Abstract
This perspective article highlights recent progress and emerging challenges in understanding the formation and function of membraneless organelles (MLOs). A long-term goal in the MLO field is to identify the sequence-encoded rules that dictate the formation of compositionally controlled biomolecular condensates, which cells utilize to perform a wide variety of functions. The molecular organization of the different components within a condensate can vary significantly, ranging from a homogeneous mixture to core-shell droplet structures. We provide many examples to highlight the richness of the observed behavior and potential research directions for improving our mechanistic understanding. The tunable environment within condensates can, in principle, alter enzymatic activity significantly. We examine recent examples where this was demonstrated, including applications in synthetic biology. An important question about MLOs is the role of liquid-like material properties in biological function. We discuss the need for improved quantitative characterization tools and the development of sequence-structure-dynamics relationships.
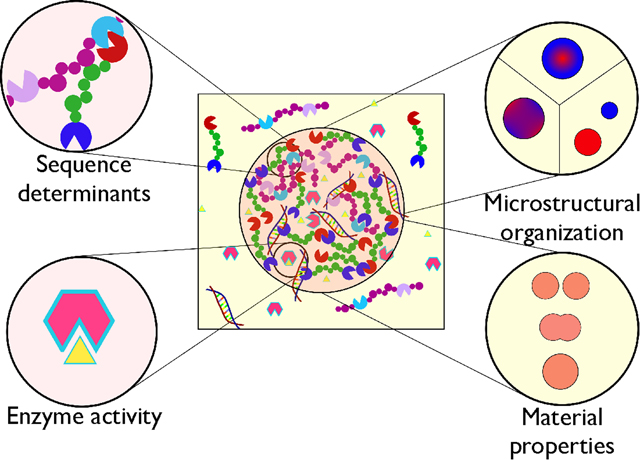
Graphical Abstract
Introduction
The many biochemical processes that take place inside a cell are highly coordinated and occur with a great degree of spatiotemporal control. Classic organelles like the endoplasmic reticulum and mitochondria use lipid bilayer membranes to organize biological molecules and activities, but there are additionally many organelles without such membranes, which also function to concentrate various required molecules for cellular pathways. These are termed membraneless organelles (MLOs) or biomolecular condensates.1 Some examples are the nucleolus, Cajal bodies, stress granules, and germ granules. MLOs have recently emerged as a central player in numerous cellular processes,2 including gene regulation, DNA repair, cellular signaling, and stress response.3
Formation of MLOs involves the selective incorporation of several biomolecules like proteins, nucleic acids, and in some cases lipids, due to liquid-liquid phase separation (LLPS).1,2,4 This process is expected to be carefully regulated in the cellular context to maintain function. Loss of function can arise from aberrant phase separation behavior, leading to excessive or insufficient condensate formation, altered material properties, and/or loss of specificity in recruiting desired constituents to the condensate. Altering the phase behavior of proteins involved in the formation of MLOs may lead to numerous diseases such as Alzheimer’s, amyotrophic lateral sclerosis (ALS), frontotemporal dementia, and cancer.5,6
The assembly of MLOs is sequence-encoded. Many MLOs have a multilayered or ‘multiphase’ architecture with distinct subcompartmentalization, reminiscent of composite materials. The composition of each subcompartment can be quite different, which may be necessary for achieving spatial control over function, e.g., in the case of the nucleolus.7 Interestingly, the material properties and dynamics within these compositionally distinct phases can also be remarkably different, e.g. in the case of P granules,8 further providing control over kinetic processes such as enzyme-catalyzed reactions. These issues and examples will be further discussed herein.
As MLOs are now being recognized as essential for cellular compartmentalization, we must obtain a better understanding of their formation to unlock the rules of life. Given the scientific excitement about this new way of thinking about critical cellular processes and the complexity of the underlying phenomena, scientists from diverse backgrounds have converged to work on this problem.9 This has brought a wide-ranging set of experimental and computational techniques10–12 to the field13, leading to an explosion of new discoveries, and in turn raising yet more questions. In this article, we discuss recent progress and future opportunities related to various thermodynamic, structural, functional, and dynamical aspects of MLOs. We note that the selection of systems and techniques is mostly guided by our interests instead of a comprehensive survey of the field.
Sequence determinants of phase separation
Intrinsically disordered regions (IDRs) have been identified as important drivers of phase separation for many proteins that underlie biomolecular condensate formation (such as FUS,14 TDP-43,15,16 LAF-1,17,18 hnRNPA2,19,20 etc.). In particular, phase-separating proteins often feature low-complexity (LC) domains or prion-like domains (PLDs), which are subtypes of IDRs. Even though a combination of experimental and computational studies has corroborated the importance of IDRs, the role of folded domains is also shown to be important for phase separation (e.g., TDP-43 NTD,21 PcG SAM domain,22 and engineered complex coacervates23 and multivalent systems such as polySUMO+polySIM or polySH3+polyPRM24). More work is certainly needed to identify the relative contributions of the disordered and folded domains (Fig. 1), and in vitro laboratory experiments and in silico computational studies can play an essential role in this.
Figure 1.
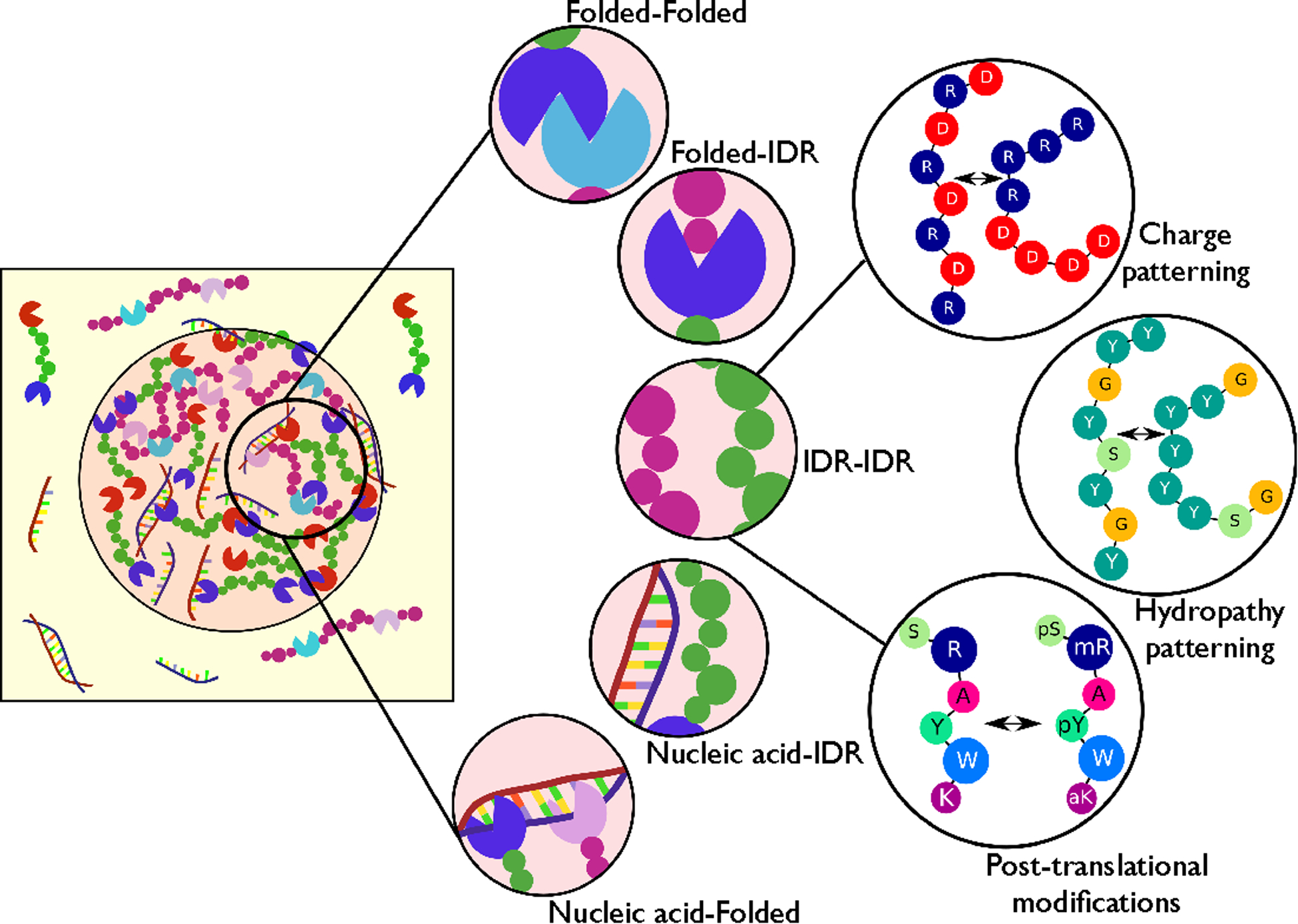
Molecular determinants of phase separation underlying the formation of biomolecular condensates. (Left) Cartoon representation of a dense droplet phase in co-existence with a dilute phase of proteins and nucleic acids. (Middle) The condensed phase can be stabilized by interactions between different protein domains [intrinsically disordered regions (IDRs) or folded domains] and interactions with nucleic acids (DNA/RNA). (Right) The strength of multivalent interactions between IDRs is determined by standard atomic interaction modes and their patterning, and it is tunable via post-translational modifications to regulate phase separation in cells.
