CONSPECTUS:
In vitro-transcribed RNAs are emerging as new biologics for therapeutic innovation, as exemplified by their application recently in SARS-CoV-2 vaccinations. RNAs prepared by in vitro transcription (IVT) allow transient expression of proteins of interest, conferring safety over DNA- or virus-mediated gene delivery systems. However, in vitro-transcribed RNAs should be used with caution because of their immunogenicity, which is in part triggered by double-stranded RNA (dsRNA) byproducts during IVT. Cellular innate immune response to dsRNA byproducts can lead to undesirable consequences, including suppression of protein synthesis and cell death, which in turn can detrimentally impact the efficacy of mRNA therapy. Thus, it is critical to understand the nature of IVT byproducts and the mechanisms by which they trigger innate immune responses.
Our lab has been investigating the mechanisms by which the innate immune system discriminates between “self” and “nonself” RNA, with the focus on the cytoplasmic dsRNA receptors retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated 5 (MDA5). We have biochemically and structurally characterized critical events involving RNA discrimination and signal transduction by RIG-I or MDA5. We have used in vitro-transcribed RNAs as tools to investigate RNA specificity of RIG-I and MDA5, which required optimization of the IVT reaction and purification processes to eliminate the effect of IVT byproducts. In this Account, we summarize our current understanding of RIG-I and MDA5 and IVT reactions and propose future directions for improving IVT as a method to generate both research tools and therapeutics. Other critical proteins in cellular innate immune response to dsRNAs are also discussed. We arrange the contents in the following order: (i) innate immunity sensors for nonself RNA, including the RIG-I-like receptors (RLRs) in the cytosol and the toll-like receptors (TLRs) in the endosome, as well as cytoplasmic dsRNA-responding proteins, including protein kinase R (PKR) and 2′,5′-oligoadenylate synthetases (OASes), illustrating the feature of protein–RNA binding and its consequences; (ii) the immunogenicity of IVT byproducts, specifically the generation of dsRNA molecules during IVT; and (iii) methods to reduce IVT RNA immunogenicity, including optimizations of RNA polymerases, reagents, and experimental conditions during IVT and subsequent purification.
Graphical Abstract
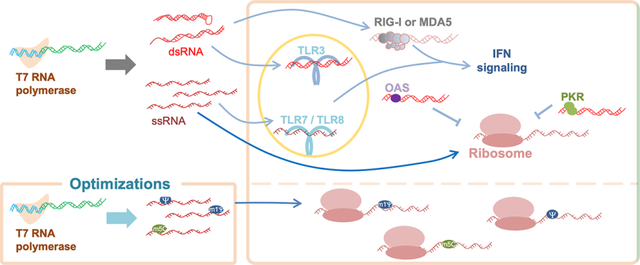
1. INTRODUCTION
Messenger RNA (mRNA)-based therapy can be used to replace endogenous malfunctioning genes or deliver antigens either from pathogens or tumors to the immune system.5 As our understanding of RNA biology and technology increases, mRNA-based therapy has become a promising direction for treatment. mRNA vaccines against SARS-CoV-2 are the most well-known example of its application to date.6
Therapeutic mRNAs are produced through in vitro transcription (IVT) catalyzed by DNA-dependent RNA polymerases derived from bacteriophages, such as T3, T7, or SP6.7 Of these, T7 RNA polymerase is the most extensively studied and widely used.8 These RNA polymerases selectively recognize the promoter region in DNA templates to synthesize RNA transcripts based on downstream template sequences. The resultant transcripts are naked RNA molecules—uncapped at the 5′-end and non-polyadenylated [poly(A)] at the 3′-end. To mimic naturally occurring mRNAs, extra steps are taken during and after IVT to cap the 5′-end (linking of N7-methylguanosine to the first nucleotide through a 5′–5′ triphosphate bond), optimize the sequences in the 5′- and 3′-untranslated regions (UTRs) and the coding sequence (CDS), and add the poly(A) tail.9
Although these modifications improve the stability and translational efficiency of in vitro-transcribed RNAs, they are challenged by the host innate immune system, which recognizes nonself molecules frequently found in invading pathogens or under pathological conditions. The innate immune response impedes therapeutic efficacy, as it not only affects treatment safety because of aberrant immune activation but also reduces the translation efficiency as part of cellular stress responses.10 Advances in innate immunity and IVT research have led us to believe that the purity and nucleotide composition of in vitro-transcribed RNA contribute to its immunogenicity. In this Account, we focus on our progress in our understanding of innate immune response to nonself RNA and IVT RNA.
2. CELLULAR INNATE IMMUNITY
Nonself molecules are sensed by host-encoded pattern recognition receptors (PRRs). PRRs generally do not recognize specific sequences from DNA, RNA, or protein but rather the molecular structures or kinds. Ligand-bound PRRs activate type-I interferon (IFN-I, such as IFNα or IFNβ) and inflammatory nuclear factor kappa B (NF-κB) signaling pathways to direct and modulate innate and adaptive immunity.11,12 Aberrant activation elicited by, for example, mutations in critical genes results in inflammation and immune diseases.13 Cellular functions, including gene expression, metabolism, proliferation, and differentiation, are under stress in cells involved in the innate immune response.14,15 In such cases, the translation of in vitro-transcribed mRNAs is negatively affected.
We will discuss PRRs that recognize nonself RNAs, most notably cytosolic RIG-I-like receptors (RLRs) and endosomal toll-like receptors (TLRs). Other double-stranded RNA (dsRNA)-binding proteins that shape cellular stress responses to dsRNA will also be discussed.
2.1. RLRs
Upon RNA virus infection, dsRNAs are formed intracellularly and recognized by RLRs, including retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated 5 (MDA5).16,17 They have similar amino acid sequences and domain architectures, with two tandem N-terminal caspase activation and recruitment domain (CARD) motifs, a central DExD/H box helicase domain, and a C-terminal domain (CTD) (Figure 1a).16 RIG-I and MDA5 use the same downstream adaptors, kinases, and transcription factors to activate IFN signaling.16 Thus, the study of one receptor provides insights into the other.
Figure 1.
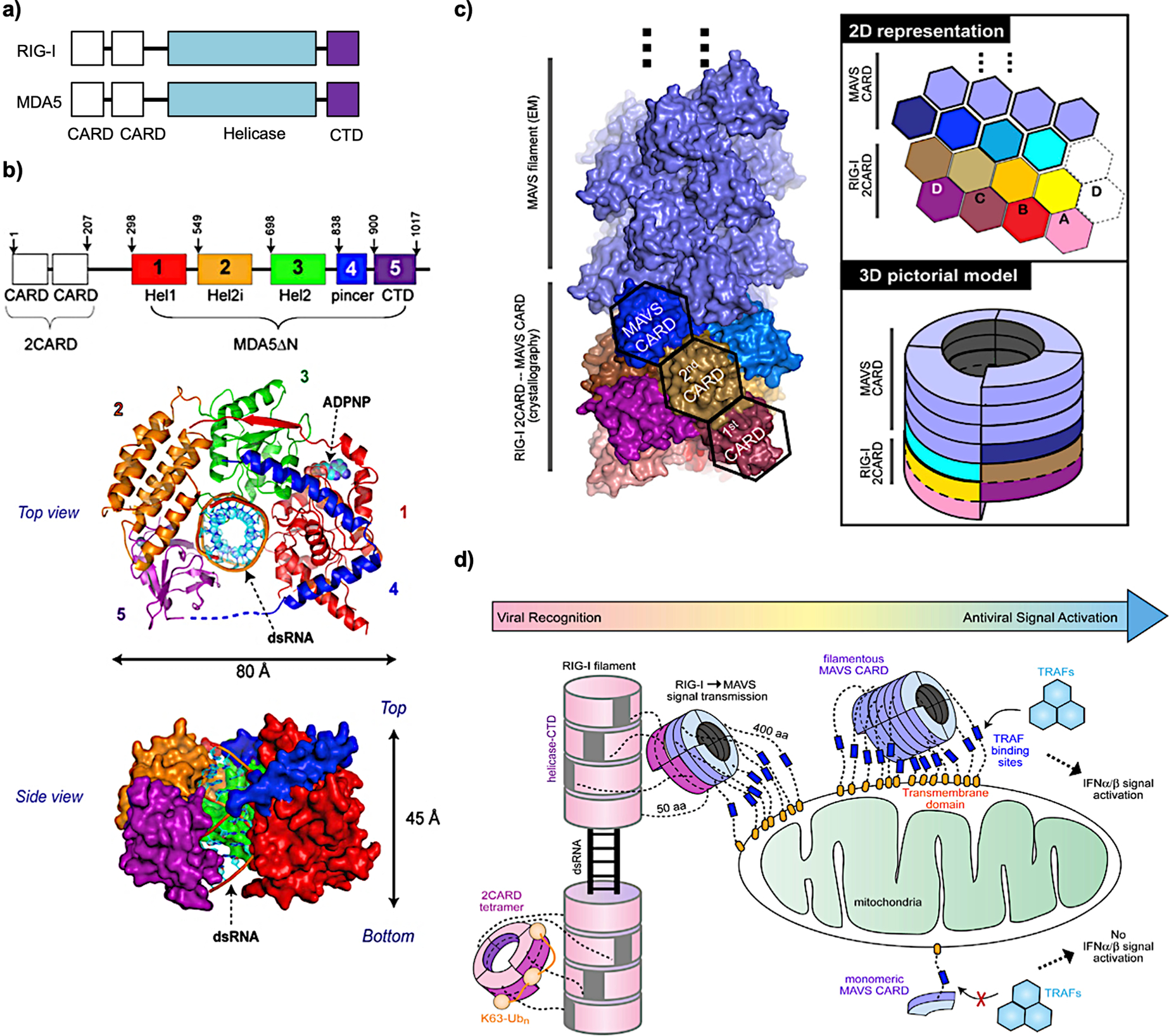
(a) Schematic of the domain organization of RIG-I and MDA5. (b) Top and side views of MDA5 (without 2CARDs): 12 bp dsRNA complex structure in the presence of the ADP analogue ADPNP. (c) Structures of MAVS CARD filament nucleated by RIG-I 2CARD oligomers. 2D and 3D models are shown on the right. (d) Cartoon model of filamentous RLRs nucleating MAVS to form filaments to activate downstream factors for IFN signaling. (b) Reprinted with permission from ref 1. Copyright 2013 Elsevier Inc. (c, d) Reprinted with permission from ref 2. Copyright 2014 Elsevier Inc.
