Abstract
Many studies have examined the impact of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants on neutralizing antibody activity after they have become dominant strains. Here, we evaluate the consequences of further viral evolution. We demonstrate mechanisms through which the SARS-CoV-2 receptor binding domain (RBD) can tolerate large numbers of simultaneous antibody escape mutations and show that pseudotypes containing up to seven mutations, as opposed to the one to three found in previously studied variants of concern, are more resistant to neutralization by therapeutic antibodies and serum from vaccine recipients. We identify an antibody that binds the RBD core to neutralize pseudotypes for all tested variants but show that the RBD can acquire an N-linked glycan to escape neutralization. Our findings portend continued emergence of escape variants as SARS-CoV-2 adapts to humans.
Evasive maneuvers by the spike protein
Throughout the course of the COVID-19 pandemic, variants have arisen in the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus that increase infectivity or reduce its susceptibility to existing antibodies. Nabel et al. focus on mutations in the spike protein, which is found on the viral surface and is responsible for binding and entering host cells, and show that the structure exhibits plasticity in resisting neutralizing antibodies. Working with pseudoviruses, the authors identify combinatorial mutations that might lead to resistance. They identify an antibody that neutralizes a pseudotype with a highly mutated spike, but also show that SARS-CoV-2 can acquire a glycan to escape this neutralization. Understanding the consequences of changes in the antigenic landscape of the spike protein is important if we are to rapidly respond to new variants of concern. —VV
As it evades antibodies, the SARS-CoV-2 spike protein receptor-binding domain can acquire multiple mutations and a glycan.
INTRODUCTION
As severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) replicates under selective pressure from natural and vaccine-induced immunity, variants of concern (VOCs) continue to emerge. Through adaptative evolution, these variants acquire mutations in the spike protein receptor binding domain (RBD) that binds the cellular receptor angiotensin-converting enzyme 2 (ACE2). The effects of spike protein mutations on immune responses make it important to monitor viral variants. While previously studied VOCs contain one to three RBD mutations that at times overlap, the potential for composite variants that contain larger numbers of mutations is being closely monitored.
RATIONALE
As parts of the world continue to face waves of infection and mitigation strategies are relaxed, viral replication in human hosts under antibody selective pressure continues to shape the antigenic landscape of the SARS-CoV-2 spike protein. As variants containing composite mutations begin to emerge, proactive approaches examining the impact of variants before they become dominant strains are critical.
RESULTS
We determined the x-ray crystal structure of human ACE2 in complex with a SARS-CoV-2 RBD that contains six substitutions that arose during persistent infection of an immunocompromised individual. We found that structural plasticity at the RBD–ACE2 interface allowed the RBD to tolerate a large number of mutations while retaining ACE2 affinity. We generated a panel of pseudotypes bearing composite RBD mutations (up to seven) from immunocompromised host-derived sequences and VOCs. Composite variants more adeptly evaded therapeutic antibody neutralization than did previously studied VOCs. After first immunization but before the second dose of an mRNA vaccine, we observed a loss in vaccine recipient serum neutralizing activity for all variants tested, although the severity differed depending on the variant. However, sampling after the second immunization revealed detectable neutralizing activity against all variants in the serum of vaccine recipients, including against a pseudotype that contains seven composite RBD mutations [denoted receptor binding mutant-2 (RBM-2)]. To identify evolutionary barriers that restrict neutralization breadth, we used the SARS-CoV spike protein to isolate a neutralizing antibody from a COVID-19 convalescent donor. Through structural analysis and functional assays, we show that N-linked glycan acquisition by the SARS-CoV-2 RBD confers pseudotype resistance to neutralization by the isolated cross-reactive antibody and at least one other antibody that binds a similar, otherwise highly conserved epitope. Therefore, acquisition of an N-linked glycan on the SARS-CoV-2 RBD is an additional means through which the virus could continue to evade immune responses.
CONCLUSION
We find that accumulation of large numbers of RBD mutations is facilitated by structural plasticity at the RBD–ACE2 interface and further erodes the activity of therapeutic antibodies and serum from vaccine recipients. Furthermore, acquisition of an N-linked glycan on the SARS-CoV-2 RBD is an additional neutralization escape pathway that should be closely monitored during viral antigenic drift.
Immune escape at the SARS-CoV-2 spike protein RBD.
Structural plasticity accommodates the accumulation of composite substitutions in the RBD ACE2 binding site and allows the RBD to adeptly escape therapeutic antibodies. Cross-neutralizing antibodies bind the RBD core, but acquisition of an N-linked glycan at RBD residue Asn370 (N370) drives further neutralization escape. Single-letter abbreviations for the amino acid residues are as follows: D, Asp; E, Glu; F, Phe; H, His; K, Lys; L, Leu; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; and Y, Tyr. LC, light chain; HC, heavy chain.
As severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continues to replicate in humans under selective pressure from natural and vaccine-induced immunity, variants of concern (VOCs) with increased transmissibility or virulence continue to emerge (1). Through adaptive evolution, these variants acquire mutations in the spike (S) protein receptor binding domain (RBD) that binds the cellular receptor human angiotensin-converting enzyme 2 (ACE2) (1–3). Many of these mutations are within the RBD receptor binding motif (RBM), a hypervariable loop that mediates most of the ACE2 contacts (2, 3). The RBD is the primary target of neutralizing antibodies in naturally acquired or vaccine-elicited humoral immunity (4, 5). The spike protein N-terminal domain (NTD) is also a target of neutralizing antibodies, and VOCs have NTD mutations that include deletions at an antigenic supersite for neutralizing antibody binding (6, 7). The effects of spike protein mutations on immune responses (8–13) make it important to monitor viral variants.
While previously studied VOCs contain one to three RBD mutations that at times overlap (1), the potential for composite variants is being closely monitored. For example, the B.1.617.2 (Delta) variant can acquire the K417NRBD mutation (Lys417→Asn) found in the B.1.351 (Beta) variant, generating the Delta AY.2 variant, for a total of three RBD mutations (Fig. 1A). Similarly, as shown in recently deposited sequences from samples collected in Angola, the Beta variant can acquire the L452RRBD mutation found in the Delta and B.1.429/427 (Epsilon) variants, for a total of four RBD mutations (Fig. 1A and table S1). Further complicating matters, variant monitoring efforts are still undersampling viral evolution. For example, a virus recently sequenced from travelers returning from Tanzania contained a previously undocumented combination of RBD mutations (E484KRBD, T478RRBD, and R346KRBD) with NTD deletions that would likely alter the spike protein antigenic surface and result in antibody escape (table S1).
Fig. 1. Structure of intrahost evolved RBD bound to human ACE2.
(A) Key RBD substitutions discussed in the text and the SARS-CoV-2 variants that contain them. Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr. (B) Day 146* RBD–ACE2 ectodomain x-ray crystal structure. RBD residues that are mutated in variants discussed in the text are shown. Boxed residues are mutated in the day 146* RBD as compared with the Wuhan-Hu-1 (wild-type) SARS-CoV-2 RBD. The Delta +3 variant contains an additional RBD mutation that is not shown in the schematic diagram (see table S2). (C) Wild-type RBD–ACE2 contacts near N501RBD [Protein Data Bank (PDB) ID 6M0J] (2). (D) Day 146* RBD contacts near Y501RBD. (E) Wild-type SARS-CoV-2 RBD–ACE2 interactions near Q493RBD. (F) Day 146* RBD interactions near K493RBD. (G) Cryo-EM structure of the SARS-CoV-2 RBD bound to the C1C-A3 antibody Fab. RBD residues discussed in the text are labeled. LC, light chain; HC, heavy chain.
Here, we investigate the structural plasticity of the SARS-CoV-2 spike protein RBD and its capacity to evade neutralizing antibodies.
Results
Structure of an evolved receptor binding domain–ACE2 complex
We previously generated two SARS-CoV-2 spike proteins that each contain six RBD changes that were detected during persistent infection of an immunocompromised individual infected with a SARS-CoV-2 strain containing the D614GS mutation (14–16). This individual received treatment with REGN-COV2 (17, 18), but several of the RBD substitutions had occurred even before administration of this therapeutic antibody cocktail (14–16). Lentivirus pseudotypes bearing these spike proteins, denoted day 146* and day 152* (Fig. 1A and table S2), were refractory to neutralization by VH3-53 heavy chain gene–derived neutralizing antibodies, a potent class of neutralizing antibodies that have been repeatedly isolated from convalescent donors (19–25). These pseudotypes were also resistant to neutralization by components of REGN-COV2 (17, 18) and by polyclonal immunoglobulin G (IgG) purified from the serum of COVID-19 convalescent donors (14). Substitutions in the day 146* and day 152* spike proteins, noted in samples sequenced from this individual in the spring and summer of 2020, foreshadowed those in currently circulating VOCs at three positions: N501RBD, E484RBD, and T478RBD (Fig. 1A). The day 146* and day 152* spike proteins also contain substitutions that are not in current dominant strains but could have serious effects if acquired. For example, the S494PRBD substitution is a therapeutic antibody (LY-CoV555) escape mutation (26) that, as of 27 September 2021, was present in more than 12,000 human-derived SARS-CoV-2 sequences on public research databases (GISAID) (27). Additionally, the Q493KRBD mutation, which is found in more than 100 human-derived SARS-CoV-2 sequences on GISAID as of 27 September 2021, confers resistance to multiple therapeutic antibodies [REGN10933, CB6 (LY-CoV016), and LY-CoV555] and VH3-53 gene–derived antibodies (14, 16, 17, 28).
To determine the impact of their combined mutations on human ACE2 binding, we generated recombinant RBDs for the day 146* and day 152* spike protein mutants. The affinity of the day 152* mutant monomeric RBD for the monomeric ACE2 ectodomain was substantially lower (binding affinity, Kd, of 2.4 μM) than that of wild-type (Wuhan-Hu-1) RBD (54 nM, consistent with other reports) (3, 29), suggesting that its mutations compromise ACE2 binding (fig. S1 and table S3). For comparison, the affinity we measured of the SARS-CoV RBD for human ACE2 was 0.26 μM, about ninefold higher than the affinity for the day 152* RBD (fig. S1 and table S3). The affinity of the day 152* RBD for ACE2 is comparable to that of the RBDs of some bat coronaviruses that are closely related to SARS-CoV-2 and bind human ACE2 (e.g., RaTG13 virus RBD affinity of 3.9 μM) (30). The day 146* RBD, however, had a similar affinity (Kd of 46 nM) for ACE2 as that of the Wuhan-Hu-1 SARS-CoV-2 RBD (fig. S1 and table S3).
We determined the x-ray crystal structure of the day 146* RBD bound to the human ACE2 ectodomain (Fig. 1B, fig. S2, and table S4). This structure is similar to previously determined structures of ACE2–SARS-CoV-2 RBD complexes (2, 3), except we observed contacts between two N-linked glycans on ACE2 (attached to N53ACE2 and N90ACE2) and the RBD (fig. S3). Removing the N90ACE2 glycan, which interacts with the RBD in both copies of the crystal asymmetric unit (fig. S3), increased Wuhan-Hu-1 SARS-CoV-2 and day 146* RBD affinity for ACE2, although the effect was modest (fig. S1 and table S3). This finding is consistent with prior work implicating the N90ACE2 glycan, which is removed in a human polymorphism (T92IACE2), as a barrier to SARS-CoV-2 RBD binding to ACE2 (31, 32).
