Abstract
The EtfAB components of two bifurcating flavoprotein systems, the crotonyl-CoA-dependent NADH:ferredoxin oxidoreductase from the bacterium Megasphaera elsdenii and the menaquinone-dependent NADH:ferredoxin oxidoreductase from the archaeon Pyrobaculum aerophilum, have been investigated. With both proteins, we find that removal of the electron-transferring flavin adenine dinucleotide (FAD) moiety from both proteins results in an uncrossing of the reduction potentials of the remaining bifurcating FAD; this significantly stabilizes the otherwise very unstable semiquinone state, which accumulates over the course of reductive titrations with sodium dithionite. Furthermore, reduction of both EtfABs depleted of their electron-transferring FAD by NADH was monophasic with a hyperbolic dependence of reaction rate on the concentration of NADH. On the other hand, NADH reduction of the replete proteins containing the electron-transferring FAD was multiphasic, consisting of a fast phase comparable to that seen with the depleted proteins followed by an intermediate phase that involves significant accumulation of FAD⋅−, again reflecting uncrossing of the half-potentials of the bifurcating FAD. This is then followed by a slow phase that represents the slow reduction of the electron-transferring FAD to FADH−, with reduction of the now fully reoxidized bifurcating FAD by a second equivalent of NADH. We suggest that the crossing and uncrossing of the reduction half-potentials of the bifurcating FAD is due to specific conformational changes that have been structurally characterized.
Keywords: electron bifurcation, electron-transferring flavoprotein, rapid-reaction kinetics, electron paramagnetic resonance
Abbreviations: bcd, butyryl-CoA dehydrogenase; BF-FAD, bifurcation FAD; EPR, electron paramagnetic resonance; ETF, electron-transferring flavoprotein; ET-FAD, electron-transferring FAD; FAD, flavin adenine dinucleotide; HQ, hydroquinone; KBr, potassium bromide; PDA, photodiode array; Q, quinine; SQ, semiquinone; TB, terrific broth
Electron-bifurcating flavoproteins are evolutionarily ancient energy-conserving systems that play essential roles in the bioenergetics of many extant anaerobic bacteria and archaea (1, 2, 3, 4). Bifurcation involves the use of a pair of reducing equivalents from a medium-potential donor to deliver individual high- and low-potential electrons to appropriate acceptors in a way that is overall thermodynamically favorable. The low-potential electrons can be used to drive processes such as CO2 and N2 fixation, H2 production, methanogenesis, and acetogenesis, depending on the organism. Several of the presently known bifurcating systems utilize an electron-transferring flavoprotein (ETF) component as the central bifurcating element. These ETFs differ from the long-studied nonbifurcating ETFs from a variety of both vertebrate and microbial organisms in possessing a second equivalent of flavin adenine dinucleotide (FAD) in place of the AMP found in the nonbifurcating proteins. This second FAD, unique to the bifurcating systems, is the site of electron bifurcation (the BF-FAD); the FAD common to both types of ETFs is referred to as the electron-transferring FAD (ET-FAD). Median-potential electron donors for these bifurcating flavoproteins are typically pyridine nucleotides, NADH or NADPH. High-potential acceptors include crotonyl-CoA, caffeyl-CoA, pyruvate, menaquinone, and ubiquinone, whereas ferredoxins and flavodoxins generally serve as the low-potential acceptors (1, 2, 3, 4).
Electron bifurcation takes advantage of the fact that the isoalloxazine ring is capable of both one- and two-electron processes, with an intermediate semiquinone (SQ, as either the neutral FADH⋅ or anionic FAD⋅−) between the oxidized FAD (quinine [Q]) and two-electron reduced FADH2 (hydroquinone [HQ], which ionizes to FADH⋅− at neutral pH). The essential aspect of a bifurcating flavin is that the half-potentials of its Q/SQ and SQ/HQ couples are highly crossed, meaning that the half-potential for the Q/SQ couple is very low and that for the SQ/HQ couple very high, with the result that the SQ oxidation state is thermodynamically extremely unstable. Subsequent to its two-electron reduction by NAD(P)H, the first electron out of the bifurcating flavin leaves with a high potential, leaving behind a very low-potential electron as the thermodynamically unstable SQ that is rapidly transferred to the low-potential acceptor. The two processes are tightly coupled in the course of turnover, with little leakage of the low-potential electron to the high-potential acceptor (1, 5, 6). The central question regarding bifurcation has to do with why this is so, that is, why the low-potential electron generated after the first electron transfer event out of the bifurcating site does not simply follow the first electron to the high-potential acceptor, a process that must be strongly thermodynamically favorable.
By way of identifying the features common to flavin-based bifurcating systems, we have investigated the EtfAB proteins from two ETF-containing complexes. The first is the crotonyl-CoA-dependent NADH:ferredoxin oxidoreductase, termed EtfAB–bcd (for butyryl-CoA dehydrogenase), from the bacterium Megasphaera elsdenii, which, in addition to the BF-FADs and ET-FADs of the EtfAB component has a third equivalent of FAD in its bcd component that is the site of reduction of the high-potential acceptor crotonyl-CoA (7, 8). The low-potential acceptor ferredoxin is reduced directly at the BF-FAD once the low-potential FAD⋅− is generated. The crystal structures of the closely related EtfAB from Acidaminococcus fermentans (8) and the complete EtfAB–bcd complex from Clostridium difficile (9) provide a structural context to understand bifurcation. It is clear from these structures that the domain possessing the ET-FAD has considerable conformational mobility and is capable of rotating some 60° relative to the remainder of the complex to alternately position the flavin close to the BF-FAD or the crotonyl-CoA-reducing FAD (9), a feature particularly relevant to the present work. The second system is the EtfABCX menaquinone-dependent NADH:ferredoxin oxidoreductase from the archaeon Pyrobaculum aerophilum, which has a 2x[4Fe-4S]-containing EtfX and an FAD-containing EtfC subunit in addition to the two FADs of its EtfAB. The FAD of EtfC is the site of menaquinone reduction, once reducing equivalents have been transferred via the ET-FAD and iron–sulfur clusters from the BF-FAD. The 2.9 Å cryoelectron microscopy structure of the closely related EtfABCX from Thermotoga maritima has been determined (10), and it is evident that the domain of the EtfAB component containing the ET-FAD is similarly mobile to the corresponding domain in the bacterial EtfAB systems. These proteins have extensive sequence and structural similarities, yet are from organisms from different domains of life (Bacteria and Archaea, respectively). The intention here is that by comparing and contrasting their behavior it will be possible to identify those characteristics that are most fundamental to bifurcation.
Results
Reductive titrations of EtfAB with NADH and sodium dithionite
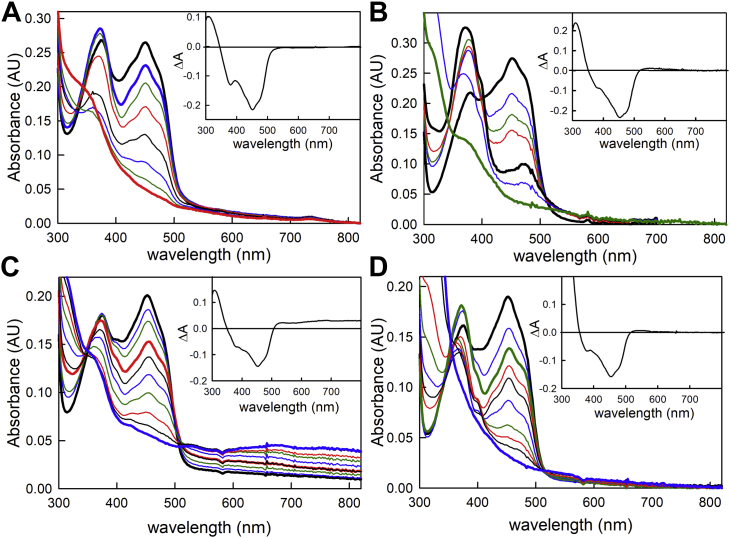
We first undertook reductive titrations of replete M. elsdenii and P. aerophilum EtfAB (i.e., proteins containing both the bifurcating and ET-FADs) with either NADH or dithionite. With NADH, a transient increase in the 360 to 377 nm region is observed with both proteins that is consistent with formation of anionic SQ, FAD⋅− (Fig. 1, A and C); FAD⋅− accumulation is even greater in the course of titrations with sodium dithionite, as shown in Figure 1, B and D, which reflects partial reduction of the ET-FAD. The M. elsdenii protein does not form a long-wavelength absorbing charge–transfer complex between the reduced protein and NAD+, but the P. aerophilum clearly does. These results are consistent with previous reports with both the M. elsdenii (11, 12, 13, 14) and P. aerophilum (15) proteins and underscore the fact that formation of a charge–transfer complex is not critical to the bifurcation process.
