Abstract
Amyotrophic lateral sclerosis (ALS) is the most prominent motor neuron disease in humans. Its etiology consists of progressive motor neuron degeneration resulting in a rapid decline in motor function starting in the limbs or bulbar muscles and eventually fatally impairing central organs most typically resulting in loss of respiration. Pathogenic variants in 4 main genes, SOD1, TARDBP, FUS, and C9orf72, have been well characterized as causative for more than a decade now. However, these only account for a small fraction of all ALS cases. In this review, we highlight many additional variants that appear to be causative or confer increased risk for ALS, and we reflect on the technologies that have led to these discoveries. Next, we call attention to new challenges and opportunities for ALS and suggest next steps to increase our understanding of ALS genetics. Finally, we conclude with a synopsis of gene therapy paradigms and how increased understanding of ALS genetics can lead us to developing effective treatments. Ultimately, a consolidated update of the field can provide a launching point for researchers and clinicians to improve our search for ALS-related genes, defining pathogenic mechanisms, form diagnostics, and develop therapies.
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disorder. Progressive degeneration of upper and lower motor neurons in the motor cortex, brainstem, and spinal cord in ALS leads to various patterns of spasticity and flaccid limb weakness as well as dysarthria and dysphagia. Up to 50%–65% of patients with ALS may have a range of cognitive impairment, thus reflecting pathology in additional cortical regions.1 The incidence is estimated to be 2–3 per 100,000.2 About 10% of ALS cases have a first-degree relative with the disorder, which have helped uncover Mendelian causes of disease, termed familial ALS (FALS). Sporadic ALS (SALS) cases also have genetic risk factors, as evidenced by twin studies that indicate 61% heritability of SALS, yet only a fraction of these factors have actually been resolved.3 From a phenotypic standpoint, familial and SALS cases are similar in presentation with the major difference that the mean age at onset is about 10 years later for individuals with SALS relative to FALS.4 Males have a ∼1.5-fold increased risk of developing SALS,5 although this increased incidence is not observed in familial cases.4 Early death occurs due to aspiration and respiratory failure in 2–5 years after the first recognition of symptoms,6 although specific clinical presentations of motor dysfunction can be quite heterogeneous.
There is not one biological test to confirm the diagnosis of ALS; the diagnosis is made clinically based on a specific phenotypic presentation to distinguish it from the broader classification of motor neuron diseases (MNDs). Classical ALS is defined as involving both upper and motor neurons. Less common variant forms are categorized along the broader diagnosis of ALS/MND. Approximately 3% of patients with ALS/MND will have isolated upper motor neuron involvement manifesting as pure spasticity over a longer duration of time and absence of other cognitive symptoms, which is termed primary lateral sclerosis.7 On the other end of the motor neuron spectrum, there are patients who only develop lower motor neuron dysfunction leading to weakness and amyotrophy termed progressive muscular atrophy (PMA). PMA is also rare, accounting for 3%–10% of ALS/MND.8 Clinical diagnosis requires precision as there are many other disease processes that mimic ALS including mechanical, infectious, neoplastic, and autoimmune that affect motor neurons and corticospinal tract function. To address the complexity in diagnosis, there have been widely accepted clinical and electrodiagnostic criteria established that derive from the definition that classical ALS involves multiple spinal regions as well as both upper and lower motor neuron involvement. To be given a diagnosis of clinically definite ALS, a patient must have both upper and lower MNDs across 3 spinal regions or bulbar and 2 spinal regions. Alternatively, patients can be diagnosed with clinically probable ALS if criteria are satisfied by upper and lower motor neuron involvement in 2 regions. Electrodiagnostic studies can also provide evidence of lower MND even in the absence of clinically evident signs.9
