Summary
The plant hormone strigolactones (SLs) are secreted by plant roots to act as rhizospheric signals. Here, we present a protocol for characterizing plant-released SLs. We first outline all necessary steps required for collection, processing, and analysis of plant root exudates using the C18 column for SL extraction, followed by liquid chromatography-mass spectrometry (LC-MS) for SL quantification. We then describe image processing by SeedQuant, an open-source artificial-intelligence-based software, for measuring the biological activity of SLs in inducing root parasitic plant seed germination.
For complete details on the use and execution of this protocol, please refer to Wang et al. (2019) and Braguy et al. (2021).
Subject areas: Metabolism, Metabolomics, Model Organisms, Plant sciences, Mass Spectrometry
Graphical abstract
Highlights
-
•
Protocol for identifying strigolactones (SLs) from exudates of plant roots
-
•
Step-by-step instructions to analyze SLs by LC-MS and seed bioassay
-
•
Can be applied to most of the plant species grown hydroponically or in sand
The plant hormone strigolactones (SLs) are secreted by plant roots to act as rhizospheric signals. Here, we present a protocol for characterizing plant-released SLs. We first outline all necessary steps required for collection, processing, and analysis of plant root exudates using the C18 column for SL extraction, followed by liquid chromatography-mass spectrometry (LC-MS) for SL quantification. We then describe image processing by SeedQuant, an open-source artificial-intelligence-based software, for measuring the biological activity of SLs in inducing root parasitic plant seed germination.
Before you begin
Strigolactones (SLs) are evolutionarily conserved apocarotenoid metabolites (Walker et al., 2019; Wang et al., 2021) that originate from carotenoids cleavage (Alder et al., 2012). SLs regulate plant architecture (Gomez-Roldan et al., 2008; Umehara et al., 2008; Al-Babili and Bouwmeester, 2015), and, particularly in nutrient deficiency conditions, under which they are exuded by plant roots for establishing arbuscular mycorrhizal symbiosis (Al-Babili and Bouwmeester, 2015; Fiorilli et al., 2019). However, SLs are released at extremely low concentrations (∼picoMolar per liter of root exudate), even under phosphate starvation, and are relatively unstable, which makes their quantification difficult (Boutet-Mercey et al., 2018). The protocol below describes specifically and in a stepwise manner: the SL extraction from root exudates of rice (Wang et al., 2019, 2020) and pearl millet - when cultured in hydroponic and sand system - and their subsequent analysis using (1) the detection and quantification of SLs by LC-MS, as well as (2) the evaluation of their efficacy as seed germination stimulant of the root parasitic plant Striga hermonthica, using SeedQuant (Braguy et al., 2021). Our protocol is also suitable for many other plant species, such as Arabidopsis (Ablazov et al., 2020) and tomato; however, it may need some modifications with respect to the parameters of the MS analysis. In addition, testing the germination-inducing bioactivity may require changes in the seed preconditioning step (Matúšová et al., 2004) if other parasitic weeds are used.
Preparation of growth mediums
Timing: ∼2 h
-
1.Preparation of ½ MS medium.
-
a.Dissolve 2.21 g Murashige & Skoog (MS) Basal Medium (without vitamins) in 1 L Milli-Q water and adjust the pH to 5.8.
-
b.Autoclave ½ MS medium for further use.
-
a.
-
2.Preparation of half-strength modified Hoagland nutrient solution.
-
a.To make +Pi solution (normal growth condition), add 1 mL of each stock solution, except 10 mL of iron EDTA (FeSO4), (Table 1) to 900 mL of Milli-Q water. Thereafter, add extra Milli-Q water to make the final volume 1 L.Note: Preparation of Hoagland nutrient solution stocks is listed in Table 2.Note: For the low Pi or -Pi conditions, the solution is made with 0.01 mL/L of 0.4 mM K2HPO4 or without 0.4 mM K2HPO4, respectively.
-
b.Adjust the pH to 5.8.Note: The pH of the half-strength modified Hoagland nutrient solution should be re-adjusted to 5.8 every time before using. The growth medium (½ MS medium and half-strength modified Hoagland nutrient solution) can be kept at room temperature (23°C–25°C) up to 1 month.
 CRITICAL: All the chemicals are dissolved in the following 7 stock solutions to avoid any precipitation. All the stock solutions should be stored at 4°C.
CRITICAL: All the chemicals are dissolved in the following 7 stock solutions to avoid any precipitation. All the stock solutions should be stored at 4°C. CRITICAL: The stock solutions can be kept at 4°C up to 3 month.
CRITICAL: The stock solutions can be kept at 4°C up to 3 month.
-
a.
Table 1.
Preparation of 1 L half-strength modified Hoagland nutrient solution
| Reagent | Stock concentration | Final concentration | Amount |
|---|---|---|---|
| 1. Ammonium nitrate (NH4NO3) | 5.6 mM | 5.6 μM | 1 mL |
| 2. Magnesium sulfate heptahydrate (MgSO4∗7H2O) | 0.8 mM | 0.8 μM | 1 mL |
| 3. Iron EDTA (FeSO4) | 0.18 mM | 1.8 μM | 10 mL |
| 4. Calcium chloride dihydrate (CaCl2.2H2O) | 1.6 mM | 1.6 μM | 1 mL |
| 5. Potassium nitrate (KNO3) | 0.8 mM | 0.8 μM | 1 mL |
| 6. Potassium phosphate dibasic trihydrate (K2HPO4.3H2O) | 0.4 mM | 0.4 μM | 1 mL |
| 7. Micronutrients∗∗ | n/a | n/a | 1 mL |
| ddH2O | n/a | n/a | 984 mL |
| Total | n/a | n/a | 1,000 mL |
| ∗∗Micronutrient | ||
|---|---|---|
| Reagent | Stock concentration | Final concentration |
| Boric Acid (H3BO4) | 0.023 mM | 11.4974 μM |
| Manganese chloride (MnCl2∗4H2O) | 0.0045 mM | 2.2484 μM |
| Copper Sulfate (CuSO4∗5H2O) | 0.0003 mM | 0.1481 μM |
| Zinc Chloride (ZnCl) | 0.0015 mM | 0.75 μM |
| Sodium molybdate (Na2MoO4∗2H) | 0.0001 mM | 0.0495 μM |
Table 2.
Stocks of half-strength modified Hoagland nutrient solution
| Stock no. | Chemicals name | Concentration (mM) | Molecular weight (g/mol) | Mass (g/L) |
|---|---|---|---|---|
| 1. | NH4NO3 | 5.6 | 80.04 | 224.1 |
| 2. | MgSO4.7H2O | 0.8 | 246.48 | 98.6 |
| K2SO4 | 0.8 | 174.2 | 70.0 | |
| 3. | FeSO4.7H2O | 0.18 | 278.0 | 2.55 |
| Na2EDTA.2H2O | 0.02 | 372.2 | 1.86 | |
| 4. | CaCl2.2H2O | 1.6 | 147.02 | 117.6 |
| 5. | KNO3 | 0.8 | 101.11 | 40.4 |
| 6. | K2HPO4.3H2O | 0.4 | 228.2 | 50.0 |
| 7. | Micronutrients | |||
| H3BO3 | 0.023 | 61.84 | 0.711 | |
| MnCl2.4H2O | 0.0045 | 197.91 | 0.445 | |
| CuSO4.5H2O | 0.0003 | 249.68 | 0.037 | |
| ZnCl2 | 0.0015 | 136.32 | 0.102 | |
| Na2MoO4.2H2O | 0.0001 | 241.95 | 0.012 | |
1. NH4NO3; 2. K2HPO4.3H2O; 3. MgSO4+ K2SO4; 4. FeSO4.7H2O and Na2EDTA.2H2O; 5. CaCl2; 6. KNO3; 7. Micro nutrient.
Plant growth
Timing: Rice (Oryza sativa L.) as an example: 21 days for hydroponic; 42 days for sand
-
3.Seed sterilization.
-
a.Place the rice seeds (Oryza sativa L.) in a 50 mL falcon tube containing 50% commercial bleach (sodium hypochlorite) solution (Milli-Q water + commercial bleach; 1:1 v/v).
-
b.Continuously shake the tube in a rotator for 10 min to sterilize the surface of the seeds.Note: An addition of 2 drops of Tween-20 to the 50% sodium hypochlorite solution can improve the seed sterilization step.Optional: For the sterilization of pearl millet (Pennisetum glaucum) seeds, do not add Tween-20 and sterilize for only 5 min.
-
c.In a sanitized laminar flow cabinet, wash the seeds successively 5–6 times with sterilized Milli-Q water.