The N501YRBD substitution is found in multiple VOCs (Fig. 1A); once it surfaced in the immunocompromised individual, it was retained at later time points (14–16). As also shown in a cryo–electron microscopy (cryo-EM) structure of the SARS-CoV-2 spike protein containing the N501YRBD substitution bound to ACE2 (33), the side chain of Y501RBD interacts with Y41ACE2 and K353ACE2 with no notable structural change (Fig. 1, C and D). E484RBD is a critical target of antibodies against SARS-CoV-2 and is mutated in several variants (12, 34, 35). In structures of Wuhan-Hu-1 SARS-CoV-2 RBD bound to ACE2, E484RBD is near but does not directly contact the receptor (Fig. 1E). In the day 146* RBD–ACE2 complex structure, the K493RBD side chain reaches over the RBD surface to recruit the E484RBD side chain to form a new salt bridge with K31ACE2 (Fig. 1F). The nearby Y489HRBD mutation, which removes a polar contact with ACE2, better accommodates repositioning of E484RBD because the histidine is smaller than the tyrosine side chain and would avoid potential steric clashes with E484RBD in this binding mode (Fig. 1, E and F). A second rotamer for residue H34ACE2 forms additional RBD contacts to fill a gap created by the reorganization of local interactions (Fig. 1, E and F). This structural plasticity may explain how the RBD tolerates an unexpectedly large number of mutations during intrahost evolution yet retains the ability to bind ACE2 tightly. It is also consistent with the large sequence divergence in the RBD residues that contact ACE2 among SARS-related coronaviruses that share this cellular receptor.
Neutralization escape of therapeutic antibodies
RBD-targeting antibodies can be categorized into classes on the basis of whether they bind an overlapping footprint with ACE2 and recognize only an open or both an open and a closed RBD on the spike protein trimer (36). CB6 (equivalent to LY-CoV016 or etesevimab) is a class 1, VH3-66–derived antibody that blocks ACE2 binding and can only bind the RBD when it is open, and LY-CoV555 (bamlanivimab) is a class 2 antibody that blocks ACE2 binding but can bind both open and closed RBDs (21, 37). LY-CoV016 and LY-CoV555 are used together as a cocktail and bind epitopes that partially overlap on the RBM such that both cannot bind simultaneously (21, 37). REGN10933 is a class 1 antibody, and REGN10987 is a class 3 antibody that sterically blocks ACE2 binding but binds the RBM outside the main ACE2 binding site; both are used as a cocktail (REGN-COV2) (17, 18).
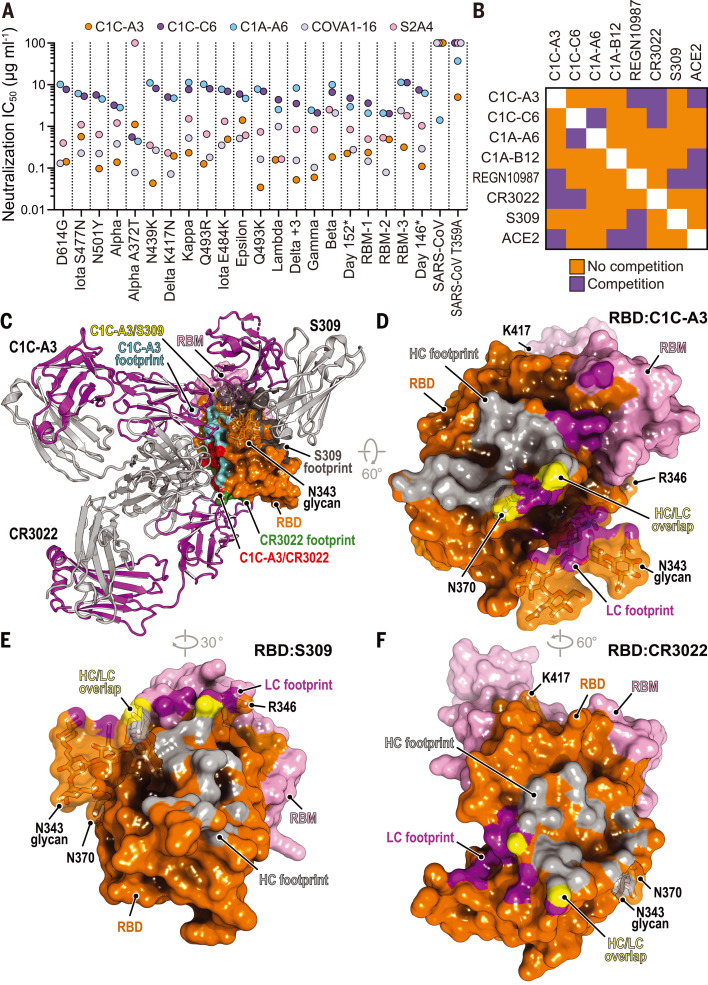
Structural plasticity at the RBD–ACE2 interface suggests that the RBD could tolerate many more mutations than found in current VOCs. We next generated pseudotypes for spike protein variants that contain composite mutations. The Delta variant, which contains the L452RRBD and T478KRBD substitutions, has become a dominant strain across the globe (38). We generated a pseudotype for the Delta AY.2 variant, which contains the K417NRBD mutation that is usually found in the Beta variant, and a Delta variant containing the N501YRBD, E484KRBD, and F490SRBD mutations usually found in the Beta, P.1 (Gamma), and C.37 (Lambda) variants (referred to here as “Delta +3”) (Fig. 1A and tables S1 and S2). The set of RBD mutations for the latter strain occurred in deposited sequences from samples collected in Turkey (table S1). We also generated pseudotypes in which we combined spike protein substitutions detected in the immunocompromised host with mutations found in the Beta variant, which we chose because this VOC is highly resistant to antibody neutralization (10, 12, 39). Starting with a day 146* spike protein sequence, which contains an NTD deletion, we incorporated either one (E484KRBD) or two (E484KRBD and K417NRBD) additional substitutions; these are referred to as receptor binding mutant-1 (RBM-1) and RBM-2, respectively (Fig. 1A and table S2). Additionally, starting with the Beta variant spike protein sequence, we generated a variant pseudotype that contains two additional mutations associated with immune evasion (L452RRBD and N439KRBD) (40, 41). This pseudotype is referred to as RBM-3 (Fig. 1A and table S2). An ACE2-Fc fusion protein neutralized RBM-1, RBM-2, and RBM-3 pseudotypes, suggesting that all entered cells by binding ACE2 (Fig. 2B and fig. S4A).
Fig. 2. Neutralization escape from therapeutic antibodies and mRNA vaccine–elicited serum.
(A) Summary of neutralization IC50 values for lentivirus pseudotypes with the indicated monoclonal antibodies. (B) Tabulated IC50 values for lentivirus pseudotypes with the indicated monoclonal antibodies and an ACE2-Fc fusion protein (ACE2). (C) Mean ID50 neutralization titers for the indicated variant pseudotypes at the time of the second immunization but before vaccination (“dose 1”), or 28 days after second immunization (“dose 2”) with mRNA-1273 or BNT162b2. The fold change of the mean ID50 neutralization titer with respect to D614GS pseudotype is shown in each panel. Each experiment was performed twice independently in triplicate (n = 6). Wilcoxon matched-pairs signed rank test; ****P < 0.0001. (D) Tabulated fold change of mean ID50 neutralization titers for the indicated pseudotypes as compared with D614GS pseudotype.
We tested the activity of therapeutic antibodies against Delta AY.2, Delta +3, RBM-1, RBM-2, RBM-3, and additional variant pseudotypes with known resistance profiles to serve as comparators in the same assay (Fig. 2, A and B, and fig. S4A). LY-CoV555 was the most affected by escape mutations, followed by CB6 (from which LY-CoV016 is derived) (Fig. 2, A and B, and fig. S4A). The Q493KRBD mutation conferred absolute resistance to LY-CoV555, generated 80-fold resistance to CB6, and also compromised REGN10933 activity, consistent with previous reports (Fig. 2, A and B, and fig. S4A) (14, 16, 17, 26). In addition to the expected loss of activity of LY-CoV555 and CB6 against Beta and Gamma variants (9, 11, 12, 42), LY-CoV555 and CB6 lost all activity against day 146*, day 152*, RBM-1, RBM-2, and RBM-3 pseudotypes (Fig. 2, A and B, and fig. S4A). Whereas the Delta variant is known to resist neutralization by LY-CoV555 but retain sensitivity to neutralization by CB6/LY-CoV016 (38), the Delta AY.2 pseudotype was resistant to both agents (Fig. 2, A and B, and fig. S4A). This is expected because CB6/LY-CoV016 is derived from a VH3-66 antibody (21), and the additional mutation the Delta AY.2 variant contains with respect to Delta (K417NRBD) confers resistance to CB6/LY-CoV16 and other members of the VH3-53 and VH3-66 class of neutralizing antibodies (9, 14, 16, 26, 43). The Delta +3 pseudotype, which despite containing six RBD mutations does not contain the K417NRBD substitution, only escaped neutralization by LY-CoV555 (Fig. 2, A and B; fig. S4A; and table S2). Although the distribution of LY-CoV016 and LY-CoV555 was paused in the United States in the summer of 2021 as the prevalence of Gamma and Beta VOCs increased, the distribution of this antibody cocktail has since been resumed with the rise of Delta as the predominant strain. However, our findings emphasize the importance of close monitoring of Delta AY.2 and of other Delta variants for acquisition of the K417NRBD mutation.
Although REGN10933 lost substantial activity against the Beta variant, which is consistent with other reports (9, 12, 42), it still had a median inhibitory concentration (IC50) value of <1 μg ml−1 in our assays (Fig. 2, A and B, and fig. S4A). However, resistance markedly worsened with the day 146*, day 152*, RBM-1, RBM-2, and RBM-3 pseudotypes, with 800- to 1900-fold loss of neutralizing activity (IC50 values ranging from 20 to 47 μg ml−1). REGN10987 potently neutralized many of the variant pseudotypes we examined. While we observed the expected resistance to REGN10987 neutralization by variants containing the N439KRBD or the adjacent N440DRBD substitutions (14, 16), we also observed some loss of activity against Epsilon and B.1.617.1 (Kappa), which was not expected because none of their substitutions fall within the REGN10987 RBD footprint (Fig. 1A and Movie 1). Nonetheless, other reports have also noted varying degrees of modest in vitro resistance of Epsilon and Kappa variants to REGN10987 neutralization (39, 42). Notably, the day 146* and RBM-3 pseudotypes were the only ones to gain resistance to both antibodies in REGN-COV2, because they contain substitutions in the REGN10933 (e.g., Q493KRBD, or E484KRBD and K417NRBD) and the REGN10987 binding sites (N439KRBD or N440DRBD) (Fig. 2, A and B; fig. S4A; and Movie 1) (14). We observed on GISAID instances of “day 146*–like” viruses that would be expected to resist neutralization by LY-CoV555, LY-CoV016, REGN10933, and REGN10987, because they contain the Q493KRBD and N439KRBD substitutions. One strain contains the N501YRBD, Q493KRBD, and N439KRBD mutations (sequenced once in South Africa), and the other contains the N501YRBD, Q493KRBD, L452RRBD, N439KRBD, and N440FRBD mutations (sequenced once in the United Kingdom) (table S1).
Movie 1. Antibody footprints on an evolving SARS-CoV-2 RBD.
Antibodies are classified according to Barnes et al. (36). PDB IDs are listed in parentheses. Key RBD residues discussed in the main text are highlighted.
The broadly neutralizing antibody S309 (44), a class 3 antibody that binds the RBD but does not interfere with ACE2 binding and from which the therapeutic antibody sotrovimab is derived, was active against all variants we tested (fig. S4A). However, we could not calculate reliable neutralization IC50 values because of variable non-neutralizable pseudotype fractions (fig. S4A). The presence of a non-neutralizable fraction is unexplained but has been noted in other reports when human cells overexpressing ACE2, as opposed to African green monkey (Vero) target cells, were used to examine S309 neutralizing activity (45, 46).
