Abstract
In 2011, a pathogenic hexanucleotide repeat expansion in the C9ORF72 gene was discovered to be the leading genetic cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). Before this, the C9ORF72 gene and its protein were unknown. The repeat expansion was found to cause both haploinsufficiency and gain of toxicity through aggregating RNA products and dipeptide repeat proteins. A worldwide effort was then initiated to define C9ORF72 ALS/FTD and unravel the pathogenic mechanism for the development of therapeutic options. A decade later, C9ORF72 genetic testing is readily available. There is now an increasing appreciation that C9ORF72 not only is the leading genetic cause of ALS/FTD but may contribute to a spectrum of disorders. This article reviews what is currently known about the C9ORF72 expansion and how C9ORF72 expansion manifests in ALS, FTD, psychiatric disorders, and movement disorders. With therapeutic strategies fast approaching the clinic, earlier recognition of possible C9ORF72 expansion related disorders is even more paramount to improve patient care.
Before the identification of the C9ORF72 gene in 2011, it had been known for some time that a gene or genes associated with frontotemporal dementia (FTD)-amyotrophic lateral sclerosis (ALS) resided on human chromosome 9 using genetic linkage analysis in pedigrees segregating ALS or FTD or both phenotypes.1,2 Mok et al.3 described a ∼140-kb risk haplotype on chromosome 9p21 that was shared by chromosome 9p-linked families and showed significant association with FTD and ALS in several populations. The presence of the same haplotype across multiple families suggested that most individuals carried the same pathogenic variant. Finding the haplotype in individuals with ALS or FTD also supported the notion that the same mutation could cause disparate phenotypes.
Two groups identified the disease-causing chromosome 9p mutation using different approaches and succeeded simultaneously. The collaborative group led by Rademakers performed detailed sequencing of the C9ORF72 gene and identified a polymorphic GGGGCC (G4C2) hexanucleotide repeat (HRE) located between the 2 noncoding C9ORF72 exons 1a and 1b.4 Analysis of this HRE in samples from members of a family with autosomal dominant ALS/FTD showed an aberrant segregation pattern with all affected individuals appearing homozygous. They suspected that lack of segregation was due to the presence of an unamplifiable repeat expansion and developed a repeat-primed PCR (RP-PCR) method (see below) designed to detect expanded G4C2 repeats. The presence of repeat expansions was detected by RP-PCR in all affected family members, but not in unaffected relatives, which was subsequently confirmed by Southern blot analysis. The maximum size of the repeat in controls was 23 units, whereas in affected individuals, it was estimated at 700 or more repeats.
The collaborative group led by Traynor used an approach that emphasized enrichment of the chromosome 9p region followed by massive parallel sequencing.5 Enrichment techniques encompassed flow sorting of chromosome 9 or enrichment of the target region using 43,000 custom oligonucleotide baits covering a total of 2.58 MB in the chromosome 9p FTD/ALS locus. Subsequent manual alignment of sequence reads in the target region identified the HRE in the C9ORF72 gene. Defining a pathogenic expansion as >30 repeats, they found that 113 of 402 ALS cases and 2 of 478 controls from Finland had an expanded repeat using rp-PCR analysis. None of the African samples carried >15 G4C2 repeats (average: 3; range: 0–15) providing the first suggestion that repeat alleles may differ in specific populations. In Finish familial ALS cases, 52 (46.4%) had the expansion. Independently confirming the pathogenic role of the G4C2 repeat in FTD, they also found a 29% occurrence of expanded repeats in Finnish FTD cases. Indeed, C9ORF72-ALS frequency varies widely based on ethnicity and geographic region and is most common in European and North American populations, accounting for about 40% of ALS cases with a family history of ALS and 8% of ALS without a family history6; in contrast, it is rarely found in Asian populations.7-10 A recent study in 140 South Africans with sporadic ALS identified 10 cases (7%) of pathogenic C9ORF72 expansions: 4 White and 6 Cape mixed Africans—ancestry derived from European settlers, Indigenous tribes, and Indian Islanders, among others.11 This regional variance, increased risk among those with a family history—along with the common haplotype—suggests a single common founder, who originated from Finland.12,13 Modeling estimates have dated this founder to around 500 AD, which closely coincides with the beginning of the Viking invasions from Finland. Alternatively, a competing hypothesis is that the identified common haplotype contributes instability to the C9ORF72 region; thus, the region itself predisposes to the creation of expansion.14 A recent study of 593 patients with ALS in India discovered the presence of C9 expansion in 3.2% (19/593) of patients with ALS where 47.4% (9/19) positive cases belonged to the eastern region of India. The common haplotype was present in 11 of the 19 cases.15 Although pathogenic C9 expansions appear to be most prevalent in individuals of European ancestry, recent work supports the need for increased testing in more diverse populations.11,15,16
Pathogenesis of C9ORF72 Expansion
C9ORF72 Expansion and Morphologic Findings
Mechanisms of neuronal dysfunction in the setting of C9ORF72 expansion are currently not clear. Pathologically, in most, but not all, patients with C9ORF72 ALS/FTD, aggregation of TAR DNA-binding protein 43 (TDP-43) occurs in clinically correlated cortical and spinal motor neurons and the frontal cortex.17 The histopathologically unique TDP-43 subtypes A and B are the predominant inclusions found in C9ORF72 patients. TDP-43 is primarily nuclear; however, in patients with ALS/FTD, posttranslationally altered TDP-43 proteins are found to aggregate the cytoplasm. A pathologic link between abnormal TDP-43 and C9ORF72 expansion is currently under extensive investigation. The collaborative study group led by Rothstein recently provided evidence that nuclear accumulation of CHMP7, a critical mediator of nuclear pore complex (NPC) function, is sufficient to cause TDP-43 mislocalization to the cytoplasm and subsequent dysfunction. Impaired nuclear export of CHMP7 led to reduced expression of NPC components and contributed to TDP-43 mislocalization, dysfunction, and downstream deficits in neuronal survival. CHMP7 expression was also found to be elevated in postmortem neuronal tissue of patients with C9ORF72 ALS.18
Approximately 50%–60% of patients with ALS will develop frontotemporal dysfunction, ranging from dementia (FTD) to one or more problems with neuropsychological, language, or speech function. These are collectively known at the frontotemporal spectrum disorders of ALS and are discussed in greater detail below. Anatomically, sporadic FTD and C9ORF72-FTSD have similar morphologic changes in the brain, with atrophy of the anterior insula, anterior cingulate, amygdala, and striatum; those with C9ORF72-FTSD, however, display more involvement of the medial pulvinar thalamus. Increased cerebellar atrophy has also been shown in C9ORF72-FTSD and ALS, though less consistently.17
Pathogenesis and Allele Size
There is debate surrounding both the pathogenic allele size for C9ORF72-ALS and the correlation of size of the expansion and disease severity. Pathologic expansions are typically hundreds to thousands of repeats in length, although disease has been seen with smaller expansions. Alleles with greater than 30 repeats are generally considered pathogenic for C9ORF72-ALS5; yet, although relatively rare, intermediate-length alleles of 20–30 repeats may increase the risk for C9ORF72-ALS19-21 and parkinsonism.22 C9ORF72 expansion size has been suggested to correlate with age at onset of ALS23,24—an association common in many neurologic repeat expansion diseases—other studies, however, have failed to demonstrate this.10,25 One of the confounding factors for both of these debates is the somatic instability of the expansion, specifically that the repeat size is highly variable from tissue to tissue, often with a significant increase in size in the CNS in comparison to blood-derived DNA, which most studies are based on.26 A second factor is the variety of testing modalities that make determining the size of the expansion often unreliable or incomparable.