The first obvious question about the phase separation of IDRs is the role of protein sequence, which can be further separated into two parts: (a) the amino acid composition and (b) their specific arrangement in the primary sequence. IDRs involved in phase separation are typically LC and rich in polar residues with interspersed aromatic residues (most commonly Tyrosine) and repetitive motifs (such as [S/N/G]Y[S/N/G], RGG/RG, FG, etc).25 Many IDRs have a significant fraction of Glycine and Proline residues that may promote disorder,26 though these residues may also be important for controlling the phase separation or co-localization of other biomolecules. Also, the presence of charged residues, especially Arginine, has been deemed important for driving LLPS, though the molecular interactions responsible for Arginine’s potency are still being debated.18,27,28
Consistent with fundamental biochemistry knowledge, hydrophobic residues are expected to drive protein self-assembly via non-specific interactions (including π-π interactions between aromatic sidechains), while polar residues may help control phase separation and maintain liquid-like condensates.29,30 Charged residues may provide additional stabilizing interactions via electrostatic attraction between oppositely charged Asp/Glu and Arg/Lys pairs.18,31,32 Several intriguing observations from mutagenesis experiments have prompted researchers to invoke interaction modes such as cation-π and sp2-π as being equally or more important drivers of phase separation than commonly described primary interaction modes in biomolecular assembly.27,28 For example, Arg-to-Lys or Tyr-to-Phe mutations can cause a significant decrease in LLPS propensity.18,27 As it has been difficult to explain the observed behavior in terms of electrostatic or hydrophobic interactions, it has been proposed that these two mutations may disrupt either cation-π interactions or sp2-π interactions. Accordingly, a ‘molecular grammar’ based on cation- π interactions between Arg-Tyr residue pairs has been proposed to explain the LLPS of PLDs.27 In another study, the importance of sp2-π interactions was emphasized and presented in the form of a propensity score (PS) predictor.28 More recent studies have also highlighted the role of hydrogen bonding abilities of specific amino acids, including Arg and Tyr, to help drive LLPS.18,33,34
Despite the lack of mechanistic clarity on which interaction modes are responsible, the contributions of different amino acids to LLPS are much clearer, and it is possible to design protein sequences with tunable phase behavior.35 Some of this design is helped by complementing the experimental results with simple transferable coarse-grained (CG) computational models that can capture effective interactions between different amino acids and can be used to study the sequence-dependent LLPS of proteins.10,36,37 Most of these CG models require further improvements to become completely predictive,30 which ultimately requires a better understanding of molecular interactions within the condensate.38 The efforts towards this goal so far use existing all-atom classical force fields that can provide faithful structural and dynamical properties of disordered proteins11 but are not necessarily able to distinguish between different interaction modes that are inherently quantum-mechanical. This is starting to be recognized by the force field development community,39 and new developments will likely emerge soon.
In addition to amino acid composition, the linear arrangement of amino acids in the protein sequence is also important in controlling LLPS. Significant previous research has demonstrated that the placement of charged residues within an IDR sequence tunes its conformational properties,40 leading to a predictable change in the phase separation behavior.18,41,42 Much less appreciated is the impact of how other amino acids are arranged, though some recent studies have shown that the patterning of Tyrosine residues within PLDs can tune the phase behavior.43 Some biologically relevant IDRs (e.g. LAF-1 RGG, FUS LC) appear to have a specific bias towards the presence of repetitive short motifs in their sequence, rather than a random arrangement of residues with the same amino acid composition (e.g., the LAF-1 RGG domain has 7 repeats of the RGG motif). It would be helpful to understand the context in which a specific bias in the IDR sequence is necessary for phase separation, and if not, which other factors might explain the repetitive short motifs.
Recent studies have identified simple order parameters based on charge and hydropathy patterning that can be used to assess conformational changes in IDRs associated with changes in protein sequence for a fixed composition.40,44–46 Of course, these simple order parameters fail to capture every single case, requiring further work in this direction.47 One of the current limitations in improving these parameters is the lack of experimental data on the systematic variation of protein sequences. We believe this to be an area of opportunity where collaboration among experimental and computational groups may be beneficial.
A conceptual framework based on the mapping of real protein sequences to a sticker-spacer architecture has been used to study the role of “sticker” residues and their arrangement within the sequence, using a highly optimized lattice-based simulation engine.12 The binary classification scheme of assigning a residue to either the sticker or spacer category may appear limited at first and at odds with the prevalence of weak, non-specific interactions between most residue pairs. Yet the framework was used successfully in recent studies48 to propose a predictive strategy for the phase behavior of many proteins in the FET family27,43 and highlight the relative role of different protein residues in the LLPS of the UBQLN2 protein.49 The use of this framework raises an important fundamental question – how does the specification of a “sticker” amino acid depend on its neighboring residues and the overall protein composition and sequence?
Despite immense progress in the past several years, many important issues remain concerning the sequence determinants of LLPS. Many IDRs contain short secondary structure elements that may facilitate intermolecular interactions to promote self-assembly. For instance, LLPS of the TDP-43 C-terminal domain (CTD) can be perturbed by point mutations within a small helical segment,15,16 which can then be related to changes in the helical propensity of this region. Given the correlation between disease-associated point mutations and aberrant phase behavior for IDRs such as the TDP-43 CTD, more work is needed to better understand how subtle changes in the protein sequence can have enormous implications for LLPS and biological function. Another example is the enhanced aggregation propensity of FUS LC due to a known ALS disease mutation (G156E);50 this aberrant liquid-to-fiber transition is difficult to explain mechanistically with the available knowledge. In many such cases, the altered LLPS behaviors are due to changes in the secondary structure. Most current computational models are not able to account for such changes in a transferable manner, though input from all-atom simulations and experiments can be used to test hypotheses about the role of secondary structure in assembly.16
The folded domains in proteins involved in the formation of condensates usually have a recognized role, e.g., RNA recognition motifs (RRMs), DNA binding domains (DBDs), homo- or hetero-dimerization domains, etc. Experimental challenges such as protein aggregation often preclude clarity on a folded domain’s contribution to the LLPS of full-length proteins. Regardless of the reason, interactions between the folded domains or between the IDRs and the folded domains are often not characterized experimentally, thereby limiting the ability to model these regions properly in simulations. As the self-assembly and the biological function of a protein may be tightly coupled to these details, an integrative approach combining various experimental and modeling tools should be a powerful strategy to move the field forward.
Multicomponent phase separation and microstructural organization
Biomolecular condensates in cells are comprised of many interacting molecules that in some cases are well-mixed and in other cases assemble into distinct, coexisting phases (Fig. 2). For example, in the case of P granules,8 MEG-3 forms a stable phase that recruits hundreds of RNAs, which coexists with a separate liquid-like phase comprised of PGL proteins and other IDPs such as LAF-1 and GLH-1.8,51 What factors control the composition of a functional biomolecular condensate and the relative arrangement of various macromolecules?
Figure 2.
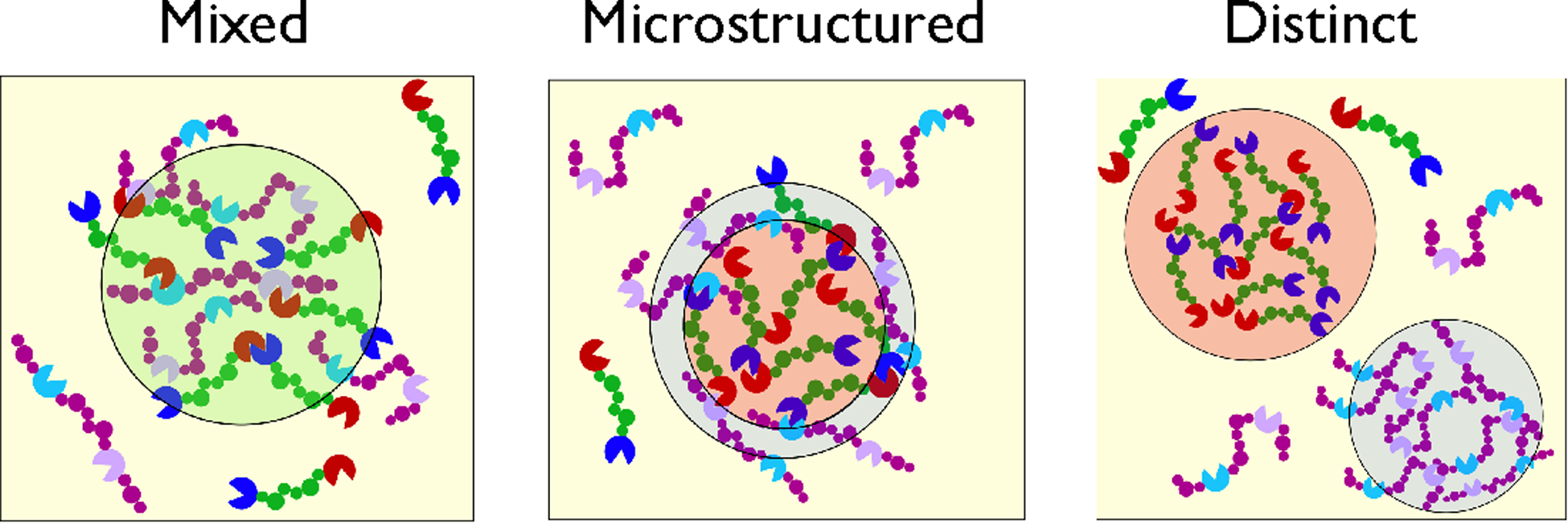
Schematic of possible scenarios in multicomponent protein phase separation. (Left) Mixing of different proteins due to similar homotypic and heterotypic interactions. (Middle) Formation of condensates with subcompartments distinct in protein composition (and possibly dynamics) due to differences in homotypic and heterotypic interactions. (Right) Formation of separate protein droplets of distinct composition when heterotypic interactions are much weaker.
