Kanbar et al. show that the long noncoding RNA Malat1 impacts the differentiation of effector and memory CD8+ T cell subsets by virtue of preferential epigenetic repression of memory-associated genes in terminal effector cells through an interaction with Ezh2.
Abstract
During an immune response to microbial infection, CD8+ T cells give rise to short-lived effector cells and memory cells that provide sustained protection. Although the transcriptional programs regulating CD8+ T cell differentiation have been extensively characterized, the role of long noncoding RNAs (lncRNAs) in this process remains poorly understood. Using a functional genetic knockdown screen, we identified the lncRNA Malat1 as a regulator of terminal effector cells and the terminal effector memory (t-TEM) circulating memory subset. Evaluation of chromatin-enriched lncRNAs revealed that Malat1 grouped with trans lncRNAs that exhibit increased RNA interactions at gene promoters and gene bodies. Moreover, we observed that Malat1 was associated with increased H3K27me3 deposition at a number of memory cell-associated genes through a direct interaction with Ezh2, thereby promoting terminal effector and t-TEM cell differentiation. Our findings suggest an important functional role of Malat1 in regulating CD8+ T cell differentiation and broaden the knowledge base of lncRNAs in CD8+ T cell biology.
Graphical Abstract
Introduction
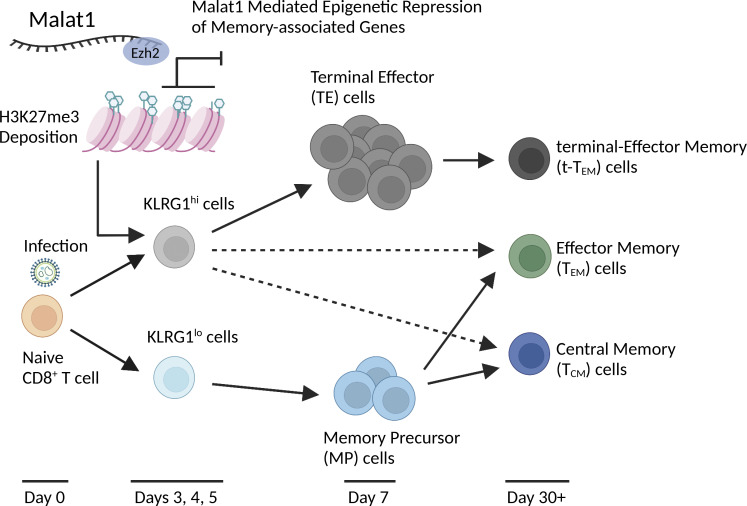
During an immune response to microbial infection, CD8+ T cells give rise to effector cells that mediate acute host defense and memory cells that provide long-lived protection (Chang et al., 2014). Effector cells have been broadly subdivided into terminal effector (TE) cells, defined by high expression of KLRG1 and low expression of CD127 (KLRG1hiCD127lo), which undergo apoptosis after pathogen clearance, and memory precursor (MP) cells, which are characterized by low expression of KLRG1 and high expression of CD127 and can give rise to long-lived memory cells (Joshi et al., 2007). It has been well-established that there is substantial heterogeneity among circulating effector and memory cells (Kakaradov et al., 2017; Kurd et al., 2020). CD62LhiCD127hi central memory (TCM) cells, home to secondary lymphoid structures, proliferate rapidly upon microbial rechallenge and possess the greatest plasticity to differentiate into secondary effector cells (Sallusto et al., 1999; Masopust et al., 2014). Effector memory (TEM) cells have recently been subdivided into conventional CD62LloCD127hi TEM cells, which circulate through nonlymphoid tissues and provide immediate effector response while maintaining multipotent capacity (Sallusto et al., 1999; Masopust et al., 2014), and TE memory (t-TEM) cells, characterized by low expression of CD62L and low expression of CD127 (CD62LloCD127lo), which exhibit limited multipotency and recall response, decreased lymphoid tissue presence, but potent cytotoxicity (Kurd et al., 2020; Olson et al., 2013; Milner et al., 2020). Another subset of memory cells, termed tissue-resident memory (TRM) cells, does not recirculate and localizes to peripheral organs and tissues, providing the first line of response to infection (Schenkel et al., 2014). Critical regulators of CD8+ T cell differentiation have been previously characterized, with transcription factors T-bet, Blimp1, Zeb2, and Id2 promoting the formation of effector cells, and Tcf1, Eomes, Foxo1, and Id3 regulating memory cell formation (Joshi et al., 2007; Kallies et al., 2009; Omilusik et al., 2015, 2018; Zhou and Xue, 2012; Banerjee et al., 2010; Michelini et al., 2013; Ji et al., 2011). However, the role of other potential mechanisms regulating CD8+ T cell differentiation, such as DNA methylation, post-translational modifications, and noncoding RNAs, remain poorly understood.
Long non-coding RNAs (lncRNAs) are a class of RNA molecules longer than 200 bp that are not translated into protein but are transcribed by RNA Polymerase II, capped at the 5′ end, and polyadenylated (Kopp and Mendell, 2018). It has been estimated in mouse and human CD8+ T cells that 25% of the transcriptome encodes for lncRNAs (Hudson et al., 2019). To date, few lncRNAs have been functionally characterized in CD8+ T cells despite published observations that lncRNA expression profiles can distinguish naive, effector, and memory subsets, suggesting that lncRNAs may play important roles in CD8+ T cell fate decisions (Hudson et al., 2019; Kotzin et al., 2019; Wang et al., 2015; Sharma et al., 2011). Our previous work raised the possibility that the lncRNA Malat1 might be involved in CD8+ T cell differentiation in response to acute infection (Kakaradov et al., 2017). Malat1 was first identified in a screen for markers of early-stage non-small cell lung cancer metastasis, with mutations in Malat1 and transcriptional dysregulation subsequently confirmed in various cancer types (Ji et al., 2003; Kim et al., 2018; Gutschner et al., 2013b; a). Malat1 localizes to nuclear speckles that sequester various proteins involved in RNA processing, transcription, and epigenetic regulation (Spector and Lamond, 2011). Nuclear speckles also contain a high density of RNA Polymerase II that associates with multiple DNA regions forming interchromosomal contacts at sites of active transcription (West et al., 2014; Quinodoz et al., 2018). In germline knockout models, Malat1 has been shown to be dispensable for normal mouse development and physiology (Zhang et al., 2012; Eißmann et al., 2012; Nakagawa et al., 2012; Yao et al., 2018). By contrast, acute knockdown of Malat1 has resulted in significant functional changes affecting cellular proliferation, motility, and differentiation (Tripathi et al., 2010; Tano et al., 2010; Bernard et al., 2010), suggesting that acute knockdown models may yield disparate phenotypes compared to genetic models.
In this study, we performed a functional genetic knockdown screen that suggested Malat1 as a regulator of CD8+ T cell differentiation. Malat1 knockdown significantly reduced TE cell differentiation at the peak of infection and t-TEM cell formation by 30 d after infection. Analyses of secondary recall immune responses revealed that t-TEM cells were not dependent on Malat1 to give rise to secondary t-TEM cells; by contrast, TEM and TCM cells were dependent on Malat1 to give rise to secondary t-TEM and TEM cells. Malat1 knockdown resulted in increased expression of a number of memory cell-associated genes in cells destined to become TEs. Examination of chromatin interactions with Malat1 revealed significant enrichment at gene promoters and gene bodies indicating an active role in transcription; furthermore, Malat1-interacting regions were correlated with a selective accumulation of the epigenetic repressive mark H3K27me3 compared with the epigenetic activation marks H3K4me3, H3K4me1, and H3K27ac. Malat1, in part through an interaction with the epigenetic repressor Ezh2, significantly increased H3K27me3 deposition on many genes associated with memory cell differentiation. Together, these findings suggest an important functional role of Malat1 in promoting terminal differentiation in CD8+ T cells and broaden the knowledge base of lncRNAs in CD8+ T cell biology.
Results
In vivo functional genetic knockdown screen reveals the lncRNA Malat1 as a critical regulator of CD8+ T cell differentiation
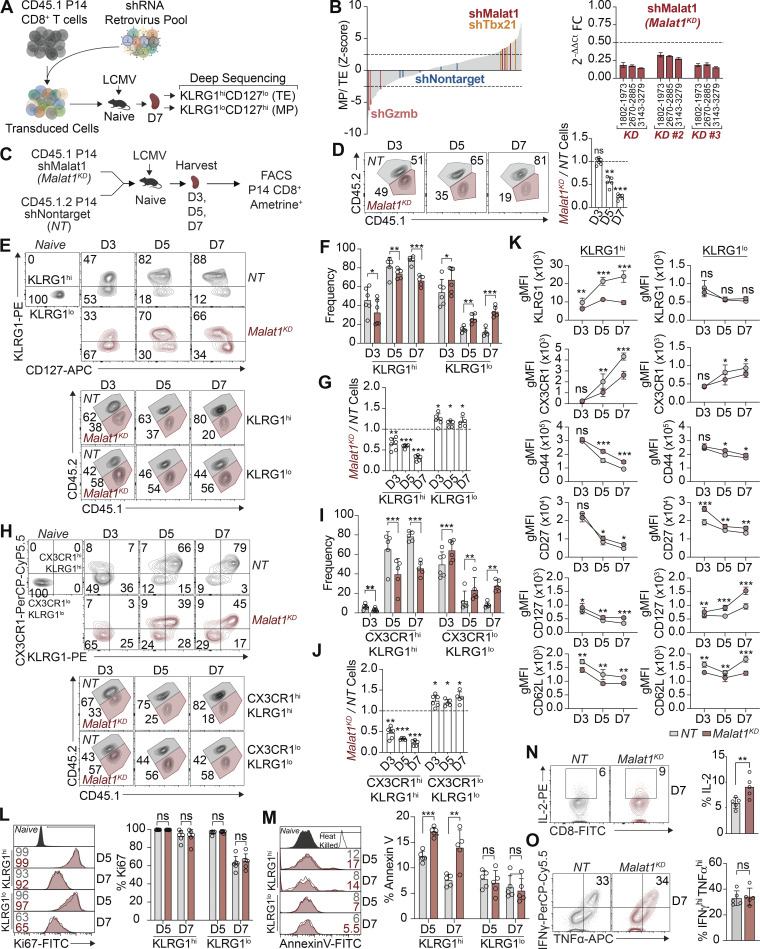
We previously observed a striking transcriptional divergence among CD8+ T cells that had undergone their first division in response to viral infection and identified a number of putative regulators of CD8+ T cell differentiation (Kakaradov et al., 2017). We, therefore, conducted a pooled shRNA screen of 365 shRNAs against 102 of these gene targets to assess their possible functional significance (Fig. 1 A) using a previously published approach (Chen et al., 2014; Milner et al., 2017). We transduced P14 CD8+ CD45.1+ T cells, which have transgenic expression of a TCR recognizing an immunodominant epitope of lymphocytic choriomeningitis virus (LCMV), with a pool of shRNA retroviruses. These cells were adoptively transferred into congenic CD45.2+ WT mice that were subsequently infected with the Armstrong strain of LCMV. On day 7 after infection, TE (KLRG1hiCD127lo) and MP (KLRG1loCD127hi) CD8+ T cells were isolated by FACS. Non-targeting shRNAs were included as a negative control, and shRNAs targeting Tbx21, the gene encoding the T-box transcription factor T-bet, were included as a positive control. Using a Z-score cutoff of ±3, three Malat1 shRNAs were observed to be differentially enriched in KLRG1loCD127hi MP cells compared with KLRG1hiCD127lo TE cells (Fig. 1 B and Table S1). The knockdown efficiencies of each Malat1 shRNA were verified using multiple quantitative PCR (qPCR) primers tiling the Malat1 locus with 83.1, 69.4, and 82.2% average knockdown (Fig. 1 B).
Figure 1.
In vivo shRNA screen reveals lncRNA Malat1 as a critical regulator of CD8+ T cell differentiation. (A) CD45.1+ P14 T cells were transduced with a shRNA pool; 7 d after infection TE- (KLRG1hiCD127lo) and MP-phenotype (KLRG1loCD127hi) cells were isolated by FACS. (B) Enrichment of shRNA construct in MP cells relative to TE cells, reported as the average Z-score from two independent screens, n = 20 mice per screen (left). Validation of three Malat1KD shRNAs in activated CD8+ T cells with locus coordinates for each Malat1 primer set (right). FC, fold change. (C) P14 CD8+ T cells were transduced with Malat1 shRNA (Malat1KD, CD45.1) or nontarget shRNA (NT, CD45.1.2) and adoptively co-transferred at a 1:1 ratio into CD45.2 recipient mice that were subsequently infected with LCMV. Splenocytes were harvested on days 3, 5, and 7. (D) Quantification of total splenic NT and Malat1KD ratios at day 3, 5, and 7 after infection. (E–G) Quantification of splenic NT and Malat1KD KLRG1hi and KLRG1lo-phenotype cells, representative flow cytometry plots (E), quantification of frequencies (F), and numeric ratio of cells (G). (H–J) Quantification of splenic NT and Malat1KD CX3CR1hiKLRG1hi and CX3CR1loKLRG1lo-phenotype cells, representative flow cytometry plots (H), quantification of frequencies (I), and numeric ratio of cells (J). (K) Quantification of key effector- and memory-associated molecules in KLRG1hi-and KLRG1lo-phenotype cells. gMFI, geometric mean fluorescence intensity. (L and M) Frequency of Ki-67+ (L) and Annexin V+ (M) cells in KLRG1hi- and KLRG1lo-phenotype cells on days 5 and 7 after infection. (N and O) Malat1KD and NT cells on day 7 after infection were cultured ex vivo in the presence of cognate gp33-41 peptide for 5 h and frequency of IL-2+ (N) or IFNγhiTNFhi (O) cells measured. All data are from one representative experiment out of two independent experiments with n = 5–6 per group; *, P < 0.05; **, P < 0.005; ***, P < 0.0005 paired t test (F, I, and K–O), one sample t test (D, G, and J) . Graphs indicate mean ± SEM, symbols represent individual mice. D, day.
