Abstract
Strengthening second-line drug-resistant tuberculosis (TB) detection is a priority. GenoType MTBDRplus VER 2.0 performance is reduced with non-recommended ramp rate usage (temperature change speed between PCR cycles); however, ramp rate’s effect on GenoType MTBDRsl VER 2.0 (MTBDRsl) performance, is unknown. Fifty-two Xpert MTB/RIF Ultra-positive rifampicin-resistant smear-negative sputa and a Mycobacterium tuberculosis dilution series were tested at a manufacturer-recommended (2.2°C/second) or suboptimal (4.0°C/second) ramp rate. M. tuberculosis–complex-DNA positivity, indeterminates, fluoroquinolone- and second-line injectable-resistance accuracy, banding differences, and, separately, inter-reader variability were assessed. Five (39%) of 13 re-surveyed laboratories did not use the manufacturer-recommended ramp rate. On sputum, 2.2°C/second improved indeterminates versus 4.0°C/second (0 of 52 versus 7 of 51; P = 0.006), incorrect drug-class diagnostic calls (0 of 104 versus 6 of 102; P = 0.013), and incorrect banding calls (0 of 1300 versus 54 of 1275; P < 0.001). Similarly, 2.2°C/second improved valid results [(52 of 52 versus 41 of 51; +21% (P = 0.001)] and banding call inter-reader variability [34 of 1300 (3%) versus 52 of 1300 (4%); P = 0.030]. At the suboptimal ramp rate, false-resistance and false-susceptible calls resulted from wild-type band absence rather than mutant band appearance, resulting in misclassification of moxifloxacin resistance level from high-to-low. Suboptimal ramp rate contributes to poor MTBDRsl performance. Laboratories must ensure that the manufacturer-recommended ramp rate is used.
In 2019, approximately 10 million individuals fell ill with tuberculosis (TB) and approximately 1.3 million individuals died.1 Drug-resistant TB is a global health problem. Approximately 465,000 individuals having multidrug-resistant TB (MDR-TB), ≥6% of whom have additional resistance to fluoroquinolones (FQs) and second-line injectables (SLIDs) (WHO Global Tuberculosis Report 2020). Worldwide in 2019, only 52% of patients with MDR-TB were tested for resistance to both these drug classes, and only 58% of those who start treatment successfully complete it (WHO Global Tuberculosis Report 2020). Phenotypic culture-based drug susceptibility testing is slow and costly, and patients need to wait up to 6 months before being placed on effective treatment, if at all.2 FQs are becoming incorporated into first-line drug regimens, which will require drastic scale-up of drug susceptibility testing. The World Health Organization (WHO) also recommends moxifloxacin for isoniazid-monoresistant TB in the newly endorsed shortened rifapentine regimen.3
GenoType MTBDRsl VER 2.0 (MTBDRsl) (Hain Lifescience, Nehren, Germany) is one of two commercially available rapid molecular WHO-endorsed assays for the detection of Mycobacterium tuberculosis complex and resistance to FQs and SLIDs.4, 5 According to the WHO, MTBDRsl should be performed directly on sputum irrespective of smear microscopy status to reduce the delay associated with culture for indirect testing.4
However, performance data for direct use on sputum are heterogeneous. In a systematic review and meta-analysis, smear-negative sensitivity estimates were imprecise: 80% [95% CI, 28–99], 80% (95% CI, 28–99), and 50% (95% CI, 1–99) for FQs, SLIDs, and extensively drug-resistant TB (XDR-TB) (using the then contemporaneous definition), respectively.6 This affected the certainty of evidence of the WHO recommendation and undermined uptake of MTBDRsl.
MTBDRsl requires thermocycling for DNA amplification. The manufacturer recommends a ramp rate of ≤2.2°C/second, which is the speed of temperature change between PCR cycles. It was previously shown that performance of GenoType MTBDRplus VER 2.0 (MTBDRplus) (Hain Lifescience), which is an assay for first-line resistance, is reduced when suboptimal thermocycler ramp rates are used, mainly on smear-negative specimens.7 These findings are incorporated into laboratory external quality assessment programs and the WHO TB laboratory training material (https://openwho.org/courses/multi-drug-resistant-tb, last accessed July 6, 2021).
If MTBDRsl is also vulnerable to this phenomenon, this would result in some of the thousands of individuals who receive this assay each day having drug resistance diagnoses missed, thereby resulting in resistance to the drugs critical to protect new regimens (eg, FQ to limit bedaquiline resistance acquisition in the oral second-line regimen) remaining delayed or undiagnosed.8,9 More broadly, this issue of ramp rate is increasingly pertinent as manufacturers are designing instruments with faster thermocycling (and hence faster ramp rates) to decrease time-to-result. Furthermore, many thermocyclers, especially those at entry level (ie, with fewer customizable settings compared with more advanced models that are typically more expensive), do not have a customizable ramp rate.
It is hypothesized that the heterogeneous and suboptimal sensitivities reported for MTBDRsl on smear-negative specimens were partly attributable to suboptimal ramp rate, and the goal was to generate empirical evidence of this theory. The current study assessed whether laboratories that reported use of suboptimal ramp rates during the authors’ previous MTBDRplus evaluation7 had switched to the manufacturer-recommended ramp rate and what the observed effect had been.
Materials and Methods
Ethics Statement
This study was approved by the Health Research Ethics Committee of Stellenbosch University (N16/04/045) and Western Cape Research Ethics Committee (WC_2016RP18_637). All methods were in accordance with relevant guidelines and regulations. Permission was granted to access anonymized residual specimens collected as part of routine diagnostic practices, and thus patient informed consent was waived.
