SUMMARY
Regulated loading of eIF3-bound 40S ribosomes on capped mRNA is generally dependent upon the translation initiation factor eIF4E; however, mRNA translation often proceeds during physiological stress, such as virus infection, when eIF4E availability and activity are limiting. It remains poorly understood how translation of virus and host mRNAs are regulated during infection stress. While initially sensitive to mTOR inhibition, which limits eIF4E-dependent translation, we show that protein synthesis in human cytomegalovirus (HCMV)-infected cells unexpectedly becomes progressively reliant upon eIF3d. Targeting eIF3d selectively inhibits HCMV replication, reduces polyribosome abundance, and interferes with expression of essential virus genes and a host gene expression signature indicative of chronic ER stress that fosters HCMV reproduction. This reveals a strategy whereby cellular eIF3d-dependent protein production is hijacked to exploit virus-induced ER stress. Moreover, it establishes how switching between eIF4E and eIF3d-responsive cap-dependent translation can differentially tune virus and host gene expression in infected cells.
Graphical abstract
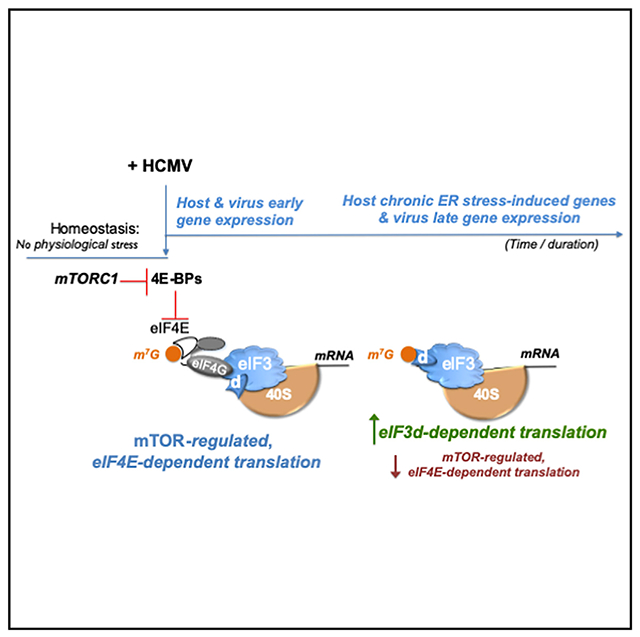
In brief
Instead of eIF4E-regulated ribosome loading, Thompson et al. show capped mRNA translation in HCMV-infected cells becomes reliant upon eIF3d. Depleting eIF3d inhibits HCMV replication, reduces polyribosomes, and restricts virus late gene and host chronic ER stress-induced gene expression. Thus, switching to eIF3d-responsive translation tunes gene expression to support virus replication.
INTRODUCTION
Gene expression rapidly responds to physiological stress like virus infection in part via the control of mRNA translation (Mohr and Sonenberg, 2012). This encompasses a vast array of strategies that range from global suppression of host mRNA translation to more nuanced tactics supporting both virus and host cell protein synthesis. The consequences are profound for all virus lifecycles, which mandate viral polypeptide production by host ribosomes, and cell intrinsic host defenses, many of which restrict infected cell protein synthesis (Stern-Ginossar et al., 2019). Precisely how this is achieved and the underlying molecular mechanisms that support infected cell protein synthesis are incompletely understood.
Regulated ribosome loading onto the 5′-end of m7GTP-capped mRNAs is a fundamental step that regulates protein synthesis in eukaryotes (Pelletier and Sonenberg, 2019). This typically is executed by eukaryotic translation initiation factors (eIFs) including the cap-binding protein eIF4E, which recognizes the 5′-terminal cap structure and assembles a multi-subunit initiation factor complex to load the eIF3-bound 40S ribosome. Assembly of this canonical initiation factor complex reliant upon cap-recognition by eIF4E is responsive to environmental and physiological cues that control mechanistic target of rapamycin complex-1 (mTORC1) activation, which regulates eIF4E binding to the eIF4E-binding protein (4E-BP) family of translational repressors and thereby adjusts 40S ribosome recruitment to capped mRNAs. To maximize translation initiation on capped virus and host mRNAs in infected cells, virus-encoded functions often stimulate mTORC1 to overcome repression by 4E-BPs. Nevertheless, the suppression of translation by 4E-BP sequestering eIF4E or mTOR kinase active-site inhibitors remains incomplete at best in both infected and uninfected cells (Chuluunbaatar et al., 2010; Feldman et al., 2009; Thoreen et al., 2009). This could reflect variable stoichiometry of eIF4E and 4E-BP in different conditions (Alain et al., 2012) or cell types or reduced reliance upon eIF4E for 40S loading onto capped mRNAs (Pelletier and Sonenberg, 2019).
Delivery of 40S ribosome subunits to a specialized eIF4E-containing initiation factor complex assembled upon the mRNA capped 5′-terminus is mediated by eIF3. Long viewed as a general translation factor required to bind, load, and recycle ribosomes, eIF3 has more recently been shown to regulate translation of a subset of mRNAs (Lamper et al., 2020; de la Parra et al., 2018; Lee et al., 2015, 2016; Pulos-Holmes et al., 2019). Assembled from 13 distinct protein subunits (Sun et al., 2011), human eIF3 contains six functional core (a, b, c, e, f, h) subunits required for generalized translation initiation activity (Masutani et al., 2007), one of which, eIF3e, also regulates phosphorylation of the cap-binding protein eIF4E (Walsh and Mohr, 2014). Among human eIF3 non-core (d, g, i, j, k, l, m) subunits dispensable for translation initiation (Masutani et al., 2007), eIF3d selectively regulates mRNA translation by serving as an alternative cap-recognition subunit that can mediate 40S loading independent of the canonical cap-binding protein eIF4E (Lee et al., 2016). This allows cap-dependent translation of select mRNAs to proceed while eIF4E is repressed, such as during nutrient stress induced by prolonged glucose starvation (Lamper et al., 2020). How eIF3 responds to virus infection and how non-canonical translation initiation mechanisms including those involving non-core subunits like eIF3d might impact infected cell protein synthesis and virus infection biology is not understood.
Although innocuous in most healthy individuals, the β-herpesvirus human cytomegalovirus (HCMV) is a widespread opportunistic pathogen responsible for severe disease among the immunocompromised and remains a major source of congenital morbidity and mortality among newborns (Boeckh and Geballe, 2011; Britt, 2008; Cannon et al., 2010; Ljungman et al., 2011; Manicklal et al., 2013; Razonable and Humar, 2013). During the productive growth cycle of this large DNA virus that replicates within the nucleus, more than 200 ORFs encoded by the genome are coordinately expressed, and different capped, polyadenylated virus-encoded mRNA populations accumulate at distinct times in the infectious program (Murphy et al., 2003a, 2003b; Stern-Ginossar et al., 2012). Since HCMV infection, unlike many viruses, does not globally restrict host protein synthesis, it provides a powerful model to investigate how a dynamic transcriptome comprised of host and virus mRNAs competes for ribosome access to remodel the proteome. While assembling an eIF4E-containing translation initiation factor complex (Kudchodkar et al., 2004; McKinney et al., 2012; Walsh et al., 2005) and remodeling the host translational landscape (McKinney et al., 2014; Tirosh et al., 2015) regulate HCMV productive replication, the sensitivity of infected cell protein synthesis to inhibitors of mTOR, which repress cap-dependent translation reliant upon eIF4E, or the eIF4A helicase activity associated with eIF4E-containing initiation factor complexes is reduced. Specifically, HCMV protein accumulation is dependent upon mTOR early in infection but becomes insensitive to mTOR and eIF4A inhibitors later in the virus life cycle (Clippinger et al., 2011; Lenarcic et al., 2014). Precisely how HCMV-infected cell protein synthesis might proceed with reduced requirements for components and/or biochemical activities needed for canonical cap-dependent translation remains unexplained, and roles for alternative cap-recognition factors like eIF3d have yet to be investigated.
Here we show that the overall abundance of eIF3 protein subunits, including the cap-interacting subunit eIF3d, increases in response to HCMV infection. While overall eIF3d mRNA levels remain relatively unchanged by HCMV infection, eIF3d protein accumulation was dependent upon host mTOR activity, consistent with post-transcriptional control of its expression. In addition to reducing accumulation of an essential HCMV late protein and interfering with infected cell protein synthesis, eIF3d depletion selectively inhibited HCMV reproduction and spread. Finally, eIF3d depletion globally remodeled the infected cell translational landscape, reducing expression of critical virus genes and interfering with a host eIF3d-dependent gene expression program that supports productive HCMV replication. This establishes that eIF3d plays an unexpected role in regulating HCMV gene expression and productive growth. Furthermore, it provides evidence that a switch favoring eIF3d-dependent mRNA translation plays a vital role remodeling the infected cell translation landscape to support HCMV productive growth.
RESULTS
Regulation of eIF3 subunit abundance in response to HCMV infection
To investigate how eIF3 is impacted by acute HCMV infection, total protein from HCMV-infected primary human fibroblasts (normal human dermal fibroblasts, NHDFs) was isolated at various times post-infection and the overall abundance of representative human eIF3 functional core (eIF3a, b, e, h), and non-core (eIF3d, g, l) subunits evaluated. Remarkably, compared to uninfected cells, protein levels of each eIF3 subunit evaluated (eIF3a, b, d, e, g, h, l) were elevated by 24 h post-infection (hpi) and continued to increase until 72 hpi (Figure 1A). Virus protein accumulation was next measured to delineate when eIF3 subunit accumulation increases relative to representative HCMV-encoded immediate-early (IE), early (E), and late (L) proteins produced with different kinetics. Detection as early as 3–6 hpi of HCMV IE proteins (IE1/2), which encode master transactivators that regulate transcription, and the E protein (UL44) required for DNA synthesis preceded virus-induced accumulation of core and non-core eIF3 subunits. By contrast, accumulation of the essential L protein pp28 was not readily observed until 48 hpi, subsequent to virus-induced eIF3 subunit accumulation (Figure 1A). Levels of GAPDH, a control cellular antigen, did not detectably increase in response to HCMV infection. To understand how this protein accumulation was coordinated, the abundance of mRNAs encoding each eIF3 subunit was examined by qRT-PCR. Core subunit eIF3b mRNA level increased nearly 5-fold by 24 hpi (Figure 1B), suggesting that elevated eIF3b transcript levels possibly resulting from transcriptional stimulation or decreased decay account for increased eIF3b protein abundance. Indeed, eIF3b protein levels relative to GAPDH (Figure 1A) increase from 2-fold at 12 hpi, to 3.3-fold at 48 hpi, to 4.8 fold at 72 hpi, mirroring the RNA abundance increase (Figure 1B). By contrast, mRNAs encoding eIF3a, d, g, and h remained at similar levels detected in uninfected cells, fluctuating less than 2-fold (Figure 1B), and levels of eIF3e and l mRNAs declined by nearly 2-fold over time despite accumulation of their encoded protein products (Figure 1B). With the exception of eIF3b, this raised the possibility that increased eIF3 subunit protein levels resulted from a post-transcriptional regulatory process triggered by infection.
Figure 1. Regulation of eIF3 subunit accumulation in response to HCMV infection.
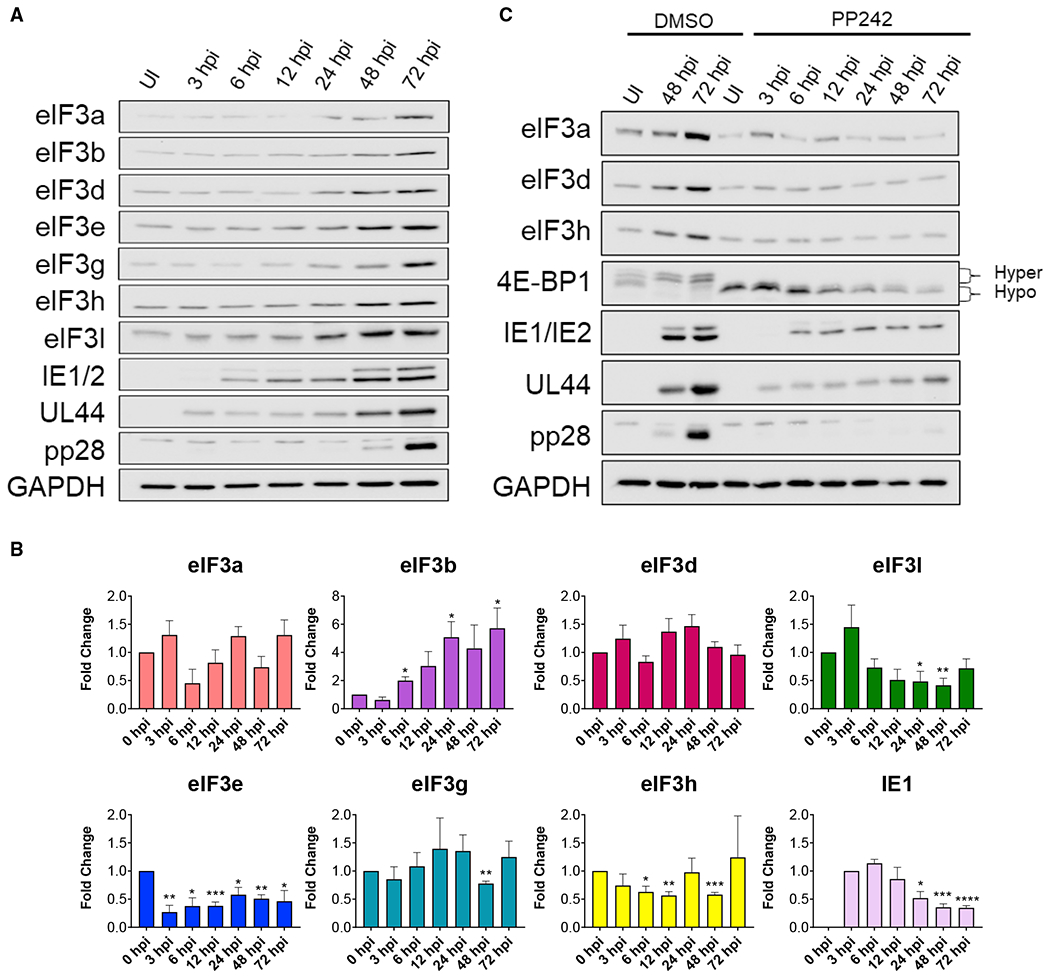
(A) NHDFs were mock (uninfected [UI]) or HCMV infected (AD169 strain; MOI = 3). At each indicated time post-infection total protein was collected, fractionated by SDS-PAGE, and analyzed by immunoblotting using the antibodies shown. GAPDH served as the loading control.