Figure 1.
Reductive titrations of the replete EtfAB.A, titration of oxidized (bold black) Megasphaera elsdenii EtfAB with NADH at 25 °C. Bold blue and red spectra correspond to maximum FAD⋅− and reduced spectra, respectively, in the course of the titration. Inset, difference spectrum, reduced minus oxidized. Negative features indicate flavin reduction. B, titration of oxidized (bold black) M. elsdenii EtfAB with sodium dithionite at 25 °C. Lower bold black and green spectra correspond to maximum FAD⋅− and reduced spectra, respectively. Inset, difference spectrum, reduced minus oxidized. Negative features indicate flavin reduction. C, titration of oxidized (bold black) Pyrobaculum aerophilum EtfAB with NADH at 10 °C. Bold red and blue spectra correspond to maximum FAD⋅− and reduced spectra, respectively. In addition, there is an accumulation of a charge–transfer complex between NAD+ and the reduced BF-FAD in the long-wavelength region. Inset, difference spectrum, reduced minus oxidized. Negative and positive features indicate flavin reduction and accumulation of charge–transfer complex, respectively. D, titration of oxidized (bold black) P. aerophilum EtfAB with sodium dithionite at 10 °C. Bold green and blue spectra correspond to maximum FAD⋅− and reduced spectra, respectively. Inset, difference spectrum, reduced minus oxidized. Negative features indicate flavin reduction. BF-FAD, bifurcation FAD; ETF, electron-transferring flavoprotein; FAD, flavin adenine dinucleotide.
We next examined the behavior of the two proteins that have been depleted of the ET-FAD by incubation with potassium bromide (KBr). Neither protein exhibited any FAD⋅− in the course of titration with NADH, as shown in Figure 2, A and C, which is to be expected since reaction of NADH with the remaining BF-FAD is expected to proceed directly to the fully reduced flavin HQ. As with the replete proteins, the P. aerophilum but not the M. elsdenii protein exhibits strong charge–transfer absorbance at longer wavelengths. Surprisingly, and contrary to expectation if the half-potentials of the BF-FAD are highly crossed, there is substantial accumulation of FAD⋅− at intermediate points in the course of titrations of both depleted proteins with sodium dithionite (Fig. 2, B and D). Based on the extinction coefficient for the FAD⋅− state in the M. elsdenii system of 17 mM−1 cm−1 at 365 nm (16), the amount of FAD⋅− accumulation at maximum in the course of the titration is approximately 80% (the remainder of the protein presumably having been fully reduced on to the HQ). This behavior has been reported previously with the M. elsdenii EtfAB and is seen here to be a property in common with the P. aerophilum protein. It is indicative of the uncrossing of the half-potentials of the BF-FAD. It is noteworthy that, no evidence for formation of sulfite adducts (17, 18) is found upon incubation of either EtfAB with 100 mM sulfite (data not shown).
Figure 2.
Reductive titrations of EtfAB depleted of the ET-FAD.A, titration of oxidized (bold black) Megasphaera elsdenii EtfAB with NADH at 25 °C. Bold red spectrum corresponds to the reduced spectrum. Inset, difference spectrum, reduced minus oxidized. Negative features indicate flavin reduction. B, titration of oxidized (bold black) M. elsdenii EtfAB reduced with sodium dithionite at 25 °C. Upper bold green spectrum corresponds to maximum accumulation of FAD⋅−. Inset, difference spectrum, reduced minus oxidized. Negative features indicate flavin reduction. C, titration of oxidized (bold black) Pyrobaculum aerophilum EtfAB with NADH at 10 °C. Bold blue spectrum corresponds to the reduced spectrum. In addition, accumulation of the charge–transfer complex between NAD+ and the reduced BF-FAD can be seen in the long-wavelength region. Inset, difference spectrum, reduced minus oxidized. Negative and positive features indicate flavin reduction and accumulation of charge–transfer complex, respectively. D, titration of oxidized (bold black) P. aerophilum EtfAB with sodium dithionite at 10 °C. Bold red spectrum corresponds to maximum accumulation of FAD⋅−. Inset, difference spectrum, reduced minus oxidized. Negative and positive features indicate flavin reduction and accumulation of FAD⋅−, respectively. BF-FAD, bifurcation FAD; ETF, electron-transferring flavoprotein; ET-FAD, electron-transferring FAD; FAD, flavin adenine dinucleotide.
Kinetics of reduction of EtfAB with NADH
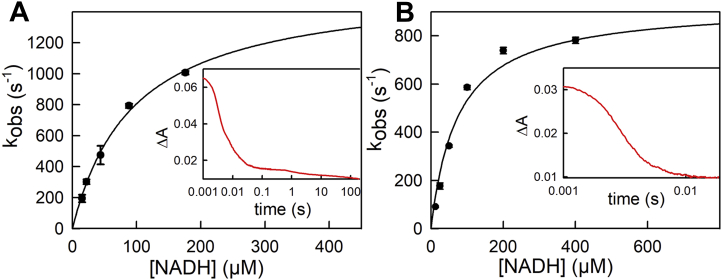
We next examined the reductive half-reaction of the two EtfABs by stopped-flow spectrophotometry, beginning with the depleted forms that had only the BF-FAD so as to examine the intrinsic kinetics of reduction of the BF-FAD without the complication of subsequent electron transfer events to the ET-FAD. Following the reaction at 450 nm, ∼90% of the total absorbance change seen with the M. elsdenii protein (Fig. 3A) and essentially 100% of the change seen with the P. aerophilum protein (Fig. 3B) occurs in a single phase, with a rate constant that is hyperbolically dependent on [NADH]. Hyperbolic fits to plots of kobs versus [NADH] yield a Kd of 96 μM and limiting kred of 1600 s−1 for the M. elsdenii protein at 5 °C and 75 μM and 930 s−1, respectively, for the P. aerophilum protein at 25 °C. We note that this value is obtained by extrapolation in the plot of rate constant versus [NADH] to infinite [NADH] and that the fastest experimentally determined rate constant is 1000 s−1. This remains at the limit of modern stopped-flow devices, but as we show in Fig. S1, there remains sufficient observed absorbance change at the highest NADH concentrations used to reliably determine that rate constant for reduction. That the transients seen at progressively lower [NADH] extrapolate back to the known initial absorbance of the oxidized enzyme used indicates that the only dead-time spectral changes in the instrument are due to the observed kinetic phase and that there is no faster phase taking place.
Figure 3.
Reduction of depleted EtfAB by NADH.A, reaction of 7 μM depleted Megasphaera elsdenii EtfAB with increasing NADH concentrations monitored at 450 nm at 5 °C. The Kd and limiting kred from hyperbolic fits to the data are 96 μM and 1600 s−1, respectively. The inset shows a representative transient out to 200 s. B, reaction of 4 μM depleted Pyrobaculum aerophilum EtfAB with increasing NADH concentrations monitored at 450 nm at 25 °C. Kd and limiting kred are 75 μM and 930 s−1, respectively. The inset shows a representative transient out to 50 ms. ETF, electron-transferring flavoprotein.
