Summary
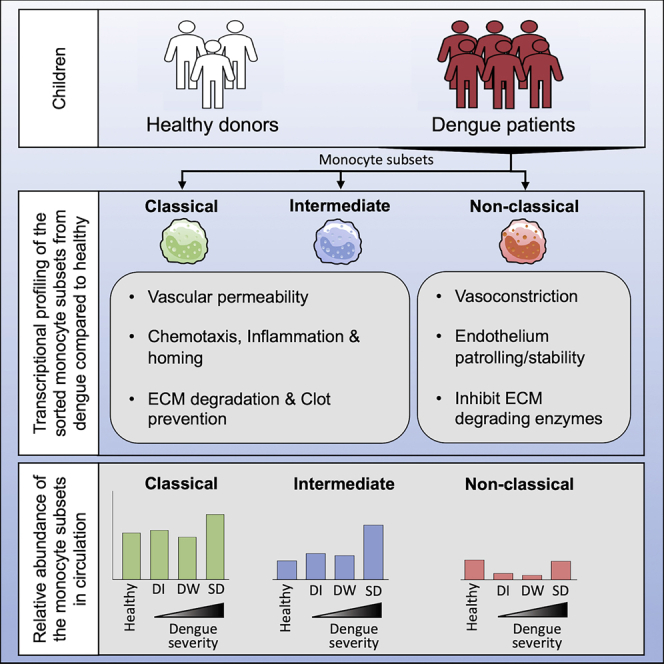
Monocytes are known to play a critical role in dengue pathophysiology. However, which monocyte subset expresses what inflammatory mediator(s) and what transcriptional features distinguish each of the monocyte subset in vivo remain poorly understood. In this study we provide a detailed transcriptional analysis of the three human monocyte subsets in healthy children and in children with dengue febrile illness. Notably, we found that the CD14+ CD16high intermediate monocyte subset from dengue patients highly upregulated key genes involved in mediating inflammation, endothelial dysfunction, vascular permeability, tissue extravasation, and clot prevention compared to healthy children. The CD14+CD16low classical monocytes shared some of these features. These two subsets increased massively in patients with severe dengue. By contrast, the CD14−CD16high nonclassical monocyte subset upregulated key genes involved in vasoconstriction, endothelial barrier stability, and are involved in endothelial patrolling while showing a significant decline from circulation. These findings improve our understanding of monocyte responses in dengue.
Subject areas: Immunology, Omics, Virology
Graphical abstract
Highlights
-
•
CM/IM in dengue express genes involved in inflammation/vascular permeability
-
•
NCM express genes related to endothelial patrolling/ stability and vasoconstriction
-
•
CM/IM expand in severe dengue whereas the NCM decline in mild dengue
Immunology; Omics; Virology
Introduction
Dengue is a mosquito-borne human viral disease of public health importance resulting in over 100 million clinical cases worldwide yearly (Bhatt et al., 2013). All the four dengue virus serotypes continue to spread globally and are currently present in over 150 countries, with India contributing to nearly a third of the global dengue disease burden (Bhatt et al., 2013; Murhekar et al., 2019; WHO, 2012; Wilder-Smith and Rupali, 2019). The clinical spectrum of dengue can range from mild febrile illness to more complex forms of the disease, including dengue hemorrhagic fever (DHF) or dengue shock syndrome (DSS) which can be fatal, especially among children (Bhatt et al., 2013; WHO, 2009, 2011).
Earlier studies suggest that dengue is primarily an inflammatory disease with vascular permeability and plasma leakage playing a central role in its pathophysiology (Stephen J Thomas, Alan L Rothman, 2021). The clinical manifestations of the dengue appear to be multifactorial in nature influenced by a complex spectrum of viral and host factors, including viral serotypes, host immune status, gender, genetic composition, and age (Boonnak et al., 2013; Guzman et al., 2002; Stephens, 2010; Vejbaesya et al., 2009; WHO, 2009, 2011; Wilder-Smith et al., 2019; Yung et al., 2015). Monocytes have been historically considered as one of the key players in this complex puzzle (Castillo et al., 2019; Wan et al., 2018).
Under steady-state conditions, monocytes constitute approximately 5% of the blood circulating nucleated cells (Emmady and Espinoza, 2021). Although the earliest studies identified these cells as CD14 expressing, later studies recognized that there exists an additional monocyte population that is negative for CD14 but positive for CD16 (Passlick et al., 1989; Ziegler-Heitbrock et al., 1988) – and beginning from the 2010s, researchers started to subdivide monocytes into three major subpopulations: the CD14+CD16low classical monocytes (CM), the CD14+CD16+ intermediate monocytes (IM), and the CD14lowCD16+ nonclassical monocytes (NCM) (Ziegler-Heitbrock, 2000). In vivo deuterium labeling studies suggested that a common stem cell precursor from bone marrow first gives rise to CM, which enters circulation and either dies within a day or two or undergoes the sequential transition to IM and NCM phenotypes (Patel et al., 2017). The IM subset has a life span of about 4-5 days whereas the NCM subset has the longest life span of around seven days. Many researchers attempted to gain a functional understanding of the features of these three subsets using in vitro systems- but it is recognized that these in vitro functional readouts can be confounded by the plasticity of these cells to rapidly change phenotypes and functions simply by culturing on tissue culture plastic (Ong et al., 2019). Ex vivo gene expression profiles of the sorted CM, IM, and NCM subsets from healthy adults suggest that while CM subset is generally enriched for genes related to phagocytosis, carbohydrate metabolism, and LPS signaling; IM subset is enriched for genes related to antigen presentation (Wong et al., 2011). In contrast, the NCM subset is enriched for endothelial patrolling genes (Wong et al., 2011). Interestingly, so far, gene expression analysis of each of the three monocyte subsets under the setting of infection with any pathogen is limited. There were studies from HIV or malaria-infected humans or rheumatoid arthritis, but these studies did not subdivide the three monocyte subsets (Gekonge et al., 2012; Loughland et al., 2020; Smiljanovic et al., 2018; Van den Bergh et al., 2010; Wampler Muskardin et al., 2020).
Monocytes are generally considered a major target of infection by Dengue virus either by direct infection or through antibody-dependent enhancement of infection (ADE) (Azeredo et al., 2010; Durbin et al., 2008; Jessie et al., 2004; Kou et al., 2008; Kwissa et al., 2014; Naranjo-Gomez et al., 2019; Singla et al., 2016; Sun et al., 2011). Some in vitro studies suggest that dengue virus-infected monocytes may cause vascular endothelial damage either directly or indirectly (Bosch et al., 2002; Kanlaya et al., 2009; Kelley et al., 2012; Wong et al., 2012). On the other hand, studies suggest that the monocytes when stimulated with dengue virus, can upregulate CD16 and enhance differentiation and expansion of dengue-specific antibody-secreting cells (Kwissa et al., 2014). Interestingly, it was found that IM subset increases in dengue patients whereas NCM subset decreases (Azeredo et al., 2010; Duyen et al., 2017; Kwissa et al., 2014; Naranjo-Gomez et al., 2019; Singla et al., 2016). Similar trends were reported in patients with a different hemorrhagic fever-causing virus, Ebola (Ludtke et al., 2016; McElroy et al., 2020). What transcriptional changes occur in each of the three monocyte subsets under the conditions of dengue febrile illness, which monocyte subset expresses what inflammatory mediator(s), and whether the relative abundance of individual monocyte subsets differs depending on disease severity remain unknown. This study seeks to address these knowledge gaps by performing RNA-seq based transcriptional profiling of the sorted CM, IM, and NCM subsets from dengue febrile children in India and evaluating the relative abundance of each monocyte subsets depending on disease severity.
Results
Analysis of the global transcriptional profiles of the CM, IM, and NCM subsets under steady-state and dengue patients
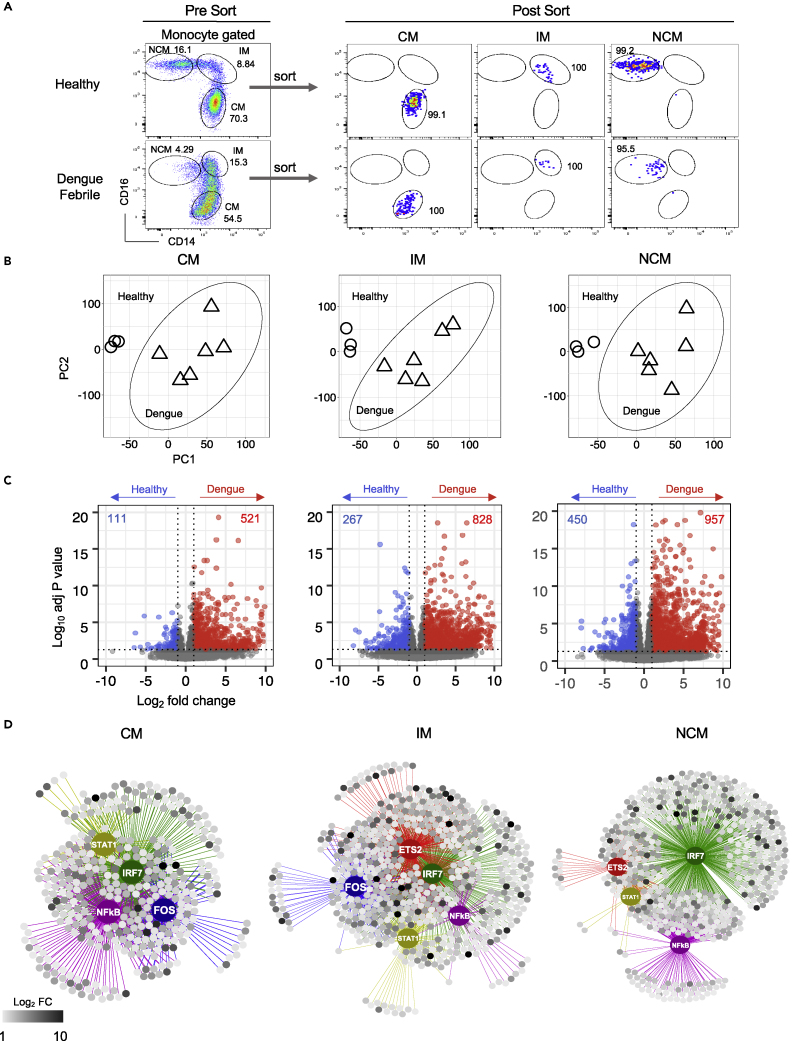
To determine global transcriptional profiles, we sorted the CM, IM, and NCM subsets from six dengue confirmed febrile children. The characteristics of these patients are shown in Table S1. These children were 7-14 years of age recruited during the febrile phase of dengue illness at 3-6 days post-onset of symptoms. Four of these six children were classified as secondary dengue infection using the standard capture ELISA assays using IgM/IgG ratios (PanBiO). Three of the six children were manifesting clinical symptoms indicative of hemorrhage at the time of recruitment. Disease severity classification by WHO 2009 guidelines indicated four of the six children had dengue with warning signs (DW), one had severe dengue (SD) and the other had dengue infection without warning signs (DI) at the time of recruitment. As shown in the gating strategy (Figure S1A), we first excluded T/B cells, CD16+ NK cells, and neutrophils by gating out CD3, CD19, CD20, CD56, CD66b, and NKp30 positive cells and then gated on HLA-DR positive population to identify individual monocyte subsets based on surface expression of CD16 and CD14. For comparison with steady-state conditions, we included sorted CM, IM, and NCM subsets from three healthy children. Consistent with previous reports, we found an increase in the frequency of the IM subset in dengue patients compared to steady-state (Azeredo et al., 2010; Kwissa et al., 2014; Singla et al., 2016); and a decrease in the frequency of the NCM subset in dengue patients compared to steady-state (Duyen et al., 2017; Kwissa et al., 2014; Naranjo-Gomez et al., 2019) (Figure 1A, left, Table S1). Although many historical studies showed that monocytes are one of the major targets of infection with dengue virus in vitro and in vivo (Durbin et al., 2008; Jessie et al., 2004; Kou et al., 2008; Lin et al., 2002; Naranjo-Gomez et al., 2019; Schmid and Harris, 2014; Singla et al., 2016; Sun et al., 2011; Zanini et al., 2018) the analysis of infection in individual monocyte subsets in vivo is only beginning to emerge (Azeredo et al., 2010; Naranjo-Gomez et al., 2019; Singla et al., 2016). A recent study showed that all the three monocyte subsets are susceptible for dengue virus infection in vivo with the frequencies of the infected cells ranging from as low as 1% to as high as 80% (Naranjo-Gomez et al., 2019). Consistent with these previous reports, we found that all the three monocyte subsets can be infected by the dengue virus in vivo (Figure S1B and Table S1). We then performed flow cytometric cell sorting to obtain a highly purified population of each of the three monocyte subsets from each of the dengue febrile children and healthy children as shown in Figure 1A, right. These purified cell populations were then processed for transcriptional analysis using bulk RNA sequencing as detailed in the methodology.