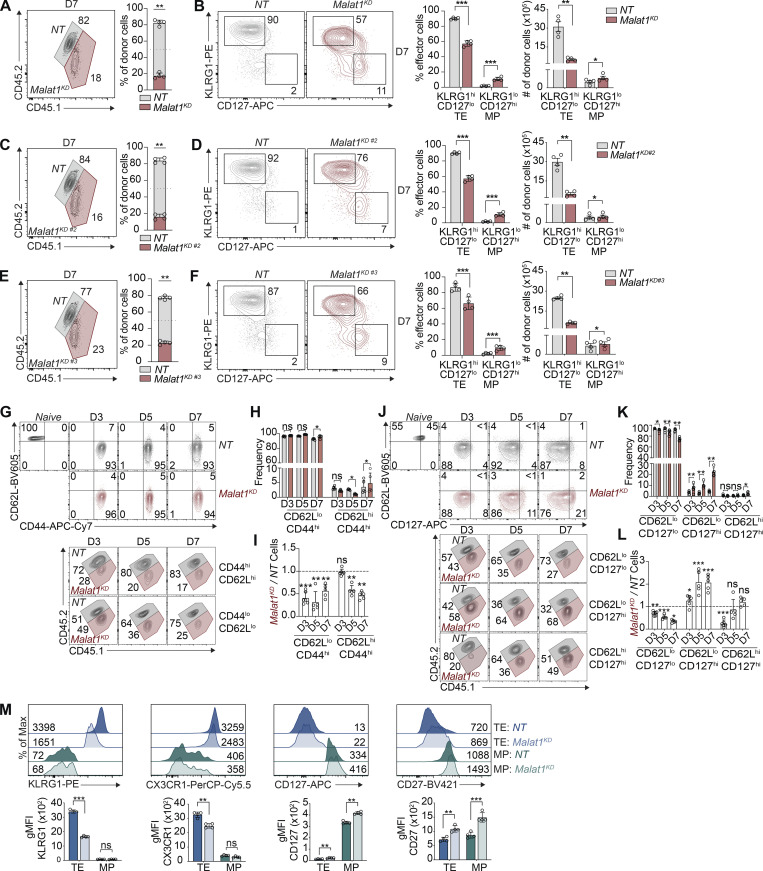
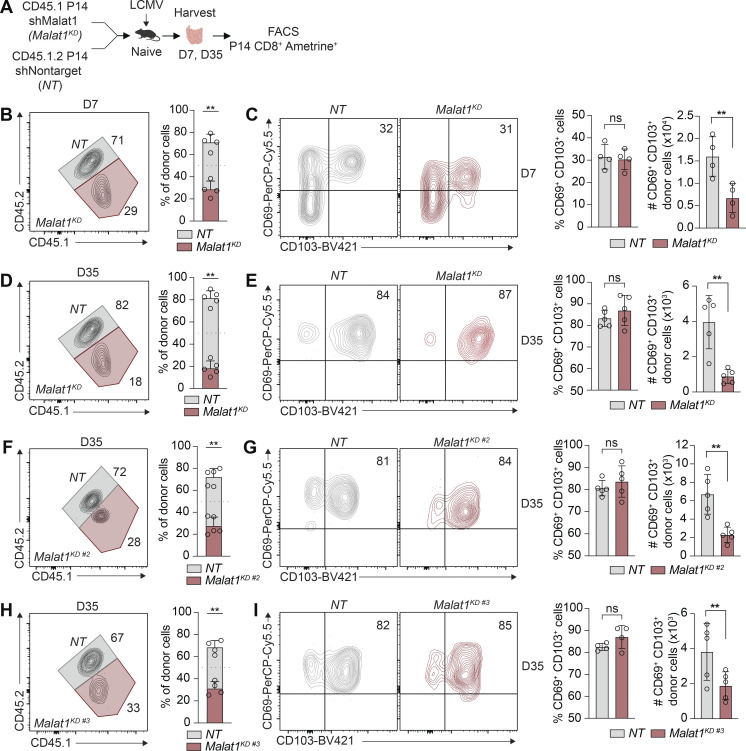
We next evaluated the functional consequences of Malat1 knockdown during CD8+ T cell differentiation. Congenically distinct P14 CD8+ T cells were transduced with Malat1 shRNA (Malat1KD, CD45.1) or nontarget shRNA (NT, CD45.1.2) and adoptively co-transferred into CD45.2 recipient mice (Fig. 1 C). On day 7 after infection, Malat1KD cells showed a marked numerical decrease as compared to NT cells (Fig. S1, A, C, and E). Moreover, Malat1KD TE cells were decreased relative to NT TE cells, whereas Malat1KD MP cells were increased relative to NT MP cells (Fig. S1, B, D, and F).
Figure S1.
lncRNA Malat1 regulates CD8+ T cell differentiation. P14 CD8+ T cells were transduced with Malat1 shRNA (Malat1KD, CD45.1) or nontarget shRNA (NT, CD45.1.2) and adoptively co-transferred at a 1:1 ratio into CD45.2 recipient mice that were subsequently infected with LCMV (see Fig. 1 C). (A, C, and E) Quantification of splenic NT and Malat1KD (A), Malat1KD #2 (C), or Malat1KD #3 (E) ratios at day 7. (B, D, and F) Representative flow cytometry plots of TE- and MP-phenotype cells (left) and quantification (right) among co-transferred cells. (G–I) Quantification of splenic NT and Malat1KD CD62LhiCD44hi and CD62LloCD44hi-phenotype cells, representative flow cytometry plots (G), quantification of frequencies (H), and numeric ratio of cells (I) at days 3, 5, and 7. (J–L) Quantification of splenic NT and Malat11KD CD127hiCD62Lhi-, CD127hiCD62Llo-, and CD127loCD62Llo-phenotype cells, representative flow cytometry plots (J), quantification of frequencies (K), and numeric ratio of cells (L) at days 3, 5, and 7. (M) Representative flow cytometry plots and quantification of key TE- and MP-associated molecules at day 7. All data are from one representative experiment out of two independent experiments with n = 5–6 per group; *, P < 0.05; **, P < 0.005; ***, P < 0.0005, paired t test (A–F, H, K, and M), one sample t test (I and L). Graphs indicate mean ± SEM, symbols represent individual mice. D, day.
We next tested whether Malat1 knockdown impaired the early differentiation of TE cells. Compared with NT cells, Malat1KD cells analyzed on days 5 and 7 after infection exhibited a marked numerical decrease (Fig. 1 D). On day 3 after infection, although the total Malat1KD pool did not exhibit a numerical decrease, the frequencies and ratios of KLRG1hi Malat1KD cells were already reduced with corresponding increases in KLRG1lo Malat1KD cells compared with their NT cell counterparts (Fig. 1, E–G). Moreover, analysis of expression of CX3CR1, another marker associated with TE cells, revealed that the frequencies and ratios of Malat1KD CX3CR1hiKLRG1hi cells were reduced compared with Malat1KD CX3CR1loKLRG1lo cells as early as day 3 after infection (Fig. 1, H–J). Additionally, relative to control cells, Malat1KD KLRG1hi cells exhibited decreased expression of KLRG1 and CX3CR1, whereas Malat1KD KLRG1lo cells exhibited increased expression of CD127 and CD27, phenotypic markers associated with memory cells (Figs. 1 K and S1 M). Notably, the majority of Malat1KD cells exhibited a CD44hi and CD62Llo phenotype (Fig. S1, G–L), indicating that the reduction in Malat1KD TE cells was not due to a failure of T cell activation.
KLRG1hi and KLRG1lo Malat1KD and NT cells exhibited similar expression of the proliferation marker Ki-67 on days 5 and 7 after infection, suggesting that Malat1 knockdown did not affect proliferation several days after T cell activation (Fig. 1 L). However, expression of the membrane-bound apoptotic marker Annexin V was modestly increased in KLRG1hi, but not in KLRG1lo Malat1KD cells on days 5 and 7 after infection, suggesting the numerical deficiency observed in Malat1KD KLRG1hi cells may be partly attributed to increased apoptosis (Fig. 1 M). Analyses of functional parameters demonstrated that Malat1KD cells expressed more IL-2, a cytokine that promotes homeostatic proliferation; however, Malat1KD and NT cells were similarly polyfunctional with equivalent frequencies of IFNγhiTNFhi cells (Fig. 1, N and O). Taken together, these results demonstrate that Malat1 plays a critical role in TE cell differentiation.
Malat1 is a critical regulator of CD8+ t-TEM cell differentiation and plays a role in the generation of secondary memory T cells
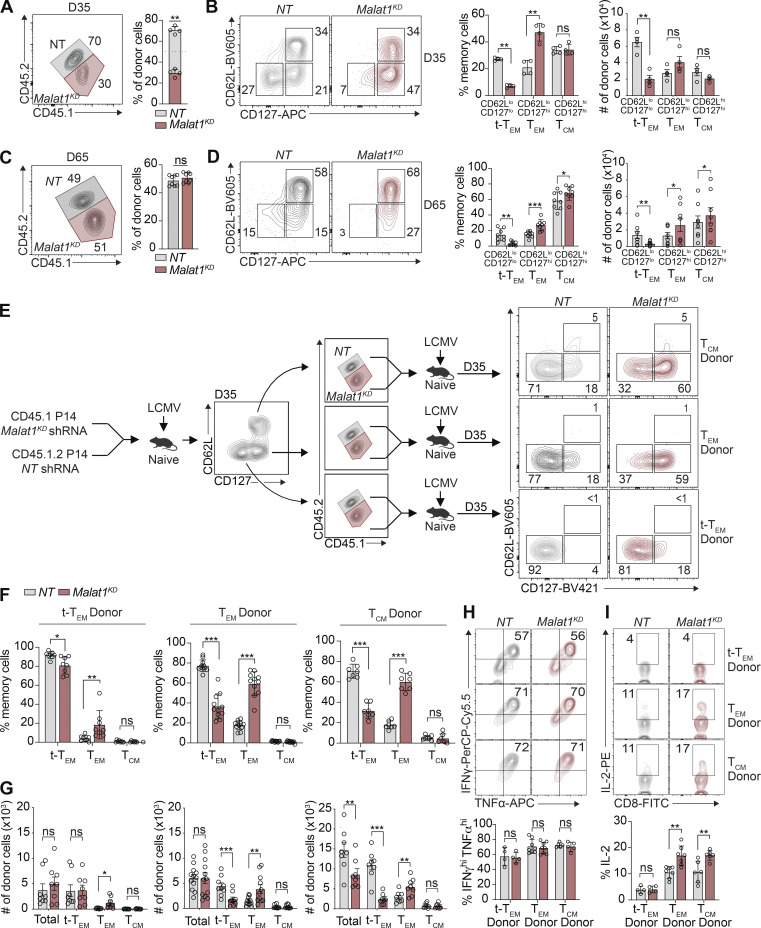
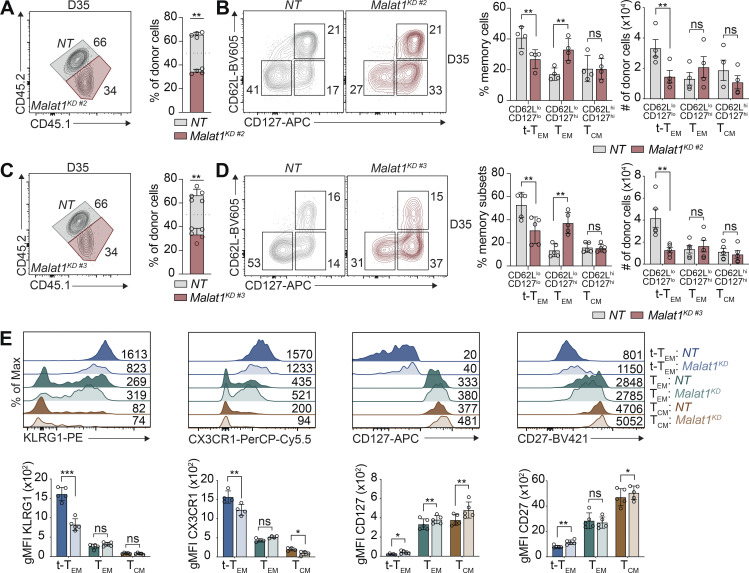
Assessment of total circulating memory cells 35 d after infection revealed decreased proportions of Malat1KD cells compared with NT cells (Figs. 2 A and S2, A and C). Compared with control cells, the proportion and absolute number of Malat1KD CD62LloCD127lo (t-TEM) cells were decreased, with a corresponding increase in the proportion, but not absolute number, of Malat1KD CD62LloCD127hi TEM cells on day 35 after infection (Figs. 2 B and S2, B and D). No changes were observed in the proportion or absolute number of Malat1KD CD62LhiCD127hi TCM cells on day 35 after infection. However, by day 65 after infection, we observed an increase in the numbers and frequencies of Malat1KD TCM and TEM cells along with decreased numbers and frequencies of t-TEM cells (Fig. 2, C and D). Moreover, Malat1KD t-TEM cells exhibited decreased expression of effector cell-associated markers KLRG1 and CX3CR1, along with increased expression of memory cell-associated markers CD127 and CD27 (Fig. S2 E). Compared with control cells, Malat1KD TEM and TCM cells also exhibited increased expressions of CD127 and CD27 (Fig. S2 E). Lastly, the proportions of Malat1KD CD69+CD103+ CD8+ T cells in the small intestine intra-epithelial (siIEL) compartment was similar to that of control cells (Fig. S3, B–I).
Figure 2.