-
d.For imbibition, keep the seeds in the falcon tube with 30 mL sterilized Milli-Q water overnight (∼12 h) in an incubator, in the dark at 30°C.Note: Pearl millet seeds can be directly placed in the Petri dish without imbibition (see step 4).
-
a.
-
4.
After imbibition, spread 10 rice seeds on a 100 mm × 15 mm Petri dish, containing two filter papers moistened with 5 mL of ½ MS medium.
Note: Manipulate under sterile conditions in a laminar flow cabinet.
Optional: For pearl millet, spread around 20 seeds per 100 mm × 15 mm Petri dish, containing two filter papers moistened with 5 mL of ½ MS medium.
-
5.
Seal the Petri dishes with parafilm, and wrap them in aluminum foil to avoid exposure of light to the seeds.
-
6.
Place the Petri dishes in a 30°C incubator for 2 days.
-
7.
Remove the aluminum foil and the parafilm, and transfer the germinated seeds to a growth chamber with day/night temperature of 28/22°C and a 12 h photoperiod, 200 μmol photons m−2 s−1 for 5 days.
Note: Check the seeds on the third day and add ½ MS medium if needed.
-
8.Grow rice seedlings under hydroponic conditions.
-
a.Make one hole in the center of the cap of 50 mL tube.
-
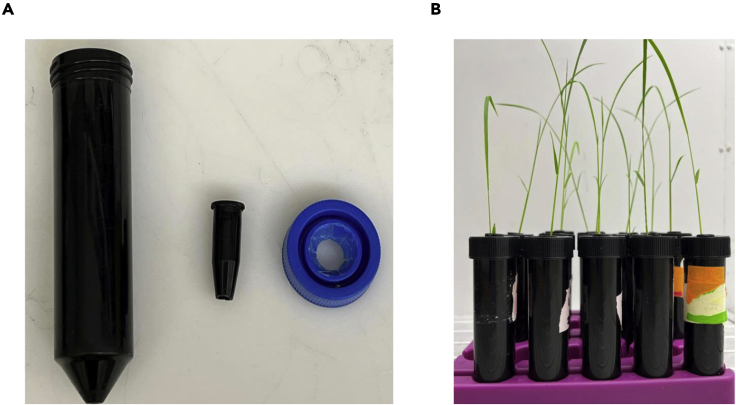
b.Cut the bottom of a 1.5 mL Eppendorf tube, remove the lid if it has one, and place it in the hole of the 50 mL tube’s cap (Figure 1A).
-
c.Fill the 50 mL black tube with +Pi Hoagland nutrient solution.
-
d.Transfer one-week-old rice seedlings (1–2) through the 1.5 mL bottomless Eppendorf tube in the center of each cap and fix the cap into 50 mL black tubes (Figure 1B).
-
e.Keep the tubes with seedlings in a growth chamber for 1 week (+P) and refresh the nutrient solution two times per week.
-
a.
Optional: For pearl millet, transfer seven germinated seedlings to a 2 L pot filled with silver sand and apply 1 L +Pi half-strength modified Hoagland nutrient solution twice per week for 4 weeks.
-
9.
Trigger the SL production and exudation by applying (–Pi) half-strength modified Hoagland nutrient solution for another 1 week (Wang et al., 2019).
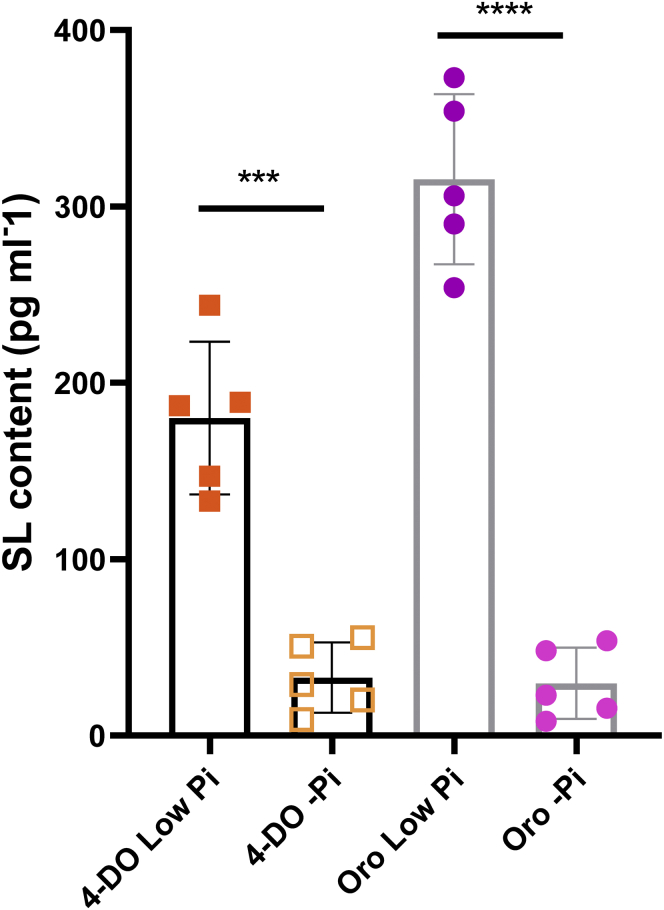
Note: Rice seedlings can be directly grown under low Pi conditions for two weeks, which generally showed higher SL content compared to -Pi conditions (Figure 2).
Figure 1.
The setup for the hydroponic system of 50 mL falcon tube
(A) Make one hole in the center of the cap of the 50 mL tube. Cut the bottom of a 1.5 mL lid-removed black Eppendorf tube and place it in the hole of the 50 mL tube’s cap.
(B) A represented picture of rice seedlings grown under hydroponic system.
Figure 2.
SL quantification of rice grown under Low Pi and -Pi conditions
Data represent mean ± SD. n=5. Statistical analysis was performed using two-tail student t-test. Different letters denote significant differences (∗p < 0.05, ∗∗p< 0.01, ∗∗∗p< 0.001, ∗∗∗∗p< 0.0001). 4-DO, 4-deoxyorobanchol; Oro, Orobanchol.
Preconditioning of Striga hermonthica seeds
Timing: ∼10 days
The bioactivity of root-released SLs can be evaluated on the parasitic seeds of the Orobanchaceae family, as seed germination stimulants. However, to be responsive to external chemical signals, the parasitic seeds require pre-conditioning steps (Jamil et al., 2012) mimicking a humid and warm environment (see steps below).
-
10.Cleaning of Striga seeds.
-
a.Place 1 g Striga seeds in a 50 mL falcon tube with 40 mL tap water. Soak the seeds in water and agitate them on a rotator for 30 min.
-
b.In a new 50 mL falcon tube, add 15 mL of 60% sucrose, followed by 10 mL of 40% sucrose solution added drop-by-drop.
 CRITICAL: Make sure to not disturb the sucrose gradient.
CRITICAL: Make sure to not disturb the sucrose gradient. -
c.Transfer gently 20 mL of the water-soaked-seeds to the sucrose-containing tube, without disturbing the gradient.
-
d.Centrifuge at 1,356 × g for 10 min and thereafter collect the Striga seeds from the inner phase with the help of pipette in a vacuum assembly.
-
e.Remove the sucrose from the seeds by 5–6 times successive washings with sterilized Milli-Q water. Let the wet seeds entirely dry in a laminar flow cabinet.Note: This drying step takes 12–42 h.
 Pause point: Keep the cleaned seeds in the Petri dish at room temperature (23°C–25°C) for long-term storage.
Pause point: Keep the cleaned seeds in the Petri dish at room temperature (23°C–25°C) for long-term storage.
-
a.
-
11.Pre-conditioning.
-
a.Place the cleaned Striga seeds (∼5 mg/12 glass fiber filter discs) in a 50 mL tube.
-
b.Add 20 mL 50% bleach solution (sterilized Milli-Q water+commercial bleach; 1:1 v/v) into the tube.
-
c.Shake the tube in a rotator for 7 min.
 CRITICAL: Please do not keep more than 7 min because a long-term exposure can be lethal for the seeds.
CRITICAL: Please do not keep more than 7 min because a long-term exposure can be lethal for the seeds. -
d.Transfer the seeds to a vacuum assembly to wash away the bleach by 5–6 successive washings with sterilized Milli-Q water in a laminar flow cabinet and dry the seeds entirely.
-
e.Place twelve 9 mm sterilized glass fiber filter paper discs on a round glass Petri dish plate and spread evenly ∼50–100 Striga seeds on each disc.
-
f.Put a sterilized 9 cm diameter Whatman filter paper in a 9 cm round plastic Petri dish plate and add 3 mL sterilized Milli-Q water on it.
