynoghevirus phages might have been particularly abundant in the wastewater that we sampled, and that they are able to infect genetically diverse P. aeruginosa isolates.
Figure 2.
Genomic similarity among isolated P. aeruginosa bacteriophages and other publicly available phage genomes
Phages are organized by family and genus, which are labeled at the top of the figure. Black outline indicates phages belonging to the same family or genus. Phage genomes were compared with one another using nucleotide BLAST to determine sequence coverage and nucleotide identity for each pairwise comparison. Coverage and identity values were multiplied to calculate the “sequence similarity” for each comparison. Similarity values range from 1 to 100%, and are shown with red shading (1% or less = white, 100% = red). Phages isolated in this study are shaded blue. Phages used previously for phage therapy are shaded yellow.
Phage susceptibility of P. aeruginosa isolates and bacteriophage infectivity
To examine the phage susceptibilities of our P. aeruginosa isolates as well as the infectivity profile of each phage, we performed a lytic activity screen of the 15 bacteriophages studied here against all 23 bacterial isolates (Figure 3). Serial dilutions of each phage were spotted onto top agar lawns of each bacterial isolate, and individual plaques were counted to determine the titer of each phage against each isolate. Three of the P. aeruginosa isolates we tested (413, 414, and 729) were resistant to all phages tested, however the other 20 isolates (87% of all isolates tested) were susceptible to at least one phage (Figure 3). Phage susceptibility profiles of the isolates were highly variable, with the exception of isolate pairs 418/423 and 427/466; these pairs contained isolates belonging to the same ST, which were more genetically similar to one another than to the other isolates in the study. Although phages were found to infect between 9 and 19 different isolates, activity of the same phage was often variable against different isolates. For example, phage PSA07/PB1 displayed titers varying from 102 to 1010 PFU/mL against different P. aeruginosa isolates (Figure 3). Finally, compared to Myoviridae and Siphoviridae phages, the Podoviridae phages we isolated were able to infect more isolates and had higher average infectivity against the isolates tested here.
Figure 3.
Infectivity of isolated phages against genetically diverse P. aeruginosa isolates
Bacterial isolates are ordered according to the core genome phylogeny in Figure 1. Infectivity is shown as the log10 titer (PFU/mL) of each phage against each isolate. Boxed white values indicate the P. aeruginosa isolate that each phage was isolated and propagated on. Blue shading corresponds to phage titer, with darker shading indicating higher titer. White shading indicates no phage activity.
Genomic and phenotypic differences of phage-resistant mutants
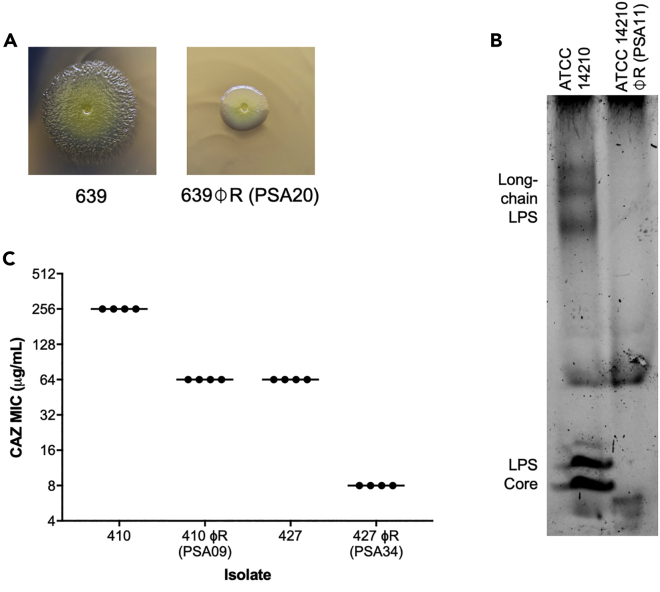
Within host-associated microbial communities, bacteriophages and their bacterial prey co-evolve with each other, and with their host (Koskella and Brockhurst, 2014). Phage predation exerts strong selective pressure on bacterial hosts, and often gives rise to resistance-conferring mutations in the bacterial genome. During the course of phage propagation, we isolated single colonies of phage-resistant mutants for four phages: PSA09, PSA11, PSA20, and PSA34 (Figure 4). Phage-resistant mutant isolates were tested to confirm their resistance, and were then subjected to whole-genome sequencing. Sequencing reads were mapped to the hybrid assembled genome of the corresponding phage-susceptible parent isolate, and protein-altering mutations in each resistant mutant were identified (Table 2). Each phage-resistant mutant genome encoded one or two protein-altering mutations. Based on the annotation of each mutated gene, we were able to identify putative phage resistance-conferring mutations in each mutant isolate genome. A phage-resistant mutant in the 639 isolate background that was resistant to phage PSA20 was found to have a Thr278Pro mutation in the Type IV pilus protein PilB (Table 2). Because Type IV pili are involved in twitching motility, we compared the twitching motility of the 639 P. aeruginosa parent isolate and the PSA20-resistant mutant, and found that the resistant mutant showed diminished twitching motility (Figure 4A). A PSA11-resistant mutant in the ATCC 14210 background encoded two mutations, one of which resided in a RfaB-like glycosyltransferase. Suspecting that this resistant mutant might have alterations in its lipopolysaccharide (LPS), we performed lipid A and O-antigen profiling on the parent and resistant mutant isolates (Figures 4B and S2). Lipid A profiles were similar between isolates, however the core and long-chain O-antigen regions were markedly absent in the resistant mutant compared to the parent isolate (Figure 4B). These findings suggest that phage PSA11 uses LPS as a receptor for infection, similar to other P. aeruginosa phages (Huszczynski et al., 2019; Pan et al., 2016).
Figure 4.
Phenotypic consequences of phage resistance
(A) Twitching motility differences between 639 and 639φR, a phage-resistant mutant raised against phage PSA20 that harbors a mutation in the Type IV pilus protein PilB.
(B) O-antigen profiles of ATCC 14210 and ATCC 14210φR, a phage-resistant mutant raised against phage PSA11 that harbors a mutation in a RfaB-like glycosyltransferase.