We determined the structure of MDA5 bound to dsRNA and found that MDA5 binds dsRNA in a sequence-independent manner by interacting with the backbone of the RNA duplex (Figure 1b).1 Upon dsRNA binding through the helicase domain and CTD, proteins oligomerize along the length of the dsRNA and form a filamentous structure.18 The two CARD motifs (2CARDs) oligomerize upon filament formation,19 activating the downstream adaptor mitochondrial antiviral signaling protein (MAVS) in the mitochondria. We used RIG-I 2CARD oligomers to study their interaction with MAVS and found that they interact with the CARD motif of MAVS, triggering MAVS to form prion-like aggregates.2 It was shown that this MAVS–MAVS interaction is mediated through its CARD–CARD binding2 in the way that 2CARD oligomers from the RIG-I filament nucleate MAVS assembly (Figure 1c).2 This work uncovered how signals are transduced from RLR–dsRNA to MAVS. Activated MAVS then recruits tumor necrosis factor (TNF) receptor-associated factors (TRAFs), TANK-binding kinase 1 (TBK1), and interferon regulatory factor 3 (IRF3) to activate IFN signaling (Figure 1d).2,20
Despite sharing similarities, RIG-I and MDA5 have different substrate preferences and modes of filament formation. We and others have shown that RIG-I senses 5′-triphosphate (5′-ppp) or 5′-diphosphate (5′-pp) dsRNA with minimal lengths of ~20 base pairs (bp) and 40–150 bp for optimal signal transduction efficiency.21–24 First, it binds to the 5′-end of dsRNA and translocates to the inner side of dsRNA by hydrolyzing adenosine triphosphates (ATPs). A second RIG-I molecule then binds to the exposed dsRNA end and processes it in the same manner.19 MDA5 prefers longer dsRNA molecules (normally >1000 bp), independent of cap structure. It binds to the inside of dsRNA to nucleate protein association for filament formation.2,18 It was found that nucleation is a rate-limiting step compared with filament elongation. Hydrolyzing ATP leads to dissociation of MDA5 from the filament end and new MDA5s then assemble to fill the gap. Long dsRNA is preferred because a certain length of filament can be maintained under such assembly-and-disassembly conditions.18,22 The ATPase activities of both RIG-I and MDA5 play important roles in discrimination of nonself from self RNA substrates.18,25,26 The sensitivity of RLRs needs to be balanced with the specificity. The gain-of-function mutations of MDA5 showed more IFN-activating capacity at the cost of self-recognition,3 leading to the onset of autoimmune diseases. RLRs were found to be involved in in vitro-transcribed RNA-mediated activation of IFN signaling.27,28
2.2. TLRs
Viruses invading cells through endocytosis enter the endosomes for uncoating and release of their genomic materials into the cytosol. Several types of TLRs localize and face toward the inside of the endosome.29 Recognition of dsRNAs with a minimal length of ~40 bp by TLR330 leads to TLR3 dimerization and subsequently activates the downstream adaptor protein Toll or interleukin-1 receptor (TIR) domain-containing adapter-inducing interferon-beta (TRIF). TRIF then recruits TRAF3 and TRAF6 for transcriptional activation of IFN and NF-κB activation.29,31 Structural studies have revealed that TLR3 binds to the ribose–phosphate backbone and not individual bases of dsRNA.32
Single-stranded RNAs (ssRNAs) are also recognized by TLR7 or TLR8 in the endosome. TLR7 dimerizes upon RNA binding, whereas TLR8 exists as a dimer before RNA recognition.33,34 Once bound to their ligands, TLR7 and TLR8 recruit the downstream adaptor myeloid differentiation primary response 88 (MyD88), which in turn associates with interleukin-1 receptor-associated kinase 4 (IRAK4) and IRAK1/IRAK2 to form the Myddosome. The Myddosome recruits TRAFs to activate IFN and NF-κB signaling.35–38 Human TLR8 prefers GU-rich ssRNAs.39 The immunogenicity of U-rich stretches was demonstrated using mouse and human cells in a study showing that uridine stretches are strong ligands for TLR7.40 Consistently, structural analysis revealed that both TLR7 and TLR8 bind to uridine in a uracil- and ribose-dependent manner (Figure 2a for TLR7 and Figure 2b for TLR8).33,34 It is noteworthy that the binding sites on the uracil base are still present in chemically modified versions of uridine, such as pseudouridine (ψ) (Figure 2c), which will be discussed in more detail later in this Account. Several studies have confirmed that in vitro-transcribed RNAs stimulate TLR3, TLR7, and TLR8.41–43
Figure 2.
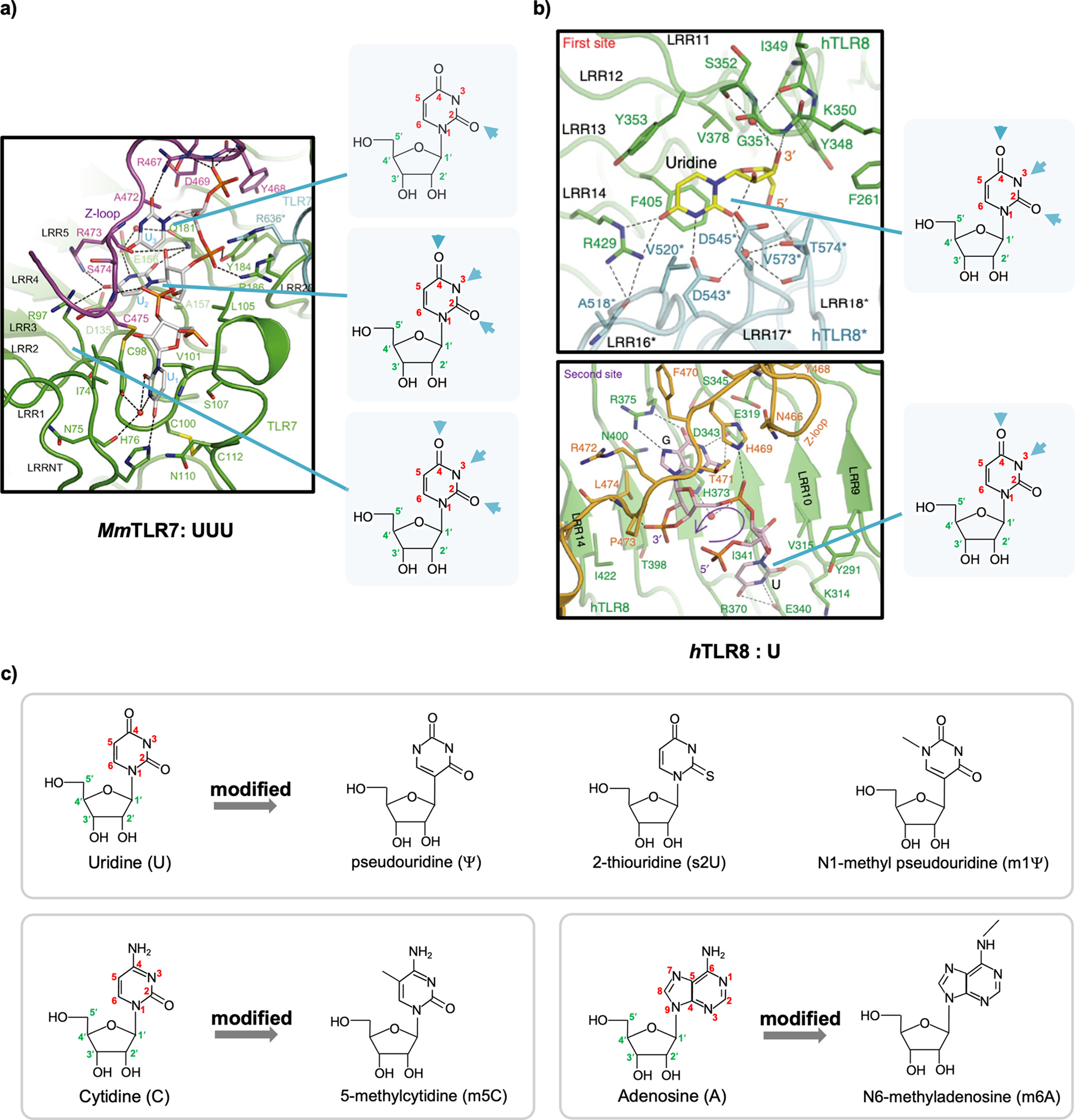
(a) View of UUU recognized by Macaca mulatta TLR7 (MmTLR7). (b) View of U recognized by human TLR8 (hTLR8). The structural formulas of uridine are highlighted to the right of each panel, and interacting atoms in the uracil base are indicated by arrows. (c) Structures of unmodified uridine, cytidine, and adenosine and their chemically modified versions. The carbon and nitrogen atoms in the base ring are numbered in red. The carbon atoms in the ribose ring are numbered in green and labeled with a prime (′). (a) Reprinted with permission from ref 33. Copyright 2016 Elsevier Inc. (b) Reprinted with permission from ref 34. Copyright 2015 Nature Publishing Group.
2.3. Other dsRNA-Binding Proteins
When protein kinase R (PKR) binds to dsRNAs with a minimal length of ~33 bp, it dimerizes and gets activated by autophosphorylation,17,44 which leads to the phosphorylation of the alpha subunit of eukaryotic translation initiation factor 2 (eIF2α). Phosphorylated eIF2α blocks global translation initiation and subsequently leads to activation of NF-κB-mediated apoptosis.45,46 Similar to RLRs, PKR binds to dsRNA in a sequence-independent manner.47,48 In vitro-transcribed RNAs have been reported to trigger PKR activation in vitro and in cells.49,50
2′,5′-Oligoadenylate synthetase (OAS) binds to dsRNA to produce 2′,5′-oligoadenylate (2–5A), which serves as a secondary messenger to activate RNase L by inducing its dimerization or oligomerization.51 Activated RNase L then globally degrades cellular RNAs52 and actively arrests global translation.53–55 Interestingly, defense mRNAs such as IFNβ are not cleaved and translate during this process. All of these reprogram the cellular environment toward making IFNs.53–55 In humans, the OAS family comprises OAS1, OAS2, OAS3, and OAS-like protein (OASL); OASL is enzymatically inactive.56 OAS3 is more potent in responding to dsRNAs than OAS1 and OAS2 and is the primary activator of RNase L.57 In vitro-transcribed RNAs activate the OAS–RNase L pathway and inhibit their own translation.58
A schematic view of cellular innate immunity is shown in Figure 3.