The secondary structure of the C9ORF72 expansion has been shown to contribute to expansion and contraction of repeat lengths27 and cause cell-to-cell transmission of dipeptide repeat proteins (DPRs),28 which are found in aggregates throughout the CNS of C9ORF72 patients.29,30 Conceivably, then, just a small population of cells with pathogenic C9ORF72 expansions could be present in the CNS and initiate the disease process. Such somatic variations would be missed by routine blood DNA testing that evaluates germline cells. A recent report found no evidence of C9ORF72 expansion in spinal cord tissue of patients with ALS negative for C9ORF72 expansions31 providing further support that normal C9ORF72 expansions are stable26 and expansion instability occurs only when restricted to expanded alleles.32 Repeat length variability in tissues of patients with very large C9ORF72 expansions has been reported,26 but as these studies suggest pathogenic expansions likely occur in most if not all cells. It remains unclear whether an exact number of repeats trigger disease or whether other factors are more important than repeat length.
Pathophysiology of C9ORF72 Expansion
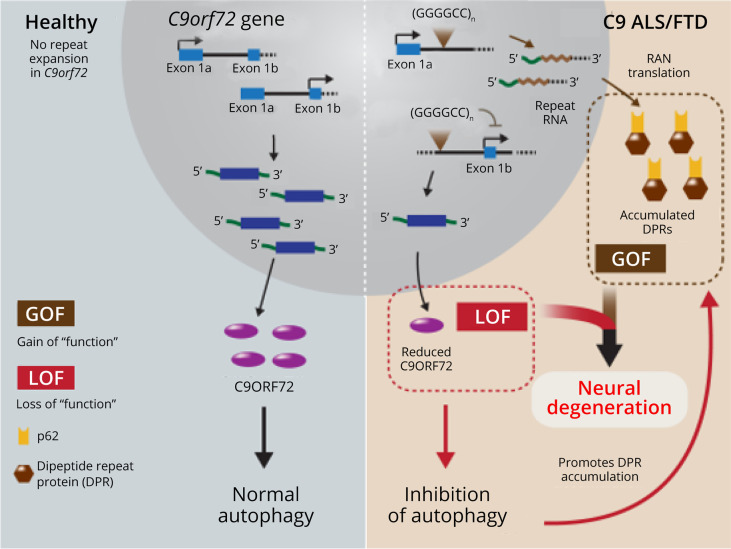
Several mechanisms are hypothesized to explain C9ORF72 expansion pathogenesis, with RNA toxicity and repeat-associated non-ATG (RAN) translation being the 2 major ones (Figure 1B and Figure 1C), and loss of C9ORF72 function likely of lesser importance (Figure 1D). C9ORF72 transcript levels and protein expression are decreased in patients with C9ORF72 ALS/FTD,4,33,34 yet neurodegenerative features are not found in loss of function models.35-37 C9ORF72 expansion occurs within an intron and therefore undergoes canonical transcription. Toxic gain of function occurs when expansion RNA transcripts block the transcription of other genes by sequestering RNA-binding proteins.38-41 Products from RAN translation of C9ORF72 expansion can also exert deleterious effects on cellular processes. First discovered in spinocerebellar ataxia type 8 and myotonic dystrophy (DM1), RAN translation is now known to occur in many microsatellite expansion–related diseases, including C9ORF72-ALS/FTD.29,42-44 RAN translation begins in hairpin forming expansion regions independent of the canonical ATG start codon. Multiple RAN products can be produced from different reading frames as expansion mutations are often bidirectionally transcribed.42,45 RAN proteins can cause cytoplasmic mislocalization and block splicing of genes related to mitochondrial, neuronal, and pre-mRNA splicing function.46 In addition, they can disrupt the ubiquitin proteosome system, form aggregates trapping macromolecular complexes, and cause vesicle and endoplasmic reticulum membrane deformation.47,48 The contribution of individual RAN proteins to disease is unclear. C9ORF72-RAN protein expression is toxic in zebrafish49 and induces neurodegeneration in mice.47 In particular, the arginine-containing proteins (polyPR and polyGR) show the greatest toxicity.38,50 These C9ORF72-RAN toxicity studies used repeat lengths of fewer than 90 units, whereas human C9ORF72 disease often involves repeat lengths in the hundreds to thousands. Continued investigation is necessary to better define the structure and pathogenesis of C9ORF72-RAN proteins.
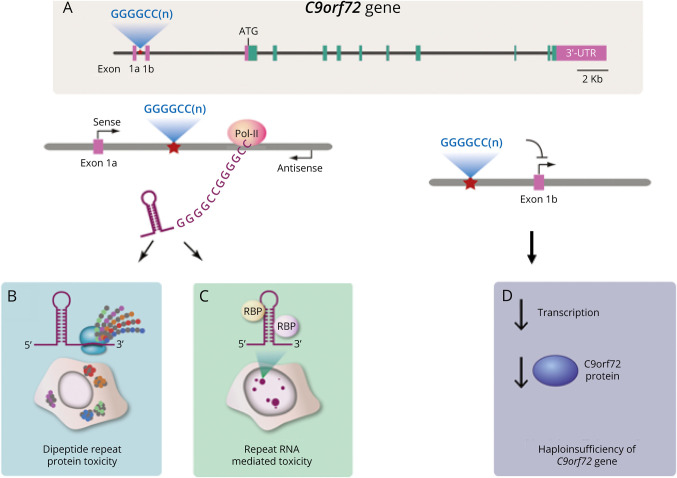
Figure 1. Proposed Mechanisms of C9ORF72 Expansion.