(C) Ceftazidime (CAZ) susceptibilities of two pairs of multidrug-resistant clinical isolates and corresponding phage-resistant mutants, both of which harbor mutations in genes impacting LPS biosynthesis.
Table 2.
Protein-altering mutations identified in phage-resistant P. aeruginosa mutants
| Isolate | Phage | Locationa | Mutationb | Description |
|---|---|---|---|---|
| ATCC 14210 φR | PSA11 | 4,104,560 | Tyr64fs | Transcriptional activator protein LasR |
| 5,912,880 | Arg237fs | RfaB-like glycosyltransferase | ||
| 410 φR | PSA09 | 5,582,688 | Asp279Gly | RfaB-like glycosyltransferase |
| 427 φR | PSA34 | 5,953,386 | Trp202fs | dTDP-4-dehydrorhamnose reductase RfbD |
| 639 φR | PSA20 | 5,758,428 | Thr278Pro | Type IV pilus protein PilB |
Genome coordinates in the parent P. aeruginosa genome.
fs = frameshift.
As many of the bacterial isolates we used for phage screening were multidrug-resistant, we also generated phage-resistant mutants from two isolates that were originally resistant to the third-generation cephalosporin ceftazidime, an antibiotic that is used to treat P. aeruginosa infections (Nguyen et al., 2018) (Table S1). Both parent isolates encoded known cephalosporin resistance-associated mutations in the Pseudomonas-derived cephalosporinase (PDC, also called AmpC). The two phage-resistant mutants we isolated encoded mutations in genes predicted to impact LPS biosynthesis, including the same RfaB-like glycosyltransferase mutated in ATCC 14210 φR, as well as the dTDP-4-dehydrorhamnose reductase RfbD (Table 2). We compared the ceftazidime susceptibilities of both phage-resistant mutants to their corresponding parent isolates, and found that the mutants showed either 4-fold or 8-fold sensitization (Figure 4C). In both cases, LPS-altering mutations were the only changes detected in the resistant mutant genomes. Overall, these data indicate that phage resistance-conferring alterations to LPS in these mutants also increased their susceptibility to a cell wall-targeting antibiotic.
Phage-mediated killing of bacterial biofilms in vitro and ex vivo
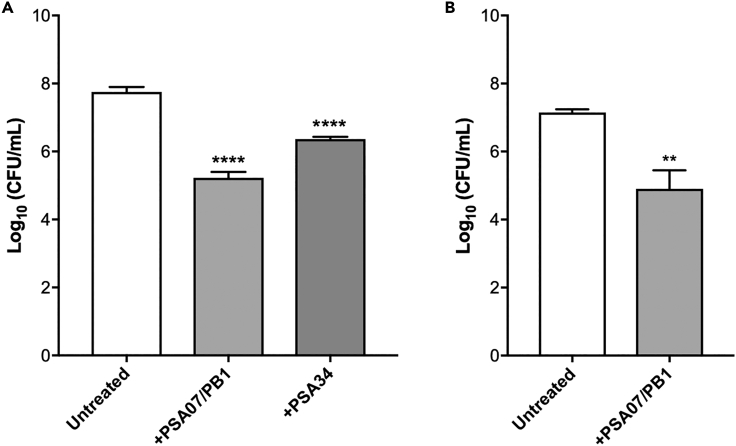
Because P. aeruginosa that causes infections frequently grows in biofilms (Maurice et al., 2018), we tested whether phages that were active against bacteria in our top agar lawn-based activity assays could also kill bacteria grown in biofilms (Figure 5). We first tested the ability of the PSA07/PB1 and PSA34 phages to kill the multidrug-resistant isolate 427 when grown in biofilms in vitro. Biofilms were grown for 24 h, planktonic cells were removed, biofilms were washed, and then phages were applied and plates were incubated for an additional 24 h. PSA07/PB1 treatment resulted in >100-fold bacterial killing, and PSA34 treatment resulted in >10-fold bacterial killing (Figure 5A). Next, we tested the ability of the PSA07/PB1 phage to kill the 427 isolate grown in association with human CF airway epithelial cells. Bacteria were incubated with epithelial cells for 8 h, then phage was added and incubated for additional 16 h before cell-associated bacteria were collected and quantified for viability. We found that similar to the in vitro assay, PSA07/PB1 treatment resulted in >100-fold bacterial killing (Figure 5B), suggesting that phages can also kill P. aeruginosa grown in association with epithelial cells and under conditions that more closely mimic infection in humans.
Figure 5.
Phage-mediated killing of P. aeruginosa grown in biofilms in vitro and ex vivo
(A) Bacterial viability measured after P. aeruginosa isolate 427 biofilms were grown in vitro and then treated with either fresh media (Untreated), or with phages PSA07/PB1 or PSA34.
(B) Viability after phage PSA07/PB1 treatment of P. aeruginosa isolate 427 biofilms grown on human-derived CF airway epithelial cells. Bars show mean values of at least three biological replicates, and error bars show standard deviation. Viable bacteria were quantified as CFU/mL, and phage-treated conditions were compared to the untreated condition using two-tailed t-tests. ∗∗p < 0.01, ∗∗∗∗p < 0.0001.
Discussion
The objective of this study was to isolate and characterize lytic bacteriophages from hospital wastewater with activity against clinical P. aeruginosa isolates representing the types of isolates we might wish to target with phage therapy. By screening wastewater samples against a genetically and phenotypically diverse panel of P. aeruginosa bacterial isolates, we were able to isolate a diverse group of P. aeruginosa-targeting phages representing the Myoviridae, Siphoviridae, and Podoviridae families. In testing our panel of P. aeruginosa isolates for susceptibility to the isolated phages, we found a broad range of phage activities. In addition, our analysis of phage-resistant mutants showed that evolving phage resistance conferred a decrease in production of virulence factors like pili and LPS, as well as an increase in antibiotic susceptibility. Finally, two of the phages we isolated were able to kill P. aeruginosa grown in biofilms, suggesting that they have therapeutic utility for the treatment of P. aeruginosa infections.