(B) As in (A) except total RNA was isolated at the indicated times. qRT-PCR analysis was performed for each indicated gene (n = 3). The HCMV IE1 gene provided a control for HCMV infection. IE1 RNA levels peaked around 6 hpi and then decreased as infection progressed as described (Stamminger et al., 1991; Stenberg and Stinski, 1985). n = 3 for each gene. Error bars indicate SEM. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001 by Student’s t test.
(C) As in (A) except cells were treated with DMSO or the mTOR active-site inhibitor, PP242 (added at 1.5 hpi; used at 2.5 μM). The change in 4E-BP1 migration from hyperphosphorylation (slow migrating) to hypophosphorylation (fast migrating) confirms PP242 activity.
Many host mRNAs whose translation is stimulated by HCMV are regulated by mTORC1 (McKinney et al., 2014; Tirosh et al., 2015), which is activated by the HCMV UL38 protein (Moorman et al., 2008). Accordingly, treatment of infected cells with the mTOR active-site inhibitor PP242 obviated the virus-induced accumulation of eIF3 subunits and severely restricted representative HCMV IE and E protein accumulation while nearly precluding detection of the L protein pp28 (Figure 1C). The overall abundance of eIF3a, d, and h in PP242-treated, HCMV-infected cells was not, however, reduced below levels detected in uninfected cells (Figure 1C). Similar findings were observed using NHDFs infected with either AD169 laboratory (Figure 1C) or clinical (TB40/E) HCMV-based strains (Figure S1). These results establish that cellular eIF3 subunits increase in response to HCMV infection in an mTOR-dependent manner. As mTORC1 regulates eIF4E-dependent translation, these findings are consistent with eIF3 subunit abundance being controlled translationally. While PP242 treatment prevented 4E-BP1 hyperphosphorylation and is expected to impair cap-dependent translation reliant upon eIF4E, HCMV protein accumulation was reduced but not eliminated. This residual accumulation of representative IE and E proteins in PP242-treated, HCMV-infected cells (Figures 1C and S1) might result from incomplete mTOR inhibition, which could support eIF4E-dependent translation, or from an alternative initiation process related to eIF3 subunit accumulation. It further agrees with earlier studies showing that HCMV mRNA translation late in infection is insensitive to inhibition of cap-dependent translation reliant upon eIF4E, and translation of capped virus mRNAs still occurs independent of eIF4E (Clippinger et al., 2011; Lenarcic et al., 2014). Moreover, it raises the question of whether HCMV-infected cell protein synthesis might be responsive to alternative forms of cap-dependent translation that are insensitive to mTOR inhibition and not contingent upon eIF4E.
Selective reliance of HCMV productive replication and gene expression upon eIF3d
Cap-dependent ribosome loading and translation reliant upon eIF3d does not require the canonical cap-recognition protein eIF4E whose capacity to promote translation initiation is restricted when mTOR is inhibited (Alain et al., 2012; de la Parra et al., 2018; Lee et al., 2015, 2016). This enables alternative regulatory mechanisms, some of which are triggered by metabolic stress like prolonged glucose starvation (Lamper et al., 2020), to shape gene expression. To investigate whether the eIF3 cap-binding subunit eIF3d regulates productive HCMV replication, NHDFs treated with non-silencing, control siRNA or eIF3d-specific siRNA were infected with HCMV and infectious virus production quantified. Compared to non-silencing control siRNA, eIF3d depletion using two different siRNAs specific for eIF3d decreased infectious virus production by 44- or 116-fold, respectively (Figure 2A). Replication of the clinical HCMV strain TB40/E was similarly reduced by approximately 90-fold by eIF3d depletion (Figure 2B). Reduced HCMV replication following eIF3d depletion was also observed in the more clinically relevant ARPE-19 epithelial cell line (Figure S2A). By contrast, eIF3d depletion did not cause a significant, detectable decrease in the productive replication of herpes simplex virus 1 (HSV-1), a related herpesvirus from a different virus subfamily, relative to non-silencing, control siRNA-treated cultures (Figure 2C). Each of two eIF3d siRNAs depleted the eIF3 subunit target effectively and to a similar extent in NHDFs (Figure 2D). This indicates that depletion of eIF3d, a non-core eIF3 subunit, selectively reduces HCMV replication in NHDFs without simply disrupting general eIF3 complex functions or translation such as to non-specifically compromise replication of the related herpesvirus HSV-1.
Figure 2. Depletion of eIF3d selectively interferes with HCMV replication and inhibits virus protein accumulation.
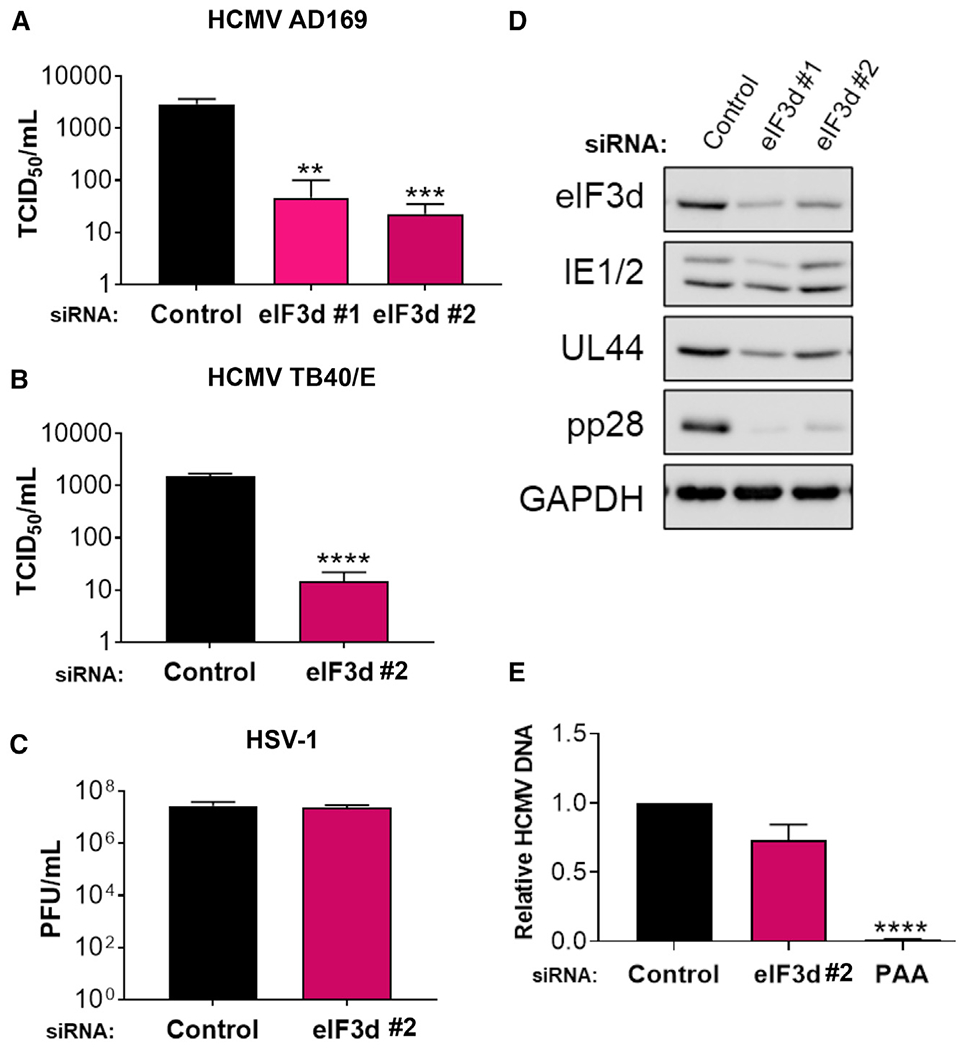
(A) NHDFs transfected with non-silencing control or eIF3d-specific siRNAs were infected with HCMV (AD169 strain; MOI = 0.1). Infectious virus was quantified from supernatants collected 5 days post-infection (dpi) using a TCID50 assay (n = 3). Error bars indicate SEM. **p ≤ 0.01; ***p ≤ 0.001 by Student’s t test.
(B) As in (A) except NHDFs were infected with HCMV (TB40/E strain; MOI = 0.1). n = 3 for each sample. Error bars indicate SEM. ****p ≤ 0.0001 by Student’s t test.
(C) As in (A) except NHDFs were infected with HSV-1 (Patton strain; MOI = 0.001). Supernatants were collected 72 hpi and virus titer was determined by plaque assay (n = 3). Error bars indicate SEM.
(D) NHDFs were infected with HCMV (AD169 strain; MOI = 3) as in (A). At 72 hpi, total protein was collected, fractionated by SDS-PAGE, and the accumulation of viral proteins encoded by representative IE (IE1/2), E (UL44), and L (pp28) genes analyzed by immunoblotting. GAPDH served as the loading control.
(E) NHDFs transfected with control or eIF3d siRNA #2 were infected with HCMV as in (D). At 72hpi, total DNA was collected, and HCMV DNA abundance was quantified by qPCR. Untransfected cells treated in parallel with PAA following inoculation, which prevents virus DNA synthesis, provide a control. Error bars indicate SEM (n = 3). ****p ≤ 0.0001 by Student’s t test.
Progressively greater reductions in HCMV L protein levels upon eIF3d depletion were observed at 72 hpi with limited effects on representative IE (IE1/2) and E (UL44) proteins. While only modest reductions in IE1/2 and UL44 levels were observed, accumulation of the essential L protein pp28 was most severely restricted in infected NHDFs (Figure 2D) and ARPE-19 cells (Figure S2B). By contrast, the abundance of mRNAs encoding IE1, IE2, UL44, and pp28 in eIF3d-depleted cultures varied no more than 50% compared to control, non-silencing siRNA-treated cultures (Figure S3). Thus, inhibition of HCMV replication in response to eIF3d depletion was accompanied by a reduction in virus-encoded protein levels, with the strongest decline in a representative L protein being most pronounced, while overall levels of their respective mRNAs fluctuated less than 2-fold.
While eIF3d depletion preferentially reduced HCMV pp28 protein accumulation (Figure 2D), the corresponding UL99 late mRNA, which encodes the pp28 protein, decreased by approximately 50% (Figure S3). This raised the possibility that eIF3d depletion might indirectly reduce pp28 gene expression by interfering with virus DNA synthesis, which is required for full expression of viral L genes including pp28. Notably, virus DNA levels were reduced by less than 30% in HCMV-infected NHDFs treated with eIF3d-siRNA compared to control, non-silencing siRNA, and this difference (p > 0.05) did not reach statistical significance (Figure 2E). By contrast, treatment of infected cultures with the virus DNA polymerase inhibitor phosphonoacetic acid (PAA) resulted in a statistically significant (p ≤ 10−4) 98% reduction of HCMV DNA (Figure 2E). Thus, eIF3d depletion does not significantly interfere with virus DNA synthesis, an obligate requirement for L gene expression. It excludes the possibility that eIF3d depletion indirectly regulates HCMV L gene expression by interfering with virus DNA synthesis and is consistent with a direct role of eIF3d selectively regulating HCMV L genes.
Consistent with other reports (Wagner et al., 2016), eIF3d depletion selectively reduced eIF3d levels without detectably perturbing overall levels of eIF3 subunits a, b, e, g, h, and l (Figure 3A). By contrast, depletion of eIF3 subunits a, b, and g in HCMV-infected cells using two different siRNAs for each eIF3 subunit target had pleotropic effects, reducing levels of other core and non-core subunits including eIF3d (Figure 3B, compare lane 1 to lanes 2, 3, 4, 5, 9, 10). Representative HCMV IE (IE1/2) and E (UL44) protein accumulation was inhibited to a much greater extent by depleting eIF3 subunits a, b, or g compared to eIF3d or e (Figure 3B, compare lanes 1, 6, 7, 8 to lanes 2, 3, 4, 5, 9, 10), whereas L protein pp28 accumulation was reduced substantially by depleting human eIF3 functional core (a, b, e) or non-core (d, g) subunits (Figure 3B). Besides interfering with virus protein accumulation, depletion of eIF3a or e reduced infectious virus production (Figure S4), as did depletion of eIF3b or g (Figure S4) (Song et al., 2019). The requirement of human eIF3 functional core subunits for HCMV protein accumulation and replication is not surprising, given their essential role loading 40S ribosomes on host and virus mRNAs during canonical and non-canonical cap-dependent translation initiation, respectively reliant upon eIF4E and eIF3d (Cate, 2017; Lee et al., 2015, 2016). Additionally, the human functional core subunit eIF3e regulates eIF4E phosphorylation, which is required for normal levels of productive HCMV replication (Walsh et al., 2005) and impacts eIF4E-driven regulatory processes (Walsh and Mohr, 2014). Only depletion of the non-core subunit eIF3l did not detectably interfere with accumulation of other eIF3 subunits, representative HCMV proteins (Figure 3B), or virus replication (Song et al., 2019). While the impact of individual eIF3 core subunit-depletion upon other core subunits and eIF3d makes it impossible to ascribe any phenotype to a single hypomorphic eIF3 core subunit, the differential impact of eIF3d depletion primarily on L protein accumulation is striking.
Figure 3. eIF3 subunits differentially impact HCMV protein accumulation.