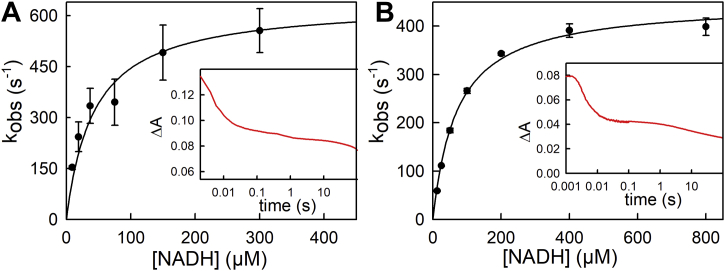
Having thus characterized the rate of reduction of the BF-FAD alone, the reaction of the replete EtfABs with NADH was next examined. Unfortunately, following any transient accumulation of FAD⋅− in the 360 to 377 nm region, as seen in the equilibrium titrations, was not possible owing to the strong absorbance of NADH (which was always present in pseudo–first order excess). As expected, however, the transients seen at 450 nm were considerably more complex than those seen with the depleted systems. With the M. elsdenii protein, three phases are apparent: a very fast phase that is complete within 20 ms; an intermediate phase that extends out to 1 s; and a slow phase that occurs on the 100 s timescale. The fastest phase is [NADH]-dependent and accounts for approximately half of the total absorbance change at 450 nm. This is attributed to the initial reduction of the BF-FAD, with a Kd of 30 μM and limiting kred of 590 s−1 at 5 °C (Fig. 4A). The intermediate phase accounts for only ∼15% of the total absorbance change at 450 nm but as shown below correlates with accumulation of a significant amount of FAD⋅−, as discussed further. The slowest phase of the reaction results in full reduction of the protein, again consistent with the equilibrium titration with NADH. With the P. aerophilum protein, only two kinetic phases are readily distinguished: a [NADH]-dependent fast phase that accounts for over half of the total spectral change and is complete within 20 ms; and a much slower (and possibly multiphasic) process that occurs over the next 10 to 100 s (Fig. 4B). Again, the total absorbance change observed is consistent with eventual full reduction of the protein and consistent with the results of equilibrium titration. A hyperbolic fit of a plot of kobs versus [NADH] yielded Kd of 71 μM and limiting kred of 450 s−1 at 25 °C (Fig. 4B); as with the M. elsdenii protein, this fastest phase is attributed to the initial reduction of the BF-FAD by NADH, with the subsequent process(es) leading to full reduction of the protein proceeding with rate constants too close to be resolved.
Figure 4.
Reduction of replete EtfAB with NADH.A, reaction of 7 μM replete Megasphaera elsdenii EtfAB with increasing NADH concentrations monitored at 450 nm at 10 °C. The Kd and limiting kred from hyperbolic fits to the data are 30 μM and 590 s−1, respectively. The inset shows a representative transient out to 100 s. B, reaction of 4 μM replete Pyrobaculum aerophilum EtfAB with increasing NADH concentrations monitored at 450 nm at 25 °C. Kd and limiting kred from hyperbolic fits to the data are 71 μM and 450 s−1, respectively. The inset shows a representative transient out to 100 s. ETF, electron-transferring flavoprotein.
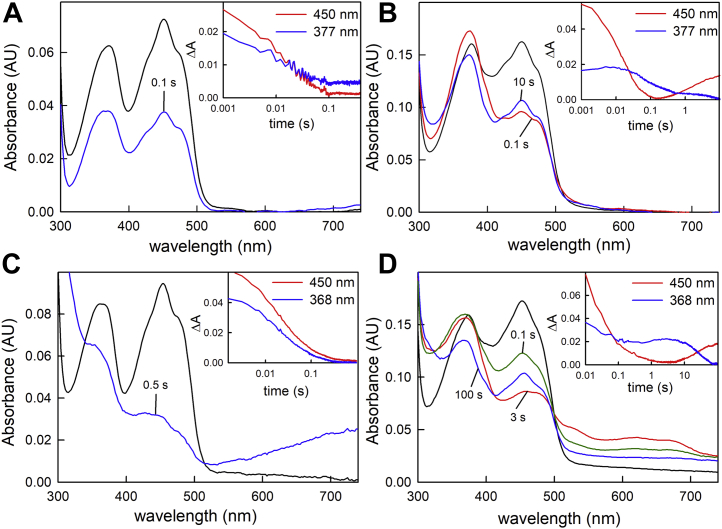
In order to follow absorbance changes that might be associated with FAD⋅− accumulation in the course of the reaction, it was necessary to perform the stopped-flow kinetics at greatly reduced concentrations of NADH. While the reaction will not be exponential under conditions that are not pseudo–first order, it is nevertheless possible to examine the time scale on which various processes occur. Figure 5 shows the results of experiments in which either M. elsdenii (panels A and B) or P. aerophilum (panels C and D) EtfAB is reacted with one equivalent of NADH. With the proteins depleted of the electron-transferring FAD (panels A and C), the reactions are monophasic and take place on the 10 to 100 ms timescale, with no accumulation of SQ. With the replete proteins (panels B and D), the kinetics are again substantially more complex. As with the depleted protein, the fast phase of the transient observed at 450 nm for M. elsdenii occurs on the same 10 to 100 ms time; for the P. aerophilum protein, an additional intermediate phase is observed that is complete by 3 s; this phase likely reflects the higher apparent Kd for the P. aerophilum protein relative to the M. elsdenii protein (compare Fig. 4, A and B). The transients seen at 377 and 368 nm (for M. elsdenii and P. aerophilum, respectively) exhibit absorbance increases on comparable timescales to reduction at 450 nm because of the accumulation of FAD⋅− (in the case of the P. aerophilum protein also yielding maximum charge–transfer complex of NAD+ with the reduced BF-FAD). At the end of the fast phase, the one equivalent of NADH is consumed. On a longer timescale, both proteins exhibit a decrease in the absorption because of FAD⋅− as judged by the decrease of absorbance at 368 or 377 nm; this is accompanied by an increase in absorbance at 450 nm. In the case of the M. elsdenii protein, this is likely a consequence of the intramolecular electron transfer between a pair of SQs in a single protomer ultimately yielding oxidized bifurcating flavin and reduced electron-transferring flavin (intermolecular electron transfer occurs on a much longer timescale with the M. elsdenii protein; Fig. 6B). This intramolecular electron transfer necessarily entails the transfer of the (nominally) low-potential electron into the high-potential pathway (which must be slow for successful bifurcation). For the P. aerophilum protein, the last phase is much slower and likely entails intermolecular as well as intramolecular electron transfer (Fig. 6D). The final spectrum seen with the P. aerophilum protein also reflects partial decay of the charge–transfer complex consistent with this interpretation.
Figure 5.
The reaction of EtfAB with stoichiometric concentrations of NADH.A, stopped-flow spectra of 7.5 μM of depleted Megasphaera elsdenii EtfAB reacted with 7.5 μM of NADH at 10 °C show that in the pre-steady state there is no accumulation of FAD⋅− seen in the first 200 ms of the reaction with the oxidized spectrum shown in black and 100 ms spectrum in blue. Inset, the 450 and 377 nm time courses extracted from the full-wavelength spectra out to 200 ms. B, stopped-flow spectra of 3 μM replete M. elsdenii EtfAB reacted with 3 μM of NADH at 10 °C shows that in the pre-steady state there is rapid accumulation of FAD⋅− seen in the first 200 ms of the reaction with the oxidized spectrum shown in black and 100 ms spectrum in red. Blue spectrum indicates loss of FAD⋅− and reoxidation of the bifurcating flavin at 10 s. Inset, the 450 and 377 nm time courses extracted from the full-wavelength spectra out to 10 s. C, stopped-flow spectra of 8 μM of depleted Pyrobaculum aerophilum EtfAB with 8 μM NADH at 25 °C. Oxidized spectrum is shown in black and 500 ms spectrum in blue. Inset, the 450 and 368 nm time courses extracted from the full-wavelength spectra out to 200 ms. D, stopped-flow spectra of 8 μM of replete EtfAB with 8 μM of NADH at 25 °C with oxidized (black), 0.1 s (green), 3 s (red), and 100 s (blue) spectra shown. Inset, the 450 and 368 nm time courses extracted from the full-wavelength spectra out to 100 s. ETF, electron-transferring flavoprotein; FAD, flavin adenine dinucleotide.
Figure 6.