Provided an accurate clinical diagnosis, the neuropathologic features are similar across most cases. Although the majority of ALS brains show no significant gross abnormalities, degeneration coalesces as atrophy in the anterior nerve roots of the spinal cord as well as occasional atrophy in the precentral gyrus.10 In patients who have clinical symptoms suggestive of frontotemporal dementia (FTD), this atrophy may extend to the frontal and temporal lobes. Microscopically, the degeneration of motor neurons is often observed as a reduction in neurons with associated astrogliosis in the anterior horn of the spinal cord and lower cranial motor nuclei in the brainstem as well as a more mild loss of motor neurons in primary motor cortex.10 Particularly in the cortex, some nonspecific features of neurodegeneration, such as microvacuolation or spongiosis, may also appear.11
Initial Discoveries and Classical Approaches to Detect ALS Genes
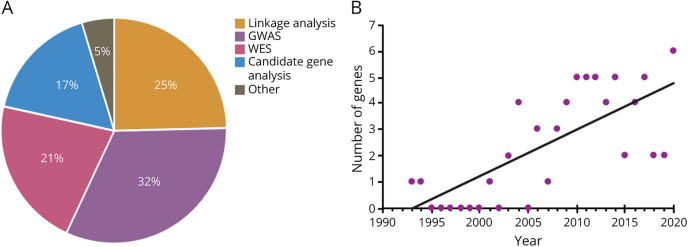
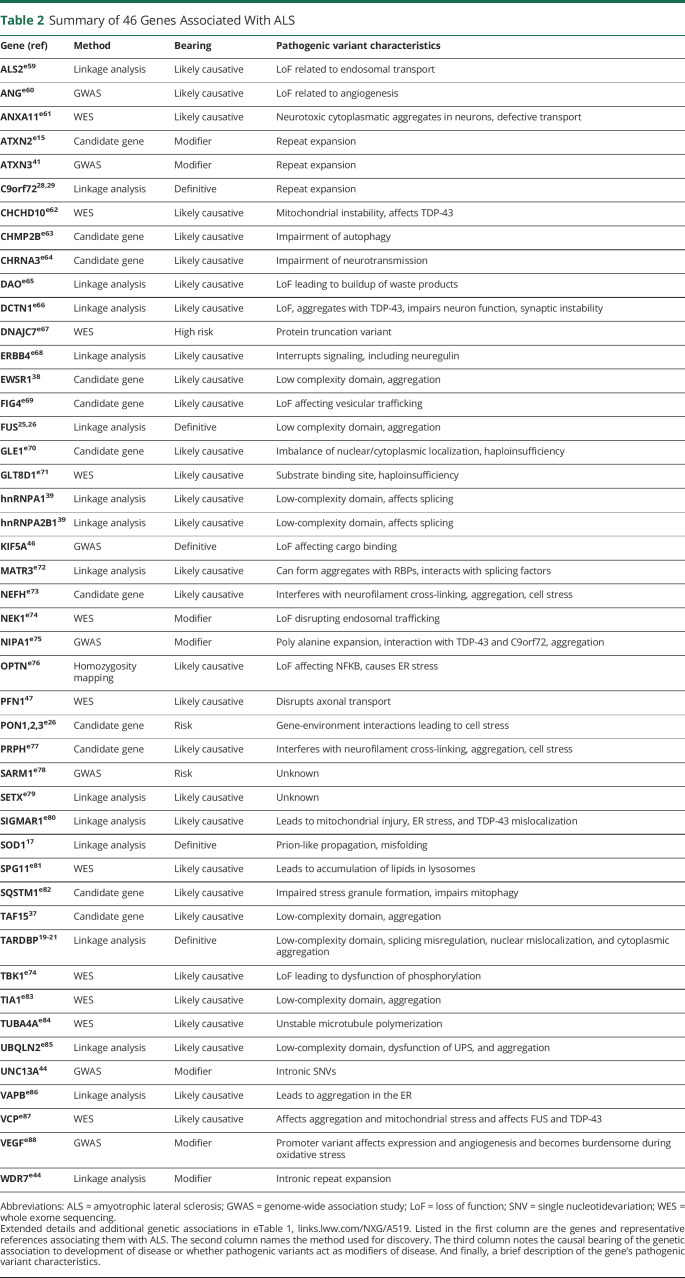
Our current toolkit for investigating genetic causes of ALS includes linkage analysis, candidate gene analysis, genome-wide association studies (GWASs), whole-exome sequencing (WES), and whole-genome sequencing (WGS). Each of these methods have individual strengths and weaknesses, and are optimized for asking specific kinds of genetic questions and have a unique use among the various discovery strategies for ALS-related genes. We highlight the kinds of studies in which each of these technologies excel as well as their weaknesses that need to be considered when designing and interpreting investigations in Table 1. Linkage studies rely on models of Mendelian inheritance and have been the most effective at identifying causal genes. Linkage analysis has accounted for about 25% of implicated genes (Figure 1A). Following these discoveries, the same genes can be identified to be associated with SALS cases using other approaches. Therefore, linkage analysis provides a robust launching point toward diagnosing ALS. With the knowledge acquired from linkage studies, the approach of candidate gene analysis can be leveraged to investigate hypotheses derived from the elucidation of associated pathways and genes that operate in tandem with, and similarity to, previously implicated genes. Candidate gene investigations contribute to 17% of ALS gene discoveries to date. Finally, GWAS and WES can be used more broadly to identify variants within or near genes that are associated with a risk of SALS incidence. These techniques have accounted for 32% and 21% of discoveries, respectively. Since 1993, many genes have been implicated as either causal or contributing risk, and the accelerating pace of technological advancement has led to an ever-increasing number of discoveries per year of postulated genetic contributions to ALS (Figure 1B). We highlight 46 genes with strong evidence of disease causation or association in Table 2 (along with several more putative ALS genes in eTable 1, links.lww.com/NXG/A519), and we explore the technologies contributing to their discovery. Many of these genes have been the subject of excellent recent reviews,12-16 and here, we address them in the context of technological advancement and implications for future investigations.
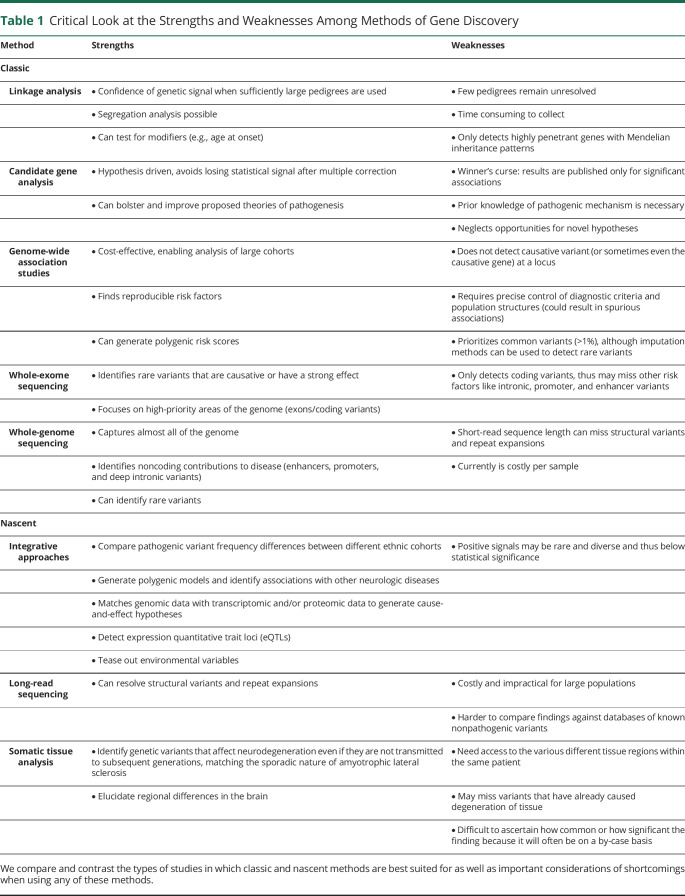
Table 1.
Critical Look at the Strengths and Weaknesses Among Methods of Gene Discovery
Figure 1. Discovery Methods and Frequency (per Year) of ALS-Associated Genes.
(A) The percent contribution of various gene discovery methods to the current repertoire of 64 ALS implicated genes that we highlight. (B) The number of novel ALS gene discoveries per year. GWAS = genome-wide association study; WES = whole-exome sequencing.
Table 2.