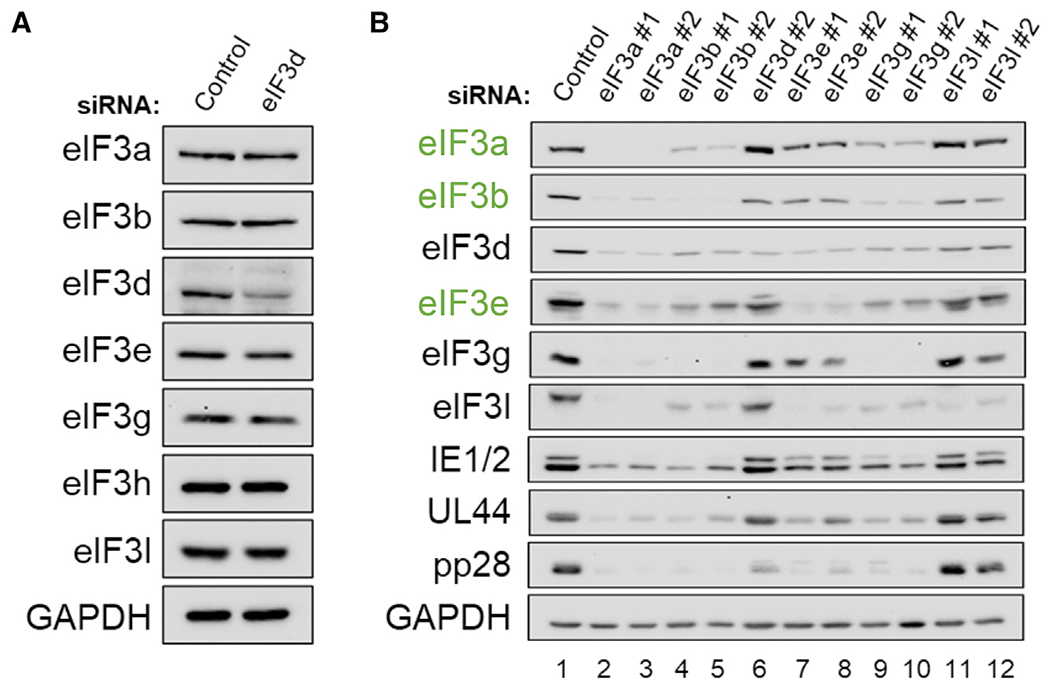
(A) NHDFs (asynchronous) were transfected with non-silencing control or eIF3d-specific siRNA (eIF3d#2). After 72 h, total protein was collected, fractionated by SDS-PAGE, and analyzed by immunoblotting using the antibodies shown. GAPDH served as the loading control.
(B) As in (A) except NHDFs were transfected with non-silencing control siRNA or siRNAs targeting the indicated eIF3 subunits. Transfected, growth-arrested cells were infected with HCMV (AD169 strain; MOI = 3), and total protein collected at 72 hpi was analyzed by immunoblotting using the indicated antibodies. Core eIF3 subunits are labeled in green text.
Control of HCMV-infected cell protein synthesis by eIF3d
To examine whether eIF3d influenced new protein synthesis, uninfected or HCMV-infected NHDFs treated with non-silencing control or eIF3d-specific siRNA were metabolically pulse-labeled with 35S-amino acids (aa) at various times post-infection. Compared to NHDFs treated with non-silencing, control siRNA, eIF3d depletion reduced the overall amount of radiolabel incorporated into polypeptides by approximately 25% (Figure 4A) in good agreement with studies performed in established cell lines (de la Parra et al., 2018; Lee et al., 2015, 2016). While statistically significant differences in total protein synthesis were not detected in HCMV-infected cells exposed to non-silencing or eIF3d-specific siRNA at 6 hpi, the greater sensitivity of infected cell protein synthesis to eIF3d depletion was apparent by 12 hpi, where protein synthesis was reduced by approximately 50% (Figures 4A and 4B). Much of the HCMV-induced increase in infected cell protein synthesis detected beginning at 24 hpi and continuing through 48 hpi was dependent upon eIF3d, as a greater than 50% reduction in infected cell protein synthesis was observed upon eIF3d depletion compared to non-silencing, control siRNA (Figures 4A and 4B). Despite differences in overall profile and quantity of proteins produced at 72 hpi, reduced new protein synthesis in infected cells was nevertheless observed in eIF3d-depleted cultures (Figures 4A and 4B). At the resolution of one-dimensional SDS-PAGE, the profile of proteins synthesized by infected cells was not radically altered by eIF3d depletion (Figure 4B). While eIF3d depletion impacted global rates of infected cell protein synthesis, the overall accumulation of representative HCMV IE (IE1/2) and E (UL44) proteins was not substantially reduced by eIF3d depletion (compared to non-silencing siRNA treatment) at the time points examined (Figure 4C). However, overall accumulation of the essential L protein pp28 was substantially impaired beginning at 48 hpi and continuing through 72 hpi (Figure 4C).
Figure 4. Protein synthesis in HCMV-infected cells and virus protein accumulation are eIF3d dependent.
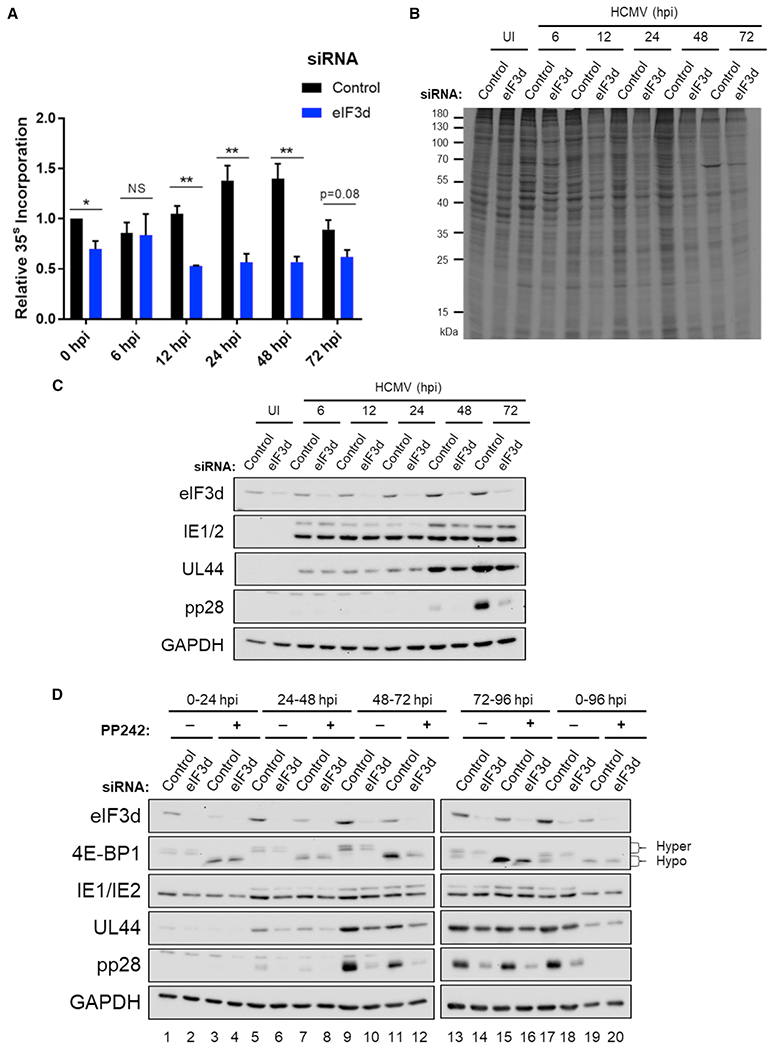
(A) NHDFs transfected with non-silencing control or eIF3d siRNA #2 were mock (uninfected [UI]) or infected with HCMV (AD169 strain; MOI = 3). At each indicated time post-infection, cells were metabolically radiolabeled with 35S-amino acids for 30 min. Total protein was collected and acid insoluble radioactivity quantified by liquid scintillation counting (n = 3). Error bars indicate SEM. *p ≤ 0.05; **p ≤ 0.01 by Student’s t test.
(B) As in (A) except total protein was fractionated by SDS-PAGE and visualized by autoradiography.
(C) As in (A) except total protein was fractionated by SDS-PAGE and analyzed by immunoblotting using the antibodies shown. GAPDH served as the loading control.
(D) NHDFs transfected with non-silencing control or eIF3d-specific siRNA #2 were infected with HCMV (AD169 strain; MOI = 3). Cultures were treated with DMSO or PP242 (2.5 μM) for the indicated time range (0–24, 24–48, 48–72, 72–96, or 0–96 hpi). Total protein was collected at 24, 48, 72, or 96 hpi, fractionated by SDS-PAGE, and analyzed by immunoblotting using the antibodies shown. The change in 4E-BP1 migration from hyperphosphorylation (slow migrating) to hypophosphorylation (fast migrating) was used to confirm PP242 activity.
To better define how mTOR and eIF3d-regulated translation contribute to HCMV protein accumulation during the virus reproductive cycle, infected cell cultures treated with non-silencing control siRNA or eIF3d specific siRNA were exposed to PP242 continuously throughout a 96-h infection or during discrete 24-h intervals. Significantly, as the time of PP242 addition progressed from 0 to 24, 24 to 48, 48 to 72, to finally 72 to 96 hpi, UL44 and pp28 accumulation became less sensitive to PP242 (Figure 4D, compare lane 5 to 7; 9 to 11; 13 to 15), but they remained sensitive to eIF3d depletion. By contrast, IE1/2 accumulation was relatively insensitive to eIF3 depletion (Figure 4D) and, in agreement with published reports, relatively insensitive to treatment with an active-site mTOR inhibitor (Clippinger et al., 2011; Lenarcic et al., 2014; Moorman and Shenk, 2010). In addition, although PP242 exposure for any 24-h interval prior to 72 hpi restricted eIF3d accumulation, similar eIF3d levels were detected in HCMV-infected cultures treated with PP242 during 72–96 hpi compared to untreated cultures (Figure 4D, lanes 13, 15). Finally, PP242 addition at 24–48, 48–72, or 72–96 hpi did not detectably further reduce UL44 or pp28 levels beyond those observed in eIF3d-depleted cultures that were not exposed to PP242. In all instances, mTOR activity was inhibited as PP242 prevented hyperphosphorylated 4E-BP1 accumulation and continuous PP242 exposure over 96 h (compare lane 17 to 19) interfered with accumulation of representative IE, E, and L virus proteins and eIF3d in response to HCMV infection (Figure 4D). A greater reduction in HCMV protein levels was achieved in eIF3d-depleted cultures continuously exposed to PP242 for 96 hpi, which likely reflects in part the abrogation of mTOR-dependent eIF3d accumulation (Figure 4D). This establishes that HCMV protein accumulation switches over time from initially being responsive to mTOR to being increasingly reliant upon eIF3d. It supports prior work showing that mTOR activity regulates virus protein accumulation early in infection, while exerting far less effect on virus protein accumulation at late times (Clippinger et al., 2011). Our findings also agree with earlier reports of reduced reliance upon cap-dependent translation requiring eIF4E and provide a plausible mechanism for understanding how cap-dependent translation might proceed in HCMV-infected cells by capitalizing upon eIF3d.
Global remodeling of host and HCMV mRNA translation by eIF3d
To comprehensively understand host and virus mRNA targets regulated by eIF3d, NHDFs treated with non-silencing, control siRNA or eIF3d-specific siRNA were mock infected or infected with HCMV and RNAs associated with ribosomes and polyribosomes identified. As eIF3d-dependent translation increases as HCMV infection progresses, cytoplasmic cell-free lysates were prepared at 72 hpi and fractionated by sucrose gradient sedimentation to separate ribosome subunits, 80S monosomes, and polyribosomes. The experiment was performed at 72 hpi because of the pronounced difference in pp28 accumulation observed (Figure 4C) and the relative insensitivity of pp28 accumulation to PP242 treatment (Figure 4D). Absorbance profiles (A254 nm) across gradient fractions revealed a slight decrease in light (two to three ribosomes) and heavy (greater than or equal to four ribosomes) polyribosome abundance in mock-infected samples along with a modest increase in 40S/60S subunits and 80S ribosomes upon eIF3d depletion (Figure 5A). These observations are consistent with eIF3d depletion resulting in reduced initiation, as evidenced by increased 80S and ribosome subunits, and the majority of mRNAs being translated in an eIF3d-independent manner reliant upon eIF4E-driven ribosome loading and cap recognition (de la Parra et al., 2018; Lee et al., 2015, 2016). Since HCMV induces ribosome biogenesis and stimulates infected cell protein synthesis (Bianco and Mohr, 2019; McKinney et al., 2014), ribosome subunit, monosome, and polysome peaks are increased, and the extent to which light and heavy polysome levels are reduced upon eIF3d depletion is greater than in mock-infected NHDFs (Figures 5A and 5B). These results agree with metabolic labeling data (Figures 4A and 4B) and demonstrate that protein synthesis in HCMV-infected cells is more sensitive to eIF3d depletion than mock-infected cells.
Figure 5. Regulation of polysome abundance and genome-wide HCMV mRNA translation by eIF3d.