-
g.Transfer the above-mentioned (f) glass fiber filter discs with Striga seeds (12 discs per plate) and seal the plate with parafilm.
-
h.Cover the plate(s) with aluminum foil and place the plate in an inverted position (keep the seeds upside down) in the incubator at 30°C for 10 days.
 CRITICAL: As water exposure will disturb Striga seed germination, please keep the plate inverted.
CRITICAL: As water exposure will disturb Striga seed germination, please keep the plate inverted.
-
a.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Methanol | VWR (≥99.9% (by GC), HiPerSolv CHROMANORM® for LC-MS) | Cat#83638 |
| Water | VWR (HiPerSolv CHROMANORM® for LC-MS) | Cat#83645 |
| Milli-Q Water | Merck (Quantum® Polishing Cartridge) | QTUM0TEX1 |
| Ethyl Acetate | VWR (≥99.8%, HiPerSolv CHROMANORM® for HPLC) | Cat#83621 |
| Acetonitrile | Fisher Scientific (Optima™ LC/MS Grade) | Cat#A955 |
| Formic acid | Merck (98%–100% for LC-MS LiChropur®) | Cat#533002 |
| 4-deoxyorobanchol | OlChemIm | Cat#0257141 |
| Orobanchol | OlChemIm | Cat#0256701 |
| rac-GR24 | Strigolab | https://strigolab.eu/ |
| D6-5-deoxystrigol | The University of Tokyo | https://www.u-tokyo.ac.jp/focus/en/people/people001154.html |
| Ammonium nitrate | Fisher Scientific US | Cat#A676212 |
| Magnesium sulfate heptahydrate | GOLD BIOTECH | Cat#M-020-1 |
| Iron(II) sulfate heptahydrate, plant cell culture tested | Sigma-Aldrich | Cat#F8263-1KG |
| Ethylenediaminetetraacetic acid disodium salt dihydrate (EDTA) | Sigma-Aldrich | Cat#E6635-1KG |
| Calcium chloride dihyd 99% acs | Sigma-Aldrich | Cat#223506-500G |
| Potassium nitrate | Fisher Scientific US | Cat#MK6715212 |
| Potassium phosphate dibasic trihydrate 2ReagentPlus(R), >=99.0% | Sigma-Aldrich | Cat#P5504-5KG |
| Boric Acid | Sigma-Aldrich | Cat#B6768 |
| Manganese chloride tetrahydrate | ACROS ORGANICS | Cat#205895000 |
| Copper Sulfate pentahydrate | Sigma-Aldrich | Cat#C8027 |
| Zinc Chloride | Sigma-Aldrich | Cat#793523 |
| Sodium molybdate | ACROS ORGANICS | Cat#206375000 |
| Murashige & Skoog (MS) Basal Medium | Sigma-Aldrich | Cat#M5524-50L |
| Grade 4 qualitative filter papers, Whatman™, 90 mm, 1 cm thickness | A-VWR, Part of Avantor | Cat#512-1026 |
| Plastic round Petri dish (PETRI DISH 90 × 16.2 MM) | VWR INTERNATIONAL, LTD-UK | Cat#391-0443 |
| Silver sand | Hanson | https://www.hanson-packedproducts.co.uk/en/products/base-aggregates/silver-sand |
| Pierce™ FlexMix™ Calibration Solution | Thermo Scientific | Cat#A39239 |
| 50 mL black falcon tube | Heathrow Scientific | Cat#518520 |
| 1.5 mL Eppendorf tube | Eppendorf, North America | Cat#022363204 |
| 1.5 mL Black Eppendorf tube | Argos Technologies™ | Cat#5087954 |
| 8 mL brown glass vial | VWR North America | Cat#548-0889 |
| Cap of 8 mL glass vial | VWR North America | Cat#548-0862 |
| 1.5 mL glass autosampler vial | VWR North America | Cat#VWRI548-0030 |
| Cap of 1.5 mL glass autosampler vial | VWR North America | Cat#89239-018 |
| 0.22 μm filter | Thermo Scientific | Cat#00215484 |
| Commercial bleach (sodium hypochlorite) | Clorox® Bleach | https://www.cloroxarabia.com/en/products/clorox-bleach/original/ |
| Tween-20 | Thermo Scientific | Cat# 85113 |
| Experimental models: Organisms/strains | ||
|
Oryza sativa: Nipponbare 3-week-old seedlings |
KAUST | Butt et al. (2018)https://doi.org/10.1186/s12870-018-1387-1 |
|
Oryza sativa: d17 (background Nipponbare) 3-week-old seedlings |
KAUST | Butt et al. (2018)https://doi.org/10.1186/s12870-018-1387-1 |
|
Pennisetum glaucum: 29Aw 7-week-old plants |
International Crops Research Institute for the Semi-Arid Tropics (ICRISAT) | Dayou et al. (2021)https://doi.org/10.1017/wsc.2021.12 |
| Software and algorithms | ||
| Xcalibur™ Software | Thermo Fisher Scientific | OPTON-30965 |
| LAS-EZ-V3-0 software | Leica Microsystems | https://www.leica-microsystems.com/products/microscope-software/p/leica-las-ez/ |
| SeedQuant | KAUST | https://braguyjm.github.io/SeedQuant2/ |
| MultiQuant 2.1 | SCIEX | https://sciex.com/products/software/multiquant-software |
| Other | ||
| Hypersil GOLD™ C18 Selectivity HPLC Columns | Thermo Scientific | 25003-032130 |
| SPE C18-Fast (500 mg/3 mL) | SEClute™ | 5138758 |
| Leica LED3000 R mounted with a CCD camera (Leica Microsystems) | Leica Microsystems | https://www.leica-microsystems.com/products/microscope-accessories/p/leica-led3000-bli/ |
| UHPLC-Orbitrap ID-X Tribrid Mass Spectrometer | Thermo Fisher Scientific | https://www.thermofisher.com/sa/en/home/industrial/mass-spectrometry/liquid-chromatography-mass-spectrometry-lc-ms/lc-ms-systems/orbitrap-lc-ms/orbitrap-tribrid-mass-spectrometers/orbitrap-iq-x.html |
| HPLC-triple quadrupole/linear ion trap instrument (QTRAP5500) | AB Sciex | https://sciex.com/products/mass-spectrometers/qtrap-systems/qtrap-5500-system |
| UHPLC- Triple-Stage Quadrupole Mass Spectrometer (TSQ-AltisTM) | Thermo Fisher Scientific | https://www.thermofisher.com/order/catalog/product/TSQ02-10002 |
Step-by-step method details
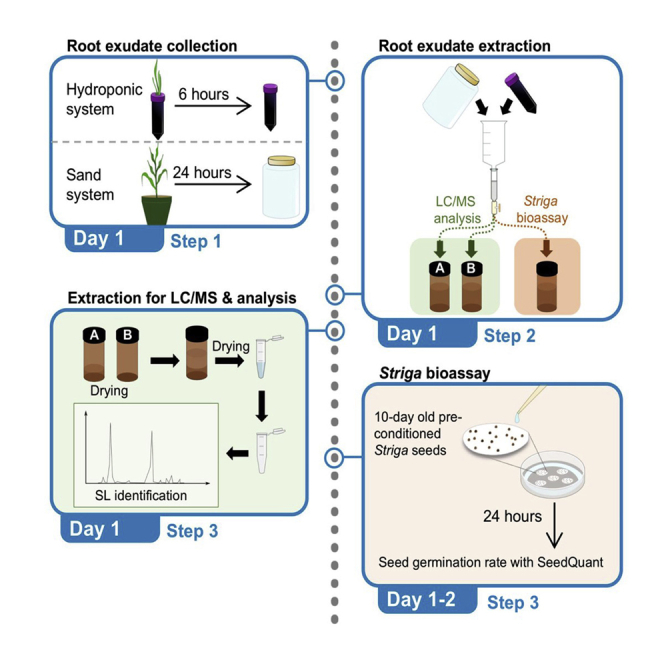
SL collection and extraction
Timing: ∼10 h
Under phosphate starvation, roots of hydroponically grown plants continuously release SLs into the medium. Therefore, we recommend to let SLs accumulate in the medium for 6 h and then collect it (Wang et al., 2019): we usually refresh the solution in the early morning and collect the medium for SL extraction 6 h later.
Optional: For plants grown in the sand system, we recommend to collect the exudate 24 h after pre-washing the pots with 1 L –Pi half-strength modified Hoagland solution (Jamil et al., 2014).
-
1.Root exudates collection.
-
a.In the early morning, prepare fresh –Pi/Low Pi half-strength modified Hoagland solution with pH adjusted to ∼5.8.