Experimental Design
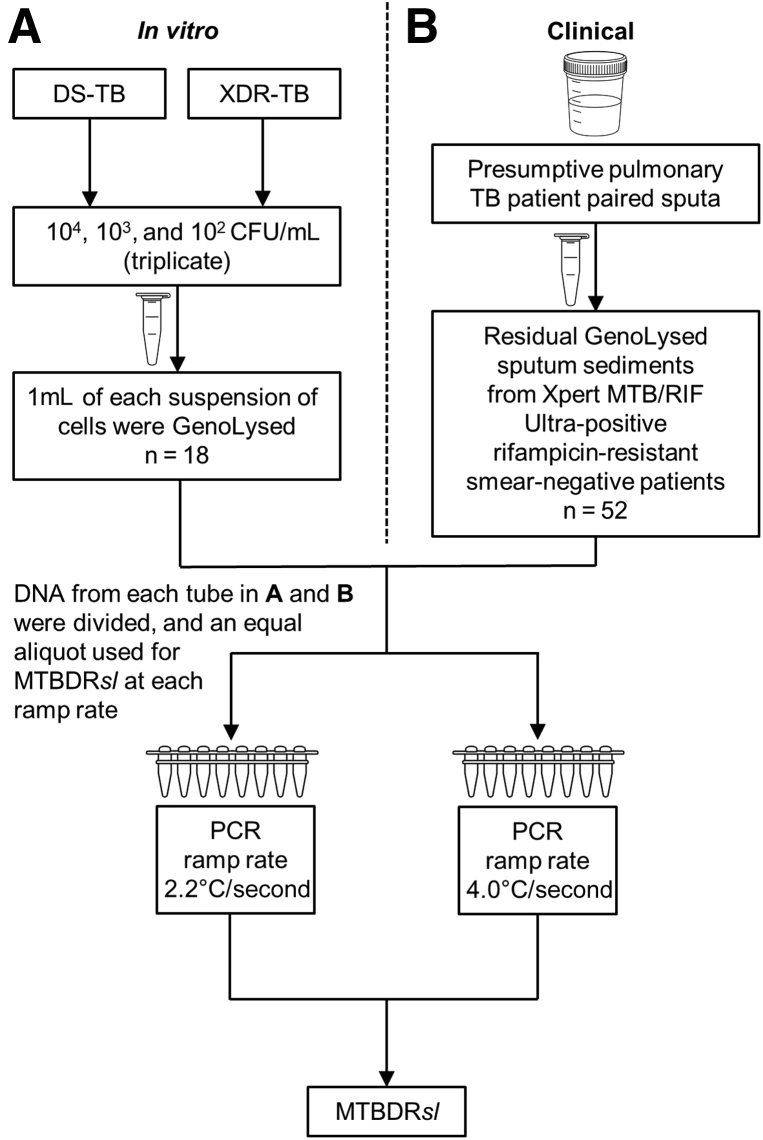
Ramp rate assessment was performed in both an in vitro dilution series and clinical sputa (Figure 1). DNA extracted from dilution series and clinical specimens were split and compared head-to-head at the manufacturer-recommended ramp rate of 2.2°C/second or the most common suboptimal ramp rate of 4.0°C/second identified previously in a survey.7 MTBDRsl was performed on all amplified DNA per manufacturer’s instructions for use (Hain Lifescience) [kit lot #39B (expiry date September 2, 2019); strip lot #ABB0117A161 (expiry date September 18, 2019)]. All experiments for this study were performed before the kits’ expiration dates. Strips were interpreted by using the WHO-endorsed Global Laboratory Initiative line probe assay interpretation guide (GLI, http://www.stoptb.org/wg/gli/assets/documents/LPA_test_web_ready.pdf; WHO, https://openwho.org/courses/multi-drug-resistant-tb/items/49CT8rhOFxxXzbJYsIIZlK, last accessed October 19, 2021) and the authors agree with the recommendations in these guidelines. For sputa, programmatic MTBDRsl results (performed at the recommended ramp rate) were compared. All equipment is annually calibrated and serviced.
Figure 1.
Study flow diagram for an in vitro [a dilution series of cells (104, 103, and 102 colony-forming units per milliliter [CFU/mL])] experiment (A) and clinical experiment (sputa) (B) to assess the impact of thermocycler ramp rate on GenoType MTBDRsl VER 2.0 (MTBDRsl). DNA extracted from the dilution series and clinical specimens was split and MTBDRsl compared head-to-head at the manufacturer-recommended ramp rate of 2.2°C/second or 4.0°C/second. DS-TB, drug-susceptible tuberculosis; TB, tuberculosis; XDR-TB, extensively drug-resistant tuberculosis.
MTBDRsl Calls and Result Definitions
Conjugate Control Band
The conjugate control (CC)-band must be present for a strip to be valid as it indicates that hybridization occurred.
Amplification Control Band
The amplification control (AC)-band is present when the assay is performed correctly. Per the manual (GenoType MTBDRsl Instructions for Use IFU-317A-04; Hain Lifescience), there are rare cases in which the AC-band disappears due to competition during the amplification reaction. In this scenario, an absent AC-band in combination with M. tuberculosis–complex-DNA (TUB-band) and locus control bands is still a valid result.
Locus Control Bands (gyrA, gyrB, rrs, and eis)
The locus control bands (gyrA, gyrB, rrs, and eis) need to be present for a call from that locus to not be indeterminate.