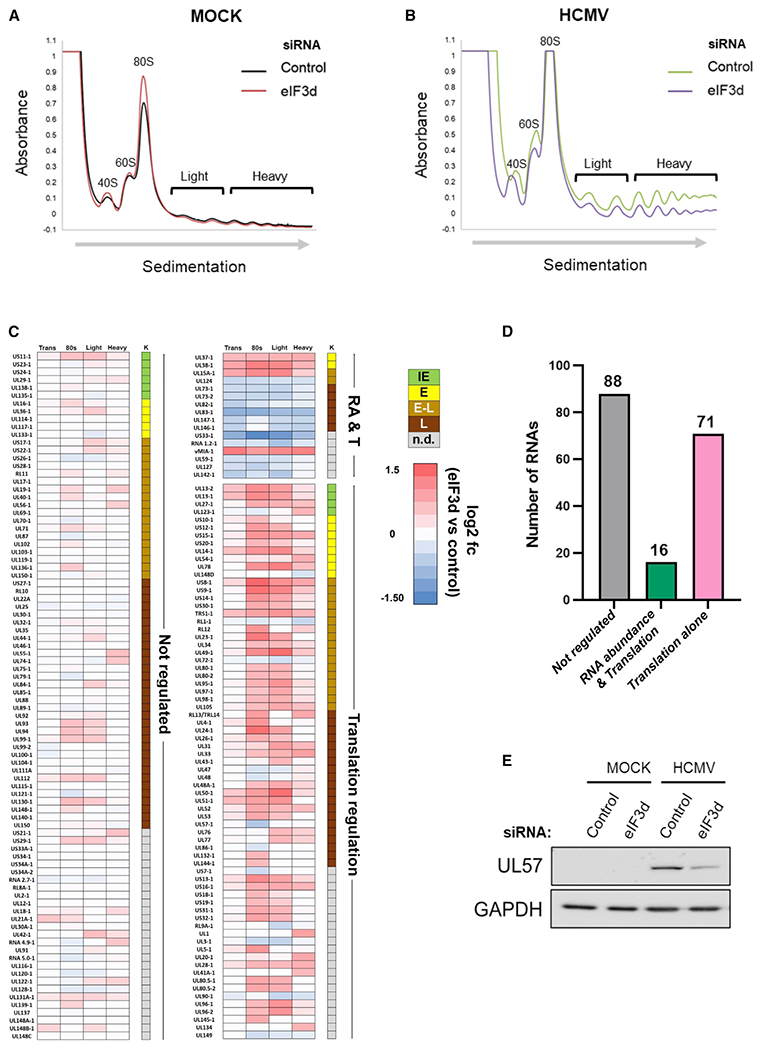
(A and B) Absorbance tracing (A254nm) comparing sucrose gradient sedimentation profiles of cytoplasmic lysates from siRNA-treated (non-silencing control versus eIF3d-specific siRNA #2) mock-infected (A) or (B) HCMV-infected (TB40/E strain; MOI = 3) NHDFs harvested at 72 hpi. Migration of ribosome subunits (40S, 60S) 80S monosomes and polysomes (light, heavy) are shown. The top of the gradient is on the left, and the direction of sedimentation indicated by an arrow.
(C) RNA sequencing was performed on total RNA and RNA isolated from pooled sucrose gradient fractions representing 80S (monosome), light polyribosome (two to three ribosomes), and heavy polysome peaks (greater than or equal to four ribosomes) with three biological replicates per condition. The heatmap illustrates the response of individual HCMV genes in each pooled fraction to eIF3d depletion. HCMV genes are classified either as not regulated, regulated by RNA abundance and translation (RA and T), or translation alone. Each row represents a distinct HCMV RNA, while numbered columns represent log2fc differences in RNA abundance between eIF3d-depleted and control datasets with values for overall transcript levels (trans), 80S ribosome (one monosome), light polysomes (two to three ribosomes), and heavy polysomes (greater than or equal to four ribosomes). Each data point represents the log2fc difference in RNA abundance between eIF3d-depleted and control datasets. Colored boxes represent statistically significant differences (adjusted p value < 0.05) with positive values (red) indicating increased abundance in eIF3d-depleted samples and negative values (blue) indicating a decreased abundance. Temporal HCMV gene classification is shown (K) representing IE (green), E (yellow), early-late (E-L, tan), L (brown), or not determined (n.d., gray).
(D) Graph representing the number and classification of regulated HCMV RNA transcripts in each category.
(E) After 72h, total protein was isolated from mock- or HCMV-infected (TB40/E strain; MOI = 3) NHDFs treated with siRNA as indicated in (A), fractionated by SDS-PAGE, and analyzed by immunoblotting using the specified antibodies. GAPDH provides a loading control.
To identify mRNA targets in HCMV-infected cells whose distribution on ribosomes is regulated by eIF3d, RNA isolated from pooled 80S monosomes, light polysome peaks comprising poorly translated mRNAs, and heavy polysome peaks representing well-translated mRNAs was subjected to short read Illumina RNA sequencing. Total polyadenylated RNA isolated in parallel served as a control for changes in overall RNA abundance in response to HCMV infection and/or eIF3d depletion. In all cases, three biological replicates were generated per condition resulting in 24 sequencing libraries that were sequenced to a depth of 20–40 million paired-end reads per library. Resulting analysis of the sequence data showed the proportion of read pairs aligning to HCMV was reduced in eIF3d-depleted samples relative to non-silencing, control siRNA-treated samples (Figure S5 and Table S4). Responses of polyadenylated RNAs to eIF3d depletion or non-silencing control siRNA were determined by pseudoalignment of sequence reads against the human transcriptome (GENCODE v37) and subsequent analysis using RIVET (Ernlund et al., 2018). This approach offers superior sensitivity as it allows quantification of individual transcript isoforms as opposed to summing counts to the level of individual genes (Bray et al., 2016).
RNAs were categorized as not detectably regulated (“not regulated”) or changed in RNA abundance alone (“RNA abundance alone”), RNA abundance and translation (both positively or negatively changed, “RNA abundance & translational regulation”), or translation alone (“translational regulation”). RNAs were considered translationally regulated if (1) their distribution and abundance within the 80S, light polysome, or heavy polysome pooled fractions changed (≥ 1.5-fold; p < 0.05) between at least one fraction pool (80S, light, and/or heavy) upon eIF3d depletion, and (2) overall RNA abundance within total, unfractionated polyadenylated RNA varied less than 1.5-fold. Inversely regulated mRNAs, where transcripts were restricted on polysomes while their abundance increased or enriched on polysomes while their abundance declined (“inverse regulation”), were also considered translationally regulated. Among eIF3d-responsive mRNAs in HCMV-infected cells, the majority of observed changes in transcript distribution and movement between the 80S monosome, light polysome, and heavy polysome fractions resulted from translational regulation (Table S1) consistent with the role of eIF3d in specialized translation.
Analysis of HCMV polysome-associated RNAs showed that out of 175 that were annotated to the TB40/E reference genome, 88 were eIF3d independent, while 87 were regulated in an eIF3d-dependent manner (Figures 5C, 5D and Table S2). Of the 87 eIF3d-dependent RNAs, 16 were regulated at the level of RNA abundance and translation, while 71 were regulated by translation alone (Figure 5D and Table S2). The heatmap (Figure 5C) shows that most virus-encoded RNAs regulated by eIF3d are translationally upregulated (63) compared to downregulated (24). Most of the RNAs annotated encode proteins that have an early-late (E/L) or late (L) classification and are thus consistent with the result that most genes regulated by eIF3d belong to an E/L or L temporal class. Surprisingly, we were unable to identify the RNA that encodes pp28, UL99, among the list of eIF3d-responsive, translationally regulated HCMV genes derived from our polysome analysis dataset (Figure S6) even though our results show pp28 is encoded by an eIF3d-responsive mRNA (Figures 2D, 3B, 4C, and 4D). We attribute this to the complexity of the UL97/UL98/UL99 gene cluster as the two RNA isoforms encoding UL99 overlap entirely with the RNAs encoding UL97 and UL98, which prevents short read sequencing from accurately distinguishing between these RNA isoforms (Figure S6).
Significantly, 37 out of the 87 RNAs regulated by eIF3d (Figure 5C) were previously shown to encode proteins essential for virus productive growth in human fibroblasts or display a substantial growth defect when deleted (Dunn et al., 2003). Furthermore, our virus genome-wide analysis identifies at least four known essential viral genes, UL48, UL57 (ICP8), UL86, and UL90, translationally repressed by eIF3d depletion (Figure 5C). This was further validated in the case of UL57, as eIF3d depletion restricted UL57 protein accumulation in HCMV-infected cells (Figure 5E). Reduced expression of multiple essential viral genes or HCMV genes required for productive virus growth (Figures 5C and 5D) likely accounts in part for the replication defect we observe upon eIF3d depletion (Figures 2A, 2B, and S2). For example, eIF3d depletion could impair translation of select L mRNAs or interfere with translation of certain E mRNAs at late times post-infection, as herpesvirus E genes continue to be expressed at late times. Besides protein coding genes, the abundance of one HCMV long non-coding (nc) RNA (RNA1.2) on polysomes is reduced by eIF3d depletion (Figure 5C). Although annotated as a long RNA lacking a canonical ORF, RNA1.2 ribosome occupancy has been demonstrated and unique peptides encoded by RNA1.2 detected (Stern-Ginossar et al., 2012). Reduced RNA1.2 expression reportedly results in NF-kB activation and could impact virus replication by regulating host genes (Lau et al., 2020). Finally, eight transcripts encoded by TB40/E, but not present in the AD169 laboratory strain, were regulated by eIF3d. Since there are 13 genes missing in the lab-adapted strain that are required for infection in non-fibroblast cell types (Wilkinson et al., 2015), eIF3d-dependent regulation of HCMV gene expression might impact additional aspects of the HCMV life cycle including productive growth in myeloid or endothelial cells or latency.
Analysis of host mRNA in infected NHDFs revealed that most (46,249) are not detectably responsive to eIF3d depletion and are unregulated, while a subset of 10% are eIF3d dependent (4,919) (Figure 6A). The relative number of cellular mRNAs exhibiting dependence upon eIF3d for their translation agrees nicely with the proportion of eIF3d-regulated genes identified by others (de la Parra et al., 2018; Lee et al., 2015, 2016; Lamper et al., 2020). Among eIF3d-regulated genes (Figures 6A and 6B), greater than 80% (4,020) were translationally regulated (translation + inverse regulation), whereas the remainder were regulated by RNA abundance and translation (11.6%, 568 genes) or transcription alone (6.7%, 331 genes). Translationally stimulated eIF3d-dependent mRNAs were statistically enriched in pathways including mRNA splicing major pathway (FDR < 0.000001), metabolism of RNA (FDR < 0.0004), mRNA3′ end processing (FDR < 0.0005), unfolded protein response (FDR < 0.002), XBP-1 (S) activates chaperone genes (FDR < 0.01), IRE1 alpha activates chaperone genes (FDR < 0.01), and transcriptional regulation by E2F6 (FDR < 0.05) (Figures 6C and S7A). By contrast, no significantly regulated pathways were found using REACTOME (Jassal et al., 2020) to profile mRNAs translationally downregulated by eIF3d. Additionally, significant differences in 5′-UTR length distribution and GC content in mRNAs translationally regulated by eIF3d compared to eIF3d-independent mRNAs were not observed (Figures S7B and S7C). Host genes translationally regulated by eIF3d in HCMV-infected cells were enriched for small upstream ORFS (uORFs) (Figures S7D and S7E). Furthermore, gene set enrichment analysis (GSEA) comparing host genes translationally regulated by eIF3d in HCMV-infected cells with cellular genes regulated in response to an integrated stress response (ISR) induced by chronic ER stress (Guan et al., 2017), which involves eIF3d-dependent translation, revealed a substantial statistically significant (p < 1e-08 by hypergeometric test) overlap (Figure 6D and Table S3). The majority of coinciding genes shared were concentrated in GO categories encompassing translational regulation and critical aspects of the unfolded protein response (UPR).
Figure 6. Remodeling the global host mRNA translational landscape in HCMV-infected cells by eIF3d.
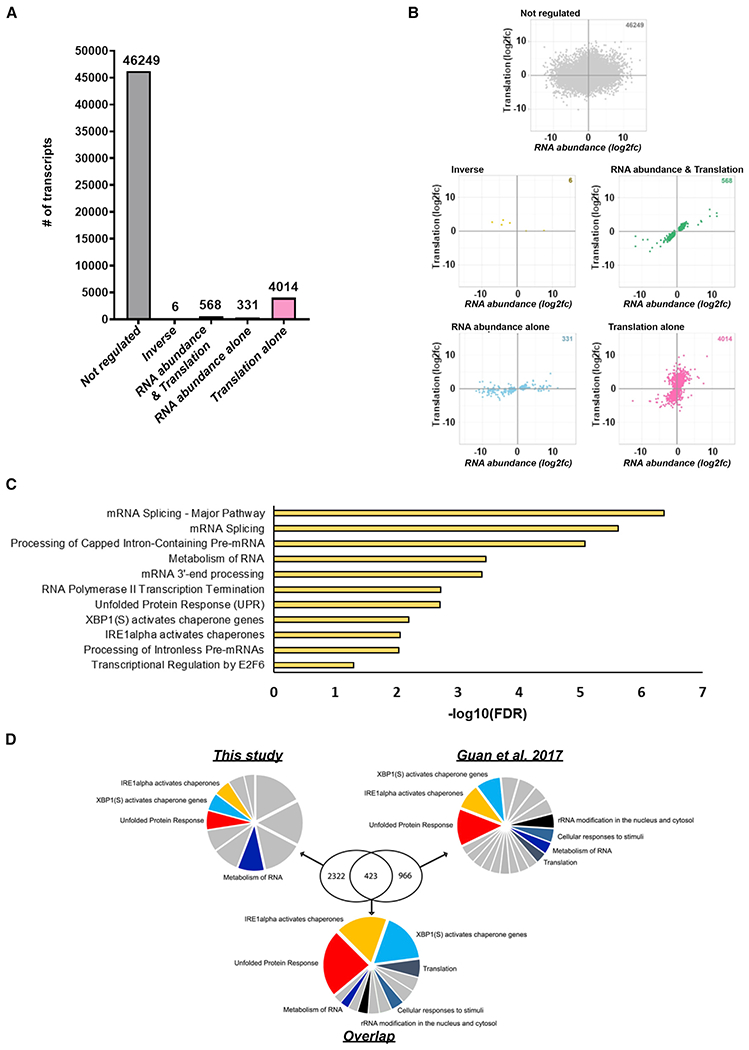
(A) The response of individual host genes in HCMV-infected NHDFs to eIF3d depletion was classified as not regulated or regulated by RNA abundance alone, translation alone, or RNA abundance and translation (direct and inverse) as described in the main text. The number of host RNAs regulated by eIF3d depletion in each category is shown by the graph.
(B) Scatterplots comparing log2-fold changes in RNA abundance and translation of host RNAs in HCMV-infected NHDFs treated with non-silencing control siRNA or eIF3d-specific siRNA. The gray, gold, green, blue, and pink colored dots represent no regulation, inverse regulation (between RNA abundance and translation), regulation by both RNA abundance and translation, regulation by RNA abundance alone, and translational regulation alone, respectively.
(C) GSEA was performed using REACTOME (Jassal et al., 2020) for genes encoding translationally upregulated RNAs in any or all fractions (80S, light, heavy). No pathways were reported in analysis of genes encoding translationally downregulated RNAs in any condition.