Figure 1.
Global transcriptional analysis of the three monocyte subsets in dengue patients versus healthy children
(A) Pre-sort and post-sort analysis of the three monocyte subsets from healthy children and dengue febrile patients.
(B) PCA score plot obtained from normalized counts of 60,448 genes from CM, IM, and NCM subsets of dengue febrile and healthy children. Genes with no counts in all the samples for each subset were excluded. Ward.D2 was used as a clustering method. ○ represents healthy; Δ represents dengue.
(C) Volcano plots showing DEGs in CM of dengue compared to CM of healthy (left); IM of dengue compared to IM of healthy (middle); NCM of dengue compared to NCM of healthy (right). Scattered dots represent genes. Red dots are genes that are significantly up-regulated, and blue dots represent significantly down-regulated genes. Gray dots are either non-significant, non-differential, or both. (For significant: adjusted P-value < 0.05 and P-value < 0.01; for differential: log2 fold change ≥ 1 or =<-1).
(D) Regulatory networks for upregulated genes in dengue versus healthy comparisons for each subset showing the putative major transcription factors (TFs). Regulons of each major TF are shown in different edge colors, with the regulating TF as a bigger node of the same color. Targets are shown as smaller circles where the gradient of high to low log2 fold change is indicated from black to white color.
All the previous reports compared transcriptional profiles of individual monocyte subsets in steady-state (i.e., in healthy individuals) were from adults. Of these, one study was by microarray analysis and the others were by single-cell RNA seq (Monaco et al., 2019; Uhlen et al., 2019). So far, there is no information on comparison transcriptional profiles of individual monocyte subsets in steady state from children. Hence, we first compared transcriptional profiles between healthy children’s sorted CM, IM, and NCM subsets. We found 1467 differentially expressed genes (DEGs) between the CM, IM and NCM subsets from healthy children (Figure S2A and Table S2A). These DEGs showed significant association with select biological processes in a subset specific and subset overlapping manner (Figure S2B and Table S2B). Over 40 of the DE-Gs from our RNA seq results matched well with the expected flow cytometric staining patterns reported in literature for individual monocyte subsets, thus further validating our RNA seq analysis (Figure S3A). Moreover, the significant DEGs previously reported by Monaco et al. in each of the three monocyte subsets from healthy adults correlated well with the top 500 DEGs in each of the three monocyte subsets from healthy children in our study (Figure S3B).
We next asked what transcriptional changes occur in the CM, IM, and NCM subsets from dengue patients compared to steady-state. Principal component analysis (PCA) showed that while there were substantial differences in the global transcriptional profiles between the CM, IM and NCM subsets under steady-state (i.e., in healthy children) (Figure S4A), the differences in the global transcriptional profiles between the CM and IM subsets became less distinct in dengue (Figure S4B, left). Hierarchical clustering analysis showed that while the CM, IM, and NCM subsets from the dengue patients continue to cluster separately within the CM, IM, and NCM subsets, there was no distinct clustering of the samples from patients with secondary dengue infection or samples from patients with hemorrhage or samples from patients with DI/DW/SD (Figure S4B, right). PCA score plot for all the six subsets (i.e., for CM, IM and NCM subsets from healthy children and dengue febrile children) showed that while the differences in the global transcriptional profiles between the CM and IM subsets become less distinct in dengue, there were substantial differences in the global transcriptional profiles between dengue versus healthy in all the three subsets (Figure S4C). Very few of the top 100 dengue versus healthy PC1 or PC2 genes were commonly shared between the CM, IM and NCM subsets suggesting that the transcriptional changes induced in the dengue patients were not necessarily uniform for all the three subsets (Figure S4D and Table S3A). Hence, we analyzed significant global transcriptional differences between healthy and dengue in each of the three monocyte subsets separately (Figure 1B). In the CM, 632 genes showed differential expression in dengue compared to healthy, of which 521 were upregulated in dengue (Figure 1C, left and Table S3B). In the IM, 1095 genes showed differential expression in dengue compared to healthy, of which 828 were upregulated in dengue (Figure 1C, middle and Table S3C). In the NCM, 1407 genes showed differential expression in dengue compared to healthy, of which 957 were upregulated in dengue (Figure 1C, right and Table S3D). Analysis of the 100 most DEGs in dengue versus healthy showed that a vast majority of these were either qualitatively or quantitatively different between the three subsets (Table S3E). Analysis of genes involved in transcriptional regulation showed that over 80 transcription factors differed in expression levels between the CM, IM, and NCM subsets or between the dengue versus steady states (i.e., healthy) in one or more subsets (Figure S5). Motif2TF search (iRegulon) for binding motifs of these transcription factors for the upregulated DEGs to predict transcriptional regulatory network (TRNs) indicated that while some TRN nodes were altered significantly in all the three monocyte subsets in dengue compared to steady-state (e.g., STAT-1, IRF7, and NFκβ), some were quantitatively preferential to the CM and IM subsets (e.g., FOS). Some others were quantitatively preferential to the IM and NCM subsets (e.g., ETS-2) (Figure 1D and Table S3F). Consistent with this finding, we found that while some biological processes such as inflammation, chemotaxis, and interferon responses were upregulated in all the three monocyte subsets from dengue patients compared to steady-state, many other processes were preferentially upregulated in one or two subsets (Figure S6 and Table S4). Because this pathway analysis did not account for qualitative and quantitative differences between the subsets and in dengue versus steady state, we performed a side-by-side comparison of gene expression between the CM, IM, and NCM subsets and in dengue versus steady-state for select immunological categories (Figure S7). The notable findings are elaborated in sections below.
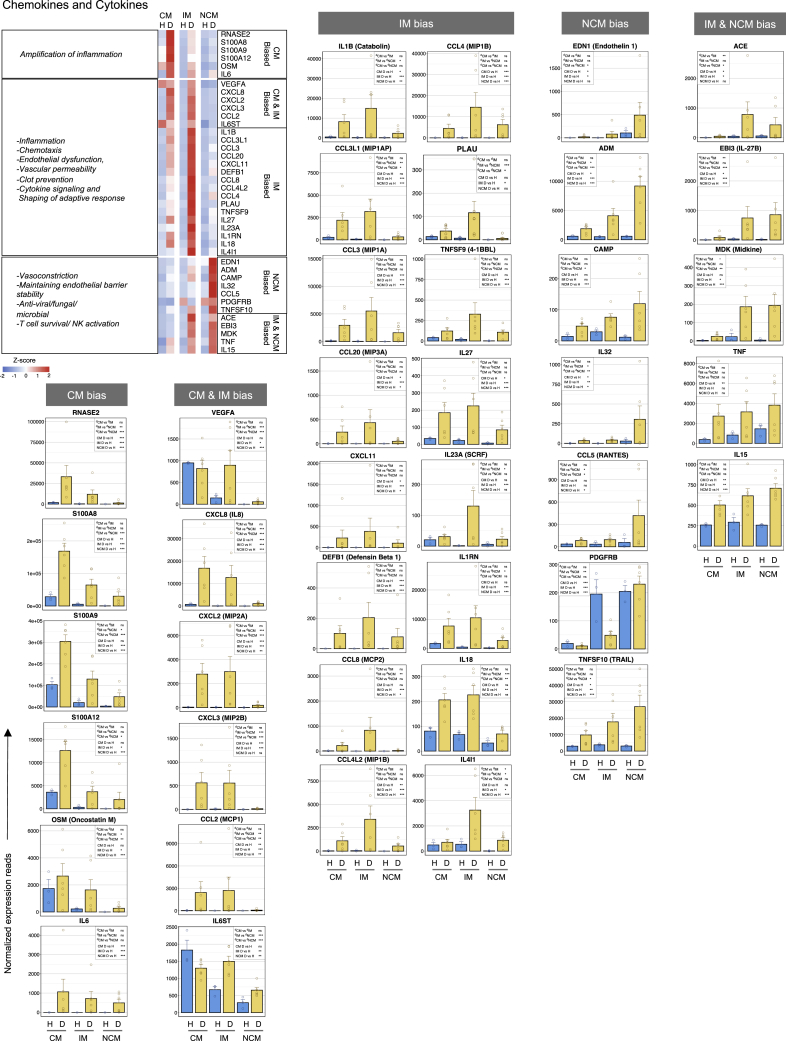
Changes in expression of chemokines/cytokines genes
Heatmap and expression counts of selected cytokines/chemokine genes are shown in Figure 2. Salient points are highlighted below.
Figure 2.
The contrasting behavior in the expression of chemokines/cytokines between the CM, IM, and NCM subsets in dengue
Heatmap of select genes on the top left. A gradient of high to low gene expression based on Z-score calculated from the average normalized counts for healthy subjects and dengue patients in each monocyte subset is shown from red to blue, respectively. Panels of bar graphs show a side-by-side comparison of normalized expression reads in the CM, IM, and NCM subsets from healthy children and dengue patients. Each dot represents the normalized counts for the individual subject (healthy, n = 3; dengue, n = 6). Bars represent average reads in a given subset for a given condition. Blue: healthy children; Yellow: dengue febrile children. Error bars represent the SEM. For each bar graph, significant and non-significant differences within subsets from dengue patients (dCM, dIM and dNCM) and for dengue versus healthy comparison (D vs H) in each subset (CM, IM and NCM) is highlighted in a box, where ∗ denotes adjusted P-value <0.05; ∗∗ denotes adjusted P-value <0.01; ∗∗∗ denotes adjusted P-value <0.001; and “ns” denotes non-significant (also mentioned in Table S3G). The adjusted p values were obtained from DeSeq2 analysis.
We found that the CM subset from dengue patients were highly biased toward expressing key genes involved in amplifying inflammation. Notable among these include RNASE2, S100A8/A9/A12, OSM, and IL-6. RNASE2 is a well-known alarmin that amplifies inflammation. S100A8/A9/A12 is also known to amplify inflammation via regulating cytokine induction, leukocyte recruitment and trans-endothelial migration. Oncostatin M (OSM) is an IL-6 cytokine family member known to activate endothelial cells and regulate their ability to produce other inflammatory cytokines such as IL-6, G-CSF, and GM-CSF.
Both CM and IM subsets from dengue patients showed a high bias toward expressing key genes involved in chemotaxis, endothelial dysfunction, leukocyte extravasation, and angiogenesis. Notable among these include VEGFA, CXCL8, CXCL2, CXCL3, CCL2, and IL6ST. VEGFA is a well-known vascular permeability factor involved in both physiological and pathological angiogenesis. In the CM subset, its expression was high under both steady-state and dengue- but in the IM subset, its expression was high, specifically in dengue. CXCL2, CXCL3, and CXCL8 are the three well--known ELR motif-containing CXC chemokines that function through the CXCR1/CXCR2 axis, mediating endothelial dysfunction, vascular dysfunction extravasation, and angiogenesis; and CCL2 is known to provide a potent chemotactic signal via CCR2/CCR4 axis. IL6ST is a well-known signal transducer for several other pro-inflammatory cytokines such as IL6, oncostatin, ciliary neurotrophic factor, and leukemia inhibitory factor. In dengue, its expression was marginally down in the CM, highly upregulated in the IM and was moderately upregulated in the NCM.