One useful framework for understanding the compositions of MLOs is to classify their components as either scaffold molecules that condense to form the MLO or clients that are recruited into the MLO.24 Clients may be incorporated into condensates due to either non-specific interactions (usually via IDRs) or specific binding modes.20,24,52,53 Regarding the mixing of different components within a condensate, studies highlight a wide set of possibilities, ranging from the formation of separate unmixed condensates that are composed of different biomolecules to complete mixing, and everything in between.7,54–58 In a binary mixture, for ex m ample, one may expect a well-mixed dense phase if the homotypic and heterotypic interactions are similar. On the other hand, if heterotypic interactions are weaker than self-interactions, these condensates may form distinct subcompartments rich in either of the two components.
The formation of such subcompartments can be crucial for providing more specialized control over the biochemical processes occurring inside the MLOs. The nucleoli’s three separately identifiable regions are enriched in RNA Pol I, fibrillarin, and nucleophosmin, which are essential for performing successive steps in generating mature pre-ribosomal particles.7 This behavior and other similar observations can be explained in terms of interfacial thermodynamics of liquid phases of different components,59 though a mechanistic understanding in terms of molecular interactions is still a work in progress. In this regard, various factors outlined in the previous section regarding sequence determinants of LLPS based on homotypic interactions, such as amino acid composition and patterning, and the presence of folded domains, should also apply to multicomponent phase separation.
Indeed, recent experimental studies on synthetic and natural protein sequences have begun to shed light on the balance of homotypic and heterotypic interactions underpinning multicomponent phase separation60. In phase separating RNA-protein mixtures,58,61 the morphology and composition of the condensate is dictated by a balance of homotypic (protein-protein and RNA-RNA) and heterotypic (protein-RNA) interactions. LLPS is enhanced in the presence of RNA at low concentrations due to favorable heterotypic interactions, whereas phase separation is inhibited at higher RNA concentrations due to repulsive interactions between RNA molecules. Such interactions, when modified by post-translational modifications (PTMs), can change the co-localization of proteins in a condensate. For instance, FMRP and CAPRIN1 are IDRs involved in translational regulation. Phosphorylation of either FMRP or CAPRIN1, but not both, promotes co-phase separation of the two proteins. Furthermore, inclusion of RNA resulted in striking differences in sub-compartmentalization depending on whether FMRP or CAPRIN1 was phosphorylated.62 A recent study presented a network model based on patchy-colloids theory to explain the assembly of stress granules (SGs) with attached P-bodies (PBs) in vivo.54 In the patchy-colloids framework, a system of interacting particles will phase separate into a connected, system-spanning network only if the particles have valence (number of binding sites) of three or more. G3BP plays a critical role in formation of SG/PB condensates upon RNA influx, with the G3BP oligomerization domain and RNA-binding domain both necessary for the high valence of G3BP. Together, these studies highlight the role of multivalency of intermolecular interactions, mixture composition, and stoichiometry. Furthermore, these examples illustrate that IDRs and folded domains can both play important roles in the intermolecular interactions underlying co-phase separation, with PTMs capable of tuning the interaction strength.
As multicomponent phase separation is dependent on a competition between homotypic and heterotypic interactions between molecules (including with solvent species), even a simple theory based on two-body interactions (ignoring multi-body effects) will require enumeration of these interactions between all possible pairs of molecules. Even if such information becomes available, it will be rather cumbersome to use it to develop multicomponent phase diagrams,29,63 which will still require untested approximations in the underlying thermodynamic mixture model.64 We believe that results of computer simulations based on theoretical models with increasing level of complexity can help identify the limitations of different approaches that are currently feasible. The phase behavior of complex polymer solution mixtures has been studied extensively due to its relevance to many industrial problems, and researchers should leverage the existing literature on that topic to make further progress in biomolecular condensates.65
Emerging experimental techniques will continue to advance our understanding of multicomponent phase separation. Proximity labeling approaches can identify unknown components of biomolecular condensates; one recent study identified ~150 human stress granule components using ascorbate peroxidase proximity labeling.66 Super-resolution microscopy has revealed micro-phases that may be difficult or impossible to resolve using diffraction-limited microscopy.54,67 Also regarding microscopy, conventional fluorescent tagging strategies used to visualize multicomponent phase separation may alter the sensitive balance of intermolecular interactions. An emerging approach particularly useful for in vivo studies is the development of phase separation biosensors, which partition into condensates without themselves phase separating or perturbing native LLPS.68
MLOs can regulate enzyme activity
What is the function of membraneless organelles? In many cases, it may be to regulate the rates of biochemical reactions (Fig. 3). Data is emerging to support this hypothesis, but many questions remain, and the field is wanting for additional quantitative evidence. There are a host of reasons why enzyme-catalyzed reactions may change upon incorporation into condensates. First, and perhaps most straightforwardly, enzyme and substrate concentrations may be altered in condensates depending on the extent of their recruitment or partitioning. Second, enzyme activity is sensitive to the environment. Data indicates that the interior of condensates may have polarity closer to that of the interior of proteins or polar organic solvents than to that of the bulk aqueous phase.69–72 Other important liquid properties that impact enzyme activity can be altered in biomolecular condensates, including higher viscosities and lower diffusion coefficients, altered effective pH, and reduced ionic strength.70,71 Third, enzyme structural changes can occur as enzymes move between environments, resulting in altered activity and stability.73 When these considerations are taken together, it seems highly likely that enzymatic reaction rates will change when an enzyme moves from the bulk phase to a condensate. Condensates can form and dissolve rapidly in response to stimuli, so condensate formation provides a mechanism for highly sensitive enzyme activity regulation. Determining the combination of enzyme characteristics and condensate composition that will result in increased or decreased enzyme activity is of high priority for advancing our understanding of the biochemical functions of membraneless organelles.