Intermolecular electron transfer of depleted and replete EtfAB. From Megasphaera elsdenii (A and B) and Pyrobaculum aerophilum (C and D). A, spectra of 9 μM of depleted M. elsdenii EtfAB reacted with 4.5 μM of NADH at 25 °C. The spectra shown are oxidized (black), after mixing (green), and after 4 h (blue). Inset, difference spectrum, 4 h minus after mixing. Positive feature in the 377 nm region corresponds to accumulation of FAD⋅−. B, spectra seen in the course of reaction of 7.5 μM of replete M. elsdenii EtfAB, pre-reduced with sodium dithionite, with 7.5 μM of oxidized protein at 25 °C. The spectra shown are after mixing (black) and after 500 s (blue). Inset, the increases in absorption at 377 nm (blue) and 450 nm (red) over the course of 500 s. C, spectra of 13 μM of P. aerophilum depleted EtfAB reacted with 6.5 μM of NADH at 25 °C. The spectra shown are oxidized (black), 5 min after mixing (green), and after 3 h (blue). Inset, difference spectrum, 3 h minus 5 min after mixing. D, spectra seen in the course of reaction of 4 μM of replete P. aerophilum EtfAB, pre-reduced with sodium dithionite, with 4 μM of oxidized protein at 25 °C. The spectra shown are after mixing (black) and after 60 s (blue). Inset, the increases in absorption at 368 nm (blue) and 450 nm (red) over the course of 120 s. ETF, electron-transferring flavoprotein; FAD, flavin adenine dinucleotide.
The accumulation of FAD⋅− on intermediate timescales in the reaction of the EtfAB component of the A. fermentans EtfAB–bcd system, highly homologous to that from M. elsdenii studied here, has been observed previously (19) and attributed to intermolecular electron transfer from two-electron reduced protein to oxidized protein to yield two equivalents of one-electron reduced enzyme in which the FAD⋅− would arise from the ET-FAD, which is known to form a stable anionic SQ. The timescale on which FAD⋅− accumulates in both our and the previous study would require that such an intermolecular electron transfer process would have to occur at a rate near the diffusion-controlled limit. In order to examine rates of intermolecular electron transfer, two different experiments were performed. In the first, EtfAB (from both organisms, depleted of the ET-FAD) was reacted with a substoichiometric concentration of NADH. As shown in Figure 6, intermolecular electron transfer with the depleted occurs over the course of hours for the M. elsdenii (panel A) or not at all in the case of the P. aerophilum (panel C). In a second experiment, fully reduced replete EtfAB was reacted with an equal concentration of fully oxidized protein, and the kinetics were followed by stopped-flow spectrophotometry. The ET-FAD is known to have a high Q/SQ half-potential (13), and the expectation is that eventually the system must equilibrate via intermolecular electron transfer to yield the FAD⋅− of the ET-FAD. In this case, intermolecular electron transfer involving either flavin could be followed. As shown in the insets to Figure 6, B and D, intermolecular electron transfer, as reflected in formation of FAD⋅−, occurs on a timescale of 100 s with proteins from both sources.
Electron paramagnetic resonance confirmation of FAD⋅− formation
In order to confirm the formation of FAD⋅−, electron paramagnetic resonance (EPR) spectra were obtained under various conditions. First, EtfABs depleted of their ET-FAD were titrated with dithionite to maximum absorbance at 377 nm or 368 nm and the EPR recorded. As shown in Figure 7, A and C, strong SQ signals were observed with both the M. elsdenii and P. aerophilum proteins when compared against a standard sample of 20 μM flavodoxin. The integrated intensity of the signals amounted to 0.6 spins per BF-FAD in the case of the depleted M. elsdenii EtfAB and 0.5 spins per BF-FAD in the case of the depleted P. aerophilum EtfAB. The replete EtfABs were reduced with either stoichiometric or excess of NADH, with the EPR sample frozen approximately 1 s after addition of substrate. The results are most clear-cut with the P. aerophilum protein, where significant amounts of anionic SQ signal are observed (Fig. 7D, lower spectra). In the case of the M. elsdenii protein (Fig. 7B, lower spectra), reduction of the replete protein with NADH does result in substantial FAD⋅− formation, although to a lesser extent than is seen with the P. aerophilum protein. The EPR signals in Figure 7B integrate to ∼0.2 spins per replete EtfAB. When NADH was mixed with the depleted EtfAB from either source, there was essentially no FAD⋅− signal observed in the first second of turnover, consistent with the lack of any increase in absorbance of ∼370 nm in the course of the reaction.
Figure 7.
X-band EPR spectra of the anionic semiquinone, FAD⋅−.A, spectra of depleted Megasphaera elsdenii EtfAB (upper, black) and replete EtfAB (lower), both titrated to maximum accumulation of semiquinone, as well as the spectrum of flavodoxin from Azotobacer vinelandii used as the quantification standard (upper, blue). The integrated intensity amounted to ∼0.6 spins/BF-FAD with the depleted EtfAB and 0.4 spins per replete EtfAB. B, spectra of depleted M. elsdenii EtfAB reacted with 30× excess of NADH (upper) and replete EtfAB reacted with stoichiometric amount (lower, black) or 30× excess of NADH (lower, red); the integrated intensity with the replete EtfAB amounted to ∼0.2 spins/EtfAB. C, spectra of depleted Pyrobaculum aerophilum EtfAB (upper, black) and replete EtfAB (lower), both titrated to maximum accumulation of semiquinone. The integrated signal intensity amounted to ~0.5 spins/BF-FAD with the depleted protein and ∼0.3 spins/EtfAB with the replete protein. D, spectra of depleted P. aerophilum EtfAB reacted with 25× excess of NADH (upper) and replete EtfAB reacted with stoichiometric amount (lower, black) or 25× excess of NADH (lower, red); the integrated signal intensities amounted to ∼0.4 spins/EtfAB for the red spectrum and ∼0.2 spins/EtfAB for the lowerblack spectrum. All spectra were collected at 150 K with 10 μW microwave power and 4 G modulation amplitude. All spectra are scaled to reflect the same protomer concentration, except for the lower spectra in (B), which were enlarged four-fold for clarity. BF-FAD, bifurcation FAD; EPR, electron paramagnetic resonance; ETF, electron-transferring flavoprotein; FAD, flavin adenine dinucleotide.
That any FAD⋅− is generated within ∼1 s upon reduction of both EtfABs with NADH is significant. With intermolecular electron transfer shown to be slow, as described previously, the only way for SQ to be formed at all in the two-electron reduced protein generated after reaction with a first equivalent of NADH is by transfer of a single electron from the now reduced BF-FAD to the ET-FAD, meaning that both flavins in the replete protein must be present as FAD⋅− in equal amounts. This in turn implies that, to the extent that FAD⋅− accumulates, the half-potentials of the BF-FAD must have become uncrossed. As is shown in Figure 6, the time for intermolecular electron transfer to occur is much too slow to result in the amount of FAD⋅− measured during the first 1 s of turnover. This is further supported by the lack of any significant FAD⋅− seen on the same timescale with the EtfAB depleted of the electron-transferring FAD. Our results indicate that this occurs in approximately 20% of the two-electron reduced M. elsdenii protein and 40% of the P. aerophilum protein. Thus, as with the EtfABs depleted of the ET-FAD, it appears that uncrossing of the half-potentials of the BF-FAD also occurs in the replete M. elsdenii protein, although to a lesser extent than is seen in the depleted proteins.†
Discussion
Although the rate constants for specific processes can be quite different in the two EtfAB systems investigated here, and while only the P. aerophilum protein forms a charge–transfer complex with pyridine nucleotide in the course of the reductive half-reaction and the M. elsdenii protein does not, the behavior of the two systems is otherwise quite similar. The ET-FADs of both proteins is easily removed, and the reduction of the remaining BF-FADs in the depleted proteins exhibits a hyperbolic dependence of the observed rate constant on [NADH]. As seen previously in both the M. elsdenii (11) and A. fermentans (18) systems, we see no indication of SQ formation in the course of the kinetics of reduction by NADH or in titrations with NADH with the depleted proteins (as expected, given that NADH obligatorily reduces the remaining BF-FAD directly to the HQ in a two-electron process). It is important to recognize that the absence of SQ formation in these experiments does not mean that the BF-FAD half-potentials have remained highly crossed in the depleted proteins, only that its midpoint potential remains sufficiently high that it is effectively reduced by NADH directly to the HQ. On the other hand, titration of the depleted ETFs with sodium dithionite, a one-electron donor, results in significant accumulation of FAD⋅−, as reflected in both the UV–visible absorption spectra and observation of the expected EPR signals. The stable accumulation of FAD⋅− on removal of the ET-FAD in both proteins must reflect that the half-potentials of the BF-FAD have become uncrossed, and significantly so. The similarities seen in the two systems examined here, one from a bacterium that grows optimally (Topt) near 37 °C, the other from an archaeon with Topt ∼100 °C, are significant and appear to reflect a fundamentally common and evolutionarily ancient mechanism.