(D) Gene symbol lists derived from host RNAs translationally regulated by eIF3d in HCMV-infected cells (this study) and those regulated in response to chronic ER stress (Guan et al., 2017) were submitted for GSEA using REACTOME. A GSEA was also performed on the subset of translationally regulated RNAs that overlapped between the two studies. Pie charts representing all significant enriched pathways (FDR < 0.05) are shown for each dataset and the overlap. The relative size of each slice is representative of the −log10(FDR) value (i.e., larger slices indicate smaller FDR values). Pathways overlapping between analyses are color coded and identified. Remaining pathways are shown in gray and are detailed in Table S3.
Host eIF3d-responsive genes impact HCMV gene expression and productive replication
To evaluate whether host eIF3d-responsive gene expression impacts HCMV infection biology, the accumulation of proteins encoded by several genes predicted to be translationally regulated by eIF3d depletion in HCMV-infected cells was examined. Seven cellular genes that function in physiological stress responses and encode mRNAs that either increased (DDX21, VCP) or decreased (SLC3A2, INPP4B, MLST8, WIPI2, RAD23A) on heavy polysomes in response to eIF3d depletion were prioritized for study. The solute carrier SLC3A2 and inositol polyphosphate 4-phosphatase type II INPP4B are ATF4-responsive genes and components of the UPR and ISR (Han et al., 2013; Harding et al., 2003), the transitional ER ATPase VCP and RAD23A regulate proteostasis respectively through ER-associated protein degradation and ubiquitin-proteasome pathway (Bertolaet et al., 2001; Elsasser et al., 2002; Hiyama et al., 1999; Meyer and Weihl, 2014), and mTORC subunit LST8 (MLST8) and WD repeat domain phosphoinositide-interacting protein 2 (WIPI2) control autophagy (Liu and Sabatini, 2020; Proikas-Cezanne et al., 2015), which is an ISR outcome (Kroemer et al., 2010; Pakos-Zebrucka et al., 2016). By contrast, the nucleolar RNA helicase 2 protein DDX21 is responsive to nucleolar stress and nucleic acid sensing/infection stress (Calo et al., 2018; Zhang et al., 2011). In accordance with enrichment of their cognate mRNAs on polysomes in HCMV-infected cells even during eIF3d depletion, overall DDX21 and VCP protein levels increased in response to HCMV infection (Table S1). Depletion of eIF3d, however, did not result in detectably greater DDX21 or VCP accumulation (Figure 7A). Studies by others have shown that depleting either DDX21 or VCP reduces productive HCMV replication (Hao et al., 2019; Lin et al., 2017). By contrast, polysome association of mRNAs encoding SLC3A2, INPP4B, MLST8, WIPI2, or RAD23A was diminished by eIF3d depletion in HCMV-infected cells (Table S1). Significantly, protein levels of SLC3A2, MLST8, INPP4B, WIPI2, and RAD23A all increased in HCMV-infected cells and were dependent on eIF3d (Figure 7A). Cellular protein GAPDH levels were not detectably responsive to eIF3d depletion in HCMV-infected cells, providing a control for specificity (Figure 7A). This establishes that these mRNA targets identified and selected by their eIF3d-dependent enrichment on polyribosomes in HCMV-infected cells encode proteins whose accumulation is eIF3d dependent.
Figure 7. Regulation of HCMV replication by host eIF3d-responsive genes.
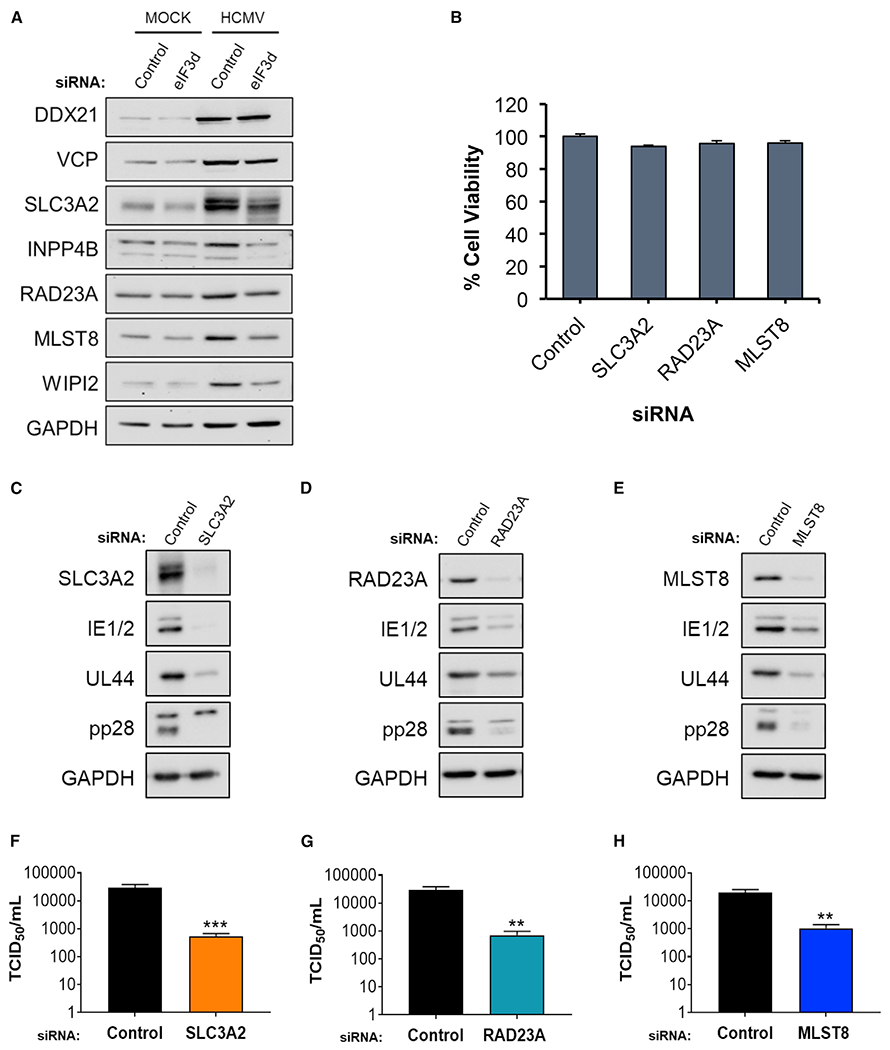
(A) Total protein from mock- or HCMV-infected (TB40/E strain; MOI = 3) NHDFs treated with the indicated siRNA (non-silencing controlor eIF3d-specific siRNA #2) was collected at 72 hpi, fractionated by SDS-PAGE, and analyzed by immunoblotting using the antibodies shown. GAPDH provides a loading control.
(B) Viability of NHDFs transfected with siRNAs specific for the indicated gene (n = 3). Error bars indicate SEM.
(C–E) As in (A) except total protein from HCMV-infected (TB40/E strain; MOI = 0.1) NHDFs transfected with the specified siRNA was collected at 5 dpi.
(F–H) As in (C)–(E) except infectious virus was quantified from supernatants collected 5 dpi using a TCID50 assay (n = 3). Error bars indicate SEM. **p ≤ 0.01; ***p ≤ 0.001 by Student’s t test.
Individual depletion of representative eIF3d-dependent gene products encoded by SLC3A2, RAD23A, or MLST8 did not significantly reduce NHDF viability (Figure 7B) and is predicted to target multiple isoforms of each transcript including eIF3d-regulated isoforms (Table S1). To determine how SLC3A2, RAD23A, and MLST8 depletion influenced HCMV gene expression, viral protein abundance was measured by immunoblotting. Compared to HCMV-infected cultures treated with non-silencing, control siRNA, depletion of SLC3A2, RAD23A, or MLST8 reduced virus protein accumulation (Figures 7C–7E). Levels of master regulators IE1/2, the E protein UL44, and the critical L protein pp28 all declined upon depletion of host eIF3d-dependent target genes (Figures 7C–7E). These broad effects on virus gene expression raised the possibility that expression of host eIF3d-dependent genes might impact productive HCMV replication and promote virus growth and spread. Compared to cultures exposed to non-silencing, control siRNA, depletion of either SLC3A2, RAD23A, or MLST8 interfered with HCMV reproduction, reducing infectious virus production by 55-, 40-, or 18-fold, respectively (Figures 7F–7H). This demonstrates that disrupting expression of select eIF3d-dependent target genes interferes with HCMV protein accumulation and productive replication. It further establishes that the virus-induced eIF3d-dependent host gene expression program regulates productive HCMV growth.
DISCUSSION
Differential loading of eIF3-bound 40S ribosomes plays a critical role controlling cap-dependent mRNA translation in response to physiological stress like infection. Although mTORC1 signaling regulates eIF4E-containing, cap-recognition initiation factor complex activity, cap-dependent translation is not completely reliant upon eIF4E. Here, we establish that HCMV-infected cell protein synthesis exhibits a greater reliance upon eIF3d, a non-core eIF3 subunit that mediates an alternative form of cap-dependent translation, in contrast with uninfected cells. Overall eIF3 subunit levels including eIF3d accumulate in an mTOR-dependent manner in response to HCMV infection. A switch from mTOR-sensitive to eIF3d-dependent protein accumulation subsequently transpires. Depleting eIF3d reduced infected cell protein synthesis and selectively inhibited HCMV L gene expression and virus replication. Moreover, eIF3d depletion reduced polyribosome abundance and remodeled the global translational landscape in HCMV-infected cells, interfering with expression of essential virus genes and abrogating an eIF3d-dependent host gene expression signature that fosters HCMV productive growth. This reveals a new, unexpected strategy whereby infection stress remodels the infected cell proteome, bypassing mTOR-regulated translational control and harnessing eIF3d-dependent protein production. It further illustrates how switching between eIF4E and eIF3d cap-dependent mRNA translation within a developmental gene expression program can be appropriated to differentially regulate gene expression in infection biology.
With the exception of being enriched for small uORF content in the 5′-UTR of host mRNAs, shared characteristics among mRNAs encoded by host or virus eIF3d-responsive genes in HCMV-infected cells were not evident at the primary sequence level, predicted secondary structure, or 5’-UTR length. While a stem-loop structure within the 5′-UTR has been shown to determine whether eIF3d activates or represses translation of two target genes (Lee et al., 2015, 2016; Lamper et al., 2020), additional sequence and/or structural elements together with subcellular transcript localization could also impact eIF3d responsiveness under varied physiological conditions in different cell types. A 40-nt sequence derived from the HCMV pp28 gene 5′UTR reportedly repressed translation of a cis-linked reporter gene incorporated into a recombinant HCMV (Kerry et al., 1997). Although it was suggested that this regulatory element might contain a putative stem-loop structure, the GC-rich content of herpesvirus genomes confounds accurate secondary structure prediction by computational methods alone. Further study is required to investigate whether this genetic element adopts a genuine structure and its contribution, if any, to the eIF3d-dependent HCMV gene expression program we describe. As eIF3d can activate and repress gene expression, genetic elements might differentially tune eIF3d responsiveness from repression to activation temporally and spatially. This could allow a virus-encoded target gene like pp28 to shift reliance from eIF4E- to eIF3d-dependent translation over the virus life cycle.
While eIF3d reportedly regulates translation of mRNAs involved in energy metabolism in the yeast S. pombe (Shah et al., 2016) and HCMV infection modulates cell metabolism to support virus replication (Rodríguez-Sánchez et al., 2019), host eIF3d-responsive genes were not found to significantly cluster within these GO classification terms. This could reflect species or transcriptome differences. Instead, genome-wide polysome profiling revealed expression of cellular genes controlling activation of chaperones by IRE1 and XBP1(s), the UPR, and ISR to be eIF3d dependent in HCMV-infected cells (Figures 6C and S7A). Significantly, HCMV activates and regulates the UPR throughout infection in part to stimulate lipogenesis (Isler et al., 2005; Yu et al., 2013), and depleting PKR-like ER kinase (PERK) restricts HCMV gene expression and attenuates virus productive growth (Yu et al., 2013). This switch to an eIF3d-dependent initiation mechanism during HCMV infection resembles an ISR program induced by chronic ER stress characterized by eIF3d-dependent mRNA recruitment and partial translational recovery in uninfected cells (Guan et al., 2017). The latter allows for sustained mRNA translation while preventing ER overload. Thus, instead of interfering with the UPR/ISR, our data suggest HCMV exploits this eIF3-responsive UPR/ISR host gene expression program associated with chronic ER stress for its benefit. Moreover, this eIF3-responsive chronic ER stress pathway is likely harnessed to regulate expression of HCMV capped mRNAs, thereby avoiding regulatory translation repression mechanisms reliant upon eIF4E as infection progresses and cumulative physiological stress on the infected cell increases. Mimicking chronic ER stress in part may have facilitated adoption of the lengthy HCMV productive replication cycle by providing a means to sustain protein synthesis during chronic infection stress.
Ongoing host protein synthesis in HCMV-infected cells in part enables HCMV to capitalize upon an infection-induced cellular gene expression program. Instead of precluding ER stress response activation, beneficial aspects such as lipogenesis control can be harnessed, while others can be tuned by differential translation or relying upon eIF3d for HCMV mRNA translation. In contrast, infection with the related herpesvirus subfamily member HSV-1 or many other RNA or DNA viruses, which typically have shorter replication cycles and impair host protein synthesis in part by accelerating mRNA decay, destroy much of the host transcriptome and limit UPR/ISR induction (Mulvey et al., 2007; Stern-Ginossar et al., 2019). Competition with host mRNAs for ribosomes is reduced, and the need to exploit a host eIF3d-dependent translation program is obviated. This does not preclude exploitation of eIF3d for virus mRNA translation in infected cells where host protein synthesis is impaired. Indeed, a recent genome-wide screen reported that eIF3d is a host factor that regulates coronavirus replication (Lee et al., 2021).