-
b.Refresh the medium (50 mL) of each tube containing rice seedlings and wait for 6 h.
-
c.After 6 h, remove the plants from the tubes and store the collected medium with 50 mL tubes on ice.
-
a.
Alternatives: When using the sand system, we recommend to collect 500 –1,000 mL root exudates by adding –Pi/Low Pi half-strength modified Hoagland solution on the top of each pot and collect the flow-through, from the bottom of the pot, in 1 L plastic bottle.
Note: During the collection, protect the collected root exudates from light to avoid SL degradation. We also recommend to extract the collected samples as soon as possible without any delay, as SLs degrade quickly in the water when exposed to light and high temperature.
Optional: Transfer the seedlings to a new 50 mL tubes containing fresh half-strength modified Hoagland solutions for a second collection in case something goes wrong during the extraction.
Optional: At this stage, plant tissues can be also collected for SL or transcript analysis. They should be immediately frozen in liquid nitrogen and stored at −80°C.
-
2.SL extraction from root exudates.
-
a.Keep the collected root exudates on ice. In the lab, spike each sample, collected both from hydroponic and sand, with 0.672 ng of D6-5-deoxystrigol - used as internal standard (IS).Alternatives: The internal standard can be replaced by using the SL analog GR24 (1 ng).Note: If the collected exudate is to be applied to parasitic seeds, do not add internal standard. The latter will trigger high germination!
-
b.Pre-condition Fast SPE C18 column (500 mg/3 mL; GracePure), connected with SPE Vacuum Manifold with 3 mL of methanol, followed by 3 mL of Milli-Q water.Note: After conditioning, keep the columns wet with water and do not let them dry before sample loading! The pre-conditioning step enables a condition to enable the analyte adsorption. Wetting the adsorbent with a suitable solvent ensures reproducibility.
-
c.Attach a 50 mL syringe to the pre-conditioned C18 column and load the collected root exudate samples (before you begin step 9) onto them. After enrichment, wash the C18 column with 3 mL Milli-Q water to remove impurities.Optional: To get better MS resolution and sensitivity for SL identification, we suggest to pool at least 500 mL–1,000 mL root exudate.Note: If the volume of root exudate exceeds 200 mL, we strongly advice to add a pre-step, using glass fiber filter paper through a vacuum assembly, to remove impurities and plant debris.
-
d.Elute SLs twice, into two separate 8 mL brown glass vials: use 2 mL acetone to collect the first fraction A, followed by 3 mL acetone for the fraction B.Note: For the parasitic seeds bioassay, elute the collected exudate with 3 mL acetone only, as the parasitic weeds are much more sensitive than LC-MS.
 Pause point: Seal the collected samples with caps in the brown glass vials and keep the acetone extracted samples in −20°C refrigerator for 2-week storage or at −80°C for long-term storage.
Pause point: Seal the collected samples with caps in the brown glass vials and keep the acetone extracted samples in −20°C refrigerator for 2-week storage or at −80°C for long-term storage. CRITICAL: Try to avoid direct light exposure during the extraction process.
CRITICAL: Try to avoid direct light exposure during the extraction process.
-
a.
-
3.SL re-extraction for LC-MS.
-
a.Fully dry the fraction B (from SL collection and extraction step 2d; containing mostly acetone) under vacuum.
-
b.Add 1 mL ethyl acetate into each vial containing the dried fraction B, and gently vortex for 5 s.
-
c.Dry the fraction A (from SL collection and extraction step 2d; containing a mix of acetone and water) under vacuum, which results in ∼300 μL partially dry (acetone-water).Optional: If space allows it, please vacuum-dry Fraction A and B together.
-
d.Transfer 1 mL of re-suspended fraction B in ethyl acetate to fraction A, and gently vortex for 5 s. Two layers with ethyl acetate (up) and water (down) will appear.
-
e.Centrifuge the samples at 1,356 × g for 2 min at room temperature.
-
f.Carefully transfer 750 μL from the upper-layer (SLs enriched organic phase) to a new 1.5 mL Eppendorf tube.
 CRITICAL: Do not touch the lower layer to avoid water contamination.
CRITICAL: Do not touch the lower layer to avoid water contamination. -
g.Fully dry the samples using a speed vacuum at room temperature.
 Pause point: Keep the extracts in −20°C refrigerator if you do not want to process them immediately.
Pause point: Keep the extracts in −20°C refrigerator if you do not want to process them immediately. -
h.Dissolve the dried samples in 100 μL of acetonitrile:water (25:75, v:v) and filter them gently through a 0.22 μm filter into a glass autosampler vial.
-
i.Close the vial with the cap before LC-MS/MS analysis.
-
j.For better detection, gently tap the bottom of the vial to remove the gases in the samples.
 Pause point: Keep the filtered samples in −20°C refrigerator for storage.
Pause point: Keep the filtered samples in −20°C refrigerator for storage.
-
a.
SL identification by LC-MS/MS
Timing: 23 min per sample
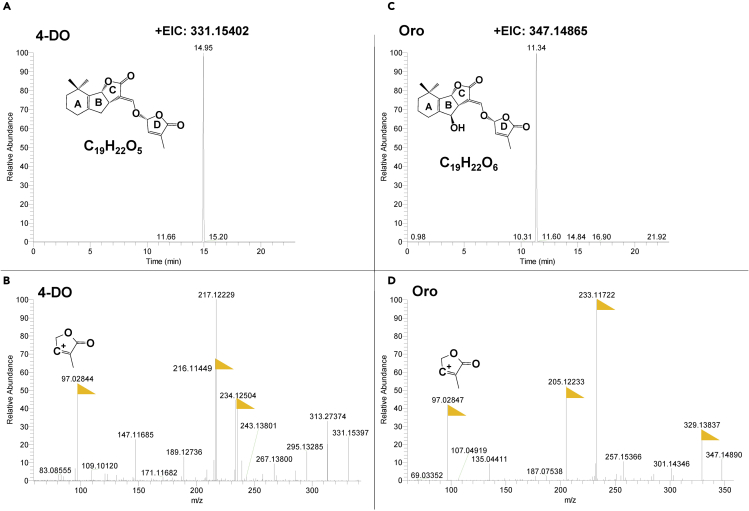
Although SLs are divided into canonical and non-canonical SLs (Wang et al., 2021), they are generally traced and identified using the same methods. In our case, we use the IDX-Qrbitrap high-resolution Mass Spectrometry instrument, including the MS and MS/MS scans. The full scan MS analysis revealed the presence of the SL (4-deoxyorobanchol) at m/z 331.15402 (RT: 14.95 min). The MS/MS scan confirmed the presence of the common D-ring moiety fragment ion at m/z 97.02823, which corresponds to the conserved diagnostic product ion of SLs (Figures 3 and 4).
Note: The IDX Orbitrap Mass Spectrometer is an analytical technique that contains three mass analyzers used for determining the m/z of small and big molecules. The Orbitrap IDX spectrometer could reach a high resolution (> 120,000) and reliable mass accuracy (<3 ppm mass error). The Mass spectrometer was calibrated using a purchasable “Calibration Mix ESI (Thermo Scientific)” and by following the manufacturer’s guidelines. Electrospray ionization in positive mode (ESI+) was applied for the studied compounds, using the following parameters: vaporized temperature = 100°C, voltage = 3,500 V, sheath gas = 30, auxiliary gas: 15, ion source fragmentation = 35 V, capillary temperature = 300°C. 10 μl of each sample were injected through a loop injection to a C18 column using an independent UPLC pump.
Note: The separation of the studied metabolites was performed on a Hypersil GOLD™ C18 Selectivity HPLC Columns (150 × 4.6 mm; 3 μm) maintained at 35°C.
Note: The mobile phases consisted of (A) 100% LC-MS grade water + 0.1% formic acid and (B) 100% Acetonitrile + 0.1% formic acid (Table 3). A gradient elution method was used (Table 4).
Note: Identification of SLs is performed using an UHPLC system coupled with an Orbitrap ID-X Tribrid Mass Spectrometer, which runs in positive and negative modes. The MS parameters are listed in Table 5 and the parameters MS/MS are listed in Table 6.
Note: Overall cycle time per sample is 23 min.
-
4.
Prepare solvents as described in the Table 3.
-
5.
Install the running solvent lines into the UHPLC solvent reservoirs.
-
6.
Purge the solvent lines for 5 min.
-
7.
Equilibrate the Orbitrap ID-X Tribrid system as shown in Tables 5 and 6.
-
8.
Create a batch table.
-
9.
Analyze samples in both positive and negative ion modes.