The bacteriophages we isolated in this study were similar in terms of phage family, genus, and other genome characteristics to P. aeruginosa-targeting phages isolated previously (Farlow et al., 2020; Kwiatek et al., 2015; Latz et al., 2017; Oliveira et al., 2020). This could be because of the fact that these prior studies also isolated phages from sewage, similar to what we did in this study. The phages we isolated also resembled P. aeruginosa-targeting phages that have been recently used for phage therapy (Aslam et al., 2019, 2020; Khatami et al., 2021; Onsea et al., 2019; Rubalskii et al., 2020). Our findings are consistent with the idea that phages active against P. aeruginosa mirror the abundant genetic and phenotypic diversity of their hosts. Although it has been noted that newly isolated phages do not often represent novel phylogenetic lineages (Latz et al., 2017), sampling and screening from more diverse sources could potentially uncover a broader range of phage genetic diversity.
Although the vast majority of P. aeruginosa isolates we screened were susceptible to one or more of the phages we isolated, three bacterial isolates were resistant to all phages studied here. These three isolates (413, 414, and 729) were genetically distinct from one another, and no clear trends emerged to explain their resistance to phage infection. For example, all three isolates lacked functional CRISPR-Cas systems, and they did not have a higher relative abundance of prophages compared to phage-susceptible isolates. We did note that isolate 729 grew very poorly, and was predicted to be a hypermutator because of a frameshift mutation in the DNA mismatch repair gene mutS. It is unknown whether hypermutators in P. aeruginosa are more resistant to phage infection; this would be a worthwhile avenue of future investigation. Nonetheless, the specific mechanism(s) conferring phage resistance in the clinical isolates we studied here remain unclear.
During the course of phage propagation, we isolated four phage-resistant P. aeruginosa mutants and studied them further. From whole-genome sequencing of these mutants, we identified two kinds of mutations that lead to measurable phenotypic changes. First, in the phage-resistant mutant of isolate 639, disruption of the Type IV pilus protein PilB appears to have also caused a reduction in twitching motility. Because the Type IV pilus has been previously described as a surface receptor used by P. aeruginosa phages for infection (Bradley and Pitt, 1974), we suspect that the phage PSA20, and also perhaps the other Yuavirus phages we isolated, use the Type IV pilus as a receptor for infection. Second, we identified mutations in genes affecting LPS biosynthesis in phage-resistant mutants of isolates ATCC 14210, 410 and 427. Bacterial LPS is also a well-known surface receptor used for phage infection in P. aeruginosa, and also contributes to bacterial virulence (Huszczynski et al., 2019). We also observed that two of our phage-resistant mutants showed increased susceptibility to ceftazidime, a cell-wall targeting antibiotic. Taken together, these findings are consistent with the notion that the development of phage resistance is often coupled with collateral effects like decreased bacterial virulence or increased antibiotic susceptibility (Chan et al., 2016). This has potentially promising implications for the treatment of P. aeruginosa infections using phages, where a tradeoff between phage resistance and antibacterial resistance or virulence could be exploited.
When we tested whether two different P. aeruginosa phages could kill bacteria grown in biofilms, we observed reductions in viable bacteria upon phage treatment of biofilms both in vitro and ex vivo. Although application of phage did not completely eradicate bacteria growing in the biofilms, it did substantially decrease the bacterial loads measured in both assays to similar levels to levels obtained after antibiotic treatment (Zemke et al., 2017). This finding is in agreement with other studies that have also documented phage-mediated reductions in P. aeruginosa biofilm density in vitro (Fong et al., 2017; Fu et al., 2010; Oliveira et al., 2020). Here we have extended these in vitro findings to test the ability of phages to infect bacteria grown on human CF airway epithelial cells, a setting that more closely mimics bacterial growth in the CF airway (Cornforth et al., 2020; Hendricks et al., 2016). Testing of phage efficacy in a context that includes eukaryotic cells is an important feature of this study. How bacteriophages interact with eukaryotic cells, and how this interaction may impact phage activity, is a focus on ongoing work by us and others (Van Belleghem et al., 2018).
Taken together, the genomic and phenotypic data presented here have promising implications for the therapeutic potential of P. aeruginosa-targeting bacteriophages. It is clear that within the tripartite relationship between phages, bacteria, and mammalian hosts, phages can be drivers of bacterial genetic and phenotypic change. We are yet to fully explore the diversity of ways in which phages could be utilized within host-associated microbial communities to increase human health and improve disease states. Nonetheless, this study provides a valuable addition to the growing literature documenting the abundance and diversity of P. aeruginosa phages, and demonstrates how systematic characterization can aid in the development of phages for clinical use as precision antibiotics.