Figure 3 –
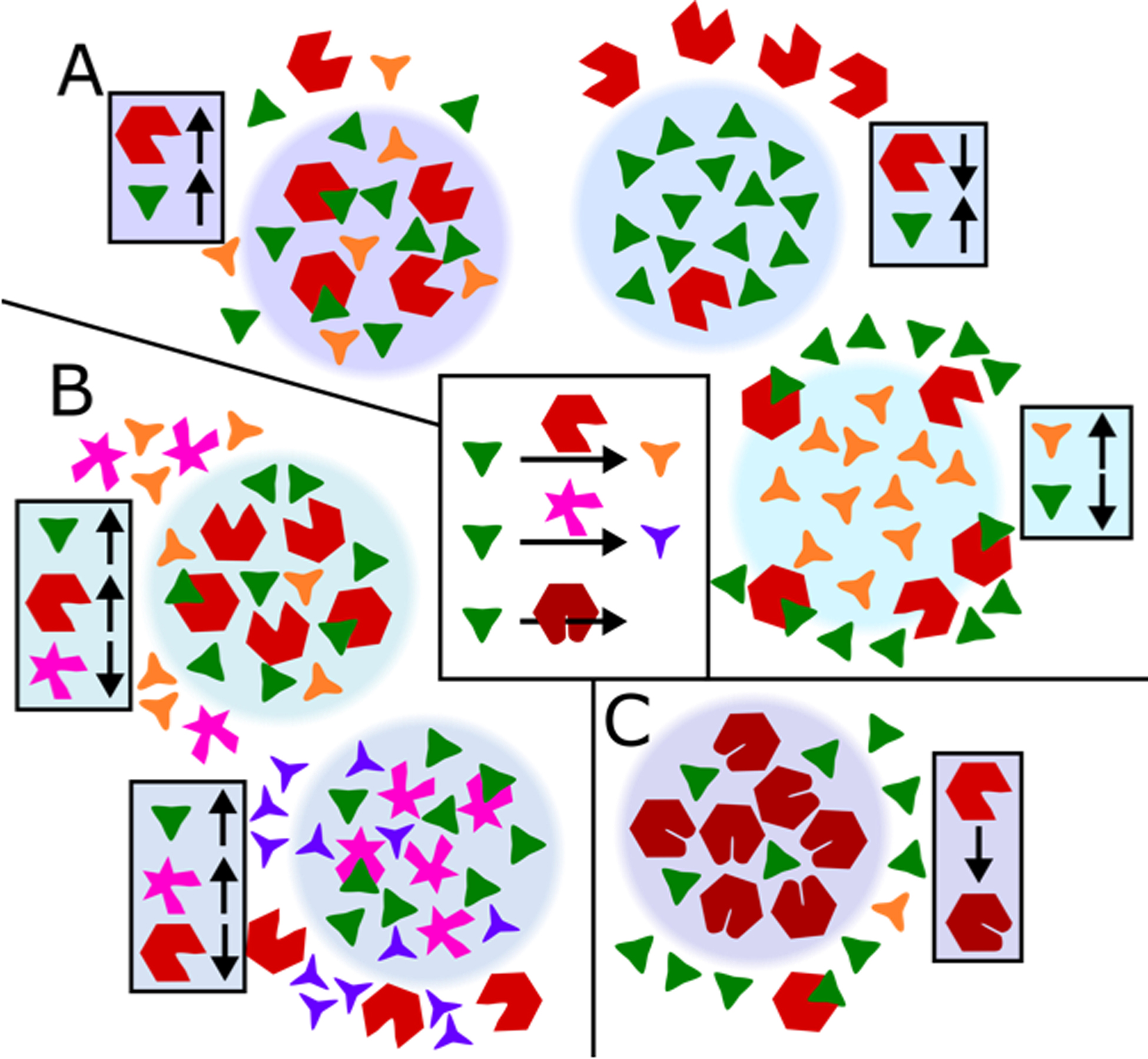
Modulation of enzyme activity by biomolecular condensates. Six outcomes are illustrated, highlighting different partitioning into condensates of enzymes, substrates, and products. A) Enzymes and substrates partitioned together or separately have been shown to enhance or reduce enzyme activity, respectively. Condensates which preferentially sequester unstable or toxic biproducts may localize the responsible enzymes at the condensate interface. B) Condensates can influence biochemical pathways by preferential incorporation of one enzyme over the other to alter reaction selectivity. C) Enzyme incorporation into the condensate environment could induce enzyme structural changes that alter enzyme kinetics.
Several recent studies in both synthetic and natural systems examined enzymatic reaction rate changes upon compartmentalization into biomolecular condensates.3 Michaelis-Menten kinetics and increased concentrations are frequently invoked to explain the changes, but as just discussed, there are numerous additional considerations. It is therefore challenging to predict enzyme activity changes upon compartmentalization. One might expect not only instances of enhanced reaction rates, but also instances of unchanged or attenuated activity. In one recent report, β-galactosidase fused to an IDP exhibited increasing enzyme activity as the phase-separating IDP increased in length, demonstrating a correlation between increased phase separation propensity and enhanced enzyme activity.35 Contrarily, another recent report describes a cascade of two enzymes for converting L-aspartate 4-semialdehyde into 1,3-diaminopropane that, once fused to a phase separating IDP, results in slower catalysis than the free enzymes.74 The discordant results of these two reports highlight that further investigation into the physical chemistry of biomolecular condensates will underpin progress towards rational engineering of artificial organelle reaction chambers.
One naturally occurring biomolecular condensate with catalytic function is the Bre1-Lge1 condensate, which promotes ubiquitination of histone H2B in gene-body nucleosomes in yeast. Lge1 forms a core surrounded by a shell of Bre1 that together recruit nucleosome core particles and enzymes required for ubiquitination. Notably, the core-shell architecture of this condensate was found to be necessary for accelerating H2B ubiquitination.75 As a second example, purinosomes colocalize enzymes involved in the de novo purine biosynthesis pathway. A recent study demonstrated that purinosome enrichment yields increased purine biosynthesis. Purinosome-enriched cells contained increased concentration of the purine nucleotide precursor IMP and exhibited enhanced metabolic flux of the de novo purine biosynthesis pathway.76 These studies reveal the impact of biomolecular condensates on enzyme activity, in both synthetic and natural systems. However, insight is lacking into the mechanisms responsible for the changes in catalysis, and future experimental and computational research will seek to dissect these mechanisms.
Studying enzymes in membraneless organelles is complex and highly dependent on the reaction in question. In classical enzymology experiments, catalysis occurs homogeneously in an aqueous solution. In the case of enzyme activity involving biomolecular condensates, even single enzyme-substrate reactions become biphasic or multiphasic experiments where enzymes, substrates, and products separate between the liquid phases. This presents challenges and opportunities for experimental and theoretical research studying enzyme kinetics involving biomolecular condensates. Enzyme activity calculations must consider at least three different locations – the condensate, the dilute phase (bulk solvent), and the interface between the two phases. If the substrate and enzyme both partition into the condensate, catalysis may occur predominantly in the condensed phase. Or, enzymes that localize to or act at the interface may play a larger catalytic role. For instance, in one recent study, phase-separated DNA droplets underwent degradation when a SmaI restriction enzyme was applied. The enzyme activity was confined to the droplet surface (phase interface) when the system was far below the dissolution temperature. However, SmaI cleaved the DNA throughout the droplet at higher temperatures, because higher temperatures altered the droplet’s permeability, allowing for increased penetration of the restriction enzyme into the condensate.77 In both of these scenarios, droplet size (and consequently the surface area to volume ratio) is an important variable, which will impact diffusion times, partitioning, and interfacial catalysis. Finally, in LLPS systems, phase-separating proteins (e.g. an enzyme-IDP fusion) will be present in the dilute phase at the saturation concentration, and so the dilute phase should not be neglected as a possible location for catalysis.
Kinetics studies must also consider enzyme structure. Protein structure rearrangements frequently occur as proteins move between environments, e.g. nucleus, cytosol, and extracellular.78 Similar structural or functional shifts should be expected of enzymes as they transition from the cytosol into condensates, resulting in Michaelis constant Km and catalytic rate constant kcat varying depending on the location of the enzyme. Specific enzyme structural characteristics will dictate enzyme activity upon condensate sequestration, and condensate properties will affect sequestered enzymes. Both aspects require investigation. Recent work on synthetic droplet reactors describes condensate properties that can be characterized in purified systems, including condensate polarity, partition coefficients, enzyme concentration, and localized enzyme activity.71 Most enzyme-condensate systems are not as well characterized, especially in vivo. We will later describe methods for determining condensate rheology, which impacts the diffusion-limited on-rate constant for enzyme-substrate binding, but these and other liquid properties, including proticity, ionicity, and polarity, will vary and impact reaction kinetics. Condensate-sequestered enzyme conformational changes could be characterized by cryogenic electron tomography and other high-resolution techniques.78,79 Molecular dynamics simulations could examine how enzymes interact with different condensate environments. Bioinformatics methods could examine conserved features of enzymes found in biomolecular condensates.80 Our understanding of condensate-enzyme interactions is still in the early stages of development, as are the methods. Exploring how enzymes interact with condensates will inform our understanding of their biological functions and potential applications in designer synthetic organelles.
Having discussed several examples and physicochemical considerations of enzymatic reactions localized to condensates, we now speculate on how enzymes may have evolved to harness LLPS for functional regulation. As discussed earlier, colocalizing enzymes to enhance activity is of great interest, but diminished activity upon colocalization to a condensate is at least equally important biologically. Membraneless organelles that sequester substrates away from enzymes can reduce reaction rates, as is the case for mRNA sequestered in stress granules away from the translational machinery.81,82 As a second example, proteins (e.g. TDP-43) that localize to stress granules may be shielded from stress-activated kinases, safeguarding the proteins from forming disease-related phosphorylated aggregates.83 Enzymes such as these kinases could have evolved to be excluded from condensates – partitioning dissimilarly from their substrates – to allow for stimulus-responsive attenuation of enzyme activity. In other scenarios, enzymes and condensate proteins could perhaps coevolve to favor partitioning, as in the case of the Lge1-Bre1 condensate.75 These regulatory mechanisms that evolved in cells have provided inspiration for synthetic biologists. One recent study developed a synthetic condensate in yeast that, under control of an optogenetic switch, colocalized enzymes from the deoxyviolacein pathway to direct flux towards the desired product, resulting in product enrichment of up to 18-fold.76
Finally, we note that recruitment of proteins into biomolecular condensates can also regulate the kinetics of non-enzymatic processes – e.g. branching microtubule nucleation, carboxysome formation, and tau fibril aggregation.84–87 These assembly events are not enzyme-based, but similarly to the examples discussed thus far, exhibit enhanced kinetics due to condensate colocalization. These natural systems could serve as inspiration for harnessing condensate colocalization to template the assembly of engineered bionanomaterials.