Positive for M.tuberculosis–Complex-DNA
The TUB-band indicates the presence of M. tuberculosis–complex-DNA.
Strip Banding Call
For a band to be classified as present, it must be equal or darker than the AC-band. Overall, there are 27 possible strip bands on MTBDRsl. When only the CC- and AC-bands are present, this represents a valid TUB-negative result.
Drug Class Diagnostic Call
Band presence or absence in a gene region determines whether the result is classified as susceptible or resistance to a drug class (two drug class diagnostic calls possible for MTBDRsl: FQs or SLIDs).
(In)determinate for a Gene Region and/or Drug Class
For a specific gene region and/or drug class to be determinate, locus control band(s) must be present. A strip was called indeterminate for a drug class if at least one gene locus control was absent.
Valid Result
TUB-band–positive strip determinate for all gene locus controls and thus has diagnostic calls for both drug classes (eg, TUB-band–positive, FQ-resistant, SLID-susceptible).
Additional Amikacin Resistance (rrs C1402T and eis C-14T)
These are new guidelines released by the WHO indicating resistance to amikacin. rrs C1402T translates to rrs WT1 band not binding and eis C-14T translates to the eis MUT1 band binding.10 The MTBDRsl will need to be updated.
Impact of Thermocycler Ramp Rate on MTBDRsl Performance on a Dilution Series
A phenotypically and genotypically resistant clinical XDR strain (gyrA D94N, gyrB wild type, rrs A1401G, and eis wild type) and a drug-susceptible strain (H37Rv, ATCC 25618) were grown to mid-exponential phase in Middlebrook 7H9 media (Becton Dickinson, Franklin Lakes, NJ) supplemented with Middlebrook Oleic Albumin Dextrose Catalase (Becton Dickinson) and adjusted to a McFarland 1.0 standard [approximately 108 colony-forming units per milliliter (CFU/mL)] (GLI Mycobacteriology Laboratory Manual, http://www.stoptb.org/wg/gli/assets/documents/gli_mycobacteriology_lab_manual_web.pdf, last accessed July 23, 2021). Serial dilutions in phosphate buffer supplemented with 0.025% Tween 80 (Merck, Sandton, South Africa) were inoculated onto Middlebrook 7H10 solid media (Becton Dickinson) and incubated for 21 days at 37°C for CFU calculations. These experiments were performed in biological triplicate. One milliliter of the 104, 103, and 102 CFU/mL suspensions were GenoLysed (Hain Lifescience) and MTBDRsl performed per the manufacturer’s instructions (Hain Lifescience). The two lower dilutions approximate to smear-negative disease (<10,000 CFU/mL),11 expected to be most affected by a suboptimal ramp rate. DNA was amplified with the CFX96 thermocycler (Bio-Rad Laboratories, Sandton, South Africa) at ramp rates of 2.2°C/second and 4.0°C/second. Two experienced readers recorded bands in a blinded manner. Accuracy analyses for TUB-band positivity, indeterminate rates, incorrect banding calls, and incorrect drug class diagnostic calls were done.
Impact of Thermocycler Ramp Rate on MTBDRsl Performance on Clinical Specimens
GenoLysed samples (n = 52) remaining after programmatic line probe assay test results were collected from a TB laboratory in Cape Town, South Africa. These samples were, per the national algorithm, derived from the paired sputum specimen of a presumptive pulmonary TB patient who received Xpert MTB/RIF Ultra (Ultra) (on separate sputum), MGIT 960 culture, and Auramine O microscopy (on the same sputum before being GenoLysed). All sputa were smear-negative and Ultra-positive rifampicin-resistant. Smear-positive specimens were not included as it was previously shown that ramp rate had no effect on MTBDRplus performance on smear-positive specimens.7 Residual GenoLysed samples were stored at −20°C.
Samples were categorized by using programmatic line probe assay results as: 17 MDR-TB, 24 pre-XDR, and 11 XDR-TB. For the experiment, DNA was amplified by using a CFX96 thermocycler (Bio-Rad, Hercules, CA) at 2.2°C/second (manufacturer-recommended) and 4.0°C/second. MTBDRsl was performed per the manufacturer’s instructions (Hain Lifescience), and two experienced readers recorded bands in a blinded fashion. Accuracy analyses for TUB-band positivity, indeterminate rates, incorrect banding calls, and incorrect drug class diagnostic calls were done.
Calculation of Laboratory Savings from an Improvement in MTBDRsl Performance on Smear-Negative Specimens Stemming from Ramp Rate
Calculations were performed on how much the routine laboratory, from which GenoLysed remnants were received, would save if the proportional increase, which was found in valid results when the optimal versus the suboptimal ramp rate was used, was applied. This cost savings calculation was based on the average number of MTBDRsl tests performed indirectly on cultured isolates per month (which would now be reduced due to direct testing on smear-negative specimens having improved performance) and the cost of each test (including consumables, labor, and overheads; the sum is pre-calculated and supplied by the laboratory provider).
Inter-reader Agreement
An additional three experienced readers, independent of the aforementioned two readers, read all strips from the dilution series and clinical specimens at either ramp rate independently from one another and blinded to each other’s calls as well as any other information regarding the specimens or strains used. Banding calls were assessed between readers, as well as resultant differences in final drug class diagnostic calls. Excluding the CC-bands and AC-bands, and including the TUB-band, gene locus control-bands, and gene-specific wild type- and mutant-bands, there are 25 possible bands per MTBDRsl strip. There are hence 450 possible bands total for the 18 samples in the dilution series and 1300 possible bands for the 52 clinical isolates. Each strip results in two drug class diagnostic calls, and there are hence 36 possible drug class diagnostic calls in total for 18 samples in the dilution series and 104 possible drug class diagnostic calls in total for the 52 clinical isolates.