Limitations of the study
This study had several limitations. Many of the phages we isolated showed variable activity, and their activity was generally diminished against isolates other than the host isolate used for their initial isolation and propagation. Although we attempted to be unbiased in our phage isolation methods, we observed some redundancy in isolated phages within the Podoviridae family, suggesting a possible enrichment of our wastewater source with Podoviridae phages. In addition, we only isolated and studied four different phage-resistant mutants, and we did not confirm that any of the resistance-associated mutations identified were indeed the cause of phage resistance, for example through genetic complementation. Furthermore, in the biofilm assays we used centrifugation before CFU counting, which may not have been sufficient to fully separate phages from bacteria. Finally, all work performed in this study was conducted in vitro or ex vivo, thus we are unable to conclude that any of the phages we isolated would be useful therapeutic candidates without additional testing, for example in relevant animal models of P. aeruginosa infection.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| P. aeruginosa ATCC 14210 | American Type Culture Collection | ATCC 14210 |
| P. aeruginosa ATCC 15692 | American Type Culture Collection | ATCC 15692 |
| P. aeruginosa 410 | CF sputum | 410 |
| P. aeruginosa 411 | CF sputum | 411 |
| P. aeruginosa 412 | CF sputum | 412 |
| P. aeruginosa 413 | CF sputum | 413 |
| P. aeruginosa 414 | CF sputum | 414 |
| P. aeruginosa 415 | CF sputum | 415 |
| P. aeruginosa 417 | CF sputum | 417 |
| P. aeruginosa 418 | CF sputum | 418 |
| P. aeruginosa 419 | CF sputum | 419 |
| P. aeruginosa 421 | CF sputum | 421 |
| P. aeruginosa 422 | CF sputum | 422 |
| P. aeruginosa 423 | CF sputum | 423 |
| P. aeruginosa 425 | CF sputum | 425 |
| P. aeruginosa 426 | CF sputum | 426 |
| P. aeruginosa 427 | CF sputum | 427 |
| P. aeruginosa 428 | CF sputum | 428 |
| P. aeruginosa 429 | CF sputum | 429 |
| P. aeruginosa 466 | CF sputum | 466 |
| P. aeruginosa 639 | CF sputum | 639 |
| P. aeruginosa 729 | CF sputum | 729 |
| P. aeruginosa 779 | CF sputum | 779 |
| Chemicals, peptides, and recombinant proteins | ||
| Calcium chloride dihydrate | Millipore Sigma | CaCl2 |
| Magnesium chloride hexahydrate | Millipore Sigma | MgCl2 |
| Trizma hydrochloride | Millipore Sigma | Tris-HCl |
| Sodium chloride | Millipore Sigma | NaCl |
| Magnesium sulfate | Millipore Sigma | MgSO4 |
| Chloroform | Millipore Sigma | Chloroform |
| 1-Octanol | Millipore Sigma | Octanol |
| Phenol:chloroform:isoamyl alcohol (25:24:1) | Fisher Scientific | Phenol:chloroform:isoamyl alcohol (25:24:1) |
| Sodium acetate | Millipore Sigma | Sodium acetate |
| Acetic acid | Thermo Fisher | Acetic acid |
| Crystal violet | Thermo Fisher | Crystal violet |
| Phosphate buffered saline | Millipore Sigma | PBS |
| Citric acid | Millipore Sigma | Citric acid |
| Tri-sodium citrate dihydrate | Millipore Sigma | Trisodium citrate |
| Methanol | Millipore Sigma | Methanol |
| Critical commercial assays | ||
| Nextera library preparation kit | Illumina Technologies | Nextera XT/Nextera kit |
| SQK-RBK004 | Oxford Nanopore Technologies | Rapid barcoding kit |
| Pro-Q Emerald 300 LPS Gel Stain Kit | Molecular Probes | Pro-Q Emerald 300 LPS Gel Stain Kit |
| Deposited data | ||
| P. aeruginosa genomes | NCBI BioProject: PRJNA610040 | P. aeruginosa bacterial host genomes |
| Phage genomes | NCBI BioProject: PRJNA721956 | Phage genomes |
| Software and algorithms | ||
| Guppy | Oxford Nanopore Technologies, Oxford, UK | Guppy |
| Unicycler | Wick et al., 2017 | unicycler |
| Prokka | Seemann, 2014 | prokka |
| Roary | Page et al., 2015 | Roary |
| RAxML | Stamatakis, 2014 | RAxML |
| CRISPRCasFinder | Couvin et al., 2018 | CRISPRCasFinder |
| DefenseFinder | Abby et al., 2014 | DefenseFinder |
| PHASTER | Arndt et al., 2016 | PHASTER |
| SPAdes v3.13.0 | Bankevich et al., 2012 | SPAdes |
| Mauve | Darling et al., 2004 | Mauve |
| CLC Genomics Workbench v11.0.1 | Qiagen | CLC Genomics Workbench |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Daria Van Tyne (vantyne@pitt.edu).
Materials availability
Bacterial isolates, isolated phages, and phage-resistant mutants are available by request from the Lead Contact under the conditions of a material transfer agreement (MTA).
Experimental model and subject details
Bacterial isolates
P. aeruginosabacterial isolates were collected from patients treated at the University of Pittsburgh Medical Center (UPMC) (n = 21), or were purchased from the American Type Culture Collection (ATCC) (n = 2). Collection of UPMC isolates was conducted with Institutional Review Board Approval (protocol #PRO12060302). Of the UPMC isolates, 20 were collected from people with CF and one was a clinical isolate from sputum collected from a non-CF patient. Both ATCC isolates were of clinical origin. All isolates were cryopreserved in brain heart infusion (BHI) media with 16.7% glycerol and stored at −80°C.
Method details
Hospital wastewater collection
Wastewater effluent was sampled from the main sewer outflow of a Pittsburgh area hospital, at a point before the outflow joined with the municipal sewer system. A total of four water samples were collected over a six-month period. Each effluent sample was centrifuged at 4,000 rpm for 20 min to pellet solid debris, the supernatant was filtered through a 0.22 μm filter, and the sample was then concentrated by centrifugation using an Amicon 100kDa filter unit (MilliporeSigma, Burlington, MA) at 4,000 rpm for 15 min.
Phage isolation
Lytic bacteriophages were identified with a soft agar overlay assay. Briefly, bottom agar plates were prepared containing BHI media with 1.5% agar, 1 mM CaCl2 and 1 mM MgCl2. Bacterial isolates were inoculated into BHI media and grown overnight at 37°C. 100 μL of stationary phase culture (OD > 1.0) was added to a tube containing 100 μL of filtered concentrated wastewater for 5–10 min at room temperature, and then 10 mL of top agarose (BHI with 0.5% agarose, 1 mM CaCl2, and 1 mM MgCl2) cooled to 55°C was added and the mixture was plated onto two bottom agar plates. Plates were incubated overnight at 37°C and were examined the following day to identify lytic phage plaques. Phages were passaged by sequential picking and plating of individual plaques grown on the same bacterial isolate. Phages were picked from individual plaques using a pipette tip and were placed into 100 μL of SM buffer (50 mM Tris-HCl pH 7.5, 100 mM NaCl, 8 mM MgSO4) and incubated overnight at 37°C. The following day, serial 10-fold dilutions were made in SM buffer, and 3 μL of each dilution was spotted onto a plate containing 5 mL of top agarose mixed with 50 μL of stationary phase bacterial culture (OD > 1.0) and layered on top of a bottom agar plate. After overnight incubation at 37°C, an individual plaque was picked and passaged again. Each phage was passaged at least twice before the generation of high-titer stocks. To generate high-titer stocks, a single plaque was picked into 100 μL of SM buffer and incubated overnight. Then, 100 μL of overnight bacterial culture (OD > 1.0) was added and the mixture was incubated for 5–10 min at room temperature, followed by addition of 10 mL of top agarose and plating onto two bottom agar plates. Plates were incubated overnight at 37°C, and then plates with high plaque density were flooded with 5 mL of SM buffer and incubated at 37°C for at least 1 h to elute phages from the top agar. SM buffer was removed from each plate, pooled, spun down at 4,000 rpm for 20 min, and filtered through a 0.22 μm filter. Phage-containing lysates were extracted with 0.1 volumes of chloroform followed by 0.4 volumes of 1-octanol, and were stored at 4°C.