Summary of 46 Genes Associated With ALS
Linkage Studies
Classical genetics approaches including genetic linkage studies have been successful in identifying pathogenic variants in ∼50%–60% of patients with a strong family history of ALS and 10%–20% of SALS.16 The first gene discovered to show linkage to familial inheritance of ALS was superoxide dismutase 1 (SOD1) in 1993.17 Pathologic variants are primarily missense, with a few translation termination variants appearing near the end of the protein, which account for 5%–10% of individuals with FALS. Although it was first hypothesized that SOD1 contributed to disease by loss of the ability to eliminate reactive oxygen species, it is now generally accepted that this loss of function has a very minimal impact and that SOD1 pathogenic variants actually lead to errors in the stability of the protein. This can cause it to fold incorrectly, not unlike prions, which can propagate misfolding among other SOD1 proteins ultimately leading to aggregation.18
Identification of pathogenic variants in the RNA-binding protein (RBP) TDP-43, encoded by the gene TAR DNAbinding protein (TARDBP), gave us more detailed insight into ALS pathology and its relationship to RBPs and splicing regulation.19-22 Normally, TDP-43 is localized to the nucleus, but in patients with TARDBP variants and in SALS cases, it shifts to the cytoplasm and builds up as insoluble aggregates.23 This cytoplasmic redistribution prevents TDP-43 from serving its normal function regulating mRNA splicing and leads to aberrant splicing patterns for several genes.24 A heightened focus of the field to examine RBPs led to the identification of pathogenic variants in FUS RNA binding protein (FUS), a related heterogeneous ribonuclear protein, detected in 1%–3% of individuals with FALS.25-27 A revolutionary finding for the field informing us of an additional pathologic mechanism was pedigrees with ALS, FTD, or both found to carry hexanucleotide repeat expansions in C9orf72-SMCR8 complex subunit (C9orf72).28,29 Repeat expansions have been detected in about 40% of FALS and surprisingly 3%–7% of sporadic individuals as well.30-32 C9orf72 repeat expansions have also been identified in FTD, and about 15% of ALS cases also have FTD that can arise before the diagnosis of ALS.33
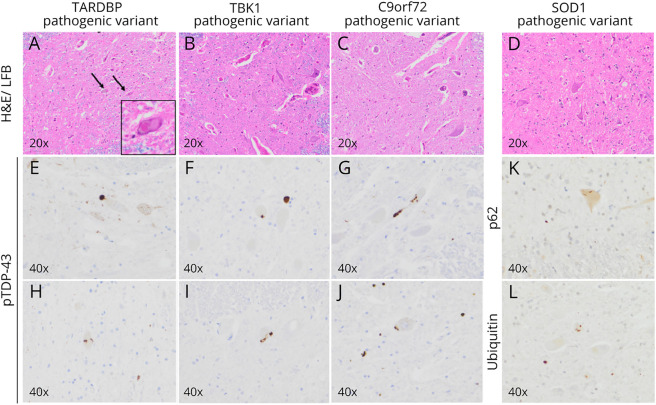
These 4 genes are the most well-defined causative genes for ALS and their pathologic consequences are reflected among the molecular hallmarks of disease. All forms of ALS demonstrate ubiquitin-positive neuronal cytoplasmic inclusions in degenerating motor neurons.34 With some notable exceptions, these ubiquitinated inclusions are comprised of TDP-43, p62, and other proteins. The abnormal TDP-43 inclusions observed in ALS can have varied morphology, including fine skeins, coarse skeins, dot-like inclusions, and dense round inclusions10 (Figure 2). In contrast, the motor neurons of patients with SOD1 pathogenic variants contain abnormal ubiquitinated neuronal inclusions, but in contrast to most other forms of ALS, these ubiquitinated inclusions are comprised of accumulated SOD1 protein and not TDP-43.35 Although these TDP-43-negative/SOD1-positive inclusions are characteristic of SOD1 ALS neuropathology, abnormal SOD1 inclusions have been rarely observed in sporadic forms of ALS.35 Similarly, patients with ALS related to pathogenic variants in FUS often demonstrate ubiquitinated neuronal inclusions, which are comprised of mutant FUS protein—and not TDP-4310—although there are still cases in which the 2 still colocalize demonstrating that better characterization of molecular pathologies is still needed.36
Figure 2. Common Neuropathologic Findings in ALS Subtypes.
(A–D) Hematoxylin and eosin (H&E)/Luxol fast blue (LFB)-stained sections demonstrating reduction of motor neurons and astrogliosis in the anterior horn of the spinal cord. Occasionally, eosinophilic intracellular inclusions termed Bunina bodies (TARDBP pathogenic variant, H&E/LFB with inset) are identified. (E–J) Ubiquitinated intracellular inclusions comprised predominantly of phosphorylated TDP-43 (pTDP-43) present in patients with a TARDBP pathogenic variant, TBK1 pathogenic variant, and a C9orf72 repeat expansion. These inclusions are morphologically heterogeneous and can take the form of coarse or fine skeins, dot-like inclusions, or dense round inclusions. (K–L) Immunohistochemistry demonstrating inclusions that are positive for p62 and ubiquitin in individuals with SOD1 pathogenic variants that have similar morphologies to the TDP-43 inclusions of the aforementioned genetic subtypes but are themselves negative for pTDP-43 immunoreactivity. Listed is the magnification in each panel. ALS = amyotrophic lateral sclerosis.
Case/Control Analysis of Candidate Genes
Candidate genes can be hypothesized to be involved in ALS based on their similarity with previously discovered genes or pathogenic mechanisms. An example of a candidate gene approach is a study that was initiated by a yeast functional screen of RBPs to find proteins that show the most similar aggregation pathology as TDP-43. Subsequently, the genes TATA-box binding protein associated factor 15 (TAF15) and EWS RNA binding protein 1 (EWSR1) were selected to be sequenced among many cases and controls, which identified variants associated with disease.37,38 Similarly, the identification of TARDBP and FUS ignited an interest to focus on RBPs among other familial cases. Most notable is the linkage of heterogeneous nuclear ribonucleoproteins A1 and A2b1 (HNRNPA1) and (HNRNPA2b1) within autosomal dominant families in which investigators specifically narrowed their genetic search to proteins harboring prion-like domains.39 However, there is an important consideration for candidate gene approaches. The method excels at bolstering prior hypotheses of pathogenesis, yet as a consequence, it discriminates against findings that may refute these hypotheses and neglects the development of novel hypotheses.
Genome-Wide Association Studies
ALS researchers contributed to some of the early successful GWAS findings. These studies point to several significant loci including at ACSL5, C21orf2, DPP6, GPX3-TNIP1, MOBP, SCFD1, SARM1, UNC13A, and others.40-45 Many GWAS investigations have been able to identify several genes per study owing to robust data acquisition, and this has led the way in uncovering variants associated with sporadic cases.14 Furthermore, subsequent studies that screen new populations frequently replicate previous findings including those found from linkage analysis and candidate gene analysis. Obtaining convincing genetic findings from GWASs will always be a function of the sample size, population structure and composition, and the genetic effect of individual loci. More recent GWAS investigations leverage novel algorithms and imputation methods to improve sensitivity.14
The drawback of GWAS's broad approach is that significant gene variant associations can only be considered risk alleles, and subsequent studies will need to delve further into identifying actual causal variants and their contribution to disease pathology. Despite this, GWAS design is improving and still remains a practical technique leading to some of the most recent findings. Another improvement that has yet to be thoroughly incorporated is more stringent selection of phenotypes. Given the heterogeneity of ALS pathology, more careful consideration of specific phenotypes may generate novel signals.
Whole-Exome Sequencing
WES takes an approach that attempts to identify an enrichment of coding variants that are more likely to be causal as opposed to GWAS, which is mostly only capable of identifying risk genes. WES can still be used in conjunction with the latest GWAS investigations to home-in on the relevance of significant GWAS signals. Using large data sets, researchers can identify variants enriched among patients with ALS. WES following a significant GWAS hit identified pathogenic variants in kinesin family member 5A (KIF5A).46 The identification of KIF5A variants clustering at the C-terminal cargo-binding domain in ALS is particularly compelling because pathogenic variants in other regions of the gene including the N-terminal motor domain cause hereditary spastic paraplegia (HSP). Although both ALS and HSP are motor neuron disorders, the phenotypes are quite distinct. HSP arises from a dying back of upper motor neurons and typically has an earlier age at onset with longer disease duration. One proposed model for how 2 similar, but distinct, disorders can have variants in the same gene is that in HSP, the pathogenic changes broadly restrict anterograde cargo transport, whereas in ALS, they act on specific cargo proteins. WES can also be used to elucidate genes in FALS that have failed to be identified by linkage analysis as with PFN1.47 In many cases, unaffected individuals also have nonsynonymous variants in genes nominated by WES approaches, and it is the increased burden of variants that can be damaging to protein function in ALS cases that ultimately contributes to disease. As more samples undergo WES (and WGS), a better resolution of the critical regions of genes and their functional implications can be established.