Neutralization escape of mRNA vaccine recipient sera
Messenger RNA (mRNA)–based vaccines encoding the SARS-CoV-2 spike protein elicit robust neutralizing antibody responses (47–49). We directly compared immune evasion of the day 146*, day 152*, and RBM-2 pseudotypes to the B.1.1.7 (Alpha), Beta, and Gamma pseudotypes in sera obtained from individuals who had received a two-dose series of an mRNA vaccine (BNT162b2 or mRNA-1273) (Fig. 2, C and D, and fig. S5). In addition to RBD substitutions, day 146*, day 152*, RBM-1, and RBM-2 spike proteins all contain NTD deletions spanning residues 141 to 144, which are positioned near NTD mutations in Alpha, Beta, and Delta in a key antigenic supersite (table S2) (6, 7). As similar NTD deletions found in Alpha, Beta, and Delta prevent binding of some neutralizing antibodies (6, 7, 46), they would be expected to escape neutralization by some NTD-targeting antibodies in addition to escaping neutralization by RBD-targeting antibodies. After initial immunization but before the second dose, we observed a loss in neutralizing activity for all variants, although the severity of this loss varied. Variants that contain any substitution at E484RBD combined with an NTD deletion (Beta, day 152*, and RBM-2) were more effective at evading antibody responses than variants that had an E484RBD substitution without an NTD deletion (Gamma) or an NTD deletion but no E484RBD substitution (day 146*) (Fig. 2, C and D; fig. S5; and table S2). These findings are consistent with the role of E484RBD as a major driver in neutralization escape of polyclonal antibody responses to SARS-CoV-2 (35) and observations that Beta more robustly escapes antibody neutralization than Gamma (9, 13). They further suggest that variants that have an NTD supersite deletion and an E484RBD substitution are the most concerning when it comes to resistance to polyclonal antibodies.
One-quarter of sampled individuals had no detectable activity against the Beta and RBM-2 pseudotypes after a single immunization (Fig. 2, C and D). However, sampling at 7 and 28 days after the second immunization revealed detectable neutralizing activity against all variants in all vaccine recipients, including against the RBM-2 pseudotype, which contains seven RBD mutations (Fig. 2, C and D, and fig. S5). Thus, repeated administration of an mRNA vaccine encoding constructs of the SARS-CoV-2 spike protein used in current formulations may provide sufficient neutralizing antibody breadth and potency to yield baseline serum neutralizing activity against variants that are more extensively mutated than the current dominant strains.
Identification of SARS-CoV cross-reactive antibodies
The RBD is also the major target of neutralizing antibodies against SARS-CoV, which caused a small outbreak of viral pneumonia from 2003 to 2004, although with a much higher case fatality rate (50, 51). Polyclonal antibody responses against SARS-CoV-2 poorly cross-neutralize SARS-CoV (52, 53). To identify barriers that restrict neutralization breadth, we performed single memory B cell sorting with the SARS-CoV spike protein to mine the memory B cell repertoire of a COVID-19 convalescent individual (“C1”). Polyclonal IgG from C1 plasma neutralized SARS-CoV-2 pseudotype but had weak activity against SARS-CoV pseudotype (fig. S6A). From C1 peripheral blood mononuclear cells, using a prefusion stabilized SARS-CoV spike protein (S2P) (54), we cloned 17 cross-reactive antibodies. Of these, 11 antibodies bound both the SARS-CoV and the SARS-CoV-2 spike protein in an enzyme-linked immunosorbent assay (ELISA) (fig. S6C and table S5). Only two RBD-binding antibodies, C1C-A3 (“A3”) and C1C-C6 (“C6”), neutralized SARS-CoV-2 pseudotypes in our assays (Figs. 2B and 3A and fig. S6F). Despite binding to the SARS-CoV spike protein and RBD by ELISA, A3 and C6 did not neutralize SARS-CoV pseudotype (fig. S6, F and G). We also included C1A-A6 (“A6”) in these assays, a SARS-CoV-2 neutralizing antibody we previously isolated from the C1 donor using prefusion stabilized SARS-CoV-2 S2P in single B cell sorting experiments (14). Unlike A3 and C6, A6 neutralized SARS-CoV pseudotypes (Figs. 2B and 3A and fig. S6F). We determined Fab RBD binding affinities using biolayer interferometry (BLI) (fig. S7 and table S3) and confirmed A3 and A6 activity against infectious SARS-CoV-2 in a plaque reduction neutralization assay (fig. S4B).
Fig. 3. Neutralization of SARS-CoV-2 variants by an RBD core–targeting antibody.
(A) Summary of neutralization IC50 values for pseudotypes and the indicated antibodies. (B) Summary of the results of BLI-based competition assays. (C) Superposition of the CR3022 (PDB ID 6W41) (55) and S309 (PDB ID 6WPS) (44) structures onto the C1C-A3–bound RBD structure. Antibody Fabs are shown as ribbon diagrams, and the RBD is shown in surface representation. Antibody footprints are shown on the RBD surface. (D) RBD footprint of C1C-A3. (E) RBD footprint of S309 (PDB ID 6WPS) (44). (F) RBD footprint of CR3022 (PDB ID 6W41) (55). In panels (D) to (F), key RBD residues discussed in the main text are highlighted.
A3 neutralized almost all SARS-CoV-2 variant pseudotypes with a neutralization IC50 value of <1 μg ml−1, including Beta, Gamma, Delta AY.2, Delta +3, RBM-1, RBM-2, and RBM-3 pseudotypes; the Epsilon variant was the only exception, with an IC50 value of 1.9 μg ml−1 (Figs. 2B and 3A and fig. S4A). C6 and A6 also broadly neutralized variants, but with higher baseline IC50 values, even against D614GS pseudotypes (ranging from 2.0 to 11.4 μg ml−1) (Figs. 2B and 3A and fig. S4A).
To determine where on the RBD A3, C6, and A6 bind, we carried out competition studies with C1A-B12 (14), a class 1 antibody; REGN10987 (17, 18) and S309 (44), two class 3 antibodies; and CR3022 (55), a class 4 antibody (Fig. 3B, fig. S8, and Movie 1). A3 competed with CR3022 and REGN10987 for RBD binding, C6 competed with CR3022, and C6 and A6 competed with each other (Fig. 3B and fig. S8). A6 did not compete with any of the other antibodies tested. Among A3, C6, and A6, only A3 competed with binding of an ACE2-Fc fusion protein, suggesting that A3 blocks cellular attachment.
Antibody C1C-A3 binds the conserved RBD core
We determined the 3.1-Å cryo-EM structure of the A3 Fab bound to the SARS-CoV-2 spike protein ectodomain (Fig. 4A, figs. S9 and S10, and table S6). A3 binds the RBD core with the spike protein trapped in the three open RBD conformation (Fig. 4A). In agreement with competition assays (Fig. 3B), A3 interacts with RBD residues that overlap significantly with those of CR3022 (Fig. 3, C, D, and F, and Movie 1). A3 is therefore a class 4 antibody, a class that includes CR3022, S2A4, S304, S2X35, H014, COVA1-16, S2X259, and DH1047 (4, 56–59) (Movie 1 and fig. S11). Although the A3 and S309 footprints on the RBD do not overlap, and S309 (a class 3 antibody) can bind the closed spike protein trimer (44), both antibodies contact the N-linked glycan attached to N343RBD but approach it from different faces (Fig. 3C and Movie 1).
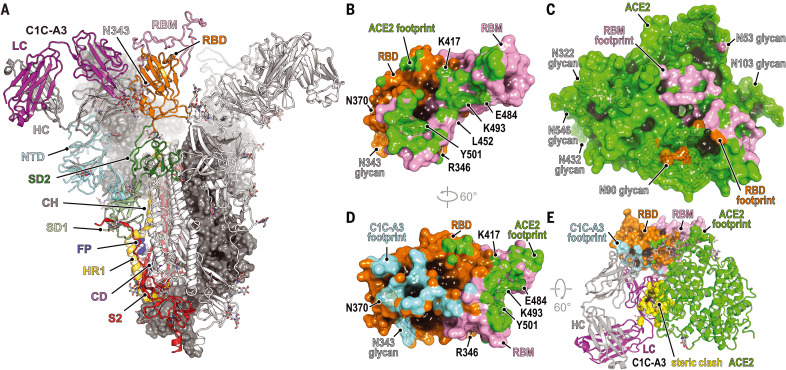
Fig. 4. Structural basis for C1C-A3 neutralization.
(A) Cryo-EM structure of the C1C-A3–Fab SARS-CoV-2 spike protein complex. Two of the three spike protein protomers are shown in surface representation. One protomer is shown as a ribbon diagram with labeled subdomains. The trimer model shown was generated by superposition of an RBD–C1C-A3 Fab model generated by subparticle classification of the RBD region onto the coordinates of the trimeric spike protein–C1C-A3 Fab complex (see materials and methods). SD1, subdomain 1; SD2, subdomain 2; FP, fusion peptide; HR1, heptad repeat 1; CD, connector domain; S2, additional portions of S2 subunit. (B) Surface representation of the SARS-CoV-2 day 146* RBD showing the ACE2 footprint, including surfaces contacted by ACE2 N-linked glycans. Key RBD positions discussed in the text are labeled. (C) Surface representation of ACE2, showing the day 146* RBD and RBM footprints. (D) Surface representation of the RBD highlighting C1C-A3 Fab and ACE2 footprints. (E) Overlay of the C1C-A3 Fab–RBD complex with the day 146* RBD–ACE2 complex. Atoms within 1.54 Å of each other are shown in yellow surface representation to highlight steric clashes. Key RBD residues discussed in the text are labeled in (B) and (D).
The A3 Fab avoids the RBD–ACE2 interface, which contains the majority of key antibody escape mutations, but, like other class 4 antibodies, nonetheless binds the RBD in a manner that would sterically interfere with ACE2 binding (Fig. 4, B to E, and fig. S11). On the basis of its epitope, in addition to retaining activity against all variants we tested, A3 would also have activity against emergent and preemergent SARS-CoV-2 variants; these include a variant sequenced from travelers from Tanzania that contains the E484KRBD, T478RRBD, and R346KRBD mutations, and B.1.621 (Mu), a variant detected early in 2021 in Colombia that has since spread internationally and contains the E484KRBD, N501YRBD, and R346KRBD mutations (Figs. 1G and 4D and table S1). The R346KRBD mutation falls within the RBD core and is in the S309 binding site but is not within A3’s footprint (Fig. 3, D and E, and Movie 1). However, S309 would likely retain activity against SARS-CoV-2 variants that contain the R346KRBD mutation, as the residue that is at the position analogous to SARS-CoV R346RBD is a lysine in SARS-CoV, and S309 neutralizes both SARS-CoV and SARS-CoV-2 (44, 60).
RBD core glycan addition drives neutralization escape
Despite A3’s breadth against SARS-CoV-2 variant pseudotypes (Figs. 2B and 3A), A3 does not neutralize SARS-CoV pseudotype (fig. S6, F and G). The A3 epitope is highly conserved between SARS-CoV-2 and SARS-CoV; however, N370RBD is a site of N-linked glycosylation in SARS-CoV (N357RBD in SARS-CoV numbering) and in animal coronaviruses but not in SARS-CoV-2 (Fig. 5, A to C and F) (61). An N-linked glycan attached to SARS-CoV-2 N370RBD would introduce steric clashes with the A3 antibody heavy and light chains (Fig. 5D). Furthermore, calculations of antibody-accessible surface areas using molecular dynamic trajectories of a fully glycosylated SARS-CoV-2 spike protein with a modeled N370RBD glycan suggest that its addition would restrict A3 epitope access and could also affect binding of other class 4 antibodies (fig. S12) (61, 62).
Fig. 5. Structural basis for immune evasion of a RBD core–targeting antibody.
(A and B) C1C-A3 antibody contacts with the SARS-CoV-2 RBD core. (C) C1C-A3 contacts with the N343RBD glycan with structural superposition of the SARS-CoV RBD (PDB ID 6NB6) (78). N-linked glycans found at N330RBD and N357RBD in SARS-CoV and the analogous N343RBD and N370RBD positions in SARS-CoV-2 are highlighted. (D) Superposition of the C1C-A3 Fab–SARS-CoV-2 RBD structure with the SARS-CoV RBD (PDB ID 6NB6) (78) showing that a glycan attached at SARS-CoV N357RBD may interfere with antibody binding. The SARS-CoV-2 RBD is not shown for clarity. (E) Superposition of the C1C-A3–SARS-CoV-2 RBD with the RaTG13 virus RBD (PDB ID 7CN4) (79) showing that a glycan attached at RaTG13 virus N370RBD would be more readily accommodated because the helix that contains it would be rotated away from the Fab. The SARS-CoV-2 RBD is omitted for clarity. (F) Sequence alignment of the RBD core region contacted by C1C-A3. SARS-CoV-2 numbering is shown at the top of the alignment, and SARS-CoV numbering is shown at the bottom. Circles indicate antibody contacts. (G) C1C-A3 neutralization curves for the indicated lentivirus pseudotypes. Data are plotted as the mean ± standard deviation of the mean. The experiment was performed twice in triplicate (n = 6). For some data points, error bars are smaller than symbols. (H) Tabulated neutralization IC50 values for the indicated pseudotypes.