In addition, the IM subset from dengue patients showed a very high bias toward expressing several other cytokines/chemokines. Notable among these include those that are known to drive inflammatory response (IL-1β), those that provide chemotaxis to various immune cells (CCL3L1, CCL3, CCL20, CXCL11, DEFB1, CCL8, CCL4L2, and CCL4), those that prevent clot formation (PLAU), and those that modulate functions or production of other cytokines or T-cell effector functions (TNFSF9, IL27, IL23A, IL1RN, IL18, and IL4L1). IL-1β is one of the most potent pro-inflammatory cytokines involved in various inflammatory reactions, including prostaglandin synthesis, leukocyte influx and activation, T/B cell activation and their cytokine production, fibroblast proliferation, collagen production, VEGF production, vascular permeability, angiogenesis, and pyrogenicity. CCL3L1, CCL3, CCL20, CXCL11, DEFB1, CCL8, CCL4L2, and CCL4 are known to provide strong chemotaxis to a wide variety of immune cells through several shared CCR receptors that usually overlap in expression between a variety of immune cell populations (Cui et al., 2020; Hughes, 2018). The plasminogen activator urokinase (PLAU) catalyzes the formation of plasmin which in turn degrades many extracellular matrix proteins, including fibrin-thereby contributing to hemorrhage via prevention of clot formation. TNFSF9 (41BBL) is a well-known bidirectional signal transducer that promotes T cell proliferation and effector functions. IL-27 is a potent immunomodulator of adaptive responses. IL23a and IL18 are potent pleiotropic cytokines that synergize with IL-12 to promote IFN-γ production from activated T cells. IL1RN is known to regulate IL-1 action by competing with its binding to IL1R1. IL4IL1 is a pleiotropic cytokine that is known to be involved in tryptophan catabolism and regulating adaptive responses.
In marked contrast to the highly biased expression of genes related to inflammation, endothelial dysfunction, vascular permeability, and clot prevention seen in the IM subset, we found that the NCM subset from dengue patients was highly biased toward expression of genes involved in vasoconstriction and endothelial stability (EDN1, ADM, CAMP, PDGFRB, and ACE). Endothelin-1 (EDN1) is a well-known vasoconstrictive peptide. Adrenomedullin (ADM) is an antimicrobial peptide known to play a key role in stabilizing endothelial barrier, thereby preventing endothelial permeability. Cathelicidin (CAMP) is also an antimicrobial peptide that increases endothelial stiffness, thereby decreasing vascular permeability. ACE is known to cleave angiotensin-1 to generate a highly potent vasoconstrictor form of angiotensin.
Interestingly, ACE showed high expression in both the IM and NCM subsets from dengue patients. Platelet-derived growth factor receptor B (PDGFRB) is known to play a key role in developing blood vessels, promoting proliferation, migration, and recruitment of pericytes and smooth muscle cells to endothelial cells, and in the formation of scar tissue at vascular injury sites. Interestingly, we found that while the IM subset decreased expression of PDGFRB in dengue compared to steady-state, the NCM maintained its expression.
In addition, the NCM from dengue patients showed a bias toward the expression of CCL5 (RANTES), which, in addition to being anti-viral, is known to mediate activation of basophils and release of histamines from eosinophils IL-32, an emerging novel cytokine with both pro- and anti-inflammatory properties; TNFSF10, which is known to mediate killing of abnormal cells by inducing apoptosis; and IL-27 subunit beta (EBI3), which, when combined with IL27 subunit alpha, is known to exert both pro and anti-inflammatory effects. Interestingly, we found that the IL-27 B expression was relatively high in the dengue NCM and IM subsets, whereas the IL-27A was relatively high in the dengue CM and IM subsets, suggesting that the generation of fully functional IL27 is likely to be most efficient in the dengue IM. The IM and NCM subsets from dengue patients also showed a bias toward the expression of Midkine (MDK), a multifaceted factor involved in inflammation and tissue regeneration/repair after injury or trauma.
Interestingly, all the three monocyte subsets from dengue patients showed high expression of IL-15 with a slight bias toward NCM. IL-15 is generally considered the pro-survival factor for different adaptive immune cell subsets and is implicated in activating T cells and NK cells. A similar pattern of expression was found with TNF-α.
Taken together, these results indicated that while the CM subset from dengue patients are potently equipped with gene expression machinery involved in amplifying inflammation, the IM subset is additionally equipped with gene expression machinery involved in mediating inflammation, vascular permeability, and clot prevention. By contrast, the NCM subset is potentially equipped with machinery involved in mediating vasoconstriction and endothelial barrier stability.
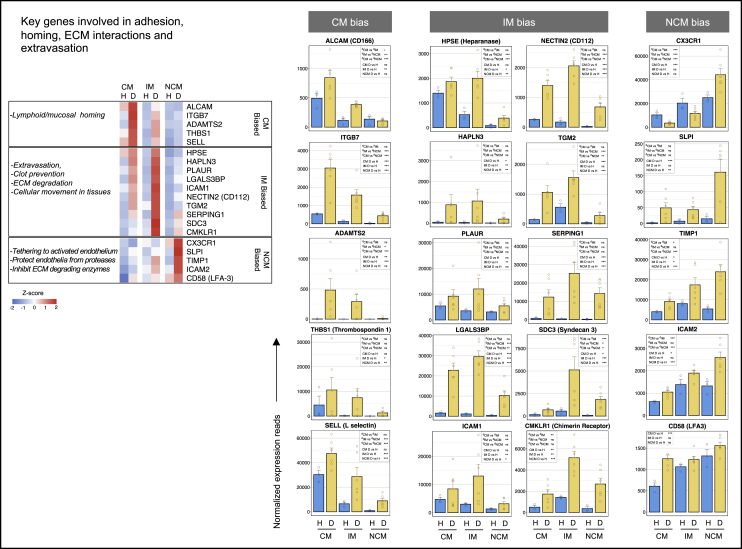
Changes in expression of genes involved in adhesion and extracellular matrix interactions
Heatmap and individual reads of select genes of interest involve adhesion, homing, extracellular matrix (ECM) interactions, and extravasation is shown in Figure 3.
Figure 3.
The contrasting behavior between the CM, IM, and NCM subsets in the expression of genes involved in adhesion, homing, extracellular matrix (ECM) interactions, and tissue extravasation
Heatmap of select genes is shown on the top left. A gradient of high to low gene expression based on Z-score calculated from the average of normalized counts for healthy subjects and dengue patients in each monocyte subset, is shown from red to blue. Panels of bar graphs showing normalized expression read in CM, IM, and NCM subsets from healthy children and dengue patients and are grouped as per their bias toward CM, IM, or NCM. Each dot represents the normalized counts for the individual subject (healthy, n = 3; dengue, n = 6). Bars represent average reads in a given subset for a given condition. Blue: healthy children; Yellow: dengue febrile children. Error bars represent the SEM. For each bar graph, significant and non-significant differences within subsets from dengue patients (dCM, dIM and dNCM) and for dengue versus healthy comparison (D vs H) in each subset (CM, IM and NCM) is highlighted in a box, where ∗ denotes adjusted P-value <0.05; ∗∗ denotes adjusted P-value <0.01; ∗∗∗ denotes adjusted P-value <0.001; and “ns” denotes non-significant (also mentioned in Table S3G). The adjusted p values were obtained from DeSeq2 analysis.
We found that the dengue CM is generally biased toward expression genes involved in adhesion, extracellular matrix interactions, and mucosal/lymphoid homing. Notable among these include ALCAM, a potent homophilic and heterophilic adhesion molecule; integrin subunit beta 7 (ITGB7), that facilitate mucosal homing; procollagen N endopeptidase (ADAMTS2), a metalloproteinase that cleaves type I/type II collagens to facilitate cellular movement through extracellular matrix; thrombospondin (THBS1) that connect cells with extracellular matrix; and L-selectin (SELL), that facilitates leukocyte migration to lymphoid organs and inflammatory sites. The hierarchy of expression of these were in the order of CM > IM > NCM.
Noticeably, the IM subset from dengue patients showed a general bias towards expression of genes involved in cellular interactions, adhesion, and movement through tissue extracellular matrix. Notable among these include heparinase (HPSE), that cleaves heparan sulfate proteoglycans to facilitate cellular movement through extracellular matrix; hyaluronan proteoglycan link protein 3 (HAPLN3) that facilitates extracellular matrix binding; PLAUR, that localizes and promotes formation of plasmin thereby inhibiting clot formation and degrading extracellular matrix proteins; galectin-3 binding protein (LGALS3BP), that promotes integrin-mediated cell adhesion; ICAM-1, that promotes cell: cell or cell:ECM interactions; NECTIN-2, an adhesion molecule which, in addition to serving as receptor for several viruses, may facilitate T cell responses; transglutaminase (TGM-2), which is known to facilitate protein cross-linking; SERPING1, that regulates complement cascade and also inhibits blood coagulation, fibrinolysis or generation of bradykinins via its regulatory effects on factor XIIa and C1; syndecan-3 (SDC), which plays a role in the organization of cell shape via regulating actin cytoskeleton; and CMKLR, which, in addition to serving as a receptor for several viruses, is involved in chemotaxis toward fat tissue by adipocyte secreted chemokine chemerin.
On the other hand, the NCM from dengue patients showed a high bias toward expressing several mediators involved in tethering and protecting the activated endothelial cells from protease-mediated damage and mediators involved in the inhibition of extracellular matrix-degrading enzymes. Notable among these include CX3CR1, which mediates a strong tethering to activated endothelial cells via binding fractalkine; secretory leukocyte peptidase inhibitor (SLPI), that protects epithelial cells from damage by proteases and is involved in tissue repair and wound healing; tissue inhibitor of metalloproteinases 1 (TIMP-1), that prevents tissue extravasation by inhibiting several extracellular matrix-degrading matrix metalloprotease enzymes.
Taken together, these results indicated that while the CM subset from dengue patients was highly biased toward the expression of key genes involved in lymphoid/mucosal homing, both the CM and IM subsets were highly biased toward the expression of key genes involved in the extracellular matrix degradation, clot dissolution, and tissue extravasation. These findings are consistent with the high bias seen toward the expression of genes involved in inflammation and vascular permeability in these cell subsets. The finding that the NCM are biased towards expression of key genes involved in tethering and protecting the activated endothelial cells is consistent with the high bias seen in these cells toward expression of genes involved in vascular constriction and maintaining endothelial barrier stability.
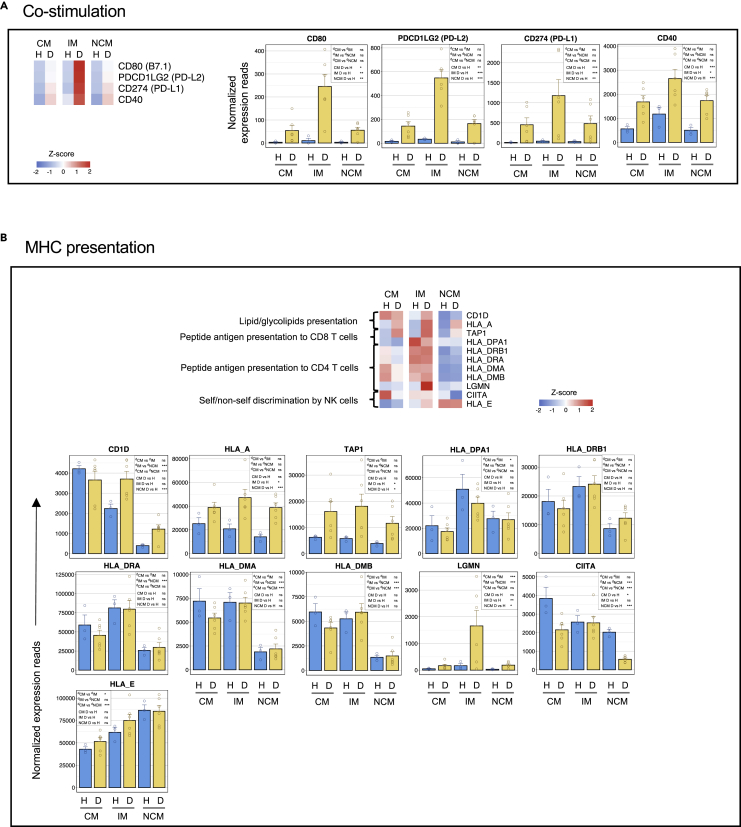
Changes in expression of genes involved in MHC presentation and co-stimulation
The expression of key genes involved in co-stimulation and MHC presentation are shown in Figure 4. We found that the IM subset from dengue patients expressed very high levels of several costimulatory molecules involved in regulating adaptive responses (CD80, CD40, CD274, and PDCDL2). Consistent with this, they also showed a moderate bias toward the expression of classical MHC class-I (HLA-A) and class-I presentation (TAP1). They also showed a bias toward the expression of several HLA class-II genes (HLADPA1, HLADRB1, HLA-DRA, HLADMA, and HLADMB); class-II trans-activator (CIITA), which is a positive regulator of MHC class-II gene transcription; and legumain (LGMN), which likely plays a role in lysosomal/endosomal presentation by class-II. Both the CM and IM, on the other hand, showed a high bias toward the expression of CD1D, which is involved in presenting lipid/glycolipid antigens of self or microbial origin to T cells.