(A) The C9ORF72 gene contains a polymorphic hexanucleotide (GGGGCC) repeat in a noncoding region. (B) RNA transcripts with the C9ORF72 repeat expansions are produced by both sense and antisense transcription, resulting in the accumulation of nuclear and cytoplasmic aggregation of both sense and antisense repeat-containing RNA. Repeat-containing RNA can cause sequestration of essential RNA-binding proteins leading to defects in pre-mRNA splicing. (C) Expansion repeat is a substrate for non–ATG-dependent translation event that generates dipeptide repeat proteins that cause toxicity through aggregation and altering multiple essential cellular functions. (D) Repeat expansion can interfere with transcription and cause downregulation of C9ORF72 gene expression and loss of C9ORF72 protein function. Adapted from Gitler et al. There has been an awakening: Emerging mechanisms C9orf72 mutations in FTD/ALS. Brain Res. 2016;1647.
C9ORF72 Expansion and Nuclear Transport
Compositional changes to the NPC occur early in C9ORF72 disease with associated reduction in NPC expression and cytoplasmic mislocalization of componentry.51 Critical in maintaining overall cellular function, disruption of the NPC and nuclear export influences neuronal viability and is a common feature in neurodegenerative disorders. C9ORF72 expansion disrupts nucleocytoplasmic transport either via direct interaction of C9ORF72 RNA fragments with nuclear pore proteins or through RAN protein disruption of the NPC. PolyPR-containing RAN proteins bind to regions enriched in phenylalanine/glycine repeats within the complex, thereby plugging the pore and blocking nuclear export.52 Whether nuclear transport deficits are a cause or consequence of other cellular events in C9ORF72 disease is actively being investigated.
C9ORF72 Testing Methods
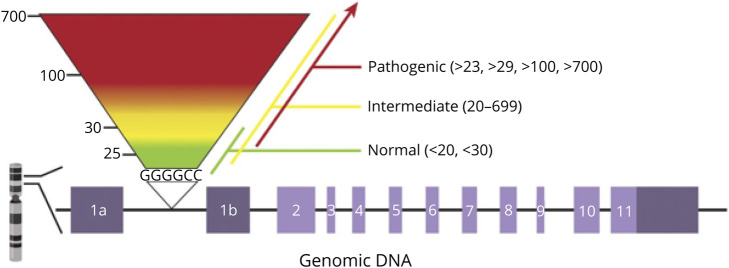
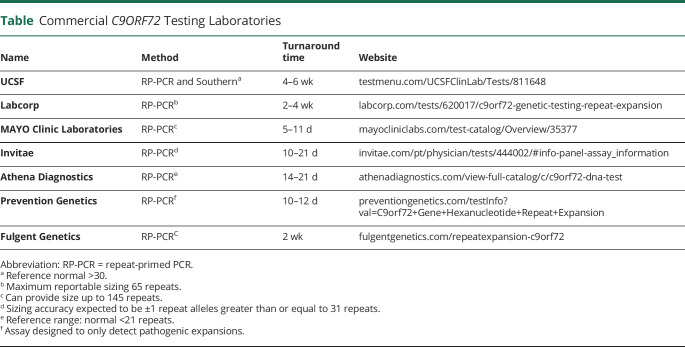
Clinical testing for C9ORF72 has been available since 2012.53 Reliable and consistent analysis of the C9ORF72 expansion across multiple laboratories, however, has proven to be problematic with a high incidence of both false-positive and negative test results.54 The characterization of the C9ORF72 expansion is complex due to its GC-rich DNA region and the presence of sequence variations at the 3′ end of the repeat region.55 This creates a challenge for utilization of conventional PCR-based fragment analysis, and therefore, at a minimum, a combination of gene-specific and RP-PCR is needed for the assessment of large repeat expansions. RP-PCR was first developed in 1996 as an approach to detect the CAG repeat expansion in the DMPK gene that causes DM1.56 RP-PCR is essentially a 3-primer PCR, where one primer flanks the repeat, another is complementary to the repeat with a tail, and a third binds within the repeat region. This generates a heterogeneous mixture of fragments differing in size by 1 repeat that are then analyzed via capillary electrophoresis (CE) (Figure 2). Because of CE size constraints, the maximum fragment size that can be analyzed by PCR is 145 repeats in length, anything larger must be evaluated with Southern blot. The capability to size larger repeats makes Southern blot the advocated gold standard; however, it is may not detect small or intermediate expansion, and few laboratories offer it.57 Interpretation of expansion size is inconsistent among laboratories with cutoffs for normal, intermediate, and expanded alleles varying significantly.58 Most laboratories do not report allele sizes within the normal range due to inability to detect fewer than 25, and what constitutes a pathogenic allele size may vary from lab to lab. (Figure 3). Amplicon-length analysis using primers flanking the repeat motif can resolve ambiguous cases where sequence variants interfere with PCR results. Combining amplicon-length analysis and bidirectional 5′ and 3′ RP-PCR is recommended for greatest sensitivity and specificity.54,55 Several laboratories offering certified C9ORF72 testing are listed in the Table.
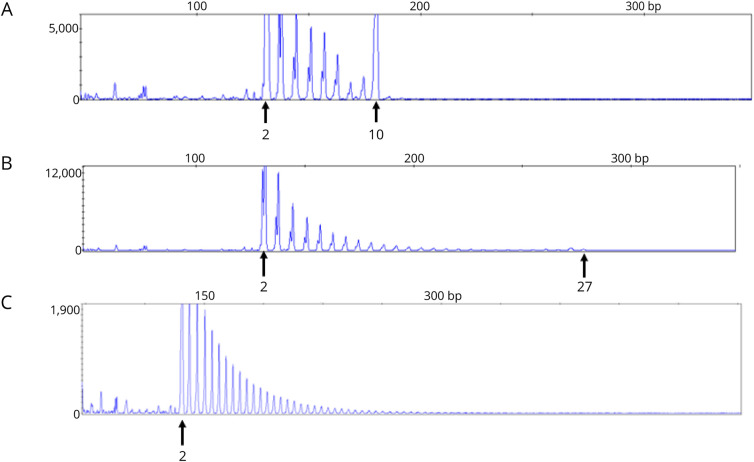
Figure 2. Variable.