Follow-Up Survey of TB Diagnostic and Research Laboratories
Prior respondents to the initial MTBDRplus-focused survey7 were re-surveyed (n = 29) to gather information on the current MTBDRsl conditions. Other laboratories newly known to us as performing MTBDRsl on smear-negative specimens (n = 11) were also surveyed for the first time, and initial nonresponders were re-contacted at least twice. Survey questions included whether ramp rate changed and impact on nonvalid results (Supplemental Appendix S17). Permission to use data in an anonymized manner was received from the Faculty of Medicine and Health Sciences Human Research Ethics Committee of Stellenbosch University (N16/04/045).
Statistical Analyses
Data were analyzed using Stata version 15 (StataCorp, College Station, TX) and GraphPad Prism version 8.0.1 (GraphPad Software, La Jolla, CA) using two-sided t-tests with α = 0.05. McNemar’s test was used to calculate differences for paired data (ie, the same DNA tested at both ramp rates). The two-sample proportion test was used for comparisons between proportions.
Results
MTBDRsl on the Dilution Series at Different Ramp Rates
Overall, there were no differences between ramp rates of 2.2°C/second and 4.0°C/second for TUB-band detection [16 of 18 (89%) versus 17 of 18 (94%); P = 0.547], indeterminate results [2 of 16 (13%) versus 3 of 17 (18%); P = 0.680], incorrect banding calls [22 of 400 (6%) versus 33 of 425 (8%); P = 0.193)], or incorrect drug resistance calls [2 of 32 (6%) versus 2 of 34 (6%); P = 0.950] (Table 1). Therefore, valid results did not differ significantly [14 of 16 (88%) versus 14 of 17 (82%); P = 0.680].
Table 1.
MTBDRsl Performance on a Dilution Series of Drug-Susceptible-TB and XDR-TB Strains (104, 103, and 102 CFU/mL) at Ramp Rates of 2.2°C/second (Manufacturer-Recommended) or 4.0°C/second (3 Replicates in Triplicate for Each Ramp Rate; 18 Total MTBDRsl Results)
| Ramp rate (°C/second) | TUB-band–positive | TUB-band–positive |
|||
|---|---|---|---|---|---|
| Indeterminate for any gene locus | Incorrect banding call | Incorrect drug class diagnostic call | Valid result | ||
| 2.2 | 16/18∗ (89) | 2/16† (13) | 22/400‡ (6) | 2/32§ (6) | 14/16† (88) |
| 4.0 | 17/18∗ (94), P = 0.547 | 3/17† (18), P = 0.680 | 33/425¶ (8), P = 0.193 | 2/34‖ (6), P = 0.950 | 14/17† (82), P = 0.680 |
Data are expressed as n/N (%). Accuracy for M. tuberculosis–complex-DNA (TUB-band) and then further analysis of indeterminate rates, incorrect banding calls, and incorrect drug class diagnostic calls were done. No significant differences were seen between ramp rates using dilution series. P values are for within-column comparisons between different ramp rates. CFU, colony-forming units; Incorrect banding call, the presence or absence of a band deviating from the true banding call; Incorrect drug class diagnostic call, the presence or absence of banding patterns resulting in deviation of the true susceptibility to a drug class; Indeterminate, one or more gene locus control is absent; MTBDRsl, GenoType MTBDRsl VER 2.0; TB, tuberculosis; TUB-band–positive, positive for Mycobacterium tuberculosis–complex-DNA; Valid result, TUB-band–positive, determinate for all gene locus controls, thus having diagnostic calls for both drug classes; XDR, extensively drug resistant.
Two strains × 3 replicates × 3 dilutions.
TUB-positive strips.
Sixteen TUB-band–positive strips × 25 bands per strip.
Sixteen TUB-band–positive strips × 2 drug class diagnostic calls.
Seventeen TUB-band–positive strips × 25 bands per strip.
Seventeen TUB-band–positive strips × 2 drug class diagnostic calls.
MTBDRsl on Clinical Sputa at Different Ramp Rates
No TUB-band detection differences were seen at 2.2°C/second versus 4.0°C/second [52 of 52 (100%) versus 51 of 52 (98%); P = 0.315; one MDR-TB patient was TUB-negative only at 4.0°C/second]. However, indeterminate rates improved at 2.2°C/second [0 of 52 (0%) versus 7 of 51 (14%); P = 0.006], as did the proportion of bands that appeared incorrectly [0 of 1300 (0%) versus 55 of 1275 (4%); P < 0.001)] and drug-resistance calls [0 of 104 (0%) versus 6 of 102 (6%); P = 0.013] (Table 2). The proportion of patients with a valid result was therefore 52 (100%) of 52 versus 41 (80%) of 51. In other words, the patients who successfully received testing for FQs and SLIDs thus improved 21% (95% CI, 8–34; P < 0.001).
Table 2.