Figure 4.
The CM and IM subsets from dengue patients are biased toward the expression of key genes involved in glycolipid presentation; the IM are additionally biased classical MHC class-I/class-II presentation and co-stimulation, whereas the NCM are biased toward nonclassical MHCs
(A) Heatmap and bar graphs of genes relating to co-stimulation.
(B) Heatmap and bar graphs of genes relating to MHC presentation. For heatmaps, a gradient of high to low gene expression based on a Z-score calculated from the average of normalized counts for healthy subjects and dengue patients in each monocyte subset is shown red to blue. Panels of bar graphs showing normalized expression read in CM, IM, and NCM subsets from healthy children and dengue patients are grouped according to their bias toward CM, IM, or NCM. Each dot represents the normalized counts for the individual subject (healthy, n = 3; dengue, n = 6). Bars represent average reads in a given subset for a given condition. Blue: healthy children; Yellow: dengue febrile children. Error bars represent the SEM. For each bar graph, significant and non-significant differences within subsets from dengue patients (dCM, dIM and dNCM) and for dengue versus healthy comparison (D vs H) in each subset (CM, IM and NCM) is highlighted in a box, where ∗ denotes adjusted P-value <0.05; ∗∗ denotes adjusted P-value <0.01; ∗∗∗ denotes adjusted P-value <0.001; and “ns” denotes non-significant (also mentioned in Table S3G). The adjusted p values were obtained from DeSeq2 analysis.
Interestingly, the expression of the CD1D was high on the CM in both steady-state and dengue. Its expression was relatively lower in the IM under state but further upregulated, reaching levels similar to what is seen in CM in dengue. By contrast to the expression pattern seen in the CM and IM, the NCM showed a bias toward expressing nonclassical MHC class-Ib (HLA-E) and nonclassical MHC class-Ib (HLA-E) in both steady-states and dengue. HLA-E presents leader sequences derived from other MHC molecules to NK inhibitory receptors, providing an important mechanism for self-non-self-discrimination by NK cells.
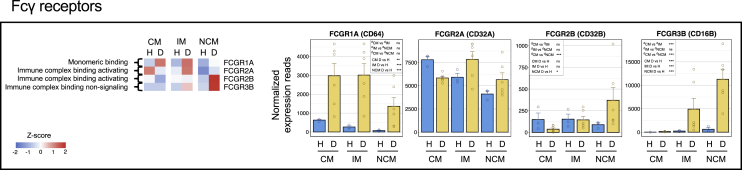
Changes in expression of Fcγ receptor genes
Fc receptors play a key role in monocyte responses by coupling cellular responses to antibody effector functions (Bournazos et al., 2020; Nimmerjahn and Ravetch, 2006; Smith and Clatworthy, 2010; Vogelpoel et al., 2015). These include responses such as cytokine production, antibody-dependent cellular cytotoxicity (ADCC), and endocytosis. They are also involved in mediating antibody-dependent enhancement of infection (ADE) when bound to IgG complexed viral particles. In humans, there are three well-characterized classes of Fcγ receptors: FcγRI (CD64), FcγRII (CD32), and FcγRIII (CD16). Expression patterns of different Fcg receptor genes are shown in Figure 5.
Figure 5.
The IM from dengue patients is biased toward FcγRIIa, whereas the NCM are biased toward the FcγRIIb and FcγRIII
Heatmap of the FC receptor genes with a gradient of high to low gene expression based on Z-score calculated from the average normalized counts for healthy subjects and dengue patients in each monocyte subset is shown from red to blue. Panels of bar graphs showing normalized expression read in CM, IM, and NCM subsets from healthy children and dengue patients are grouped according to their bias toward CM, IM, or NCM. Each dot represents the normalized counts for the individual subject (healthy, n = 3; dengue, n = 6). Bars represent average reads in a given subset for a given condition. Blue: healthy children; Yellow: dengue febrile children. Error bars represent the SEM. For each bar graph, significant and non-significant differences within subsets from dengue patients (dCM, dIM and dNCM) and for dengue versus healthy comparison (D vs H) in each subset (CM, IM and NCM) is highlighted in a box, where ∗ denotes adjusted P-value <0.05; ∗∗ denotes adjusted P-value <0.01; ∗∗∗ denotes adjusted P-value <0.001; and “ns” denotes non-significant (also mentioned in Table S3G). The adjusted p values were obtained from DeSeq2 analysis.
Consistent with previous studies (Sampath et al., 2018; Wong et al., 2011), we found that, under steady-state, the expression of FcγRIa, which is known to bind monomeric IgG because of its high affinity, was biased toward CM. Interestingly, we found that its expression was highly upregulated in all the three monocyte subsets in dengue, with the highest expression being in the CM and IM subsets.
The FcγRII class showed a very complex expression pattern. This class comprises two distinct receptors: an activating receptor, FcγRIIa (CD32a), and an inhibitory receptor, FcγRIIb (CD32b). The binding of immune complexes to the inhibitory receptor FcγRIIb is known to counterbalance collaborating signals through other activating receptors thus serving as a key immune checkpoint mechanism for counterbalancing excessive damage by unwanted immune responses (Kerntke et al., 2020; Rosales, 2017; Vogelpoel et al., 2015). It is shown that even a minute shift in the balance between the expression of activating versus inhibitory receptors toward inhibitory receptor in vitro can result in a massive decrease of inflammatory cytokine responses by innate immune cells (Blom et al., 2003; Boruchov et al., 2005; Clynes et al., 1999; Dhodapkar et al., 2007)- thus making it an attractive target for monoclonal antibody-based therapeutic applications (Teige et al., 2019). We found that the expression of the activating FcγRIIa (CD32a) was biased toward the CM subset under steady-state. In dengue, while its expression was down-regulated in the CM subset, it's expression was upregulated in the IM and NCM subsets, with expression being highest in the IM subset. Interestingly, we found that the expression of the inhibitory receptor FcγRIIb (CD32b) was biased toward the CM and IM subsets under steady-state. In dengue, its expression was down-regulated in the CM, remained unaltered in the IM, and was highly up-regulated in the NCM. These findings, together, indicate that while the IM subset from the dengue patients is biased toward the expression of machinery involved in transmitting activating signals upon binding immune complexes, the NCM subset from dengue patients is biased toward the expression of the machinery involved in transmitting inhibitory signals upon binding immune complexes.
It is known that the FcγRIII class comprises two distinct receptors: FcγRIIIa (CD16, which is used for defining monocyte subsets by flow cytometry) and FcγRIIIb (CD16b). Although the FcγRIIIa/CD16 is known to give activation signals, the FcγRIIIb is generally considered a unique GPI-linked Fcγ receptor that lacks signaling motif and thus implicated in trapping excessive immune complexes or antibody-dependent cellular cytotoxicity (ADCC). Interestingly, we found that the expression of the FcγRIIIb was highly upregulated in IM and NCM subsets from dengue patients, with the expression being highest in the NCM subset.
Taken together, the expression patterns of the Fcγ receptors that we found in the dengue patients suggests that while all the three monocyte subsets are equipped with one or more activating Fcγ receptors, the NCM are preferentially biased toward the expression of Fcγ receptors that are implicated in either negative signaling (FcγRIIb) or signal neutral/trapping of immune complexes or ADCC (FcγRIIIb).
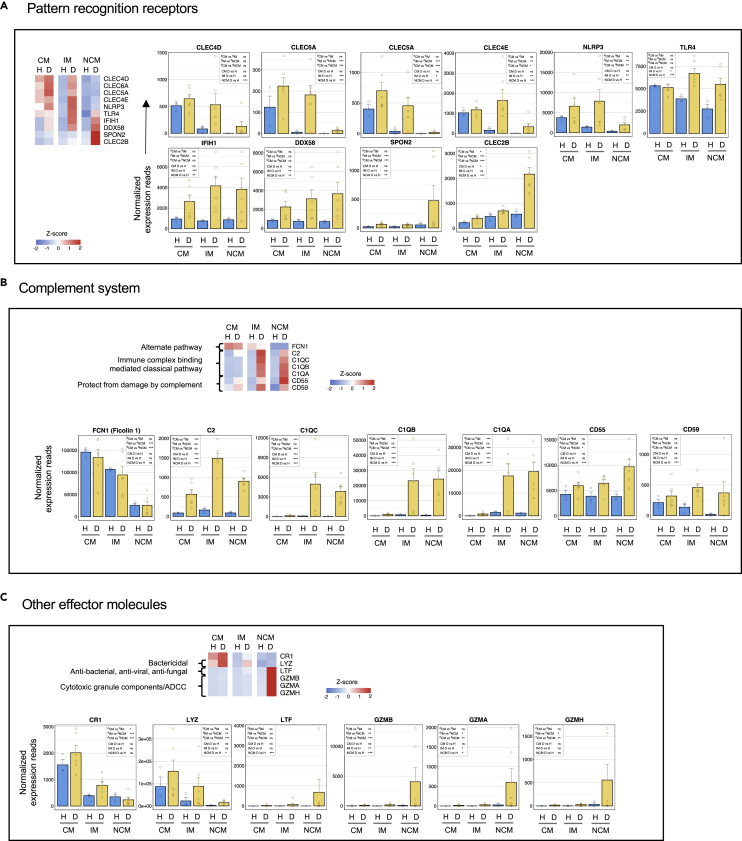
Changes in genes involved in pattern recognition
Select genes of interest encoding molecules involved in innate sensing and microbial pattern recognition is shown in Figure 6A. The CM and IM subsets from dengue patients showed a bias toward expressing various genes involved in microbial carbohydrate pattern recognition, cell adhesion, and inflammation (CLEC4D, CLEC6A, CLEC5A, and CLEC4E), and NLRP3 that pays a key role in the formation of an inflammasome. Consistent with previous studies, the expression of TLR4 was biased toward the CM subset under steady-state, but we found that its expression was upregulated in all the three monocyte subsets in dengue, with the highest expression being in the IM subset. Expression of RIG-I (DDX58) and IFIH1 (MDA5), the two key intracellular pattern recognition molecules in inducing inflammation upon sensing double-stranded viral RNA, was upregulated on all the three monocyte subsets from dengue patients with a bias in expression toward both the IM and NCM subsets. By contrast, the NCM form dengue patients showed a strong bias toward the expression of Spondin-2 (SPON2) and CLEC2B. Limited data available from literature so far suggests that SPON2 acts as a unique pattern recognition molecule in the extracellular matrix that binds pathogens and functions as an opsonin by flagging them for phagocytosis (He et al., 2004). Emerging literature suggests that CLEC2B can mediate monocyte-NK cross-talk by binding killer cell lectin-like receptor subfamily F member 1 (KLRF1/NKp80) that is expressed virtually on all human NK cells (and subsets of effector CD8 T cells or gamma/delta cells) (Welte et al., 2006).
Figure 6.
The CM and IM subsets from dengue patients are biased toward an expression of key genes involved in microbial carbohydrate pattern recognition and inflammation, the IM and NCM are biased toward intracellular double-stranded RNA recognition – whereas the NCM are biased toward pattern recognition in the extracellular matrix and monocyte-NK cross talk
(A) Select genes related to pattern recognition.
(B) Select genes related to complement.