(A) RP-PCR/CE representative profile for a heterozygote normal range repeats denoted by the arrows indicate repeat with corresponding size below (2 and 10). (B) RP-PCR/CE fragment representative of a normal expanded repeats of (2 and 27), note the diminishing signal peak as the repeat becomes larger. (C) RP-PCR/CE representative of an expanded repeat greater than 30.
Figure 3. Variable Interpretations of the C9orf72 Hexanucleotide Repeat Number Among Commercial Laboratories.
Jennifer Roggenbuck. Neurol Genet. 2021;7:e542.
Table.
Commercial C9ORF72 Testing Laboratories
Recommendations for C9ORF72 Genetic Testing
Clinician surveys reveal a growing consensus (∼93%) to offer genetic testing to patients with familial ALS.57 Whether to extend testing in cases of sporadic ALS is less clear. With the growing availability of clinical trials for C9ORF72-ALS, and very recently FTD, testing should be offered to any patient diagnosed with either condition, particularly of European ancestry.57 Recent studies identified the C9ORF72 expansion as accounting for 75%–79% of pathogenic variants in ALS. It is becoming increasingly evident, therefore, that at least in US clinics, the prevalence of pathogenic variants in genes other than C9ORF72 may be much lower than originally thought.16 Presymptomatic testing may be considered for adult relatives of patients with C9ORF72 disease, but only when combined with extensive genetic counseling and psychological assessment. The limitations of testing should be emphasized as the penetrance of C9ORF72-related disease remains unclear. A few studies report a penetrance of 80%–90% by age 80 years, yet others report a much lower rate.6,59,60 Clinical guidelines for C9ORF72 testing in both sporadic and familial ALS/FTD might allow us to better understand the incidence of C9ORF72 expansion in our diverse population and guide decision making for testing affected persons of non-European descent, in which the incidence rates are likely significantly lower.
C9ORF72 ALS
Clinically, patients with C9ORF72 ALS are relatively indistinguishable from nonexpanded patients, with a few exceptions. One of the most obvious is the increased risk among patients with a family history of ALS or FTD, particularly in those of Caucasian ethnicity. A second exception is the coexistence of cognitive and behavioral impairment—most commonly behavioral variant FTD (FTDbv), which is seen in about 50%.e1-e3 There is a reported increased risk of bulbar-onset ALS (up to 40%),e2,e3 although this has not been supported in all cohorts.10,e4 The age at onset ranges from the 2nd to the 9th decades, with a peaked age-related prevalence in the 5th decade.59 Penetrance is high, although incomplete, and is estimated to be 80%–90% by age 80 years.6,e5 Duration of disease (from onset to death) is comparable to that of patients with ALS without C9ORF72 expansions, although on average, death occurs earlier by 1.8 years.e3
Nonmotor Symptoms in ALS
The most common nonmotor symptom in patients with ALS is bvFTD. The other FTD subtypes, semantic and progressive aphasia, are uncommon in patients with ALS.e6 FTDbv is diagnosed clinically based on established criteria and is characterized by progressive deterioration of personality, social norms, and cognition.e7 Ultimately, FTD is caused by frontotemporal lobar degeneration (FTLD), which is a pathologic diagnosis based on the presence of proteinaceous inclusions containing either tau or TDP-43 proteins found in neural tissue at autopsy. Frontotemporal lobe spectrum disorder (FTSD) represents a lesser degree of symptoms, manifesting as apathy, disinhibition, compulsivity, loss of empathy, and executive dysfunction.e8,e9 Screening tests for elements of FTD/FTSD have been developed for use in ALS clinics.e10,e11
Among patients who present with dementia and fulfill the criteria for FTD, about 15% also have motor features consistent with ALS or develop such over a short time.e12 Among patients with ALS, a lesser percentage fulfill the criteria for FTD, and a larger number have elements of FTSD—in 274 patients with ALS assessed with a clinical screening tool, 6.5% had a score consistent with FTD and 54.2% consistent with FTSD.e13 FTSD symptoms usually begin a few years before onset of weakness but are frequently unrecognized by the family.
Patients with ALS can develop psychiatric disease including mood (13.7%) and neurotic (6.2%) disorders and rarely personality disorders and schizophrenia.e14 Schizophrenia in the form of a single, late-onset psychotic episode occurring 1–5 years before onset of ALS although has been observed in population-based studies. Other symptoms include drug use/abuse and suicide. All are present a long period before onset of weakness.e15 Similarities also exist between symptoms of bvFTD and those in autism spectrum disorder (ASD), including social isolation, obsessive compulsion, inflexibility, and stereotypy of speech.e16
Whether there is an increased incidence of nonmotor symptoms in patients with ALS and relatives with C9ORF72 expansions, particularly psychiatric symptoms, is difficult to answer with precision. Although C9ORF72 expansion is the most common genetic abnormality associated with ALS, the percentage is only moderately high and varies among populations; thus, the number of patients and families available for study is relatively small.e14,e17 C9ORF72 expansion size varies among patients with ALS. There is also variable expression of expansion size within different tissues from a patient (blood and brain). There does not appear to be a phenotypic correlation with expansion number and ALS or FTD/FTSD.17
When investigating the incidence of dementia in family members, the type of dementia is not commonly assessed rigorously for type. In both patients and family members, psychiatric symptoms can be missed as they are frequently not diagnosed. Furthermore, the incidence of any psychiatric disorder in the general population is relatively high, and methods of query for these disorders vary, making comparisons among studies and robust generalizations difficult.
C9ORF72 Expansions in Patients With ALS and FTD/FTSD and Families
Nonmotor symptoms occur in patients with ALS with and without C9ORF72 expansions. In a study of 60 patients with ALS and FTD, ∼20% had repeat expansions, and 80% had normal repeat numbers.e18 The associations below are considered statistically significant.