Note: Injection volumes can vary from 5 to 10 μL, depending on sample concentrations.
CRITICAL: To prevent a shift in the retention time(s), the same batch of samples should be analyzed within 48 h in the same run.
-
10.
Data processing by using Xcalibur™ Software.
-
11.
Elemental composition to Calculate the theoretical accurate mass, e.g., 331.15400 for 4-DO, and search from the mass data using Xcalibur™ Software with 5 ppm mass tolerance (Figure 3).
-
12.
Check MS/MS data to make sure to identify with the authentic standard (Figure 3).
Note: We provide the MS/MS spectrum of authentic 4-DO, Oro, and GR24 which can be used as references.
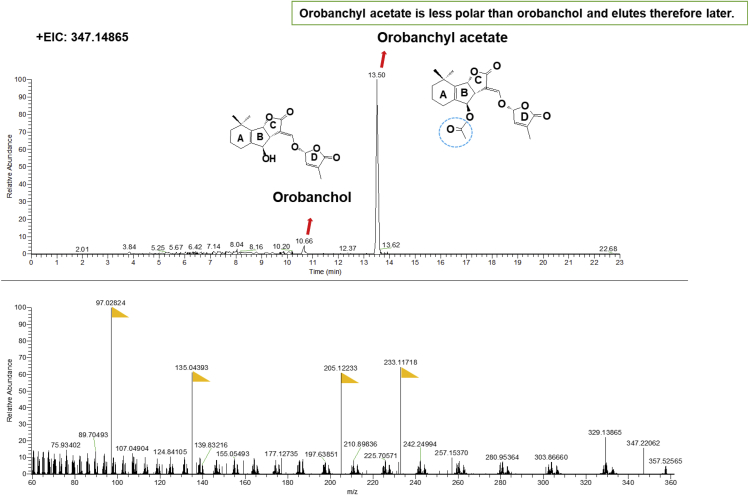
Note: In Figure 5, the Oro and Orobanchyl acetate showed the same MS/MS, which could be distinguished by retention time.
Figure 3.
The chromatography of SL identification from authentic standards
(A) Accurate mass of 4-deoxyorobanchol (4-DO).
(B) MS/MS fragmentation of 4-DO (Seto et al., 2014).
(C) Accurate mass of Orobanchol (Oro).
(D) MS/MS fragmentation of Oro (Seto et al., 2014). The yellow flags point out the major fragments. +EIC = extracted ion chromatogram of positive mode.
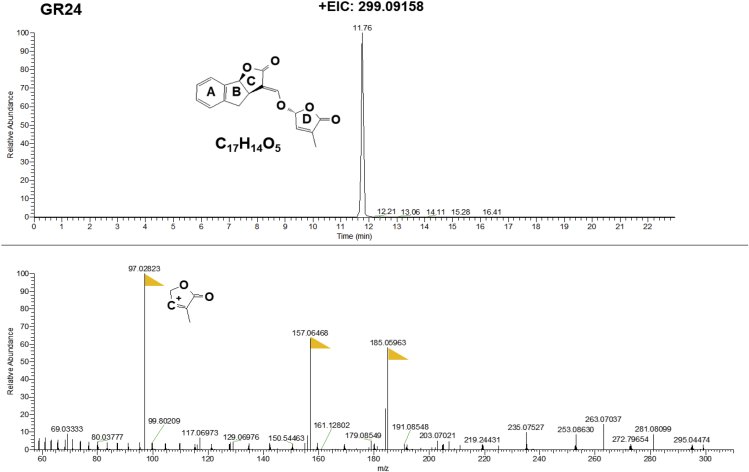
Figure 4.
The chromatography of SL identification from authentic standard GR24
Accurate mass of GR24 (up). MS/MS fragmentation of GR24 (down) (Rebecca et al., 2020). The yellow flags point out the major fragments. +EIC = extracted ion chromatogram of positive mode.
Table 3.
Solvent composition
| Reagent | Final concentration | Amount |
|---|---|---|
| (A) Water | N/A | 500 mL |
| (B) Acetonitrile | N/A | 500 mL |
| Formic acid | 0.1% (V/V) |
Table 4.
LC gradient condition for SL identification
| Time | Flow (mL/min) | %A | %B |
|---|---|---|---|
| 0 | 0.5 | 75 | 25 |
| 15 | 0.5 | 0 | 100 |
| 20 | 0.5 | 0 | 100 |
| 21 | 0.5 | 75 | 25 |
| 23 | 0.5 | 75 | 25 |
Table 5.
General settings of MS
| Parameters | Values |
|---|---|
| Ionization | Heated Electrospray Ionization |
| Spray Voltage | Static |
| Positive Ion (V) | 3,500 |
| Negative Ion (V) | 2,500 |
| Gas Mode | Static |
| Sheath Gas (Arb) | 60 |
| Aux Gas (Arb) | 15 |
| Sweep Gas (Arb) | 2 |
| Ion Transfer Tube Temperature (°C) | 350 |
| Vaporized Temperature (°C) | 400 |
| Detector Type | Orbitrap |
| Orbitrap Resolution | 120000 |
| Use Quadrupole Isolation | TRUE |
| Scan Range (m/z) | 50–500 |
| RF Lens (%) | 60 |
| AGC Target | Standard |
| Microscans | 1 |
| Data Type | Centroid |
Table 6.
General settings of MS/MS
| Parameters | Values |
|---|---|
| Isolation Mode | Quadrupole |
| Isolation Window (m/z) | 1.6 |
| Isolation Offset | Off |
| Activation Type | HCD |
| Collision Energy Mode | Assisted |
| HCD Assisted Collision Energies (%) | 10,20,30,40 |
| Detection Type | Orbitrap |
| Orbitrap Resolution | 60,000 |
| Scan Range Mode | Auto |
| AGC Target | Standard |
| Microscans | 1 |
| Data Type | Centroid |
Figure 5.
The chromatography of SL identification from pearl millet exudate
Accurate mass of Orobanchol (up). MS/MS fragmentation of Orobanchyl acetate (down). The yellow flags point out the major fragments. +EIC= Extracted ion chromatogram of positive mode.
SL quantification by LC-MS/MS
Timing: 25 min per sample
A highly specific and sensitive method has been accomplished to selectively quantify compounds within complex mixtures by quadrupole MS analyzer. Quantification of SLs was performed by the UHPLC-TSQ-Altis Mass spectrometer using Multiple Reaction Monitoring (MRM) experiment (Figure 6A).
Note: The TSQ Altis is a triple stage quadrupole Mass Spectrometer that determines the m/z of small molecules and which provides the best transmission and peak shape. This instrument has an electrospray ionization source and is capable of scan functions, such as Selective Ion Monitoring (SIM), Selective/Multiple Reaction Monitoring (S/MRM), and precursor ion scanning. The Mass spectrometer was calibrated using a purchasable “Calibration Mix ESI (Thermo Scientific)” by following the manufacturer’s guidelines.
Note: The separation of metabolites is on a Hypersil GOLD™ C18 Selectivity HPLC Columns (150 × 4.6 mm; 3 μm) maintained at 35°C.
Note: The mobile phases consist of (A) 100% LC-MS grade water + 0.1% formic acid and (B) 100% Acetonitrile plus 0.1% formic acid are used for gradient elution (Tables 7 and 8).
Note: Identification of SLs is performed using an UHPLC system coupled with a Triple-Stage Quadrupole Mass Spectrometer (TSQ-AltisTM), which runs in positive mode. The general MS parameters of MRM are listed in Table 9, and the ion transitions of SLs are listed in Table 10.
Note: Overall cycle time per sample is 25 min.
Optional: The MRM parameters of HPLC-triple quadrupole/linear ion trap instrument (QTRAP5500) are listed in Table 11 that shown in Figure 6B while the settings of ion transitions are the same as in Table 10.
-
13.
Prepare solvents as described in the Table 7.
-
14.
Install the running solvent lines into the UHPLC solvent reservoirs.
-
15.
Purge the solvent lines for 5 min.
-
16.
Equilibrate the Triple-Stage Quadrupole Mass Spectrometer (TSQ-AltisTM) system as shown in Table 9.
-
17.
Create a batch table and insert pooled QC samples once every ten times.
Optional: Insert authentic standard samples, if having, in the beginning and end of running.
-
18.
Analyze samples in positive ion modes.
Note: Injection volumes can be changed from 5 to 10 μL, depending on the sample concentrations.
CRITICAL: To prevent retention time-shifting, the same batch of samples should be analyzed within 48 h in the same run.
-
19.
Analyze data in Xcalibur™ Software.
-
20.
Create a processing method based on the MRM results before batch analysis.