Figure 3.
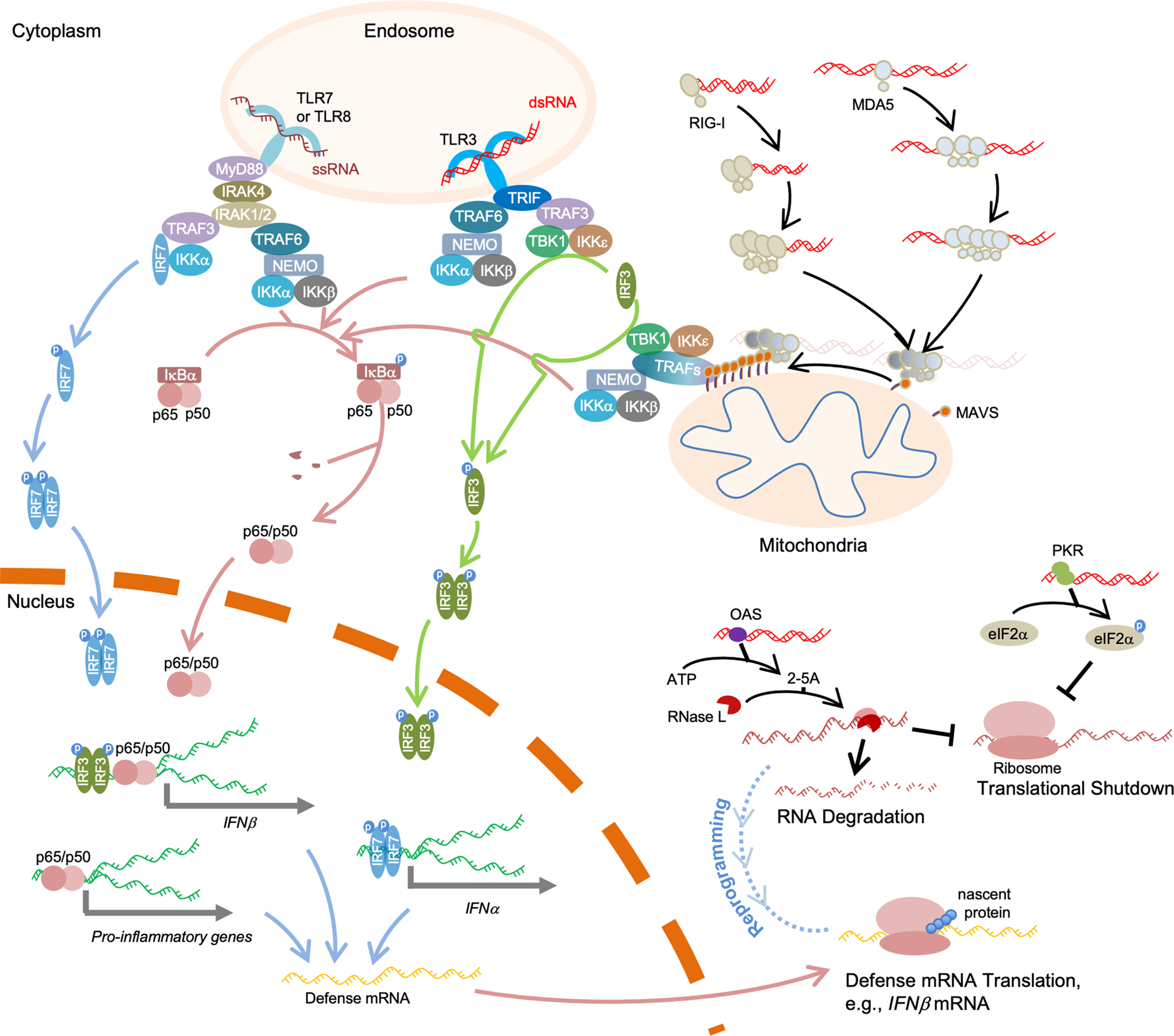
In the cytoplasm, RIG-I and MDA5 form filaments along the length of dsRNA to activate MAVS, whose aggregation functions as a platform to recruit TRAF proteins for IFN-I and NF-κB signaling activation. dsRNAs are also recognized by PKR to trigger global translation repression and by OAS for RNA degradation and translation shutdown. OAS–RNase L reprograms cellular translation to allow defense mRNAs, such as IFNβ mRNAs, to translate normally. In the endosome, dsRNAs are recognized by TLR3, and such binding activates the downstream adaptor TRIF. ssRNAs in the endosome bind to TLR7 or TLR8, activating MyD88, which further recruits IRAKs to form the Myddosome. Activated TRIF or Myddosome then recruits TRAFs for downstream activation. Red-colored strands represent RNA molecules, and green strands represent DNA molecules. “p” denotes phosphorylation.
It is noteworthy that theoretically a potential consequence of in vitro-transcribed RNAs applied in vivo is the raising of anti-RNA antibodies. These RNA-recognizing antibodies are evident in the case of autoimmune diseases like systemic lupus erythematosus.59,60 Studies have shown that TLR7 in B cells contributes to such antibody production and that RNA antigens can be RNA epitopes on small nuclear RNAs (snRNAs), 28S rRNA (rRNA), certain tRNAs (tRNAs), and even some mRNAs.59–62 Although reports on anti-IVT RNA antibodies are still lacking, one might consider examining mRNAs for therapeutics, as they themselves do not induce anti-RNA antibodies.
3. RNA IMMUNOGENICITY DUE TO IVT REACTION
IVT RNAs are often used to investigate the RNA specificity of RNA sensors in the innate immune system. IVT can be easily performed using commercial kits or homemade T7 RNA polymerase. However, IVT yields not only the desired full-length ssRNA but also several types of byproducts, including short transcripts due to abortive transcription63 and dsRNAs.4,64,65 Such complexity can be exemplified by the observation that RNA produced by IVT activated OAS while that with the same sequence produced by chemical synthesis did not.66 Understanding how dsRNA byproducts are formed during IVT will help reduce unwanted immunogenicity.
dsRNA byproducts are mainly produced through the 3′-extension of the ssRNA transcript, caused by annealing of the 3′-end to complementary sequences in cis or trans and continued transcription (Figure 4a).64,65 Additionally, we identified that dsRNA byproducts that were resistant to ssRNA-specific RNase I digestion but sensitive to dsRNA-targeting RNase III treatment (Figure 4b) were composed of two separate reverse complementary RNA strands (Figure 4c). The sense strand in the dsRNA corresponds to the desired ssRNA product, whereas the antisense strand was a full-length RNA molecule transcribed from the 3′-end of the template to the promoter region (Figure 4c), indicating that the antisense strand is transcribed in a promoter-independent, 3′-end extension-free manner and anneals to the sense strand to form a duplex (Figure 4d). This full-length dsRNA byproduct activates both RIG-I and MDA5 for IFN signaling (Figure 4e).4 The promoter-free transcription from the 3′-end of the DNA template is dependent on the sequence of the 3′-end in the DNA template.4,67
Figure 4.
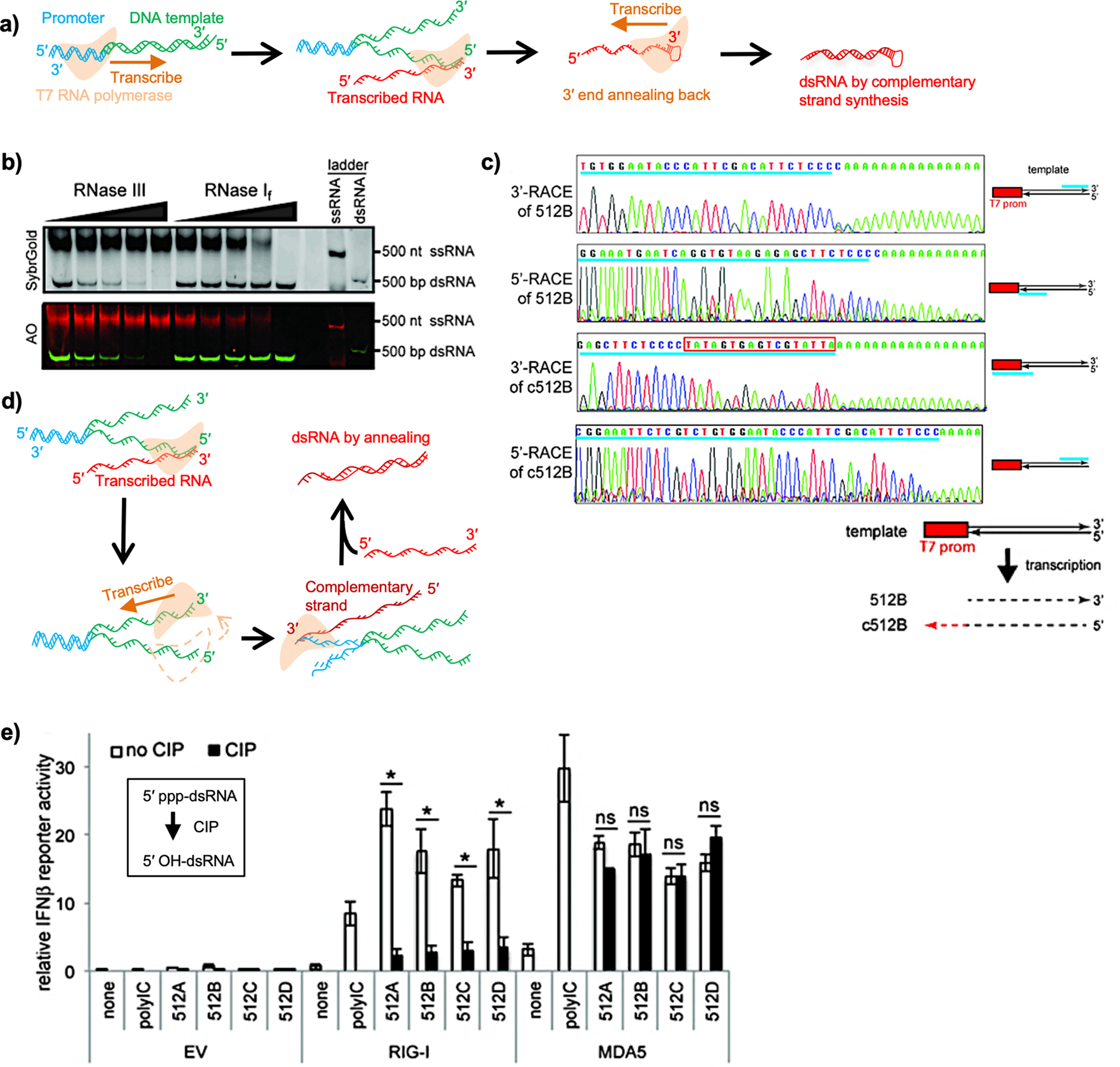
(a) Schematic view of 3′-end extension-mediated dsRNA production by T7 RNA polymerase. The promoter region in the template DNA is shown in blue. Downstream sequences are shown in green. The transcribed RNAs are shown in red. The orange arrow indicates the direction of transcription. (b) Transcripts were challenged with RNase III or RNase I treatment and subjected to native PAGE analysis. The RNAs were visualized using SybrGold and AO. In AO staining, dsRNA is stained in green and ssRNA in red. (c) Start and end sites of sense (512B) and antisense (c512B) strands of transcripts were determined by 3′ and 5′ rapid amplification of cDNA ends (RACE). The schematic illustration of the sequencing results is shown at the bottom. (d) Schematic view of dsRNA production by the mechanism of annealing of sense and antisense strands. After RNA transcription by the sense strand, the RNA polymerase initiates RNA transcription by the antisense strand. The orange arrow indicates the direction of transcription. (e) A luciferase assay was done in HEK293T cells by cotransfection of RIG-I- or MDA5-expressing plasmids with luciferase reporters driven by IFNβ promoter, followed by transfection of stimulant RNAs indicated. CIP: calf intestinal phosphatase. (b), (c), and (e) Reprinted with permission from ref 4. Copyright 2018 Oxford University Press.