Partial occupancy of the glycan attached to SARS-CoV N357RBD in recombinant protein preparations may explain why we observed spike protein and RBD binding but a lack of SARS-CoV pseudotype neutralization (fig. S6, F and G). In surface plasmon resonance binding assays, A3 IgG bound tightly to the SARS-CoV RBD only when the RBD was enzymatically deglycosylated (fig. S13 and table S7). Consistent with the role of the SARS-CoV N357RBD N-linked glycan as a barrier to A3 neutralization, introducing a substitution that would prevent its addition (T359ARBD) sensitized SARS-CoV pseudotypes to A3 neutralization (IC50 value of 5 μg ml−1) (Fig. 2B and fig. S4A).
The A372S/TRBD mutations, which would introduce an N-linked glycosylation motif and allow for modification of N370RBD in the SARS-CoV-2 spike protein, are found in human-derived SARS-CoV-2 sequences (GISAID) (27), including on sequences for VOCs Alpha and Delta, without apparent geographic restriction (48 sequence counts as of 10 October 2021 and detected in at least 14 countries) (Fig. 5F and table S8). Although the mutations are currently rare, their presence in sequence databases suggests that SARS-CoV-2 strains containing these mutations can replicate in humans. To confirm that an N-linked glycan could be added to N370RBD, we conducted glycan analysis on recombinant SARS-CoV-2 RBD containing the A372SRBD substitution and observed 90% occupancy of an N-linked glycan at position N370RBD (fig. S13B).
Because acquisition of a putative N-linked glycan at N370RBD was the most frequent on the Alpha variant at the time of our initial analysis, we generated an Alpha pseudotype that contains the A372TRBD substitution (Alpha A372T). We tested the effect of this substitution on three class 4 antibodies: A3, the antibody we isolated here; S2A4, an antibody that does not cross-react with the SARS-CoV RBD (4); and COVA1-16, an antibody that has weak cross-neutralizing activity against SARS-CoV (57). The mutation resulted in eightfold greater resistance to A3 neutralization (IC50 value of 1.1 μg ml−1, as compared with 0.14 μg ml−1 with the Alpha pseudotype) and complete resistance to S2A4 neutralization (Figs. 2B, 3A, and 5, G and H, and fig. S4A). S2A4 and COVA1-16 neutralized variants with potency that was overall comparable to A3 in most cases (Figs. 2B and 3A and fig. S4A). COVA1-16, probably because it has some activity against SARS-CoV [above the limit of detection in our assays, but 29 μg ml−1 as reported by Liu et al. (57)], retained activity against Alpha A372T pseudotype (Figs. 2B, 3A, and 5H, and fig. S4A). The Fab binding pose of certain class 4 antibodies, therefore, may allow them to avoid steric hindrance from an N-linked glycan attached to N370RBD (S2X259 is one such antibody) (Movie 1) (56).
Antibody C1C-A3 neutralizes a related coronavirus
Coronaviruses that circulate in animals and have spike protein RBDs that can bind human ACE2 are a continued threat. RaTG13 virus, which is closely related to SARS-CoV-2 phylogenetically, is one example (63). The RaTG13 virus spike protein contains a threonine at RBD position 372, which would allow for N370RBD glycosylation (Fig. 5F). Despite the presence of the N-linked glycan, A3 potently neutralized RaTG13 virus pseudotype (neutralization IC50 value of 21 ng ml−1), suggesting that A3 neutralization breadth extends to preemergent coronaviruses that are closely related to SARS-CoV-2 (Fig. 5, G and H). Structural superposition reveals that the N370RBD glycan on the RaTG13 RBD is positioned in a manner that may not block A3 epitope access but could interfere with binding of other antibodies that bind nearby epitopes on the RBD core (Fig. 5E).
Discussion
As variants containing composite mutations begin to emerge, continued SARS-CoV-2 immune evasion will remain a big concern. We found that accumulation of large numbers of RBD mutations, which mimics accelerated spike protein evolution occurring in a persistently infected immunocompromised host (14–16), is facilitated by structural plasticity at the ACE2–RBD interface (Fig. 1, B to F). The severity of the phenotypes we observed in vitro suggests that further evolved variants will more adeptly escape therapeutic antibody neutralization than currently circulating VOCs, with potential resistance to two-component antibody cocktails (Fig. 2, A and B).
After two mRNA vaccine immunizations and as early as 7 days after the second dose, all mRNA vaccine recipients had detectable neutralizing activity against pseudotypes containing an NTD supersite deletion and RBDs with six to seven mutations (e.g., day 146*, day 152*, and RBM-2), with mean neutralization ID50 values decreased by 2.3- to 6.1-fold (Fig. 2, C and D, and fig. S5). While the precise epitopes targeted by this residual vaccine-elicited serum neutralizing activity remain to be determined, we surmise that antibodies targeting the RBD core (e.g., those that bind away from the RBM) at least in part account for some of this activity. As the RBD is a major target of vaccine-elicited and naturally acquired humoral immunity to SARS-CoV-2 (4, 5), and the RBM is a critical site of potent neutralizing antibody binding (19, 21–25, 64) that is the most antibody-accessible and the least masked by glycan and conformational shielding (fig. S12), continued RBM evolution may guide antibody responses toward more conserved neutralizing epitopes on the RBD core.
We mined genome sequences in the GISAID database for substitutions that would introduce additional N-linked glycans onto the RBD. This analysis identified D364NRBD as an additional mutation that would introduce a putative N-linked glycosylation site in a surface-exposed loop in the footprint of some class 4 antibodies (Movie 1). The independent acquisition of N-linked glycosylation sites (through the A372S/TRBD and D364NRBD substitutions) on the same surface of the RBD core, but not on other RBD sites, suggests that this region may be a target of immune selective pressure.
While glycan addition may allow neutralization escape, this change could come at a cost to viral fitness and infectivity. Indeed, the A1114G:T372A mutation that removed the glycan in the SARS-CoV-2 RBD appeared under selective pressure, and addition of the glycan decreases viral replication in human lung epithelial (Calu-3) cells by more than 60-fold (65). A recent molecular dynamics study suggests that introducing the glycan at N370RBD in SARS-CoV-2 would favor the closed conformation with the N370RBD glycan stabilizing the closed RBD structure on the trimeric spike protein (66). A lack of a glycan at position N370RBD, therefore, may increase SARS-CoV-2 ACE2-binding and infectivity by favoring the open state but may also make SARS-CoV-2 more vulnerable to neutralizing antibodies that can only bind the RBD in the open conformation.
Although addition of the N370RBD glycan may be associated with a cost to viral fitness, should the selective immune pressure be considerable at this site over a long enough time scale, this may also afford the virus an opportunity to acquire permissive secondary mutations during evolution that restore viral fitness, as is observed in influenza virus drug resistance (67). Such compensatory mutations would be ones that promote ACE2 binding and RBD opening; for example, the D614GS mutation (68), which favors the open conformation, and the N501YRBD mutation, which introduces more favorable interactions with ACE2 (Fig. 1D).
As parts of the world continue to face waves of infection and mitigation strategies are relaxed, viral replication in human hosts under antibody selective pressure will continue to shape the antigenic landscape of the SARS-CoV-2 spike protein. With vigorous variant monitoring efforts underway to help design next-generation antibody-based therapeutics, and with mRNA- or DNA-based vaccines that can be updated to rapidly adapt to new variants, proactively examining the consequences of further viral evolution before the next highly antibody resistant strain emerges is of utmost importance.
Materials and methods summary
We isolated monoclonal antibodies from the blood of a COVID-19 convalescent individual using single B cell sorting with prefusion-stabilized SARS-CoV spike protein ectodomain as bait and using established protocols (14, 54). We obtained venous blood samples from healthy mRNA-1273 and BNT162b2 vaccine recipients. We produced recombinant glycoproteins and antibodies or Fabs in transiently transfected mammalian cells grown in suspension culture and purified these proteins using affinity-based methods. We used ELISAs to measure antibody binding and BLI or surface plasmon resonance to determine kinetic parameters of binding. We packaged lentivirus pseudotypes by transient transfection of HEK293T cells, as previously described (14). We used HEK293T cells expressing human ACE2 in pseudotype neutralization assays or Vero E6 cells in plaque reduction neutralization tests, as previously described (14). We collected x-ray diffraction data on crystals of a day 146*–SARS-CoV-2 RBD complex at the Advanced Photon Source (APS, Argonne, IL) NE-CAT beamline and used established procedures for data processing, molecular replacement, atomic model building, and refinement (69–73). We used mass spectrometry to perform glycopeptide analysis. After data collection on a Titan Krios cryo–electron microscope equipped with a Gatan K3 camera, we used single-particle cryo-EM to determine the structure of a prefusion-stabilized SARS-CoV-2 spike protein ectodomain (7) complexed with C1C-A3 Fab complex using established procedures for image processing, atomic model building, and refinement (72–77).
Supplementary Material
Acknowledgments
This work is based on research conducted at the Northeastern Collaborative Access Team (NE-CAT) beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P30 GM124165). The Pilatus 6M detector on 24-ID-C beamline is funded by a NIH-ORIP HEI grant (S10 RR029205). This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract DE-AC02-06CH11357. We thank the staff at NE-CAT for assistance with x-ray data collection. Cryo-EM data were collected at the Harvard Cryo-Electron Microscopy Center for Structural Biology. We thank Harvard Cryo-EM Center staff members for assistance with cryo-EM data collection. We acknowledge support for COVID-19–related structural biology research at Harvard from the Nancy Lurie Marks Family Foundation. We thank B. Chen for providing a human ACE2 ectodomain–expressing cell line, and H. Choe for providing cDNA encoding the SARS-CoV spike protein.
Funding: This work was funded in part by the Massachusetts Consortium on Pathogen Readiness (MassCPR) (J.A.). J.A. was also supported by the Star-Friedman Award Challenge for Promising Scientific Research. L.R.B. is supported by the Harvard Clinical and Translational Science Center, from the National Center for Advancing Translational Science (1UL1TR002541-01); by Barbara and Amos Hostetter; and by the Chleck Family Foundation. S.E.T., R.C.L., E.T.R., and R.C.C. receive grant support from the Centers for Disease Control and Prevention (U01CK000490). K.G.N., A.C., and H.H.V. are supported by award T32GM007753 from the National Institute of General Medical Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Author contributions: Conceptualization: K.G.N., S.A.C., S.S., J.P., L.E.C., P.Y., A.Co., L.G.A.M., N.V.T., M.D., S.E.T., S.K., A.C.S., A.D., C.T., J.F.C.-S., A.T.D., D.T., A.Cl., Y.F., A.G., I.R.C., J.S., L.R.B., R.C.C., and J.A. Methodology: K.G.N., S.A.C., S.S., J.P., L.E.C., P.Y., A.Co., L.G.A.M., H.H.V., N.V.T., G.Z., M.D., S.E.T., S.K., A.C.S., A.D., R.C.L., E.T.R., C.T., J.F.C.-S., A.T.D., D.T., A.Cl., Y.F., A.G., I.R.C., J.S., L.R.B., R.C., and J.A. Investigation: K.G.N., S.A.C., S.S., J.P., L.E.C., P.Y., A.C., L.G.A.M., H.H.V., V.B., N.V.T., S.E.T., S.K., M.J., A.C.S., C.T., J.F.C.-S., A.T.D., D.T., A.C., Y.F., A.G., I.R.C., J.S., L.R.B., R.C.C., and J.A. Visualization: K.G.N., S.A.C., S.S., J.P., L.E.C., P.Y., A.Co., L.G.A.M., H.H.V., N.V.T., G.Z., M.D., S.E.T., S.K., A.C.S., C.T., J.F.C.-S., A.T.D., D.T., A.Cl., Y.F., A.G., I.R.C., J.S., L.R.B., R.C.C., and J.A. Funding acquisition: R.C.L., E.T.R., R.C.C., and J.A. Project administration: K.G.N., S.A.C., S.S., J.P., N.V.T., M.D., S.E.T., S.K., A.C.S., A.G., I.R.C., J.S., L.R.B., R.C.C., and J.A. Supervision: A.D., J.F.C.-S., Y.F., A.G., I.R.C., J.S., L.R.B., R.C.C., and J.A. Writing–original draft: J.A. Writing–review & editing: K.G.N., S.A.C., S.S., J.P., L.E.C., A.C., L.G.A.M., H.H.V., G.Z., M.D., S.E.T., S.K., A.C.S., R.C.L., E.T.R., C.T., J.F.C., A.T.D., D.T., A.C., Y.F., A.G., I.R.C., J.S., L.R.B., R.C.C., and J.A.