Whole-Genome Sequencing
The rapid decline in sequencing costs has enabled WGS technology to supplant WES. WGS techniques bypass some of the challenges of WES including probe coverage and can provide a comprehensive profile of variants across the genome. As many genes with coding variants have presumably already been identified in ALS, WGS represents an additional layer of detail to examine some of the noncoding contributors to disease such as those that may lie in promoters, enhancers, or introns. One such strategy is to look for accumulations of variants in bins across the genome.48 The same strategy can be used near ALS candidate genes to identify regions far from coding exons. For example, using linkage analysis coupled with RNA analysis, we identified a deep intronic variant in SOD1, which led to the inclusion of a cryptic exon in intron 4.49 Variants at or around the cryptic SOD1 exon can now be identified with WGS techniques, along with other sites at genes critical for development of ALS. Despite the vast capability of WGS to scan almost the entire content of the genome, there are important technical limitations. For instance, WGS has limited capacity in identifying tandem repeat expansions because the length of short reads is often shorter than the length of repeats. The complete sequence coverage afforded by WGS enables regions to be reanalyzed to identify variants in regions such as promoter/enhancer elements and protein binding sites as additional genetic and molecular discoveries are made.
Consolidating Our Knowledge of ALS Genetics to Generate Theories of Pathology
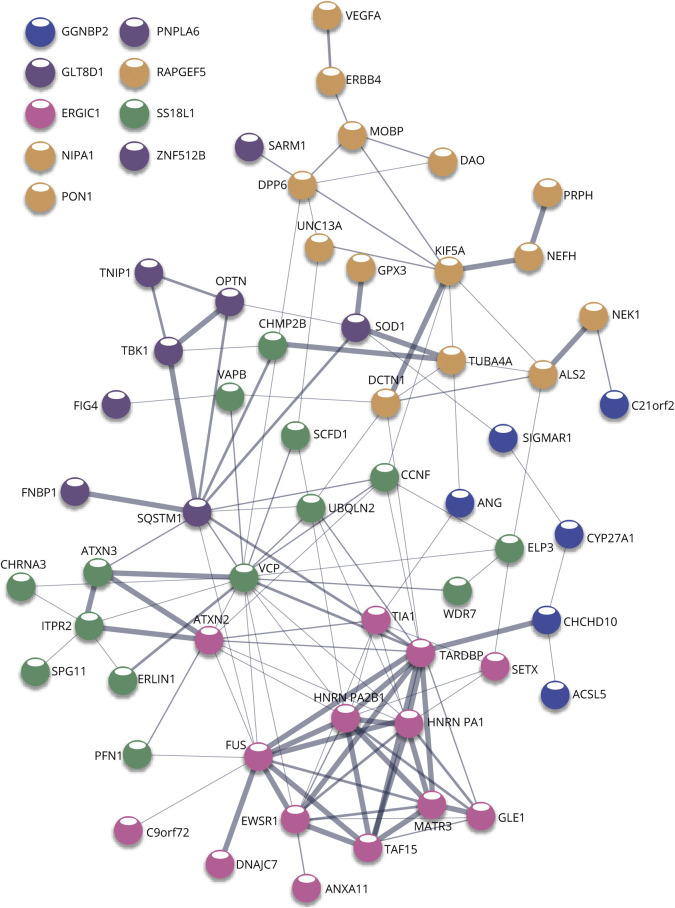
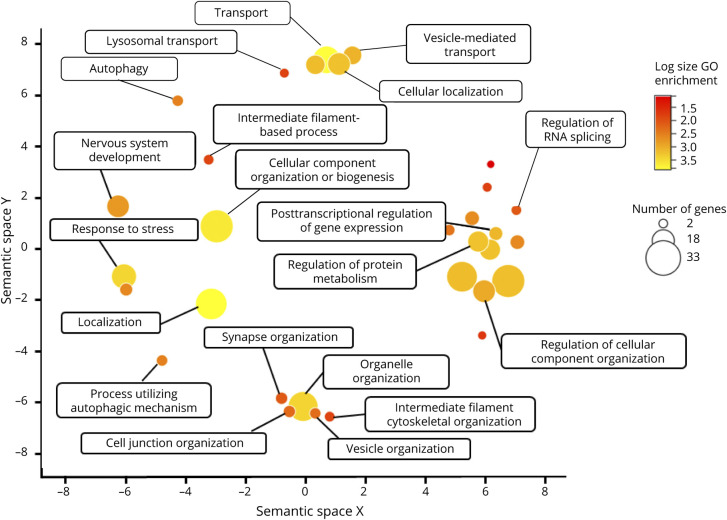
Based on the genes identified for ALS so far, we can draw important conclusions regarding common pathways implicated and convergence toward ALS phenotypes and/or related diseases. The finding that TDP-43 mislocalization to cytoplasmic aggregates present in the overwhelming majority of patients with ALS50 indicates a central pathology from which we can work backward to understand genetic and environmental triggers to disease progression. One step we can take toward understanding ALS is to take a holistic view of how the vast array of ALS-indicated genes is related to one another. One strategy is to construct network clusters based on the confidence of experimental evidence of functional associations using resources such as STRING (Figure 3).51 A second strategy can leverage our understanding of these genes' ontological roles to hypothesize additional genetic candidates involved in these central processes that can be screened for among patients (Figure 4).
Figure 3. STRING Network Analysis Reveals Distinctive Clusters of Interactions Among Implicated Genes.
Node colors represent 5 groups from k-means clustering. Edge thickness represents a confidence score of gene-gene interaction based on data collected from experimental and database records (thinnest is < 0.15, thickest is > 0.9).51 Orphan nodes are listed in the top left, colors reflecting associated clusters.
Figure 4. Revigo Gene Ontology (GO) Summary Based on the GO Panther Biological Processes Complete Statistical Overrepresentation Teste57,e58.
Axes reflect 2D semantic space similarity between GO terms. Color represents the number and variety of lower-level GO terms encompassed by the broader category. Size of each point represents the total number of ALS-implicated genes included in the GO category.