Genome sequencing
Bacterial genomic DNA from each P. aeruginosa host isolate was extracted from 1 mL overnight cultures grown in BHI media using a Qiagen DNeasy Blood and Tissue Kit (Qiagen, Germantown, MD) following the manufacturer’s protocol. Illumina sequencing libraries were prepared with a Nextera XT or Nextera kit (Illumina, San Diego, CA), and libraries were sequenced on a MiSeq using 300-bp paired-end reads, or on a NextSeq using 150-bp paired-end reads. Genomic DNA was also used to construct long-read sequencing libraries using a rapid barcoding kit (SQK-RBK004, Oxford Nanopore Technologies, Oxford, UK), and libraries were sequenced on a MinION device. Base-calling of nanopore reads was performed with Guppy. Phage genomic DNA was extracted from 500 μL of phage lysate using phenol-chloroform, followed by ethanol precipitation. Briefly, 500 μL phenol:chloroform:isoamyl alcohol (25:24:1) was added to each lysate, samples were vortexed and then centrifuged at 14,000 rpm for 1 min. The upper aqueous phase was transferred to a new tube and 500 μL of chloroform was added. Samples were vortexed and centrifuged again at 14,000 rpm for 1 min, and the upper aqueous phase was again transferred to a new tube. Then 1 μL glycogen, 0.1x volume 3 M sodium acetate, and 2.5x volume 100% ethanol were added and samples were incubated overnight at −20°C. The next day samples were centrifuged at 14,000 rpm for 30 min at 4°C, then the supernatant was removed and the DNA pellet was washed with 150 μL 70% ethanol. DNA pellets were resuspended in 100 μL nuclease-free water, and DNA was quantified with a Qubit fluorimeter (Thermo Fisher Scientific, Waltham, MA). Illumina sequencing libraries were prepared with a Nextera XT or Nextera kit (Illumina, San Diego, CA), and libraries were sequenced on a MiSeq using 300-bp paired-end reads, or on a NextSeq using 150-bp paired-end reads.
Phenotyping and phage-resistant mutants
When phage-resistant mutants were observed in the course of preparing high-titer phage lysates, they were saved for additional characterization. Individual bacterial colonies were picked and restreaked onto BHI agar. Phage resistance was confirmed by titering the same phage lysate on both parent and resistant mutant isolates and confirming high titer (>1 × 105 PFU/mL) on the parent isolate but no phage activity against the resistant mutant. Resistant mutants were also spotted onto a lawn of the parent bacterial isolate to confirm that they were not lysogens. Genomic DNA was extracted from each phage-resistant mutant, sequenced on the Illumina platform, and analyzed using CLC Genomics Workbench v11.0.1. Raw sequencing reads were mapped to the annotated genome of the corresponding parent isolate, and variants were identified with a read depth cut-off of 10 and a variant frequency cut-off of 80%.
Biofilm assays were performed following a previously published protocol (O'Toole, 2011). Briefly, bacteria were first inoculated into LB media and incubated overnight at 37°C. Cultures were then diluted 1:100 into M63 media. Diluted cultures were aliquoted into vinyl 96-well plates (100 μL per well) sealed, and incubated at 37°C for 24 hours. After incubation, planktonic cells were removed by inverting the plates and shaking liquid out into a sterilization tub. Plates were then submerged in water and rinsed twice to remove unattached cells. Wells were stained with 0.1% crystal violet and incubated at room temperature for 15 min. Plates were rinsed three times with water and shaken out vigorously, then allowed to dry completely. Crystal violet stain was solubilized with 30% acetic acid. Absorbance was read in each well at 550 nm using a BioTek Synergy H1 microplate reader with GenMark software (BioTek, Winooski, VT). Two biological replicates, each containing 24 technical replicates, were run for each isolate. To test phage activity against bacteria grown in biofilms, biofilms were inoculated into 96-well plates as above and incubated for 24 h at 37°C. Planktonic cells were removed and biofilms were washed with sterile 1xPBS using a multichannel pipettor, and either fresh M63 media or phage at 1 × 1012 PFU/mL in M63 media was added to each well. Plates were incubated for 24 hours at 37°C, then bacteria in each well were resuspended and serial 10-fold dilutions were tracked onto BHI agar plates to determine the number of colony-forming units per mL (CFU/mL) in each condition.
Swimming motility was measured by spotting 2.5 μL from an overnight bacterial culture grown in BHI media onto plates containing LB + 0.3% agar. Plates were incubated overnight at 37°C and colony diameters were measured the following day. Twitching motility was assessed by inserting a pipet tip coated in overnight bacterial culture completely through a BHI agar plate to create a small hole in the agar, and then incubating for 48 hours at 37°C. Mucoidy was measured by assessing the morphology of each isolate when grown on both LB agar and Pseudomonas isolation agar (PIA) plates. Each isolate was struck onto each agar type, then incubated at 37°C overnight, followed by a 48-hour incubation at room temperature.
Antibiotic susceptibility testing was performed by the Kirby-Bauer disk diffusion method (aztreonam, cefepime, cefiderocol, ceftazidime, ceftazidime-avibactam, ciprofloxacin, gentamicin, imipenem-relebactam, levofloxacin, meropenem, piperacillin-tazobactam, tobramycin), or Etest (ceftolozane-tazobactam). Clinical and Laboratory Standards Institute (CLSI) interpretive criteria was used to determine susceptibility (CLSI, 2019). Ceftazidime susceptibility was also determined for selected parent and phage-resistant mutant isolates using broth microdilution in Mueller-Hinton Broth. Serial two-fold dilutions of ceftazidime were tested and the minimum inhibitory concentration (MIC) was recorded as the lowest antibiotic concentration that inhibited bacterial growth by visual inspection.