Uncrossing also appears to occur in the replete systems as well, as reflected in the transient absorption increases at ca. 377 nm (M. elsdenii) and 368 nm (P. aerophilum) seen on the tens of milliseconds to seconds timescale in the course of reduction with NADH, again reflecting formation of FAD⋅−. This transient accumulation of FAD⋅− has previously been observed with the EtfAB from A. fermentans (19) but was attributed solely to one-electron reduction of the ET-FAD subsequent to intermolecular electron transfer from NADH-reduced protein. Given the protein concentrations that were used, it was recognized that the observed rates of FAD⋅− formation required that this (nonphysiological) intermolecular electron transfer must occur at a bimolecular rate near the diffusion-controlled limit. In our hands, intermolecular electron transfer as determined in two different types of experiments is, in fact, extremely slow in both systems, occurring much more slowly than the transient accumulation of FAD⋅− in the reaction with NADH. Given that NADH introduces reducing equivalents in pairs, the only partially reduced form that accumulates in the course of the reaction with NADH is the two-electron reduced EtfAB2e− (one-electron and three-electron reduced forms being formed only as a result of intermolecular electron transfer). The possible distributions of the two reducing equivalents within EtfAB2e− are:
meaning that to the extent that one of the FADs is present as FAD⋅−, the other must be as well. Furthermore, for this distribution to be thermodynamically stable, the half-potentials for the BF-FAD must have again become uncrossed, this in the replete proteins. On the basis of quantification of the SQ, it appears that during the course of reduction of the replete protein, only some 20% of the M. elsdenii EtfAB and 40% of the P. aerophilum protein exist in the two-electron reduced form with two SQs, meaning that uncrossing is incomplete and suggesting that an equilibrium exists between conformational states in which the half-potentials of the BF-FAD are either crossed or uncrossed.
There is, in fact, evidence in the literature that the half-potentials of the BF-FAD are not always highly crossed in a bifurcating EtfAB. Whitfield and Mayhew (12), for example, showed that the FADs of EtfAB from M. elsdenii (not recognized at the time as being part of a bifurcating system) could be quantitatively converted to FAD⋅− upon photoreduction and observed a transient increase in absorbance at ∼377 nm in the course of titrations with NADH that reflected accumulation of the SQ state. Previous work with the P. aerophilum EtfAB by Schut et al. (15) has also indicated the transient formation of anionic SQ in the course of titration of the replete EtfAB and EtfABCX with NADH. The FAD⋅− that was observed was suggested to be due to the ET-FAD, generated by intermolecular electron transfer. It was also shown that Ti(III)citrate reduction of both EtfAB and EtfABCX gave rise to a stable FAD⋅− signal, as reflected in the absorbance increase at ∼370 nm (15). Based on the work presented here, this can now be attributed to an uncrossed BF-FAD, and it is clear that both the BF-FAD− and ET-FADs contribute to the transient SQ formation. It has not previously been suggested that the half-potentials of a BF-FAD might uncross under specific circumstances.
The kinetics of NADH reduction of both replete EtfABs examined here are similarly complex. An initial fast phase of the reaction is [NADH]-dependent and generally similar to the reduction of the depleted protein and is attributed to the initial reduction of the BF-FAD by NADH. This is followed by the aforementioned intermediate phase that reflects accumulation of FAD⋅− in the intermediate EtfAB2e− state that accumulates in the course of the reaction. As discussed further, we attribute this to a reorientation of the domain containing the ET-FAD that results in uncrossing of the half-potentials of the BF-FAD. This is followed by an extremely slow phase of the reduction by NADH that ultimately results in full reduction to EtfAB4e−. This requires that the BF-FAD become fully reoxidized, meaning that the second nominally high-potential electron must pass to the ET-FAD (i.e., “leak” into the high-potential pathway in a nonphysiological process that defeats bifurcation), followed by rapid re-reduction of the BF-FAD by a second equivalent of NADH.
We note that several aspects of the kinetics of the M. elsdenii and P. aerophilum EtfABs seen here differ significantly from what has been reported for the A. fermentans EtfAB. Under conditions similar to ours (although we note that we have worked at 10 °C with the M. elsdenii EtfAB, and 25 °C with the P aerophilum protein, Sucharitakul et al. (19) worked at 5 °C), the initial reduction was reported to be too fast to be observed, and the fastest process that could be seen was not [NADH]-dependent. As discussed previously, the slower accumulation of FAD⋅− was observed but attributed to accumulation of reducing equivalents on the ET-FAD subsequent to intermolecular electron transfer. Again, our results indicate that intermolecular electron transfer is very slow, meaning that the transient accumulation of FAD⋅− on the seconds timescale upon reduction with NADH must be intramolecular and entail uncrossing of the half-potentials of the BF-FAD.
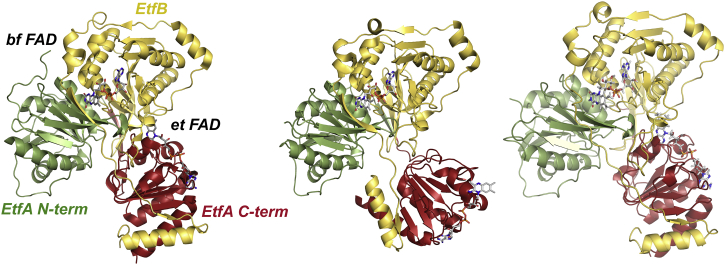
Some comment as to the structural basis for the uncrossing of the half-potentials of the BF-FAD seen here is appropriate. As illustrated in Figure 8, a comparison of the X-ray crystal structures of the A. fermentans EtfAB subcomplex and the EtfAB–bcd holoenzyme from C. difficile provides clear evidence that the domain containing the ET-FAD is quite mobile. The recent cryoelectron microscopy of the EtfABCX complex from T. maritima (homologous to the P. aerophilum system) suggests that the domain with the ET-FAD is similarly mobile in this system. Such mobility has, in fact, also been observed in nonbifurcating ETFs (20) and appears to be a general property of these systems. Significantly, the C terminus of the BF-FAD–containing EtfB subunit of the bacterial systems (which, somewhat confusingly, corresponds to the EtfA subunit of the archaeal system) loops into the C-terminal domain of EtfA that contains the ET-FAD. As illustrated in Figure 8, this loop extends into the final strand of β-sheet in the body of EtfB that crosses directly over the dimethylbenzene ring of the BF-FAD. Motion of the ET-FAD–containing domain could thus easily alter the environment of the BF-FAD to the point that its half-potentials become uncrossed by altering the orientation of this loop region. Removal of the ET-FAD could similarly perturb the environment of the BF-FAD. What seems most plausible is that the configuration seen in the C. difficile structure, with the ET-FAD oriented away from the BF-FAD, is that in which the half-potentials of the BF-FAD have become uncrossed, whereas that seen in the A. fermentans structure, with the ET-FAD in close proximity to the BF-FAD, has crossed potentials. Site-directed mutagenesis studies targeting the loop region of EtfB are presently under way to test this hypothesis.
Figure 8.