(C) Select genes related to other effector functions. For heatmaps, the gradient of high to low gene expression based on Z-score calculated from the average of normalized counts for healthy subjects and dengue patients in each monocyte subset is shown from red to blue. Panels of bar graphs showing normalized expression read in CM, IM, and NCM subsets from healthy children and dengue patients are grouped as prerecording to their bias toward CM, IM, or NCM. Each dot represents the normalized counts for the individual subject (healthy, n = 3; dengue, n = 6). Bars represent average reads in a given subset for a given condition. Blue: healthy children; Yellow: dengue febrile children. Error bars represent the SEM. For each bar graph, significant and non-significant differences within subsets from dengue patients (dCM, dIM and dNCM) and for dengue versus healthy comparison (D vs H) in each subset (CM, IM and NCM) is highlighted in a box, where ∗ denotes adjusted P-value <0.05; ∗∗ denotes adjusted P-value <0.01; ∗∗∗ denotes adjusted P-value <0.001; and “ns” denotes non-significant (also mentioned in Table S3G). The adjusted p values were obtained from DeSeq2 analysis.
Changes in genes involved in complement
Select genes of interest encoding molecules involved in the complement are shown in Figure 6B. We found that the expression of ficolin-1 (FCN1), which is involved in recognizing microbial carbohydrate patterns and initiation of the alternate pathway of complement, was highly biased toward the CM subset in both steady-state and dengue. Expression of key genes involved in recognizing the immune complexes and initiation of the classical complement pathway (C2, C1QC, C1QB, and C1QA) was relatively low in all the subsets under steady conditions steady-state but highly upregulated in both the IM and NCM subsets from dengue patients. Expression of complement decay-accelerating factor (CD55) and complement membrane attack inhibitory protein (CD59), the two key genes involved in protecting the cells from complement-mediated damage, however, was upregulated on all the three monocyte subsets from dengue patients with a moderate bias in expression toward the IM and NCM subsets.
Although the CM from dengue patients were very low in the expression of proteins involved in the initiation of the classical pathway of complement, they showed very high expression of CR1, a key receptor involved in recognition and phagocytosis of particles coated by opsonins C3b/C4b that are released during complement cascade; and Lysozyme (LYZ), a potent bacteriolytic agent that is also involved in facilitating phagocytosis (Figure 6C). By contrast, the NCM from dengue patients were highly biased toward the lactoferrin (LTF) expression, a broad spectrum antimicrobial and anti-inflammatory peptide. They also expressed very high granzyme genes (GZMB, GZMA, and GZMH), which are likely to play a key role in ADCC activity. It is important to note that granzymes are implicated in several other functions, including serving as a the a bridge between complement proteins and apoptosis by facilitating generation of C3a and C5a and regulating endothelial stability and vascular homeostasis (Garzon-Tituana et al., 2020; Hendel et al., 2014).
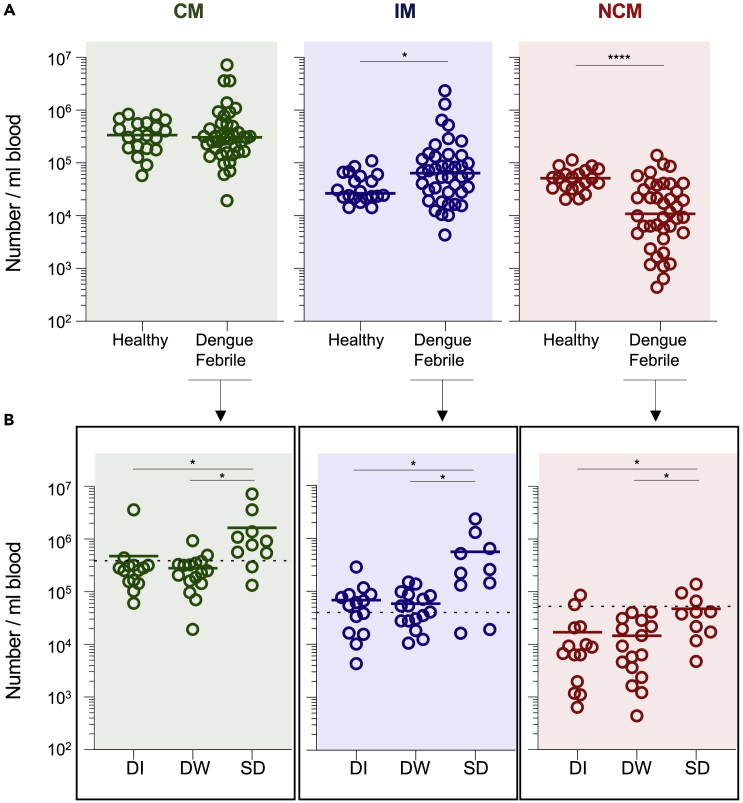
Changes in the relative abundance of the monocyte subsets depending on disease severity
Given the contrasting behavior between the individual monocyte subsets in gene expression profiles, we next asked whether the relative abundance of each monocyte subset differs depending on disease severity. For this, we analyzed the relative abundance of the CM and IM subsets in a total of 41 dengue febrile patients. Of these, 14 patients had mild dengue febrile illness (DI), 16 patients had dengue with warning signs (DW) and 10 patients had severe dengue (SD) at the time of clinical presentation (Table S5). Consistent with previous studies (Azeredo et al., 2010; Kwissa et al., 2014; Naranjo-Gomez et al., 2019; Singla et al., 2016; Welte et al., 2006), we found that while the IM increased moderately, the NCM subset decreased massively in the overall dengue patient group compared to healthy (Figure 7A). Interestingly, by further subdividing the dengue patient group based on disease severity, we found that the CM and IM subsets increased significantly in SD cases compared to DI or DW cases (Figure 7B). By contrast, the decrease of the NCM was less pronounced in the SD cases than DW and DI cases.
Figure 7.
Characterization of individual monocytes subsets in dengue patients with different grades of disease severity
(A) Scatterplots comparing the number per mL blood of the CM, IM, and NCM subsets in healthy children (n= 20) and dengue febrile patients (n= 40).
(B) Scatterplots comparing number per mL blood of CM, IM, and NCM in dengue febrile samples (n= 40) with different grades of disease severity, i.e., (DI, n= 14; DW, n= 16; and SD, n= 10). Mean values are indicated. Significance values between two groups were calculated using two-tailed Mann-Whitney’s U test for non-parametric datasets. Significance values are calculated using Kruskal-Wallis one-way ANOVA followed by Dunn’s multiple comparison test for non-parametric datasets. Significance indicated by: ∗ = p < 0.05, ∗∗ = p < 0.01, ∗∗∗ = p < 0.001 and ∗∗∗∗ = p < 0.0001.
Discussion
In summary, our study provides a comprehensive description of transcriptional profiles for the three human monocyte subsets in children under steady state and in dengue. We show several important differences between the three monocyte subsets in steady state, in dengue, and in dengue versus steady state. A notable finding is that while the CM and IM subsets from dengue patients were highly biased toward the expression of key genes involved in inflammation, vascular permeability, lymphoid homing, extracellular matrix degradation, clot prevention, and tissue extravasation– the NCM were biased toward vasoconstriction, maintenance of vascular tone, endothelial tethering, complement fixation, inhibition of immune complex-mediated signaling and protection from extracellular matrix-degrading enzymes. Although the global transcriptional profiles of the individual monocyte subsets from dengue patients did not cluster separately depending on disease severity or the hemorrhage it is important to note that the CM and IM subsets showed a significantly higher expansion in patients with severe dengue compared to those with dengue infection with or without warning signs. By contrast, the NCM subset showed a decline from circulation in all dengue patients and this decline was less pronounced in patients with severe disease. These findings highlight the contrasting and somewhat opposing behavior between the CM/IM versus NCM in dengue pathophysiology. This knowledge has implications for developing treatment strategies against vascular permeability disorders such as dengue and sepsis as discussed below.
It is important to note that many of the inflammatory mediators that we found upregulated by the CM and IM subsets in dengue have been historically shown to be elevated in the plasma of the dengue patients, especially those that show vascular leakage and or severe dengue (e.g., VEGF, IL-1, IL-6, IL-8, MCP-1 MIP1β) - and hence considered potential biomarkers of dengue disease severity (Patro et al., 2019; Priyadarshini et al., 2010; Singla et al., 2016). Our finding that the CM and IM are not only biased toward the expression of several of these inflammatory mediators- but that these two subsets also expand massively in patients with severe dengue, provide a possible mechanistic source for these inflammatory mediators in dengue.
Based on our finding that the CM and IM subsets robustly express many other genes involved in homing, extracellular matrix degradation, and tissue extravasation and hence likely to home to lymphoid organs or tissues, it is conceivable that the actual expansion of these cells in severe dengue cases might be much higher than what we have estimated in the blood circulation alone.
Many studies suggest that monocytes can make IL-10 upon in vitro culture with various stimuli (Bekkering et al., 2016; Skrzeczynska-Moncznik et al., 2008). Our study surprised us that none of the blood circulating monocyte subsets from dengue patients showed notable IL-10 transcription. This finding warrants further studies to identify in vivo source (s) of IL-10, a major anti-inflammatory cytokine generally elevated in dengue patients (Patro et al., 2019; Singla et al., 2016). Emerging studies suggest that a subpopulation of dengue-specific CD4 T cells can produce IL-10. It is also possible that other myeloid cells, such as myeloid-derived suppressor cells, macrophages, and dendritic cells, could produce IL-10 (Park et al., 2018; Saraiva and O'Garra, 2010; Yaseen et al., 2020).
Our finding that the CM and IM from dengue patients, in addition to expressing key genes involved in vascular permeability and extracellular matrix degradation, also expressed genes involved in clot prevention or clot dissolution – in addition to expanding massively in severe dengue cases, suggest that these two monocyte subsets might contribute to hemorrhage-the hallmark of dengue pathophysiology.
On the other hand, our finding that the NCM expresses an opposing set of genes involved in vasoconstriction, maintenance of endothelial barrier stability, vascular homeostasis and vascular tone, and protection from extracellular matrix-degrading enzymes suggests that these cells might be protecting against vascular leakage.
In addition, the reason why NCM decreases from the blood circulation remains to be addressed. We hypothesize that the decrease of the NCM from blood of the dengue patients may be because of their strong binding to activated endothelial cells. This hypothesis is consistent with their endothelial patrolling behavior that has been extensively reported under steady-state (Thomas et al., 2015); and further supported by our observation that the NCM from dengue patients – but not the CM or IM-strongly expressed CX3CR1 that is involved in tethering to activated endothelial cells.
Intriguingly, we found that the decrease of the NCM from blood was most pronounced in patients with non-severe dengue. This finding raises an interesting question on whether endothelial tethering along with their ability to express vasoconstrictive and endothelial stabilizing factors may be opposing the vascular permeability effects by CM/IM in some patients and thus protecting them from severe dengue. In this regard, it is important to note that numerous antagonists of key molecules involved in vasoconstriction that we found expressed by the dengue NCM (i.e., endothelin, ACE) have been in clinical use for successful treatment of hypertension disorders for several decades (Borek et al., 1989; Raja, 2010).
Thus, our study raises the question whether activators of vasoconstriction (instead of antagonists), either alone or in conjunction with other inhibitors of vascular permeability (e.g., VEGF blockade), may be useful for treating hemorrhagic disorders such as dengue.
In this context, it is interesting to note that NCM shows a decrease in the circulation of other hemorrhagic disorders such as Ebola (Ludtke et al., 2016; McElroy et al., 2020) – thus, further strengthening our hypothesis. Future studies, thus, are needed for a detailed understanding of the qualitative and quantitative differences in the fine balance between individual monocyte subsets in lymphoid, vascular, and tissue compartments and how these events determine disease severity in dengue or other hemorrhagic fevers.
Limitations of the study
Our study is limited to analysis of the monocyte subsets in blood circulation. Given our finding that the dengue-patient derived classical and intermediate subsets express high levels of genes encoding lymphoid/mucosal tissue homing receptors whereas the nonclassical subset express high levels of activated endothelium binding receptor CX3CR1, further studies are needed to address the transcriptional and or functional analysis of these cells in different anatomical locations.