Based on the vast amount of functional evidence gathered from investigating the effects of genetic variants in model systems, a few general understandings and theories of pathology have been popularized.52 Most of the genes involved can be segregated into a few overarching categories: RNA binding, proteostasis, cytoskeletal, and metabolism/redox (eTable 1, links.lww.com/NXG/A519). Ontologically, there is an enrichment of more diverse cellular processes including stress response, endomembrane system organization, localization, and RNA splicing regulation (Figure 4). An intriguing observation is that the most confident and interconnected cluster among our network of genes (highlighted in pink) is most strongly enriched for Gene Ontology terms related to RNA splicing (Figure 3). This supports theories of pathogenesis that centralize on one of the main consequences of TDP-43 mislocalization being aberrant splicing of many target genes. Of course, caution must be taken when making these interpretations because they could simply be a result of a greater investigative depth of these genes among the scientific community.
Harking back to attempting to understanding why TDP-43 and RNA regulation is a central pathology, many studies have shown molecular similarities among the proteins involved and their pathogenic characteristics (Table 1). An essential component of RNA regulation relies on the mechanisms by which RNA molecules interact with proteins. Highly active and ubiquitous RBPs often rely on a condensation mechanism like liquid-liquid phase separation mediated by low-complexity domains.53 This mechanism must be dynamic and versatile. Because RNA regulation is spread among many proteins, any disruption to this fine-tuned mechanism can create bottlenecks in global homeostasis that are chronically exacerbated. Given that other critical functions are wrapped into this interaction network, this leads to additional imbalances in splicing regulation, nuclear export, and cytoskeletal trafficking, as well as microRNA expression. In the case of non-RBPs, pathologic aggregation can attract components of the proteostasis network and reduce its overall efficiency. RNA aggregates, including tandem repeats, can also misappropriately sequester RBPs from performing their proper function or synthesize peptide repeats that also interfere with proteostasis machinery. Overall, motor neurons, among other affected cell types, are extremely sensitive to disruptions in cellular homeostasis that chronically accumulate beyond cells' capacity to manage.54
Challenges in ALS Genetics and Exploring Emerging Solutions
Larger Data Sets, Integrated Approaches, and Novel Computational Strategies
We have witnessed a recent push to have central repositories with an organized structure of hundreds or thousands of samples. What is becoming apparent is that single gene identifications may become increasingly infrequent, and a combination of genetic factors likely contributes to ALS susceptibility. Project MinE is at the forefront of these approaches, with efforts to sequence the genome and connect findings with transcriptome, epigenome, and noncoding genome findings.55,56 Other data sets are also taking integrative approaches and providing resources for researchers at large for straightforward access to sample data. The Answer ALS consortium is one such approach, which provides whole genome data along with induced pluripotent stem cell lines and their corresponding transcriptome data, thereby saving researchers precious time and resources in trying to generate their own version of these lines. Collectively, these powerful resources will propel novel gene discoveries, especially in combination with state-of-the-art genomic methods and technologies.
Using integrative approaches that link TDP-43 binding sites with transcriptional changes has helped uncover places where loss or cytoplasmic redistribution of TDP-43 leads to improper activation of cryptic exons, for example, in the middle of intron 1 of stathmin 2 (STMN2).57,58 This activation leads to a prematurely truncated STMN2 protein product, which is detrimental to neuronal outgrowth. More recently, another aberrant splice product creating a cryptic exon was identified in unc-13 homolog A (UNC13A).59 Intriguingly, UNC13A is a well-established GWAS hit for ALS,44 and one of the lead single nucleotide variations (formerly single nucleotide polymorphisms) at this locus is directly in the cryptic exon that is generated.
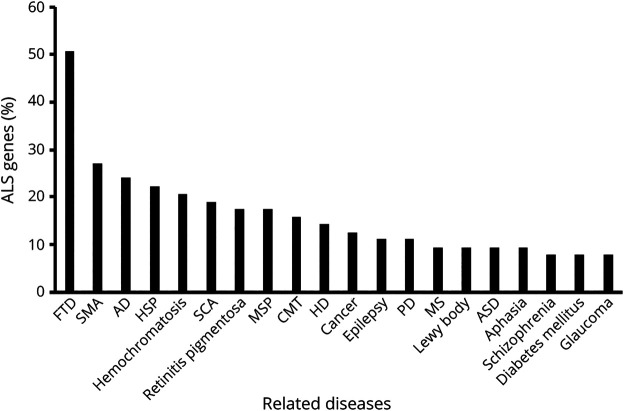
With larger and more complex genomics data becoming available, the ability to process the data by individual researchers is becoming increasingly challenging. Novel machine learning and artificial intelligence applications are helping resolve some of the complexity of data analysis revealing connections between genetic variants that are not immediately obvious. These strategies have been used successfully for imaging analysis60 and are a promising direction in the context of ALSe1. Recently, novel computational approaches have attempted to incorporate protein interactions and pathway data into machine learning modelse2. An accurate ALS diagnosis is important when curating increasingly larger collections of cases as there is considerable overlap between genetic associations of ALS and associations with other related disorders (Figure 5).
Figure 5. Percentage of the Listed ALS-Indicated Genes Associated With Other Diseases.
For each non-ALS disease listed horizontally, the percentage of ALS-indicated genes that also have relevance to that disease is plotted. AD = Alzheimer disease; ALS = amyotrophic lateral sclerosis; ASD = autism spectrum disorder; CMT = Charcot-Marie-Tooth; FTD = frontotemporal dementia; HD = Huntington disease; HSP = hereditary spastic paraplegia; MS = multiple sclerosis; MSP = multisystem proteinopathy; PD = Parkinson disease; SCA = spinocerebellar ataxia; SMA = spinal muscular atrophy.