MTBDRsl Performance on Smear-Negative Sputa at Ramp Rates of 2.2°C/second (Manufacturer-Recommended) or 4.0°C/second (52 Isolates)
| Ramp rate (°C/second) | TUB-band–positive | TUB-band–positive |
|||
|---|---|---|---|---|---|
| Indeterminate for any gene locus | Incorrect banding call | Incorrect drug class diagnostic call | Valid result | ||
| 2.2 | 52/52∗ (100) | 0/52† (0) | 0/1300‡ (0) | 0/104§ (0) | 52/52† (100) |
| 4.0 | 51/52∗ (98), P = 0.315 | 7/51† (14), P = 0.006 | 54/1275¶ (4), P < 0.001 | 6/102‖ (6), P = 0.013 | 41/51† (80), P = 0.001 |
Data are expressed as n/N (%). Accuracy for Mycobacterium tuberculosis–complex-DNA, and then further analysis of indeterminate rates, incorrect banding calls, and incorrect drug class diagnostic calls were done. The number of valid results [52 of 52 (100%) versus 41 of 51 (80%)] improved by 21% (95% CI, 8–34; P < 0.001). P values are for within-column comparisons between different ramp rates. Significant P values are marked in bold. Incorrect banding call, the presence or absence of a band deviating from the true banding call; Incorrect drug class diagnostic call, the presence or absence of banding patterns resulting in deviation of the true susceptibility to a drug class; Indeterminate, one or more gene locus control is absent; MTBDRsl, GenoType MTBDRsl VER 2.0; TB, tuberculosis; TUB-band–positive, positive for Mycobacterium tuberculosis–complex-DNA; Valid result, TUB-band–positive, determinate for all gene locus controls, thus having diagnostic calls for both drug classes.
Total number of clinical specimens.
TUB-positive strips.
Fifty-two TUB-band–positive strips × 25 bands per strip.
Fifty-two TUB-band–positive strips × 2 drug class diagnostic calls.
Fifty-one TUB-band–positive strips × 25 bands per strip.
Fifty-one TUB-band–positive strips × 2 drug class diagnostic calls.
Programmatic Ultra semi-quantitative data were available for 41 (79%) of 52 sputa. When bacterial load in sputa that gave a valid result at 2.2°C/second was compared versus sputa that gave a valid result at 4.0°C/second, there were no differences [median (interquartile range) minimum cycle threshold (CTmin), 18.7 (17.7–19.9) versus 18.8 (18.0–19.9); P = 0.899]. It was expected that 2.2°C/second would result in an improved limit of detection in MTBDRsl (better ability to detect higher CTmin and therefore fewer bacilli); however, no differences were detected.
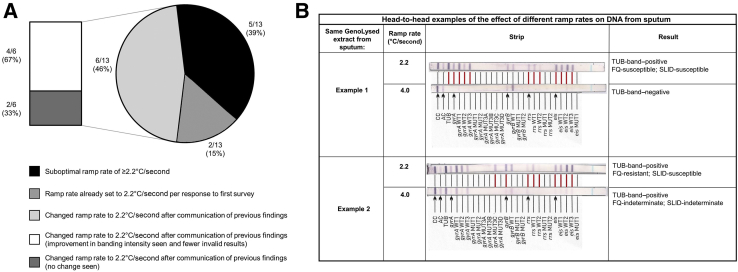
Head-to-head examples of the effect of different ramp rates on DNA from sputum are provided in Figure 2B.
Figure 2.
A: Follow-up survey results summarizing thermocycler ramp rates for GenoType MTBDRsl VER 2.0. Two (15%) of 13 initially surveyed laboratories already had their ramp rate set to 2.2°C/second, and five (39%) of 13 were still using a suboptimal ramp rate of ≥2.2°C/second upon resurveying. Six (46%) of 13 laboratories had, since the first survey on GenoType MTBDRplus VER 2.0, changed the GenoType MTBDRsl VER 2.0 ramp rate to the recommended ramp rate. Of these, four (67%) of six reported an improvement in banding intensity and fewer invalid results. B: An illustrative example of differences in banding patterns (and consequences for patient diagnoses) caused using suboptimal ramp rate. In example 1, at the suboptimal ramp rate (4.0°C/second), no tuberculosis or drug susceptibility information would be generated. In example 2, at the suboptimal ramp rate (4.0°C/second), again no drug susceptibility information would be generated, but, in this case, it would lead to a missed diagnosis of fluoroquinolone (FQ) resistance. Different banding patterns between strips are shown with a red line. SLID, second-line injectables; TUB-band–positive, positive for Mycobacterium tuberculosis–complex-DNA.
Banding patterns from both the dilution series and clinical sputa are listed in Supplemental Tables S1 and S2. For the dilution series (Supplemental Table S1), irrespective of ramp rate, MTBDRsl did not classify the XDR-TB strain correctly at 102 CFU/mL across all replicates (Table 1). Overall, for dilution series (both strains, all dilutions), the overall effect was missed resistance due to a TUB-negative, indeterminate, or a missing gene-specific band, or false-resistance due to an erroneously absent wild-type band. For clinical sputa (Supplemental Table S2) at the suboptimal ramp rate, there was worse detection of the TB and locus control bands and, when TB was detected and the locus control bands present, gene-specific bands that should have been present were absent. In the dilution series, one replicate (XDR-TB, 10 to 2 dilution) missed amikacin resistance at the suboptimal ramp rate. In clinical specimens, two samples (RR2-31 and RR2-38) with high-level moxifloxacin resistance were incorrectly classified at the suboptimal ramp rate as low-level resistant (RR2-38) or susceptible (RR2-31). At the suboptimal ramp rate of 4.0°C/second, 55 gene locus bands were erroneous. The breakdown is as follows: gyrA, 14 of 55 (25%); gyrB, 5 of 55 (9%); rrs, 28 of 55 (51%); and eis, 8 of 55 (15%).