Accession numbers
The GEO accession number for the RNA-Seq data reported in this paper is GEO: GSE176079.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD14 (M5E2) | BioLegend | Cat# 301830; RRID: AB_10959324 |
| CD16 (3G8) | BioLegend | Cat# 302048; RRID: AB_2562085 |
| HLA-DR (L243) | BioLegend | Cat# 307654; RRID: AB_2563646 |
| Lineage Cocktail (CD3-UCHT1, CD19-HIB19, CD20-2H7, CD56-5.1H11) | BioLegend | Cat# 363601; RRID: AB_2916117 |
| CD66b (G10F5) | BioLegend | Cat# 305118; RRID: AB_2566607 |
| NKp30 (P30-15C) | BioLegend | Cat# 325210; RRID: AB_2149449 |
| Mouse Anti-Dengue Complex, clone D3 -2H2-9-21 (1mg) | Merck Millipore | Cat# MAB-8705-K; RRID: AB_1586985 |
| Fixable Viability dye e-Flour 780 | E-Bioscience | Cat# 65-0865-18; RRID: AB_2916118 |
| Biological samples | ||
| Dengue samples | All India Institute of Medical Sciences, New Delhi | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| IC Fixation Buffer | E-Bioscience | 00-8222-49 |
| Permeabilization Buffer | E-Bioscience | 00-8333-56 |
| Critical commercial assays | ||
| Dengue NS1 ELISA | J.Mitra | 1R031096 |
| Dengue IgM ELISA | PanBio | 01PE20/ 01PE21 |
| Dengue IgG ELISA | PanBio | 01PE10 |
| ab102903 – PE/Cy7® Conjugation Kit - Lightning-Link® | abcam | ab102903 |
| Rneasy Micro Kit | Qiagen | 74004 |
| RLT Buffer | Qiagen | 79216 |
| Clontech SMART-Seq v4 Ultra Low Input RNA kit | Takara Bio | 634891 |
| Nextera XT DNA Library Preparation kit | Illumina | FC-131-1096 |
| Deposited data | ||
| RNA seq data | This study | NCBI GEO: GSE176079 |
| RNA seq data | Monaco et al., 2019 | NCBI GEO: GSE107011 |
| Software and algorithms | ||
| FlowJo V10 software | Becton Dickinson | N/A |
| GraphPad Prism (8.4.3) | N/A | N/A |
| R studio version 3.6.0 | The R Foundation for Statistical Computing | https://www.rstudio.com/ |
| Cytoscape v3.8.2 | Shannon.et al., 2003 | https://www.cytoscape.org |
| RTA version 2.7.7 | Illumina pipeline | https://www.illumina.com/ |
| Bcl2fastq version 2.20.0.422 | Illumina pipeline | https://www.illumina.com/ |
| Subread version 1.6.4 | Liao et al., 2014 | http://subread.sourceforge.net/ |
| DESeq2 version 1.26.0 | Love et al., 2014 | https://github.com/mikelove/DESeq2 |
| DAVID Bioinformatics Resources v6.7 | Huang da et al., 2009a, Huang da et al., 2009b | https://david.ncifcrf.gov/ |
| iRegulon v1.3 | Janky et al., 2014 | https://apps.cytoscape.org/apps/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead contact Author, Murali Krishna Kaja (murali.kaja@emory.edu).
Materials availability
This study did not generate new materials.
Experimental model and subject details
Ethics statement
The study was approved by institutional ethical boards, and informed consent or assent was obtained before inclusion in the study.
Patient recruitment
Dengue patients were recruited at the All-India Institute of Medical Sciences (AIIMS), New Delhi. Blood samples obtained from children who do not have any recent infection, allergies or vaccination are recruited at either AIIMS or Emory University, Atlanta, are included as healthy controls and are referred as steady-state in the entire paper. Dengue was confirmed by Dengue NS1 ELISA (J.Mitra, Cat # 1R031096) and/or Dengue IgM (PanBio, Cat # 01PE20, 01PE21) and/or Dengue RT-PCR. The RT-PCR was performed as described in our previous studies (Singla et al., 2016). Patients in this study were enrolled between the years 2017-2019.
Information about sex and median age of the analyzed dengue patients is specified in Table S5. The influence of gender in this study on the results were not explicitly measured.
Disease classification
Based on extensive clinical laboratory tests and evaluation, the attending physicians classified the dengue disease grade based on the WHO 2009 guidelines as dengue infection without warning signs (DI), dengue with warning signs (DW), and severe dengue (SD) at the time of recruitment (WHO, 2009).
Characterization of primary and secondary infections
Serum levels of Dengue IgM and IgG were quantified using the standard Pan-Bio capture ELISAs. As per WHO criteria, those who show levels of dengue-specific IgM greater than those of IgG by a ratio of 1.2 are considered to have primary dengue infections. Patients who show IgG only or IgM levels lower than IgG by a ratio of 1.2 are considered to have secondary dengue infection (WHO, 2009).
Method details
PBMC and plasma isolation
A blood sample was collected in Vacutainer CPT tubes (Becton Dickenson, Cat# 362761). The PBMC’s and plasma were separated as described by the manufacturer protocol. The PBMC’s were washed extensively in 1% complete RPMI (RPMI containing 1% FCS, 1X Pen/Strep, 1X Glutamine) and used for analytical flow cytometry.
Analytical flow cytometry
Fresh cells (i.e., without cryopreservation) were washed and stained for 30 min in ice-cold PBS containing 0.25% FBS. The following anti-human antibodies were used - CD14 (M5E2) (301830, BioLegend), CD16 (3G8) (302048, BioLegend) HLA-DR (L243) (307654, BioLegend). To exclude lineage positive cells, a lineage cocktail (CD3− UCHT1, CD19-HIB19, CD20-2H7, CD56-5.1H11) (363601, BioLegend), CD66b- G10F5 (305118, BioLegend), NKp30-P30-15C (325210, BioLegend) was used. The fixable viability dye e-Fluor 780 (E-Bioscience; 65-0865-18) was used to exclude dead cells during the analysis. For intracellular dengue virus staining cells are fixed after surface staining with IC fixation Buffer (e-bioscience, 00-8222-49) buffer for 10-15 minutes followed by permeabilization with perm buffer (e-bioscience, 00-8333-56) and then incubated for 45-60 minutes with anti-dengue antibody (MERCK Millipore, MAB-8705-K) that were diluted in the perm wash buffer. Then washed once with 1X perm buffer and twice with 0.25% facs buffer. Flow cytometry data acquisition was performed on a BD FACSCanto II or BD LSR-II or BD LSRFortessaTM X-20 (Becton Dickinson), and the data was analyzed using FlowJo V10 software (BD).
Fluorescence-activated cell sorter (FACS) sorting of monocyte subsets
PBMCs isolated from the peripheral whole blood of dengue patients were stained with the relevant antibodies at 4°C for 30 min, washed thoroughly, suspended in PBS containing 2% FCS, and sorted on a BD FACS Aria III (BD) with lineage negative and HLA-DR positive cells to identify monocytes. HLADR+Lineage-CD14+CD16− (classical monocytes, CM), HLADR+Lineage-CD14+CD16+ (intermediate monocytes, IM) and HLADR+Lineage-CD14−CD16+ (non-classical monocytes, NCM), cells were sorted with purity of more than 95%.
RNA isolation and library preparation
The cells were sorted using select markers for monocytes and thoroughly washed. For resuspension RLT buffer (Qiagen) was added to the cell pellet. All samples were processed simultaneously. Monocytes sorted from each patient was processed separately for RNA isolation using RNeasy Micro kit (Qiagen) with on-column DNase digestion. Quality of RNA was assessed through Agilent Bioanalyzer. For cDNA synthesis, Clonetech SMART-Seq v4 Ultra Low Input RNA kit (Takara Bio) was used as per the manufacturer’s instruction with 500pg of total RNA as input, which was kept constant for all the samples to ensure that the difference of cell numbers do not affect the analysis. We also confirmed several genes that are conventionally considered housekeeping (e.g., ACTB, ACTG1, ARF1, ATP5PB, GUSB, HPRT1, PGK1, PSMB4, PTPRC, RPS18, TBP, UBB, UBC, UBE2D2) did not score as the significantly DEGs in our analysis. cDNA library was prepared using Nextera XT DNA Library Preparation kit (Illumina) with dual indexed barcodes appended to the fragmented amplified cDNA, followed by validation through capillary electrophoresis on an Agilent 4200 TapeStation. The libraries were pooled at equimolar concentrations and sequenced on Illumina HiSeq3000 at 100SR. Approximately 20 million reads were obtained per sample.
RNA seq data analysis
RTA version 2.7.7 was used for base calling. Demultiplexing was performed by using bcl2fastq version 2.20.0.422 software. Human genome GRCh38 was used as reference for mapping the sequenced reads. Quality of the sequences were monitored by using FastQC followed by trimming of the adapter sequences. Read summarization was performed using feature counts option of Subread package version 1.6.4 (Liao et al., 2014). Raw counts were normalized and analyzed for differential expression using the DESeq2 package (Love et al., 2014). Low count genes (44,167) with counts < 10 in more than two healthy subjects and more than three dengue subjects were filtered out.
Further, genes with low average normalized counts of more than 100 were included in the differentially expressed gene list. PCA plots were generated from all remaining genes after excluding genes with 0-row sums for all subsets. Z-score (a function of standard deviation from the mean zero of all samples) transformation of the normalized counts was done for plotting the heatmaps. Hierarchical clustering was created using Euclidean distance with Ward.D2 method. All analysis was performed in R studio software.
Open representation analysis
Overexpressed genes from each differential expression analysis were functionally annotated to GO biological processes (BP) using DAVID v6.7 (Huang da et al., 2009a; b). BP terms with adjusted P-value (BH method) < 0.05 were considered as significant.
Transcription factor prediction
iRegulon (Janky et al., 2014) plug-in of the Cytoscape (v 3.8.2) was used to predict transcription factors based on motif2TF search. Genes overexpressed compared to Dengue versus Healthy for CM, IM, and NCM were used as input. The genomic regions for TF-motif search were limited to 10 kb centered around the transcription start site. 6K motif collection was used, and a minimum NES score > 3 was chosen to filter the predicted TFs. Further, TFs of known relevance to monocytes were chosen and constructed a network among each cluster.
Quantification and statistical analysis
Statistical analyses were performed using GraphPad Prism 8.4.3. Statistical significance between multiple groups was assessed by one-way analysis of variance (ANOVA) followed by Turkey’s multiple comparison test if data passed normality tests (parametric data sets). For the non-parametric data sets, Kruskal-Wallis one-way analysis of variance (ANOVA) followed by Dunn’s multiple comparison test was performed. For comparison between two groups, a two-tailed unpaired t-test with Welch’s correction was used for parametric data sets, and a two-tailed Mann–Whitney U test was used for non-parametric data sets. Significance values are indicated by: ∗ = p < 0.05, ∗∗ = p < 0.01, ∗∗∗ = p < 0.001, ∗∗∗∗ = p < 0.0001.
Acknowledgments
This work was supported by US National Institutes of Health grant ICIDR 1UO1A/115654; Government of India Department of Biotechnology grant DBT BT/PR5132/MED/15/85/2012; NIH-DBT, Human immunology Project Consortium (HIPC) grant BT/PR30260/MED/15/194/2018. We thank Mr. Satendra Singh, Mr. Ajay Singh, ICGEB, New Delhi, for technical support, and Aditya Rathi (ICGEB-TACF) for cell sorting.
Author contributions
E.A., R.L., and S.K.K. were involved in human subject recruitment. D.M., K.S., P.S., K.N., C.A., Y.C., M.K., S.G., K.G., C.S.E., R.N., and K.M. were involved in experimental work and analysis. D.M., K.S., M.S., G.M., R.A., A.C., and K.M.K. were involved in study design, analysis, interpretations, and/or manuscript preparation. All authors are involved in manuscript editing.
Declaration of interests
The authors declare no competing interests.
Published: June 17, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104384.
Contributor Information
Anmol Chandele, Email: chandeleanmol@gmail.com.
Kaja Murali-Krishna, Email: murali.kaja@emory.edu.
Supplemental information
(A) Differentially expressed genes between the CM, IM and NCM subsets in healthy children, related to Figure 1 and Figure S2A. (B) The significant biological processes associated with Differentially expressed genes between the CM, IM and NCM subsets in healthy children, related to Figure 1 and Figure S2B.