Lipid A analysis of parent and phage-resistant mutant isolates was performed by first growing each isolate overnight in LB media and then pelleting bacteria. Bacterial pellets were then applied to a target location on a stainless steel MALDI plate along with 1 μL of FLAT extraction buffer (0.2 M citric acid, 0.1 M trisodium citrate). The target plate was incubated in a humidified chamber for 30 min at 110°C. The MALDI plate was washed with deionized water from a squeeze bottle, allowed to air dry, then 1 μL of norharmane matrix solution was applied (10 mg/mL in 2:1, v/v chloroform/methanol) to each target location. Following the method of Leung et al. (2017), spectra were acquired from target locations in negative ion mode using a Microflex LRF MALDI-TOF MS (Bruker, Billerica, MA) in reflectron mode with a limited mass range of 1,000–2,400 m/z. Mass range was further narrowed to focus on the regions of interest. O-antigen profiling was performed by extracting bacterial pellets from 10 mL overnight cultures grown in LB media with 3 mL Tri-Reagent (Molecular Research, Albany, NY). After incubation at room temperature for 15 min, 600 μL chloroform was added, samples were vortexed, incubated at room temperature for 15 min, and were then centrifuged at 4,200 rpm for 20 min. The aqueous phase was removed and the organic phase was washed with 3 mL water, vortexed, incubated at room temperature for 15 min, centrifuged at 4,200 rpm for 20 min, and then the aqueous phase was again removed and combined with the prior aqueous phase. Samples were frozen, lyophilized, resuspended in Laemmli sample buffer, and boiled for 10 min. SDS-PAGE was performed using precast Tris-Tricine 10–20% gradient gels (Invitrogen, Waltham, MA) with separate cathode (0.1M Tris, 0.1M Tricine, 0.1% SDS, pH 8.25) and anode (0.2M Tris, pH 8.9) buffers. Gels were fixed in 60% methanol/10% acetic acid overnight at room temperature, and were then stained with a Pro-Q Emerald 300 LPS Gel Stain Kit (Molecular Probes, Eugene, OR) following manufacturer instructions.
Ex vivo biofilm assay
Immortalized human bronchial epithelial cells isolated from a ΔF508/ΔF508 CF patient (CFBE41o- cells) (Bruscia et al., 2002) were cultured on Transwell inserts (Corning, Corning, NY) and grown for 7–10 days at the air-liquid interface, as described (Hendricks et al., 2021). Basolateral medium was replaced with minimum essential medium (MEM) supplemented with 2 mM L-glutamine 24 hours before bacterial inoculation. P. aeruginosa isolate 427 was inoculated into the apical compartment as described previously (Zemke et al., 2014), with the following modifications: bacteria were inoculated at a multiplicity of infection of 0.015 CFU/cell in MEM and were incubated at 37°C for 1 hour, followed by inoculum removal and addition of MEM supplemented with 2 mM L-glutamine and 23 mM L-arginine. P. aeruginosa phage PSA07/PB1 was added to the apical compartment 8 hours post-bacterial inoculation at a concentration of 8 × 106 PFU/mL and incubated at 37°C for 16 hours. Following washing of the apical and basolateral media, biofilms were harvested, single bacterial cells were isolated with a Triton X-100 wash and vortexing, and bacterial counts were enumerated. CFUs were quantified by dilution plating onto LB agar.
Quantification and statistical analysis
Genome sequence processing
P. aeruginosa genomes were hybrid assembled with unicycler (Wick et al., 2017), annotated with prokka (Seemann, 2014), and were compared to one another with Roary (Page et al., 2015). A core genome phylogenetic tree was generated using RAxML with the GTRCAT substitution model and 1,000 iterations (Stamatakis, 2014). Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) loci in each bacterial isolate were identified with CRISPRCasFinder (Couvin et al., 2018) and DefenseFinder (Abby et al., 2014). CRISPR loci were called present in a genome if Cas proteins were also found; results matched between the two tools used in all cases. Prophages were identified in each bacterial genome using PHASTER (Arndt et al., 2016). Prophages of any length that were predicted to be intact or questionable by PHASTER were included. Prophage sequences were compared to one another with nucleotide BLAST, and clusters of similar prophage sequences were identified as those sharing >90% sequence coverage and >90% sequence identity. Phage genome sequences were assembled with SPAdes v3.13.0 (Bankevich et al., 2012), and were annotated with prokka (Seemann, 2014). Phage genomes were compared to one another and to other available phage genomes using BLAST (Altschul et al., 1990), PHASTER (Arndt et al., 2016), and Mauve (Darling et al., 2004).
Statistical analyses
Two-tailed t-tests were used to assess the significance of prophage differences between CRISPR+ and CRISPR- isolates, and differences in bacterial cell density in biofilm killing experiments. p-values are indicated on the corresponding figures; results were considered significant if p-values were less than 0.05.
Acknowledgments
We gratefully acknowledge all members of the Van Tyne lab, and in particular Shu-Ting Cho, for helpful input during the preparation of this manuscript. We also thank Carlos Guerrero-Bustamante and Catherine Armbruster for helpful contributions to the study. Research reported in this publication was supported in part by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number UM1AI104681. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work was also supported by grants BOMBER19R0, BOMBER21P0, VANTYN21GO, and ZAMORA20F0 from the Cystic Fibrosis Foundation, and by the Department of Medicine at the University of Pittsburgh School of Medicine. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
H.R.N. and D.V.T. conceived the study. M.H.Y., R.K.S., and Y.D. provided wastewater and bacterial isolates for screening. H.R.N., D.R.E., A.G.F., and K.J.W. screened and isolated phages. H.R.N., D.R.E., and D.V.T. performed genomic and phenotypic analyses. C.E.H., A.P., P.F.Z., A.I., R.K.E., and J.M.B. assisted with phenotypic characterization of resistant mutants and biofilm-targeting assays. H.R.N. and D.V.T. wrote the manuscript. All authors reviewed and edited the manuscript.
Declaration of interests
J.B. is a consultant for BiomX, Inc. The other authors declare no competing interests.
Published: June 17, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104372.