Whereas protein synthesis in uninfected cells largely results from eIF4E-responsive cap-dependent translation, a smaller (approximately 20%–30%) but pervasive amount of eIF3d-dependent translation is insensitive to mTORC1 repression. This is reflected in the initial reliance upon HCMV-infected cell protein synthesis on mTOR and eIF4E and impacts both virus and host protein accumulation. Constitutive activation of mTORC1 by the HCMV UL38 protein in part drives this process, remodeling the host translational landscape to enhance translation of mRNAs containing 5′-terminal oligopyrimidine (TOP) motifs including PABP (McKinney et al., 2012, 2014; Perez et al., 2011) and eIF3 subunits like eIF3d. Overtime, the dependence of protein accumulation upon an mTOR-sensitive process diminishes likely in part due to an increase in the hypophosphorylated 4E-BP1 (Walsh et al., 2005), which represses eIF4E. Infected cell protein synthesis subsequently becomes reliant upon eIF3d, impacting viral protein accumulation and remodeling host translation to support HCMV productive growth. Whether the rise in eIF3d alone is sufficient to drive this process or acts in concert with another regulatory mechanism like eIF3d dephosphorylation (Lamper et al., 2020) is an exciting possibility. Production of transcription factors, such as JUN, ATF6, or FOXP1, regulated by eIF3d could further remodel the transcriptome and add additional layers of regulation. Taken together, successive waves remodeling the host translational landscape, first via an mTORC1-sensitive process followed by an eIF3d-driven process, occur during the HCMV life cycle, tuning and orchestrating a dynamic host translational program that regulates virus infection.
Limitations of the study
We discover an unexpected strategy whereby virus infection capitalizes upon an eIF3d-dependent switch associated with ER stress, which remodels the host cell translational landscape and drives virus L gene expression to promote virus reproduction. While eIF3d is depleted prior to infection in our experiments, it is difficult to distinguish whether the reliance upon eIF3d-dependent translation in infected cells increases as infection progresses or if HCMV proteins potentiate the penetrance of the eIF3d depletion phenotype. The overlapping nature of HCMV transcripts, the quality of existing virus genome annotations, and constraints associated with short read RNA-seq also imposed limitations on our virus transcriptome analysis. Understanding how switching to an eIF3d-dependent gene expression program might be regulated by HCMV, its relationship to ER stress, and whether eIF3d cap-binding is required warrant further investigation.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ian Mohr (ian.mohr@med.nyu.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
All sequencing datasets generated during this study have been deposited at the European Nucleotide Archive (ENA) and are publically available as of the publication date. The accession number is listed in the key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-eIF3a | Cell Signaling | cat# 3411S |
| anti-eIF3b | Bethyl | cat# A301-761A |
| anti-eIF3d | Proteintech | cat# 10219-1-AP |
| anti-eIF3e | Bethyl | cat# A302-985A |
| anti-eIF3g | Bethyl | cat# A301-757A |
| anti-eIF3h | Cell Signaling | cat# 3413S |
| anti-eIF3l | GeneTex | cat# GTX120119 |
| anti-4EBP1 | Bethyl | cat# A300-501A |
| anti-GAPDH | Cell Signaling | cat# 2118S |
| anti-IE1/IE2 | Millipore | cat# MAB810 |
| anti-UL44 | Virusys | cat# CA006 |
| anti-pp28 | Virusys | cat# CA004-100 |
| anti-UL57/ICP8 | Novus | cat# NB110-57369 |
| anti-DDX21 | Proteintech | cat# 10528-1-AP |
| anti-VCP | Proteintech | cat# 10736-1-AP |
| anti-SLC3A2/CD98 | Proteintech | cat# 15193-1-AP |
| anti-INPP4B | Cell Signaling | cat# 4039S |
| anti-RAD23A | Cell Signaling | cat# 24555S |
| anti-MLST8/GBL | Cell Signaling | cat# 3274S |
| anti-WIPI2 | Cell Signaling | cat# 8567S |
| anti-rabbit IgG HRP | GE Healthcare | cat# NA934-1ML |
| anti-mouse IgG HRP | GE Healthcare | cat# NA931-1ML |
| Bacterial and virus strains | ||
| HCMV AD169-GFP | Dong Yu | N/A |
| HCMV TB40/E | Domenico Tortorella (Mount Sinai, New York, NY, USA) | N/A |
| HCMV TB40/E-GFP | Eain Murphy (SUNY, Syracuse, NY, USA) | N/A |
| HSV-1-GFP-Us11 | Dr. Ian Mohr (NYU Schiool of Medicine, New York, NY, USA) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| DMEM | Corning | cat# 10-013-CV |
| DMEM without L-Glutamine/L-Methionine/L-Cystine | Corning | cat# MT17204CI |
| 25mM HEPES | Corning | cat# 25060CI |
| 100 U/mL penicillin-100μg/mL streptomycin | Corning | cat# MT-30-002-Cl |
| pp242 | Invivogen | cat# inh-pp242 |
| RNAimax | Invitrogen | cat# 13778075 |
| Opti-MEM | Gibco | cat# 31985070 |
| TRIzol | Invitrogen | cat# 15596018 |
| GlycoBlue | Invitrogen | cat# AM9515 |
| nuclease-free water | Invitrogen | cat# AM9937 |
| qScript XLT cDNA SuperMix | QuantaBio | cat# 95161-500 |
| SsoAdvanced Universal SYBR Green Supermix | Bio-Rad | cat# 1725275 |
| Phosphonoacetic acid | Sigma | cat# P6909 |
| EasyTag Express 35S Labeling Mix | Perkin Elmer | cat# NEG772007MC |
| CytoScint ES scintillation fluid | MP Biomedicals, Inc | cat# 0188245301 |
| Cycloheximide | Sigma | cat# C7698 |
| RiboLock RNase Inhibitor | ThermoFisher Scientific | cat# EO0381 |
| cOmplete mini EDTA-free Protease Inhibitor Cocktail tablet | Roche | cat# 05892791001 |
| Critical commercial assays | ||
| Allprep DNA Kit | Qiagen | cat# 80204 |
| RNA Clean and Concentrator-5 kit | Zymo Research | cat# R1014 |
| NEBNext® Ultra II Directional RNA Library Prep Kit for Illumina | New England Biolabs | cat# E7760S |
| Deposited data | ||
| RNA sequencing datasets | European Nucleotide Archive (ENA) | BioProject ID PRJEB45749 |
| Experimental models: Cell lines | ||
| Normal human dermal fibroblasts (NHDF) | Lonza | cat# CC-2509 |
| African green monkey cells (Vero) | ATCC | cat# CCL-81 |
| Human retinal pigment epithelial cells (ARPE-19) | ATCC | cat# CRL-2302 |
| Oligonucleotides | ||
| DNA oligonucleotides for PCR | This paper and other sources | Table S5 |
| siRNAs | Qiagen, Sigma | Table S5 |
| Software and algorithms | ||
| GraphPad Prism (Version 9) | Graphpad | RRID: SCR_002798 |
| RIVET | Ernlund et al., 2018 | N/A |
| Kallisto | Bray et al., 2016 | N/A |
| ComBat-seq | Zhang et al., 2020 | N/A |
| Rstudio using ggplot2 | Wickham, 2016 | N/A |
| REACTOME | Jassal et al., 2020 | N/A |
| TrimGalore | https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ | N/A |
| STAR | Dobin et al., 2013 | N/A |
| SAMtools | Li et al., 2009 | N/A |
| BEDtools | Quinlan, 2014 | N/A |
| biomaRt | Durinck et al., 2005; Durinck et al., 2009 | N/A |
| Other | ||
| Nitrocellulose membranes | GE Healthcare | cat# 10600002 |
| Whatman Grade GF/C Glass Microfiber Filters | GE Healthcare | cat# 18822-025 |
| thinwall polypropylene ultracentrifuge tube | Beckman Coulter | cat# 331372 |
| SW41Ti rotor | Beckman Coulter | cat# 331362 |
| density gradient fractionation system | Brandel | cat# BR-188 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cells, viruses, and chemicals
Normal human dermal fibroblasts (Lonza; CC-2509) were cultured in Dulbecco’s modification of Eagle’s medium (DMEM) (Corning; 10-013-CV) supplemented with 5% (v/v) fetal bovine serum (FBS) and 100 U/mL penicillin-100μg/mL streptomycin (Corning; MT-30-002-Cl) at 37°C in 5% CO2. Unless specifically indicated otherwise, NHDFs growth-arrested by serum-deprivation were used for all experiments. Briefly, NHDFs were grown to confluency, washed twice with 1X PBS and cultured in DMEM containing 0.2% FBS for 72 h (Walsh et al., 2005). Under these conditions, >98% of the cells were in the G1 phase of the cell cycle, as revealed by propidium iodide staining followed by flow cytometry (Walsh and Mohr, 2004). Vero cells were obtained from ATCC and cultured in DMEM supplemented with 5% (v/v) calf serum and 100 U/mL penicillin-100μg/mL streptomycin. ARPE-19 cells were obtained from ATCC and cultured in DMEM-F12 media supplemented with 10% (v/v) FBS, 0.5mM sodium pyruvate, 15 mM HEPES, and 100 U/mL penicillin-100μg/mL streptomycin. HCMV AD169-GFP was a gift from Dong Yu and propagated in NHDF cells, as previously described (Bianco and Mohr, 2017). Virus was concentrated by centrifugation at 20,000 RPM in a SW28 rotor for 90 min. at 18°C followed by resuspension in 1.5% bovine serum albumin (BSA) in DMEM. Virus stock titers were determined using a plaque assay. For HCMV replication experiments, cells were infected (MOI = 0.1) for 5 d. EGFP-expression was used to measure HCMV spread and visualized under 5x magnification using a Zeiss Axiovert 200 fluorescent microscope, as previously described (McKinney et al., 2014). Supernatants were collected and virus titers were determined using a 50% tissue culture infectious dose (TCID50) assay, as previously described (Bianco and Mohr, 2017). HCMV TB40/E was kindly provided by the laboratory of Domenico Tortorella, Icahn School of Medicine at Mount Sinai, NY. HCMV TB40/E-GFP (Procter et al., 2018) was kindly provided by E. Murphy (SUNY Upstate Medical School). HSV-1 (Patton strain) expressing a GFP-Us11 fusion protein was described previously (Benboudjema et al., 2003). PP242 was used at 2.5μM.
METHOD DETAILS
siRNA transfections
siRNAs were transfected at a final concentration of 20nM each using Lipofectamine RNAimax as previously described (Burgess and Mohr, 2015). NHDFs were seeded in a 12-well plate the day prior to transfection. The day of transfection cell medium was replaced. 2μL RNAimax was mixed with 50μL Opti-MEM. 1 μL siRNA was mixed with 50μL Opti-MEM. RNAimax and siRNA mixtures were then combined and incubated for 20 min. at RT. 100μL RNAimax siRNA complexes were then added to each designated well containing cells. For polysome experiments reactions were scaled up based on volume. 48 h after transfection, cells were growth arrested by serum-deprivation for 72 h and then either mock-infected or infected with virus.
Cell viability assay
NHDFs seeded in 96-well black walled plates were transfected with the indicated siRNAs as described in the preceding section. After 3d, cells were fixed in 4% PFA in PBS for 30 min, permeabilized in 0.5% triton-X-100 for 15 min. and stained with DAPI for 2h. After washing in PBS, cells were identified using a Thermo Cellinsight CX7 LZR high-content screening platform, which collected 9 images at 4x magnification, and quantified using HCS Navigator software. Technical duplicates were performed for each independent experiment.
RNA isolation, cDNA synthesis, and quantitative PCR
RNA was isolated from cells using TRIzol according to the manufacturer’s protocol. Chloroform (100μL) was added to samples in 500μL of TRIzol, shaken vigorously by hand for 15 sec., incubated at RT for 2 min. and centrifuged at 12,000 x g (15 min, 4°C). After recovering the aqueous phase, 1 μL of GlycoBlue and isopropanol (250μL) were added and the samples were incubated for 10 min. at RT. Following centrifugation (12,000 x g, 10 min. 4°C), the RNA pellet was washed with 75% ethanol (1 mL), air dried and resuspended in nuclease-free water. cDNA was synthesized from 250 ng of purified RNA using qScript XLT cDNA SuperMix according to the manufacturer’s protocol. Quantitative PCR (qPCR) was performed in a Bio-Rad C1000 Touch Thermal Cycler using SsoAdvanced Universal SYBR Green Supermix with a 58°C annealing temperature, 44 cycles, and 25μL final volume. Primers specified in key resources table and Table S5 were used at 0.5μM.
DNA isolation and qPCR
Total DNA was isolated from HCMV-infected NHDFs using an Allprep DNA Kit according to the manufacturer’s instructions. Virus DNA was subsequently quantified by qPCR as described (Srinivas et al., 2021). Briefly, relative amounts of HCMV DNA were determined by qPCR using UL44 forward and Reverse primers. cT values were normalized to human RPL19 DNA levels detected with RPL19 Forward and RPL19-Reverse primers. Treatment of infected cells with 300 μg/mL phosphonoacetic acid (PAA) was used to validate that the qPCR DNA signal was dependent upon HCMV DNA synthesis.
Immunoblotting and antibodies
Total cellular protein was collected by lysis in sample buffer (62.5 mM Tris-HCl pH 6.8, 2% SDS, 10% glycerol, 0.7M β-mercaptoethanol) followed by boiling for 4 minutes. Lysates were fractionated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. Membranes were blocked in 5% non-fat milk in TBST and incubated in primary antibody overnight at 4°C. Primary antibodies were detected using either anti-rabbit IgG HRP or anti-mouse IgG HRP secondary antibodies and visualized by chemiluminescent detection using an Invitrogen iBright FL1000 Imaging System.
Metabolic labeling with 35S amino acids, detection and quantification
Cells were incubated with EasyTag Express 35S Labeling Mix in DMEM (77 μCi/mL) without L-Glutamine/L-Methionine/L-Cystine (supplemented with 25mM HEPES. After 30 min, cells were lysed in 1X sample buffer and boiled for 3 min. To quantify radioactive amino acid incorporation, trichloroacetic acid (TCA) was added to 10% (v/v) final concentration and the samples held on ice for 30 min. Acid insoluble radioactivity was collected on Whatman Grade GF/C Glass Microfiber Filters (washed twice with 100% ethanol, and quantified by counting in CytoScint ES scintillation fluid.