Surface tension and rheology are critical properties of condensates, but challenging to quantify
How do the material properties of biomolecular condensates, such as viscosity and surface tension, affect biochemical and biological function? This important question is just beginning to be explored. Surface tension and viscosity of condensates are essential for subcompartment formation,7,55,88 gene regulation,89,90 and condensate-membrane interactions.4 Another recent report found that Ape1 condensates must be liquid, not solid, for selective autophagy in yeast.91 Apart from normal cell physiology, many aging-related diseases are linked to pathological fibrils that form when cells lose control over the liquidity of corresponding protein condensates.50,92–94 Detailed understanding of material properties is therefore central to understanding biomolecular condensate biology and pathology, and furthermore, will guide the design of medical and biotechnological applications.95,96
Surface tension and viscosity are two main parameters that define the material state of a liquid. Surface tension is a static property, whereas viscosity describes the motion of a liquid under external force. Both originate from intermolecular forces, and in macromolecular systems, intramolecular forces also play an important role through their effect on biopolymer structure, radius of gyration, and flexibility.97 Nonetheless, the relation between surface tension and viscosity is highly nontrivial, where only empirical correlations exist within certain families of liquids.98,99 Therefore, surface tension and viscosity are often considered as two independent liquid properties.100 Indeed, among the limited data of surface tension and viscosity for biomolecular condensates, they have been almost exclusively measured using separate techniques.7,101–103 The micro-scale nature of biomolecular condensates means that traditional rheometers or tensiometers are rarely applicable when studying the properties of protein/RNA condensates,92 and instead microscopy-based measurements predominate.
So far, two techniques have been mainly applied to quantify the viscosity of condensates: most commonly fluorescence recovery after photobleaching (FRAP), and to a lesser extent, single particle tracking (SPT). Both techniques take advantage of the Stoke-Einstein relation,104 where the viscosity of a liquid is inferred from the Brownian motion of spherical particles within it, (η is the liquid viscosity, D and a are the diffusion coefficient and the radius of the chosen particle, respectively). Apart from several technical complications,105 FRAP is intrinsically limited in that the shape of the monitored fluorescent molecule can significantly deviate from a sphere, and that the hydrodynamic radii of molecules are often unknown. Therefore, often only an apparent viscosity can be extracted from the recovery of bleached molecules, limiting the comparability of FRAP results using different tracer molecules. Additionally, biomolecular condensates are often not pure liquids, and an elastic component can make the FRAP data nontrivial to quantify.92,106 SPT solves the first issue by using well-defined spherical particles. For the second issue, a generalized Stokes-Einstein relation107,108 can be applied to SPT data to extract both viscous and elastic moduli. However, to apply SPT in different systems, the surface chemistry of particles often must be finely tuned to avoid particle aggregation inside the condensate.109 Both techniques can in principle be conducted on in vitro or in vivo condensates, but in vivo experiments are more difficult to perform and quantitatively interpret, due to the small size of intracellular condensates and the complexity of the cellular environment. Additionally, both FRAP and SPT are limited to measuring passive viscosity under zero applied force, a likely oversimplification compared to the heterogeneous mechanical environment that condensates experience in the cell.110 More recently, two more approaches based on microfluidic shear stress103 and oscillatory stretch through dual optical traps106,111 have been applied to study the active rheology of condensates. Albeit challenging on their own rights, these new approaches promise a more accurate description of the viscoelastic behavior of biomolecular condensates. Other active microrheology techniques, such as magnetic twisting,112 are promising candidates to be applied in biomolecular condensate studies.
Compared to viscosity measurements, quantifying the surface tension of condensates is more obscure. (We use the term surface tension to be consistent with the MLO literature, although elsewhere the term surface tension is reserved for gas-liquid interfaces while the more general term interfacial tension is used for gas-liquid, liquid-liquid, or solid-liquid interfaces.) The only widely applied technique to measure condensate surface tension relies on the scaling between the fusion time and the length scale of two coalescing droplets, from which only the ratio of surface tension and viscosity can be extracted.101 One of the first direct surface tension measurements was achieved by adapting the sessile drop technique. Side-view morphology of condensates, which represents a balance between surface tension and gravity, was used to quantify surface tension when the density of condensates was known.7 More recently, shape fluctuations of the nucleolus were monitored to report on its surface tension,102 resembling the use of flickering spectroscopy to measure membrane tension of giant lipid vesicles.113 So far, all measurements of biomolecular condensates report a very low surface tension, ~ 10−3 mN/m,7,101,102,106 less than ten thousandth of the surface tension of oil droplets in water. Is the low surface tension a unique feature of biomolecular condensates? For entropy-driven colloidal condensates in a polymer solution, surface tension of the condensates scales as , leading to a low surface tension for condensates of protein-size particles (γ~10−2 mN/m, for a ~10 nm colloid).90,114 However, biomolecules can have direct enthalpic attractions towards each other, potentially producing high surface tension condensates that are waiting to be discovered.
When quantifying condensate rheology, it is tempting to treat condensates as a bulk material. To be sure, the bulk rheology is salient when considering whether a condensate as a whole behaves as a liquid or gel, which contributes to the condensate’s fusion rate and how it may interact with other organelles and cellular structures. As the field looks to the future, we suggest that rheological studies should also consider the molecular microstructure of condensates. Condensates may be thought of as a porous material, with a mesh size on the order of nanometers, dependent on the intermolecular interactions and crosslinks in the condensed phase.95,115 Small molecules can access the interstices between the biopolymers, which are expected to be a watery, low-viscosity environment. A small molecule in a condensate may therefore experience a viscosity that is orders of magnitude smaller than would a probe particle larger than the mesh size of the condensate.115,116 This nanoscale rheology may be particularly relevant to understanding the dynamics and biochemical activities of small drug molecules or native metabolites that partition into membraneless organelles.96 Many of the abovementioned techniques are most suitable for bulk rheology measurements, but FRAP of small molecule fluorophores or fluorescence correlation spectroscopy (FCS) can provide information about nanoscale rheology.115
To more fully understand the physical chemistry of biomolecular condensates, future work must unravel the sequence determinants of material properties. Important progress has been made in understanding the sequence determinants of phase separation, but much less is known about how sequence encodes droplet rheology and surface tension. This information would inform how mutations and post-translational modifications alter material properties, and how changes to rheology and surface tension may execute biological function or cause disease. In recent work, we found that, surprisingly, two charge-patterned variants of the LAF-1 RGG domain did not significantly alter droplet dynamics.18 In contrast, several other recent papers identified sequence perturbations that did alter material properties. In hnRNPA1, clustering of aromatic residues resulted in amorphous aggregates rather than spherical liquid.43 In FUS, G to A mutation reduced droplet dynamics, whereas Q to G mutation prevented droplet hardening over time.27 In the UBQLN2 C-terminal region, certain point mutations that increased hydrophobicity slowed droplet fusion.49 Condensates of poly-R show ~100 fold greater viscosity than comparable poly-K condensates and ~10-fold greater surface tension.88 Future work will aim to ascertain the molecular basis of such effects and expand our understanding of what sequence features encode material properties, ultimately aiming to develop a predictive framework for relating sequence to rheology and surface tension in biomolecular condensates.
As researchers continue to elucidate the relationship between condensate rheology and function, there is much room for development of new techniques and analysis, both for in vitro and in vivo condensates. Non-invasive approaches, such as genetically-encoded multimeric nanoparticles117 and optical trap-based microrheology106,111 are particularly appealing for their potential to be applied to live samples. Another desirable feature of future technology is high-sensitivity tracing of the change in condensate properties over time. This is important because biomolecular condensates in cells are rarely at thermodynamic equilibrium: material properties of condensates can not only change with applied force, but also change with time. For instance, this is manifested in the glass-like aging of PGL-3 condensates111 or the disease-related liquid-to-solid fibrilization of FUS bearing the G156E mutation found in some ALS patients.50 New in vitro tools must minimize the effects of potential artifacts due to adhesion between condensates and the substrate (e.g. the coverglass). There is also need for higher throughput approaches to measure condensate material properties. Large-scale mutagenesis studies and IDP libraries have been employed to uncover sequence determinants of phase separation and IDP toxicity,35,118 but measurements of membraneless organelle material properties have been comparatively low throughput, and higher-throughput approaches may help advance the field.
Conclusion/Outlook
Tremendous research effort in recent years has begun to shed light on the biophysical chemistry of membraneless organelles. Although the field may no longer be in its infancy, it is far from maturity, with many open questions remaining to be tackled. Building upon insights into the sequence determinants of phase separation of individual proteins, experimental and computational studies are now investigating the complex interactions that give rise to multi-component and multi-phase condensates, which may be the rule rather than the exception in biology. The ultimate goal of this research is to predictively understand how biopolymer sequence and molecular interactions give rise to the emergent properties and functions of membraneless organelles. One such function is the regulation of enzymatic activity through sequestration of enzymes, substrates, or both into biomolecular condensates. Central to this function, and many other biological functions and dysfunctions, are the material properties of MLOs. New techniques are emerging that enable improved measurements of the rheology and surface tension of biomolecular condensates, promising to help resolve key questions about the time- and context-dependence of material properties and their biochemical consequences. In the coming years, progress towards these fundamental questions will also translate into advances in biomaterials, engineered cells and protocells, and novel therapeutic approaches for disease processes linked with biomolecular phase separation.