Laboratory Savings
If the improvement in FQ and SLID testing due to optimal ramp rate usage is applied, there would be a 21% decrease in the number of tests required to be performed indirectly (which would require culture and a second MTBDRsl). At a local reference laboratory, approximately 320 MTBDRsl assays, initially attempted on smear-negative sputa, are performed per month and are subsequently repeated on culture isolates. Hence, in a scenario in which this laboratory was using an incorrect ramp rate and changed to the correct rate, they would perform approximately 67 fewer indirect MTBDRsl assays per month. At a total per test cost of US$60 (6% per annum inflation),12 this translates to a savings of US$48,240 per year (only factoring in pure laboratory costs).
Inter-reader Agreement
In the dilution series, diagnostic calls did not differ between the three readers at either ramp rate. All readers incorrectly classified the XDR-TB strain (as either TUB-band–negative or indeterminate) at all 102 CFU/mL replicates and the drug-susceptible–TB strain (as indeterminate) at one of the three replicates at 102 CFU/mL (Table 3). The proportion of disagreement between readers (banding calls) did not differ at suboptimal versus optimal ramp rates [for the drug-susceptible (1 of 225 versus 0 of 225; P = 0.317) or the XDR (3 of 225 versus 1 of 225; P = 0.313)] strain.
Table 3.
Comparison of Banding and Drug Class Diagnostic Calls Done on a Dilution Series of DS-TB and XDR-TB Strains and Clinical Specimens Interpreted by Three Experienced Readers
| Ramp rate (°C/second) | DS-TB strain |
XDR-TB strain |
Clinical specimens |
|||
|---|---|---|---|---|---|---|
| Different banding call between readers | Different drug class diagnostic call between readers | Different banding call between readers | Different drug class diagnostic call between readers | Different banding call between readers | Different drug class diagnostic call between readers | |
| 2.2 | 0/225∗ (0) | 0/18† (0) | 1/225∗ (0.4) | 0/18† (0) | 34/1300‡ (3) | 5/104§ (5) |
| 4.0 | 1/225∗ (0.4), P = 0.317 | 1/18† (6), P = 0.311 | 3/225∗ (1), P = 0.313 | 0/18† (0), P > 0.999 | 52/1300‡ (4), P = 0.030 | 8/104§ (8), P = 0.390 |
Data are expressed as n/N (%). Differences in banding calls or drug class diagnostic calls did not differ between the three readers at either ramp rate for the dilution series of cells, neither did the drug class diagnostic calls in the clinical specimens; however, significant difference between readers for banding calls on the clinical sputa occurred. P values are for within-column comparisons between different ramp rates. Significant P values are marked in bold. banding call, the presence or absence of a band deviating from the true banding call; diagnostic call, the presence or absence of banding patterns resulting in deviation of the true susceptibility to a drug class; DS-TB, drug-susceptible tuberculosis; XDR-TB, extensively drug-resistant tuberculosis.
One strain × 3 replicates × 3 dilutions × 25 bands per strip.
One strain × 3 replicates × 3 dilutions × 2 drug class diagnostic calls.
Fifty-two clinical specimens × 25 bands per strip.
Fifty-two clinical specimens × 2 drug class diagnostic calls.
In clinical sputa, however, although the disagreement in drug class diagnostic calls did not differ between readers at the optimal versus suboptimal ramp rate [5 of 104 (5%) versus 8 of 104 (8%); P = 0.390], banding calls did differ [34 of 1300 (3%) versus 52 of 1300 (4%); P = 0.030].
Additional Survey
Twenty-nine follow-up surveys were sent to the original respondents and 11 to new laboratories. Thirteen total responses were received (45%), including four from new respondents (Figure 2A). Two (15%) of 13 respondents already had their ramp rate at 2.2°C/second (per their response to the first survey), and six (46%) of 13 had subsequently changed their ramp rate to 2.2°C/second after the previous findings were communicated.7 Concerningly, five (39%) of 13 had not changed, for which varied reasons were offered (Table 4). Of the laboratories who changed to 2.2°C/second, four (67%) of six reported that this resulted in an improvement in banding intensity and fewer nonvalid results for MTBDRplus and MTBDRsl.
Table 4.
Laboratories That Indicated Their Ramp Rate Had Not yet Changed to the Manufacturer-Recommended Ramp Rate of ≤2.2°C/second Since the Last Survey, the Reason Why, and Total Number of Line Probe Assays Performed per Month
| Country | Reason given | No. of line probe assays performed per month by this respondent laboratory |
|---|---|---|
| Kenya | Do not know | 240 |
| South Africa | Ramp rate change was not necessary as MTBDRplus assays are performed on cultured isolates only and no MTBDRsl assays are performed, as well as any changes to a standard operating procedure requires a validation process | 40 |
| Belarus | Ramp rate change in a standard operation procedure is not permitted without a prior approval process | 155 |
| Denmark | Ramp rate was not changed due to the run time of the original amplification protocol being faster | 25 |
| Spain | The thermocycler did not permit a ramp rate change | 12 |
These laboratories perform either GenoType MTBDRplus VER 2.0 (MTBDRplus), GenoType MTBDRsl VER 2.0 (MTBDRsl), or both on smear-negative specimens, but data on the subtotals for each assay were not collected.