Double-stranded structures can also be formed through annealing of complementary sequences intra- or intermolecularly.68 Therefore, checking in vitro-transcribed RNA for the presence of dsRNA byproducts even after RNA purification is critical. Traditional purification methods, including phenol/chloroform extraction, lithium chloride precipitation, and column-based purification methods, cannot distinguish dsRNA from ssRNAs, and short abortive RNAs are not always removed. We used acridine orange (AO) to stain RNAs on native PAGE because AO helps distinguish between ssRNA and dsRNA (Figure 4b). Moreover, analysis of RNA products using different types of RNases is helpful (Figure 4b).
4. METHODS TO REDUCE IMMUNOGENICITY
Many groups, including ours, have attempted to eliminate the dsRNA byproduct during transcription or purification. We have arranged this section based on operational order, from during IVT to after IVT. During IVT, optimizations are performed on RNA polymerases, chemically modified NTPs, reaction conditions, and template sequences. For the subsequent purification steps, chromatography-based methods are effective.
4.1. RNA Polymerases
IVT can be performed at higher temperatures (51–55 °C) by using an engineered thermostable T7 RNA polymerase to yield ssRNA products lacking 3′-extension-derived dsRNA byproducts. Higher temperatures are believed to decrease the efficiency of either polymerase binding to the 3′-end of RNA or 3′-end priming for antisense synthesis.67 Interestingly, this high-temperature condition could not reduce the formation of the dsRNA byproduct formed by the annealing of sense and antisense RNAs. However, introduction of the poly(A) tail encoded by the template reduced antisense byproduct formation at higher temperatures. Thus, a combination of high temperature and poly(A) tailing was proposed to reduce immunogenicity67
RNA polymerases from other bacteriophages have also been studied. An RNA polymerase from the psychrophilic phage VSW-3 was shown to be more effective than T7 RNA polymerase in maintaining the yield of ssRNA products and decreasing the yield of dsRNA byproducts. Reactions can be performed at ≤25 °C, which is reportedly beneficial for the stability of RNA products.69
4.2. Chemically Modified Nucleotides
DNA molecules undergo modifications such as cytosine methylation on nucleosides, not to affect base paring but to influence protein binding.70 Similarly, mammalian RNAs, including mRNAs, are modified post-transcriptionally. Chemically modified nucleosides include pseudouridine (ψ), N6-methyladenosine (m6A), 5-methylcytidine (m5C), etc.71 Other common modified nucleosides include 2-thiouridine (s2U) and N1-methylpseudouridine (m1ψ) (Figure 2c). Such modifications are suggested to mark the RNAs as self and protect them from innate immune recognition.42 Extensive efforts have been made to investigate the protective role of modified nucleosides toward in vitro-transcribed mRNAs. Studies in dendritic cells (DCs) from peripheral blood have found that m5C and m6A present immunostimulatory effects comparable to those with unmodified bases, whereas ψ, s2U, and ψ/m6A are nonstimulatory.42 This suggests a dominant role of ψ (over m6A) in protecting RNAs from recognition and that mRNAs containing m1ψ are less immunogenic.72,73 Consistently, in mice, modified nucleosides such as s2U/m5C, ψ/m5C, and ψ reduce the immunogenicity of in vitro-transcribed mRNA.43,74,75
These chemically modified nucleotides containing in vitro-transcribed RNAs may lose efficient binding affinity to innate immune receptors. We reported that the presence of ψ, s2U, or m6A in the RNA duplex suppresses RIG-I from forming filaments on dsRNA.19 Consistently, Gehrke et al. observed that m6A protects in vitro-transcribed RNAs from binding to RIG-I, whereas ψ, m1ψ, and m5C prevent RIG-I conformational change after binding.28 A combination of m5C and s2U protects mRNAs from binding to TLR3, TLR7, TLR8, and RIG-I.43
Studies have shown that for IVT mRNA, translation efficacy is higher with modified nucleotides than with normal nucleotides.43,72,73,75 For example, in vitro-transcribed mRNAs containing ψ or m5C exhibited a dramatic increase in translation, which was immunogenicity-independent for in vitro-transcribed mRNAs containing ψ.75 Comparison of in vitro-transcribed yeast tRNA and natively isolated tRNA showed that the IVT product is a strong agonist of PKR activation and that nucleoside modifications, including ψ, m5C, and s2U, abolished this activation.76 Consistently, tests with in vitro-transcribed mRNAs have shown that ψ, m1ψ, and s2U are responsible for protection against PKR recognition.49,50,77 Moreover, replacement of U with ψ protects the RNA molecule from activating OAS and the downstream RNase L-mediated RNA cleavage event.58 Mounting evidence from mice studies supports the beneficial role of modified nucleosides (ψ and m1ψ) in mRNA translation.58,72,73,75,78–80 However, it should be noted that exactly why certain modifications are more effective than others in evading translational suppression is unclear. Not all modifications are beneficial to translation, even if they contribute to reducing PKR binding. Additionally, different cells may exhibit different sensitivities to modified nucleosides. For example, in vitro-transcribed mRNAs containing s2U and m6A in HEK293 cells (an immortal human embryonic kidney cell line) and murine monocyte-derived dendritic cells (MDDCs) are not translated.75 Furthermore, ψ and m1ψ could not improve translation of in vitro-transcribed mRNAs in HeLa cells (an immortal human cervical cancer cell line) or keratinocytes (an epidermal cell line) but could enhance the translation of in vitro-transcribed mRNA in C2C12 cells (an immortalized mouse myoblast cell line).72
These observations suggest that the effect of RNA modification on immunogenicity and translation suppression may not be entirely due to the direct effect of modified nucleosides on the RNA sensors. In fact, our study found that modified nucleotides in the IVT reaction also alter the behavior of the RNA polymerase and consequently the synthesized RNA products. We tested selected modified nucleotides to replace original nucleotides for IVT and found that antisense-mediated dsRNA byproducts were reduced when ψ, m1ψ, or m5C was used but not when m6A was used (Figure 5a, 512B transcript).4 A similar observation was made by Karikó et al. when ψ was used in place of U.78 Intriguingly, the MDA5-mediated IFN signaling efficacy was unaffected by gel-purified dsRNA containing modified nucleotides (Figure 5b, purified 512B:c512B), consistent with the notion that MDA5 binds to the dsRNA backbone rather than interacting with bases, where modification often occurs. Our data suggest that modified nucleotides in part reduce the immunogenicity of in vitro-transcribed RNA by inhibiting the production of dsRNA byproducts, thus improving RNA purity. It is also possible that modified nucleotides in the IVT reaction also suppress other immunogenic byproducts besides the dsRNA byproduct as described above, which would be an interesting area of future research.
Figure 5.