-
21.
Output automatic integrative peak area results as a CSV file for calculation (see quantification and statistical analysis section).
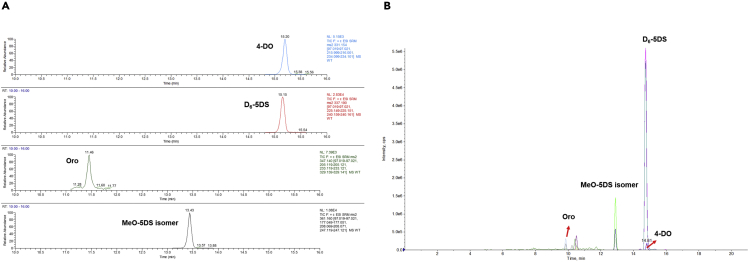
Figure 6.
The representative UHPLC–MS/MS chromatograms.
Overview of MRM chromatography from (A) TSQ-Altis and (B) QTRAP5500 Mass spectrometer, respectively. 4-DO, 4-deoxyorobanchol; Oro, Orobanchol; MeO-5DS, methoxy-5-deoxystrigol; D6-5DS, isotopically labeled (D6)-5-deoxystrigol.
Table 7.
Solvent composition
| Reagent | Final concentration | Amount |
|---|---|---|
| (A) Water | N/A | 500 mL |
| (B) Acetonitrile | N/A | 500 mL |
| Formic acid | 0.1% (V/V) |
Table 8.
LC gradient condition for MRM analysis
| Time | Flow (mL/min) | %A | %B |
|---|---|---|---|
| 0 | 0.5 | 75 | 25 |
| 15 | 0.5 | 0 | 100 |
| 20 | 0.5 | 0 | 100 |
| 21 | 0.5 | 75 | 25 |
| 25 | 0.5 | 75 | 25 |
Table 9.
MS parameter settings of SRM analysis
| Parameters | Values |
|---|---|
| Ionization | Heated Electrospray Ionization |
| Spray Voltage | Static |
| Positive Ion (V) | 4,000 |
| Negative Ion (V) | 3,500 |
| Sheath Gas (Arb) | 40 |
| Aux Gas (Arb) | 15 |
| Sweep Gas (Arb) | 5 |
| Ion Transfer Tube Temperature (°C) | 350 |
| Vaporized Temperature (°C) | 350 |
| Cycle Time (Sec) | 0.8 |
| Q1 Resolution (FWHM) | 0.2 |
| Q3 Resolution (FWHM) | 0.2 |
| CID Gas (mTorr) | 2 |
| Chromatographic Peak Width (Sec) | 6 |
Table 10.
The MRM detection parameters of SLs
| Strigolactones | Monitoring transitions (m/z; [M+H] +) |
||
|---|---|---|---|
| Precursor ion | Diagnostic product ion | Confirming product ion | |
| D6-5DS | 337.19 | 97.02 | 240.16 | 222.15 |
| GR24 | 299.08 | 97.02 | 157.06 | 185.05 |
| 4DO | 331.15 | 97.02 | 234.1 | 216.0 |
| Oro | 347.14 | 97.02 | 329.14 | 233.12 | 205.12 |
| MeO-5DS isomer | 361.16 | 97.02 | 247.12 | 208.07 | 177.05 |
| Oro-Acetate∗ | 389.15 | 97.02 | 329.14 | 233.12 | 205.12 |
| 411.1∗ ∗[M+Na] + |
97.02 | 351.1 | 254.1 | |
Table 11.
The MRM parameters of HPLC-triple quadrupole/linear ion trap instrument (QTRAP5500)
| Parameters | Values |
|---|---|
| Ionization | Turbo Spray |
| Spray Voltage | 5,500 |
| Curtain Gas (CUR) | 20 |
| Collision Gas (CAD) | Medium |
| Temperature (TEM) | 400 |
| Ion Source Gas 1 (GS1) | 80 |
| Ion Source Gas 2 (GS2) | 70 |
| Declustering Potential (DP) | 60 |
| Entrance Potential (EP) | 12 |
| Collision Energy (CE) | 16 |
| Collision Cell Exit Potential (CXP) | 15 |
SL bioactivity on Striga seed germination
Timing: ∼30 h
-
22.Preparation of collected SLs for bioassays.
-
a.Take 400 μL of each SL-containing sample eluted by acetone (from SL collection and extraction step 2d) into 2 mL Eppendorf tube and add 400 μL sterilized Milli-Q water (1:1 dilution).
-
b.Remove the acetone by speed vacuum for 60 min at room temperature (23°C–25°C) or preferably lower temperature.
-
a.
CRITICAL: As acetone can affect the germination, make sure to keep enough time for evaporation. One can make a mark on the tube corresponding to 400 μL volume, to ensure the total evaporation of acetone in the sample.
Optional: For the SL-containing samples collected from 500 mL to 1,000 mL, try to dilute the sample into a 1:3 to 1:9 ratio for the bioassays.
-
23.Striga germination bioassays.
-
a.Dry the 10-day-preconditioned Striga discs (from the section of preconditioning of Striga hermonthica seeds) in a laminar flow cabinet.
-
b.Place a filter paper ring (∼1 cm wide) cut from a 9 cm diameter Whatman filter paper in the 9 cm plastic Petri dish plate.
-
c.Transfer six Striga discs to each Petri dish plate in the middle of the ring.
 CRITICAL: Avoid touching the filter paper ring, as water will interfere with the seed germination.
CRITICAL: Avoid touching the filter paper ring, as water will interfere with the seed germination. -
d.Apply 50 μL of aforementioned SL-containing sample (1:1 dilution) on each Striga disc and humidify the filter paper ring with 900 μL sterilized Milli-Q water.
-
e.Seal the plates with parafilm and cover with aluminum foil. Incubate at 30°C for 24 h to germinate.
 CRITICAL: Please include a positive control (50 μL of 1 μM GR24) and negative control (50 μL of sterilized H2O) to ensure the quality of the experiment.
CRITICAL: Please include a positive control (50 μL of 1 μM GR24) and negative control (50 μL of sterilized H2O) to ensure the quality of the experiment.
-
a.
-
24.Scanning and counting.
-
a.Take the incubated plates out of the incubator and open them to dry for 15 min.Note: If the discs are not dry, there will be reflected when the discs will be imaged.
-
b.Capture the images of seed-containing discs individually with a Leica LED3000 R adjusted to 50% medium light, mounted with a CCD camera (Leica Microsystems) and store the images in a laptop.Note: The images were saved in 8-bit, 2,592 × 1,944 pixels and exported in JPEG format using the LAS-EZ-V3-0 software for image acquisition.
-
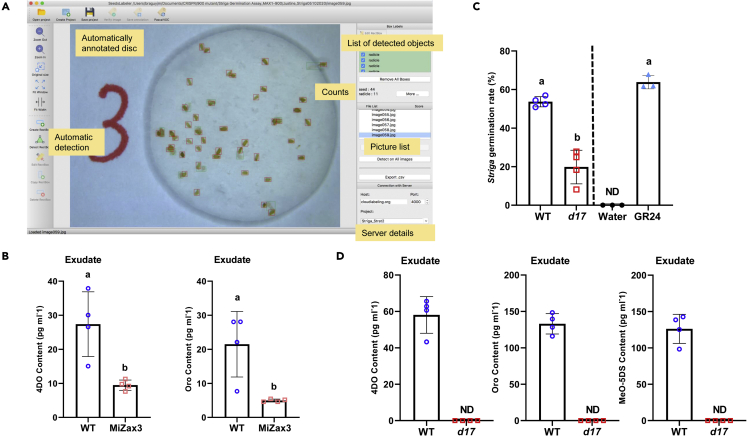
c.Detect and count the germinated and non-germinated seeds with SeedQuant (Braguy et al., 2021) (Figure 7A).
-
d.Calculate the germination percentage by dividing germinated seeds over total seeds to evaluate the bioactivity of the plant released SLs.
-
a.
Figure 7.
Expected outcome obtained from this protocol
(A) Overview and annotation of SeedQuant software interface.
(B) Evaluation of MiZax effect by SL quantification.
(C) Striga bioassay results processed by SeedQuant.