Figure 4.
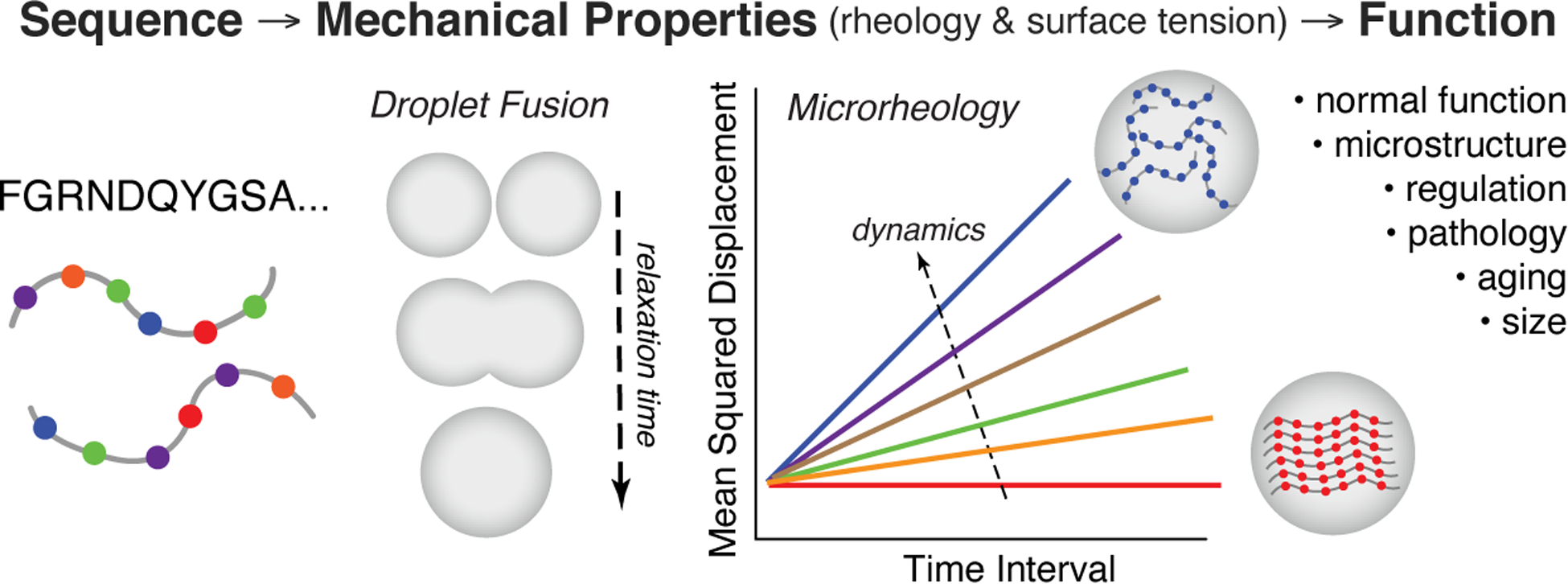
The material properties of biomolecular condensates arise from sequence-encoded information and impart biological function. A growing suite of techniques for measuring condensate surface tension and rheology is beginning to shed light on how condensate material properties relate to native biological processes, as well as how material properties change with aging or disease.
Acknowledgement
This work was supported in part by the National Institutes of Health R01NS116176 (J.M.), R01GM120537 (J.M.), National Science Foundation grant DMR2004796 (J.M.), CBET1929237 (S.D.K.).
Biographies
Benjamin S. Schuster is an assistant professor at Rutgers University in the Department of Chemical and Biochemical Engineering. His lab studies the biophysics of protein liquid-liquid phase separation (LLPS) and bioengineering applications of LLPS. He joined Rutgers in 2019 following a postdoctoral fellowship at the University of Pennsylvania, graduate studies at Johns Hopkins University, and undergraduate studies at the University of Minnesota.
Roshan M. Regy is a Ph.D student advised by Professor Jeetain Mittal at the Chemical and Biomolecular Engineering Department, Lehigh University. His work focuses on investigating sequence-specific phase separation drivers of Intrinsically Disordered Proteins using computational methods. He started his Ph.D. in Chemical Engineering at Lehigh University in 2018 after finishing his undergraduate studies in Chemical Engineering in 2017 from B.I.T.S. Pilani, India.
Elliott M. Dolan is a PhD Candidate in the Khare Lab at Rutgers University in the Department of Chemistry and Chemical Biology. His research centers on designing stimulus-responsive proteins including protein assemblies and photo-controllable enzyme catalysts. He began his doctoral work in 2016 following his Masters studies in Biomedical Engineering at Rutgers University and undergraduate studies in Chemical Engineering at The Ohio State University.
Aishwarya Kanchi Ranganath is a Master’s student working in the Schuster lab at Rutgers University in the Department of Chemical and Biochemical Engineering. Her work is mainly focused on understanding the sequence determinants and biophysics of liquid-liquid phase separation (LLPS) using techniques like turbidity assays and fluorescence microscopy. She joined Rutgers in 2018 following her undergraduate studies in Chemical Engineering from SRMIST, India.
Nina Jovic has a Bachelors in Chemical Engineering from the University of Florida. She joined the Chemical and Biomolecular Department at Lehigh University in 2018 for her PhD and is working with Professor Jeetain Mittal. One of her areas of study is investigating molecular determinants of protein phase separation using atomistic simulations.
Sagar D. Khare is an Associate Professor in the Department of Chemistry and Chemical Biology at Rutgers University. His lab is developing computational and experimental tools to dissect and design proteins. Current projects include the design of enzyme stability, stimulus responsiveness, substrate specificity and supramolecular structures for applications in therapeutics and industrial biocatalysts. He was an undergraduate student at the Indian Institute of Technology Delhi, a graduate student at The University of North Carolina at Chapel Hill, and a postdoctoral fellow at the University of Washington, Seattle, prior to starting his lab at Rutgers in 2012.
Zheng Shi is an assistant professor at Rutgers University in the department of Chemistry and Chemical Biology. His lab studies the mechanics of cell membranes and biomolecular condensates. Before joining Rutgers, Zheng was a postdoc at Harvard University. He got his Ph.D. in Chemistry at the University of Pennsylvania and Bachelor’s degree in Physics at the University of Science and Technology of China.
Jeetain Mittal is currently Ruth H. and Sam Madrid Endowed Chair Professor of Chemical and Biomolecular Engineering at Lehigh University. He joined Lehigh in 2009 following a postdoctoral fellowship at the National Institutes of Health and Ph.D. at the University of Texas at Austin. His group is developing predictive physics-based computational tools to identify the fundamental rules that govern structural and compositional ordering in a wide variety of systems with a specific focus on the following active research projects: (1) biomolecular phase separation and (2) nanoparticle superlattice engineering by DNA-mediated interactions.