Malat1 regulates CD8+ T cell memory formation and represses generation of secondary TEM cells. (A and C) Quantification of splenic NT and Malat1KD ratios on days 35 and 65 after infection. (B and D) Representative flow cytometry plots of t-TEM, TEM, and TCM cells (left) and quantification (right) among co-transferred cells. (E) P14 CD8+ T cells were transduced with Malat1 shRNA (Malat1KD, CD45.1) or nontarget shRNA (NT, CD45.1.2) and adoptively co-transferred at a 1:1 ratio into CD45.2 recipient mice, which were then infected with LCMV. 35 d after primary infection, Malat1KD and NT cells were sorted from t-TEM, TEM, or TCM subsets, mixed at a 1:1 (5,000 Malat1KD cells/5,000 NT cells) ratio, and adoptively transferred into naive CD45.2 recipient followed by infectious challenge with LCMV (secondary infection). (F) Frequency of secondary memory populations derived from transferred t-TEM (left), TEM (middle), and TCM (right) donor cells was assessed at 30 d after secondary LMCV infection. (G) Quantification of secondary memory subsets derived from t-TEM (left), TEM (middle), and TCM (right) donor populations. (H and I) Malat1KD and NT secondary t-TEM, TEM, and TCM cells were cultured ex vivo in the presence of cognate gp33-41 peptide for 5 h and frequency of IFNγhiTNFhi (H) or IL-2+ (I) cells measured. All data are from one representative experiment out of two independent experiments with n = 4–7 (A–D), n = 9–10 (E–G), or n = 4–6 (H and I) mice per group; *, P < 0.05; **, P < 0.005; ***, P < 0.0005, paired t test. Graphs indicate mean ± SEM, symbols represent individual mice. D, day.
Figure S2.
lncRNA Malat1 regulates memory CD8+ T cell differentiation. P14 CD8+ T cells were transduced with Malat1 shRNA (Malat1KD #2 or Malat1KD #3, CD45.1) or nontarget shRNA (NT, CD45.1.2); cells were adoptively co-transferred at a 1:1 ratio into CD45.2 recipient mice that were subsequently infected with LCMV. (A and C) Quantification of splenic NT and Malat1KD #2 (A) or Malat1KD #3 (C) ratios at day 35 after infection. (B and D) Representative flow cytometry plots of t-TEM, TEM, and TCM cells (left) and quantification (right) among co-transferred cells. (E) Representative flow cytometry plots and quantification of t-TEM, TEM, and TCM cell-associated molecules. All data are from one representative experiment out of two independent experiments with n = 4–5 per group; *, P < 0.05; **, P < 0.005; ***, P < 0.0005, paired t test. Graphs indicate mean ± SEM, symbols represent individual mice. D, day.
Figure S3.
lncRNA MALAT1 knockdown reduces siIEL TRM cell differentiation. (A) P14 CD8+ T cells were transduced with Malat1 shRNA (Malat1KD, Malat1KD #2, or Malat1KD #3, CD45.1) or nontarget shRNA (NT, CD45.1.2) and adoptively co-transferred at a 1:1 ratio into CD45.2 recipient mice that were subsequently infected with LCMV. (B) Quantification of siIEL NT and Malat1KD ratios at day 7 after infection. (C) Representative flow cytometry plots of CD69+CD103+-phenotype cells (left) and quantification (right) at day 7 after infection among co-transferred cells. (D, F, and H) Quantification of siIEL NT and Malat1KD (D), Malat1KD #2 (F), or Malat1KD #3 (H) ratios at day 35 after infection. (E, G, and I) Representative flow cytometry plots of TRM cells (left) and quantification (right) at day 35 among co-transferred cells. All data are from one representative experiment out of two independent experiments with n = 4–5 mice per group; **, P < 0.005, paired t test. Graphs indicate mean ± SEM, symbols represent individual mice. D, day.
Having established a role for Malat1 in differentiation of CD8+ T cells during primary infection, we next sought to determine whether Malat1 also plays a role in the generation of secondary circulating memory T cell subsets upon infectious rechallenge. 35 d after primary LCMV infection, Malat1KD and NT t-TEM, TEM, or TCM cells were FACS isolated, mixed in equal proportions, and transferred into new naive recipient mice which were then infected with LCMV (Fig. 2 E). 35 d later, the numbers and proportions of secondary circulating memory T subsets were assessed (Fig. 2, F and G). Donor Malat1KD and NT t-TEM cells gave rise to equal numbers of total and t-TEM cells in Malat1KD and NT cells (Fig. 2, F and G), suggesting that Malat1 does not play a critical role in the ability of primary t-TEM cells to give rise to secondary t-TEM cells upon rechallenge. By contrast, compared with donor NT cells, donor Malat1KD t-TEM cells gave rise to increased numbers and proportions of secondary TEM cells. Moreover, compared with donor NT cells, donor Malat1KD TEM and TCM cells gave rise to reduced numbers and proportions of secondary t-TEM cells, but increased numbers and proportions of secondary TEM cells (Fig. 2, F and G). Secondary memory T cells derived from donor Malat1KD and NT cells were similarly polyfunctional with equivalent frequencies of IFNγhiTNFhi cells (Fig. 2 H), though secondary memory cells derived from Malat1KD TEM and TCM donors tended to express more IL-2 compared with those derived from NT donors (Fig. 2 I). Taken together, these results suggest that Malat1 plays a role in repressing the generation of secondary TEM cells derived from t-TEM, TEM, and TCM cells, in addition to its role in regulating primary t-TEM cell differentiation.
Single-cell RNA-seq (scRNA-seq) analyses reveal that Malat1 depletion upregulates factors associated with memory cell differentiation
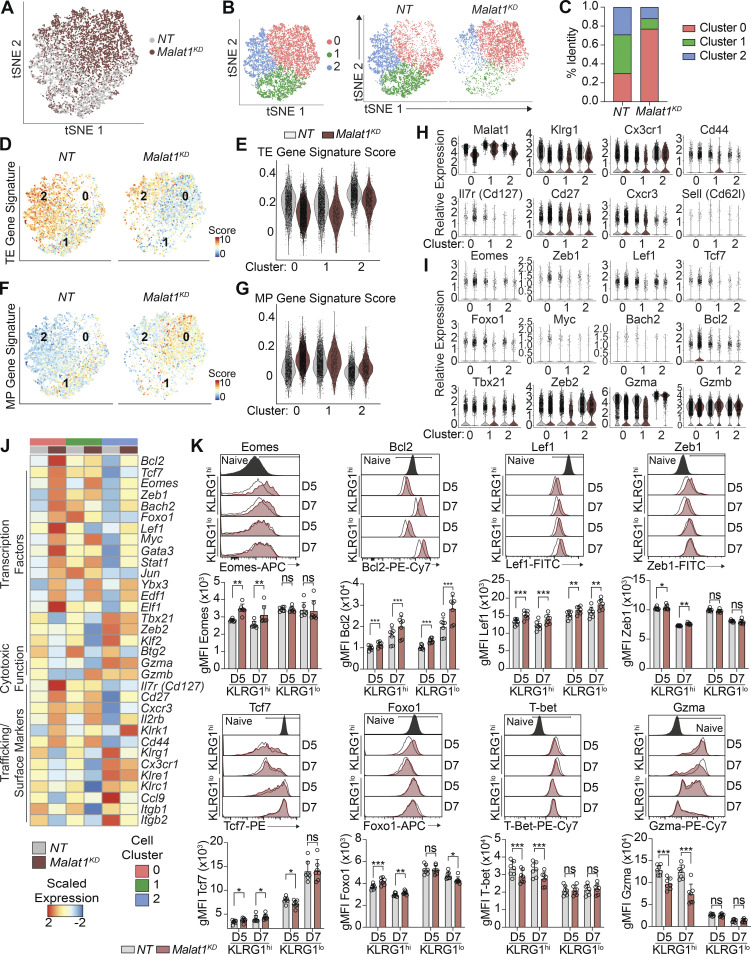
To begin to investigate the mechanisms underlying the role of Malat1 in CD8+ T cell differentiation, we performed scRNA-seq of FACS-sorted Malat1KD and NT cells responding to viral infection. P14 CD8+ T cells were transduced with Malat1 shRNA (Malat1KD, CD45.1) or nontarget shRNA (NT, CD45.1.2), adoptively co-transferred into CD45.2 recipient mice and isolated on day 7 after infection. Unsupervised t-distributed stochastic neighborhood embedding (tSNE) analysis revealed separation of the majority of Malat1KD and NT cells (Fig. 3 A). Clustering analysis yielded three distinct clusters, with 76% of Malat1KD cells in Cluster 0 and the remaining Malat1KD cells distributed between Clusters 1 (10%) and 2 (14%; Fig. 3, B and C). NT cells were distributed in nearly equal proportions among Clusters 0 (30%), 1 (37%), and 2 (33%).
Figure 3.
scRNA-seq reveals that Malat1 depletion upregulates memory-associated factors. (A) tSNE analysis of Malat1KD and NT cells on day 7 after LCMV infection. (B) Clustering analysis of Malat1KD and NT cells as one plot (left) or separated by sample type (right). (C) Bar graph quantifying proportion of Malat1KD and NT cells among each cluster type. (D and E) TE signature enrichment of all Malat1KD and NT cells displayed on tSNE plots (D) or split by scRNA-seq clusters (E). (F and G) MP signature enrichment of all Malat1KD and NT cells displayed on tSNE plots (F) or split by scRNA-seq clusters (G). (H and I) scRNA-seq expression profiles of genes relevant to CD8+ T cell trafficking (H) and effector and memory differentiation (I) in Malat1KD and NT cells split by scRNA-seq clusters. (J) Average expression profiles of genes relevant to CD8+ T cell differentiation, cytotoxicity, and trafficking in Malat1KD and NT cells split by scRNA-seq clusters. (K) Representative flow cytometry plots and quantification of protein expression of memory-associated genes and TE-associated genes in Malat1KD and NT KLRG1hi and KLRG1lo cells at days 5 and 7 after infection. All data are from one representative experiment out of two independent experiments with n = 5–7 mice per group; *, P < 0.05; **, P < 0.005; ***, P < 0.0005, paired t test. Graphs indicate mean ± SEM, symbols represent individual mice. D, day.
Evaluation of the transcriptional profile of each scRNA-seq cluster revealed that among NT cells, Cluster 1 cells expressed the lowest levels of Klrg1 and Cx3cr1 and were enriched for the MP gene signature (Fig. 3, F–H and Table S2); conversely, Clusters 0 and 2 cells expressed higher levels of Klrg1 and Cx3cr1 and were enriched for the TE gene signature (Fig. 3, D, E, and H; and Table S2). Among Malat1KD cells, Cluster 0 and 2 cells were enriched for the MP gene signature (Fig. 3, F–H); consistent with this observation, Malat1KD Cluster 0 cells exhibited higher expression of memory-associated molecules (Tcf7, Eomes, Foxo1, Zeb1, Lef1, Bach2, Bcl2, Il7r, Cd27, and Cxcr3) compared with their NT counterparts (Fig. 3, H–J and Table S3). Moreover, Malat1KD Cluster 0 cells exhibited reduced expression of TE-associated molecules (Tbx21, Zeb2, Gzma, Gzmb, Klrg1, and Cx3cr1) compared with their NT Cluster 0 counterparts (Fig. 3, H–J), consistent with the diminished TE gene signature observed in these cells (Fig. 3 E). Flow cytometry was performed in Malat1KD and NT cells on days 5 and 7 after infection to confirm the observations revealed by scRNA-seq. Indeed, compared with their NT KLRG1hi counterparts, Malat1KD KLRG1hi cells exhibited increased expression of memory-associated molecules such as Eomes, Bcl2, Lef1, Zeb1, Tcf7, and Foxo1 (Fig. 3 K). Taken together, these results suggest that Malat1 may act specifically in cells destined to become TEs and reduce the expression of genes that promote memory formation.