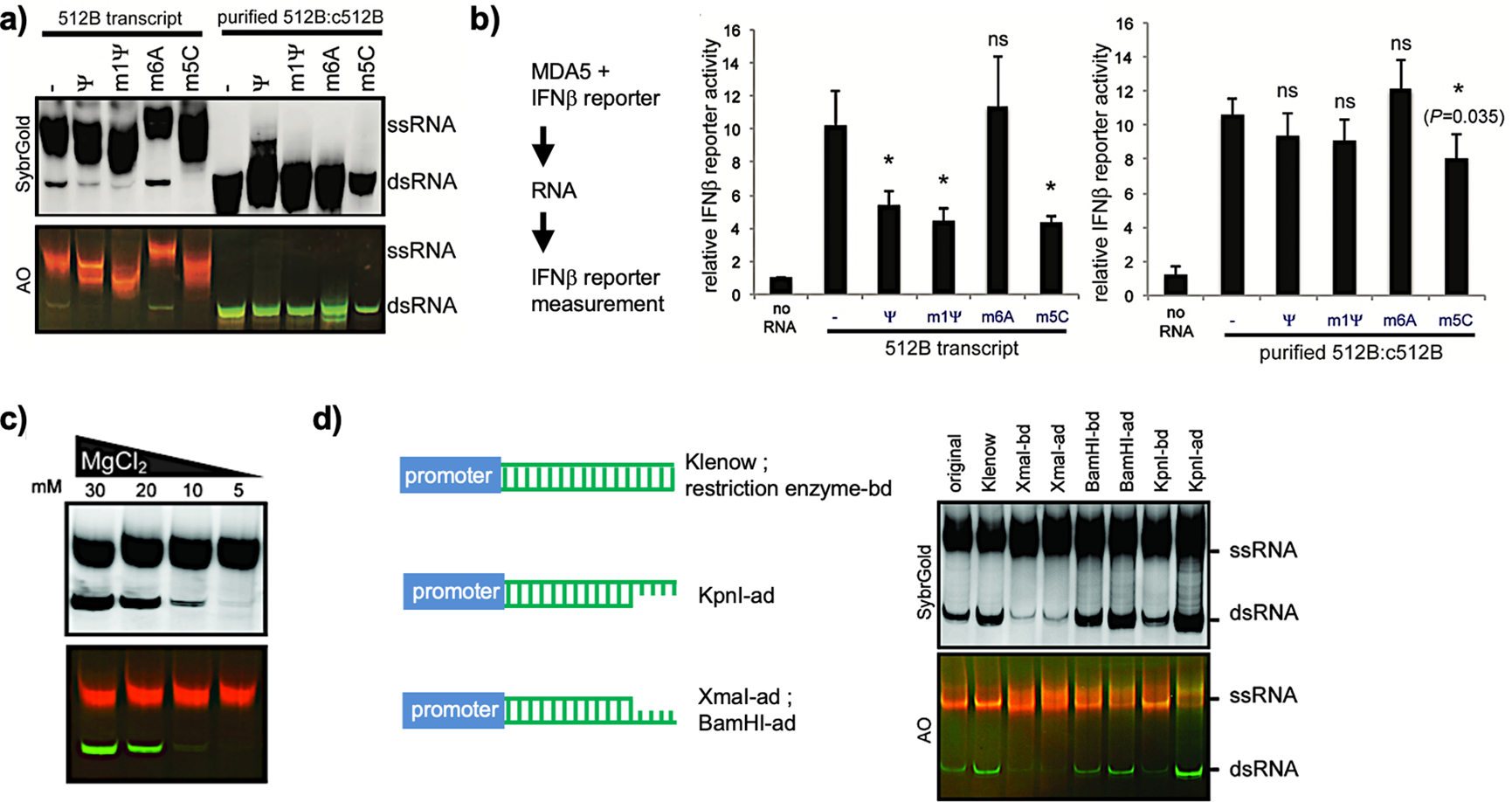
(a) Native PAGE analysis of transcripts using unmodified or modified nucleotides for IVT. The purified dsRNA 512B:c512B was initiated by incubation of IVT products with RNase I to digest ssRNAs. (b) A luciferase assay was done in HEK293T cells by cotransfection of MDA5-expressing plasmids and luciferase reporters driven by IFNβ promoter, followed by unpurified (512B transcript) or purified (purified 512B:c512B) RNA transfection, respectively. (c) Native PAGE analysis of transcribed RNAs at 512 nucleotides (nt) using different Mg2+ concentrations for IVT. dsRNA byproducts were validated by AO staining. (d) Native PAGE analysis of transcribed RNAs with different 3′-end sequences and structures. Templates before (original) and after the Klenow reaction (Klenow), derivatives with indicated restriction sites at the 3′-end before digestion (-bd) and after digestion (-ad) are shown on the left and were used for IVT on the right. (a–d) Reprinted with permission from ref 4. Copyright 2018 Oxford University Press.
4.3. Transcription Conditions
We tested different IVT conditions, such as the concentrations of RNA polymerase, template, NTPs, and NaCl, to decrease dsRNA formation and found that lowering the Mg2+ concentration (from 30 to 5 mM) reduced the amount of dsRNA byproducts generated by the promoter-free antisense RNA synthesis mechanism (Figure 5c) independent of template length.4 Although the reason for this improvement is unclear, we speculate that the T7 RNA polymerase in the promoter-free antisense RNA synthesis maintains its conformation in the elongation phase to stay transcriptionally active and that a high concentration of Mg2+ (30 mM) promotes maintenance of such a conformation.4
4.4. Template Sequences
Sequence manipulation can reduce the immunogenicity of in vitro-transcribed mRNA. By using different restriction sites to change the 3′-end sequence in the IVT template and digestion of these sites to create different structures, we noticed that the 3′-end sequence and structures impact the promoter-free antisense transcription (Figure 5d).4 Other studies have shown that using uridine-depleted sequences lowers the immunogenicity of in vitro-transcribed Cas9 mRNA containing modified nucleosides by an unknown mechanism,81 supporting the notion that TLR7 and TLR8 bind U-containing ssRNAs.
4.5. Purification of In Vitro-Transcribed RNAs
On the basis of RNA size and structure, native PAGE or agarose gel electrophoresis can identify the correct ssRNA products on a small scale.82 For larger amounts of in vitro-transcribed RNAs, chromatographic purification effectively separates IVT byproducts, such as short abortive transcripts, nucleotides, and dsRNAs with distinct conformations.83–85 High-performance liquid chromatography (HPLC) was reported to effectively remove dsRNA contaminants; however, the resultant in vitro-transcribed mRNA, composed of unmodified nucleotides, remained immunostimulatory—DCs maintained high levels of TNF-α and IFN-α. By contrast, HPLC-purified ψ-, m5C- or ψ/m5C- modified mRNAs did not induce cytokine release.85 Findings from a study using m1ψ were consistent.86 Thus, using a combination of modified nucleotides for IVT and HPLC purification can produce non-immunostimulatory mRNAs.
Another approach to remove dsRNA contaminants takes advantage of the fact that dsRNA prefers to bind to cellulose in ethanol-containing buffer78 and is utilized in the study of microorganisms.87,88 Cellulose-bound dsRNA is separated in the spin column from the ssRNA flowthrough. Nucleoside-modified RNAs (m1ψ) can be effectively purified using this method. Results from in vivo assays suggested that mRNAs purified using this method are comparable with HPLC-purified mRNAs in terms of translation efficiency.78 Consistent with results from HPLC purification, cellulose-purified in vitro-transcribed mRNAs containing unmodified nucleosides were immunostimulatory, whereas m1ψ-containing in vitro-transcribed mRNAs were not, further highlighting the protective role of modified nucleosides.
A summary of methods to reduce immunogenicity is illustrated in Figure 6. Purification of in vitro-transcribed mRNAs to discard dsRNA contaminants or abortive short transcripts is not effective enough to keep the in vitro-transcribed mRNAs from being immunogenic. For now, a combination of modified nucleotide usage for transcription and chromatographic purification appears to be promising for in vitro-transcribed mRNA preparation.
Figure 6.
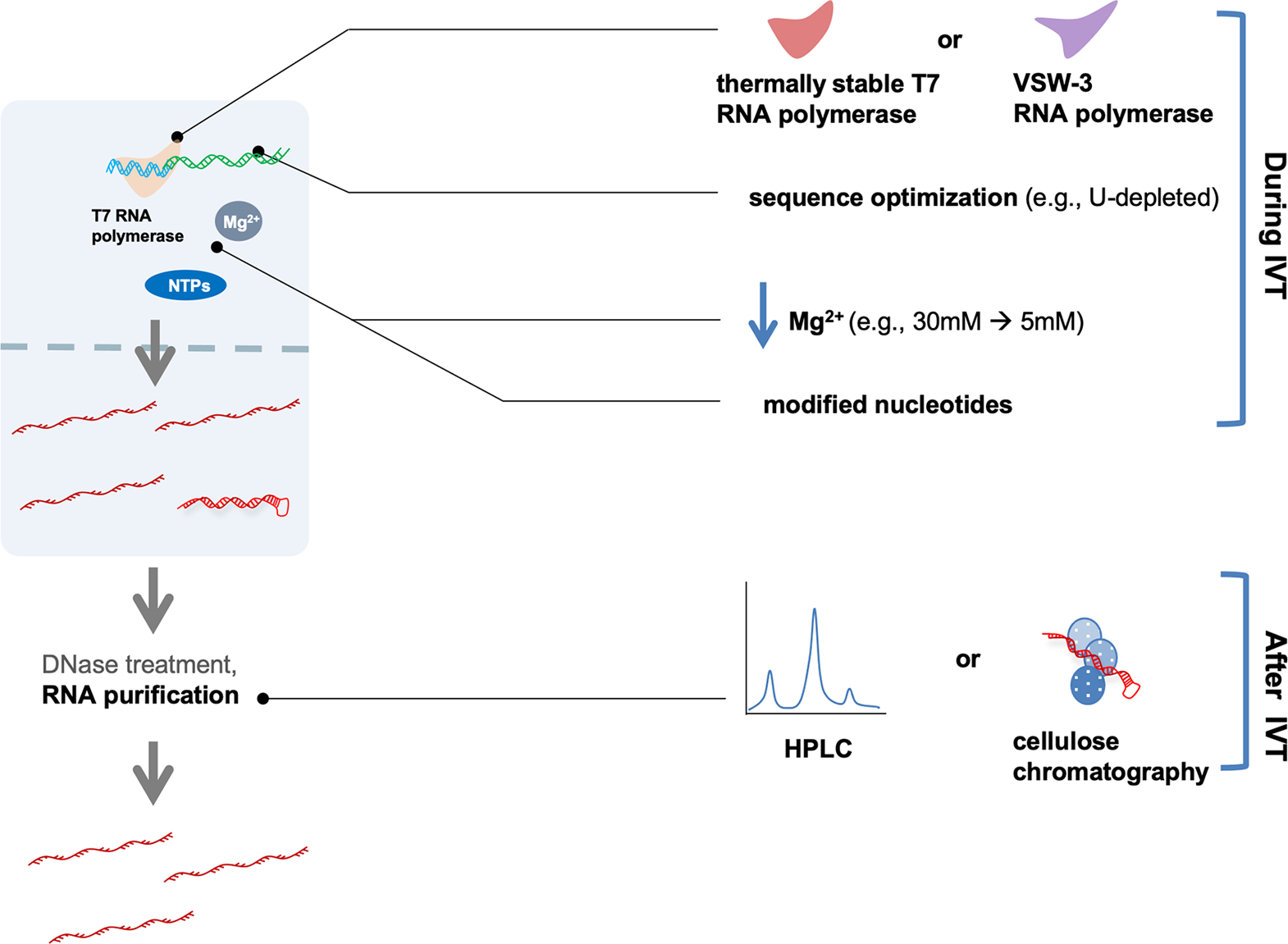
Optimizations to reduce dsRNA contaminants include the use of different RNA polymerases, changes in experimental conditions (e.g., Mg2+ concentration or the use of selected modified nucleotides), and the use of manipulated-sequence-containing templates (e.g., deletion of U-rich sequence) during IVT. In the purification steps, HPLC or cellulose chromatography can be applied.