Ethnic Differences
Identifying genetic risk factors in multiethnic populations is currently a priority for researchers and represents an emerging yet still understudied feature of ALS.41,42,e3-e5 This research is important because additional genetic risk factors can be identified leading to potential breakthroughs in our understanding of ALS pathobiology. For example, the frequency of pathogenic variants in ALS-causing genes is considerably different in China compared with Europe and North America, with a less prominent role for C9orf72 repeat expansions.e6
Using samples from diverse ethnic groups has been successful at identifying or independently validating certain risk factors for African American individuals, who have a greater prevalence of Alzheimer disease (AD).e7,e8 Not only are some of the GWAS hits different in AD,e9 but relative risk at known loci for AD can be dramatically different, such as increased risk for ATP binding cassette subfamily A member 7(ABCA7) variants in African Americanse10 compared with in non-Hispanic Whites.e11,e12 Examining local ancestry around a GWAS hit can also reveal that variants that have a stronger effect in certain populations.e13
Oligogenic (or Polygenic) Model
Two or more sequence variants that on their own have a low probability of causing disease may have an additive or synergistic role in tandem, as has been identified in a proportion (1%–4%) of FALS and SALS cases.e14 Some modifying genes may have no bearing on disease susceptibility but could influence the age at onset and progression of ALS. An example is the ataxin 2 gene (ATXN2),e15 where intermediate CAG repeat expansions numbering 27–33 (but not >34, which causes spinocerebellar ataxia 2)e16 are enriched in patients with ALS.e17 Modifier genes can also explain how genes that are ubiquitously expressed (as in the case with SOD1, which also accounts for as much as 1% of brain proteine18) proceed to elicit only a motor neuron-specific phenotype. Despite several risk genes having been reported from GWAS, there is still a lack of understanding about the functional significance of these variants. This presents an opportunity for RNA sequencing and use of disease models to interpret the potential contributions of these variants. Computational approaches have attempted to elucidate polygenic explanations and epistatic mechanisms. However, such approaches are computationally demanding, and the resources are not yet available to make groundbreaking discoveries.e1
Gene and Environment Interactions
Several environmental risk factors have been proposed and interrogated for ALS. Many of these candidates originated due to increased prevalence of ALS in a particular occupation or geographical region. An unusual cluster of ALS cases was observed in Guam,e19 and although several hypotheses have been proposed regarding the specific neurotoxic agent, especially L-beta-N-methylamino-L-alanine, the connection is still a subject of debate.e20 Increased clusters of ALS have also been observed in the Kii peninsula of Japan.e21 More recently, false morel mushrooms (Gyromitra gigas) have been proposed as a risk factor for a cluster of ALS cases in the French Alps.e22 Military veterans are also at increased risk for developing ALS, especially those that served in the Middle East, perhaps due to repeated head trauma or exposure to neurotoxins.e23-e25 A unique discovery in this regard is risk variants in paraoxonases 1, 2, and 3 appeared to render soldiers more vulnerable to toxins that would trigger ALS pathology often associated with Gulf War syndrome.e26 Similarly, paraoxonase variants have also been connected to ALS and other neurologic disease in populations frequently exposed to pesticides.e27,e28 It is likely that variants in additional candidate genes predispose individuals to toxic damage on exposure to chemical agents. Traumatic brain injury among veterans and sports players may also increase the probability of developing ALS or modify the severity of disease in addition to genetic causes.e29,e30 And there is evidence that sterile alpha and TIR motif containing 1 (SARM1) deletion modifies axon injury response.e31,e32
Mendelian randomization models are a powerful emerging tool that enable the discovery of causal genetics and environmental risk factors through use of summary statistics data resulting from GWASs. Although results have been mixed, there are suggestions that ALS susceptibility can be linked to type 2 diabetes, smoking, cholesterol, and physical activity.e33,e34 Improvement in phenotypic measurements and controlling for possible confounding variables could make this approach more practical.
Somatic Tissue Analysis for Pathogenic Variants and Repeat Expansions
Work in brain samples from individuals with autism has revealed somatic mutation events in single cells that may have been challenging to detect using bulk tissuee35 as well as somatic variants from high-density (∼×250) coverage DNA sequencing.e36 A similar scenario may exist for ALS, although one of the challenges of single-cell studies is that if a cell with an offending variant dies and is cleared, the variant will not be picked up from genomic analyses. It is notable that in the same individual, C9orf72 expansion carriers can have a much longer repeat length in the frontal cortex vs cerebellum.e37 This is in line with other repeat disorders, where expansions are unique to the affected brain region, such as huntingtin (HTT) somatic mutations in the striatum.e38 It follows that sensitive and mutation-prone loci may precipitate pathology by somatic mutation events, which could explain a failure to identify a single genetic cause in some patients.
Long-Read Sequencing for Repeat Expansions and Structural Changes
A substantial fraction of the missing genetic contribution to ALS may come from tandem repeat expansions, 40 of which have been found to cause neurodegenerative diseases.e39,e40 Our ability to identify such repeats is limited from the technical limitations of WES and WGS. The advent of long-read sequencing technology represents an emerging strategy to uncover the contribution of repeat expansions to ALS. Increasing examples of repeat expansions with a much larger internal sequence length (in contrast to short repeats such as the C9orf72 expansion) associated with disease are being found to have associations with AD and schizophrenia/bipolar disorder.e41-e43 Recently, expansions of a much larger 69-nt repeat in an intron of WD repeat domain 7 (WDR7) were found to be enriched in length in several ALS cohorts using long-read sequencing and length estimates from WGS data.e44 Repeat expansions such as those ATXN2 and WDR7 commonly appear insufficient on their own to cause disease, but the prevalence of their expansions in the general population is much higher than the number of individuals with ALS. Therefore, these repeats should be treated as potential modifiers of ALS or risk factors similar to how we interpret GWAS hits. In the case of WDR7, the expansion event can be coupled with a FUS variant to elicit disease.e44
Copy number variants have also been explored in the context of ALS, and some examples have been identified.e45-e47 By performing long-read sequencing, the relative frequency of these changes can be compared with background population rates, and we can detect whether there are certain chromosomal regions and breakpoints enriched in ALS. Moreover, the approach will help detect other tandem repeats throughout the genome in an unbiased manner to reveal their relative contribution to ALS.
Conclusions
Like many neurodegenerative disorders, understanding the contributions of the many genes that are implicated in ALS remains a challenge. Classical techniques of gene discovery remain effective, and although no single approach can capture a complete picture of ALS genetics, each has strengths and weaknesses and geared toward answering specific genetics questions. The tremendous breadth of genetic, clinical, and neuropathologic information acquired thus far have aided the field in proposing a more complete model of disease. Using the most recent knowledge of common pathways can guide more refined hypotheses and experimental designs to identify additional candidate genes either causative on their own or subtle modifiers and risk loci that increase or decrease predisposition.
One of the ultimate objectives in uncovering the genetics of ALS is to develop accurate gene therapy solutions to correct pathogenic variants. The gene therapy field is accelerating, and recent successes give hope that we can leverage our understanding of ALS genetics to develop effective treatments. The Food and Drug Administration approval of recombinant adeno-associated virus (rAAV) delivery of retinoid isomerohydrolase RPE65 (RPE65) for the treatment of retinal degeneration and rAAV survival of motor neuron 1, telomeric (SMN1) for spinal muscular atrophye48,e49 are model examples. The approval was the culmination of decades of research into designing a safe and potent delivery vector. Progress in bioengineering and isolation of capsids with enhanced properties or tropism, including the ability to cross the blood-brain barrier and transduce neurons, afford the possibility of targeted delivery to specific neuronal subtypes and enhancing delivery throughout the nervous system.e50-e53
Examples of promising gene therapy solutions for ALS include the delivery of antisense molecules targeting SOD1 and C9orf72, which now are progressing to clinical trials.e54-e56 The advantage of using genes with known pathogenic variants is that therapies conceivably can be commenced before symptom onset, as is often the case for rAAV-SMN1 delivery. This has the advantage of restoring motor neuron function before decline. Although therapies targeting familial inherited pathogenic variants are arguably the most promising for ALS, they only apply to a subset of patients, which minimizes their utility for treating the greater majority of ALS cases. However, given that up to 97% of individuals with ALS are positive for TDP-43 inclusions,50 there may be an opportunity for gene therapy targeting TDP-43 to restore functionality, which could benefit many more patients.