An Australian study involving 89 patients with the diagnosis of FTD and ALS and 1,414 first- and second-degree relatives found the following associations: among patients with FTD, 32% had a family member with ASD, whereas only 9% with ALS had an affected family member. Comparing other psychiatric disorders, in patients with FTD or ALS, there were no differences in the percentages of family members with a psychotic, mood, or bipolar disorder or risk of suicide. Family members of patients with FTD and patients with ALS were more likely to have any psychiatric disorder if the family had C9ORF72 expansions.e19
In a study of 127 patients with ALS and 2,116 first- and second-degree relatives from an Irish ALS registry, the following were noted: 61% of patients with ALS reported greater than one first- and second-degree relative with psychiatric symptoms, in contrast to 38.6% in a control population. Among families with greater than 3 affected family members, the diagnostic breakdown was schizophrenia, suicide, autism, and alcoholism. All patients with ALS with a C9ORF72 expansion had 1 family member with psychiatric diagnoses, and in those with 3 or more affected members with psychiatric diagnoses, only 20.7% had an expansion. The data support a strong connection between ALS and psychiatric disorders, but the presence of a C9ORF72 expansion did not associate with greater numbers of affected family members. Of note, depression was not overrepresented in patients with ALS.e16
A study based on a Scottish national motor neuron disease (ALS) registry used a uniform 4-generation family history to assess dementias and psychiatric disorders in the patient and family and also included assessment for frontotemporal lobe dysfunction.e9 Determination of C9ORF72 repeat expansion was available for 84.9% (256/305), and 9.3% had disease-causing expansions. Among the patients with ALS, FTD was identified in 5.6% and FTSD in 51.1%, with apathy the most common behavioral change in 29.7%. A family history of dementia was reported in 29% (76/266). The above associations were not linked with C9ORF72 status, and cognitive and psychiatric features in patients and family members were felt to represent a general overlap or continuum with these conditions and ALS.
Movement Disorders
Initial studies of patients with C9ORF72 ALS/FTD noted the presence of additional motor symptoms with parkinsonian features, suggesting that C9ORF72 expansion could cause a more heterogeneous syndrome.e20-e22 Movement disorders occur frequently in patients with C9ORF72 expansion and can precede the diagnosis of ALS/FTD. Movement disorders can also be the initial or sole manifestation of C9ORF72 expansion.e23 Parkinsonism is a prevalent (∼40%) feature of C9ORF72-ALS/FTD, particularly in those with bvFTD where prevalence can increase to 75%.e20 C9ORF72 expansion–associated movement disorders are clinically heterogeneous and frequently found in combination. Tremor and parkinsonism were the most frequent movement disorders reported in a recent cohort study.e23 Although there are no consensus guidelines, several studies provide support for testing for C9ORF72 expansion in persons presenting with parkinsonism, tremor, distal myoclonus, cervical dystonia, and chorea with orofacial involvement in the setting of familial neurodegenerative conditions.
C9ORF72 Expansion Carriers, Typical Parkinson Disease, and Parkinsonism
There is no evidence that C9ORF72 positivity (as defined by >60 repeat units) contributes a significant risk for Parkinson disease (PD). Yet, several studies have shown an increased risk associated with intermediate repeat expansions (20–30 units).e24,e25 The clinical significance of this finding is limited due to the relatively small number of cases, and further studies are needed to explore this correlation. Likely, there are disease-modifying genetic and environmental factors present that may partially explain the inconclusive link between C9ORF72 expansion number and PD. Of interest, patients with PD with intermediate expansions appear to be quite Dopa responsive, which is not the case in most C9ORF72 patients harboring larger expansions and displaying parkinsonian features as part of a more complex phenotype.e20,e24,e26
Patients with C9ORF72-FTD/ALS who develop parkinsonism often present with the typical symptoms of asymmetric-onset akinetic rigidity with prominent bradykinesia and little or no tremor.e20,e28 On average, patients with C9ORF72-FTD/ALS with parkinsonism present at a younger age (∼52 years), with parkinsonian signs typically manifesting in the first 2 years of disease and even earlier in familial FTD/ALS cases.4,e20,e28 In a review of 45 C9ORF72 patients with parkinsonism, the syndrome consisted of hypokinetic rigidity without resting tremor (61%) with both asymmetric (59%) and symmetric (41%) distributions. Additional features included upper motor neuron signs (60%), lower motor neuron signs (36%), cognitive dysfunction (85%), behavior/personality changes (55%), and psychiatric symptoms (29%). Family history yielded evidence of ALS (31%) and FTD (21%).e28
Among the few histopathologically confirmed studies, the presence of parkinsonism in C9ORF72 patient correlated with TDP-43 and p62 pathology within the basal ganglia. Across 2 large studies of autopsy-proven PD (pooled sample size >800 patients), just 1 C9ORF72 expansion carrier (>30 units) had both typical PD (diffuse Lewy bodies throughout the neocortex and TDP-43 pathology with FTLD) and C9ORF72-mediated pathology consisting of p62-positive, TDP-43–negative neuronal cytoplasmic inclusions in the hippocampus and cerebellar granule cells.e26,e29 The distinct pathology in the substantia nigra in these cases suggests a unique mechanism for C9ORF72-mediated parkinsonism.e29,e30
C9ORF72 and Atypical Parkinsonian Syndromes
The prevalence of C9ORF72 expansion in atypical parkinsonian syndromes is difficult to determine based on only a few case reports and small cohort studies. C9ORF72 expansion has been found in patients with dementia with Lewy bodies, corticobasal syndrome, and progressive supranuclear palsy. It has been suggested, however, that these cases constitute incomplete diagnoses as ALS/FTD signs are often absent in the beginning.e27,e28,e31
C9ORF72 expansion is now recognized as the leading genetic cause of Huntington-like (Huntington disease [HD]-like) disease, which is clinically indistinguishable from HD but without the diagnostic CAG repeat expansion in the HTT gene.e32,e33 Further neuropathologic studies elucidating whether C9ORF72-mediated pathology occurs in HD-related brain regions are needed; however, there does not appear to be a causal relationship between C9ORF72 expansion and HD. That C9ORF72 expansion can cause HD-like disease does support the findings that atypical movement disorders including chorea, dystonia, tremor, myoclonus, and rigidity can be present among C9ORF72 expansion carriers.e33 With limited cases, it is unclear if there is age-related penetrance or disease anticipation. It is feasible that even intermediate-length C9ORF72 expansions can interact with repeats of other genes, thereby altering the phenotypic presentation of HD and other repeat disorders.e34-e37 Conversely, the repeats of other disease-causing expansions can interact with and influence the C9ORF72 phenotype.e36
C9ORF72 Mouse Models
There are several mouse models probing the pathogenic mechanism of C9ORF72 expansion. As discussed previously, the pathogenesis of C9ORF72 expansion is thought to be due to loss of C9ORF72 function, gain of toxicity from expansion RNA, or via products of RAN translation. Likely, there is a synergetic effect among these processes, which may also be disease or tissue specific given the heterogeneity that is observed across similar repeat lengths.