Malat1 interacts in trans at promoters and gene bodies
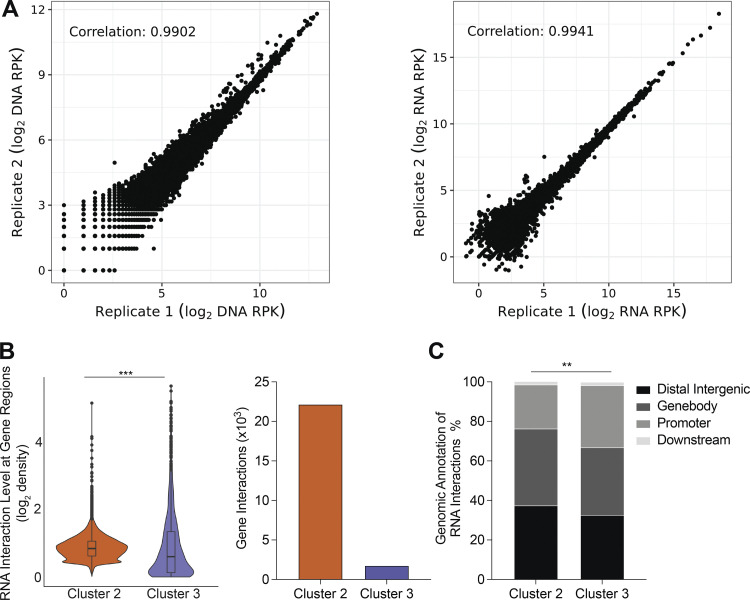
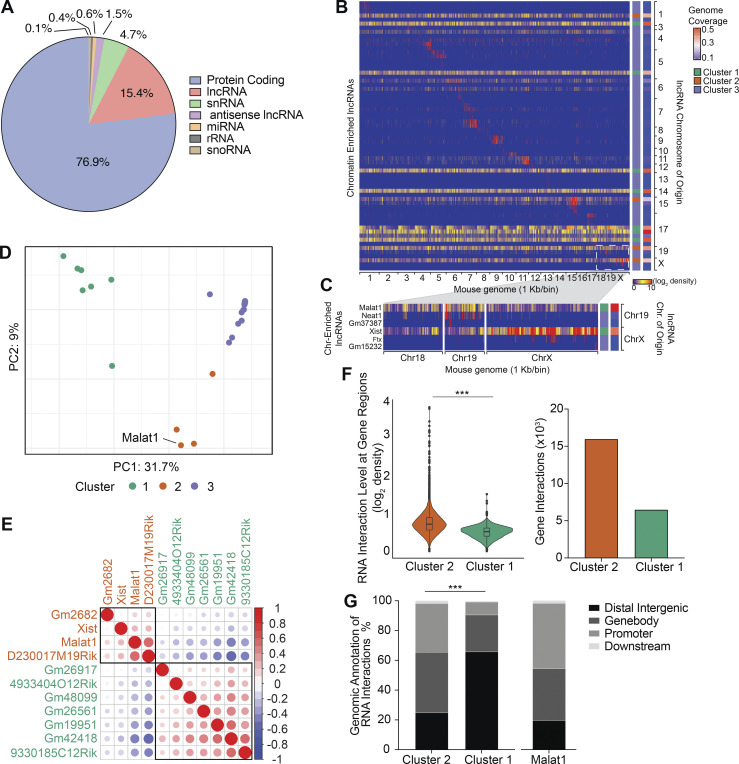
To begin to elucidate the transcriptional influence of Malat1 on gene expression, we performed GRID-seq (global RNA interactions with DNA by deep sequencing) with a specific focus on lncRNAs (Li et al., 2017). Replicate libraries of P14 CD8+ T cells were cultured in the presence of cognate gp33-41 peptide for 5 d in vitro (Fig. S4 A). Evaluation of the CD8+ T cell transcriptome revealed that a substantial proportion of RNA chromatin interactions were represented by lncRNAs (15.4%), in line with previous reports of the lncRNA abundance in activated CD8+ T cells (Fig. 4 A; Hudson et al., 2019). Chromatin-enriched lncRNAs analysis revealed that although a majority of lncRNAs interact within their chromosome of origin, there was a separate group of lncRNAs that engage in highly trans interactions throughout the mouse genome (Fig. 4 B). Increased representation of lncRNAs originating from chromosomes 19 and X confirmed a high level of Xist (X-inactive specific transcript) enrichment on chromosome X (Fig. 4 C). Malat1 and Neat1 were both enriched on chromosome 19; however, in contrast to Neat1, Malat1 engaged in trans interactions beyond its chromosome of origin (Fig. 4 C). Principal component analysis (PCA) and k-means clustering of all 66 chromatin-enriched lncRNAs resulted in three clusters of lncRNAs, thereby grouping lncRNAs by the similarity of their chromatin interaction patterns across the entire mouse genome. (Fig. 4, B and D). Clusters 1 and 2 separated 11 highly trans-interacting lncRNAs, each with genomic coverage >20%, with Malat1 grouped with the Cluster 2 lncRNAs (Fig. 4, B and D). Cluster 3 grouped all lncRNAs with <12% genomic coverage with interactions predominantly within their chromosome of origin (Fig. 4, B and D). We further evaluated the 11 highly trans-interacting lncRNAs by performing Spearman pairwise correlation analyses and hierarchical clustering, which further separated these lncRNAs into two additional sub-clusters, suggesting distinct genomic interaction features between these two groups of trans-interacting lncRNAs (Fig. 4 E). To test this hypothesis, we averaged the RNA interaction levels at each 1 kb genomic bin for Cluster 1 and Cluster 2 lncRNAs to analyze their differential RNA interaction patterns across the entire genome. Compared to Cluster 1 lncRNAs, Cluster 2 lncRNAs exhibited greater RNA interaction levels on genomic regions annotated to genes (Fig. 4 F). This trend was similarly observed when comparing Cluster 2 and Cluster 3 lncRNAs (Fig. S4 B). Genomic feature annotation of differential RNA chromatin interactions demonstrated that Cluster 2 lncRNAs were associated with promoters and gene bodies with greater frequency, whereas Cluster 1 lncRNAs were associated with distal intergenic regions with greater frequency, highlighting the distinct interaction features of these two clusters of highly trans-interacting lncRNAs (Figs. 4 G and S4 C). Taken together, these results indicate that Malat1 associates with a cluster of trans-interacting lncRNAs that have RNA interactions preferentially at promoters and gene bodies.
Figure S4.
Characterizing lncRNAs in activated CD8+ T cells. (A) Reproducibility of GRID-seq libraries for expression of all RNA enriched chromatin (reads per kilobase, 1-kb binned genome; left) and DNA interaction level of all chromatin interacting RNA (reads per kilobase, 1-kb binned genome; right). (B) Differential lncRNA chromatin interaction regions between Clusters 2 and 3 lncRNAs displayed by direct comparison in a violin plot (left). Number of unique gene interactions between Clusters 2 or 3 lncRNAs (right). (C) Distribution of genomic annotations from differential lncRNA chromatin interaction regions between Clusters 2 and 3. Statistical significance was determined in by Student’s t test, ***, P < 0.0005 (B) and Pearson’s Chi-squared test, **, P < 0.005 (C).
Figure 4.
Malat1 clusters with trans lncRNAs that focus chromatin interactions on gene promoters and gene bodies. (A) Distribution of genome-wide RNA chromatin interactions in P14 CD8+ T cells 4 d after activation. snRNA, small nuclear RNA; miRNA, microRNA; rRNA, ribosomal RNA; snoRNA, small nucleolar RNA. GRID-seq analyses was performed in duplicate and samples pooled together for analysis. (B) Heatmap of chromatin-enriched lncRNA across the murine genome. Rows represent chromatin-enriched lncRNAsand columns represent the murine genome binned at 1-kb resolution. (C) Enlarged representative region of lncRNAs from chromosomes Chr 19 and X and their chromatin interactions on chromosomes 18, 19, and X at 1-kb resolution. (D) PCA plot and k-means clustering of all 66 lncRNAs colored by cluster groups. PCA vectors used were the entire mouse genome binned into 1-kb segments removing bins with zero interactions. lncRNAs with similar global genome chromatin interaction patterns clustered together. (E) Spearman correlation matrix plot and hierarchical clustering of 11 highly trans lncRNAs with rectangles surrounding each cluster. lncRNA gene names are color-coded to match colors of k-means clusters in D. (F) Differential lncRNA chromatin interaction regions between Cluster 2 and 1 lncRNAs, displayed by direct comparison in a violin blot with box-plot denoting median, 25th, and 75th percentile (left). Number of unique gene interactions between Cluster 2 and 1 lncRNAs (right). (G) Distribution of genomic annotations from differential lncRNA chromatin interaction regions of Cluster 2 and 1 lncRNAs (left) and Malat1 alone (right). Statistical significance was determined by Student’s t test, ***, P < 0.0005 (F) and Pearson’s Chi-squared test, ***, P < 0.0005 (G).
Malat1-chromatin enrichment correlates with high coverage of H3K27me3 on genes associated with memory cell differentiation
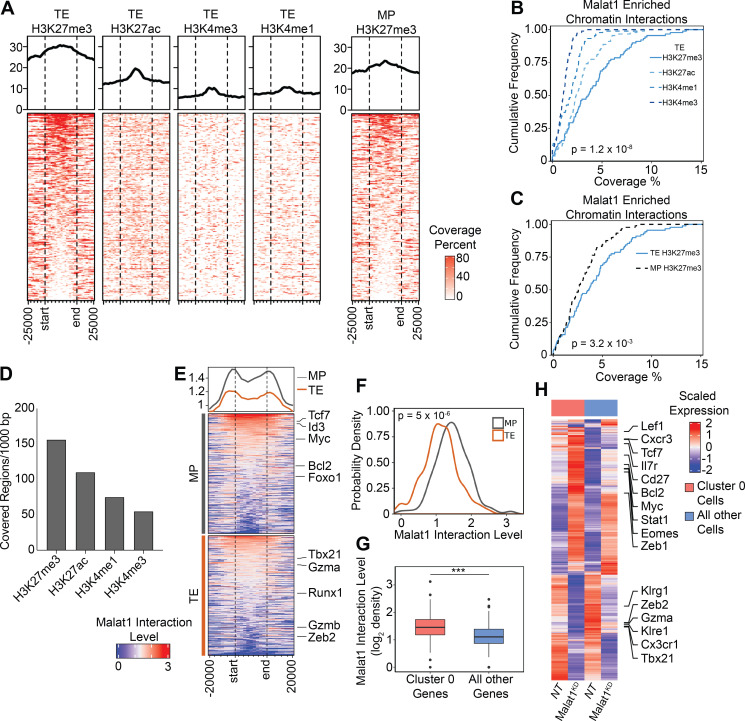
Owing to the previously reported association of Malat1 and Ezh2, the functional enzymatic component of the polycomb repressive complex 2, which deposits repressive methyl groups to histone H3 at lysine 27 (Wang et al., 2015; Kim et al., 2017), we explored epigenetic regulation at Malat1-interacting regions, utilizing publicly available chromatin immunoprecipitation sequencing (ChIP-seq) datasets in CD8+ T cells for several histone marks from TE cells, H3K27me3 and H3K27ac (GSE72408: Gray et al., 2017), and H3K4me3 and H3k4me1 (GSE95237: Yu et al., 2017). The repressive histone mark H3K27me3 demonstrated the highest proportions of coverage and covered regions at Malat1 interaction regions, as compared with active histone marks H3K4me3, H3K27ac, and H3K4me1 (Fig. 5, A, B, and D; and Table S4). We next tested Malat1 interaction regions of H3K27me3 marks from either TE or MP cells (GSE72408: Gray et al., 2017) and observed higher coverage of H3K27me3 marks in TE cells as compared with MP cells, demonstrating preferential interaction of Malat1 at H3K27me3 regions enriched in TE cells (Fig. 5, A and C; and Table S4). Focusing on genes known to play a role in CD8+ T cell differentiation, we next evaluated Malat1 interaction levels at gene regions harboring H3K27me3 (Yu et al., 2017) and found increased Malat1 enrichment at genes associated with MP cells as compared with those associated with TE cells (Fig. 5, E and F; Table S5; GSE157072; Milner et al., 2020). Furthermore, Malat1 enrichment was increased at genes that were differentially expressed in scRNA-seq Cluster 0 (Fig. 3 C) compared with all other single-cell clusters (Fig. 5 G). These genes included memory cell–associated genes Tcf7, Eomes, Zeb1, Lef1, and Bcl2, which were all upregulated as a consequence of Malat1 knockdown (Fig. 5 H and Table S6), raising the possibility of Malat1-mediated transcriptional suppression. Taken together, these results suggest that Malat1 may play a role in repressing genes associated with memory cell differentiation.
Figure 5.
Malat1 enriches on chromatin marked by the epigenetic repressive histone mark H3K27me3. (A) Coverage heatmap of H3K27me3, H3K27ac, H4K3me3, and H3K4me1 epigenetic marks from TE cells (left) and H3K27me3 from MP cells (right) at Malat1-interacting genomic regions ±25 kb. (B and C) Cumulative distribution of coverage of each epigenetic mark from TE cells (B) and only the H3K27me3 mark from TE and MP cells (C) within Malat1-interacting regions at 100-kb resolution. (D) Normalized covered regions per 1,000 bp of each epigenetic mark at Malat1-interacting genomic regions. (E) Heatmap of Malat1 interaction level on gene bodies of TE- and MP-associated genes. (F) Probability density distribution of Malat1 interactions from E. (G) Malat1 RNA interaction level of differentially expressed genes in Cluster 0 cells compared to all other cells. (H) Heatmap representing expression of selected genes in Cluster 0 Malat1KD and NT cells compared to all other cells. Statistical significance was determined in by Student’s t test, ***, P < 0.0005 (B, C, F, and G).
Malat1 interacts with Ezh2 to maintain H3K27me3 deposition at genes associated with memory cell differentiation
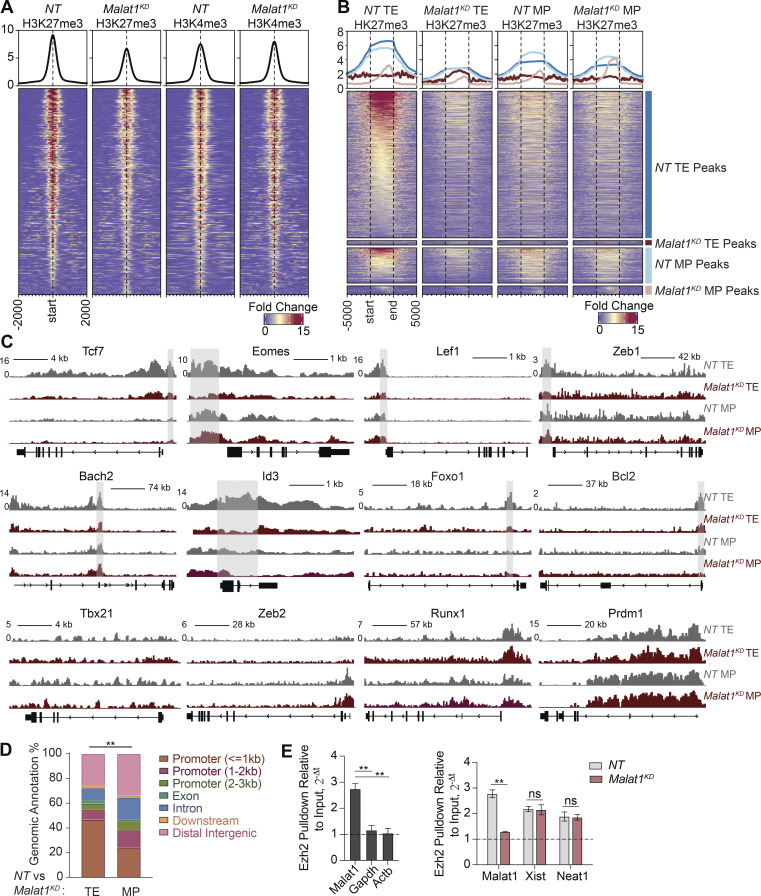
Having observed upregulated expression of memory cell–associated genes in Malat1KD cells, we explored whether H3K27me3-mediated epigenetic suppression was coordinately attenuated in Malat1KD cells. We performed H3K27me3, H3K4me3, and Ezh2 ChIP-seq on FACS-purified Malat1KD and NT cells isolated 7 d after infection. We identified 5,012 differentially methylated regions (DMRs) due to loss of H3K27me3 deposition (Fig. 6 A and Table S7); by contrast, H3K4me3 DMRs were not found in these regions. Furthermore, Ezh2 deposition in Malat1KD cells was reduced at DMRs, suggesting a role for Malat1 in maintaining Ezh2 activity (Fig. S5 A). Genomic annotation of H3K27me3 DMRs revealed that >85% of these DMRs were located within 2–3 kb of transcription start sites and gene bodies, strengthening the notion of transcriptional dysregulation in Malat1KD cells (Fig. S5 B). Consistent with a Malat1-mediated role in transcriptional repression, correlation of RNA-seq data with H3K27me3 DMRs demonstrated H3K27me3 loss with concurrent upregulation in gene expression (Fig. S5 C).