Competing interests: S.A.C., L.E.C., and J.A. are inventors on a provisional patent application that covers antibodies described in this publication filed by the president and fellows of Harvard University. S.E.T. receives monetary compensation from UpToDate, which provides clinical decision support.
Data and materials availability: C1C-A3 antibody heavy chain and light chain variable region gene sequences have been deposited in GenBank under accession codes OL621229 and OL621230, respectively. Protein Data Bank (PBD) IDs for the day 146* RBD human ACE2 ectodomain complex, the unliganded C1C-A3 Fab, the C1C-A3 Fab SARS-CoV-2 spike protein ectodomain complex, and focused refinement of the C1C-A3 Fab/RBD region are 7SN0, 7SN1, 7SN3, and 7SN2, respectively. Electron Microscopy Data Bank (EMDB) accession numbers for the C1C-A3 Fab SARS-CoV-2 spike protein ectodomain complex and focused refinement of the C1C-A3 Fab/RBD region are EMD-25210 and EMD-2509, respectively. All materials are available upon request. This work is licensed under a Creative Commons Attribution 4.0 International (CC BY 4.0) license, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. To view a copy of this license, visit https://creativecommons.org/licenses/by/4.0/. This license does not apply to figures/photos/artwork or other content included in the article that is credited to a third party; obtain authorization from the rights holder before using such material.
Supplementary Materials
This PDF file includes:
Other Supplementary Material for this manuscript includes the following:
MDAR Reproducibility Checklist
References and Notes
- 1.Harvey W. T., Carabelli A. M., Jackson B., Gupta R. K., Thomson E. C., Harrison E. M., Ludden C., Reeve R., Rambaut A., Peacock S. J., Robertson D. L.; COVID-19 Genomics UK (COG-UK) Consortium , SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 19, 409–424 (2021). 10.1038/s41579-021-00573-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X., Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581, 215–220 (2020). 10.1038/s41586-020-2180-5 [DOI] [PubMed] [Google Scholar]
- 3.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F., Structural basis of receptor recognition by SARS-CoV-2. Nature 581, 221–224 (2020). 10.1038/s41586-020-2179-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Piccoli L., Park Y.-J., Tortorici M. A., Czudnochowski N., Walls A. C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L. E., Bowen J. E., Acton O. J., Jaconi S., Guarino B., Minola A., Zatta F., Sprugasci N., Bassi J., Peter A., De Marco A., Nix J. C., Mele F., Jovic S., Rodriguez B. F., Gupta S. V., Jin F., Piumatti G., Lo Presti G., Pellanda A. F., Biggiogero M., Tarkowski M., Pizzuto M. S., Cameroni E., Havenar-Daughton C., Smithey M., Hong D., Lepori V., Albanese E., Ceschi A., Bernasconi E., Elzi L., Ferrari P., Garzoni C., Riva A., Snell G., Sallusto F., Fink K., Virgin H. W., Lanzavecchia A., Corti D., Veesler D., Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell 183, 1024–1042.e21 (2020). 10.1016/j.cell.2020.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greaney A. J., Loes A. N., Gentles L. E., Crawford K. H. D., Starr T. N., Malone K. D., Chu H. Y., Bloom J. D., Antibodies elicited by mRNA-1273 vaccination bind more broadly to the receptor binding domain than do those from SARS-CoV-2 infection. Sci. Transl. Med. 13, eabi9915 (2021). 10.1126/scitranslmed.abi9915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCarthy K. R., Rennick L. J., Nambulli S., Robinson-McCarthy L. R., Bain W. G., Haidar G., Duprex W. P., Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science 371, 1139–1142 (2021). 10.1126/science.abf6950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCallum M., De Marco A., Lempp F. A., Tortorici M. A., Pinto D., Walls A. C., Beltramello M., Chen A., Liu Z., Zatta F., Zepeda S., di Iulio J., Bowen J. E., Montiel-Ruiz M., Zhou J., Rosen L. E., Bianchi S., Guarino B., Fregni C. S., Abdelnabi R., Foo S. C., Rothlauf P. W., Bloyet L.-M., Benigni F., Cameroni E., Neyts J., Riva A., Snell G., Telenti A., Whelan S. P. J., Virgin H. W., Corti D., Pizzuto M. S., Veesler D., N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell 184, 2332–2347.e16 (2021). 10.1016/j.cell.2021.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Supasa P., Zhou D., Dejnirattisai W., Liu C., Mentzer A. J., Ginn H. M., Zhao Y., Duyvesteyn H. M. E., Nutalai R., Tuekprakhon A., Wang B., Paesen G. C., Slon-Campos J., López-Camacho C., Hallis B., Coombes N., Bewley K. R., Charlton S., Walter T. S., Barnes E., Dunachie S. J., Skelly D., Lumley S. F., Baker N., Shaik I., Humphries H. E., Godwin K., Gent N., Sienkiewicz A., Dold C., Levin R., Dong T., Pollard A. J., Knight J. C., Klenerman P., Crook D., Lambe T., Clutterbuck E., Bibi S., Flaxman A., Bittaye M., Belij-Rammerstorfer S., Gilbert S., Hall D. R., Williams M. A., Paterson N. G., James W., Carroll M. W., Fry E. E., Mongkolsapaya J., Ren J., Stuart D. I., Screaton G. R., Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera. Cell 184, 2201–2211.e7 (2021). 10.1016/j.cell.2021.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dejnirattisai W., Zhou D., Supasa P., Liu C., Mentzer A. J., Ginn H. M., Zhao Y., Duyvesteyn H. M. E., Tuekprakhon A., Nutalai R., Wang B., López-Camacho C., Slon-Campos J., Walter T. S., Skelly D., Costa Clemens S. A., Naveca F. G., Nascimento V., Nascimento F., Fernandes da Costa C., Resende P. C., Pauvolid-Correa A., Siqueira M. M., Dold C., Levin R., Dong T., Pollard A. J., Knight J. C., Crook D., Lambe T., Clutterbuck E., Bibi S., Flaxman A., Bittaye M., Belij-Rammerstorfer S., Gilbert S. C., Carroll M. W., Klenerman P., Barnes E., Dunachie S. J., Paterson N. G., Williams M. A., Hall D. R., Hulswit R. J. G., Bowden T. A., Fry E. E., Mongkolsapaya J., Ren J., Stuart D. I., Screaton G. R., Antibody evasion by the P.1 strain of SARS-CoV-2. Cell 184, 2939–2954.e9 (2021). 10.1016/j.cell.2021.03.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Planas D., Bruel T., Grzelak L., Guivel-Benhassine F., Staropoli I., Porrot F., Planchais C., Buchrieser J., Rajah M. M., Bishop E., Albert M., Donati F., Prot M., Behillil S., Enouf V., Maquart M., Smati-Lafarge M., Varon E., Schortgen F., Yahyaoui L., Gonzalez M., De Sèze J., Péré H., Veyer D., Sève A., Simon-Lorière E., Fafi-Kremer S., Stefic K., Mouquet H., Hocqueloux L., van der Werf S., Prazuck T., Schwartz O., Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat. Med. 27, 917–924 (2021). 10.1038/s41591-021-01318-5 [DOI] [PubMed] [Google Scholar]
- 11.Wang P., Casner R. G., Nair M. S., Wang M., Yu J., Cerutti G., Liu L., Kwong P. D., Huang Y., Shapiro L., Ho D. D., Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe 29, 747–751.e4 (2021). 10.1016/j.chom.2021.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang P., Nair M. S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P. D., Graham B. S., Mascola J. R., Chang J. Y., Yin M. T., Sobieszczyk M., Kyratsous C. A., Shapiro L., Sheng Z., Huang Y., Ho D. D., Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 593, 130–135 (2021). 10.1038/s41586-021-03398-2 [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Beltran W. F., Lam E. C., St. Denis K., Nitido A. D., Garcia Z. H., Hauser B. M., Feldman J., Pavlovic M. N., Gregory D. J., Poznansky M. C., Sigal A., Schmidt A. G., Iafrate A. J., Naranbhai V., Balazs A. B., Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell 184, 2523 (2021). 10.1016/j.cell.2021.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark S. A., Clark L. E., Pan J., Coscia A., McKay L. G. A., Shankar S., Johnson R. I., Brusic V., Choudhary M. C., Regan J., Li J. Z., Griffiths A., Abraham J., SARS-CoV-2 evolution in an immunocompromised host reveals shared neutralization escape mechanisms. Cell 184, 2605–2617.e18 (2021). 10.1016/j.cell.2021.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi B., Choudhary M. C., Regan J., Sparks J. A., Padera R. F., Qiu X., Solomon I. H., Kuo H.-H., Boucau J., Bowman K., Adhikari U. D., Winkler M. L., Mueller A. A., Hsu T. Y.-T., Desjardins M., Baden L. R., Chan B. T., Walker B. D., Lichterfeld M., Brigl M., Kwon D. S., Kanjilal S., Richardson E. T., Jonsson A. H., Alter G., Barczak A. K., Hanage W. P., Yu X. G., Gaiha G. D., Seaman M. S., Cernadas M., Li J. Z., Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N. Engl. J. Med. 383, 2291–2293 (2020). 10.1056/NEJMc2031364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Starr T. N., Greaney A. J., Addetia A., Hannon W. W., Choudhary M. C., Dingens A. S., Li J. Z., Bloom J. D., Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science 371, 850–854 (2021). 10.1126/science.abf9302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baum A., Fulton B. O., Wloga E., Copin R., Pascal K. E., Russo V., Giordano S., Lanza K., Negron N., Ni M., Wei Y., Atwal G. S., Murphy A. J., Stahl N., Yancopoulos G. D., Kyratsous C. A., Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science 369, 1014–1018 (2020). 10.1126/science.abd0831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen J., Baum A., Pascal K. E., Russo V., Giordano S., Wloga E., Fulton B. O., Yan Y., Koon K., Patel K., Chung K. M., Hermann A., Ullman E., Cruz J., Rafique A., Huang T., Fairhurst J., Libertiny C., Malbec M., Lee W. Y., Welsh R., Farr G., Pennington S., Deshpande D., Cheng J., Watty A., Bouffard P., Babb R., Levenkova N., Chen C., Zhang B., Romero Hernandez A., Saotome K., Zhou Y., Franklin M., Sivapalasingam S., Lye D. C., Weston S., Logue J., Haupt R., Frieman M., Chen G., Olson W., Murphy A. J., Stahl N., Yancopoulos G. D., Kyratsous C. A., Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science 369, 1010–1014 (2020). 10.1126/science.abd0827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan M., Liu H., Wu N. C., Lee C. D., Zhu X., Zhao F., Huang D., Yu W., Hua Y., Tien H., Rogers T. F., Landais E., Sok D., Jardine J. G., Burton D. R., Wilson I. A., Structural basis of a shared antibody response to SARS-CoV-2. Science 369, 1119–1123 (2020). 10.1126/science.abd2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rogers T. F., Zhao F., Huang D., Beutler N., Burns A., He W. T., Limbo O., Smith C., Song G., Woehl J., Yang L., Abbott R. K., Callaghan S., Garcia E., Hurtado J., Parren M., Peng L., Ramirez S., Ricketts J., Ricciardi M. J., Rawlings S. A., Wu N. C., Yuan M., Smith D. M., Nemazee D., Teijaro J. R., Voss J. E., Wilson I. A., Andrabi R., Briney B., Landais E., Sok D., Jardine J. G., Burton D. R., Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 369, 956–963 (2020). 10.1126/science.abc7520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., Gao G., Hu X., Zhang Y., Tong Z., Huang W., Liu W. J., Wu G., Zhang B., Wang L., Qi J., Feng H., Wang F.-S., Wang Q., Gao G. F., Yuan Z., Yan J., A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 584, 120–124 (2020). 10.1038/s41586-020-2381-y [DOI] [PubMed] [Google Scholar]