Preparation of cell-free extracts for polysome isolation
Per replicate, eight 15-cm dishes of NHDFs were transfected with either AllStars Negative Control or eIF3d siRNA, growth-arrested, and then mock or HCMV-infected (1 × 107 cells/dish; MOI = 3). At 72 hpi, cells were incubated with 100μg/mL cycloheximide in complete media for 10 min. at 37°C in 5% CO2 and then washed twice with 1X PBS containing 100 μg/mL cycloheximide. Cells were then lysed on ice in polysome lysis buffer (15mM Tris-HCl pH 7.5, 0.3M NaCl, 15mM MgCl2, 1% Triton X-100, 100 μg/mL cycloheximide) containing 100 U/mL RiboLock RNase Inhibitor and cOmplete mini EDTA-free Protease Inhibitor Cocktail tablet. Cell lysates were transferred to microcentrifuge tubes, incubated on ice for an additional 10 min., and then insoluble material was removed by centrifugation (20,800 x g for 1 min. 4°C) and supernatants were recovered. 20μL of cell-free lysate was combined with 20μL of 2X protein sample buffer for immunoblotting and 100μL of cell-free lysate was reserved for total RNA isolation.
Gradient fractionation and polysome isolation
For each gradient, 5.5mL 50% sucrose solution in 1X polysome buffer (15mM Tris-HCl pH 7.5, 0.3M NaCl, 15mM MgCl2, 100 μg/mL cycloheximide) was pipetted into a thinwall polypropylene ultracentrifuge tube followed by 5.5mL 10% sucrose solution in 1X polysome buffer, which was layered onto the 50% sucrose solution. Each gradient was sealed with parafilm and placed horizontally overnight at 4°C. The next day, cell free lysates were layered onto 11mL 10-50% sucrose gradients and sedimented at 38,000 RPM for 2 h 15 min. in a SW41Ti rotor at 4°C. Gradients were fractionated and absorbance profiles were produced by pumping the gradients through a flow cell while measuring RNA absorbance at 254nM using a density gradient fractionation system (Brandel; BR-188).
Polysome RNA isolation
Total RNA was isolated from 100μL of cell-free lysate as described in the preceding RNA isolation section with modifications (300μL TRIzol, 80μL chloroform, and 200μL isopropanol were used). RNA was isolated from 500μL gradient fractions in 2mL microcentrifuge tubes. Following TRIzol (1mL) addition to fractions and incubation for 5 min. at RT, samples were extracted with chloroform (300μL) and centrifuged at 12,000 x g for 15 min at 4°C. The aqueous phase was recovered and 2μL of GlycoBlue along with 750μL of isopropanol was added to each sample to precipitate RNA overnight at −20°C. After centrifugation (14,000 x g; 15 min, 4° C), RNA pellets were washed with 70% ice-cold ethanol (1 mL), air dried, and resuspended in nuclease-free water. Fractions representing 80S (1 ribosome), light polysomes (2-3 ribosomes), or heavy polysomes (4 + ribosomes) were pooled per replicate. Total RNA and pooled polysome fractions were purified using the RNA Clean and Concentrator-5 kit according to the manufacturer’s protocol.
Library preparation and sequencing
For polysome profiling-sequencing, a total of 24 RNA-seq libraries were prepared from RNA using the NEBNext® Ultra II Directional RNA Library Prep Kit for Illumina according to the manufacturer’s protocol. Three biological replicates were generated for each polysome fraction (80S, Light, Heavy) and the total RNA in each condition (eIF3d-depletion, non-silencing control) (Table S4). Resulting libraries were sequenced by the staff at the New York University Genome Technology Center (NYU GTC) on an Illumina NovaSeq 6000 in paired-end mode using a S1 100 cycle kit version, yielding 25-50 million paired-end reads per sample.
RNA-seq analysis (host)
Sequence reads derived from the polysome profiling were pseudoaligned against a Homo sapiens transcriptome database obtained from Gencode v37 and comprising protein-coding (n = 104,760) and lncRNA transcripts (n = 48,741). Pseudoalignments were generated using Kallisto (Bray et al., 2016) with bootstrapping set to 100. Raw transcript counts were corrected for batch effects (Figure S8) using ComBat-seq (Zhang et al., 2020), aggregated, normalized (Limma) and parsed using RIVET (Ernlund et al., 2018) to quantify RNA abundance (total mRNA fraction) and translational (80S, Light polysome, Heavy polysome pooled peak fractions) changes. RNA abundance and translational changes were classified as inert if (i) the adjusted p value >0.05; or (ii) if the adjusted p value <0.05 but the change in RNA abundance/translation was <1.5-fold (i.e. −0.58 > log2fc < 0.58). Graphical outputs of the RIVET analyses were generated in Rstudio (https://www.rstudio.com) using ggplot2 (Wickham, 2016) Gene set enrichment analysis was performed with REACTOME (Jassal et al., 2020) using gene lists derived from translationally regulated RNAs.
RNA-seq analysis (virus)
In the absence of a high quality HCMV transcriptome annotation, sequence reads were trimmed using TrimGalore (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) (–paired –length 30 –quality 30) and aligned against a hybrid genome comprising HG38 and the HCMV TB40/BAC4 genome (EF999921.1) using STAR (Dobin et al., 2013) with the –outFilterMultimapN-max 1 parameter. Post-alignment processing was performed using SAMtools (Li et al., 2009) and BEDtools (Quinlan, 2014) to generate Bedgraph files. Graphical outputs of the HCMV analyses were also generated in Rstudio using ggplot2 (Wickham, 2016). Abundance counts were determined using a strand-specific sliding window analysis (250 nucleotides) that was corrected for batch effects (Figure S8) using ComBat-seq (Zhang et al., 2020) and subsequently analyzed using RIVET. Windows showing statistically significant differences in abundance counts between conditions were annotated using the BEDtools intersect function to enable identification of regulated HCMV RNAs.
UTR analysis
Translationally regulated (FC, p value) transcripts were further contrasted against non-regulated transcripts by extracting 5′-UTR sequences using biomaRt (Durinck et al., 2005, 2009) and analyzing their length distributions and %GC content using countgc.sh from the bbtools package (https://jgi.doe.gov/data-and-tools/bbtools/).
QUANTIFICATION AND STATISTICAL ANALYSIS
GraphPad Prism and Excel software were used for statistical analysis. Statistical tests performed, sample sizes (N) and significance values are indicated in each individual figure legend or the main text. Error bars represent standard error of the mean (SEM) and only p ≤ 0.05 was deemed significant.
Supplementary Material
Highlights.
mRNA translation in HCMV-infected cells becomes progressively reliant upon eIF3d
eIF3d depletion selectively restricts HCMV replication and late gene expression
Chronic ER stress-induced host genes regulated by eIF3d promote HCMV replication
An eIF3d-dependent switch tunes infected cell mRNA translation to support HCMV growth
ACKNOWLEDGMENTS
We thank members of the Mohr laboratory for helpful discussions, Hannah Burgess for critically reading the manuscript, Eain Murphy and Domenico Tortorella for sharing TB40/E viruses, and Ivan Topisirovic and Ola Larsson for sharing analyzed data sets. This work was supported by National Institutes of Health (NIH) grants to I.M. (GM056927, AI073898), I.M. and D.P.D. (AI152543), and H.M.B. (AI151436, AI166638). L.T. was supported in part by an NIH training grant (T32 AI007180).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110767.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Alain T, Morita M, Fonseca BD, Yanagiya A, Siddiqui N, Bhat M, Zammit D, Marcus V, Metrakos P, Voyer LA, et al. (2012). eIF4E/4E-BP ratio predicts the efficacy of mTOR targeted therapies. Cancer Res. 72, 6468–6476. [DOI] [PubMed] [Google Scholar]
- Benboudjema L, Mulvey M, Gao Y, Pimplikar SW, and Mohr I (2003). Association of the herpes simplex virus type 1 Us11 gene product with the cellular kinesin light-chain-related protein PAT1 results in the redistribution of both polypeptides. J. Virol 77, 9192–9203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolaet BL, Clarke DJ, Wolff M, Watson MH, Henze M, Divita G, and Reed SI (2001). UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat. Struct. Biol 8, 417–422. [DOI] [PubMed] [Google Scholar]
- Bianco C, and Mohr I (2017). Restriction of human cytomegalovirus replication by ISG15, a host effector regulated by cGAS-STING double-stranded-DNA sensing. J. Virol 91, e02483–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco C, and Mohr I (2019). Ribosome biogenesis restricts innate immune responses to virus infection and DNA. Elife 8, e49551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckh M, and Geballe AP (2011). Cytomegalovirus: pathogen, paradigm, and puzzle. J. Clin. Invest 121, 1673–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, and Pachter L (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol 34, 525–527. [DOI] [PubMed] [Google Scholar]
- Britt W (2008). Manifestations of human cytomegalovirus infection: proposed mechanisms of acute and chronic disease. Curr. Top. Microbiol. Immunol 325, 417–470. [DOI] [PubMed] [Google Scholar]
- Burgess HM, and Mohr I (2015). Cellular 5′-3′ mRNA exonuclease Xrn1 controls double-stranded RNA accumulation and anti-viral responses. Cell Host Microbe 17, 332–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E, Gu B, Bowen ME, Aryan F, Zalc A, Liang J, Flynn RA, Swigut T, Chang HY, Attardi LD, et al. (2018). Tissue-selective effects of nucleolar stress and rDNA damage in developmental disorders. Nature 554, 112–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon MJ, Schmid DS, and Hyde TB (2010). Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev. Med. Virol 20, 202–213. [DOI] [PubMed] [Google Scholar]
- Cate JH (2017). Human eIF3: from ’blobology’ to biological insight. Philos. Trans. R. Soc. Lond. B Biol. Sci 372, 20160176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuluunbaatar U, Roller R, Feldman ME, Brown S, Shokat KM, and Mohr I (2010). Constitutive mTORC1 activation by a herpesvirus Akt surrogate stimulates mRNA translation and viral replication. Genes Dev. 24, 2627–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clippinger AJ, Maguire TG, and Alwine JC (2011). The changing role of mTOR kinase in the maintenance of protein synthesis during human cytomegalovirus infection. J. Virol 85, 3930–3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Parra C, Ernlund A, Alard A, Ruggles K, Ueberheide B, and Schneider RJ (2018). A widespread alternate form of cap-dependent mRNA translation initiation. Nat. Commun 9, 3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn W, Chou C, Li H, Hai R, Patterson D, Stolc V, Zhu H, and Liu F (2003). Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. U S A 100, 14223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, and Huber W (2005). BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21, 3439–3440. [DOI] [PubMed] [Google Scholar]
- Durinck S, Spellman PT, Birney E, and Huber W (2009). Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc 4, 1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsasser S, Gali RR, Schwickart M, Larsen CN, Leggett DS, Müller B, Feng MT, Tübing F, Dittmar GA, and Finley D (2002). Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat. Cell Biol 4, 725–730. [DOI] [PubMed] [Google Scholar]
- Ernlund AW, Schneider RJ, and Ruggles KV (2018). RIVET: comprehensive graphic user interface for analysis and exploration of genome-wide translatomics data. BMC Genomics, 809. 10.1186/s12864-018-5166-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D, and Shokat KM (2009). Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 7, e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan B-J, van Hoef V, Jobava R, Elroy-Stein O, Valasek LS, Cargnello M, Gao X-H, Krokowski D, Merrick WC, Kimball SR, et al. (2017). A unique ISR program determines cellular responses to chronic stress. Mol. Cell 68, 885–900.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Back SH, Hur J, Lin YH, Gildersleeve R, Shan J, Yuan CL, Krokowski D, Wang S, Hatzoglou M, et al. (2013). ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol 15, 481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H, Han T, Xuan B, Sun Y, Tang S, Yue N, and Qian Z (2019). Dissecting the role of DDX21 in regulating human cytomegalovirus replication. J. Virol 93, e01222–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, et al. (2003). An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11, 619–633. [DOI] [PubMed] [Google Scholar]
- Hiyama H, Yokoi M, Masutani C, Sugasawa K, Maekawa T, Tanaka K, Hoeijmakers JHJ, and Hanaoka F (1999). Interaction of hHR23 with S5a: the ubiquitin-like domain of hhr23 mediates interaction with S5a subunit of 26S Proteasone. J. Biol. Chem 274, 28019–28025. [DOI] [PubMed] [Google Scholar]
- Isler JA, Skalet AH, and Alwine JC (2005). Human cytomegalovirus infection activates and regulates the unfolded protein response. J. Virol 79, 6890–6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jassal B, Matthews L, Viteri G, Gong C, Lorente P, Fabregat A, Sidiropoulos K, Cook J, Gillespie M, Haw R, et al. (2020). The reactome pathway knowledgebase. Nucleic Acids Res. 48. D498–d503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerry JA, Priddy MA, Kohler CP, Staley TL, Weber D, Jones TR, and Stenberg RM (1997). Translational regulation of the human cytomegalovirus pp28 (UL99) late gene. J. Virol 71, 981–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Mariño G, and Levine B (2010). Autophagy and the integrated stress response. Mol. Cell 40, 280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudchodkar SB, Yu Y, Maguire TG, and Alwine JC (2004). Human cytomegalovirus infection induces rapamycin-insensitive phosphorylation of downstream effectors of mTOR kinase. J. Virol 78, 11030–11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamper AM, Fleming RH, Ladd KM, and Lee ASY (2020). A phosphorylation-regulated eIF3d translation switch mediates cellular adaptation to metabolic stress. Science 370, 853–856. [DOI] [PubMed] [Google Scholar]