Gain of Toxicity Models
Transgenic mouse models carrying the intronic C9orf72 expansion have been generated by several groups.35,e38,e39 Although all develop molecular characteristics of ALS/FTD, only 2 models reported behavioral and pathologic phenotypes.35,e40 The model backgound appears to exert influential effects on phenotype. In the C57 black 6 (C57Bl/6) background C9orf72 expansion resulted in age-dependent cognitive deficits and mild hippocampal neuronal loss at 12 months. Conversely, on the Friend Virus B NIH Jackson (FVB/NJ) background C9orf72 expansion resulted in length and dose-dependent phenotypes, including motor dysfunction and ALS/FTD neurodegenerative features.e40 Consistent with human disease, there is variable penetrance in these animals, with some developing acute disease (∼30%) and others showing milder, slowly progressing phenotypes (∼40%) or no detectable phenotype (∼25%). End-stage, acutely affected mice had severe hippocampal and cortical degeneration, weakness, and paralysis of one or both hindlimbs. Motor neuron loss, neuroinflammation, gait abnormalities, decreased survival, and aggregation of RAN proteins were detectable in both acutely and slowly progressing mice.e40 These mice (FVB C9-500) were deposited at The Jackson Laboratory (JAX) in 2017 and are available to the research community. To note, follow-up studies with these mice have shown disparate phenotypes, including the absence of survival and motor deficits,e41 with this difference being ascribed to differing research methods.e42 A recent study did show a robust ALS/FTD phenotype in this JAX FVB C9-500 cohort that was prevented with an antibody against the poly-GA dipeptide RAN proteins. This study provides support not only for the relevance of this model for study but also that RAN protein pathology is a driving mechanism for C9ORF72 disease.e43 Other groups have now also reported decreased survival and similar findings in these JAX FVB C9-500.e42,e44
Loss of Function Models
Loss of C9orf72 in zebrafishe45 and Caenorhabditis eleganse46 causes motor deficits, whereas there is no neurodegenerative phenotype in mice with reduced or eliminated expression of C9ORF72. Decreased lifespan is seen in these animals resulting from age-dependent abnormalities independent of the nervous system, including splenomegaly and lymph node enlargement.35-37 The selective elimination of C9ORF72 from neurons and glial cells, at least in mice, does not cause motor neuron degeneration, motor deficits, or disease.e47 An unresolved question has been whether reduced C9ORF72 production from the affected allele synergizes with gain of toxicity mechanisms from repeat expansion to drive disease. To better understand this, one groupe48 developed a cohort of mice in which neither, one, or both endogenous C9orf72 alleles are inactivated and express transgenes encoding either 66 repeats or a 450 repeat-containing C9orf72 gene that does not encode the C9ORF72 protein. Reduction or elimination of C9ORF72 protein resulted in early accumulation of DPR proteins, cognitive deficits, and hippocampal neural degeneration. Both genotypes in the reduction or absence of C9ORF72 had reduced lifespan. These studies support the ALS/FTD C9ORF72 model that pathogenesis is a result of reduced C9ORF72 function synergizing with repeat-dependent gain of toxicity (Figure 4).
Figure 4. Synergistic Effect of C9ORF72 Gain of Toxicity and Loss of Function.
Loss of C9orf72 function exacerbates the accumulation of poly-dipeptide repeat proteins and leads to cellular toxicity. Zhu Q, et al. Reduced C9ORF72 function exacerbates gain of toxicity from ALS/FTD-causing repeat expansion in C9orf72. Nat Neurosci. 2020;23(5):615–624.
Clinical Trials
There are several compounds that have entered the pipeline and are progressing toward clinical use. Most of these therapeutics use antisense oligonucleotides (ASOs) targeting the sequences adjacent to the HRE in the first intron of the C9ORF72 pre-mRNA. Doing so can direct degradation of repeat-containing RNAs, without lowering the level of C9ORF72 protein encoding mRNA.35 A phase 1b/2a trial (FOCUS-C9) is currently investigating the safety and tolerability of an ASO selectively targeting transcriptional variants containing the HRE expansion (G4C2), thereby sparing C9ORF72 protein. The study has recently been opened to patients with C9ORF72 ALS/FTD. Preclinical work demonstrated knock down of repeat-containing transcripts in the spinal cord and cortex and knock down of 80% of DPRs in the cortex. These results persisted for at least 6 months. Similarly, Biogen has an ongoing phase 1 trial with BIIB78, another ASO shown to knock down toxic mRNA while preserving normal C9ORF72 protein. Preclinical studies showed that a single IV dose reduced toxic RNA and dipeptide aggregates and attenuated behavioral and cognitive deficits.35
Alector, Inc. recently announced that patients with C9ORF72 ALS have been dosed in a phase 2 study evaluating the safety and efficacy of AL001, a monoclonal antibody designed to elevate progranulin levels in the brain. There is also a phase 2 study enrolling symptomatic patients with C9ORF72 FTD. Progranulin is a key immune modulator in the brain, and low levels of progranulin are associated with TDP-43 aggregation. AL001 has shown encouraging results in patients with FTD who harbor progranulin mutations, increasing progranulin levels and normalizing cerebral inflammatory markers. Theoretically, if inflammation can be normalized and TDP-43 aggregation decreased in patients with C9ORF72 ALS/FTD, this may represent a novel therapeutic approach that is expansion independent.
Highlights
C9ORF72 expansion is the most common genetic cause of ALS/FTD in European and North American populations, with the haplotype likely arising from a single founder. Recent studies, however, highlight the importance of recruiting a more diverse population of affected individuals to better understand the worldwide prevalence and geographic variance.