Discussion
The current study evaluated for the first-time the impact of thermocycler ramp rates on the most widely used molecular test for second-line drug-resistant TB (MTBDRsl). This study shows: i) in sputa, valid results improved by 21% when using the optimal ramp rate, which results in significant laboratory cost savings and would decrease diagnostic delay; ii) banding call and drug susceptibility call reader disagreement worsened at the suboptimal ramp rate; and iii) several laboratory respondents had not corrected their line probe assay ramp rate but, those that had, reported fewer nonvalid results from MTBDRsl on smear-negative specimens.
In a previous study, the authors found that a suboptimal thermocycler ramp rate negatively affects the diagnostic accuracy of potentially thousands of MTBDRplus assays, especially on smear-negative sputa,7 and ramp rate monitoring was incorporated into laboratory quality control and training documentation (WHO Drug-resistant tuberculosis: how to interpret rapid molecular test results, https://openwho.org/courses/multi-drug-resistant-tb, last accessed July 6, 2021). The current study shows that a 21% increase in MTBDRsl diagnoses (valid results) in smear-negative specimens is possible through ramp rate correction. This is not a niche problem; diagnostic laboratories that still do not perform MTBDRsl correctly were identified. This correction, which this study has now provided MTBDRsl-specific empirical evidence, could reduce drug-resistant TB diagnostic care cascade gaps: a recent study found that only 65% of MDR-TB cases were evaluated for FQ resistance.13
Critically, ramp rate correction will reduce repeat MTBDRsl testing on isolates. Most directly, this will translate into substantial laboratory cost savings in high-burden countries, especially when TB services are fragile due to the COVID-19 pandemic, not to mention the myriad of other individual and population benefits that can stem from improved drug susceptibility testing14; these include reduced time to treatment, transmission, and mortality.
Most laboratories in the follow-up survey had corrected the ramp rate; however, a significant amount, including those responsible for routine diagnostic testing on smear-negative specimens, still used a suboptimal ramp rate. It should be emphasized that: i) laboratories must ensure that they are using the optimal ramp rate; ii) thermocycler ramp rate monitoring should be added to laboratory external quality assurance programs and accreditation processes for MTBDRsl; and iii) the manufacturer should make the recommended ramp rate more prominent in assay documentation. It is worth evaluating further why incorrect ramp rates continued to be used. This may be due to quality assurance lapses, a deliberate choice (eg, to potentially speed up turn-around-time) without an awareness of downsides, or a design limitation of available thermocyclers.
When a band was present at the optimal ramp rate (2.2°C/second) and not the suboptimal ramp rate (4.0°C/second), FQ and/or SLID diagnoses were missed completely due to gene locus control bands not binding. False drug class diagnostic calls for FQs and/or SLIDs (false resistance) due to the inability of a band to bind were also seen. No false resistance was observed due to the binding of mutant probes when the suboptimal ramp rate was used. However, false resistance calls due to an erroneous absence of wild-type bands occurred. It was noted that more than one-half of the incorrect bands in sputa occurred in one gene locus (rrs), which may be due to secondary structures that interfere with PCR and detection.
A more prominent performance difference was seen between ramp rates in clinical sputa than in spiked solution. Bacilli in mucus sputa matrices behave differently from bacilli spiked in in vitro experiments, and these findings illustrate potential downsides to investigating the effect of PCR parameters on molecular assays when in vitro or mock specimens are used.
The current evaluation has strengths and limitations. A wider ramp rate range or different thermocycler models were not assessed due to limited sputa and cost. The utility of additional testing when a useful (ie, valid) result failed to be generated was also not evaluated. The most frequently reported incorrect ramp rate from the previous survey was used.7 DNA from samples was not directly quantified; however, when comparing Ultra semi-quantitative (CTmin) data between valid results across ramp rates, no differences occurred. When there is an indeterminate result for a gene locus, regardless of whether that indeterminate result is caused by optimal ramp rate, it may influence the reliability of other diagnostic calls from loci with valid control bands. However, this requires a large diagnostic accuracy study to investigate, and the current work was not designed to do so.
The survey results would have also been subjected to selection, response, and reporting biases. The authors suggest that a formal survey be done by the manufacturer and/or the appropriate regulatory and oversight agency (the study survey was done independently). Savings stemming from quicker diagnosis, treatment initiation, and long-term reductions in transmission and mortality due to improved performance were not evaluated; there is already a saving in laboratory costs alone, with no downside.
In conclusion, this study found that a still incorrectly configured and innocuous technical setting (ramp rate) has a real-world negative impact on patients’ diagnoses for second-line drug resistance using MTBDRsl. Patients with smear-negative specimens, for whom early diagnosis is important to curtail transmission of drug resistance, are especially vulnerable. All stakeholders must ensure that the optimal thermocycler ramp rate for MTBDRsl is used, and the impact of this source of technical variation should be investigated for other molecular diagnostics.
Acknowledgments
We thank the laboratories that participated in the survey and provided data, the National Health Laboratory Service (Green Point, Cape Town, South Africa), and Hain Lifescience, which donated the MTBDRsl kits used for the dilution series.
Footnotes
Supported by the European and Developing Countries Clinical Trials (EDCTP2) program supported by the European Union (grant SF1401, OPTIMAL DIAGNOSIS), The Center for Innovation in Point-of-Care Technologies for HIV/AIDS at Northwestern University (National Institute of Biomedical Imaging and Bioengineering of the NIH, Award Number U54EB027049), and the National Institute of Allergy and Infectious Diseases. Research reported in this publication was also supported by the South African Medical Research Council.
Disclosures: Hain Lifescience donated the MTBDRsl kits used for the dilution series. They did not have any role in study design or result interpretation.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the South African Medical Research Council.