Supplemental information
Data and code availability
-
•
Hybrid assembled bacterial host genomes and bacteriophage genomes have been deposited at NCBI under BioProjects PRJNA610040 and PRJNA721956 and are publicly available as of the date of publication. Accession numbers are listed in Tables 1 and S1.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abby S.S., Neron B., Menager H., Touchon M., Rocha E.P.C. MacSyFinder: a program to mine genomes for molecular systems with an application to CRISPR-Cas systems. PLoS One. 2014;9 doi: 10.1371/journal.pone.0110726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arndt D., Grant J.R., Marcu A., Sajed T., Pon A., Liang Y., Wishart D.S. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslam S., Courtwright A.M., Koval C., Lehman S.M., Morales S., Furr C.L., Rosas F., Brownstein M.J., Fackler J.R., Sisson B.M., et al. Early clinical experience of bacteriophage therapy in 3 lung transplant recipients. Am. J. Transpl. 2019;19:2631–2639. doi: 10.1111/ajt.15503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslam S., Lampley E., Wooten D., Karris M., Benson C., Strathdee S., Schooley R.T. Lessons learned from the first 10 consecutive cases of intravenous bacteriophage therapy to treat multidrug-resistant bacterial infections at a single center in the United States. Open Forum Infect. Dis. 2020;7:ofaa389. doi: 10.1093/ofid/ofaa389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondy-Denomy J., Qian J., Westra E.R., Buckling A., Guttman D.S., Davidson A.R., Maxwell K.L. Prophages mediate defense against phage infection through diverse mechanisms. ISME J. 2016;10:2854–2866. doi: 10.1038/ismej.2016.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley D.E., Pitt T.L. Pilus-dependence of four Pseudomonas aeruginosa bacteriophages with non-contractile tails. J. Gen. Virol. 1974;24:1–15. doi: 10.1099/0022-1317-24-1-1. [DOI] [PubMed] [Google Scholar]
- Bruscia E., Sangiuolo F., Sinibaldi P., Goncz K.K., Novelli G., Gruenert D.C. Isolation of CF cell lines corrected at ΔF508-CFTR locus by SFHR-mediated targeting. Gene Ther. 2002;9:683–685. doi: 10.1038/sj.gt.3301741. [DOI] [PubMed] [Google Scholar]
- CDC . U.S. Department of Health and Human Services; 2019. Antibiotic Resistance Threats in the United States, 2019. [Google Scholar]
- Chan B.K., Sistrom M., Wertz J.E., Kortright K.E., Narayan D., Turner P.E. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci. Rep. 2016;6:26717. doi: 10.1038/srep26717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI . Clinical and Laboratory Standards Institute; 2019. Performance standards for antimicrobial susceptibilty testing, 29th ed. CLSI Supplement M100. [Google Scholar]
- Cornforth D.M., Diggle F.L., Melvin J.A., Bomberger J.M., Whiteley M. Quantitative framework for model evaluation in microbiology Research using Pseudomonas aeruginosa and cystic fibrosis infection as a test case. mBio. 2020;11 doi: 10.1128/mBio.03042-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvin D., Bernheim A., Toffano-Nioche C., Touchon M., Michalik J., Neron B., Rocha E.P.C., Vergnaud G., Gautheret D., Pourcel C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018;46:W246–W251. doi: 10.1093/nar/gky425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling A.C., Mau B., Blattner F.R., Perna N.T. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo-Calap P., Delgado-Martinez J. Bacteriophages: protagonists of a post-antibiotic era. Antibiotics (Basel) 2018;7:66. doi: 10.3390/antibiotics7030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evseev P.V., Gorshkova A.S., Sykilinda N.N., Drucker V.V., Miroshnikov K.A. Pseudomonas bacteriophage AN14 – a Baikal-borne representative of Yuavirus. Limnol. Freshw. Biol. 2020;5:1055–1066. doi: 10.31951/2658-3518-2020-a-5-1055. [DOI] [Google Scholar]
- Farlow J., Freyberger H.R., He Y., Ward A.M., Rutvisuttinunt W., Li T., Campbell R., Jacobs A.C., Nikolich M.P., Filippov A.A. Complete genome sequences of 10 phages lytic against multidrug-resistant Pseudomonas aeruginosa. Microbiol. Resour. Announc. 2020;9 doi: 10.1128/mra.00503-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong S.A., Drilling A., Morales S., Cornet M.E., Woodworth B.A., Fokkens W.J., Psaltis A.J., Vreugde S., Wormald P.J. Activity of bacteriophages in removing biofilms of Pseudomonas aeruginosa isolates from chronic rhinosinusitis patients. Front Cell Infect Microbiol. 2017;7:418. doi: 10.3389/fcimb.2017.00418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman N.D., Temkin E., Carmeli Y. The negative impact of antibiotic resistance. Clin. Microbiol. Infect. 2016;22:416–422. doi: 10.1016/j.cmi.2015.12.002. [DOI] [PubMed] [Google Scholar]
- Fu W., Forster T., Mayer O., Curtin J.J., Lehman S.M., Donlan R.M. Bacteriophage cocktail for the prevention of biofilm formation by Pseudomonas aeruginosa on catheters in an in vitro model system. Antimicrob. Agents Chemother. 2010;54:397–404. doi: 10.1128/AAC.00669-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks M.R., Lane S., Melvin J.A., Ouyang Y., Stolz D.B., Williams J.V., Sadovsky Y., Bomberger J.M. Extracellular vesicles promote transkingdom nutrient transfer during viral-bacterial co-infection. Cell Rep. 2021;34:108672. doi: 10.1016/j.celrep.2020.108672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks M.R., Lashua L.P., Fischer D.K., Flitter B.A., Eichinger K.M., Durbin J.E., Sarkar S.N., Coyne C.B., Empey K.M., Bomberger J.M. Respiratory syncytial virus infection enhances Pseudomonas aeruginosa biofilm growth through dysregulation of nutritional immunity. Proc. Natl. Acad. Sci. U S A. 2016;113:1642–1647. doi: 10.1073/pnas.1516979113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoiby N., Ciofu O., Bjarnsholt T. Pseudomonas aeruginosa biofilms in cystic fibrosis. Future Microbiol. 2010;5:1663–1674. doi: 10.2217/fmb.10.125. [DOI] [PubMed] [Google Scholar]