There is growing recognition that in addition to ALS/FTD, C9ORF72 expansion likely contributes to a spectrum of disorders. Movement disorders, particularly parkinsonism, are frequently observed in C9ORF72 expansion carriers and can precede the onset of ALS/FTD symptoms or present in isolation. While there does not appear to be an association between C9ORF72 pathogenic expansion and Parkinson disease, it is the most frequent phenocopy of Huntington disease.
The pathogenic mechanism of C9ORF72 expansion is actively being explored in various animal models. RNA toxicity and products of RAN translation are thought to be leading contributors; however, recent findings suggest reduction in C9ORF72 synergizes with gain of toxicity from repeat expansion to cause early accumulation of DPR proteins and neural degeneration.
With commercial testing widely available, consensus guidelines would help establish consistent repeat length interpretations and address genetic testing and counseling for patients and families with both familial and sporadic disease. As C9ORF72 expansion targeted therapies are approaching clinical approval, the offer and decision to test is even more important.
Expansion targeting therapies are expected to enter the clinic in the next few years with several studies having recently opened to patients with C9ORF72 ALS/FTD. These opportunities represent an exciting time for the study of C9ORF72 disease and highlight the importance to continue to advance our understanding of the mechanism, prevalence, and natural history of C9ORF72-related disorders.
Glossary
- ALS
amyotrophic lateral sclerosis
- ASD
autism spectrum disorder
- ASO
antisense oligonucleotide
- CE
capillary electrophoresis
- DM1
myotonic dystrophy
- DPR
dipeptide repeat protein
- FTD
frontotemporal dementia
- FTDbv
behavioral variant FTD
- FTLD
frontotemporal lobar degeneration
- FTSD
frontotemporal lobe spectrum disorder
- HD
Huntington disease
- HRE
hexanucleotide repeat
- NPC
nuclear pore complex
- PD
Parkinson disease
- RAN
repeat-associated non-ATG
- rp-PCR
repeat-primed PCR
- TDP-43
TAR DNA-binding protein 43
Appendix. Authors

Contributor Information
Sarah Breevoort, Email: sarah.breevoort@hsc.utah.edu.
Summer Gibson, Email: summer.gibson@hsc.utah.edu.
Karla Figueroa, Email: karlaf@genetics.utah.edu.
Mark Bromberg, Email: mbromberg@hsc.utah.edu.
Study Funding
Supported by grant R37NS033123 from the National Institutes of Neurological Disorders and Stroke.
Disclosure
The authors report no disclosure relevant to this manuscript. Go to Neurology.org/NG for full disclosure.
References
- 1.Vance C, Al-Chalabi A, Ruddy D, et al. Familial amyotrophic lateral sclerosis with frontotemporal dementia is linked to a locus on chromosome 9p13.2-21.3. Brain. 2006;129(pt 4):868-876. [DOI] [PubMed] [Google Scholar]
- 2.Morita M, Al-Chalabi A, Andersen PM, et al. A locus on chromosome 9p confers susceptibility to ALS and frontotemporal dementia. Neurology. 2006;66(6):839-844. [DOI] [PubMed] [Google Scholar]
- 3.Mok K, Traynor BJ, Schymick J, et al. Chromosome 9 ALS and FTD locus is probably derived from a single founder. Neurobiol Aging. 2012;33(1):209-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72(2):245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Renton AE, Majounie E, Waite A, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72(2):257-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Majounie E, Renton AE, Mok K, et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 2012;11(4):323-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ogaki K, Li Y, Atsuta N, et al. Analysis of C9orf72 repeat expansion in 563 Japanese patients with amyotrophic lateral sclerosis. Neurobiol Aging. 2012;33(10):2527-2536. [DOI] [PubMed] [Google Scholar]
- 8.Zou ZY, Li XG, Liu MS, Cui LY. Screening for C9orf72 repeat expansions in Chinese amyotrophic lateral sclerosis patients. Neurobiol Aging. 2013;34(6):1710-1716. [DOI] [PubMed] [Google Scholar]
- 9.Tsai CP, Soong BW, Tu PH, et al. A hexanucleotide repeat expansion in C9ORF72 causes familial and sporadic ALS in Taiwan. Neurobiol Aging. 2012;33(9):2232.e11–2232.e18. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y, Lin Z, Chen X, et al. Large C9orf72 repeat expansions are seen in Chinese patients with sporadic amyotrophic lateral sclerosis. Neurobiol Aging. 2016;38:217.e15–217.e22. [DOI] [PubMed] [Google Scholar]
- 11.Nel M, Agenbag GM, Henning F, Cross HM, Esterhuizen A, Heckmann JM. C9orf72 repeat expansions in South Africans with amyotrophic lateral sclerosis. J Neurol Sci. 2019;401:51-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pliner HA, Mann DM, Traynor BJ. Searching for Grendel: origin and global spread of the C9ORF72 repeat expansion. Acta Neuropathol. 2014;127(3):391-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith BN, Newhouse S, Shatunov A, et al. The C9ORF72 expansion mutation is a common cause of ALS+/-FTD in Europe and has a single founder. Eur J Hum Genet. 2013;21(1):102-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Zee J, Gijselinck I, Dillen L, et al. A pan-European study of the C9orf72 repeat associated with FTLD: geographic prevalence, genomic instability, and intermediate repeats. Hum Mutat. 2013;34(2):363-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shamim U, Ambawat S, Singh J, et al. C9orf72 hexanucleotide repeat expansion in Indian patients with ALS: a common founder and its geographical predilection. Neurobiol Aging. 2020;88:156.e1–156.e9. [DOI] [PubMed] [Google Scholar]
- 16.Roggenbuck J, Palettas M, Vicini L, Patel R, Quick A, Kolb SJ. Incidence of pathogenic, likely pathogenic, and uncertain ALS variants in a clinic cohort. Neurol Genet. 2020;6(1):e390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vatsavayai SC, Nana AL, Yokoyama JS, Seeley WW. C9orf72-FTD/ALS pathogenesis: evidence from human neuropathological studies. Acta Neuropathol. 2019;137(1):1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coyne AN, Baskerville V, Zaepfel BL, et al. Nuclear accumulation of CHMP7 initiates nuclear pore complex injury and subsequent TDP-43 dysfunction in sporadic and familial ALS. Sci Transl Med. 2021;13(604):eabe1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iacoangeli A, Al Khleifat A, Jones AR, et al. C9orf72 intermediate expansions of 24-30 repeats are associated with ALS. Acta Neuropathol Commun. 2019;7(1):115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ng ASL, Tan EK. Intermediate C9orf72 alleles in neurological disorders: does size really matter? J Med Genet. 2017;54(9):591-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaivola K, Salmi SJ, Jansson L, et al. Carriership of two copies of C9orf72 hexanucleotide repeat intermediate-length alleles is a risk factor for ALS in the Finnish population. Acta Neuropathol Commun. 2020;8(1):187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cannas A, Solla P, Borghero G, et al. C9ORF72 intermediate repeat expansion in patients affected by atypical parkinsonian syndromes or Parkinson's disease complicated by psychosis or dementia in a Sardinian population. J Neurol. 2015;262:2498-2503. [DOI] [PubMed] [Google Scholar]