- 22.Seydoux E., Homad L. J., MacCamy A. J., Parks K. R., Hurlburt N. K., Jennewein M. F., Akins N. R., Stuart A. B., Wan Y.-H., Feng J., Whaley R. E., Singh S., Boeckh M., Cohen K. W., McElrath M. J., Englund J. A., Chu H. Y., Pancera M., McGuire A. T., Stamatatos L., Analysis of a SARS-CoV-2-infected individual reveals development of potent neutralizing antibodies with limited somatic mutation. Immunity 53, 98–105.e5 (2020). 10.1016/j.immuni.2020.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robbiani D. F., Gaebler C., Muecksch F., Lorenzi J. C. C., Wang Z., Cho A., Agudelo M., Barnes C. O., Gazumyan A., Finkin S., Hägglöf T., Oliveira T. Y., Viant C., Hurley A., Hoffmann H.-H., Millard K. G., Kost R. G., Cipolla M., Gordon K., Bianchini F., Chen S. T., Ramos V., Patel R., Dizon J., Shimeliovich I., Mendoza P., Hartweger H., Nogueira L., Pack M., Horowitz J., Schmidt F., Weisblum Y., Michailidis E., Ashbrook A. W., Waltari E., Pak J. E., Huey-Tubman K. E., Koranda N., Hoffman P. R., West A. P. Jr., Rice C. M., Hatziioannou T., Bjorkman P. J., Bieniasz P. D., Caskey M., Nussenzweig M. C., Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 584, 437–442 (2020). 10.1038/s41586-020-2456-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du S., Cao Y., Zhu Q., Yu P., Qi F., Wang G., Du X., Bao L., Deng W., Zhu H., Liu J., Nie J., Zheng Y., Liang H., Liu R., Gong S., Xu H., Yisimayi A., Lv Q., Wang B., He R., Han Y., Zhao W., Bai Y., Qu Y., Gao X., Ji C., Wang Q., Gao N., Huang W., Wang Y., Xie X. S., Su X. D., Xiao J., Qin C., Structurally resolved SARS-CoV-2 antibody shows high efficacy in severely infected hamsters and provides a potent cocktail pairing strategy. Cell 183, 1013–1023.e13 (2020). 10.1016/j.cell.2020.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y., Gong Y., Xiao H., Fan Z., Tan S., Wu G., Tan W., Lu X., Fan C., Wang Q., Liu Y., Zhang C., Qi J., Gao G. F., Gao F., Liu L., A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science 368, 1274–1278 (2020). 10.1126/science.abc2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Starr T. N., Greaney A. J., Dingens A. S., Bloom J. D., Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016. Cell Rep. Med. 2, 100255 (2021). 10.1016/j.xcrm.2021.100255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elbe S., Buckland-Merrett G., Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Chall. 1, 33–46 (2017). 10.1002/gch2.1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J. C. C., Muecksch F., Rutkowska M., Hoffmann H.-H., Michailidis E., Gaebler C., Agudelo M., Cho A., Wang Z., Gazumyan A., Cipolla M., Luchsinger L., Hillyer C. D., Caskey M., Robbiani D. F., Rice C. M., Nussenzweig M. C., Hatziioannou T., Bieniasz P. D., Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife 9, e61312 (2020). 10.7554/eLife.61312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wrapp D., Wang N., Corbett K. S., Goldsmith J. A., Hsieh C.-L., Abiona O., Graham B. S., McLellan J. S., Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263 (2020). 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu K., Pan X., Li L., Yu F., Zheng A., Du P., Han P., Meng Y., Zhang Y., Wu L., Chen Q., Song C., Jia Y., Niu S., Lu D., Qiao C., Chen Z., Ma D., Ma X., Tan S., Zhao X., Qi J., Gao G. F., Wang Q., Binding and molecular basis of the bat coronavirus RaTG13 virus to ACE2 in humans and other species. Cell 184, 3438–3451.e10 (2021). 10.1016/j.cell.2021.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suryamohan K., Diwanji D., Stawiski E. W., Gupta R., Miersch S., Liu J., Chen C., Jiang Y.-P., Fellouse F. A., Sathirapongsasuti J. F., Albers P. K., Deepak T., Saberianfar R., Ratan A., Washburn G., Mis M., Santhosh D., Somasekar S., Hiranjith G. H., Vargas D., Mohan S., Phalke S., Kuriakose B., Antony A., Ustav M. Jr., Schuster S. C., Sidhu S., Junutula J. R., Jura N., Seshagiri S., Human ACE2 receptor polymorphisms and altered susceptibility to SARS-CoV-2. Commun. Biol. 4, 475 (2021). 10.1038/s42003-021-02030-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mehdipour A. R., Hummer G., Dual nature of human ACE2 glycosylation in binding to SARS-CoV-2 spike. Proc. Natl. Acad. Sci. U.S.A. 118, e2100425118 (2021). 10.1073/pnas.2100425118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu X., Mannar D., Srivastava S. S., Berezuk A. M., Demers J.-P., Saville J. W., Leopold K., Li W., Dimitrov D. S., Tuttle K. S., Zhou S., Chittori S., Subramaniam S., Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies. PLOS Biol. 19, e3001237 (2021). 10.1371/journal.pbio.3001237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen R. E., Zhang X., Case J. B., Winkler E. S., Liu Y., VanBlargan L. A., Liu J., Errico J. M., Xie X., Suryadevara N., Gilchuk P., Zost S. J., Tahan S., Droit L., Turner J. S., Kim W., Schmitz A. J., Thapa M., Wang D., Boon A. C. M., Presti R. M., O’Halloran J. A., Kim A. H. J., Deepak P., Pinto D., Fremont D. H., Crowe J. E. Jr., Corti D., Virgin H. W., Ellebedy A. H., Shi P.-Y., Diamond M. S., Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 27, 717–726 (2021). 10.1038/s41591-021-01294-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greaney A. J., Loes A. N., Crawford K. H. D., Starr T. N., Malone K. D., Chu H. Y., Bloom J. D., Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe 29, 463–476.e6 (2021). 10.1016/j.chom.2021.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barnes C. O., Jette C. A., Abernathy M. E., Dam K. A., Esswein S. R., Gristick H. B., Malyutin A. G., Sharaf N. G., Huey-Tubman K. E., Lee Y. E., Robbiani D. F., Nussenzweig M. C., West A. P. Jr., Bjorkman P. J., SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 588, 682–687 (2020). 10.1038/s41586-020-2852-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones B. E., Brown-Augsburger P. L., Corbett K. S., Westendorf K., Davies J., Cujec T. P., Wiethoff C. M., Blackbourne J. L., Heinz B. A., Foster D., Higgs R. E., Balasubramaniam D., Wang L., Zhang Y., Yang E. S., Bidshahri R., Kraft L., Hwang Y., Žentelis S., Jepson K. R., Goya R., Smith M. A., Collins D. W., Hinshaw S. J., Tycho S. A., Pellacani D., Xiang P., Muthuraman K., Sobhanifar S., Piper M. H., Triana F. J., Hendle J., Pustilnik A., Adams A. C., Berens S. J., Baric R. S., Martinez D. R., Cross R. W., Geisbert T. W., Borisevich V., Abiona O., Belli H. M., de Vries M., Mohamed A., Dittmann M., Samanovic M. I., Mulligan M. J., Goldsmith J. A., Hsieh C.-L., Johnson N. V., Wrapp D., McLellan J. S., Barnhart B. C., Graham B. S., Mascola J. R., Hansen C. L., Falconer E., The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci. Transl. Med. 13, eabf1906 (2021). 10.1126/scitranslmed.abf1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M. M., Planchais C., Porrot F., Robillard N., Puech J., Prot M., Gallais F., Gantner P., Velay A., Le Guen J., Kassis-Chikhani N., Edriss D., Belec L., Seve A., Courtellemont L., Péré H., Hocqueloux L., Fafi-Kremer S., Prazuck T., Mouquet H., Bruel T., Simon-Lorière E., Rey F. A., Schwartz O., Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 596, 276–280 (2021). 10.1038/s41586-021-03777-9 [DOI] [PubMed] [Google Scholar]
- 39.Chen R. E., Winkler E. S., Case J. B., Aziati I. D., Bricker T. L., Joshi A., Darling T. L., Ying B., Errico J. M., Shrihari S., VanBlargan L. A., Xie X., Gilchuk P., Zost S. J., Droit L., Liu Z., Stumpf S., Wang D., Handley S. A., Stine W. B. Jr., Shi P.-Y., Davis-Gardner M. E., Suthar M. S., Knight M. G., Andino R., Chiu C. Y., Ellebedy A. H., Fremont D. H., Whelan S. P. J., Crowe J. E. Jr., Purcell L., Corti D., Boon A. C. M., Diamond M. S., In vivo monoclonal antibody efficacy against SARS-CoV-2 variant strains. Nature 596, 103–108 (2021). 10.1038/s41586-021-03720-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCallum M., Bassi J., De Marco A., Chen A., Walls A. C., Di Iulio J., Tortorici M. A., Navarro M.-J., Silacci-Fregni C., Saliba C., Sprouse K. R., Agostini M., Pinto D., Culap K., Bianchi S., Jaconi S., Cameroni E., Bowen J. E., Tilles S. W., Pizzuto M. S., Guastalla S. B., Bona G., Pellanda A. F., Garzoni C., Van Voorhis W. C., Rosen L. E., Snell G., Telenti A., Virgin H. W., Piccoli L., Corti D., Veesler D., SARS-CoV-2 immune evasion by the B.1.427/B.1.429 variant of concern. Science 373, 648–654 (2021). 10.1126/science.abi7994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomson E. C., Rosen L. E., Shepherd J. G., Spreafico R., da Silva Filipe A., Wojcechowskyj J. A., Davis C., Piccoli L., Pascall D. J., Dillen J., Lytras S., Czudnochowski N., Shah R., Meury M., Jesudason N., De Marco A., Li K., Bassi J., O’Toole A., Pinto D., Colquhoun R. M., Culap K., Jackson B., Zatta F., Rambaut A., Jaconi S., Sreenu V. B., Nix J., Zhang I., Jarrett R. F., Glass W. G., Beltramello M., Nomikou K., Pizzuto M., Tong L., Cameroni E., Croll T. I., Johnson N., Di Iulio J., Wickenhagen A., Ceschi A., Harbison A. M., Mair D., Ferrari P., Smollett K., Sallusto F., Carmichael S., Garzoni C., Nichols J., Galli M., Hughes J., Riva A., Ho A., Schiuma M., Semple M. G., Openshaw P. J. M., Fadda E., Baillie J. K., Chodera J. D., Rihn S. J., Lycett S. J., Virgin H. W., Telenti A., Corti D., Robertson D. L., Snell G.; ISARIC4C Investigators; COVID-19 Genomics UK (COG-UK) Consortium , Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell 184, 1171–1187.e20 (2021). 10.1016/j.cell.2021.01.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang L., Zhou T., Zhang Y., Yang E. S., Schramm C. A., Shi W., Pegu A., Oloniniyi O. K., Henry A. R., Darko S., Narpala S. R., Hatcher C., Martinez D. R., Tsybovsky Y., Phung E., Abiona O. M., Antia A., Cale E. M., Chang L. A., Choe M., Corbett K. S., Davis R. L., DiPiazza A. T., Gordon I. J., Hait S. H., Hermanus T., Kgagudi P., Laboune F., Leung K., Liu T., Mason R. D., Nazzari A. F., Novik L., O’Connell S., O’Dell S., Olia A. S., Schmidt S. D., Stephens T., Stringham C. D., Talana C. A., Teng I.-T., Wagner D. A., Widge A. T., Zhang B., Roederer M., Ledgerwood J. E., Ruckwardt T. J., Gaudinski M. R., Moore P. L., Doria-Rose N. A., Baric R. S., Graham B. S., McDermott A. B., Douek D. C., Kwong P. D., Mascola J. R., Sullivan N. J., Misasi J., Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants. Science 373, eabh1766 (2021). 10.1126/science.abh1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan M., Huang D., Lee C. D., Wu N. C., Jackson A. M., Zhu X., Liu H., Peng L., van Gils M. J., Sanders R. W., Burton D. R., Reincke S. M., Prüss H., Kreye J., Nemazee D., Ward A. B., Wilson I. A., Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants. Science 373, 818–823 (2021). 10.1126/science.abh1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pinto D., Park Y.-J., Beltramello M., Walls A. C., Tortorici M. A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., Peter A., Guarino B., Spreafico R., Cameroni E., Case J. B., Chen R. E., Havenar-Daughton C., Snell G., Telenti A., Virgin H. W., Lanzavecchia A., Diamond M. S., Fink K., Veesler D., Corti D., Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583, 290–295 (2020). 10.1038/s41586-020-2349-y [DOI] [PubMed] [Google Scholar]