5. OUTLOOK
Progress in innate immunity has increased our understanding of the immunogenicity of in vitro-transcribed mRNAs. Mechanistic studies of the interactions between host-cell-encoded receptors and their RNA ligands have revealed details of innate immune activation, and investigations of IVT reactions have shed light on the origin of such immunogenicity. Although immunogenicity can be potentially beneficial in the case of vaccination,89 a well-controlled in vitro-transcribed mRNA product is crucial for therapeutic applications, as the goal of mRNA therapy is to express proteins of interest in target cells. The use of chemically modified nucleotides for IVT and chromatographic purification steps makes in vitro-transcribed mRNAs competent in protein translation and immunosilencing.78,85,86 New RNA polymerases, including the optimized T7 RNA polymerase, are also worth exploring.67,69
The use of chemically modified nucleotides in IVT contributes not only to reducing the immunogenicity of purified ssRNAs but also to decreasing dsRNA production.4,78 We speculate that modified nucleotides may benefit IVT by increasing transcript purity and decreasing transcript immunogenicity. Because of the complex nature of in vitro transcription, using AO staining can be a straightforward way to check the presence of dsRNA byproducts, thus reducing uncertainties about the IVT samples.
How modified nucleotides suppress dsRNA production or RNA immunogenicity is still unclear. Considering immune evasion as an example, the dsRNA-binding proteins discussed here sense RNA in a sequence-independent manner. For the ssRNA-sensing receptors TLR7 and TLR8, structural studies have shown that the receptor-bound atoms in uridine include oxygen atoms linked to C2 and C4 and N3 in uracil (Figure 2a,b), all of which are equally present in ψ or m1ψ (Figure 2c). Precisely how these modified nucleotides affect immunogenicity and translational efficiency, whether it is directly or indirectly mediated by RNA sensors, and how much of these effects are due to their impact on the IVT reaction itself remain to be investigated. We believe that studying the molecular mechanism ofn this process will improve our understanding of how innate immunity distinguishes “nonself” from “self” RNAs.
ACKNOWLEDGMENTS
This work was supported by the NIH (Grants R01AI154653, R01AI111784, and DP1AI152074 to S.H.) and the State Key Laboratory of Veterinary Etiological Biology, CAAS (Grant SKLVEB2020KFKT001 to X.M.).
ABBREVIATIONS
- AO
acridine orange
- ATP
adenosine triphosphate
- CARD
caspase activation and recruitment domain; bp, base pair
- CDS
coding sequence
- CTD
C-terminal domain
- DC
dendritic cell
- dsRNA
double-stranded RNA
- eIF2α
alpha subunit of eukaryotic translation initiation factor 2
- HPLC
high-performance liquid chromatography
- IFN
interferon
- IRAK
interleukin-1 receptor-associated kinase
- IRF3
interferon regulatory factor 3
- IVT
in vitro transcription
- MAVS
mitochondrial antiviral-signaling protein
- MDA5
melanoma differentiation-associated 5
- MDDC
monocyte-derived dendritic cell
- MyD88
myeloid differentiation primary response 88
- m1ψ
N1-methylpseudouridine
- m5C
5-methylcytidine
- m6A
N6-methyladenosine
- NF-κB
nuclear factor kappa B
- nt
nucleotides
- OAS
2′,5′-oligoadenylate synthetase
- OASL
OAS-like protein
- PAGE
polyacrylamide gel electrophoresis
- PKR
protein kinase R
- poly(A)
polyadenylated
- PRR
pattern recognition receptor
- RIG-I
retinoic acid-inducible gene I
- RLR
RIG-I-like receptor
- ssRNA
single-stranded RNA
- s2U
2-thiouridine
- TBK1
TANK-binding kinase 1
- TLR
toll-like receptor
- TNF
tumor necrosis factor
- TRAF
TNF receptor-associated factor
- TRIF
TIR-domain-containing adapter-inducing interferon-beta
- UTR
untranslated region
- ψ
pseudouridine
- 2–5A
2′,5′-oligoadenylate
Biographies
Xin Mu was born on November 26, 1982, in Tianjin, China. He received his B.S. degree from Tianjin University and his Ph.D. degree from the Institute of Biophysics, Chinese Academy of Sciences (CAS), supervised by Professor Guangxia Gao. He joined Professor Sun Hur’s lab at Boston Children’s Hospital, Harvard Medical School, in 2015 as a postdoctoral fellow, where he worked on dissecting the molecular mechanism of self and nonself discrimination by MDA5 and RIG-I. He also worked on optimizing IVT to diminish dsRNA byproducts. At the end of 2019, he left Professor Hur’s lab and established his group as an associate professor at Tianjin University. He is now studying regulation of the interferon signaling pathway.
Sun Hur was born on December 30, 1978, in Seoul, Republic of Korea. She received her B.S. degree from Ewha Womans University in Seoul and her Ph.D. degree from the University of California, Santa Barbara, supervised by Professor Thomas C. Bruice. She performed postdoctoral research under the mentorship of Professor Robert M. Stroud lab at the University of California, San Francisco. She joined Harvard Medical School and Boston Children’s Hospital as an assistant professor in 2008, working on the molecular mechanisms of self and nonself discrimination by MDA5 and RIG-I. She is currently the Oscar Schloss Professor at Harvard Medical School and Boston Children’s Hospital. Her current focus is on innate immune regulation and transcriptional regulation of T cell development.
Footnotes
The authors declare no competing financial interest.
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.accounts.1c00521
KEY REFERENCES
- Wu, B.; Peisley, A.; Richards, C.; Yao, H.; Zeng, X.; Lin, C.; Chu, F.; Walz, T.; Hur, S. Structural Basis for dsRNA Recognition, Filament Formation, and Antiviral Signal Activation by MDA5. Cell 2013, 152, 276–289.1 The structure of MDA5:dsRNA complex demonstrates that MDA5 forms filaments along the length of dsRNA and binds to dsRNA in a sequence-independent manner.
- Wu, B.; Peisley, A.; Tetrault, D.; Li, Z.; Egelman, E. H.; Magor, K. E.; Walz, T.; Penczek, P. A.; Hur, S. Molecular Imprinting as a Signal-Activation Mechanism of the Viral RNA Sensor RIG-I. Mol. Cell 2014, 55, 511–523.2 MAVS filament assembly proceeds through its CARD–CARD interaction, which is nucleated by RIG-I 2CARD oligomers, revealing how IFN signaling is transduced from receptor to adaptor.
- Ahmad, S.; Mu, X.; Yang, F.; Greenwald, E.; Park, J. W.; Jacob, E.; Zhang, C.-Z.; Hur, S. Breaching Self-Tolerance to Alu Duplex RNA Underlies MDA5-Mediated Inflammation. Cell 2018, 172, 797–810.3 The aberrant activation of MDA5 in autoimmune disease is elicited by recognition of endogenous Alu retroelements, indicating that the increased efficiency of innate immunity in sensing substrate molecules is accompanied by the danger of self-intolerance.
- Mu, X.; Greenwald, E.; Ahmad, S.; Hur, S. An Origin of the Immunogenicity of In Vitro Transcribed RNA. Nucleic Acids Res. 2018, 46, 5239–5249.4 A previously undiscovered mechanism of dsRNA byproduct formation during IVT was found, implicating the important roles of Mg2+ and modified nucleotides in T7 RNA polymerase activity.
Contributor Information
Xin Mu, School of Pharmaceutical Science and Technology, Tianjin University, Tianjin 300072, China; Tianjin University and Health-Biotech United Group Joint Laboratory of Innovative Drug Development and Translational Medicine, Tianjin University, Tianjin 300072, China.
Sun Hur, Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, Massachusetts 02115, United States; Program in Cellular and Molecular Medicine, Boston Children’s Hospital, Boston, Massachusetts 02115, United States.