The structures of ETFs in different orientations.Left, the EtfAB component seen in the EtfAB–bcd complex from Clostridium difficile (Protein Data Bank [PDB]: 5OL2), with the ET-FAD orientated away from the BF-FAD. Center, the structure of isolated EtfAB from Acidaminococcus fermentans (PDB: 4KPU), with the ET-FAD in close proximity to the BF-FAD. Right, the EtfAB component from the cryo-EM structure of Thermotoga maritima EtfABCX (PDB: 7KOE), with the ET-FAD oriented similarly to that seen in the A. fermentans structure. For the C. difficile and A. fermentans proteins, the N terminus of EtfA is in green and its C terminus in red; EtfB is in yellow. Note that because of differences in operon organization, EtfA of T. maritima (and of P. aerophilum) corresponds to EtfB of the two bacterial systems, and EtfB to EtfA. BF-FAD, bifurcation FAD; ETF, electron-transferring flavoprotein; ET-FAD, electron-transferring FAD; FAD, flavin adenine dinucleotide.
Experimental procedures
Organisms and growth conditions
Plasmids containing the M. elsdenii EtfAB genes (MELS_2126 and MELS_2127) were constructed with a C-terminal Strep II tag derived from pASG-IBA3 (IBA GmbH) as previously described by Chowdhury et al. (5). The plasmids were transformed into chemically competent Escherichia coli BL21 cells and grown as previously described (1) except that the growth media was changed to terrific broth (TB) for both preculture and growth. Precultures were grown in 10 ml of TB with 100 μl/ml of ampicillin overnight with the shaker set to 30 °C and 220 rpm. For growth, 100 μg/μl of ampicillin was added to 2 l of TB in a 6 l Erlenmeyer flask and was then inoculated with the preculture and grown to a final absorbance of 0.0625 at 600 nm. This was grown for an additional ∼6 h at 37 °C with shaking at 180 rpm until an absorbance of 0.5 at 600 nm was reached, at which point the temperature was reduced to 21 °C, and growth was continued until an absorbance of 0.8 at 600 nm was reached. Cells were then induced with 0.1 mM anhydrotetracycline and grown for an additional 24 h. Cells were harvested by centrifugation for 20 min at 5500g at 4 °C, frozen in liquid nitrogen, and stored at −80 °C until lysed (yields were typically ∼11 g cells/l of media). The plasmid containing the P. aerophilum EtfABCX genes and growth of the recombinant Pyrococcus furiosus have been described previously by Schut et al. (15).
Purification of M. elsdenii EtfAB
All steps were performed at 0 to 4 °C and under aerobic conditions unless otherwise stated, using a procedure modified from Chowdhury et al. (5) as described by Vigil et al. (21). Frozen cells (∼22 g wet weight) were thawed and suspended in 0.5 to 1 g/ml 50 mM Tris–HCl, 100 mM NaCl, pH 8.0 (buffer A1); this suspension was then supplemented with 1 mM NaF (empirically noted to be sufficient for prevention of proteolysis in this expression system), 1 mM benzamidine, 0.5 mM PMSF, and catalytic amounts of lysozyme and DNase I. After a 1 h incubation period, cells were disrupted by one to two passages through a French pressure cell at 20,000 psi. Ultracentrifugation was performed to remove cell debris at 42,000g for 60 min, and supernatant not immediately used was frozen in liquid nitrogen and stored at −80 °C until needed. For protein purification, a gravity column containing 6 ml of Strep-Tactin XT Superflow resin (IBA GmbH) was first equilibrated with five column volumes of buffer A1, and the supernatant loaded at a rate of 0.5 ml/min. Flow-through spectra were taken after every column volume to monitor binding of protein, and the absorbance at 450 nm was continuously monitored. After loading the cell lysate, buffer A1 was used to wash the column at a rate of 1 ml/min until absorbance at 280 nm was less than 0.1 (typically achieved after 30 column volumes). Elution was performed with 50 mM biotin in 50 mM Hepes (pH 7.5) and was typically completed after four column volumes. During purification, it was noted that some FAD dissociates from the protein. In fact, as-isolated EtfAB has been shown to be partially depleted of the ET-FAD (13). In order to maximize retention of the ET-FAD, as the protein is buffer exchanged into 50 mM Tris–HCl, 150 mM NaCl, and pH 7.5 (buffer A), 5× molar excess–free FAD is added during each round of concentration using an Amicon Ultra 4 (Millipore). Buffer-exchanged protein was then incubated in 5× molar excess FAD for ∼8 h in the dark. After incubation, the excess FAD was removed using an 8.3 ml PD-10 (GE Healthcare) column equilibrated in buffer A. Protein treated in this manner was typically ∼95% replete in its ET-FAD, as confirmed by SDS denaturation. This was reflected in the final absorption spectrum of the protein. Finally, the purified protein was aliquoted, flash frozen, and stored at −80 °C prior to use.
Purification of P. aerophilum EtfAB
Purification of P. aerophilum EtfAB was carried out as previously described by Schut et al. (15). In short, P. aerophilum EtfABCX was recombinantly expressed in P. furiosus with a histidine affinity tag on the EtfA subunit. EtfAB and EtfABCX were purified under anaerobic conditions by affinity chromatography followed by size exclusion. EtfAB was the main form obtained with little (<10%) of the full-length EtfABCX. Protein used in quantification experiments was approximately 74% replete in the electron-transferring flavin, as confirmed by SDS denaturation.
Preparation of EtfAB depleted of its ET-FAD
As discussed previously, the electron-transferring FAD of EtfAB was prone to dissociate. In order to fully remove the ET-FAD, protein was incubated and then washed with a chaotropic salt (KBr) similar to the method described by Sato et al. (13) but without the (NH4)2SO4 wash. For the M. elsdenii protein, samples were concentrated to minimal volume in 50 mM K-PO4, pH 6.0 (buffer A2), and then incubated in 50 mM K-PO4, pH 6.0, and 2 M KBr (buffer B2) for 2 h. The protein was then concentrated to minimal volume and washed with alternating buffers A2 and B2 until no more FAD was detectable in the flowthrough (this was typically achieved after four cycles). The protein (fully depleted of the ET-FAD) was exchanged into buffer A, aliquoted, and stored as described previously. For the P. aerophilum EtfAB, depletion was carried out as described previously but using 50 mM Hepes, 200 mM NaCl, pH 7.5 (buffer C) and 50 mM Hepes, 200 mM NaCl, pH 7.5, and 2 M KBr (buffer D). The purified protein was buffer exchanged into buffer C, aliquoted, and stored as described previously.
SDS denaturation of proteins
Denaturation and flavin quantification of replete and depleted forms of M. elsdenii EtfAB, P. aerophilum EtfAB, and Azotobacer vinelandii flavodoxin were carried out using the method previously described by Aliverti et al. (22). Proteins were kept in their respective reaction buffers, and SDS was added to a final concentration of 1%. The solutions were monitored spectroscopically until no more changes were observed at 280 and 450 nm. The final spectra were compared with the free FAD absorbance spectra, and concentration was calculated using ε450 nm = 11.3 mM−1 cm−1 (22, 23).
UV–visible absorbance measurements
All static anaerobic titrations and absorbance measurements were performed using a Hewlett-Packard 8452A diode array spectrophotometer equipped with a thermostatted cell holder. The molar extinction coefficient used for both M. elsdenii and P. aerophilum EtfAB was ε450 nm = 21.1 mM−1 cm−1 as reported by Sato et al. (13). Titrations were performed using an anaerobic quartz sidearm cuvette sealed with a rubber septum (24). M. elsdenii samples were made anaerobic using an Ar train at room temperature and contained catalytic amounts of glucose oxidase from Acidaminococcus niger and bovine liver catalase. After anaerobiosis was achieved, glucose was added to a final concentration of 2 to 10 mM to continuously scrub the reaction mixture of O2 as described by Vigil et al. (21). Because the P. aerophilum EtfAB spontaneously denatures when being made anaerobic on the Ar train, samples had to be made anaerobic by first making the reaction buffer C containing 2 to 10 mM of glucose anaerobic as described previously. The concentrated protein and a stock solution of glucose oxidase and catalase was then injected into the sealed and anaerobic sidearm cuvette using a Hamilton syringe. The gas was turned off, and the sample was then allowed to mix for ∼30 min with stirring to enable the glucose oxidase and catalase to scrub the reaction mixture of O2. The reductant (NADH or sodium dithionite) was made anaerobic and introduced into the cuvette by piercing the septum seal via a Hamilton syringe (the needle/septum interface being sealed with Apiezon N vacuum grease). The spectrum of the anaerobic oxidized protein was recorded, followed by spectra after addition of each aliquot of reductant. A first spectrum was recorded ∼5 s after each addition, followed by a second 5 to 10 min later to ensure complete reaction. Additions continued until the enzymes were fully reduced as compared with the published spectra (11, 13, 15, 25).