An accurate diagnosis in the clinic and neuropathologic characterization are required for continuing success at identifying novel and rare genetic associations. So far, our collection of ALS genetic associations point to TDP-43 and RNA regulation as one of the most likely convergent mechanisms. However, it remains to be seen how widespread and how strong of a bearing on disease alternative pathologic mechanisms may have such as gene-environmental interactions, noncoding genetic modifiers of disease, and somatic variants. Currently, techniques better suited to finding subtle and elusive contributors to disease should be brought to the forefront. Integrative analyses, somatic mutation analysis, and long-read sequencing technologies are some of the most promising new approaches for novel gene discovery beyond the scope of protein-coding variants. Computational approaches toward identifying mechanisms of genetic epistasis and pleiotropy are still in their infancy but show great potential. Importantly, ALS genetic research is accelerating, and there is hope for the future that we can create a more complete picture of disease as well as develop successful therapeutic interventions.
Glossary
- AD
Alzheimer disease
- ALS
amyotrophic lateral sclerosis
- FALS
familial ALS
- FTD
frontotemporal dementia
- GO
Gene Ontology
- GWAS
genome-wide association study
- HSP
hereditary spastic paraplegia
- MND
motor neuron disease
- PMA
progressive muscular atrophy
- rAAV
recombinant adeno-associated virus
- RBP
RNA-binding protein
- SALS
sporadic ALS
- SMA
spinal muscular atrophy
- WES
whole-exome sequencing
- WGS
whole-genome sequencing
Appendix. Authors
Contributor Information
Samuel N. Smukowski, Email: samsmuko@uw.edu.
Heather Maioli, Email: hlmaioli@uw.edu.
Caitlin S. Latimer, Email: caitlinl@uw.edu.
Thomas D. Bird, Email: tomnroz@uw.edu.
Suman Jayadev, Email: sumie@uw.edu.
Study Funding
S.N.S. is supported by a National Science Foundation Graduate Fellowship. H.M. is supported in part by the Department of Veterans Affairs Office of Academic Affiliations Advanced Fellowship Program in Mental Illness Research and Treatment and the Department of Veterans Affairs Puget Sound Mental Illness Research, Education, and Clinical Center (MIRECC). P.N.V. acknowledges research support from the Ann Arbor Active Against ALS (A2A3) foundation.
Disclosure
The authors report no disclosures relevant to the manuscript. Go to Neurology.org/NG for full disclosure.
References
- 1.Goldstein LH, Abrahams S. Changes in cognition and behaviour in amyotrophic lateral sclerosis: nature of impairment and implications for assessment. Lancet Neurol. 2013;12(4):368–380. [DOI] [PubMed] [Google Scholar]
- 2.Quinn C, Elman L. Amyotrophic lateral sclerosis and other motor neuron diseases. Continuum (Minneap MN). 2020;26(5):1323–1347. [DOI] [PubMed] [Google Scholar]
- 3.Al-Chalabi A, Fang F, Hanby MF, et al. An estimate of amyotrophic lateral sclerosis heritability using twin data. J Neurol Neurosurg Psychiatry. 2010;81(12):1324–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Camu W, Khoris J, Moulard B, et al. Genetics of familial ALS and consequences for diagnosis. J Neurol Sci. 1999;165:S21–S26. [DOI] [PubMed] [Google Scholar]
- 5.Sejvar JJ, Holman RC, Bresee JS, Kochanek KD, Schonberger LB. Amyotrophic lateral sclerosis mortality in the United States, 1979-2001. Neuroepidemiology. 2005;25(3):144–152. [DOI] [PubMed] [Google Scholar]
- 6.Verma A. Clinical manifestation and management of amyotrophic lateral sclerosis. In: Araki T, ed. Amyotrophic Lateral Sclerosis. Exon Publications; 2021. [PubMed] [Google Scholar]
- 7.Pinto WBVR, Debona R, Nunes PP, et al. Atypical motor neuron disease variants: still a diagnostic challenge in neurology. Rev Neurol (Paris). 2019;175(4):221–232. [DOI] [PubMed] [Google Scholar]
- 8.de Vries BS, Rustemeijer LMM, Bakker LA, et al. Cognitive and behavioural changes in PLS and PMA: challenging the concept of restricted phenotypes. J Neurol Neurosurg Psychiatry. 2019;90(2):141–147. [DOI] [PubMed] [Google Scholar]
- 9.Brooks BR, Miller RG, Swash M, Munsat TL. El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Mot Neuron Disord. 2000;1(5):293–299. [DOI] [PubMed] [Google Scholar]
- 10.Saberi S, Stauffer JE, Schulte DJ, Ravits J. Neuropathology of amyotrophic lateral sclerosis and its variants. Neurol Clin. 2015;33(4):855–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellison D, Love S. Neuropathology: A Reference Text of CNS Pathology. Elsevier; 2013. [Google Scholar]
- 12.Mathis S, Goizet C, Soulages A, Vallat JM, Masson GL. Genetics of amyotrophic lateral sclerosis: a review. J Neurol Sci. 2019;399:217–226. [DOI] [PubMed] [Google Scholar]
- 13.Mejzini R, Flynn LL, Pitout IL, Fletcher S, Wilton SD, Akkari PA. ALS genetics, mechanisms, and therapeutics: where are we now? Front Neurosci. 2019;13:1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rich KA, Roggenbuck J, Kolb SJ. Searching far and genome-wide: the relevance of association studies in amyotrophic lateral sclerosis. Front Neurosci. 2021;14:603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marangi G, Traynor BJ. Genetic causes of amyotrophic lateral sclerosis: new genetic analysis methodologies entailing new opportunities and challenges. Brain Res. 2015;1607:75–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chia R, Chiò A, Traynor BJ. Novel genes associated with amyotrophic lateral sclerosis: diagnostic and clinical implications. Lancet Neurol. 2018;17(1):94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosen DR, Siddique T, Patterson D, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362(6415):59–62. [DOI] [PubMed] [Google Scholar]
- 18.Hayashi Y, Homma K, Ichijo H. SOD1 in neurotoxicity and its controversial roles in SOD1 mutation-negative ALS. Adv Biol Regul. 2016;60:95–104. [DOI] [PubMed] [Google Scholar]
- 19.Rutherford NJ, Zhang YJ, Baker M, et al. Novel mutations in TARDBP (TDP-43) in patients with familial amyotrophic lateral sclerosis. PLoS Genet. 2008;4(9):e1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kabashi E, Valdmanis PN, Dion P, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40(5):572–574. [DOI] [PubMed] [Google Scholar]
- 21.Sreedharan J, Blair IP, Tripathi VB, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319(5870):1668–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Deerlin VM, Leverenz JB, Bekris LM, et al. TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis. Lancet Neurol. 2008;7(5):409–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pesiridis GS, Lee VM, Trojanowski JQ. Mutations in TDP-43 link glycine-rich domain functions to amyotrophic lateral sclerosis. Hum Mol Genet. 2009;18(R2):R156–R162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ling JP, Pletnikova O, Troncoso JC, Wong PC. TDP-43 repression of nonconserved cryptic exons is compromised in ALS-FTD. Science. 2015;349(6248):650–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwiatkowski TJ, Bosco DA, LeClerc AL, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323(5918):1205–1208. [DOI] [PubMed] [Google Scholar]