REFERENCES
- (1).Wu B; Peisley A; Richards C; Yao H; Zeng X; Lin C; Chu F; Walz T; Hur S Structural Basis for DsRNA Recognition, Filament Formation, and Antiviral Signal Activation by MDA5. Cell 2013, 152, 276–289. [DOI] [PubMed] [Google Scholar]
- (2).Wu B; Peisley A; Tetrault D; Li Z; Egelman EH; Magor KE; Walz T; Penczek PA; Hur S Molecular Imprinting as a Signal-Activation Mechanism of the Viral RNA Sensor RIG-I. Mol. Cell 2014, 55, 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Ahmad S; Mu X; Yang F; Greenwald E; Park JW; Jacob E; Zhang C-Z; Hur S Breaching Self-Tolerance to Alu Duplex RNA Underlies MDA5-Mediated Inflammation. Cell 2018, 172, 797–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Mu X; Greenwald E; Ahmad S; Hur S An Origin of the Immunogenicity of in Vitro Transcribed RNA. Nucleic Acids Res. 2018, 46, 5239–5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Sahin U; Karikó K; Türeci Ö mRNA-Based Therapeutics — Developing a New Class of Drugs. Nat. Rev. Drug Discovery 2014, 13, 759–780. [DOI] [PubMed] [Google Scholar]
- (6).Huang Q; Zeng J; Yan J COVID-19 mRNA Vaccines. J. Genet. Genomics 2021, 48, 107–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Eun H-M Enzymology Primer for Recombinant DNA Technology, 1st ed.; Elsevier, 1996. DOI: 10.1016/B978-0-12-243740-3.X5000-5. [DOI] [Google Scholar]
- (8).Wang W; Li Y; Wang Y; Shi C; Li C; Li Q; Linhardt RJ Bacteriophage T7 Transcription System: An Enabling Tool in Synthetic Biology. Biotechnol. Adv. 2018, 36, 2129–2137. [DOI] [PubMed] [Google Scholar]
- (9).Linares-Fernández S; Lacroix C; Exposito J-Y; Verrier B Tailoring MRNA Vaccine to Balance Innate/Adaptive Immune Response. Trends Mol. Med. 2020, 26, 311–323. [DOI] [PubMed] [Google Scholar]
- (10).De Beuckelaer A; Pollard C; Van Lint S; Roose K; Van Hoecke L; Naessens T; Udhayakumar VK; Smet M; Sanders N; Lienenklaus S; Saelens X; Weiss S; Vanham G; Grooten J; De Koker S Type I Interferons Interfere with the Capacity of MRNA Lipoplex Vaccines to Elicit Cytolytic T Cell Responses. Mol. Ther. 2016, 24, 2012–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Hervas-Stubbs S; Perez-Gracia JL; Rouzaut A; Sanmamed MF; Le Bon A; Melero I Direct Effects of Type I Interferons on Cells of the Immune System. Clin. Cancer Res. 2011, 17, 2619–2627. [DOI] [PubMed] [Google Scholar]
- (12).Liu T; Zhang L; Joo D; Sun S-C NF-κB Signaling in Inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Kretschmer S; Lee-Kirsch MA Type I Interferon-Mediated Autoinflammation and Autoimmunity. Curr. Opin. Immunol. 2017, 49, 96–102. [DOI] [PubMed] [Google Scholar]
- (14).Swiecki M; Colonna M Type I Interferons: Diversity of Sources, Production Pathways and Effects on Immune Responses. Curr. Opin. Virol. 2011, 1, 463–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Tam JCH; Jacques DA Intracellular Immunity: Finding the Enemy within–How Cells Recognize and Respond to Intracellular Pathogens. J. Leukocyte Biol. 2014, 96, 233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Loo Y-M; Gale M, Jr Immune Signaling by RIG-I-like Receptors. Immunity 2011, 34, 680–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Hur S Double-Stranded RNA Sensors and Modulators in Innate Immunity. Annu. Rev. Immunol. 2019, 37, 349–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Peisley A; Jo MH; Lin C; Wu B; Orme-Johnson M; Walz T; Hohng S; Hur S Kinetic Mechanism for Viral DsRNA Length Discrimination by MDA5 Filaments. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, E3340–E3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Peisley A; Wu B; Yao H; Walz T; Hur S RIG-I Forms Signaling-Competent Filaments in an ATP-Dependent, Ubiquitin-Independent Manner. Mol. Cell 2013, 51, 573–583. [DOI] [PubMed] [Google Scholar]
- (20).Sohn J; Hur S Filament Assemblies in Foreign Nucleic Acid Sensors. Curr. Opin. Struct. Biol. 2016, 37, 134–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Cadena C; Ahmad S; Xavier A; Willemsen J; Park S; Park JW; Oh S-W; Fujita T; Hou F; Binder M; Hur S Ubiquitin-Dependent and -Independent Roles of E3 Ligase RIPLET in Innate Immunity. Cell 2019, 177, 1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Kato H; Takeuchi O; Mikamo-Satoh E; Hirai R; Kawai T; Matsushita K; Hiiragi A; Dermody TS; Fujita T; Akira S Length-Dependent Recognition of Double-Stranded Ribonucleic Acids by Retinoic Acid-Inducible Gene-I and Melanoma Differentiation-Associated Gene 5. J. Exp. Med. 2008, 205, 1601–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Schlee M; Roth A; Hornung V; Hagmann CA; Wimmenauer V; Barchet W; Coch C; Janke M; Mihailovic A; Wardle G; Juranek S; Kato H; Kawai T; Poeck H; Fitzgerald KA; Takeuchi O; Akira S; Tuschl T; Latz E; Ludwig J; Hartmann G Recognition of 5′ Triphosphate by RIG-I Helicase Requires Short Blunt Double-Stranded RNA as Contained in Panhandle of Negative-Strand Virus. Immunity 2009, 31, 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Kohlway A; Luo D; Rawling DC; Ding SC; Pyle AM Defining the Functional Determinants for RNA Surveillance by RIG-I. EMBO Rep. 2013, 14, 772–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Devarkar SC; Schweibenz B; Wang C; Marcotrigiano J; Patel SS RIG-I Uses an ATPase-Powered Translocation-Throttling Mechanism for Kinetic Proofreading of RNAs and Oligomerization. Mol. Cell 2018, 72, 355–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Yu Q; Qu K; Modis Y Cryo-EM Structures of MDA5-DsRNA Filaments at Different Stages of ATP Hydrolysis. Mol. Cell 2018, 72, 999–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Wienert B; Shin J; Zelin E; Pestal K; Corn JE In Vitro-Transcribed Guide RNAs Trigger an Innate Immune Response via the RIG-I Pathway. PLoS Biol. 2018, 16, No. e2005840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Durbin AF; Wang C; Marcotrigiano J; Gehrke L RNAs Containing Modified Nucleotides Fail To Trigger RIG-I Conformational Changes for Innate Immune Signaling. mBio 2016, 7, No. e00833–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Blasius AL; Beutler B Intracellular Toll-like Receptors. Immunity 2010, 32, 305–315. [DOI] [PubMed] [Google Scholar]
- (30).Leonard JN; Ghirlando R; Askins J; Bell JK; Margulies DH; Davies DR; Segal DM The TLR3 Signaling Complex Forms by Cooperative Receptor Dimerization. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 258–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Kawai T; Akira S Signaling to NF-κB by Toll-like Receptors. Trends Mol. Med. 2007, 13, 460–469. [DOI] [PubMed] [Google Scholar]
- (32).Liu L; Botos I; Wang Y; Leonard JN; Shiloach J; Segal DM; Davies DR Structural Basis of Toll-Like Receptor 3 Signaling with Double-Stranded RNA. Science 2008, 320, 379–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Zhang Z; Ohto U; Shibata T; Krayukhina E; Taoka M; Yamauchi Y; Tanji H; Isobe T; Uchiyama S; Miyake K; Shimizu T Structural Analysis Reveals That Toll-like Receptor 7 Is a Dual Receptor for Guanosine and Single-Stranded RNA. Immunity 2016, 45, 737–748. [DOI] [PubMed] [Google Scholar]
- (34).Tanji H; Ohto U; Shibata T; Taoka M; Yamauchi Y; Isobe T; Miyake K; Shimizu T Toll-like Receptor 8 Senses Degradation Products of Single-Stranded RNA. Nat. Struct. Mol. Biol. 2015, 22,109–115. [DOI] [PubMed] [Google Scholar]
- (35).Hoshino K; Sugiyama T; Matsumoto M; Tanaka T; Saito M; Hemmi H; Ohara O; Akira S; Kaisho T IκB Kinase-α Is Critical for Interferon-α Production Induced by Toll-like Receptors 7 and 9. Nature 2006, 440, 949–953. [DOI] [PubMed] [Google Scholar]
- (36).Hornung V SnapShot: Nucleic Acid Immune Sensors, Part 2. Immunity 2014, 41, 1066–1066. [DOI] [PubMed] [Google Scholar]
- (37).Hornung V; Barchet W; Schlee M; Hartmann G RNA Recognition via TLR7 and TLR8. Handb. Exp. Pharmacol. 2008, 183, 71–86. [DOI] [PubMed] [Google Scholar]
- (38).Balka KR; De Nardo D Understanding Early TLR Signaling through the Myddosome. J. Leukocyte Biol. 2019, 105, 339–351. [DOI] [PubMed] [Google Scholar]
- (39).Heil F; Hemmi H; Hochrein H; Ampenberger F; Kirschning C; Akira S; Lipford G; Wagner H; Bauer S Species-Specific Recognition of Single-Stranded RNA via Toll-like Receptor 7 and 8. Science 2004, 303, 1526–1529. [DOI] [PubMed] [Google Scholar]
- (40).Diebold SS; Massacrier C; Akira S; Paturel C; Morel Y; ReiseSousa C Nucleic Acid Agonists for Toll-like Receptor 7 Are Defined by the Presence of Uridine Ribonucleotides. Eur. J. Immunol. 2006, 36, 3256–3267. [DOI] [PubMed] [Google Scholar]
- (41).Karikó K; Ni H; Capodici J; Lamphier M; Weissman D MRNA Is an Endogenous Ligand for Toll-like Receptor 3. J. Biol. Chem. 2004, 279, 12542–12550. [DOI] [PubMed] [Google Scholar]
- (42).Karikó K; Buckstein M; Ni H; Weissman D Suppression of RNA Recognition by Toll-like Receptors: The Impact of Nucleoside Modification and the Evolutionary Origin of RNA. Immunity 2005, 23, 165–175. [DOI] [PubMed] [Google Scholar]
- (43).Kormann MSD; Hasenpusch G; Aneja MK; Nica G; Flemmer AW; Herber-Jonat S; Huppmann M; Mays LE; Illenyi M; Schams A; Griese M; Bittmann I; Handgretinger R; Hartl D; Rosenecker J; Rudolph C Expression of Therapeutic Proteins after Delivery of Chemically Modified MRNA in Mice. Nat. Biotechnol. 2011, 29, 154–157. [DOI] [PubMed] [Google Scholar]
- (44).Husain B; Mukerji I; Cole JL Analysis of High-Affinity Binding of Protein Kinase R to Double-Stranded RNA. Biochemistry 2012, 51, 8764–8770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Srivastava SP; Kumar KU; Kaufman RJ Phosphorylation of Eukaryotic Translation Initiation Factor 2 Mediates Apoptosis in Response to Activation of the Double-Stranded RNA-Dependent Protein Kinase. J. Biol. Chem. 1998, 273, 2416–2423. [DOI] [PubMed] [Google Scholar]
- (46).Gil J; Alcamí J; Esteban M Induction of Apoptosis by Double-Stranded-RNA-Dependent Protein Kinase (PKR) Involves the α Subunit of Eukaryotic Translation Initiation Factor 2 and NF-κB. Mol. Cell. Biol. 1999, 19, 4653–4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Gleghorn ML; Maquat LE ‘Black Sheep’ That Don’t Leave the Double-Stranded RNA-Binding Domain Fold. Trends Biochem. Sci. 2014, 39, 328–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Bevilacqua PC; Cech TR Minor-Groove Recognition of Double-Stranded RNA by the Double-Stranded RNA-Binding Domain from the RNA-Activated Protein Kinase PKR. Biochemistry 1996, 35, 9983–9994. [DOI] [PubMed] [Google Scholar]