Figure 6.
Malat1 interacts with Ezh2 to maintain H3K27me3 deposition on memory-associated genes. (A) Deposition of H3K27me3 and H3K4me3 centered on DMRs ± 2 kb in Malat1KD and NT cells at day 7 after infection. (B) Deposition of H3K27me3 ± 5 kb in FACS-purified TE and MP subsets of NT and Malat1KD cells. 3,646 peaks were enriched in NT relative to Malat1KD TE cells (blue annotation); conversely, 108 peaks were enriched in Malat1KD relative to NT TE cells (dark red annotation). 875 peaks were enriched NT relative to Malat1KD MP cells (light blue annotation); conversely, 209 peaks were enriched in Malat1KD relative to NT MP cells (light red annotation). (C) Alignment tracks of H3K27me3 in Malat1KD and NT TE and MP cells for key genes associated with memory and TE differentiation. Gray highlight denotes differential peak sites observed in NT relative to Malat1KD TE cells. (D) Genomic annotations of differential peak sites observed in Malat1KD and NT TE or MP cells. (E) Ezh2 pull-down RIP-qPCR analyses of Ezh2-bound RNA in WT CD8+ T cells (left) and Ezh2-bound lncRNA in Malat1KD and NT cells (right). Statistical significance was determined by Student’s t test, **, P < 0.005 (E) and Pearson’s Chi-squared test, **, P < 0.005 (D).
Figure S5.
Impact of Malat1 knockdown on Ezh2 function and nuclear localization. (A) Deposition of Ezh2 centered on DMRs (Fig. 6 A) ± 2 kb in Malat1KD and NT cells at day 7 after infection. (B) Genomic annotations of DMRs from Fig. 6 A. (C) Log fold change of H3K27me3 deposition as a function of log fold change gene expression in Malat1KD versus NT cells at day 7 after infection. (D) Representative flow cytometry plot of H3K27me3, H3K4me3, and Ezh2 levels in Malat1KD and NT TE and MP cells at day 7 after infection. (E) Alignment tracks of H3K27me3 (Malat1KD and NT), H3K4me3 (Malat1KD and NT), Ezh2, and RNA expression (Malat1KD and NT) for key genes associated with memory cells. Gray highlight denotes H3K27me3 DMRs. (F) Ezh2 immunofluorescence in Malat1KD and NT cells 5 d after in vitro transduction (left) and bar graph quantification of area coverage of Ezh2 within the nucleus (right). Flow cytometry data are from one representative experiment out of two independent experiments with n = 4 mice per group (D); **, P < 0.005, paired t test. Graphs indicate mean ± SEM, symbols represent individual mice.
To elucidate a subset-specific role of Malat1 in the deposition of repressive H3K27me3 histone marks, we performed H3K27me3 CUT–RUN (Skene et al., 2018) experiments using FACS-purified TE and MP subsets of NT and Malat1KD cells. We observed a striking loss of 3,646 peaks in Malat1KD TE cells, with only a small set of 108 peaks gained (Fig. 6 B and Table S8). In contrast, we observed in Malat1KD MP cells a modest loss of 875 peaks and a gain of 209 peaks (Table S8). Consistent with our bulk ChIP-seq data, we observed a loss of H3K27me3 deposition on key memory-associated molecules Tcf7, Eomes, Id3, Foxo1, Bach2, and Bcl2 specifically in Malat1KD TE cells, but not in Malat1KD MP cells (Figs. 6 C and S5 E). Notably, we did not observe loss of H3K27me3 deposition on TE-associated molecules Tbx21, Runx1, Zeb2, and Prdm1 in either Malat1KD TE or MP cells (Fig. 6 C). Genomic annotation of differential peak sites between Malat1KD and NT TE and MP peaks revealed that most peak sites in Malat1KD TE cells were closer to gene promoter regions, consistent with the findings from our bulk ChIP-seq data (Figs. 6 D and S5 B).
Flow cytometry analyses of H3K37me3 expression levels on day 7 after infection confirmed global decreases within Malat1KD TE and MP subsets compared to their NT counterparts, while H3K4me3 levels remained unchanged, consistent with the ChIP-seq data (Fig. S5 D). Moreover, no changes in Ezh2 protein expression or nuclear localization pattern of Ezh2 in Malat1KD cells were observed, indicating that Malat1 does not regulate expression or localization of Ezh2 (Fig. S5, D and F). Lastly, we assessed Malat1 interaction with Ezh2 using RNA-binding protein immunoprecipitation (RIP) qPCR using in vitro activated CD8+ T cells. We observed enrichment for Malat1 as compared to housekeeping genes (Gapdh and Actb) upon Ezh2 pulldown (Fig. 6 D). Notably, knockdown of Malat1 did not impact Ezh2 interaction with other lncRNAs Xist and Neat1, demonstrating that knockdown of Malat1 specifically disrupted Ezh2–Malat1 interactions. Together, these results suggest that Malat1 maintains H3K27me3 deposition at a number of memory cell–associated genes, specifically in TE cells, through direct interactions with Ezh2.
Discussion
The molecular regulation of memory CD8+ T cell differentiation has been an area of intense investigation. Prior work in this field has focused primarily on protein-coding genes, while the role of the noncoding portion of the transcriptome in this process remains poorly understood. Here, we performed a functional genetic knockdown screen that identified lncRNA Malat1 as a regulator of CD8+ T cell differentiation. We provide functional evidence that Malat1 plays a critical role in the differentiation of the t-TEM circulating memory subset during primary infection and represses secondary TEM cell generation during infectious rechallenge. Notably, we observed that once t-TEM cells had formed, Malat1 did not play a critical role in the ability of primary t-TEM cells to give rise to secondary t-TEM cells upon infectious rechallenge, in contrast to primary TEM and TCM cells, which were dependent on Malat1 to give rise to secondary t-TEM cells. This suggests that multipotent CD8+ naive and memory TEM and TCM cells are dependent on Malat1, whereas more differentiated t-TEM cells with reduced proliferative capacity may be less so. Furthermore, these results demonstrate that Malat1 has a selective effect in promoting certain circulating memory cell subsets (t-TEM), but not others (TEM and TCM). To our knowledge, this is the one of few examples in which a regulator of CD8+ T cell differentiation selectively affects a specific circulating memory subset. Genetic deletions of known regulators of CD8+ T cell differentiation, such as Foxo1, Bcl6, and T-bet, tend to reduce t-TEM cell formation in response to LCMV infection, but also lead to reductions in the formation of TEM and TCM cells, effectively diminishing the entire pool of circulating memory cells (Milner et al., 2020).
Although a significant fraction of the CD8+ T cell coding genome is represented by lncRNAs, their roles remain poorly understood. We determined that >15% of chromatin-enriched RNAs are engaged by lncRNAs, in line with previous data showing that 25% of total poly-A captured RNAs are represented by lncRNAs in both mouse and human CD8+ T cells (Hudson et al., 2019). This underscores the importance of studying this class of molecules in CD8+ T cells in the context of microbial infection. NeST, one of the first identified lncRNA in T cells, was shown to promote expression of Ifng through an interaction of the MLL (mixed-lineage leukemia 1)/SET1 H3K4 methylase complex, thereby conferring resistance to Salmonella (Gomez et al., 2013). The lncRNA244, through its interactions with Ezh2, epigenetically represses Ifng and Tnf leading to CD8+ T cell dysfunction and increased susceptibility to Mycobacterium tuberculosis infection (Wang et al., 2015). In LCMV infection, the lncRNA Morrbid was specifically induced following Type 1 IFNγ stimulation, which in turn promoted the expression of the proapoptotic factor Bcl2l1, thereby negatively regulating CD8+ T cell expansion (Kotzin et al., 2019). A prior report using a germline deletion model demonstrated that Malat1 was dispensable for CD8+ T cell responses in LCMV infection (Yao et al., 2018), in contrast to the defects we observed in the current study using acute knockdown approaches. A possible explanation for these disparate results is that germline deletion models may have led to compensatory effects, as has been previously observed in T cells (El-Brolosy and Stainier, 2017). For example, deletion of the DNA epigenetic modifier Tet2 did not lead to any obvious defects in T cell development, but deletion of both Tet2 and Tet3 led to a massive lymphoproliferative phenotype (Tsagaratou et al., 2017; Lio and Rao, 2019), suggesting that Tet2 and Tet3 may be able to compensate for each other. As another example, T lymphocyte proliferation and immune function were unaffected by the deletion of Rbl2, a recruiter of histone methyltransferases, likely due to compensation by Rbl1 (Mulligan et al., 1998). Since lncRNAs represent a substantial fraction of chromatin-enriched RNAs, future studies elucidating chromatin-enriched lncRNAs in a Malat1 knockout model may reveal which lncRNA interactions are upregulated at sites normally occupied by Malat1.
Studies investigating mechanistic roles for Malat1 in transcriptional regulation have focused on interactions with various RNA processing enzymes, transcription factors, and epigenetic modifiers (Sun et al., 2018; Arun et al., 2020). Malat1 localizes to nuclear speckles which contain a large density of RNA Polymerase II and forms inter-chromosomal contacts, placing Malat1 in trans at regions of active transcription (Mao et al., 2011; West et al., 2014; Quinodoz et al., 2018). Our GRID-seq analysis allowed for an unbiased view of all chromatin-enriched RNAs and a direct comparison of Malat1 chromatin enrichment patterns relative to all other lncRNAs in activated CD8+ T cells. We found two clusters (Clusters 1 and 2) of highly enriched trans lncRNAs; Malat1 grouped with Cluster 2 lncRNAs, which were more highly associated with gene promoters and gene bodies, whereas Cluster 1 lncRNAs were more highly associated with distal intergenic sites. While our analysis explored highly trans-interacting lncRNAs, further investigation into locally interacting lncRNAs may shed light on the functional mechanisms of these classes of lncRNAs, many of which remain unexplored. Moreover, our GRID-seq analysis focused on lncRNAs and their genomic enrichment sites, but future studies may benefit by taking the reverse approach, focusing on genomic regions with high levels of RNA interaction and then identifying unifying groups of chromatin-enriched RNAs that contribute to these high-level interaction regions. In our study, we focused on known drivers of effector and memory CD8+ T cell differentiation and found that Malat1 has higher levels of interactions with genes associated with memory cell differentiation relative to those associated with TE cell differentiation. Extension of this analysis to our single-cell data analysis demonstrated that genes upregulated in a cluster of cells with a TE gene signature also exhibited higher levels of Malat1 interaction. Many memory-associated genes in this cell cluster were upregulated upon Malat1 knockdown, raising the possibility that Malat1 influences transcriptional regulation through epigenetic repression, potentially through a direct interaction with Ezh2. Indeed, Malat1-interaction sites had higher levels of H3K27me3 coverage as compared to other epigenetic markers H3K27ac, H3K4me4, and H3K4me1; moreover, H3K27me3 deposition was dramatically reduced upon Malat1 depletion.
In concordance with increased Malat1 interactions at gene bodies and gene promoters, 86.8% at DMRs were within the transcription start sites of gene promoters, providing further evidence of Malat1 transcriptional regulation in CD8+ T cells. Reduced H3K27me3 deposition on, and coordinately increased expression of, numerous memory cell–associated genes were reminiscent of the changes previously observed in Ezh2-deficient CD8+ T cells (Kakaradov et al., 2017; Gray et al., 2017), consistent with the idea that Malat1 acts, in part, through its actions on Ezh2. We note that Malat1KD cells exhibited upregulation of memory-associated genes along with downregulation of genes associated with TE differentiation. Since we did not observe changes in the epigenetic activation mark H3K4me3 upon Malat1 depletion, it remains possible that Malat1 depletion promotes the activity of another epigenetic repressor acting on genes associated with TE cell differentiation. One such mechanism may be through DNA methylation, which is often associated with gene silencing. Indeed, previous work has shown that naive cell activation led to DNA demethylation of many effector cell-associated loci including Prf1, Ifng, and Gzmk (Youngblood et al., 2017). Taken together, these findings demonstrate that Malat1 may promote t-TEM cell formation by repressing a transcriptional program that promotes TEM cell differentiation and advance our understanding of the functional role and underlying mechanisms by which lncRNAs may influence CD8+ T cell memory differentiation.