(D) SL quantification between two rice genotypes. Data represent mean ± SD. n=4. Statistical analysis was performed using One-way analysis of variance (ANOVA) and Tukey’s post hoc test. MiZax3, zaxinone mimics 3; WT, wild type; d17, dwarf17, a SL biosynthesis mutant. ND, not-detected
Expected outcomes
By using this protocol, we can unarguably identify endogenous SLs by comparing MS/MS pattern with authentic standards. In addition, we can evaluate the effect of a synthetic compound, such as the zaxinone and zaxinone mimics (MiZax), which regulate SL biosynthesis and release (Wang et al., 2019, 2020) (Figure 7B), as well as quantify the SL levels among different genotypes (Figure 7D). Finally, by applying collected root exudates to pre-conditioned Striga seeds – which is a much more sensitive method than LC-MS detection -, we can simply correlate bioassays (Figure 7C) with LC-MS results, and confirm the released SL’s bioactivity in the rhizosphere.
Quantification and statistical analysis
For the quantification of analytes (Table 12), we calculate endogenous amounts by dividing the analyte peak area with the one of the internal standard (IS), which is represented as Content (pg/mL) = (Analyte Peak Area/IS Peak Area)∗added IS (ng)∗1000/Sample collected Volume (mL).
Table 12.
MRM raw data of LCMS used in the SL quantification between two rice genotypes
| Sample | Analyte name | Analyte peak area (counts) | Analyte retention time (min) | IS peak name | IS peak area (counts) | IS retention time (min) | Content (pg/replicate) |
|---|---|---|---|---|---|---|---|
| d17-1 | 4-DeoxyOrobanchol | N/A | N/A | D6-5DS | 5.97E+06 | 14.86 | N/A |
| d17-2 | 4-DeoxyOrobanchol | N/A | N/A | D6-5DS | 5.88E+06 | 14.83 | N/A |
| d17-3 | 4-DeoxyOrobanchol | N/A | N/A | D6-5DS | 4.85E+06 | 14.84 | N/A |
| d17-4 | 4-DeoxyOrobanchol | N/A | N/A | D6-5DS | 7.22E+06 | 14.84 | N/A |
| WT-1 | 4-DeoxyOrobanchol | 4.74E+05 | 14.9 | D6-5DS | 7.36E+06 | 14.86 | 43.28 |
| WT-2 | 4-DeoxyOrobanchol | 6.72E+05 | 14.9 | D6-5DS | 7.45E+06 | 14.86 | 60.62 |
| WT-3 | 4-DeoxyOrobanchol | 6.96E+05 | 14.93 | D6-5DS | 7.13E+06 | 14.88 | 65.62 |
| WT-4 | 4-DeoxyOrobanchol | 6.70E+05 | 14.91 | D6-5DS | 7.17E+06 | 14.86 | 62.79 |
| d17-1 | Orobanchol | N/A | N/A | D6-5DS | 5.97E+06 | 14.86 | N/A |
| d17-2 | Orobanchol | N/A | N/A | D6-5DS | 5.88E+06 | 14.83 | N/A |
| d17-3 | Orobanchol | N/A | N/A | D6-5DS | 4.85E+06 | 14.84 | N/A |
| d17-4 | Orobanchol | N/A | N/A | D6-5DS | 7.22E+06 | 14.84 | N/A |
| WT-1 | Orobanchol | 1.41E+06 | 10.62 | D6-5DS | 7.36E+06 | 14.86 | 128.30 |
| WT-2 | Orobanchol | 1.29E+06 | 10.61 | D6-5DS | 7.45E+06 | 14.86 | 115.97 |
| WT-3 | Orobanchol | 1.57E+06 | 10.64 | D6-5DS | 7.13E+06 | 14.88 | 148.31 |
| WT-4 | Orobanchol | 1.49E+06 | 10.61 | D6-5DS | 7.17E+06 | 14.86 | 139.61 |
| d17-1 | MeO-5DS | N/A | N/A | D6-5DS | 5.97E+06 | 14.86 | N/A |
| d17-2 | MeO-5DS | N/A | N/A | D6-5DS | 5.88E+06 | 14.83 | N/A |
| d17-3 | MeO-5DS | N/A | N/A | D6-5DS | 4.85E+06 | 14.84 | N/A |
| d17-4 | MeO-5DS | N/A | N/A | D6-5DS | 7.22E+06 | 14.84 | N/A |
| WT-1 | MeO-5DS | 1.08E+06 | 13.01 | D6-5DS | 7.36E+06 | 14.86 | 98.35 |
| WT-2 | MeO-5DS | 1.39E+06 | 13.01 | D6-5DS | 7.45E+06 | 14.86 | 125.18 |
| WT-3 | MeO-5DS | 1.51E+06 | 13.03 | D6-5DS | 7.13E+06 | 14.88 | 142.65 |
| WT-4 | MeO-5DS | 1.48E+06 | 13.01 | D6-5DS | 7.17E+06 | 14.86 | 138.67 |
Alternatively, if no IS is used, simply measure the peak area of the selected analyte and compare it to the total average peak area across all the studied genotypes.
Table 13 summarizes the raw data of Figure 7C, counted by SeedQuant. To determine the Striga germination percentage, we divide the number of radicals (corresponding to the number of germinated Striga seeds) by the total number of seeds (including germinated and non-germinated Striga seeds). Usually, we apply each collected sample to four Striga discs – four technical replicates - and count their average; thereafter, the Striga germination rate for each plant of the same genotype is averaged to give the final germination rate (Figure 7C).
Table 13.
Example of SeedQuant raw data
| Sample | Replicate | Radicle | Seed | Percentage of germinated (radical/seed) | Average |
|---|---|---|---|---|---|
| GR24 | 1 | 38 | 56 | 67.85714286 | 63.82134414 |
| 2 | 29 | 47 | 61.70212766 | ||
| 3 | 39 | 63 | 61.9047619 | ||
| H2O | 1 | 0 | 74 | 0 | 0 |
| 2 | 0 | 74 | 0 | ||
| 3 | 0 | 72 | 0 | ||
| 4 | 0 | 53 | 0 | ||
| WT1 | 1 | 36 | 83 | 43.37349398 | 52.41200095 |
| 2 | 31 | 51 | 60.78431373 | ||
| 3 | 40 | 68 | 58.82352941 | ||
| 4 | 28 | 60 | 46.66666667 | ||
| WT2 | 1 | 44 | 79 | 55.69620253 | 50.83424193 |
| 2 | 37 | 79 | 46.83544304 | ||
| 3 | 29 | 56 | 51.78571429 | ||
| 4 | 25 | 51 | 49.01960784 | ||
| WT3 | 1 | 36 | 65 | 55.38461538 | 54.44710337 |
| 2 | 36 | 69 | 52.17391304 | ||
| 3 | 32 | 60 | 53.33333333 | ||
| 4 | 33 | 58 | 56.89655172 | ||
| WT4 | 1 | 52 | 93 | 55.91397849 | 56.94924783 |
| 2 | 23 | 48 | 47.91666667 | ||
| 3 | 43 | 65 | 66.15384615 | ||
| 4 | 37 | 64 | 57.8125 | ||
| d17-1 | 1 | 21 | 89 | 23.59550562 | 24.93116051 |
| 2 | 32 | 84 | 38.0952381 | ||
| 3 | 13 | 59 | 22.03389831 | ||
| 4 | 12 | 75 | 16 | ||
| d17-2 | 1 | 3 | 82 | 3.658536585 | 8.170308197 |
| 2 | 3 | 53 | 5.660377358 | ||
| 3 | 6 | 69 | 8.695652174 | ||
| 4 | 11 | 75 | 14.66666667 | ||
| d17-3 | 1 | 13 | 57 | 22.80701754 | 18.33589502 |
| 2 | 16 | 75 | 21.33333333 | ||
| 3 | 12 | 74 | 16.21621622 | ||
| 4 | 10 | 77 | 12.98701299 | ||
| d17-4 | 1 | 17 | 53 | 32.0754717 | 27.78734863 |
| 2 | 12 | 59 | 20.33898305 | ||
| 3 | 11 | 44 | 25 | ||
| 4 | 28 | 83 | 33.73493976 |
Limitations
Although this protocol can be applied to several plant species, the non-canonical SLs in the exudate are barely detected by LC-MS in general, due to the instability and lack of authentic standards. Undoubtedly, this could be partially solved by Striga bioassay on the basis of SL bioactivity. However, we cannot exclude the possibility of the presence of non-SL compounds that trigger parasitic plant seed germination, as observed in the rice d17 mutant. These germinating stimulants should be referred to as SL-independent metabolites. Another limitation is coming from SeedQuant, which the counting accuracy of Striga germination counted is around 93%, which is comparable to the error rate of a trained scientist.
Troubleshooting
Problem 1
SL cannot be isolated for MS/MS (step 9 in SL identification by LC-MS/MS).
Potential solution
We suggest adding a data-dependent MS/MS scan of the targeted SL ion after a full scan MS and to lower the threshold for the ion count to optimize the MS/MS fragmentation.