- (49).Nallagatla SR; Bevilacqua PC Nucleoside Modifications Modulate Activation of the Protein Kinase PKR in an RNA Structure-Specific Manner. RNA 2008, 14, 1201–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Anderson BR; Muramatsu H; Nallagatla SR; Bevilacqua PC; Sansing LH; Weissman D; Karikó K Incorporation of Pseudouridine into MRNA Enhances Translation by Diminishing PKR Activation. Nucleic Acids Res. 2010, 38, 5884–5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Han Y; Donovan J; Rath S; Whitney G; Chitrakar A; Korennykh A Structure of Human RNase L Reveals the Basis for Regulated RNA Decay in the IFN Response. Science 2014, 343, 1244–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Chakrabarti A; Jha BK; Silverman RH New Insights into the Role of RNase L in Innate Immunity. J. Interferon Cytokine Res. 2011, 31, 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Chitrakar A; Rath S; Donovan J; Demarest K; Li Y; Sridhar RR; Weiss SR; Kotenko SV; Wingreen NS; Korennykh A Real-Time 2–5A Kinetics Suggest That Interferons β and λ Evade Global Arrest of Translation by RNase L. Proc. Natl. Acad. Sci. U. S. A. 2019, 116, 2103–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Burke JM; Moon SL; Matheny T; Parker R RNase L Reprograms Translation by Widespread MRNA Turnover Escaped by Antiviral MRNAs. Mol. Cell 2019, 75, 1203–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Rath S; Prangley E; Donovan J; Demarest K; Wingreen NS; Meir Y; Korennykh A Concerted 2–5A-Mediated MRNA Decay and Transcription Reprogram Protein Synthesis in the DsRNA Response. Mol. Cell 2019, 75, 1218–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Sadler AJ; Williams BRG Interferon-Inducible Antiviral Effectors. Nat. Rev. Immunol. 2008, 8, 559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Li Y; Banerjee S; Wang Y; Goldstein SA; Dong B; Gaughan C; Silverman RH; Weiss SR Activation of RNase L Is Dependent on OAS3 Expression during Infection with Diverse Human Viruses. Proc. Natl. Acad. Sci. U. S. A. 2016, 113, 2241–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Anderson BR; Muramatsu H; Jha BK; Silverman RH; Weissman D; Kariko K Nucleoside Modifications in RNA Limit Activation of 2’−5′-Oligoadenylate Synthetase and Increase Resistance to Cleavage by RNase L. Nucleic Acids Res. 2011, 39, 9329–9338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Chauhan SK; Singh VV; Rai R; Rai M; Rai G Distinct Autoantibody Profiles in Systemic Lupus Erythematosus Patients Are Selectively Associated with TLR7 and TLR9 Upregulation. J. Clin. Immunol. 2013, 33 (5), 954–964. [DOI] [PubMed] [Google Scholar]
- (60).Hwang S-H; Lee H; Yamamoto M; Jones LA; Dayalan J; Hopkins R; Zhou XJ; Yarovinsky F; Connolly JE; Curotto de Lafaille MA; Wakeland EK; Fairhurst A-M B Cell TLR7 Expression Drives Anti-RNA Autoantibody Production and Exacerbates Disease in Systemic Lupus Erythematosus-Prone Mice. J. Immunol. 2012, 189, 5786–5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Giltiay NV; Chappell CP; Sun X; Kolhatkar N; Teal TH; Wiedeman AE; Kim J; Tanaka L; Buechler MB; Hamerman JA; Imanishi-Kari T; Clark EA; Elkon KB Overexpression of TLR7 Promotes Cell-Intrinsic Expansion and Autoantibody Production by Transitional T1 B Cells. J. Exp. Med. 2013, 210, 2773–2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Lipes BD; Keene JD Autoimmune Epitopes in Messenger RNA. RNA 2002, 8, 762–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Milligan JF; Groebe DR; Witherell GW; Uhlenbeck OC Oligoribonucleotide Synthesis Using T7 RNA Polymerase and Synthetic DNA Templates. Nucleic Acids Res. 1987, 15, 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Triana-Alonso FJ; Dabrowski M; Wadzack J; Nierhaus KH Self-Coded 3′-Extension of Run-off Transcripts Produces Aberrant Products during in Vitro Transcription with T7 RNA Polymerase. J. Biol. Chem. 1995, 270, 6298–6307. [DOI] [PubMed] [Google Scholar]
- (65).Gholamalipour Y; Karunanayake Mudiyanselage A; Martin CT 3′ End Additions by T7 RNA Polymerase Are RNA Self-Templated, Distributive and Diverse in Character—RNA-Seq Analyses. Nucleic Acids Res. 2018, 46, 9253–9263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Donovan J; Whitney G; Rath S; Korennykh A Structural Mechanism of Sensing Long DsRNA via a Noncatalytic Domain in Human Oligoadenylate Synthetase 3. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 3949–3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Wu MZ; Asahara H; Tzertzinis G; Roy B Synthesis of Low Immunogenicity RNA with High-Temperature in Vitro Transcription. RNA 2020, 26, 345–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Nacheva GA; Berzal-Herranz A Preventing Nondesired RNA-Primed RNA Extension Catalyzed by T7 RNA Polymerase. Eur. J. Biochem. 2003, 270, 1458–1465. [DOI] [PubMed] [Google Scholar]
- (69).Xia H; Jiang Y; Cheng R; Yu B; Lu X; Wu H; Zhu B In Vitro Transcription Using Psychrophilic Phage VSW-3 RNA Polymerase. bioRxiv 2020, DOI: 10.1101/2020.09.14.297226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Moore LD; Le T; Fan G DNA Methylation and Its Basic Function. Neuropsychopharmacology 2013, 38, 23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Zhao L-Y; Song J; Liu Y; Song C-X; Yi C Mapping the Epigenetic Modifications of DNA and RNA. Protein Cell 2020, 11, 792–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Andries O; Mc Cafferty S; De Smedt SC; Weiss R; Sanders NN; Kitada T N1-Methylpseudouridine-Incorporated MRNA Outperforms Pseudouridine-Incorporated MRNA by Providing Enhanced Protein Expression and Reduced Immunogenicity in Mammalian Cell Lines and Mice. J. Controlled Release 2015, 217, 337–344. [DOI] [PubMed] [Google Scholar]
- (73).Tusup M; French LE; Guenova E; Kundig T; Pascolo S Optimizing the Functionality of in Vitro-Transcribed MRNA. Biomed. J. Sci. Technol. Res. 2018, 7, 5845–5850. [Google Scholar]
- (74).Andries O; De Filette M; De Smedt SC; Demeester J; Van Poucke M; Peelman L; Sanders NN Innate Immune Response and Programmed Cell Death Following Carrier-Mediated Delivery of Unmodified MRNA to Respiratory Cells. J. Controlled Release 2013, 167, 157–166. [DOI] [PubMed] [Google Scholar]
- (75).Karikó K; Muramatsu H; Welsh FA; Ludwig J; Kato H; Akira S; Weissman D Incorporation of Pseudouridine into MRNA Yields Superior Nonimmunogenic Vector with Increased Translational Capacity and Biological Stability. Mol. Ther. 2008, 16, 1833–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Nallagatla SR; Jones CN; Ghosh SKB; Sharma SD; Cameron CE; Spremulli LL; Bevilacqua PC Native Tertiary Structure and Nucleoside Modifications Suppress TRNA’s Intrinsic Ability to Activate the Innate Immune Sensor PKR. PLoS One 2013, 8, No. e57905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Svitkin YV; Cheng YM; Chakraborty T; Presnyak V; John M; Sonenberg N N1-Methyl-Pseudouridine in MRNA Enhances Translation through EIF2α-Dependent and Independent Mechanisms by Increasing Ribosome Density. Nucleic Acids Res. 2017, 45, 6023–6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Baiersdörfer M; Boros G; Muramatsu H; Mahiny A; Vlatkovic I; Sahin U; Karikó K A Facile Method for the Removal of DsRNA Contaminant from In Vitro-Transcribed MRNA. Mol. Ther.–Nucleic Acids 2019, 15, 26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Karikó K; Muramatsu H; Keller JM; Weissman D Increased Erythropoiesis in Mice Injected With Submicrogram Quantities of Pseudouridine-Containing MRNA Encoding Erythropoietin. Mol. Ther. 2012, 20, 948–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Pardi N; Secreto AJ; Shan X; Debonera F; Glover J; Yi Y; Muramatsu H; Ni H; Mui BL; Tam YK; Shaheen F; Collman RG; Karikó K; Danet-Desnoyers GA; Madden TD; Hope MJ; Weissman D Administration of Nucleoside-Modified MRNA Encoding Broadly Neutralizing Antibody Protects Humanized Mice from HIV-1 Challenge. Nat. Commun. 2017, 8, 14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Vaidyanathan S; Azizian KT; Haque AKMA; Henderson JM; Hendel A; Shore S; Antony JS; Hogrefe RI; Kormann MSD; Porteus MH; McCaffrey AP Uridine Depletion and Chemical Modification Increase Cas9MRNA Activity and Reduce Immunogenicity without HPLC Purification. Mol. Ther.–Nucleic Acids 2018, 12, 530–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Peisley A; Lin C; Wu B; Orme-Johnson M; Liu M; Walz T; Hur S Cooperative Assembly and Dynamic Disassembly of MDA5 Filaments for Viral DsRNA Recognition. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 21010–21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).McKenna SA; Kim I; Puglisi EV; Lindhout DA; Aitken CE; Marshall RA; Puglisi JD Purification and Characterization of Transcribed RNAs Using Gel Filtration Chromatography. Nat. Protoc. 2007, 2, 3270–3277. [DOI] [PubMed] [Google Scholar]
- (84).Weissman D; Pardi N; Muramatsu H; Karikó K HPLC Purification of In Vitro Transcribed Long RNA. Methods Mol. Biol. 2013, 969, 43–54. [DOI] [PubMed] [Google Scholar]
- (85).Karikó K; Muramatsu H; Ludwig J; Weissman D Generating the Optimal MRNA for Therapy: HPLC Purification Eliminates Immune Activation and Improves Translation of Nucleoside-Modified, Protein-Encoding MRNA. Nucleic Acids Res. 2011, 39, No. e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Nelson J; Sorensen EW; Mintri S; Rabideau AE; Zheng W; Besin G; Khatwani N; Su SV; Miracco EJ; Issa WJ; Hoge S; Stanton MG; Joyal JL Impact of MRNA Chemistry and Manufacturing Process on Innate Immune Activation. Sci. Adv. 2020, 6, No. eaaz6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Day PR Double-Stranded RNA in Endothia Parasitica. Phytopathology 1977, 77, 1393. [Google Scholar]
- (88).Castillo A; Cottet L; Castro M; Sepúlveda F Rapid Isolation of Mycoviral Double-Stranded RNA from Botrytis Cinerea and Saccharomyces Cerevisiae. Virol. J. 2011, 8, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Edwards DK; Jasny E; Yoon H; Horscroft N; Schanen B; Geter T; Fotin-Mleczek M; Petsch B; Wittman V Adjuvant Effects of a Sequence-Engineered MRNA Vaccine: Translational Profiling Demonstrates Similar Human and Murine Innate Response. J. Transl. Med. 2017, 15, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]