Materials and methods
Mice
All mice were housed under specific pathogen–free conditions in an American Association of Laboratory Animal Care–approved facility at University of California, San Diego (UCSD), and all procedures were approved by the UCSD Institutional Animal Care and Use Committee. C57BL6/J (CD45.1.2+ or CD45.2+) and P14 TCR transgenic (CD45.1+ or CD45.1.2+ maintained on a C57BL6/J background) mice were bred at UCSD or purchased from The Jackson Laboratory. Recipient male and donor female mice used in adoptive transfer experiments were all 6–9 wk of age. No randomization or blinding was used in infection experiments, and only mice that had rejected adoptively transferred P14 CD8+ T cells were excluded.
CD8+ T cell isolation
For isolation of CD8+ T cells from spleen and peripheral lymph nodes, tissues were dissociated through 70-μm cell strainers. Cells were then treated with Red Blood Cell Lysis Buffer for 5 min. CD8+ T cells were then enriched using the CD8a+ T Cell Isolation Kit and LS MACS Columns (catalog number: 130-104-075; Miltenyi Biotec) according to the manufacturer’s protocol. For CD8+ T cells isolated from tissues, small intestines were resected, Peyer’s patches removed, and washed with PBS. Tissues were then cut into 1 cm and incubated in DTE buffer (dithioerythritol [1 μg/ml; Thermo Fisher Scientific] in 10% HBSS and 10% Hepes bicarbonate) at 37°C for 30 min. Lymphocytes were then enriched using a 44/67% Percoll density gradient. CD8+ T cells were maintained in T cell medium (TCM; Iscove’s Modification of DMEM [catalog number: 10-016-CV] supplemented with 10% FBS [vol/vol], 2 mM L-glutamine [catalog number: 25030149], 100 U/ml penicillin–streptomycin [catalog number: 15140122], and 55 mM μl 2-mercaptoethanol [catalog number: 21985023]) at 37°C.
Antibodies and flow cytometry
Surface proteins were stained for 10 min on ice in HBSS (catalog number: 21-021-CV) with the following antibodies: Vα2 (B20.1), CD8α (53-6.7), CD45.1 (A20), CD45.2 (104), KLRG1 (2F1/KLRG1), CD127 (A7R34), CD27 (LG.3A10), CX3CR1 (SA011F11), CD44 (IM7), CD62L (MEL-14), CD69 (H1.2F3), CD103 (2E7), all purchased from BioLegend. For intracellular protein staining, samples were fixed in 2% paraformaldehyde (catalog number: 15710; Electron Microscopy Services) at room temperature for 45 min. Cells were then permeabilized using the FoxP3/Transcription Factor Staining Buffer Kit (catalog number: 00-5523-00; Thermo Fisher Scientific) and stained for 8 h at 4°C with the following antibodies: Tcf7 (C63D9), Eomes (Dan11mag), Bcl2 (BCL/1064), Ezh2 (11/EZH2), Ki67 (B56), Zeb1 (E2G6Y), Lef1 (C12A5), Gzma (GzA-3G8.5), T-bet (4B10), H3K27me3 (C36B11), and K3K4me3 (C42D8), purchased from BioLegend, Cell Signaling, BD Biosciences, and Thermo Fisher Scientific.
shRNA CD8+ T cell transfers, infection, and treatments
shERWOOD-designed UltramiR sequences targeting Malat1 (Malat1KD 5′-ACGTAACATATGAACACAAATT-3′, Malat1KD #2 5′-TTCAACCGTTCATTGAGGGTTA-3′, Malat1KD #3 5′-AATGCTAGTAGAGTAGGTGAAT-3′) and nontarget control scramble (NT, 5′-GTCCGTCTTCATACGTTTCGTA-3′) in an LMP-d Ametrine vector backbone were purchased from transOMIC technologies (Table S1). To generate retroviral particles, platinum E cells were grown in 10-cm plates with full selection media (DMEM [catalog number: 11965-118], 10% FBS [vol/vol], 2 mM L-glutamine [catalog number: 25030149], 100 U/ml penicillin–streptomycin [catalog number: 15140122], 1 μg/ml puromycin and 10 μg/ml blasticidin). 18 h before transfection, selection media was replaced with antibiotic-free media (DMEM, 10% FBS [vol/vol], 2 mM L-glutamine). For each 10-cm plate, 10 μg of each shRNA and 5 μg pCL-Eco helper plasmids were mixed in Opti-MEM (catalog number: 31985062) to a volume of 700 μl. This was combined with 45 μl TransIT-LT1 Reagent and 655 μl Opti-MEM for 20 min at room temperature. The mixture was then added dropwise to each 10-cm plate. 12 h later, media was replaced with fresh antibiotic-free media, and the supernatant was subsequently harvested at 24 and 48 h. Retroviral supernatant was filtered through a 0.45-μm syringe filter and stored at −80°C.
Naive WT or P14 CD8+ T cells were plated at a density of 1 × 106 cells/ml in 24-well plates precoated with 100 μg/ml goat anti-hamster IgG (catalog number: 31115), followed by 5 μg/ml each of anti-CD3 (clone 3C11) and anti-CD28 (clone 37.51). 18 h after activation, media was removed and replaced with 1 ml of retrovirus supplemented with 8 μg/ml polybrene followed by centrifugation for 90 min at 2,000 rpm. Retroviral supernatant was removed and replaced with fresh TCM, allowing cells to rest at 37°C for 2 h.
For co-transfer experiments, Malat1KD and NT P14 cells were mixed at a 1:1 ratio and a total of 5 × 105 donor cells/mouse was adoptively transferred into CD45.2+ male recipient mice. 1 h later, mice were infected with 2 × 105 PFU LCMV-Armstrong. 3, 5, 7, 35, and 65 d after infection, mice were euthanized, spleens and small intestine intraepithelial compartments were harvested, and cells were analyzed by flow cytometry to determine the ratio of Malat1KD/NT cells.
For rechallenge assays, 35 d after infection, Malat1KD and NT P14 cells were FACS sorted into three populations: CD127loCD62lo t-TEM, CD127hiCD62llo TEM, and CD127hiCD62Lhi TCM, constituting the donor populations. Donor Malat1KD and NT cells from each memory population were mixed at a 1:1 ratio for a total of 10,000 donor cells/mouse and adoptively transferred into naive male CD45.2+ recipient mice and infected with 2 × 105 PFU LCMV. 35 d after secondary rechallenge infection, mice were euthanized to determine Malat1KD/NT ratio.
For ex vivo restimulation to assess cytokine production, P14 CD8+ T cells were plated in a 96-well plate at 5 × 106 cells/well in the presence of 1 ng/μl LCMV GP33–41 peptide (catalog number: RP20257; Genscript) and 1× Brefeldin A Solution (catalog number: 420601; BioLegend) for 6 h at 37°C. Cells were then fixed and permeabilized using BD Cytofix/Cytoperm (catalog number: 554714) and stained for IFNγ (XMG1.2), TNF-α (MP6-XT22), and IL-2 (JES6-5H4) antibodies, all purchased from BioLegend, for 30 min on ice. Flow cytometry of all samples was run on a LSRFortessa X-20 (BD Biosciences) or Novocyte (ACEA Biosciences). FACS sorting of cells was done on a FACSAria Fusion or FACSAria2 (BD Biosciences). FlowJo software (BD Biosciences) was used for analysis of flow cytometry data.
Pooled shRNA screen and validation of individual Malat1 constructs
A pooled LMP-d Ametrine plasmid library consisting of 375 shRNA constructs with replicates for 103 genes and 5 nontarget controls was purchased from transOMIC technologies (Table S1). Retroviral particles were made and stored as described above. Multiplicity of infection was determined by serially diluting the retroviral supernatant twofold and performing activation and transfection as described above. 24 h after transfection, a dilution factor, which yielded 15–25% of Ametrine+ cells, was determined. Next, 5 × 105 transfected P14 cells were adoptively transferred into naive CD45.2+ recipient mice and infected with LCMV. To determine the baseline distribution of all shRNAs in the plasmid pool, part of the adoptive transfer mixture was grown for 24 h in IL-2 (100 U/ml), and Ametrine+ cells were sorted and genomic DNAs were extracted (catalog number: K182001; Thermo Fisher Scientific). 7 d after infection, 20 mice were euthanized, spleens extracted, and Amtetrine+ CD8+ T cells were sorted into KLRG1hiCD127lo TE and KLRG1loCD127hi MP populations. Genomic DNA was extracted, and integrated shRNA constructs were amplified with two rounds of PCR, adding TruSeq indexed barcodes for deep sequencing (catalog numbers: TRP0001, TRP0002; transOMIC technologies). Libraries were sequenced on a HiSeq 4000. Sequencing reads were mapped to the reference plasmid library. TE and MP reads were normalized to the input reads followed by taking the log2 ratio of TE/MP for every unique shRNA in the library. Z-score values for all shRNAs were calculated as follows:
where TENT, TEKD denote nontarget and knockdown shRNAs in TE cells, and MPNT, MPKD denote nontarget and knockdown shRNAs in MP cells. Individual Malat1 shRNA constructs were validated for knockdown efficiency by sorting WT CD8+ T cells 5 d after in vitro transduction. Total RNA was extracted using a Qiagen miRNeasy Micro Kit according to the manufacturer’s protocol (catalog number: 217084). 200 ng of RNA was converted to cDNA using the Bio-Rad Script cDNA Synthesis Kit according to manufacturer’s protocol (catalog number: 1708890) and diluted with water for a final 1:5 dilution. qPCR was performed with 1 ng template per reaction using the Bio-Rad SsoAdvanced Universal SYBR Green Supermix according to manufacturer’s protocol (catalog number: 1725270) on a BioRad CFX. Three primer sets tiling the Malat1 locus (bp position 2670-2885: 5′-GGGTGGGGGTGTTAGGTAAT-3′, 5′-GGCAGAGGAACCAACCTTC-3′. bp position 3143-3279: 5′-TGATTTTCCTTGTGACTAAACAAGA-3′, 5′-AAGCCCACCCTCTAAAAGACA-3′. bp position 4546-4741: 5′-AGGTGGGAGATGATGGTCAG-3′, 5′-ACTCGTGGCTCAAGTGAGGT-3′) and one primer set to RPL13a (bp position 41-257: 5′-GGGCAGGTTCTGGTATTGGAT-3′, 5′-GGCTCGGAAATGGTAGGGG-3′) as a control were used. Knockdown efficiency was quantified using the method, .
Bulk RNA-seq library generation and analysis
Malat1KD and NT P14 CD8+ T cells were sorted 7 d after LCMV infection, and total RNA was extracted using a Qiagen miRNeasy Micro Kit. RNA quality was evaluated using the Agilent TapeStation, confirming all samples with RNA integrity number scores >9.8. Samples were submitted to the UCSD Institute for Genomic Medicine for TruSeq V2 mRNA library prep. Libraries were then sequenced on a HiSeq 4000. Sequencing reads were mapped to mm10 reference genome using STAR aligner (v2.7.6a) with default parameters. Mapped reads to genes were summarized using featureCounts (v1.5.3) with default parameters. This table was used as input for differential gene analysis using DeSeq2 (v1.32.0). Upregulated TE and MP gene lists were generated from GEO accession number GSE157072 (Milner et al., 2020) with reads mapped, counted, and differential gene analysis determined using DeSeq2 as described above. Genes were filtered by log2 fold change >1.
scRNA-seq library generation and analysis
Malat1KD and NT P14 CD8+ T cells were sorted 7 d after LCMV infection and resuspended in PBS + 0.04% (wt/vol) bovine serum albumin. 10,000 cells per sample were loaded into Single Cell A chips and partitioned into Gel Bead In-Emulsions in a Chromium Controller (10× Genomics). scRNA-seq libraries were prepared with 10× Genomics Chromium Single Cell 3′ Reagent Kits v2 according to the manufacturer’s protocol. Libraries were sequenced on a HiSeq 4000.
Reads from scRNA-seq were aligned to mm10 using the 10× Genomics Cell Ranger software (v 2.1.0). Reads were collapsed into unique molecular identifier counts. All samples had >2,000 cells detected with >1,000 genes per cell and with >70% of the coding genome covered. Genes that were not expressed in at least 5% of all cells were excluded. As previously described (Boland et al., 2020), replicates of single-cell libraries were normalized removing batch effects using RUVnormalize (v1.15.0). The raw unique molecular identifier matrix was scaled and input to the naiveRandRUV function with parameters coeff = 1e−3 and k = 10. 50 negative control genes were taken from a list of housekeeping genes (Eisenberg and Levanon, 2013) with least variability in all datasets. Seurat (v3.0.1) functions were used to calculate top variable genes, PCA, and tSNE with FindVariableGenes, RunPCA, and RunTSNE. The top 5,000 genes were considered as input for the PCA calculation, and only the top 25 PCs were used in tSNE. Louvain clustering was performed by Seurat’s FindClusters function based on the top 25 PCs, with resolution set to 0.9. Differentially expressed genes were performed between clusters or within clusters comparing Malat1KD and NT cells using two-sided Wilcox test and threshold of P < 0.05.
GRID-seq library generation and analysis
Spleens from P14 mice were homogenized, and 1 × 106 cells/ml in TCM were pulsed with 1 ng/μl LCMV GP33–41 peptide for 1 h at 37°C. Cells were washed once with equal volume of warm TCM and then plated in a 96-well plate at 5 × 104 cells/well. Cells were harvested 4.5 d later, and dead cells were removed using the Dead Cell Removal kit according to the manufacturer’s protocol (catalog number: 130-010-101; Miltenyi Biotec). CD8+ T cells were then enriched using the CD8a+ T Cell Isolation Kit. Cells were then crosslinked, nuclei isolated, and GRID-seq libraries prepared, as described previously (Li et al., 2017; Zhou et al., 2019). Final libraries were sequenced on HiSeq 4000.