Problem 2
SL content is lower than LC-MS detection (step 18 in SL quantification by LC-MS/MS).
Potential solution
Make sure the plants are growing under phosphate deficient conditions. Else, you can pool two to three 50 mL sample tubes or increase the sample volume as one single biological replicate to enhance the SL concentration.
Problem 3
Striga seeds did not germinate (step 23 in SL bioactivity on Striga seed germination).
Potential solution
The lack of seed germination can have different reasons. 1) An inefficient pre-conditioning that can be verified by using the positive control GR24 (1 μM), which usually leads to >50% germination. 2) The presence of residual amount of acetone in the samples, which is toxic for the seeds.
Problem 4
SeedQuant software does not respond (step 24 in SL bioactivity on Striga seed germination).
Potential solution
Reopen the software and make sure the internet connection is active. If it still does not respond, it could be that the server itself is down, please do not hesitate to contact right away Justine.braguy@kaust.edu.sa or Silvio.giancola@kaust.edu.sa and share your issue.
Problem 5
SeedQuant does not detect properly the germinated and non-germinated seeds (step 24 in SL bioactivity on Striga seed germination).
Potential solution
SeedQuant seed detection can be impaired by the presence of plant debris or a different light intensity during the discs capture. The detection algorithm has reached its limitations, but a fine-tuning for your experimental conditions is possible. Contact Justine.braguy@kaust.edu.sa.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Salim Al-Babili (salim.babili@kaust.edu.sa).
Materials availability
All materials generated in this study are available from the lead contact upon completing a Materials Transfer Agreement.
Acknowledgments
We thank Dr. Tadao Asami for providing D6-5-deoxystrigol standard. This work was supported by the Competitive Research Grants (CRG2017 and CRG2020) and baseline funding from King Abdullah University of Science and Technology, and the Bill & Melinda Gates Foundation (grant number OPP1136424) given to S. A.-B..
Author contributions
S. A-B. and J.Y.W. conceived the project. J.Y.W., G-T.E.C., M.J., J.B., and A.B. conducted the laboratory work. J.B. designed and provided artistic work. J.Y.W., S.S., and K.X.L. performed and validated the LCMS-related work. J.Y.W. and G-T.E.C. wrote the original draft. M.J., J.B., S.S., and S.A-B. reviewed and edited the manuscript. All authors revised the final version of the manuscript and approved it.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Jian You Wang, Email: jianyou.wang@kaust.edu.sa.
Salim Al-Babili, Email: salim.babili@kaust.edu.sa.
Data and code availability
No unique datasets or codes were generated in this study.
References
- Ablazov A., Mi J., Jamil M., Jia K.P., Wang J.Y., Feng Q., Al-Babili S. The apocarotenoid zaxinone is a positive regulator of strigolactone and abscisic acid biosynthesis in Arabidopsis roots. Front. Plant Sci. 2020;11:578. doi: 10.3389/fpls.2020.00578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Babili S., Bouwmeester H.J. Strigolactones, a novel carotenoid-derived plant hormone. Annu. Rev. Plant Biol. 2015;66:161–186. doi: 10.1146/annurev-arplant-043014-114759. [DOI] [PubMed] [Google Scholar]
- Alder A., Jamil M., Marzorati M., Bruno M., Vermathen M., Bigler P., Ghisla S., Bouwmeester H., Beyer P., Al-Babili S. The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science. 2012;335:1348–1351. doi: 10.1126/science.1218094. [DOI] [PubMed] [Google Scholar]
- Boutet-Mercey S., Perreau F., Roux A., Clavé G., Pillot J.P., Schmitz-Afonso I., Touboul D., Mouille G., Rameau C., Boyer F.D. Validated method for strigolactone quantification by ultra-high-performance liquid chromatography-electrospray ionization tandem mass spectrometry using novel deuterium-labeled standards. Phytochem. Anal. 2018;29:59–68. doi: 10.1002/pca.2714. [DOI] [PubMed] [Google Scholar]
- Braguy J., Ramazanova M., Giancola S., Jamil M., Kountche B.A., Zarban R., Felemban A., Wang J.Y., Lin P.Y., Haider I., Zurbriggen M. SeedQuant: a deep learning-based tool for assessing stimulant and inhibitor activity on root parasitic seeds. Plant Physiol. 2021;186:1632–1644. doi: 10.1093/plphys/kiab173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt H., Jamil M., Wang J.Y., Al-Babili S., Mahfouz M. Engineering plant architecture via CRISPR/Cas9-mediated alteration of strigolactone biosynthesis. BMC Plant Biol. 2018;18:174. doi: 10.1186/s12870-018-1387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayou O., Kibet W., Ojola P., Gangashetty P.I., Wicke S., Runo S. Two-tier witchweed (Striga hermonthica) resistance in wild pearl millet (Pennisetum glaucum) 29Aw. Weed Sci. 2021;69:300–306. doi: 10.1017/wsc.2021.12. [DOI] [Google Scholar]
- Fiorilli V., Wang J.Y., Bonfante P., Lanfranco L., Al-Babili S. Apocarotenoids: old and new mediators of the arbuscular mycorrhizal symbiosis. Front. Plant Sci. 2019;10:1186. doi: 10.3389/fpls.2019.01186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roldan V., Fermas S., Brewer P.B., Puech-Pagès V., Dun E.A., Pillot J.P., Letisse F., Matusova R., Danoun S., Portais J.C., et al. Strigolactone inhibition of shoot branching. Nature. 2008;455:189–194. doi: 10.1038/nature07271. [DOI] [PubMed] [Google Scholar]
- Jamil M., Kanampiu F.K., Karaya H., Charnikhova T., Bouwmeester H.J. Striga hermonthica parasitism in maize in response to N and P fertilisers. Field Crops Res. 2012;134:1–10. doi: 10.1016/j.fcr.201’2.03.015. [DOI] [Google Scholar]
- Jamil M., Charnikhova T., Jamil T., Ali Z., Mohamed N.E.M.A., Van Mourik T., Bouwmeester H.J. Influence of fertilizer microdosing on strigolactone production and Striga hermonthica parasitism in pearl millet. Int. J. Agric. Biol. 2014;16:935–940. [Google Scholar]
- Matúšová R., van Mourik T., Bouwmeester H.J. Changes in the sensitivity of parasitic weed seeds to germination stimulants. Seed Sci. Res. 2004;14:335–344. [Google Scholar]
- Rebecca J., Whitfield J.H., Pouvreau B., Cao D., Alexandrov K., Beveridge C.A., Vickers C.E. Rational design of novel fluorescent enzyme biosensors for direct detection of strigolactones. ACS Synth. Biol. 2020;9:2107–2118. doi: 10.1021/acssynbio.0c00192. [DOI] [PubMed] [Google Scholar]
- Seto Y., Sado A., Asami K., Hanada A., Umehara M., Akiyama K., Yamaguchi S. Carlactone is an endogenous biosynthetic precursor for strigolactones. Proc. Natl. Acad. Sci. U.S.A. 2014;111:1640–1645. doi: 10.1073/pnas.1314805111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara M., Hanada A., Yoshida S., Akiyama K., Arite T., Takeda-Kamiya N., Magome H., Kamiya Y., Shirasu K., Yoneyama K., et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature. 2008;455:195–200. doi: 10.1038/nature07272. [DOI] [PubMed] [Google Scholar]
- Walker C.H., Siu-Ting K., Taylor A., O'Connell M.J., Bennett T. Strigolactone synthesis is ancestral in land plants, but canonical strigolactone signalling is a flowering plant innovation. BMC Biol. 2019;17:70. doi: 10.1186/s12915-019-0689-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.Y., Haider I., Jamil M., Fiorilli V., Saito Y., Mi J., Baz L., Kountche B.A., Jia K.P., Guo X., et al. The apocarotenoid metabolite zaxinone regulates growth and strigolactone biosynthesis in rice. Nat. Commun. 2019;10:810–819. doi: 10.1038/s41467-019-08461-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.Y., Jamil M., Lin P.Y., Ota T., Fiorilli V., Novero M., Zarban R.A., Kountche B.A., Takahashi I., Martínez C., Lanfranco L. Efficient mimics for elucidating zaxinone biology and promoting agricultural applications. Mol. Plant. 2020;13:1654–1661. doi: 10.1016/j.molp.2020.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.Y., Lin P.-Y., Al-Babili S. On the biosynthesis and evolution of apocarotenoid plant growth regulators. Semin. Cell Dev. Biol. 2021;109:3–11. doi: 10.1016/j.semcdb.2020.07.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No unique datasets or codes were generated in this study.