(A) Top 100 PC1 and PC2 genes for CM, IM and NCM subsets in Dengue versus Healthy, related to Figure1B and Figure S4. (B) Differentially expressed genes between CM of dengue versus healthy, related to Figure 1C. (C) Differentially expressed genes between IM of dengue versus healthy, related to Figure 1C. (D) Differentially expressed genes between NCM of dengue versus healthy, related to Figure 1C. (E) Top 100 most DEGs in Dengue versus healthy in CM, IM and NCM, related to Figure 1C. (F) Direct targets of master regulators of transcription factors (TFs) for each regulatory network, related to Figure 1D. (G) List of adjusted P-values for bar graphs is shown in Figure 2,3,4,5 and 6, related to Figures 2–6.
Data and code availability
-
•
The RNA seq data of this study has been deposited in the Gene Expression Omnibus (GEO) with the accession number GEO: GSE176079.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Azeredo E.L., Neves-Souza P.C., Alvarenga A.R., Reis S.R.N.I., Torrentes-Carvalho A., Zagne S.M.O., Nogueira R.M.R., Oliveira-Pinto L.M., Kubelka C.F. Differential regulation of toll-like receptor-2, toll-like receptor-4, CD16 and human leucocyte antigen-DR on peripheral blood monocytes during mild and severe dengue fever. Immunology. 2010;130:202–216. doi: 10.1111/j.1365-2567.2009.03224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekkering S., Blok B.A., Joosten L.A.B., Riksen N.P., van Crevel R., Netea M.G. In vitro experimental model of trained innate immunity in human primary monocytes. Clin. Vaccin. Immunol. 2016;23:926–933. doi: 10.1128/CVI.00349-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt S., Gething P.W., Brady O.J., Messina J.P., Farlow A.W., Moyes C.L., Drake J.M., Brownstein J.S., Hoen A.G., Sankoh O., et al. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom A.B., van Lent P.L., Holthuysen A.E., Jacobs C., van den Berg W.B. Skewed balance in basal expression and regulation of activating v inhibitory Fcgamma receptors in macrophages of collagen induced arthritis sensitive mice. Ann. Rheum. Dis. 2003;62:465–471. doi: 10.1136/ard.62.5.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonnak K., Slike B.M., Donofrio G.C., Marovich M.A. Human FcγRII cytoplasmic domains differentially influence antibody-mediated dengue virus infection. J. Immunol. 2013;190:5659–5665. doi: 10.4049/jimmunol.1203052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borek M., Charlap S., Frishman W.H. Angiotensin-converting enzyme inhibitors in heart failure. Med. Clin. North Am. 1989;73:315–338. doi: 10.1016/s0025-7125(16)30675-7. [DOI] [PubMed] [Google Scholar]
- Boruchov A.M., Heller G., Veri M.C., Bonvini E., Ravetch J.V., Young J.W. Activating and inhibitory IgG Fc receptors on human DCs mediate opposing functions. J. Clin. Invest. 2005;115:2914–2923. doi: 10.1172/JCI24772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch I., Xhaja K., Estevez L., Raines G., Melichar H., Warke R.V., Fournier M.V., Ennis F.A., Rothman A.L. Increased production of interleukin-8 in primary human monocytes and in human epithelial and endothelial cell lines after dengue virus challenge. J. Virol. 2002;76:5588–5597. doi: 10.1128/jvi.76.11.5588-5597.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournazos S., Gupta A., Ravetch J.V. The role of IgG Fc receptors in antibody-dependent enhancement. Nat. Rev. Immunol. 2020;20:633–643. doi: 10.1038/s41577-020-00410-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo J.A., Naranjo J.S., Rojas M., Castano D., Velilla P.A. Role of monocytes in the pathogenesis of dengue. Arch. Immunol. Ther. Exp. (Warsz) 2019;67:27–40. doi: 10.1007/s00005-018-0525-7. [DOI] [PubMed] [Google Scholar]
- Clynes R., Maizes J.S., Guinamard R., Ono M., Takai T., Ravetch J.V. Modulation of immune complex-induced inflammation in vivo by the coordinate expression of activation and inhibitory Fc receptors. J. Exp. Med. 1999;189:179–186. doi: 10.1084/jem.189.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L.Y., Chu S.F., Chen N.H. The role of chemokines and chemokine receptors in multiple sclerosis. Int. Immunopharmacol. 2020;83:106314. doi: 10.1016/j.intimp.2020.106314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhodapkar K.M., Banerjee D., Connolly J., Kukreja A., Matayeva E., Veri M.C., Ravetch J.V., Steinman R.M., Dhodapkar M.V. Selective blockade of the inhibitory Fcγ receptor (FcγRIIB) in human dendritic cells and monocytes induces a type I interferon response program. J. Exp. Med. 2007;204:1359–1369. doi: 10.1084/jem.20062545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin A.P., Vargas M.J., Wanionek K., Hammond S.N., Gordon A., Rocha C., Balmaseda A., Harris E. Phenotyping of peripheral blood mononuclear cells during acute dengue illness demonstrates infection and increased activation of monocytes in severe cases compared to classic dengue fever. Virology. 2008;376:429–435. doi: 10.1016/j.virol.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyen H.T.L., Cerny D., Trung D.T., Pang J., Velumani S., Toh Y.X., Qui P.T., Hao N.V., Simmons C., Haniffa M., et al. Skin dendritic cell and T cell activation associated with dengue shock syndrome. Sci. Rep. 2017;7:14224. doi: 10.1038/s41598-017-14640-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmady P.D., Espinoza VE. Histology, Monocytes. StatPearls; 2021. [PubMed] [Google Scholar]
- Garzon-Tituana M., Arias M.A., Sierra-Monzon J.L., Morte-Romea E., Santiago L., Ramirez-Labrada A., Martinez-Lostao L., Pano-Pardo J.R., Galvez E.M., Pardo J. The multifaceted function of granzymes in sepsis: some facts and a lot to discover. Front. Immunol. 2020;11:1054. doi: 10.3389/fimmu.2020.01054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gekonge B., Giri M.S., Kossenkov A.V., Nebozyhn M., Yousef M., Mounzer K., Showe L., Montaner L.J. Constitutive gene expression in monocytes from chronic HIV-1 infection overlaps with acute Toll-like receptor induced monocyte activation profiles. PLoS One. 2012;7:e41153. doi: 10.1371/journal.pone.0041153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman M.G., Kouri G., Bravo J., Valdes L., Susana V., Halstead S.B. Effect of age on outcome of secondary dengue 2 infections. Int. J. Infect. Dis. 2002;6:118–124. doi: 10.1016/s1201-9712(02)90072-x. [DOI] [PubMed] [Google Scholar]
- He Y.W., Li H., Zhang J., Hsu C.L., Lin E., Zhang N., Guo J., Forbush K.A., Bevan M.J. The extracellular matrix protein mindin is a pattern-recognition molecule for microbial pathogens. Nat. Immunol. 2004;5:88–97. doi: 10.1038/ni1021. [DOI] [PubMed] [Google Scholar]
- Hendel A., Hsu I., Granville D.J. Granzyme B releases vascular endothelial growth factor from extracellular matrix and induces vascular permeability. Lab. Invest. 2014;94:716–725. doi: 10.1038/labinvest.2014.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hughes C.E., Nibbs R.J.B. A guide to chemokines and their receptors. FEBS J. 2018;285:2944–2971. doi: 10.1111/febs.14466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janky R., Verfaillie A., Imrichova H., Van de Sande B., Standaert L., Christiaens V., Hulselmans G., Herten K., Naval Sanchez M., Potier D., et al. iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput. Biol. 2014;10:e1003731. doi: 10.1371/journal.pcbi.1003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessie K., Fong M.Y., Devi S., Lam S.K., Wong K.T. Localization of dengue virus in naturally infected human tissues, by immunohistochemistry and in situ hybridization. J. Infect. Dis. 2004;189:1411–1418. doi: 10.1086/383043. [DOI] [PubMed] [Google Scholar]
- Kanlaya R., Pattanakitsakul S.N., Sinchaikul S., Chen S.T., Thongboonkerd V. Alterations in actin cytoskeletal assembly and junctional protein complexes in human endothelial cells induced by dengue virus infection and mimicry of leukocyte transendothelial migration. J. Proteome Res. 2009;8:2551–2562. doi: 10.1021/pr900060g. [DOI] [PubMed] [Google Scholar]
- Kelley J.F., Kaufusi P.H., Nerurkar V.R. Dengue hemorrhagic fever-associated immunomediators induced via maturation of dengue virus nonstructural 4B protein in monocytes modulate endothelial cell adhesion molecules and human microvascular endothelial cells permeability. Virology. 2012;422:326–337. doi: 10.1016/j.virol.2011.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerntke C., Nimmerjahn F., Biburger M. There is (scientific) strength in numbers: a comprehensive quantitation of fc gamma receptor numbers on human and murine peripheral blood leukocytes. Front. Immunol. 2020;11:118. doi: 10.3389/fimmu.2020.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kou Z., Quinn M., Chen H., Rodrigo W.S.I., Rose R.C., Schlesinger J.J., Jin X. Monocytes, but not T or B cells, are the principal target cells for dengue virus (DV) infection among human peripheral blood mononuclear cells. J. Med. Virol. 2008;80:134–146. doi: 10.1002/jmv.21051. [DOI] [PubMed] [Google Scholar]
- Kwissa M., Nakaya H.I., Onlamoon N., Wrammert J., Villinger F., Perng G.C., Yoksan S., Pattanapanyasat K., Chokephaibulkit K., Ahmed R., Pulendran B. Dengue virus infection induces expansion of a CD14(+)CD16(+) monocyte population that stimulates plasmablast differentiation. Cell Host Microbe. 2014;16:115–127. doi: 10.1016/j.chom.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Lin Y.W., Wang K.J., Lei H.Y., Lin Y.S., Yeh T.M., Liu H.S., Liu C.C., Chen S.H. Virus replication and cytokine production in dengue virus-infected human B lymphocytes. J. Virol. 2002;76:12242–12249. doi: 10.1128/jvi.76.23.12242-12249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughland J.R., Woodberry T., Field M., Andrew D.W., SheelaNair A., Dooley N.L., Piera K.A., Amante F.H., Kenangalem E., Price R.N., et al. Transcriptional profiling and immunophenotyping show sustained activation of blood monocytes in subpatent Plasmodium falciparum infection. Clin. Transl. Immunol. 2020;9:e1144. doi: 10.1002/cti2.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke A., Ruibal P., Becker-Ziaja B., Rottstegge M., Wozniak D.M., Cabeza-Cabrerizo M., Thorenz A., Weller R., Kerber R., Idoyaga J., et al. Ebola virus disease is characterized by poor activation and reduced levels of circulating CD16+ monocytes. J. Infect. Dis. 2016;214:S275–S280. doi: 10.1093/infdis/jiw260. [DOI] [PubMed] [Google Scholar]
- McElroy A.K., Akondy R.S., McLlwain D.R., Chen H., Bjornson-Hooper Z., Mukherjee N., Mehta A.K., Nolan G., Nichol S.T., Spiropoulou C.F. Immunologic timeline of Ebola virus disease and recovery in humans. JCI Insight. 2020;5:e137260. doi: 10.1172/jci.insight.137260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco G., Lee B., Xu W., Mustafah S., Hwang Y.Y., Carre C., Burdin N., Visan L., Ceccarelli M., Poidinger M., et al. RNA-seq signatures normalized by mRNA abundance allow absolute deconvolution of human immune cell types. Cell Rep. 2019;26:1627–1640.e7. doi: 10.1016/j.celrep.2019.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murhekar M., Joshua V., Kanagasabai K., Shete V., Ravi M., Ramachandran R., Sabarinathan R., Kirubakaran B., Gupta N., Mehendale S. Epidemiology of dengue fever in India, based on laboratory surveillance data, 2014-2017. Int. J. Infect. Dis. 2019;84:S10–S14. doi: 10.1016/j.ijid.2019.01.004. [DOI] [PubMed] [Google Scholar]