References
- 1.Banani SF, Lee HO, Hyman AA & Rosen MK Biomolecular condensates: Organizers of cellular biochemistry. Nature Reviews Molecular Cell Biology vol. 18 285–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shin Y & Brangwynne CP Liquid phase condensation in cell physiology and disease. Science vol. 357 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Lyon AS, Peeples WB & Rosen MK A framework for understanding the functions of biomolecular condensates across scales. Nature Reviews Molecular Cell Biology (2020) doi: 10.1038/s41580-020-00303-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Milovanovic D, Wu Y, Bian X & De Camilli P A liquid phase of synapsin and lipid vesicles. Science 361, 604–607 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryan VH & Fawzi NL Physiological, Pathological, and Targetable Membraneless Organelles in Neurons. Trends in Neurosciences vol. 42 693–708 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alberti S & Dormann D Liquid-Liquid Phase Separation in Disease. Annu. Rev. Genet 53, 171–194 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Feric M et al. Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell 165, 1686–1697 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Putnam A, Cassani M, Smith J & Seydoux G A gel phase promotes condensation of liquid P granules in Caenorhabditis elegans embryos. Nat. Struct. Mol. Biol 26, 220–226 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boeynaems S et al. Protein Phase Separation: A New Phase in Cell Biology. Trends in Cell Biology vol. 28 420–435 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dignon GL, Zheng W & Mittal J Simulation methods for liquid–liquid phase separation of disordered proteins. Current Opinion in Chemical Engineering vol. 23 92–98 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shea J, Best RB & Mittal J Physics-based computational and theoretical approaches to intrinsically disordered proteins. Curr. Opin. Struct. Biol 67, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi JM, Dar F & Pappu RV LASSI: A lattice model for simulating phase transitions of multivalent proteins. PLoS Comput. Biol 15, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Methods in Enzymology: Liquid-liquid Phase Coexistence and Membraneless Organelles. (Elsevier, 2021). [Google Scholar]
- 14.Burke KA, Janke AM, Rhine CL & Fawzi NL Residue-by-Residue View of In Vitro FUS Granules that Bind the C-Terminal Domain of RNA Polymerase II. Mol. Cell 60, 231–241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conicella AE, Zerze GH, Mittal J & Fawzi NL ALS Mutations Disrupt Phase Separation Mediated by α-Helical Structure in the TDP-43 Low-Complexity C-Terminal Domain. Structure (2016) doi: 10.1016/j.str.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conicella AE et al. TDP-43 α-helical structure tunes liquid–liquid phase separation and function. Proc. Natl. Acad. Sci. U. S. A 117, 5883–5894 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elbaum-Garfinkle S et al. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. U. S. A 112, 7189–7194 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schuster BS et al. Identifying sequence perturbations to an intrinsically disordered protein that determine its phase-separation behavior. Proc. Natl. Acad. Sci. U. S. A 117, 11421–11431 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryan VH et al. Mechanistic View of hnRNPA2 Low-Complexity Domain Structure, Interactions, and Phase Separation Altered by Mutation and Arginine Methylation. Mol. Cell 69, 465–479.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryan VH et al. Tyrosine phosphorylation regulates hnRNPA2 granule protein partitioning & reduces neurodegeneration. The EMBO Journal 1–38 (2020) doi: 10.1101/2020.03.15.992768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Labialle S et al. The miR- 379 / miR- 410 cluster at the imprinted Dlk 1 -Dio 3 domain controls neonatal metabolic adaptation. EMBO J. 33, 2216–2230 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seif E et al. Phase separation by the polyhomeotic sterile alpha motif compartmentalizes Polycomb Group proteins and enhances their activity. Nat. Commun 11, 1–19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yeong V, Werth EG, Brown LM & Obermeyer AC Formation of Biomolecular Condensates in Bacteria by Tuning Protein Electrostatics. ACS Cent. Sci (2020) doi: 10.1021/acscentsci.0c01146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banani SF et al. Compositional Control of Phase-Separated Cellular Bodies. Cell 166, 651–663 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alberti S, Gladfelter A & Mittag T Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell vol. 176 419–434 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quiroz FG & Chilkoti A Sequence heuristics to encode phase behaviour in intrinsically disordered protein polymers. Nat. Mater 14, 1164–1171 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J et al. A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell 174, 688–699.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vernon RMC et al. Pi-Pi contacts are an overlooked protein feature relevant to phase separation. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dignon GL, Best RB & Mittal J Biomolecular phase separation: From molecular driving forces to macroscopic properties. Annual Review of Physical Chemistry vol. 71 53–75 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Das S, Lin YH, Vernon RM, Forman-Kay JD & Chan HS Comparative roles of charge, π, and hydrophobic interactions in sequence-dependent phase separation of intrinsically disordered proteins. Proc. Natl. Acad. Sci. U. S. A 117, 28795–28805 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brady JP et al. Structural and hydrodynamic properties of an intrinsically disordered region of a germ cell-specific protein on phase separation. Proc. Natl. Acad. Sci. U. S. A 114, E8194–E8203 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Danielsen SPO, McCarty J, Shea JE, Delaney KT & Fredrickson GH Molecular design of self-coacervation phenomena in block polyampholytes. Proc. Natl. Acad. Sci. U. S. A 116, 8224–8232 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gabryelczyk B et al. Hydrogen bond guidance and aromatic stacking drive liquid-liquid phase separation of intrinsically disordered histidine-rich peptides. Nat. Commun 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murthy AC et al. Molecular interactions underlying liquid−liquid phase separation of the FUS low-complexity domain. Nat. Struct. Mol. Biol 26, 637–648 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dzuricky M, Rogers BA, Shahid A, Cremer PS & Chilkoti A De novo engineering of intracellular condensates using artificial disordered proteins. Nat. Chem 12, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dignon GL, Zheng W, Kim YC, Best RB & Mittal J Sequence determinants of protein phase behavior from a coarse-grained model. PLoS Comput. Biol 14, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perdikari TM et al. A coarse-grained model for position-specific effects of post-translational modifications on disordered protein phase separation. Biophys. J In press, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng W et al. Molecular Details of Protein Condensates Probed by Microsecond Long Atomistic Simulations. J. Phys. Chem. B 124, 11671–11679 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turupcu A, Tirado-Rives J & Jorgensen WL Explicit representation of cation-π interactions in force fields with 1/r4nonbonded terms. J. Chem. Theory Comput 16, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Das RK & Pappu RV Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues. Proc. Natl. Acad. Sci. U. S. A 110, 13392–13397 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin Y-H & Chan HS Phase Separation and Single-Chain Compactness of Charged Disordered Proteins Are Strongly Correlated. Biophys. J 112, 2043–2046 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dignon GL, Zheng W, Best RB, Kim YC & Mittal J Relation between single-molecule properties and phase behavior of intrinsically disordered proteins. Proc. Natl. Acad. Sci. U. S. A 115, 9929–9934 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin EW et al. Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science 367, 694–699 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huihui J, Firman T & Ghosh K Modulating charge patterning and ionic strength as a strategy to induce conformational changes in intrinsically disordered proteins. J. Chem. Phys 149, (2018). [DOI] [PubMed] [Google Scholar]
- 45.Sawle L & Ghosh K A theoretical method to compute sequence dependent configurational properties in charged polymers and proteins. J. Chem. Phys 143, 085101 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Zheng W, Dignon G, Brown M, Kim YC & Mittal J Hydropathy Patterning Complements Charge Patterning to Describe Conformational Preferences of Disordered Proteins. J. Phys. Chem. Lett 11, 3408–3415 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Das S, Amin AN, Lin YH & Chan HS Coarse-grained residue-based models of disordered protein condensates: Utility and limitations of simple charge pattern parameters. Phys. Chem. Chem. Phys 20, 28558–28574 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Choi JM, Holehouse AS & Pappu RV Physical Principles Underlying the Complex Biology of Intracellular Phase Transitions. Annual Review of Biophysics vol. 49 107–133 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Y, Jones HB, Dao TP & Castañeda CA Single Amino Acid Substitutions in Stickers, but Not Spacers, Substantially Alter UBQLN2 Phase Transitions and Dense Phase Material Properties. J. Phys. Chem. B 123, 3618–3629 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Patel A et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell (2015) doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 51.Lee CYS et al. Recruitment of mRNAs to P granules by condensation with intrinsically-disordered proteins. Elife 9, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schuster BS et al. Controllable protein phase separation and modular recruitment to form responsive membraneless organelles. Nat. Commun 9, 1–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Janke AM et al. Lysines in the RNA Polymerase II C-Terminal Domain Contribute to TAF15 Fibril Recruitment. Biochemistry 57, 2549–2563 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanders DW et al. Competing Protein-RNA Interaction Networks Control Multiphase Intracellular Organization. Cell 181, 306–324.e28 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaur T et al. Sequence-encoded and Composition-dependent Protein-RNA Interactions Control Multiphasic Condensate Topologies. bioRxiv 2020.08.30.273748 (2020) doi: 10.1101/2020.08.30.273748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roberts S et al. Complex microparticle architectures from stimuli-responsive intrinsically disordered proteins. Nat. Commun 11, 1342 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.H Y et al. HSP70 chaperones RNA-free TDP-43 into anisotropic intranuclear liquid spherical shells. Science (2020) doi: 10.1126/SCIENCE.ABB4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Regy RM, Dignon GL, Zheng W, Kim YC & Mittal J Sequence dependent phase separation of protein-polynucleotide mixtures elucidated using molecular simulations. Nucleic Acids Res. 48, 12593–12603 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mao S, Chakraverti-Wuerthwein MS, Gaudio H & Kosmrlj A Designing morphology of separated phases in multicomponent liquid mixtures. Phys. Rev. Lett 125, (2020). [DOI] [PubMed] [Google Scholar]