- 45.Rappazzo C. G., Tse L. V., Kaku C. I., Wrapp D., Sakharkar M., Huang D., Deveau L. M., Yockachonis T. J., Herbert A. S., Battles M. B., O’Brien C. M., Brown M. E., Geoghegan J. C., Belk J., Peng L., Yang L., Hou Y., Scobey T. D., Burton D. R., Nemazee D., Dye J. M., Voss J. E., Gunn B. M., McLellan J. S., Baric R. S., Gralinski L. E., Walker L. M., Broad and potent activity against SARS-like viruses by an engineered human monoclonal antibody. Science 371, 823–829 (2021). 10.1126/science.abf4830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu C., Ginn H. M., Dejnirattisai W., Supasa P., Wang B., Tuekprakhon A., Nutalai R., Zhou D., Mentzer A. J., Zhao Y., Duyvesteyn H. M. E., López-Camacho C., Slon-Campos J., Walter T. S., Skelly D., Johnson S. A., Ritter T. G., Mason C., Costa Clemens S. A., Gomes Naveca F., Nascimento V., Nascimento F., Fernandes da Costa C., Resende P. C., Pauvolid-Correa A., Siqueira M. M., Dold C., Temperton N., Dong T., Pollard A. J., Knight J. C., Crook D., Lambe T., Clutterbuck E., Bibi S., Flaxman A., Bittaye M., Belij-Rammerstorfer S., Gilbert S. C., Malik T., Carroll M. W., Klenerman P., Barnes E., Dunachie S. J., Baillie V., Serafin N., Ditse Z., Da Silva K., Paterson N. G., Williams M. A., Hall D. R., Madhi S., Nunes M. C., Goulder P., Fry E. E., Mongkolsapaya J., Ren J., Stuart D. I., Screaton G. R., Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell 184, 4220–4236.e13 (2021). 10.1016/j.cell.2021.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baden L. R., El Sahly H. M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S. A., Rouphael N., Creech C. B., McGettigan J., Khetan S., Segall N., Solis J., Brosz A., Fierro C., Schwartz H., Neuzil K., Corey L., Gilbert P., Janes H., Follmann D., Marovich M., Mascola J., Polakowski L., Ledgerwood J., Graham B. S., Bennett H., Pajon R., Knightly C., Leav B., Deng W., Zhou H., Han S., Ivarsson M., Miller J., Zaks T.; COVE Study Group , Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 384, 403–416 (2021). 10.1056/NEJMoa2035389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Polack F. P., Thomas S. J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J. L., Pérez Marc G., Moreira E. D., Zerbini C., Bailey R., Swanson K. A., Roychoudhury S., Koury K., Li P., Kalina W. V., Cooper D., Frenck R. W. Jr., Hammitt L. L., Türeci Ö., Nell H., Schaefer A., Ünal S., Tresnan D. B., Mather S., Dormitzer P. R., Şahin U., Jansen K. U., Gruber W. C.; C4591001 Clinical Trial Group , Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N. Engl. J. Med. 383, 2603–2615 (2020). 10.1056/NEJMoa2034577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jackson L. A., Anderson E. J., Rouphael N. G., Roberts P. C., Makhene M., Coler R. N., McCullough M. P., Chappell J. D., Denison M. R., Stevens L. J., Pruijssers A. J., McDermott A., Flach B., Doria-Rose N. A., Corbett K. S., Morabito K. M., O’Dell S., Schmidt S. D., Swanson P. A. 2nd, Padilla M., Mascola J. R., Neuzil K. M., Bennett H., Sun W., Peters E., Makowski M., Albert J., Cross K., Buchanan W., Pikaart-Tautges R., Ledgerwood J. E., Graham B. S., Beigel J. H.; mRNA-1273 Study Group , An mRNA vaccine against SARS-CoV-2 — Preliminary report. N. Engl. J. Med. 383, 1920–1931 (2020). 10.1056/NEJMoa2022483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ksiazek T. G., Erdman D., Goldsmith C. S., Zaki S. R., Peret T., Emery S., Tong S., Urbani C., Comer J. A., Lim W., Rollin P. E., Dowell S. F., Ling A.-E., Humphrey C. D., Shieh W.-J., Guarner J., Paddock C. D., Rota P., Fields B., DeRisi J., Yang J.-Y., Cox N., Hughes J. M., LeDuc J. W., Bellini W. J., Anderson L. J.; SARS Working Group , A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348, 1953–1966 (2003). 10.1056/NEJMoa030781 [DOI] [PubMed] [Google Scholar]
- 51.Chen Z., Zhang L., Qin C., Ba L., Yi C. E., Zhang F., Wei Q., He T., Yu W., Yu J., Gao H., Tu X., Gettie A., Farzan M., Yuen K. Y., Ho D. D., Recombinant modified vaccinia virus Ankara expressing the spike glycoprotein of severe acute respiratory syndrome coronavirus induces protective neutralizing antibodies primarily targeting the receptor binding region. J. Virol. 79, 2678–2688 (2005). 10.1128/JVI.79.5.2678-2688.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu Y., Yu D., Han Y., Yan H., Chong H., Ren L., Wang J., Li T., He Y., Cross-reactive neutralization of SARS-CoV-2 by serum antibodies from recovered SARS patients and immunized animals. Sci. Adv. 6, eabc9999 (2020). 10.1126/sciadv.abc9999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lv H., Wu N. C., Tsang O. T.-Y., Yuan M., Perera R. A. P. M., Leung W. S., So R. T. Y., Chan J. M. C., Yip G. K., Chik T. S. H., Wang Y., Choi C. Y. C., Lin Y., Ng W. W., Zhao J., Poon L. L. M., Peiris J. S. M., Wilson I. A., Mok C. K. P., Cross-reactive antibody response between SARS-CoV-2 and SARS-CoV infections. Cell Rep. 31, 107725 (2020). 10.1016/j.celrep.2020.107725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pallesen J., Wang N., Corbett K. S., Wrapp D., Kirchdoerfer R. N., Turner H. L., Cottrell C. A., Becker M. M., Wang L., Shi W., Kong W.-P., Andres E. L., Kettenbach A. N., Denison M. R., Chappell J. D., Graham B. S., Ward A. B., McLellan J. S., Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. U.S.A. 114, E7348–E7357 (2017). 10.1073/pnas.1707304114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yuan M., Wu N. C., Zhu X., Lee C. D., So R. T. Y., Lv H., Mok C. K. P., Wilson I. A., A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science 368, 630–633 (2020). 10.1126/science.abb7269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tortorici M. A., Czudnochowski N., Starr T. N., Marzi R., Walls A. C., Zatta F., Bowen J. E., Jaconi S., Di Iulio J., Wang Z., De Marco A., Zepeda S. K., Pinto D., Liu Z., Beltramello M., Bartha I., Housley M. P., Lempp F. A., Rosen L. E., Dellota E. Jr., Kaiser H., Montiel-Ruiz M., Zhou J., Addetia A., Guarino B., Culap K., Sprugasci N., Saliba C., Vetti E., Giacchetto-Sasselli I., Fregni C. S., Abdelnabi R., Foo S. C., Havenar-Daughton C., Schmid M. A., Benigni F., Cameroni E., Neyts J., Telenti A., Virgin H. W., Whelan S. P. J., Snell G., Bloom J. D., Corti D., Veesler D., Pizzuto M. S., Broad sarbecovirus neutralization by a human monoclonal antibody. Nature 597, 103–108 (2021). 10.1038/s41586-021-03817-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu H., Wu N. C., Yuan M., Bangaru S., Torres J. L., Caniels T. G., van Schooten J., Zhu X., Lee C. D., Brouwer P. J. M., van Gils M. J., Sanders R. W., Ward A. B., Wilson I. A., Cross-neutralization of a SARS-CoV-2 antibody to a functionally conserved site is mediated by avidity. Immunity 53, 1272–1280.e5 (2020). 10.1016/j.immuni.2020.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lv Z., Deng Y.-Q., Ye Q., Cao L., Sun C.-Y., Fan C., Huang W., Sun S., Sun Y., Zhu L., Chen Q., Wang N., Nie J., Cui Z., Zhu D., Shaw N., Li X.-F., Li Q., Xie L., Wang Y., Rao Z., Qin C.-F., Wang X., Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science 369, 1505–1509 (2020). 10.1126/science.abc5881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li D., Edwards R. J., Manne K., Martinez D. R., Schäfer A., Alam S. M., Wiehe K., Lu X., Parks R., Sutherland L. L., Oguin T. H. 3rd, McDanal C., Perez L. G., Mansouri K., Gobeil S. M. C., Janowska K., Stalls V., Kopp M., Cai F., Lee E., Foulger A., Hernandez G. E., Sanzone A., Tilahun K., Jiang C., Tse L. V., Bock K. W., Minai M., Nagata B. M., Cronin K., Gee-Lai V., Deyton M., Barr M., Von Holle T., Macintyre A. N., Stover E., Feldman J., Hauser B. M., Caradonna T. M., Scobey T. D., Rountree W., Wang Y., Moody M. A., Cain D. W., DeMarco C. T., Denny T. N., Woods C. W., Petzold E. W., Schmidt A. G., Teng I.-T., Zhou T., Kwong P. D., Mascola J. R., Graham B. S., Moore I. N., Seder R., Andersen H., Lewis M. G., Montefiori D. C., Sempowski G. D., Baric R. S., Acharya P., Haynes B. F., Saunders K. O., In vitro and in vivo functions of SARS-CoV-2 infection-enhancing and neutralizing antibodies. Cell 184, 4203–4219.e32 (2021). 10.1016/j.cell.2021.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Starr T. N., Czudnochowski N., Liu Z., Zatta F., Park Y.-J., Addetia A., Pinto D., Beltramello M., Hernandez P., Greaney A. J., Marzi R., Glass W. G., Zhang I., Dingens A. S., Bowen J. E., Tortorici M. A., Walls A. C., Wojcechowskyj J. A., De Marco A., Rosen L. E., Zhou J., Montiel-Ruiz M., Kaiser H., Dillen J. R., Tucker H., Bassi J., Silacci-Fregni C., Housley M. P., di Iulio J., Lombardo G., Agostini M., Sprugasci N., Culap K., Jaconi S., Meury M., Dellota E. Jr., Abdelnabi R., Foo S. C., Cameroni E., Stumpf S., Croll T. I., Nix J. C., Havenar-Daughton C., Piccoli L., Benigni F., Neyts J., Telenti A., Lempp F. A., Pizzuto M. S., Chodera J. D., Hebner C. M., Virgin H. W., Whelan S. P. J., Veesler D., Corti D., Bloom J. D., Snell G., SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature 597, 97–102 (2021). 10.1038/s41586-021-03807-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watanabe Y., Berndsen Z. T., Raghwani J., Seabright G. E., Allen J. D., Pybus O. G., McLellan J. S., Wilson I. A., Bowden T. A., Ward A. B., Crispin M., Vulnerabilities in coronavirus glycan shields despite extensive glycosylation. Nat. Commun. 11, 2688 (2020). 10.1038/s41467-020-16567-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grant O. C., Montgomery D., Ito K., Woods R. J., Analysis of the SARS-CoV-2 spike protein glycan shield reveals implications for immune recognition. Sci. Rep. 10, 14991 (2020). 10.1038/s41598-020-71748-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boni M. F., Lemey P., Jiang X., Lam T. T.-Y., Perry B. W., Castoe T. A., Rambaut A., Robertson D. L., Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat. Microbiol. 5, 1408–1417 (2020). 10.1038/s41564-020-0771-4 [DOI] [PubMed] [Google Scholar]