Reads were trimmed with cutadapt -l 86 --max-n 5 -o (v1.18), mapped to RNA-Linker-DNA (5′-GTTGGATTCNNNGACACAGCTCACTCCCACACACCGAACTCCAAC-3′) with bwa mem -k 5 -L 4 -B 2 -O 4 (v0.7.15) and sorted with samtools sort (v1.7). RNA and DNA reads were separated with GridTools matefq (https://github.com/biz007/gridtools). Reads were then mapped to the mm10 genome with bwa mem -k 17 -w 1 -T 1. GridTools evaluate was then used to correct against background and RNA–DNA mate read pair quality and quantity with a bin size of 1 kb and moving windows of 10. GridTools.py evaluate function uses a gene annotation gene transfer format file to ensure that the RNA end of the read always uniquely maps to its correct chromosomal location. Thus, during the analysis, only RNA reads that map back to their own locus will be retained. Moreover, the DNA end of the read will also be uniquely mapped, ensuring that both the RNA and DNA ends of each read are all unique. No read that maps to multiple locations in the genome (including repeat elements all over chromosomes) will be retained during analysis. GridTools RNA was used to identify expression levels of chromatin-enriched RNA. GridTools matrix was used to construct an interaction contact matrix of chromatin-associated RNA within specified genomic bin sizes of 1 kb or 100 kb. All lncRNAs with reads per kilobase DNA read density ≥10 on any genomic region were filtered from the interaction contact matrix. A 1-kb interaction matrix was directly visualized on a heatmap using the ComplexHeatmap package (v2.2.0). PCA, k-means clustering set to three clusters, and Pearson correlation analysis were performed on the interaction matrix with the R stats package (v3.6.2) removing all bins with zero interactions. The correlation matrix was visualized with corrplot package (v0.89) and circos plots with the circlize package (v0.42). Differential RNA chromatin interaction regions were determined by taking the average RNA interaction level of each genomic bin for all lncRNAs in a cluster. These averaged genomic bins were annotated to the transcription start site of the nearest gene using the ChIPpeakAnno. Consecutive genomic bins annotated to a single gene with greater RNA interaction for each genomic bin in one cluster relative to another cluster were considered differentially interacting. Differential RNA chromatin interaction regions were annotated with ChiPseeker.
Bedtools coverage (v2.29) was used to calculate coverage of histone marks, H3K27me3, H3K27ac, H3K4me3, H3K4me1, on Malat1 interacting chromatin regions. Malat1 interaction level was averaged over the gene body of TE genes, MP genes, and scRNA-seq cluster 0 and 2 differentially expressed genes.
ChIP-seq library generation and analysis
Malat1KD and NT P14 CD8+ T cells were sorted 7 d after LCMV infection and fixed in 1% fresh formaldehyde. Chromatin was then prepared using the EMD Millipore Magna ChIP A/G Chromatin Immunoprecipitation Kit according to manufacturer’s protocol (catalog number: 1710085) and flash-frozen in liquid nitrogen. Nuclei were sheared in Covaris microTUBES (catalog number: 520045) using the Covaris E220 (peak incident power 175W, duty factor 10%, cycles per burst 200, treatment time 600 s). For each immunoprecipitation (IP), 3 μg antibody per 1 × 106 cell equivalents were used for overnight incubation at 4°C. Antibodies used for IPs were as follows: anti-H3K27me3 (07-449; EMD Millipore), anti-H3K4me3 (ab8580; Abcam), and anti-Ezh2 (AC22, 07-449; EMD Millipore). 5% of each sample was kept as input control. Samples were then submitted for KAPA DNA Library Preparation and sequencing on a HiSeq 4000.
Libraries were filtered and mapped to the mm10 genome using ENCODE Transcription Factor and Histone ChIP-Seq processing pipeline with default parameters for histone marks (http://github.com/ENCODE-DCC/chip-seq-pipeline2). Final pooled bigwig files were used for visualization. Mapped non-duplicate read bam files for each biological replicate and overlapped optimal irreproducible discovery rate peaks were used as inputs for DiffBind (v2.0.2). DMRs between Malat1KD and NT were determined with a false discovery rate of <0.1. DMRs were annotated to their closest gene using CheapAnnoseak (v3.20.0) and genomic annotations using ChiPseeker (v1.22.0).
TE and MP H3K27me3 (GSE72408), TE H3K4me3 (GSE95237), TE H3K27ac (GSE72408), TE K3K4me1 (GSE95237), and input ChIP-seq libraries were mapped to the mm10 genome using ENCODE Transcription Factor and Histone ChIP-Seq processing pipeline with default parameters for histone marks. Optimal irreproducible discovery rate peaks for each histone mark were used as peak calls and coverage quantification in GRID-seq analysis.
CUT–RUN library generation and analysis
100,000 TE and MP Malat1KD and NT P14 CD8+ T cells were sorted 7 d after LCMV, and CUT–RUN libraries were generated using the Cell Signaling CUT–RUN Assay kit according to the manufacturer’s protocol (catalog number: 86652). Equal numbers of 100,000 cells per sample condition were isolated for isotype controls. Binding with primary anti-H3K27me3 antibody (07-449; EMD Millipore) and IgG control antibody binding was conducted overnight at 4°C. 50 pg of Spike-In Yeast DNA was added per sample after pAG-MNase activation with calcium chloride. DNA was purified and sequencing libraries were generated using SimpleChIP ChIP-seq DNA Library Prep Kit for Illumina with Dual indices according to manufacturer protocol (catalog number: 56795). Final libraries were amplified for 10 cycles and 100-bp paired end sequencing performed on a NovaSeq 6000.
Adapter sequences were trimmed with trimmomatic PE-threads 8 ILLUMINACLIP:$adapters:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:10 MINLEN:25 (v0.38). Reads were mapped either to the mouse or yeast genome with bowtie2 -p 8 --local --very-sensitive-local --no-unal --no-mixed --no-discordant --phred33 -I 10 -X 700 (v1.2.2). Mapped reads were then converted into bed files. A scale factor for spike-in normalization was calculated for each sample by dividing 1,000 from the sequencing depth of mapped yeast reads. Bed files were normalized using bedtools genomcov (v2.29.2) with the respective scaling factor calculated, as described above. Peaks calls were determined using macs2 macs2 callpeak -broad --broad-cutoff 0.1 -B --nomodel --keep-dup all -f BED (v2.1.2) with matched isotype samples as controls. Differential peak analysis was performed with Diffbind (v2.0.2) for Malat1KD and NT TE or MP cells respectively.
RIP preparation and analysis
10,000,000 Malat1KD and NT P14 CD8+ T cells were sorted 5.5 d after transduction, washed twice in cold PBS, and lysed with the EMD Millipore Magna RIP RNA-binding protein immunoprecipitation kit according to the manufacturer’s protocol (catalog number: 17-700). Lysates were then flash-frozen in liquid nitrogen. 10% of each lysate was removed as input control while the remaining lysates of each sample was immunoprecipitated with 5 μg of anti-Ezh2 (AC22) overnight at 4°C. Final bound RNA was quantified with Qubit HS RNA (catalog number: Q32852) and 200 ng of each pull-down sample with matched input were converted into cDNA (catalog number: 1708890). qPCR was performed with a primer sets against Malat1 (bp position 3143-3279), Gapdh (bp position 683-925: 5′-AGAGAGGGAGGAGGGGAAAT-3′, 5′-GATTTTCACCTGGCACTGCA-3′) and Actb (bp position 1388-1602: 5′-ACTGGGACGACATGGAGAAG-3′, 5′-ATGGGAGAACGGCAGAAGAA-3′), with fold change calculated, .
Immunofluorescence and analysis
Malat1KD and NT P14 CD8+ T cells were sorted 5.5 d after transduction. Cells were dried on a microscope coverslip at 37°C for 10 min, fixed in 3% PFA at room temperature, and then quenched for three washes with 50 mM NH4Cl. Slides were then permeabilized with 0.3% Triton X-100 in PBS followed by 1× Block treatment (5×, 0.01% sap, 0.25% fish skin gelatin, 0.02% NaN3 in PBS). Primary antibody staining anti-Ezh2 (AC22) was diluted 1:50 in PBS for 1 h at room temperature followed by five washes with 1× Block. Secondary antibody staining was performed with Alexa Fluor 488 Donkey anti-rabbit IgG (catalog number: 406417; Biolegend) diluted 1:200 in PBS for 1 h at room temperature, followed by five washes with 1× Block. Coverslips were mounted on glass slides with Prolong Glass Mounting Reagent containing DAPI (catalog number: P36981; Thermo Fisher Scientific) and left in the dark at room temperature overnight. Imaging was performed on a Leica SP8 Confocal with Lightning Deconvolution at 63× magnification. Minor adjustments of brightness and contrast were made equally to all images with ImageJ (v1.53a). Color channels were split and converted to grayscale 8-bit images. The DAPI channel was converted to a binary mask, edges of each nuclei found, then added as regions of interest. The Ezh2 channel was converted to a binary image, region of interest overlayed, and percentage area was calculated to quantify coverage of Ezh2 within each nucleus.
Online supplemental material
Fig. S1 provides supporting information validating the shRNA pooled screen. Fig. S2 provides supporting information for the role of Malat1 in memory cells. Fig. S3 evaluates the consequence of Malat1 depletion in TRM cells in the siIEL compartment. Fig. S4 explores differential interaction patterns of cis and trans lncRNAs. Fig. S5 evaluates the consequence of Malat1 depletion in Ezh2 localization and function. Table S1 provides Z-scores and shRNA sequences of constructs tested in the in vivo shRNA pooled screen. Table S2 provides TE and MP gene lists used to quantify enrichment scores in the scRNA-seq data. Table S3 lists differentially expressed genes between Malat1KD and NT cells in each scRNA-seq cluster. Table S4 lists coverage of epigenetic marks at Malat1 interaction sites. Table S5 lists Malat1 interaction levels at TE and MP genes marked by H3K27me3. Table S6 lists differentially expressed genes between scRNA-seq cluster 0 and all other clusters. Table S7 lists H3K27me3 differentially methylated regions in Malat1KD and NT cells at day 7. Table S8 lists differential peak sites in Malat1KD and NT TE and MP cells.
Supplementary Material
provides Z-scores and shRNA sequences of constructs tested in the in vivo shRNA pooled screen.
provides TE and MP gene lists used to quantify enrichment scores in the scRNA-seq data.
lists differentially expressed genes between Malat1KD and NT cells in each scRNA-seq cluster.
lists coverage of epigenetic marks at Malat1 interaction sites.
lists Malat1 interaction levels at TE and MP genes marked by H3K27me3.
lists differentially expressed genes between scRNA-seq cluster 0 and all other clusters.
lists H3K27me3 differentially methylated regions in Malat1KD and NT cells at day 7.
lists differential peak sites in Malat1KD and NT TE and MP cells.
Acknowledgments
scRNA-seq using the 10× Genomics platform was performed at the UCSD IGM Genomics Center and supported by National Institutes of Health (NIH) grants P30KC063491, P30CA023100, and S10OD026929. Microscopy was performed at the UCSD Microscopy Core and supported by NIH grant NS047101. This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases–funded San Diego Digestive Diseases Research Center (P30DK120515) and funded by grants from the NIH: AI129973 and BX005106 (J.T. Chang.); AI132122, AI123202 (G.W. Yeo and J.T. Chang); HG004659 (G.W. Yeo); GM124494 (W.J. Huang); and HG004659 and DK098808 (X.-D. Fu).
Author contributions: Conceptualization: J.N. Kanbar and J.T. Chang; investigation: J.N. Kanbar, S. Ma, N.S. Kurd, M.S. Tsai, T. Tysl, C.E. Widjaja, E.S. Kim, A.E. Limary; analysis: J.N. Kanbar, B. Yee, Z. He, Y. Hao; supervision: X.-D. Fu, G.W. Yeo, W.J. Huang, J.T. Chang. Graphical abstract created with BioRender.com.
Data availability
Data are available at the Gene Expression Omnibus under accession no. GSE203092.