- 26.Vance C, Rogelj B, Hortobágyi T, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323(5918):1208–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shang Y, Huang EJ. Mechanisms of FUS mutations in familial amyotrophic lateral sclerosis. Brain Res. 2016;1647:65–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Renton Alan E, Majounie E, Waite A, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72(2):257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeJesus-Hernandez M, Mackenzie Ian R, Boeve Bradley F, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72(2):245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahoney CJ, Beck J, Rohrer JD, et al. Frontotemporal dementia with the C9ORF72 hexanucleotide repeat expansion: clinical, neuroanatomical and neuropathological features. Brain. 2012;135(pt 3):736–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Majounie E, Renton AE, Mok K, et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 2012;11(4):323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Byrne S, Elamin M, Bede P, et al. Cognitive and clinical characteristics of patients with amyotrophic lateral sclerosis carrying a C9orf72 repeat expansion: a population-based cohort study. Lancet Neurol. 2012;11(3):232–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ringholz GM, Appel SH, Bradshaw M, Cooke NA, Mosnik DM, Schulz PE. Prevalence and patterns of cognitive impairment in sporadic ALS. Neurology. 2005;65(4):586–590. [DOI] [PubMed] [Google Scholar]
- 34.Mackenzie IR, Rademakers R. The role of transactive response DNA-binding protein-43 in amyotrophic lateral sclerosis and frontotemporal dementia. Curr Opin Neurol. 2008;21(6):693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Da Cruz S, Bui A, Saberi S, et al. Misfolded SOD1 is not a primary component of sporadic ALS. Acta Neuropathol. 2017;134(1):97–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keller BA, Volkening K, Droppelmann CA, Ang LC, Rademakers R, Strong MJ. Co-aggregation of RNA binding proteins in ALS spinal motor neurons: evidence of a common pathogenic mechanism. Acta Neuropathol. 2012;124(5):733–747. [DOI] [PubMed] [Google Scholar]
- 37.Couthouis J, Hart MP, Shorter J, et al. A yeast functional screen predicts new candidate ALS disease genes. Proc Natl Acad Sci U S A. 2011;108(52):20881–20890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Couthouis J, Hart MP, Erion R, et al. Evaluating the role of the FUS/TLS-related gene EWSR1 in amyotrophic lateral sclerosis. Hum Mol Genet. 2012;21(13):2899–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim HJ, Kim NC, Wang YD, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495(7442):467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Registry P, Group S, Registry S, et al. Genome-wide association analyses identify new risk variants and the genetic architecture of amyotrophic lateral sclerosis. Nat Genet. 2016;48(9):1043–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakamura R, Misawa K, Tohnai G, et al. A multi-ethnic meta-analysis identifies novel genes, including ACSL5, associated with amyotrophic lateral sclerosis. Commun Biol. 2020;3(1):526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benyamin B, He J, Zhao Q, et al. Cross-ethnic meta-analysis identifies association of the GPX3-TNIP1 locus with amyotrophic lateral sclerosis. Nat Commun. 2017;8(1):611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Es MA, van Vught PW, Blauw HM, et al. Genetic variation in DPP6 is associated with susceptibility to amyotrophic lateral sclerosis. Nat Genet. 2008;40(1):29–31. [DOI] [PubMed] [Google Scholar]
- 44.van Es MA, Veldink JH, Saris CGJ, et al. Genome-wide association study identifies 19p13.3 (UNC13A) and 9p21.2 as susceptibility loci for sporadic amyotrophic lateral sclerosis. Nat Genet. 2009;41(10):1083–1087. [DOI] [PubMed] [Google Scholar]
- 45.van Rheenen W, van der Spek RAA, Bakker MK, et al. Common and rare variant association analyses in amyotrophic lateral sclerosis identify 15 risk loci with distinct genetic architectures and neuron-specific biology. Nat Genet. 2021;53(12):1636–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nicolas A, Kenna KP, Renton AE, et al. Genome-wide analyses identify KIF5A as a novel ALS gene. Neuron. 2018;97(6):1268-1283, e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu CH, Fallini C, Ticozzi N, et al. Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature. 2012;488(7412):499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fujimoto A, Furuta M, Totoki Y, et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet. 2016;48(5):500–509. [DOI] [PubMed] [Google Scholar]
- 49.Valdmanis PN, Belzil VV, Lee J, et al. A mutation that creates a pseudoexon in SOD1 causes familial ALS. Ann Hum Genet. 2009;73(6):652–657. [DOI] [PubMed] [Google Scholar]
- 50.Neumann M, Sampathu DM, Kwong LK, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314(5796):130–133. [DOI] [PubMed] [Google Scholar]
- 51.Szklarczyk D, Gable AL, Nastou KC, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49(D1):D605–D612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morgan S, Orrell RW. Pathogenesis of amyotrophic lateral sclerosis. Br Med Bull. 2016;119(1):87–98. [DOI] [PubMed] [Google Scholar]
- 53.Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cel Biol. 2017;18(5):285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blokhuis AM, Groen EJ, Koppers M, van den Berg LH, Pasterkamp RJ. Protein aggregation in amyotrophic lateral sclerosis. Acta Neuropathol. 2013;125(6):777–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Project MinE ALS Sequencing Consortium. Project MinE: study design and pilot analyses of a large-scale whole-genome sequencing study in amyotrophic lateral sclerosis. Eur J Hum Genet. 2018;26(10):1537–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang S, Cooper-Knock J, Weimer AK, et al. Genome-wide identification of the genetic basis of amyotrophic lateral sclerosis. Neuron. 2022;110(6):992–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klim JR, Williams LA, Limone F, et al. ALS-implicated protein TDP-43 sustains levels of STMN2, a mediator of motor neuron growth and repair. Nat Neurosci. 2019;22(2):167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Theunissen F, Anderton RS, Mastaglia FL, et al. Novel STMN2 variant linked to amyotrophic lateral sclerosis risk and clinical phenotype. Front Aging Neurosci. 2021;13:658226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ma XR, Prudencio M, Koike Y, et al. TDP-43 represses cryptic exon inclusion in FTD/ALS gene UNC13A. Nature. 2021;603(7899):124–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Christiansen EM, Yang SJ, Ando DM, et al. In silico labeling: predicting fluorescent labels in unlabeled images. Cell. 2018;173(3):792–803, e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Final supplemental data available at: links.lww.com/NXG/A518. Additional eReferences e1-e95 available at: links.lww.com/NXG/A519.