- 60.Mammen Regy R & Mittal J Living cells as test tubes. Nat. Chem. Biol 16, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Banerjee PR, Milin AN, Moosa MM, Onuchic PL & Deniz AA Reentrant Phase Transition Drives Dynamic Substructure Formation in Ribonucleoprotein Droplets. Angew. Chemie - Int. Ed 56, 11354–11359 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim TH et al. Phospho-dependent phase separation of FMRP and CAPRIN1 recapitulates regulation of translation and deadenylation. Science 365, 825–829 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Mao S, Chakraverti-Wuerthwein MS, Gaudio H & Košmrlj A Designing morphology of separated phases in multicomponent liquid mixtures. arXiv (2020) doi: 10.1103/physrevlett.125.218003. [DOI] [PubMed] [Google Scholar]
- 64.Heidenreich M et al. Designer protein assemblies with tunable phase diagrams in living cells. Nat. Chem. Biol 16, 939–945 (2020). [DOI] [PubMed] [Google Scholar]
- 65.Koningsveld R, Stockmayer WH & Nies E Polymer Phase Diagrams. vol. 36 (Oxford University Press, 2001). [Google Scholar]
- 66.Markmiller S et al. Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules. Cell 172, 590–604.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang JT et al. Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. Elife 3, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Quiroz FG et al. Liquid-liquid phase separation drives skin barrier formation. Science 367, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Klibanov AM Improving enzymes by using them in organic solvents. Nature vol. 409 241–246 (2001). [DOI] [PubMed] [Google Scholar]
- 70.Nott TJ et al. Phase Transition of a Disordered Nuage Protein Generates Environmentally Responsive Membraneless Organelles. Mol. Cell 57, 936–947 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Küffner AM et al. Acceleration of an Enzymatic Reaction in Liquid Phase Separated Compartments Based on Intrinsically Disordered Protein Domains. ChemSystemsChem 2, (2020). [Google Scholar]
- 72.Crabtree MD et al. Repulsive electrostatic interactions modulate dense and dilute phase properties of biomolecular condensates. bioRxiv 2020.10.29.357863 (2020) doi: 10.1101/2020.10.29.357863. [DOI] [Google Scholar]
- 73.Secundo F Conformational changes of enzymes upon immobilisation. Chem. Soc. Rev 42, 6250–6261 (2013). [DOI] [PubMed] [Google Scholar]
- 74.Wei SP et al. Formation and functionalization of membraneless compartments in Escherichia coli. Nat. Chem. Biol 16, 1143–1148 (2020). [DOI] [PubMed] [Google Scholar]
- 75.Gallego LD et al. Phase separation directs ubiquitination of gene-body nucleosomes. Nature 579, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao EM et al. Light-based control of metabolic flux through assembly of synthetic organelles. Nat. Chem. Biol 15, 589–597 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Saleh OA, Jeon BJ & Liedl T Enzymatic degradation of liquid droplets of DNA is modulated near the phase boundary. Proc. Natl. Acad. Sci. U. S. A 117, 16160–16166 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.König I et al. Single-molecule spectroscopy of protein conformational dynamics in live eukaryotic cells. Nat. Methods 12, 773–779 (2015). [DOI] [PubMed] [Google Scholar]
- 79.Franzmann TM et al. Phase separation of a yeast prion protein promotes cellular fitness. Science 359, (2018). [DOI] [PubMed] [Google Scholar]
- 80.Kuechler ER, Budzyńska PM, Bernardini JP, Gsponer J & Mayor T Distinct Features of Stress Granule Proteins Predict Localization in Membraneless Organelles. J. Mol. Biol 432, 2349–2368 (2020). [DOI] [PubMed] [Google Scholar]
- 81.Anderson P & Kedersha N RNA granules. Journal of Cell Biology vol. 172 803–808 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Simon JR, Eghtesadi SA, Dzuricky M, You L & Chilkoti A Engineered Ribonucleoprotein Granules Inhibit Translation in Protocells. Mol. Cell 75, 66–75.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McGurk L et al. Poly(ADP-Ribose) Prevents Pathological Phase Separation of TDP-43 by Promoting Liquid Demixing and Stress Granule Localization. Mol. Cell 71, 703–717.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.King MR & Petry S Phase separation of TPX2 enhances and spatially coordinates microtubule nucleation. Nat. Commun 11, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Oltrogge LM et al. Multivalent interactions between CsoS2 and Rubisco mediate α-carboxysome formation. Nat. Struct. Mol. Biol 27, 281–287 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang H et al. Rubisco condensate formation by CcmM in β-carboxysome biogenesis. Nature 566, 131–135 (2019). [DOI] [PubMed] [Google Scholar]
- 87.Wegmann S et al. Tau protein liquid–liquid phase separation can initiate tau aggregation. EMBO J. 37, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fisher RS & Elbaum-Garfinkle S Tunable multiphase dynamics of arginine and lysine liquid condensates. Nat. Commun 11, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shin Y et al. Liquid Nuclear Condensates Mechanically Sense and Restructure the Genome. Cell 175, 1481–1491.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brangwynne CP, Mitchison TJ & Hyman AA Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. U. S. A 108, 4334–4339 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yamasaki A et al. Liquidity Is a Critical Determinant for Selective Autophagy of Protein Condensates. Mol. Cell 77, 1163–1175.e9 (2020). [DOI] [PubMed] [Google Scholar]
- 92.Guo L et al. Nuclear-Import Receptors Reverse Aberrant Phase Transitions of RNA-Binding Proteins with Prion-like Domains. Cell 173, 677–692.e20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mathieu C, Pappu RV & Paul Taylor J Beyond aggregation: Pathological phase transitions in neurodegenerative disease. Science vol. 370 56–60 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nedelsky NB & Taylor JP Bridging biophysics and neurology: aberrant phase transitions in neurodegenerative disease. Nature Reviews Neurology vol. 15 272–286 (2019). [DOI] [PubMed] [Google Scholar]
- 95.Bracha D, Walls MT & Brangwynne CP Probing and engineering liquid-phase organelles. Nature Biotechnology vol. 37 1435–1445 (2019). [DOI] [PubMed] [Google Scholar]
- 96.Klein IA et al. Partitioning of cancer therapeutics in nuclear condensates. Science 368, 1386–1392 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wilkes GL An overview of the basic rheological behavior of polymer fluids with an emphasis on polymer melts. J. Chem. Educ 58, 880–892 (1981). [Google Scholar]
- 98.Pelofsky AH Surface Tension-Viscosity Relation for Liquids. J. Chem. Eng. Data 11, 394–397 (1966). [Google Scholar]
- 99.Queimada AJ, Marrucho IM, Stenby EH & Coutinho JAP Generalized relation between surface tension and viscosity: A study on pure and mixed n-alkanes. in Fluid Phase Equilibria vols 222–223 161–168 (Elsevier, 2004). [Google Scholar]
- 100.Bormashenko EY Wetting of Real Surfaces. Wetting of Real Surfaces (De Gruyter, 2018). doi: 10.1515/9783110583144. [DOI] [Google Scholar]
- 101.Brangwynne CP et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732 (2009). [DOI] [PubMed] [Google Scholar]
- 102.Caragine CM, Haley SC & Zidovska A Surface Fluctuations and Coalescence of Nucleolar Droplets in the Human Cell Nucleus. Phys. Rev. Lett 121, 148101 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Taylor N et al. Biophysical characterization of organelle-based RNA/protein liquid phases using microfluidics. Soft Matter 12, 9142–9150 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Einstein A On the Motion of Small Particles Suspended in Liquids at Rest Required by the Molecular-Kinetic Theory of Heat. (1905).
- 105.Taylor NO, Wei MT, Stone HA & Brangwynne CP Quantifying Dynamics in Phase-Separated Condensates Using Fluorescence Recovery after Photobleaching. Biophys. J 117, 1285–1300 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jawerth LM et al. Salt-Dependent Rheology and Surface Tension of Protein Condensates Using Optical Traps. Phys. Rev. Lett 121, 258101 (2018). [DOI] [PubMed] [Google Scholar]
- 107.Mason TG, Ganesan K, Van Zanten JH, Wirtz D & Kuo SC Particle tracking microrheology of complex fluids. Phys. Rev. Lett 79, 3282–3285 (1997). [Google Scholar]
- 108.Squires TM & Mason TG Fluid Mechanics of Microrheology. Annu. Rev. Fluid Mech 42, 413–438 (2010). [Google Scholar]
- 109.Schuster BS, Allan DB, Kays JC, Hanes J & Leheny RL Photoactivatable fluorescent probes reveal heterogeneous nanoparticle permeation through biological gels at multiple scales. J. Control. Release 260, 124–133 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rosowski KA et al. Elastic ripening and inhibition of liquid–liquid phase separation. Nat. Phys 16, 422–425 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jawerth L et al. Protein condensates as aging Maxwell fluids. Science 370, 1317–1323 (2020). [DOI] [PubMed] [Google Scholar]
- 112.Wu P-H et al. A comparison of methods to assess cell mechanical properties. Nat. Methods 15, 491–498 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Brochard F & Lennon JF Frequency spectrum of the flicker phenomenon in erythrocytes. J. Phys 36, 1035–1047 (1975). [Google Scholar]
- 114.Aarts DGAL, Schmidt M & Lekkerkerker HNW Direct Visual Observation of Thermal Capillary Waves. Science 304, 847–850 (2004). [DOI] [PubMed] [Google Scholar]
- 115.Wei MT et al. Phase behaviour of disordered proteins underlying low density and high permeability of liquid organelles. Nat. Chem 9, 1118–1125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cai LH, Panyukov S & Rubinstein M Hopping diffusion of nanoparticles in polymer matrices. Macromolecules 48, 847–862 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Delarue M et al. mTORC1 Controls Phase Separation and the Biophysical Properties of the Cytoplasm by Tuning Crowding. Cell 174, 338–349.e20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bolognesi B et al. The mutational landscape of a prion-like domain. Nat. Commun (2019) doi: 10.1038/s41467-019-12101-z. [DOI] [PMC free article] [PubMed] [Google Scholar]