References
- Arun, G., Aggarwal D., and Spector D.L.. 2020. MALAT1 long non-coding RNA: Functional implications. Noncoding RNA. 6:22. 10.3390/ncrna6020022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee, A., Gordon S.M., Intlekofer A.M., Paley M.A., Mooney E.C., Lindsten T., Wherry E.J., and Reiner S.L.. 2010. Cutting edge: The transcription factor eomesodermin enables CD8+ T cells to compete for the memory cell niche. J. Immunol. 185:4988–4992. 10.4049/jimmunol.1002042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard, D., Prasanth K.V., Tripathi V., Colasse S., Nakamura T., Xuan Z., Zhang M.Q., Sedel F., Jourdren L., Coulpier F., et al. 2010. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 29:3082–3093. 10.1038/emboj.2010.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland, B.S., He Z., Tsai M.S., Olvera J.G., Omilusik K.D., Duong H.G., Kim E.S., Limary A.E., Jin W., Milner J.J., et al. 2020. Heterogeneity and clonal relationships of adaptive immune cells in ulcerative colitis revealed by single-cell analyses. Sci Immunol. 5:eabb4432. 10.1126/sciimmunol.abb4432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, J.T., Wherry E.J., and Goldrath A.W.. 2014. Molecular regulation of effector and memory T cell differentiation. Nat. Immunol. 15:1104–1115. 10.1038/ni.3031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, R., Bélanger S., Frederick M.A., Li B., Johnston R.J., Xiao N., Liu Y.C., Sharma S., Peters B., Rao A., et al. 2014. In vivo RNA interference screens identify regulators of antiviral CD4+ and CD8+ T cell differentiation. Immunity. 41:325–338. 10.1016/j.immuni.2014.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg, E., and Levanon E.Y.. 2013. Human housekeeping genes, revisited. Trends Genet. 29:569–574. 10.1016/j.tig.2013.05.010 [DOI] [PubMed] [Google Scholar]
- Eißmann, M., Gutschner T., Hämmerle M., Günther S., Caudron-herger M., Groß M., Schirmacher P., Rippe K., Braun T., Zörnig M., and Diederichs S.. 2012. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 9:1076–1087. 10.4161/rna.21089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Brolosy, M.A., and Stainier D.Y.R.. 2017. Genetic compensation: A phenomenon in search of mechanisms. PLoS Genet. 13:e1006780. 10.1371/journal.pgen.1006780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez, J.A., Wapinski O.L., Yang Y.W., Bureau J.F., Gopinath S., Monack D.M., Chang H.Y., Brahic M., and Kirkegaard K.. 2013. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-γ locus. Cell. 152:743–754. 10.1016/j.cell.2013.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, S.M., Amezquita R.A., Guan T., Kleinstein S.H., Kaech S.M., Gray S.M., Amezquita R.A., Guan T., Kleinstein S.H., and Kaech S.M.. 2017. Polycomb repressive complex 2-mediated chromatin repression guides effector CD8+ T cell terminal differentiation and loss of multipotency. Immunity. 46:596–608. 10.1016/j.immuni.2017.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner, T., Hämmerle M., and Diederichs S.. 2013a. MALAT1: A paradigm for long noncoding RNA function in cancer. J. Mol. Med. 91:791–801. 10.1007/s00109-013-1028-y [DOI] [PubMed] [Google Scholar]
- Gutschner, T., Hämmerle M., Eissmann M., Hsu J., Kim Y., Hung G., Revenko A., Arun G., Stentrup M., Gross M., et al. 2013b. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 73:1180–1189. 10.1158/0008-5472.CAN-12-2850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, W.H., Prokhnevska N., Gensheimer J., Akondy R., McGuire D.J., Ahmed R., and Kissick H.T.. 2019. Expression of novel long noncoding RNAs defines virus-specific effector and memory CD8+ T cells. Nat Commun. 10:196. 10.1038/s41467-018-07956-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, P., Diederichs S., Wang W., Boing S., Metzger R., Paul M., Tidow N., Brandt B., Buerger H., Bulk E., et al. 2003. MALAT-1, a novel noncoding RNA, and thymosin b 4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 22:8031–8041. 10.1038/sj.onc.1206928 [DOI] [PubMed] [Google Scholar]
- Ji, Y., Pos Z., Rao M., Klebanoff C.A., Yu Z., Sukumar M., Reger R.N., Palmer D.C., Borman Z.A., Muranski P., et al. 2011. Repression of the DNA-binding inhibitor Id3 by Blimp-1 limits the formation of memory CD8+ T cells. Nat. Immunol. 12:1230–1237. 10.1038/ni.2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi, N.S., Cui W., Chandele A., Lee H.K., Urso D.R., Hagman J., Gapin L., and Kaech S.M.. 2007. Inflammation directs memory precursor and short-lived effector CD8+ T cell fates via the graded expression of T-bet transcription factor. Immunity. 27:281–295. 10.1016/j.immuni.2007.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakaradov, B., Arsenio J., Widjaja C.E., He Z., Aigner S., Metz P.J., Yu B., Wehrens E.J., Lopez J., Kim S.H., et al. 2017. Early transcriptional and epigenetic regulation of CD8+ T cell differentiation revealed by single-cell RNA sequencing. Nat. Immunol. 18:422–432. 10.1038/ni.3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallies, A., Xin A., Belz G.T., and Nutt S.L.. 2009. Blimp-1 transcription factor is required for the differentiation of effector CD8+ T cells and memory responses. Immunity. 31:283–295. 10.1016/j.immuni.2009.06.021 [DOI] [PubMed] [Google Scholar]
- Kim, J., Piao H.L., Kim B.J., Yao F., Han Z., Wang Y., Xiao Z., Siverly A.N., Lawhon S.E., Ton B.N., et al. 2018. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat. Genet. 50:1705–1715. 10.1038/s41588-018-0252-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.H., Kim S.H., Yang W.I., Kim S.J., and Yoon S.O.. 2017. Association of the long non-coding RNA MALAT1 with the polycomb repressive complex pathway in T and NK cell lymphoma. Oncotarget. 8:31305–31317. 10.18632/oncotarget.15453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp, F., and Mendell J.T.. 2018. Functional classification and experimental dissection of long noncoding RNAs. Cell. 172:393–407. 10.1016/j.cell.2018.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotzin, J.J., Iseka F., Wright J., Basavappa M.G., Clark M.L., Ali M.A., Abdel-Hakeem M.S., Robertson T.F., Mowel W.K., Joannas L., et al. 2019. The long noncoding RNA Morrbid regulates CD8 T cells in response to viral infection. Proc. Natl. Acad. Sci. USA. 116:11916–11925. 10.1073/pnas.1819457116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurd, N.S., He Z., Louis T.L., Milner J.J., Omilusik K.D., Jin W., Tsai M.S., Widjaja C.E., Kanbar J.N., Olvera J.G., et al. 2020. Early precursors and molecular determinants of tissue-resident memory CD8+ T lymphocytes revealed by single-cell RNA sequencing. Sci. Immunol. 5:eaaz6894. 10.1126/sciimmunol.aaz6894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Zhou B., Chen L., Gou L.T., Li H., and Fu X.D.. 2017. GRID-seq reveals the global RNA-chromatin interactome. Nat. Biotechnol. 35:940–950. 10.1038/nbt.3968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lio, C.W.J., and Rao A.. 2019. TET enzymes and 5hMC in adaptive and innate immune systems. Front. Immunol. 10:210. 10.3389/fimmu.2019.00210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, Y.S., Zhang B., and Spector D.L.. 2011. Biogenesis and function of nuclear bodies. Trends Genet. 27:295–306. 10.1016/j.tig.2011.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masopust, D., Vezys V., Marzo A.L., and Lefrançois L.. 2014. Preferential localization of effector memory cells in nonlymphoid tissue. J. Immunol. 192:845–849. [PubMed] [Google Scholar]
- Michelini, R.H., Doedens A.L., Goldrath A.W., and Hedrick S.M.. 2013. Differentiation of CD8 memory T cells depends on Foxo1. J. Exp. Med. 210:1189–1200. 10.1084/jem.20130392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner, J.J., Nguyen H., Omilusik K., Reina-Campos M., Tsai M., Toma C., Delpoux A., Boland B.S., Hedrick S.M., Chang J.T., and Goldrath A.W.. 2020. Delineation of a molecularly distinct terminally differentiated memory CD8 T cell population. Proc. Natl. Acad. Sci. USA. 117:25667–25678. 10.1073/pnas.2008571117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner, J.J., Toma C., Yu B., Zhang K., Omilusik K., Phan A.T., Wang D., Getzler A.J., Nguyen T., Crotty S., et al. 2017. Runx3 programs CD8+ T cell residency in non-lymphoid tissues and tumours. Nature. 552:253–257. 10.1038/nature24993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan, G.J., Wong J., and Jacks T.. 1998. p130 is dispensable in peripheral T lymphocytes: Evidence for functional compensation by p107 and pRB. Mol Cell Biol. 18:206–220. 10.1128/MCB.18.1.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, S., Ip J.Y., Shioi G.O., Tripathi V., Zong X., Hirose T., and Prasanth K.V.. 2012. Malat1 is not an essential component of nuclear speckles in mice. RNA. 18:1487–1499. 10.1261/rna.033217.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson, J.A., McDonald-Hyman C., Jameson S.C., and Hamilton S.E.. 2013. Effector-like CD8+ T cells in the memory population mediate potent protective immunity. Immunity. 38:1250–1260. 10.1016/j.immuni.2013.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omilusik, K.D., Best J.A., Yu B., Goossens S., Weidemann A., Nguyen J.V., Seuntjens E., Stryjewska A., Zweier C., Roychoudhuri R., et al. 2015. Transcriptional repressor ZEB2 promotes terminal differentiation of CD8+ effector and memory T cell populations during infection. J. Exp. Med. 212:2027–2039. 10.1084/jem.20150194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omilusik, K.D., Nadjsombati M.S., Shaw L.A., Yu B., Milner J.J., and Goldrath A.W.. 2018. Sustained Id2 regulation of E proteins is required for terminal differentiation of effector CD8+ T cells. J. Exp. Med. 215:773–783. 10.1084/jem.20171584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinodoz, S.A., Ollikainen N., Tabak B., Palla A., Schmidt J.M., Detmar E., Lai M.M., Shishkin A.A., Bhat P., Takei Y., et al. 2018. Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell. 174:744–757.e24. 10.1016/j.cell.2018.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto, F., Lenig D., Förster R., Lipp M., and Lanzavecchia A.. 1999. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 401:708–712. 10.1038/44385 [DOI] [PubMed] [Google Scholar]
- Schenkel, J.M., Fraser K.A., Beura L.K., Pauken K.E., Vezys V., and Masopust D.. 2014. T cell memory. Resident memory CD8 T cells trigger protective innate and adaptive immune responses. Science. 346:98–101. 10.1126/science.1254536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, S., Findlay G.M., Bandukwala H.S., Oberdoerffer S., Baust B., Li Z., Schmidt V., Hogan P.G., Sacks D.B., and Rao A.. 2011. Dephosphorylation of the nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA-protein scaffold complex. Proc. Natl. Acad. Sci. USA. 108:11381–11386. 10.1073/pnas.1019711108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene, P.J., Henikoff J.G., and Henikoff S.. 2018. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat. Protoc. 13:1006–1019. 10.1038/nprot.2018.015 [DOI] [PubMed] [Google Scholar]
- Spector, D.L., and Lamond A.I.. 2011. Nuclear speckles. Cold Spring Harb. Perspect. Biol. 3:a000646. 10.1101/cshperspect.a000646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Q., Hao Q., Prasanth K.V., et al. 2018. Nuclear long noncoding RNAs: Key regulators of gene expression. Trends Genet. 34:142–157. 10.1016/j.tig.2017.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tano, K., Mizuno R., Okada T., Rakwal R., Shibato J., Masuo Y., Ijiri K., and Akimitsu N.. 2010. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 584:4575–4580. 10.1016/j.febslet.2010.10.008 [DOI] [PubMed] [Google Scholar]
- Tripathi, V., Ellis J.D., Shen Z., Song D.Y., Pan Q., Watt A.T., Freier S.M., Bennett C.F., Sharma A., Bubulya P.A., et al. 2010. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 39:925–938. 10.1016/j.molcel.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsagaratou, A., González-Avalos E., Rautio S., Scott-Browne J.P., Togher S., Pastor W.A., Rothenberg E.V., Chavez L., Lähdesmäki H., and Rao A.. 2017. TET proteins regulate the lineage specification and TCR-mediated expansion of iNKT cells. Nat. Immunol. 18:45–53. 10.1038/ni.3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Zhong H., Xie X., Chen C.Y., Huang D., Shen L., Zhang H., Chen Z.W., and Zeng G.. 2015. Long noncoding RNA derived from CD244 signaling epigenetically controls CD8+ T-cell immune responses in tuberculosis infection. Proc. Natl. Acad. Sci. USA. 112:E3883–E3892. 10.1073/pnas.1501662112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, J.A., Davis C.P., Sunwoo H., Simon M.D., Sadreyev R.I., Wang P.I., Tolstorukov M.Y., and Kingston R.E.. 2014. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell. 55:791–802. 10.1016/j.molcel.2014.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, Y., Guo W., Chen J., Guo P., Yu G., Liu J., Wang F., Liu J., You M., Zhao T., et al. 2018. Long noncoding RNA Malat1 is not essential for T cell development and response to LCMV infection. RNA Biol. 15:1477–1486. 10.1080/15476286.2018.1551705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngblood, B., Hale J.S., Kissick H.T., Ahn E., Xu X., Wieland A., Araki K., West E.E., Ghoneim H.E., Fan Y., et al. 2017. Effector CD8 T cells dedifferentiate into long-lived memory cells. Nature. 552:404–409. 10.1038/nature25144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, B., Zhang K., Milner J.J., Toma C., Chen R., Scott-Browne J.P., Pereira R.M., Crotty S., Chang J.T., Pipkin M.E., et al. 2017. Epigenetic landscapes reveal transcription factors that regulate CD8+ T cell differentiation. Nat. Immunol. 18:573–582. 10.1038/ni.3706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B., Arun G., Mao Y.S., Lazar Z., Hung G., Bhattacharjee G., Xiao X., Booth C.J., Wu J., Zhang C., and Spector D.L.. 2012. The lncRNA malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2:111–123. 10.1016/j.celrep.2012.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, B., Li X., Luo D., Lim D.H., Zhou Y., and Fu X.D.. 2019. GRID-seq for comprehensive analysis of global RNA–chromatin interactions. Nat. Protoc. 14:2036–2068. 10.1038/s41596-019-0172-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X., and Xue H.-H.. 2012. Cutting edge: Generation of memory precursors and functional memory CD8+ T cells depends on T cell factor-1 and lymphoid enhancer-binding factor-1. J. Immunol. 189:2722–2726. 10.4049/jimmunol.1201150 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
provides Z-scores and shRNA sequences of constructs tested in the in vivo shRNA pooled screen.
provides TE and MP gene lists used to quantify enrichment scores in the scRNA-seq data.
lists differentially expressed genes between Malat1KD and NT cells in each scRNA-seq cluster.
lists coverage of epigenetic marks at Malat1 interaction sites.
lists Malat1 interaction levels at TE and MP genes marked by H3K27me3.
lists differentially expressed genes between scRNA-seq cluster 0 and all other clusters.
lists H3K27me3 differentially methylated regions in Malat1KD and NT cells at day 7.
lists differential peak sites in Malat1KD and NT TE and MP cells.
Data Availability Statement
Data are available at the Gene Expression Omnibus under accession no. GSE203092.