Current address of M.d.V., Foundation for Innovative New Diagnostics, Geneva, Switzerland.
Supplemental material for this article can be found at http://doi.org/10.1016/j.jmoldx.2022.01.003.
Author Contributions
B.D., M.d.V., G.T., and R.W. conceived the experiments; T.D., N.B., and S.P. provided specimens and data from the National Health Laboratory Service; B.D. conducted the experiments and analyzed the data; S.P, Y.G., R.V., and S.M. assisted with analysis of results; J.M. provided critical input. All authors reviewed the manuscript.
Supplemental Data
References
- 1.World Health Organization . Global Tuberculosis Report 2020. WHO; Geneva, Switzerland: 2020. https://apps.who.int/iris/bitstream/handle/10665/336069/9789240013131-eng.pdf Available at: [Google Scholar]
- 2.Basu S., Friedland G.H., Medlock J., Andrews J.R., Shah N.S., Gandhi N.R., Moll A., Moodley P., Sturm A.W., Galvani A.P. Averting epidemics of extensively drug-resistant tuberculosis. Proc Natl Acad Sci U S A. 2009;106:7672–7677. doi: 10.1073/pnas.0812472106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dorman S.E., Nahid P., Kurbatova E.V., Phillips P.P.J., Bryant K., Dooley K.E., Engle M., Goldberg S.V., Phan H.T.T., Hakim J., Johnson J.L., Lourens M., Martinson N.A., Muzanyi G., Narunsky K., Nerette S., Nguyen N.V., Pham T.H., Pierre S., Purfield A.E., Samaneka W., Savic R.M., Sanne I., Scott N.A., Shenje J., Sizemore E., Vernon A., Waja Z., Weiner M., Swindells S., Chaisson R.E., AIDS Clinical Trials Group. Tuberculosis Trials Consortium Four-month rifapentine regimens with or without moxifloxacin for tuberculosis. N Engl J Med. 2021;384:1705–1718. doi: 10.1056/NEJMoa2033400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization . The use of molecular line probe assays for the detection of resistance to second-line anti-tuberculosis drugs: policy guidance. WHO; Geneva, Switzerland: 2016. https://apps.who.int/iris/handle/10665/246131 Available at: [Google Scholar]
- 5.World Health Organization . WHO consolidated guidelines on tuberculosis. Module 3: Diagnosis—Rapid diagnostics for tuberculosis detection 2021 update. WHO; Geneva, Switzerland: 2021. https://www.who.int/publications/i/item/9789240029415 Available at: [Google Scholar]
- 6.Theron G., Peter J., Richardson M., Warren R., Dheda K., Steingart K.R. GenoType® MTBDRsl assay for resistance to second-line anti-tuberculosis drugs. Cochrane Database Syst Rev. 2016;9:CD010705. doi: 10.1002/14651858.CD010705.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derendinger B., de Vos M., Nathavitharana R.R., Dolby T., Simpson J.A., van Helden P.D., Warren R.M., Theron G. Widespread use of incorrect PCR ramp rate negatively impacts multidrug-resistant tuberculosis diagnosis (MTBDRplus) Sci Rep. 2018;8:3206. doi: 10.1038/s41598-018-21458-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dowdy D.W., Theron G., Tornheim J.A., Warren R., Kendall E.A. Of testing and treatment: implications of implementing new regimens for multidrug-resistant tuberculosis. Clin Infect Dis. 2017;65:1206–1211. doi: 10.1093/cid/cix486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.World Health Organization . WHO; Geneva, Switzerland: 2019. WHO consolidated guidelines on drug-resistant tuberculosis treatment.https://apps.who.int/iris/handle/10665/311389 Available at: [PubMed] [Google Scholar]
- 10.World Health Organization . WHO; Geneva, Switzerland: 2021. Catalogue of mutations in Mycobacterium tuberculosis complex and their association with drug resistance.https://www.who.int/publications/i/item/9789240028173 Available at: [Google Scholar]
- 11.Hobby G.L., Holman A.P., Iseman M.D., Jones J.M. Enumeration of tubercle bacilli in sputum of patients with pulmonary tuberculosis. Antimicrob Agents Chemother. 1973;4:94–104. doi: 10.1128/aac.4.2.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Groessl E.J., Ganiats T.G., Hillery N., Trollip A., Jackson R.L., Catanzaro D.G., Rodwell T.C., Garfein R.S., Rodrigues C., Crudu V., Victor T.C., Catanzaro A. Cost analysis of rapid diagnostics for drug-resistant tuberculosis. BMC Infect Dis. 2018;18:102. doi: 10.1186/s12879-018-3013-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Vos E., Scott L., Voss De Lima Y., Warren R.M., Stevens W., Hayes C., da Silva P., Van Rie A. Management of rifampicin-resistant TB: programme indicators and care cascade analysis in South Africa. Int J Tuberc Lung Dis. 2021;25:134–141. doi: 10.5588/ijtld.20.0598. [DOI] [PubMed] [Google Scholar]
- 14.Dheda K., Gumbo T., Maartens G., Dooley K.E., Murray M., Furin J., Nardell E.A., Warren R.M., Lancet Respiratory Medicine drug-resistant tuberculosis Commission group The Lancet Respiratory Medicine Commission: 2019 update: epidemiology, pathogenesis, transmission, diagnosis, and management of multidrug-resistant and incurable tuberculosis. Lancet Respir Med. 2019;7:820–826. doi: 10.1016/S2213-2600(19)30263-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.