- 64.Hurlburt N. K., Seydoux E., Wan Y.-H., Edara V. V., Stuart A. B., Feng J., Suthar M. S., McGuire A. T., Stamatatos L., Pancera M., Structural basis for potent neutralization of SARS-CoV-2 and role of antibody affinity maturation. Nat. Commun. 11, 5413 (2020). 10.1038/s41467-020-19231-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kang L., He G., Sharp A. K., Wang X., Brown A. M., Michalak P., Weger-Lucarelli J., A selective sweep in the Spike gene has driven SARS-CoV-2 human adaptation. Cell 184, 4392–4400.e4 (2021). 10.1016/j.cell.2021.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harbison A. M., Fogarty C. A., Phung T., Satheesan A., Schulz B. L., Fadda E., Fine-tuning the Spike: Role of the nature and topology of the glycan shield in the structure and dynamics of SARS-CoV-2 S. Chem. Sci. 10.1039/D1SC04832E (2021). 10.1039/D1SC04832E [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bloom J. D., Gong L. I., Baltimore D., Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science 328, 1272–1275 (2010). 10.1126/science.1187816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yurkovetskiy L., Wang X., Pascal K. E., Tomkins-Tinch C., Nyalile T. P., Wang Y., Baum A., Diehl W. E., Dauphin A., Carbone C., Veinotte K., Egri S. B., Schaffner S. F., Lemieux J. E., Munro J. B., Rafique A., Barve A., Sabeti P. C., Kyratsous C. A., Dudkina N. V., Shen K., Luban J., Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell 183, 739–751.e8 (2020). 10.1016/j.cell.2020.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kabsch W., XDS. Acta Crystallogr. D 66, 125–132 (2010). 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Evans P. R., Murshudov G. N., How good are my data and what is the resolution? Acta Crystallogr. D 69, 1204–1214 (2013). 10.1107/S0907444913000061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.G. Bricogne, E. Blanc, M. Brandl, C. Flensburg, P. Keller, W. Paciorek, P. Roversi, A. Sharff, O. S. Smart, C. Vonrhein, T. O. Womack, BUSTER version 2.10.3 (Global Phasing Ltd., 2017).
- 72.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010). 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010). 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zheng S. Q., Palovcak E., Armache J.-P., Verba K. A., Cheng Y., Agard D. A., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). 10.1038/nmeth.4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). 10.1016/j.jsb.2015.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wagner T., Merino F., Stabrin M., Moriya T., Antoni C., Apelbaum A., Hagel P., Sitsel O., Raisch T., Prumbaum D., Quentin D., Roderer D., Tacke S., Siebolds B., Schubert E., Shaikh T. R., Lill P., Gatsogiannis C., Raunser S., SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2, 218 (2019). 10.1038/s42003-019-0437-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). 10.1038/nmeth.4169 [DOI] [PubMed] [Google Scholar]
- 78.Walls A. C., Park Y.-J., Tortorici M. A., Wall A., McGuire A. T., Veesler D., Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181, 281–292.e6 (2020). 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang S., Qiao S., Yu J., Zeng J., Shan S., Tian L., Lan J., Zhang L., Wang X., Bat and pangolin coronavirus spike glycoprotein structures provide insights into SARS-CoV-2 evolution. Nat. Commun. 12, 1607 (2021). 10.1038/s41467-021-21767-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xiao T., Lu J., Zhang J., Johnson R. I., McKay L. G. A., Storm N., Lavine C. L., Peng H., Cai Y., Rits-Volloch S., Lu S., Quinlan B. D., Farzan M., Seaman M. S., Griffiths A., Chen B., A trimeric human angiotensin-converting enzyme 2 as an anti-SARS-CoV-2 agent. Nat. Struct. Mol. Biol. 28, 202–209 (2021). 10.1038/s41594-020-00549-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Harcourt J., Tamin A., Lu X., Kamili S., Sakthivel S. K., Murray J., Queen K., Tao Y., Paden C. R., Zhang J., Li Y., Uehara A., Wang H., Goldsmith C., Bullock H. A., Wang L., Whitaker B., Lynch B., Gautam R., Schindewolf C., Lokugamage K. G., Scharton D., Plante J. A., Mirchandani D., Widen S. G., Narayanan K., Makino S., Ksiazek T. G., Plante K. S., Weaver S. C., Lindstrom S., Tong S., Menachery V. D., Thornburg N. J., Severe acute respiratory syndrome coronavirus 2 from patient with coronavirus disease, United States. Emerg. Infect. Dis. 26, 1266–1273 (2020). 10.3201/eid2606.200516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aricescu A. R., Lu W., Jones E. Y., A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. D 62, 1243–1250 (2006). 10.1107/S0907444906029799 [DOI] [PubMed] [Google Scholar]
- 83.Hsieh C. L., Goldsmith J. A., Schaub J. M., DiVenere A. M., Kuo H.-C., Javanmardi K., Le K. C., Wrapp D., Lee A. G., Liu Y., Chou C.-W., Byrne P. O., Hjorth C. K., Johnson N. V., Ludes-Meyers J., Nguyen A. W., Park J., Wang N., Amengor D., Lavinder J. J., Ippolito G. C., Maynard J. A., Finkelstein I. J., McLellan J. S., Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 369, 1501–1505 (2020). 10.1126/science.abd0826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Misasi J., Gilman M. S. A., Kanekiyo M., Gui M., Cagigi A., Mulangu S., Corti D., Ledgerwood J. E., Lanzavecchia A., Cunningham J., Muyembe-Tamfun J. J., Baxa U., Graham B. S., Xiang Y., Sullivan N. J., McLellan J. S., Structural and molecular basis for Ebola virus neutralization by protective human antibodies. Science 351, 1343–1346 (2016). 10.1126/science.aad6117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brochet X., Lefranc M. P., Giudicelli V., IMGT/V-QUEST: The highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 36 (suppl. 2), W503–W508 (2008). 10.1093/nar/gkn316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J., Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007). 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M., Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991). 10.1107/S0108767390010224 [DOI] [PubMed] [Google Scholar]
- 88.Williams C. J., Headd J. J., Moriarty N. W., Prisant M. G., Videau L. L., Deis L. N., Verma V., Keedy D. A., Hintze B. J., Chen V. B., Jain S., Lewis S. M., Arendall W. B. 3rd, Snoeyink J., Adams P. D., Lovell S. C., Richardson J. S., Richardson D. C., MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018). 10.1002/pro.3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pless D. D., Lennarz W. J., Enzymatic conversion of proteins to glycoproteins. Proc. Natl. Acad. Sci. U.S.A. 74, 134–138 (1977). 10.1073/pnas.74.1.134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Watanabe Y., Allen J. D., Wrapp D., McLellan J. S., Crispin M., Site-specific glycan analysis of the SARS-CoV-2 spike. Science 369, 330–333 (2020). 10.1126/science.abb9983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zivanov J., Nakane T., Forsberg B. O., Kimanius D., Hagen W. J. H., Lindahl E., Scheres S. H. W., New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). 10.7554/eLife.42166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhou T., Tsybovsky Y., Gorman J., Rapp M., Cerutti G., Chuang G.-Y., Katsamba P. S., Sampson J. M., Schön A., Bimela J., Boyington J. C., Nazzari A., Olia A. S., Shi W., Sastry M., Stephens T., Stuckey J., Teng I.-T., Wang P., Wang S., Zhang B., Friesner R. A., Ho D. D., Mascola J. R., Shapiro L., Kwong P. D., Cryo-EM structures of SARS-CoV-2 spike without and with ACE2 reveal a pH-dependent switch to mediate endosomal positioning of receptor-binding domains. Cell Host Microbe 28, 867–879.e5 (2020). 10.1016/j.chom.2020.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mitternacht S., FreeSASA: An open source C library for solvent accessible surface area calculations. F1000Res. 5, 189 (2016). 10.12688/f1000research.7931.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee B., Richards F. M., The interpretation of protein structures: Estimation of static accessibility. J. Mol. Biol. 55, 379–400 (1971). 10.1016/0022-2836(71)90324-X [DOI] [PubMed] [Google Scholar]
- 95.Novotný J., Auffray C., A program for prediction of protein secondary structure from nucleotide sequence data: Application to histocompatibility antigens. Nucleic Acids Res. 12, 243–255 (1984). 10.1093/nar/12.1Part1.243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pavlova A., Zhang Z., Acharya A., Lynch D. L., Pang Y. T., Mou Z., Parks J. M., Chipot C., Gumbart J. C., Machine learning reveals the critical interactions for SARS-CoV-2 spike protein binding to ACE2. J. Phys. Chem. Lett. 12, 5494–5502 (2021). 10.1021/acs.jpclett.1c01494 [DOI] [PubMed] [Google Scholar]
- 97.Li F., Li W., Farzan M., Harrison S. C., Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 309, 1864–1868 (2005). 10.1126/science.1116480 [DOI] [PubMed] [Google Scholar]
- 98.Mirdita M., Ovchinnikov S., Steinegger M., ColabFold - Makingprotein folding accessible to all. bioRxiv 2021.2008.2015.456425 [Preprint] (2021). 10.1101/2021.08.15.456425. [DOI]
- 99.Tortorici M. A., Beltramello M., Lempp F. A., Pinto D., Dang H. V., Rosen L. E., McCallum M., Bowen J., Minola A., Jaconi S., Zatta F., De Marco A., Guarino B., Bianchi S., Lauron E. J., Tucker H., Zhou J., Peter A., Havenar-Daughton C., Wojcechowskyj J. A., Case J. B., Chen R. E., Kaiser H., Montiel-Ruiz M., Meury M., Czudnochowski N., Spreafico R., Dillen J., Ng C., Sprugasci N., Culap K., Benigni F., Abdelnabi R., Foo S. C., Schmid M. A., Cameroni E., Riva A., Gabrieli A., Galli M., Pizzuto M. S., Neyts J., Diamond M. S., Virgin H. W., Snell G., Corti D., Fink K., Veesler D., Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science 370, 950–957 (2020). 10.1126/science.abe3354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ke Z., Oton J., Qu K., Cortese M., Zila V., McKeane L., Nakane T., Zivanov J., Neufeldt C. J., Cerikan B., Lu J. M., Peukes J., Xiong X., Kräusslich H.-G., Scheres S. H. W., Bartenschlager R., Briggs J. A. G., Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature 588, 498–502 (2020). 10.1038/s41586-020-2665-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MDAR Reproducibility Checklist