EPR spectroscopy
EPR samples containing FAD⋅− for the replete and depleted forms of M. elsdenii EtfAB were prepared in buffer A; replete and depleted forms of P. aerophilum EtfAB were prepared in buffer C. All samples for quantitation were prepared in 714 PQ Precision EPR tubes. A standard FADH⋅ sample was made using 20 μM of A. vinelandii flavodoxin SQ, whose difference between the Q/SQ and SQ/HQ is more than 234 mV, ensuring essentially quantitative formation of the SQ (26). EPR tubes were transferred into an anaerobic chamber several hours before sample preparation. Enzyme solutions and NADH were made anaerobic at room temperature in the manner described previously and transferred to the anaerobic chamber. For samples prepared with both excess and stoichiometric amounts of NADH, a small volume (typically 5 μl) of NADH was added to the EPR tube via Hamilton syringe inside the anaerobic chamber, and the tubes were then sealed with rubber septa, removed from the anaerobic chamber, and incubated at 4 °C for 5 to 10 min. Finally, 240 μl of the anaerobic enzyme solution was added to the tubes containing the NADH solution via Hamilton syringe, and the solution was frozen immediately in an ethanol/dry ice bath and transferred into liquid nitrogen. Using the reaction of azide with myoglobin (27), the total reaction time from mixing to freezing was estimated as 750 ms. EPR spectra were recorded with a Bruker ESP300 spectrometer equipped with a Bruker ER 4119HS high-sensitivity X-band cavity. Temperature control was handled using a Bruker variable temperature unit set at 150 K and a liquid nitrogen cryostat. Acquisition software, EWWIN 6.01 (Scientific Software Services), was used to baseline correct spectra for analysis and perform the double integrations of the spectra for comparison to the quantitation standard. The linewidths of the spectra acquired were compared with the values obtained by Palmer et al. (28).
Rapid reaction kinetics
The reaction of the replete oxidized EtfAB with NADH was observed using a SX-20 stopped-flow spectrophotometer (Applied Photophysics, Inc) equipped with photodiode array (PDA) and photomultiplier tube detection running ProData SX 2.2.5.6 acquisition software (Applied Photophysics, Inc). All experiments with the M. elsdenii protein were performed in buffer A and with the P. aerophilum protein in buffer C with the addition of catalytic amounts of glucose oxidase and catalase as described previously. The enzyme was made anaerobic in a tonometer prior to mounting it onto the stopped-flow apparatus in the manner described previously for M. elsdenii and P. aerophilum, respectively. Transients were collected either at a given wavelength (typically 450 nm) using a photomultiplier detector or over the wavelength range of 230 to 740 nm using a PDA detector. For the replete M. elsdenii, EtfAB traces were taken at 10 °C. Only the first 10 to 500 ms corresponding to the fast phase of FAD reduction were used for analysis. The time courses were fitted to a sum of two exponentials for the replete proteins from both M. elsdenii and P. aerophilum by nonlinear least squares regression analysis using the equation:
| (1) |
where n refers to the number of kinetic phases observed during the fast phase of reduction. The analysis of the time courses was performed using the ProData Viewer 4.2.0 software (Applied Photophysics, Inc). The observed rate constants, kobs, at 450 nm were plotted against the NADH concentrations to produce a hyperbolic curve. From this curve, the limiting rate constant for reduction, kred, and the dissociation constant, Kd, were obtained using the following equation:
| (2) |
The reaction of the depleted form of M. elsdenii EtfAB with NADH was performed and analyzed as described previously, although the temperature was lowered to 5 °C to compensate for the faster kinetics. The fit for the depleted P. aerophilum was single exponential. The reaction of both the replete and depleted forms of P. aerophilum EtfAB was done at 25 °C.
Protein was reacted with stoichiometric amounts of NADH per protomer, and full-wavelength spectra were obtained using a PDA detector in place of the photomultiplier tube in order to obtain full spectral changes during the course of reduction. To estimate the end point of the kinetic phases, the traces used were extracted at 450 nm following flavin reduction for both systems. At substoichiometric and/or stoichiometric amounts of NADH, 377 nm transients (for M. elsdenii) and 368 nm transients (for P. aerophilum) were used to monitor FAD⋅− formation. For the P. aerophilum protein, transients at 650 nm were used to follow formation of the charge–transfer complex between NAD+ and the reduced BF-FAD.
Intermolecular electron transfer experiments
To determine the rate of electron transfer between replete EtfAB molecules, fully oxidized replete EtfAB was reacted with reduced replete EtfAB in a 1:1 stoichiometry. Enzyme samples were made anaerobic as previously described and then transferred to an anaerobic chamber. Proteins were titrated with sodium dithionite and monitored spectroscopically, placed into glass syringes equipped with a three-way stopcock, and loaded onto the stopped-flow apparatus. Spectra were recorded with diode array detector at the temperatures given previously for each protein. Kinetic traces at 377 nm (for M. elsdenii), 368 nm (for P. aerophilum), and 450 nm were extracted from the set of spectra to follow FAD⋅− formation and flavin reduction, respectively.
To follow intermolecular electron transfer with the depleted EtfABs, the enzyme samples were made anaerobic in a sidearm cuvette prior to addition of sufficient NADH to give a concentration half that of the associated protomers. The reaction was followed using Hewlett–Packard 8452A diode array spectrophotometer, monitoring the absorbance at 377 nm or 368 nm to follow FAD⋅−, with spectra recorded every 5 min for approximately 3 h at room temperature.
Data availability
Data not contained in the article (protein gels, absorption spectra, and raw titration data) are available on request to the corresponding author (russ.hille@ucr.edu).
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We gratefully thank Dr Wolfgang Buckel for the recombinant M. elsdenii EtfAB system and Dr Dennis Dean for purified flavodoxin from A. vinelandii.
Author contributions
J. T., G. J. S., and X. G. methodology; W. V., J. T., and D. N. investigation; D. N., M. W. W. A, and R. H. supervision; W. V., J. T., D. N., G. J. S., X. G., M. W. W. A., and R. H. writing–original draft.
Funding and additional information
Work with the P. aerophilum EtfAB was supported by the National Institutes of Health, United States grant GM 135088 (to M. W. W. A. and R. H.); work with the M. elsdenii EtfAB was supported by the National Science Foundation, United States award CHE 2101672 (to R. H.).The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Ruma Banerjee
Footnotes
The observation of such high levels of FAD⋅− accumulation in the course of reductive titrations of the depleted EtfABs with dithionite as reported here is at variance with the results of electrochemical studies of the A. fermentans EtfAB, where it was reported that removal of the ET-FAD had no effect on the half-potentials of the BF-FAD (29). We have examined the A. fermentans protein ourselves and find the same high levels of FAD⋅− accumulation as seen with the M. elsdenii and P. aerophilum EtfABs reported here, clearly demonstrating that the half-potentials of the BF-FAD are indeed significantly perturbed on removal of the ET-FAD. We are unable to account for the failure to detect SQ accumulation in the electrochemical experiment.