- Naranjo-Gomez J.S., Castillo J.A., Rojas M., Restrepo B.N., Diaz F.J., Velilla P.A., Castano D. Different phenotypes of non-classical monocytes associated with systemic inflammation, endothelial alteration and hepatic compromise in patients with dengue. Immunology. 2019;156:147–163. doi: 10.1111/imm.13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn F., Ravetch J.V. Fcγ receptors: old friends and new family members. Immunity. 2006;24:19–28. doi: 10.1016/j.immuni.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Ong S.M., Teng K., Newell E., Chen H., Chen J., Loy T., Yeo T.W., Fink K., Wong S.C. A novel, five-marker alternative to CD16-CD14 gating to identify the three human monocyte subsets. Front. Immunol. 2019;10:1761. doi: 10.3389/fimmu.2019.01761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M.J., Lee S.H., Kim E.K., Lee E.J., Baek J.A., Park S.H., Kwok S.K., Cho M.L. Interleukin-10 produced by myeloid-derived suppressor cells is critical for the induction of Tregs and attenuation of rheumatoid inflammation in mice. Sci. Rep. 2018;8:3753. doi: 10.1038/s41598-018-21856-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passlick B., Flieger D., Ziegler-Heitbrock H.W. Identification and characterization of a novel monocyte subpopulation in human peripheral blood. Blood. 1989;74:2527–2534. doi: 10.1182/blood.v74.7.2527.bloodjournal7472527. [DOI] [PubMed] [Google Scholar]
- Patel A.A., Zhang Y., Fullerton J.N., Boelen L., Rongvaux A., Maini A.A., Bigley V., Flavell R.A., Gilroy D.W., Asquith B., et al. The fate and lifespan of human monocyte subsets in steady state and systemic inflammation. J. Exp. Med. 2017;214:1913–1923. doi: 10.1084/jem.20170355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro A.R.K., Mohanty S., Prusty B.K., Singh D.K., Gaikwad S., Saswat T., Chattopadhyay S., Das B.K., Tripathy R., Ravindran B. Cytokine signature associated with disease severity in dengue. Viruses. 2019;11:34. doi: 10.3390/v11010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyadarshini D., Gadia R.R., Tripathy A., Gurukumar K.R., Bhagat A., Patwardhan S., Mokashi N., Vaidya D., Shah P.S., Cecilia D. Clinical findings and pro-inflammatory cytokines in dengue patients in Western India: a facility-based study. PLoS One. 2010;5:e8709. doi: 10.1371/journal.pone.0008709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja S.G. REVIEW: endothelin receptor antagonists for pulmonary arterial hypertension: an overview. Cardiovasc. Ther. 2010;28:e65–e71. doi: 10.1111/j.1755-5922.2010.00158.x. [DOI] [PubMed] [Google Scholar]
- Rosales C. Fcγ receptor heterogeneity in leukocyte functional responses. Front. Immunol. 2017;8:280. doi: 10.3389/fimmu.2017.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath P., Moideen K., Ranganathan U.D., Bethunaickan R. Monocyte subsets: phenotypes and function in tuberculosis infection. Front. Immunol. 2018;9:1726. doi: 10.3389/fimmu.2018.01726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraiva M., O'Garra A. The regulation of IL-10 production by immune cells. Nat. Rev. Immunol. 2010;10:170–181. doi: 10.1038/nri2711. [DOI] [PubMed] [Google Scholar]
- Schmid M.A., Harris E. Monocyte recruitment to the dermis and differentiation to dendritic cells increases the targets for dengue virus replication. PLoS Pathog. 2014;10:e1004541. doi: 10.1371/journal.ppat.1004541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla M., Kar M., Sethi T., Kabra S.K., Lodha R., Chandele A., Medigeshi G.R. Immune response to dengue virus infection in pediatric patients in New Delhi, India--Association of viremia, inflammatory mediators and monocytes with disease severity. PLoS Negl. Trop. Dis. 2016;10:e0004497. doi: 10.1371/journal.pntd.0004497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrzeczynska-Moncznik J., Bzowska M., Loseke S., Grage-Griebenow E., Zembala M., Pryjma J. Peripheral blood CD14high CD16+ monocytes are main producers of IL-10. Scand. J. Immunol. 2008;67:152–159. doi: 10.1111/j.1365-3083.2007.02051.x. [DOI] [PubMed] [Google Scholar]
- Smiljanovic B., Radzikowska A., Kuca-Warnawin E., Kurowska W., Grun J.R., Stuhlmuller B., Bonin M., Schulte-Wrede U., Sorensen T., Kyogoku C., et al. Monocyte alterations in rheumatoid arthritis are dominated by preterm release from bone marrow and prominent triggering in the joint. Ann. Rheum. Dis. 2018;77:300–308. doi: 10.1136/annrheumdis-2017-211649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K.G., Clatworthy M.R. FcγRIIB in autoimmunity and infection: evolutionary and therapeutic implications. Nat. Rev. Immunol. 2010;10:328–343. doi: 10.1038/nri2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen J Thomas MD. Dengue virus infection: Pathogenesis (2021).
- Stephens H.A.F. HLA and other gene associations with dengue disease severity. Curr. Top Microbiol. Immunol. 2010;338:99–114. doi: 10.1007/978-3-642-02215-9_8. [DOI] [PubMed] [Google Scholar]
- Sun P., Bauza K., Pal S., Liang Z., Wu S.J., Beckett C., Burgess T., Porter K. Infection and activation of human peripheral blood monocytes by dengue viruses through the mechanism of antibody-dependent enhancement. Virology. 2011;421:245–252. doi: 10.1016/j.virol.2011.08.026. [DOI] [PubMed] [Google Scholar]
- Teige I., Martensson L., Frendeus B.L. Targeting the antibody checkpoints to enhance cancer immunotherapy–focus on FcγRIIB. Front. Immunol. 2019;10:481. doi: 10.3389/fimmu.2019.00481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G., Tacke R., Hedrick C.C., Hanna R.N. Nonclassical patrolling monocyte function in the vasculature. Arterioscler Thromb. Vasc. Biol. 2015;35:1306–1316. doi: 10.1161/ATVBAHA.114.304650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M., Karlsson M.J., Zhong W., Tebani A., Pou C., Mikes J., Lakshmikanth T., Forsstrom B., Edfors F., Odeberg J., et al. A genome-wide transcriptomic analysis of protein-coding genes in human blood cells. Science. 2019;366:eaax9198. doi: 10.1126/science.aax9198. [DOI] [PubMed] [Google Scholar]
- Van den Bergh R., Florence E., Vlieghe E., Boonefaes T., Grooten J., Houthuys E., Tran H.T.T., Gali Y., De Baetselier P., Vanham G., Raes G. Transcriptome analysis of monocyte-HIV interactions. Retrovirology. 2010;7:53. doi: 10.1186/1742-4690-7-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vejbaesya S., Luangtrakool P., Luangtrakool K., Kalayanarooj S., Vaughn D.W., Endy T.P., Mammen M.P., Green S., Libraty D.H., Ennis F.A., et al. TNF and LTA gene, allele, and extended HLA haplotype associations with severe dengue virus infection in ethnic Thais. J. Infect. Dis. 2009;199:1442–1448. doi: 10.1086/597422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelpoel L.T.C., Baeten D.L.P., de Jong E.C., den Dunnen J. Control of cytokine production by human fc gamma receptors: implications for pathogen defense and autoimmunity. Front. Immunol. 2015;6:79. doi: 10.3389/fimmu.2015.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wampler Muskardin T.L., Fan W., Jin Z., Jensen M.A., Dorschner J.M., Ghodke-Puranik Y., Dicke B., Vsetecka D., Wright K., Mason T., et al. Distinct single cell gene expression in peripheral blood monocytes correlates with tumor necrosis factor inhibitor treatment response groups defined by type I interferon in rheumatoid arthritis. Front. Immunol. 2020;11:1384. doi: 10.3389/fimmu.2020.01384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan S.W., Wu-Hsieh B.A., Lin Y.S., Chen W.Y., Huang Y., Anderson R. The monocyte-macrophage-mast cell axis in dengue pathogenesis. J. Biomed. Sci. 2018;25:77. doi: 10.1186/s12929-018-0482-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte S., Kuttruff S., Waldhauer I., Steinle A. Mutual activation of natural killer cells and monocytes mediated by NKp80-AICL interaction. Nat. Immunol. 2006;7:1334–1342. doi: 10.1038/ni1402. [DOI] [PubMed] [Google Scholar]
- WHO . WHO; 2009. Dengue Guidelines for Diagnosis, Treatment, Prevention and Control. [PubMed] [Google Scholar]
- WHO . WHO; 2011. Dengue: Call for Urgent Interventions for a Rapidly Expanding Emerging Disease. [Google Scholar]
- WHO . WHO; 2012. Dengue and Severe Dengue. [Google Scholar]
- Wilder-Smith A., Ooi E.E., Horstick O., Wills B. Dengue. Lancet. 2019;393:350–363. doi: 10.1016/S0140-6736(18)32560-1. [DOI] [PubMed] [Google Scholar]
- Wilder-Smith A., Rupali P. Estimating the dengue burden in India. Lancet Glob. Health. 2019;7:e988–e989. doi: 10.1016/S2214-109X(19)30249-9. [DOI] [PubMed] [Google Scholar]
- Wong K.L., Chen W., Balakrishnan T., Toh Y.X., Fink K., Wong S.C. Susceptibility and response of human blood monocyte subsets to primary dengue virus infection. PLoS One. 2012;7:e36435. doi: 10.1371/journal.pone.0036435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K.L., Tai J.J.Y., Wong W.C., Han H., Sem X., Yeap W.H., Kourilsky P., Wong S.C. Gene expression profiling reveals the defining features of the classical, intermediate, and nonclassical human monocyte subsets. Blood. 2011;118:e16–e31. doi: 10.1182/blood-2010-12-326355. [DOI] [PubMed] [Google Scholar]
- Yaseen M.M., Abuharfeil N.M., Darmani H., Daoud A. Mechanisms of immune suppression by myeloid-derived suppressor cells: the role of interleukin-10 as a key immunoregulatory cytokine. Open Biol. 2020;10:200111. doi: 10.1098/rsob.200111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung C.F., Lee K.S., Thein T.L., Tan L.K., Gan V.C., Wong J.G.X., Lye D.C., Ng L.C., Leo Y.S. Dengue serotype-specific differences in clinical manifestation, laboratory parameters and risk of severe disease in adults, Singapore. Am. J. Trop. Med. Hyg. 2015;92:999–1005. doi: 10.4269/ajtmh.14-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanini F., Robinson M.L., Croote D., Sahoo M.K., Sanz A.M., Ortiz-Lasso E., Albornoz L.L., Rosso F., Montoya J.G., Goo L., et al. Virus-inclusive single-cell RNA sequencing reveals the molecular signature of progression to severe dengue. Proc. Natl. Acad. Sci. U S A. 2018;115:E12363–E12369. doi: 10.1073/pnas.1813819115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler-Heitbrock H.W.L. Definition of human blood monocytes. J. Leukoc. Biol. 2000;67:603–606. doi: 10.1002/jlb.67.5.603. [DOI] [PubMed] [Google Scholar]
- Ziegler-Heitbrock H.W., Passlick B., Flieger D. The monoclonal antimonocyte antibody My4 stains B lymphocytes and two distinct monocyte subsets in human peripheral blood. Hybridoma. 1988;7:521–527. doi: 10.1089/hyb.1988.7.521. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Differentially expressed genes between the CM, IM and NCM subsets in healthy children, related to Figure 1 and Figure S2A. (B) The significant biological processes associated with Differentially expressed genes between the CM, IM and NCM subsets in healthy children, related to Figure 1 and Figure S2B.
(A) Top 100 PC1 and PC2 genes for CM, IM and NCM subsets in Dengue versus Healthy, related to Figure1B and Figure S4. (B) Differentially expressed genes between CM of dengue versus healthy, related to Figure 1C. (C) Differentially expressed genes between IM of dengue versus healthy, related to Figure 1C. (D) Differentially expressed genes between NCM of dengue versus healthy, related to Figure 1C. (E) Top 100 most DEGs in Dengue versus healthy in CM, IM and NCM, related to Figure 1C. (F) Direct targets of master regulators of transcription factors (TFs) for each regulatory network, related to Figure 1D. (G) List of adjusted P-values for bar graphs is shown in Figure 2,3,4,5 and 6, related to Figures 2–6.
Data Availability Statement
-
•
The RNA seq data of this study has been deposited in the Gene Expression Omnibus (GEO) with the accession number GEO: GSE176079.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.