- Lau B, Kerr K, Gu Q, Nightingale K, Antrobus R, Suárez NM, Stanton RJ, Wang ECY, Weekes MP, and Davison AJ (2020). Human cytomegalovirus long non-coding RNA1.2 suppresses extracellular release of the pro-inflammatory cytokine IL-6 by blocking NF-κB activation. Front. Cell. Infect. Microbiol 10, 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ASY, Kranzusch PJ, and Cate JHD (2015). eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature 522, 111–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ASY, Kranzusch PJ, Doudna JA, and Cate JHD (2016). eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature 536, 96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Lee YS, Choi Y, Son A, Park Y, Lee KM, Kim J, Kim JS, and Kim VN (2021). The SARS-CoV-2 RNA interactome. Mol. Cell 81, 2838–2850.e2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenarcic EM, Ziehr B, De Leon G, Mitchell D, and Moorman NJ (2014). Differential role for host translation factors in host and viral protein synthesis during human cytomegalovirus infection. J. Virol 88, 1473–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YT, Prendergast J, and Grey F (2017). The host ubiquitin-dependent segregase VCP/p97 is required for the onset of human cytomegalovirus replication. PLoS Pathog. 13, e1006329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GY, and Sabatini DM (2020). mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol 21, 183–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungman P, Hakki M, and Boeckh M (2011). Cytomegalovirus in hematopoietic stem cell transplant recipients. Hematol. Oncol. Clin. North Am 25, 151–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manicklal S, Emery VC, Lazzarotto T, Boppana SB, and Gupta RK (2013). The “silent” global burden of congenital cytomegalovirus. Clin. Microbiol. Rev 26, 86–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani M, Sonenberg N, Yokoyama S, and Imataka H (2007). Reconstitution reveals the functional core of mammalian eIF3. EMBO J. 26, 3373–3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney C, Perez C, and Mohr I (2012). Poly(A) binding protein abundance regulates eukaryotic translation initiation factor 4F assembly in human cytomegalovirus-infected cells. Proc. Natl. Acad. Sci. U S A 109, 5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney C, Zavadil J, Bianco C, Shiflett L, Brown S, and Mohr I (2014). Global reprogramming of the cellular translational landscape facilitates cytomegalovirus replication. Cell Rep. 6, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer H, and Weihl CC (2014). The VCP/p97 system at a glance: connecting cellular function to disease pathogenesis. J. Cell Sci 127, 3877–3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr I, and Sonenberg N (2012). Host translation at the nexus of infection and immunity. Cell Host Microbe 12, 470–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman NJ, Cristea IM, Terhune SS, Rout MP, Chait BT, and Shenk T (2008). Human cytomegalovirus protein UL38 inhibits host cell stress responses by antagonizing the tuberous sclerosis protein complex. Cell Host Microbe 3, 253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman NJ, and Shenk T (2010). Rapamycin-resistant mTORC1 kinase activity is required for herpesvirus replication. J. Virol 84, 5260–5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey M, Arias C, and Mohr I (2007). Maintenance of endoplasmic reticulum (ER) homeostasis in herpes simplex virus type 1-infected cells through the association of a viral glycoprotein with PERK, a cellular ER stress sensor. J. Virol 81, 3377–3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E, Rigoutsos I, Shibuya T, and Shenk TE (2003a). Reevaluation of human cytomegalovirus coding potential. Proc. Natl. Acad. Sci. U S A 100, 13585–13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E, Yu D, Grimwood J, Schmutz J, Dickson M, Jarvis MA, Hahn G, Nelson JA, Myers RM, and Shenk TE (2003b). Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc. Natl. Acad. Sci. U S A 100, 14976–14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, and Gorman AM (2016). The integrated stress response. EMBO Rep. 17, 1374–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, and Sonenberg N (2019). The organizing principles of eukaryotic ribosome recruitment. Annu. Rev. Biochem 88, 307–335. [DOI] [PubMed] [Google Scholar]
- Perez C, McKinney C, Chulunbaatar U, and Mohr I (2011). Translational control of the abundance of cytoplasmic poly(A) binding protein in human cytomegalovirus-infected cells. J. Virol 85, 156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procter DJ, Banerjee A, Nukui M, Kruse K, Gaponenko V, Murphy EA, Komarova Y, and Walsh D (2018). The HCMV assembly compartment is a dynamic golgi-derived MTOC that controls nuclear rotation and virus spread. Dev. Cell 45, 83–100.e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proikas-Cezanne T, Takacs Z, Dönnes P, and Kohlbacher O (2015). WIPI proteins: essential PtdIns3P effectors at the nascent autophagosome. J. Cell Sci 128, 207–217. [DOI] [PubMed] [Google Scholar]
- Pulos-Holmes MC, Srole DN, Juarez MG, Lee AS, McSwiggen DT, Ingolia NT, and Cate JH (2019). Repression of ferritin light chain translation by human eIF3. Elife 8, e48193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR (2014). BEDTools: the Swiss-Army tool for genome feature analysis. Curr. Protoc. Bioinformatics 47, 11.12.1–11.12.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razonable RR, and Humar A (2013). Cytomegalovirus in solid organ transplantation. Am. J. Transplant 13, 93–106. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Sánchez I, Schafer XL, Monaghan M, and Munger J (2019). The Human Cytomegalovirus UL38 protein drives mTOR-independent metabolic flux reprogramming by inhibiting TSC2. PLoS Pathog. 15, e1007569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah M, Su D, Scheliga JS, Pluskal T, Boronat S, Motamedchaboki K, Campos AR, Qi F, Hidalgo E, Yanagida M, et al. (2016). A transcript-specific eIF3 complex mediates global translational control of energy metabolism. Cell Rep. 16, 1891–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Lee S, Cho D-Y, Lee S, Kim H, Yu N, Lee S, and Ahn K (2019). Human cytomegalovirus induces and exploits Roquin to counteract the IRF1-mediated antiviral state. Proc. Natl. Acad. Sci. U S A 116, 18619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas KP, Depledge DP, Adebe JS, Rice SA, Mohr I, and Wilson AC (2021). Widespread remodeling of the m6A RNA modification landscape by a viral regulator of RNA processing and export. Proc. Natl. Acad. Sci. U S A 118. e2104805118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamminger T, Puchtler E, and Fleckenstein B (1991). Discordant expression of the immediate-early 1 and 2 gene regions of human cytomegalovirus at early times after infection involves posttranscriptional processing events. J. Virol 65, 2273–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenberg RM, and Stinski MF (1985). Autoregulation of the human cytomegalovirus major immediate-early gene. J. Virol 56, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Ginossar N, Thompson SR, Mathews MB, and Mohr I (2019). Translational control in virus-infected cells. Cold Spring Harb. Perspect. Biol 11, a033001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Ginossar N, Weisburd B, Michalski A, Le VT, Hein MY, Huang SX, Ma M, Shen B, Qian SB, Hengel H, et al. (2012). Decoding human cytomegalovirus. Science 338, 1088–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Todorovic A, Querol-Audí J, Bai Y, Villa N, Snyder M, Ashchyan J, Lewis CS, Hartland A, Gradia S, et al. (2011). Functional reconstitution of human eukaryotic translation initiation factor 3 (eIF3). Proc. Natl. Acad. Sci. U S A 108, 20473–20478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, and Gray NS (2009). An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem 284, 8023–8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh O, Cohen Y, Shitrit A, Shani O, Le-Trilling VT, Trilling M, Friedlander G, Tanenbaum M, and Stern-Ginossar N (2015). The transcription and translation landscapes during human cytomegalovirus infection reveal Novel host-pathogen interactions. PLoS Pathog. 11, e1005288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Herrmannová A, Šikrová D, and Valiášek LS (2016). Human eIF3b and eIF3a serve as the nucleation core for the assembly of eIF3 into two interconnected modules: the yeast-like core and the octamer. Nucleic Acids Res. 44, 10772–10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, and Mohr I (2004). Phosphorylation of eIF4E by Mnk-1 enhances HSV-1 translation and replication in quiescent cells. Genes Dev. 18, 660–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, and Mohr I (2014). Coupling 40S ribosome recruitment to modification of a cap-binding initiation factor by eIF3 subunit e. Genes Dev. 28, 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, Perez C, Notary J, and Mohr I (2005). Regulation of the translation initiation factor eIF4F by multiple mechanisms in human cytomegalovirus-infected cells. J. Virol 79, 8057–8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H (2016). ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag; ). [Google Scholar]
- Wilkinson GWG, Davison AJ, Tomasec P, Fielding CA, Aicheler R, Murrell I, Seirafian S, Wang ECY, Weekes M, Lehner PJ, et al. (2015). Human cytomegalovirus: taking the strain. Med. Microbiol. Immunol 204, 273–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Pierciey FJ Jr., Maguire TG, and Alwine JC (2013). PKR-like endoplasmic reticulum kinase is necessary for lipogenic activation during HCMV infection. PLoS Pathog. 9, e1003266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Parmigiani G, and Johnson WE (2020). ComBat-seq: batch effect adjustment for RNA-seq count data. NAR Genom. Bioinform 2, lqaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Kim T, Bao M, Facchinetti V, Jung SY, Ghaffari AA, Qin J, Cheng G, and Liu YJ (2011). DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity 34, 866–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing datasets generated during this study have been deposited at the European Nucleotide Archive (ENA) and are publically available as of the publication date. The accession number is listed in the key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-eIF3a | Cell Signaling | cat# 3411S |
| anti-eIF3b | Bethyl | cat# A301-761A |
| anti-eIF3d | Proteintech | cat# 10219-1-AP |
| anti-eIF3e | Bethyl | cat# A302-985A |
| anti-eIF3g | Bethyl | cat# A301-757A |
| anti-eIF3h | Cell Signaling | cat# 3413S |
| anti-eIF3l | GeneTex | cat# GTX120119 |
| anti-4EBP1 | Bethyl | cat# A300-501A |
| anti-GAPDH | Cell Signaling | cat# 2118S |
| anti-IE1/IE2 | Millipore | cat# MAB810 |
| anti-UL44 | Virusys | cat# CA006 |
| anti-pp28 | Virusys | cat# CA004-100 |
| anti-UL57/ICP8 | Novus | cat# NB110-57369 |
| anti-DDX21 | Proteintech | cat# 10528-1-AP |
| anti-VCP | Proteintech | cat# 10736-1-AP |
| anti-SLC3A2/CD98 | Proteintech | cat# 15193-1-AP |
| anti-INPP4B | Cell Signaling | cat# 4039S |
| anti-RAD23A | Cell Signaling | cat# 24555S |
| anti-MLST8/GBL | Cell Signaling | cat# 3274S |
| anti-WIPI2 | Cell Signaling | cat# 8567S |
| anti-rabbit IgG HRP | GE Healthcare | cat# NA934-1ML |
| anti-mouse IgG HRP | GE Healthcare | cat# NA931-1ML |
| Bacterial and virus strains | ||
| HCMV AD169-GFP | Dong Yu | N/A |
| HCMV TB40/E | Domenico Tortorella (Mount Sinai, New York, NY, USA) | N/A |
| HCMV TB40/E-GFP | Eain Murphy (SUNY, Syracuse, NY, USA) | N/A |
| HSV-1-GFP-Us11 | Dr. Ian Mohr (NYU Schiool of Medicine, New York, NY, USA) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| DMEM | Corning | cat# 10-013-CV |
| DMEM without L-Glutamine/L-Methionine/L-Cystine | Corning | cat# MT17204CI |
| 25mM HEPES | Corning | cat# 25060CI |
| 100 U/mL penicillin-100μg/mL streptomycin | Corning | cat# MT-30-002-Cl |
| pp242 | Invivogen | cat# inh-pp242 |
| RNAimax | Invitrogen | cat# 13778075 |
| Opti-MEM | Gibco | cat# 31985070 |
| TRIzol | Invitrogen | cat# 15596018 |
| GlycoBlue | Invitrogen | cat# AM9515 |
| nuclease-free water | Invitrogen | cat# AM9937 |
| qScript XLT cDNA SuperMix | QuantaBio | cat# 95161-500 |
| SsoAdvanced Universal SYBR Green Supermix | Bio-Rad | cat# 1725275 |
| Phosphonoacetic acid | Sigma | cat# P6909 |
| EasyTag Express 35S Labeling Mix | Perkin Elmer | cat# NEG772007MC |
| CytoScint ES scintillation fluid | MP Biomedicals, Inc | cat# 0188245301 |
| Cycloheximide | Sigma | cat# C7698 |
| RiboLock RNase Inhibitor | ThermoFisher Scientific | cat# EO0381 |
| cOmplete mini EDTA-free Protease Inhibitor Cocktail tablet | Roche | cat# 05892791001 |
| Critical commercial assays | ||
| Allprep DNA Kit | Qiagen | cat# 80204 |
| RNA Clean and Concentrator-5 kit | Zymo Research | cat# R1014 |
| NEBNext® Ultra II Directional RNA Library Prep Kit for Illumina | New England Biolabs | cat# E7760S |
| Deposited data | ||
| RNA sequencing datasets | European Nucleotide Archive (ENA) | BioProject ID PRJEB45749 |
| Experimental models: Cell lines | ||
| Normal human dermal fibroblasts (NHDF) | Lonza | cat# CC-2509 |
| African green monkey cells (Vero) | ATCC | cat# CCL-81 |
| Human retinal pigment epithelial cells (ARPE-19) | ATCC | cat# CRL-2302 |
| Oligonucleotides | ||
| DNA oligonucleotides for PCR | This paper and other sources | Table S5 |
| siRNAs | Qiagen, Sigma | Table S5 |
| Software and algorithms | ||
| GraphPad Prism (Version 9) | Graphpad | RRID: SCR_002798 |
| RIVET | Ernlund et al., 2018 | N/A |
| Kallisto | Bray et al., 2016 | N/A |
| ComBat-seq | Zhang et al., 2020 | N/A |
| Rstudio using ggplot2 | Wickham, 2016 | N/A |
| REACTOME | Jassal et al., 2020 | N/A |
| TrimGalore | https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ | N/A |
| STAR | Dobin et al., 2013 | N/A |
| SAMtools | Li et al., 2009 | N/A |
| BEDtools | Quinlan, 2014 | N/A |
| biomaRt | Durinck et al., 2005; Durinck et al., 2009 | N/A |
| Other | ||
| Nitrocellulose membranes | GE Healthcare | cat# 10600002 |
| Whatman Grade GF/C Glass Microfiber Filters | GE Healthcare | cat# 18822-025 |
| thinwall polypropylene ultracentrifuge tube | Beckman Coulter | cat# 331372 |
| SW41Ti rotor | Beckman Coulter | cat# 331362 |
| density gradient fractionation system | Brandel | cat# BR-188 |