- Huszczynski S.M., Lam J.S., Khursigara C.M. The role of pseudomonas aeruginosa lipopolysaccharide in bacterial pathogenesis and physiology. Pathogens. 2019;9 doi: 10.3390/pathogens9010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatami A., Lin R.C.Y., Petrovic-Fabijan A., Alkalay-Oren S., Almuzam S., Britton P.N., Brownstein M.J., Dao Q., Fackler J., Hazan R., et al. Bacterial lysis, autophagy and innate immune responses during adjunctive phage therapy in a child. EMBO Mol. Med. 2021;13 doi: 10.15252/emmm.202113936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskella B., Brockhurst M.A. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol. Rev. 2014;38:916–931. doi: 10.1111/1574-6976.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatek M., Mizak L., Parasion S., Gryko R., Olender A., Niemcewicz M. Characterization of five newly isolated bacteriophages active against Pseudomonas aeruginosa clinical strains. Folia Microbiol. (Praha) 2015;60:7–14. doi: 10.1007/s12223-014-0333-3. [DOI] [PubMed] [Google Scholar]
- Latz S., Krüttgen A., Häfner H., Buhl E.M., Ritter K., Horz H.P. Differential effect of newly isolated phages belonging to PB1-like, phiKZ-like and LUZ24-like viruses against multi-drug resistant Pseudomonas aeruginosa under varying growth conditions. Viruses. 2017;9:315. doi: 10.3390/v9110315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung L.M., Fondrie W.E., Doi Y., Johnson J.K., Strickland D.K., Ernst R.K., Goodlett D.R. Identification of the ESKAPE pathogens by mass spectrometric analysis of microbial membrane glycolipids. Sci. Rep. 2017;7:6403. doi: 10.1038/s41598-017-04793-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice N.M., Bedi B., Sadikot R.T. Pseudomonas aeruginosa biofilms: host response and clinical implications in lung infections. Am. J. Respir. Cell Mol. Biol. 2018;58:428–439. doi: 10.1165/rcmb.2017-0321TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradali M.F., Ghods S., Rehm B.H.A. Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front. Cell Infect. Microbiol. 2017;7:39. doi: 10.3389/fcimb.2017.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L., Garcia J., Gruenberg K., MacDougall C. Multidrug-resistant Pseudomonas infections: hard to treat, but hope on the horizon? Curr. Infect. Dis. Rep. 2018;20:23. doi: 10.1007/s11908-018-0629-6. [DOI] [PubMed] [Google Scholar]
- Oliveira V.C., Bim F.L., Monteiro R.M., Macedo A.P., Santos E.S., Silva-Lovato C.H., Paranhos H.F.O., Melo L.D.R., Santos S.B., Watanabe E. Identification and characterization of new bacteriophages to control multidrug-resistant Pseudomonas aeruginosa biofilm on endotracheal tubes. Front. Microbiol. 2020;11:580779. doi: 10.3389/fmicb.2020.580779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onsea J., Soentjens P., Djebara S., Merabishvili M., Depypere M., Spriet I., De Munter P., Debaveye Y., Nijs S., Vanderschot P., et al. Bacteriophage application for difficult-to-treat musculoskeletal infections: development of a standardized multidisciplinary treatment protocol. Viruses. 2019;11 doi: 10.3390/v11100891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole G.A. Microtiter dish biofilm formation assay. J. Vis. Exp. 2011;2437 doi: 10.3791/2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page A.J., Cummins C.A., Hunt M., Wong V.K., Reuter S., Holden M.T., Fookes M., Falush D., Keane J.A., Parkhill J. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Cui X., Zhang F., He Y., Li L., Yang H. Genetic evidence for O-specific antigen as receptor of Pseudomonas aeruginosa phage K8 and its genomic analysis. Front. Microbiol. 2016;7:252. doi: 10.3389/fmicb.2016.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubalskii E., Ruemke S., Salmoukas C., Boyle E.C., Warnecke G., Tudorache I., Shrestha M., Schmitto J.D., Martens A., Rojas S.V., et al. Bacteriophage therapy for critical infections related to cardiothoracic surgery. Antibiotics (Basel) 2020;9:232. doi: 10.3390/antibiotics9050232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schooley R.T., Biswas B., Gill J.J., Hernandez-Morales A., Lancaster J., Lessor L., Barr J.J., Reed S.L., Rohwer F., Benler S., et al. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant acinetobacter baumannii infection. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.00954-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- Singer A.C., Kirchhelle C., Roberts A.P. Reinventing the antimicrobial pipeline in response to the global crisis of antimicrobial-resistant infections. F1000Res. 2019;8:238. doi: 10.12688/f1000research.18302.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trend S., Fonceca A.M., Ditcham W.G., Kicic A., Cf A. The potential of phage therapy in cystic fibrosis: essential human-bacterial-phage interactions and delivery considerations for use in Pseudomonas aeruginosa-infected airways. J. Cyst. Fibros. 2017;16:663–670. doi: 10.1016/j.jcf.2017.06.012. [DOI] [PubMed] [Google Scholar]
- Van Belleghem J.D., Dabrowska K., Vaneechoutte M., Barr J.J., Bollyky P.L. Interactions between bacteriophage, bacteria, and the mammalian immune system. Viruses. 2018;11:10. doi: 10.3390/v11010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick R.R., Judd L.M., Gorrie C.L., Holt K.E. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. Plos Comput. Biol. 2017;13 doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winstanley C., O'Brien S., Brockhurst M.A. Pseudomonas aeruginosa evolutionary adaptation and diversification in cystic fibrosis chronic lung infections. Trends Microbiol. 2016;24:327–337. doi: 10.1016/j.tim.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemke A.C., Kocak B.R., Bomberger J.M. Sodium nitrite inhibits killing of Pseudomonas aeruginosa biofilms by ciprofloxacin. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.00448-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemke A.C., Shiva S., Burns J.L., Moskowitz S.M., Pilewski J.M., Gladwin M.T., Bomberger J.M. Nitrite modulates bacterial antibiotic susceptibility and biofilm formation in association with airway epithelial cells. Free Radic. Biol. Med. 2014;77:307–316. doi: 10.1016/j.freeradbiomed.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Hybrid assembled bacterial host genomes and bacteriophage genomes have been deposited at NCBI under BioProjects PRJNA610040 and PRJNA721956 and are publicly available as of the date of publication. Accession numbers are listed in Tables 1 and S1.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.