Supporting information
References
- 1.Herrmann G., Jayamani E., Mai G., Buckel W. Energy conservation via electron-transferring flavoprotein in anaerobic bacteria. J. Bacteriol. 2008;190:784–791. doi: 10.1128/JB.01422-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buckel W., Thauer R.K. Flavin-based electron bifurcation, ferredoxin, flavodoxin, and anaerobic respiration with protons (Ech) or NAD(+) (Rnf) as electron acceptors: A historical review. Front. Microbiol. 2018;9:401. doi: 10.3389/fmicb.2018.00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buckel W., Thauer R.K. Flavin-based electron bifurcation, A new mechanism of biological energy coupling. Chem. Rev. 2018;118:3862–3886. doi: 10.1021/acs.chemrev.7b00707. [DOI] [PubMed] [Google Scholar]
- 4.Buckel W., Thauer R.K. Energy conservation via electron bifurcating ferredoxin reduction and proton/Na+ translocating ferredoxin oxidation. Biochim. Biophys. Acta-Bioenerg. 2013;1827:94–113. doi: 10.1016/j.bbabio.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Chowdhury N.P., Kahnt J., Buckel W. Reduction of ferredoxin or oxygen by flavin-based electron bifurcation in Megasphaeraelsdenii. FEBS J. 2015;282:3149–3160. doi: 10.1111/febs.13308. [DOI] [PubMed] [Google Scholar]
- 6.Brereton P.S., Verhagen M., Zhou Z.H., Adams M.W.W. Effect of iron-sulfur cluster environment in modulating the thermodynamic properties and biological function of ferredoxin from Pyrococcus furiosus. Biochemistry. 1998;37:7351–7362. doi: 10.1021/bi972864b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li F., Hinderberger J., Seedorf H., Zhang J., Buckel W., Thauer R.K. Coupled ferredoxin and crotonyl coenzyme a (CoA) reduction with NADH catalyzed by the butyryl-CoA dehydrogenase/Etf complex from Clostridium kluyveri. J. Bacteriol. 2008;190:843–850. doi: 10.1128/JB.01417-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chowdhury N.P., Mowafy A.M., Demmer J.K., Upadhyay V., Koelzer S., Jayamani E., Kahnt J., Hornung M., Demmer U., Ermler U., Buckel W. Studies on the mechanism of electron bifurcation catalyzed by electron transferring flavoprotein ( Etf) and butyryl-CoA dehydrogenase (bcd) of Acidaminococcus fermentans. J. Biol. Chem. 2014;289:5145–5157. doi: 10.1074/jbc.M113.521013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demmer J.K., Chowdhury N.P., Selmer T., Ermler U., Buckel W. The semiquinone swing in the bifurcating electron transferring flavoprotein/butyryl-CoA dehydrogenase complex from Clostridium difficile. Nat. Commun. 2017;8:1577. doi: 10.1038/s41467-017-01746-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng X., Schut G.J., Lipscomb G.L., Li H.L., Adams M.W.W. Cryoelectron microscopy structure and mechanism of the membrane-associated electron-bifurcating flavoprotein Fix/EtfABCX. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2016978118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sato K., Nishina Y., Shiga K. Interaction between NADH and electron-transferring flavoprotein from Megasphaera elsdenii. J. Biochem. 2013;153:565–572. doi: 10.1093/jb/mvt026. [DOI] [PubMed] [Google Scholar]
- 12.Whitfield C.D., Mayhew S.G. Purification and properties of electron-transferring flavoprotein from Peptostreptococcus elsdenii. J. Biol. Chem. 1974;249:2801–2810. [PubMed] [Google Scholar]
- 13.Sato K., Nishina Y., Shiga K. Purification of electron-transferring flavoprotein from Megasphaera elsdenii and binding of additional FAD with an unusual absorption spectrum. J. Biochem. 2003;134:719–729. doi: 10.1093/jb/mvg199. [DOI] [PubMed] [Google Scholar]
- 14.Whitfield C., Mayhew S., Massey V. Electron transfer flavoprotein from. Peptostreptococcus Elsdenii. Fed. Proc. 1972;31:447. [Google Scholar]
- 15.Schut G.J., Mohamed-Raseek N., Tokmina-Lukaszewska M., Mulder D.W., Nguyen D.M.N., Lipscomb G.L., Hoben J.P., Patterson A., Lubner C.E., King P.W., Peters J.W., Bothner B., Miller A.-F., Adams M.W.W. The catalytic mechanism of electron-bifurcating electron transfer flavoproteins (ETFs) involves an intermediary complex with NAD(+) J. Biol. Chem. 2019;294:3271–3283. doi: 10.1074/jbc.RA118.005653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pace C.P., Stankovich M.T. Redox properties of electron-transferring flavoprotein from Megasphaera-elsdenii. Biochim. Biophys. Acta. 1987;911:267–276. doi: 10.1016/0167-4838(87)90067-7. [DOI] [PubMed] [Google Scholar]
- 17.Massey V., Muller F., Feldberg R., Schuman M., Sullivan P.A., Howell L.G., Mayhew S.G., Matthews R.G., Foust G.P. Reactivity of flavoproteins with sulfite - possible relevance to problem of oxygen reactivity. J. Biol. Chem. 1969;244:3999–4006. [PubMed] [Google Scholar]
- 18.Muller F., Massey V. Flavin-sulfite adducts and their structures. J. Biol. Chem. 1969;244:4007–4016. [PubMed] [Google Scholar]
- 19.Sucharitakul J., Buckel W., Chaiyen P. Rapid kinetics reveal surprising flavin chemistry in bifurcating electron transfer flavoprotein from Acidaminococcus fermentans. J. Biol. Chem. 2021;296:100124. doi: 10.1074/jbc.RA120.016017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toogood H.S., Leys D., Scrutton N.S. Dynamics driving function - new insights from electron transferring flavoproteins and partner complexes. FEBS J. 2007;274:5481–5504. doi: 10.1111/j.1742-4658.2007.06107.x. [DOI] [PubMed] [Google Scholar]
- 21.Vigil W., Jr., Niks D., Franz-Badur S., Chowdhury N., Buckel W., Hille R. Spectral deconvolution of redox species in the crotonyl-CoA-dependent NADH:ferredoxin oxidoreductase from Megasphaera elsdenii. A flavin-dependent bifurcating enzyme. Arch. Biochem. Biophys. 2021;701:108793. doi: 10.1016/j.abb.2021.108793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aliverti A., Curti B., Vanoni M.A. Identifying and quantitating FAD and FMN in simple and in iron-sulfur-containing flavoproteins. Flavoprotein Protoc. 1999;131:9–23. doi: 10.1385/1-59259-266-X:9. [DOI] [PubMed] [Google Scholar]
- 23.Koziol J. Academic Press; New York, NY: 1971. Fluorometric Analysis of Riboflavin and its Coennzymes. [Google Scholar]
- 24.Niks D., Hille R. Molybdenum-containing enzymes. Methods Mol. Biol. 2019;1876:55–63. doi: 10.1007/978-1-4939-8864-8_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoben J.P., Lubner C.E., Ratzloff M.W., Schut G.J., Nguyen D.M.N., Hempel K.W., Adams M.W.W., King P.W., Miller A.F. Equilibrium and ultrafast kinetic studies manipulating electron transfer: A short-lived flavin semiquinone is not sufficient for electron bifurcation. J. Biol. Chem. 2017;292:14039–14049. doi: 10.1074/jbc.M117.794214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor M.F., Boylan M.H., Edmondson D.E. Azotobacer vinelandii flavodoxin - purification and properties of the recocmbinant, dephospho form expressed in Escherichia coli. Biochemistry. 1990;29:6911–6918. doi: 10.1021/bi00481a022. [DOI] [PubMed] [Google Scholar]
- 27.Ballou D.P., Palmer G.A. Practical rapid quenching instrument for study of reaction mechanisms by electron paramagnetic resonance spectroscopy. Anal. Chem. 1974;46:1248–1253. [Google Scholar]
- 28.Palmer G., Mueller F., Massey V. Proceedings of the Third International Symposium on Flavins and Flavoproteins. University Park Press; Baltimore, MD: 1971. Flavins and flavoproteins. [Google Scholar]
- 29.Sucharitakul J., Buttranon S., Wongnate T., Chowdhury N.P., Prongjit M., Buckel W., Chaiyen P. Modulations of the reduction potentials of flavin-based electron bifurcation complexes and semiquinone stabilities are key to control directional electron flow. FEBS J. 2021;288:1008–1026. doi: 10.1111/febs.15343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data not contained in the article (protein gels, absorption spectra, and raw titration data) are available on request to the corresponding author (russ.hille@ucr.edu).