- 23.Gijselinck I, Van Mossevelde S, van der Zee J, et al. The C9orf72 repeat size correlates with onset age of disease, DNA methylation and transcriptional downregulation of the promoter. Mol Psychiatry. 2016;21(8):1112-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beck J, Poulter M, Hensman D, et al. Large C9orf72 hexanucleotide repeat expansions are seen in multiple neurodegenerative syndromes and are more frequent than expected in the UK population. Am J Hum Genet. 2013;92(3):345-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dols-Icardo O, Garcia-Redondo A, Rojas-Garcia R, et al. Characterization of the repeat expansion size in C9orf72 in amyotrophic lateral sclerosis and frontotemporal dementia. Hum Mol Genet. 2014;23(3):749-754. [DOI] [PubMed] [Google Scholar]
- 26.Nordin A, Akimoto C, Wuolikainen A, et al. Extensive size variability of the GGGGCC expansion in C9orf72 in both neuronal and non-neuronal tissues in 18 patients with ALS or FTD. Hum Mol Genet. 2015;24(11):3133-3142. [DOI] [PubMed] [Google Scholar]
- 27.Thys RG, Wang YH. DNA replication dynamics of the GGGGCC repeat of the C9orf72 gen. J Biol Chem. 2015;290(48):28953-28962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Westergard T, Jensen BK, Wen X, et al. Cell-to-Cell transmission of dipeptide repeat proteins linked to C9orf72-ALS/FTD. Cell Rep. 2016;17(3):645-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ash PE, Bieniek KF, Gendron TF, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77(4):639-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mori K, Arzberger T, Grässer FA, et al. Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol. 2013;126(6):881-893. [DOI] [PubMed] [Google Scholar]
- 31.Ross JP, Leblond CS, Catoire H, et al. Somatic expansion of the C9orf72 hexanucleotide repeat does not occur in ALS spinal cord tissues. Neurol Genet. 2019;5(2):e317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pamphlett R, Cheong PL, Trent RJ, Yu B. Can ALS-associated C9orf72 repeat expansions be diagnosed on a blood DNA test alone? PLoS One. 2013;8(7):e70007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waite AJ, Bäumer D, East S, et al. Reduced C9orf72 protein levels in frontal cortex of amyotrophic lateral sclerosis and frontotemporal degeneration brain with the C9ORF72 hexanucleotide repeat expansion. Neurobiol Aging. 2014;35(7):1779.e5–1779.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Belzil VV, Bauer PO, Prudencio M, et al. Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood. Acta Neuropathol. 2013;126(6):895-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang J, Zhu Q, Gendron TF, et al. Gain of toxicity from ALS/FTD-Linked repeat expansions in C9ORF72 is alleviated by antisense oligonucleotides targeting GGGGCC-containing RNAs. Neuron. 2016;90(3):535-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Rourke JG, Bogdanik L, Yáñez A, et al. C9orf72 is required for proper macrophage and microglial function in mice. Science. 2016;351(6279):1324-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Atanasio A, Decman V, White D, et al. C9orf72 ablation causes immune dysregulation characterized by leukocyte expansion, autoantibody production, and glomerulonephropathy in mice. Sci Rep. 2016;6:23204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Freibaum BD, Lu Y, Lopez-Gonzalez R, et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature. 2015;525(7567):129-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jovicic A, Mertens J, Boeynaems S, et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci. 2015;18(9):1226-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang K, Donnelly CJ, Haeusler AR, et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525(7567):56-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Almeida S, Gascon E, Tran H, et al. Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol. 2013;126(3):385-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zu T, Gibbens B, Doty NS, et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci U S A. 2011;108(1):260-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mori K, Weng SM, Arzberger T, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339(6125):1335-1338. [DOI] [PubMed] [Google Scholar]
- 44.Zu T, Liu Y, Bañez-Coronel M, et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci U S A. 2013;110(51):E4968-E4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Batra R, Charizanis K, Swanson MS. Partners in crime: bidirectional transcription in unstable microsatellite disease. Hum Mol Genet. 2010;19(R1):R77-R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yin S, Lopez-Gonzalez R, Kunz RC, et al. Evidence that C9ORF72 dipeptide repeat proteins associate with U2 snRNP to cause mis-splicing in ALS/FTD patients. Cel Rep. 2017;19(11):2244-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang YJ, Gendron TF, Grima JC, et al. C9ORF72 poly(GA) aggregates sequester and impair HR23 and nucleocytoplasmic transport proteins. Nat Neurosci. 2016;19(5):668-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bäuerlein FJB, Saha I, Mishra A, et al. In situ architecture and cellular interactions of PolyQ inclusions. Cell. 2017;171(1):179–187.e10. [DOI] [PubMed] [Google Scholar]
- 49.Ohki Y, Wenninger-Weinzierl A, Hruscha A, et al. Glycine-alanine dipeptide repeat protein contributes to toxicity in a zebrafish model of C9orf72 associated neurodegeneration. Mol Neurodegener. 2017;12(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wen X, Tan W, Westergard T, et al. Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron. 2014;84(6):1213-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coyne AN, Zaepfel BL, Hayes L, et al. G4C2 repeat RNA initiates a pom121-mediated reduction in specific nucleoporins in C9orf72 ALS/FTD. Neuron. 2020;107(6):1124–1140.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi KY, Mori E, Nizami ZF, et al. Toxic PRn poly-dipeptides encoded by the C9orf72 repeat expansion block nuclear import and export. Proc Natl Acad Sci U S A. 2017;114(7):E1111–E17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fong JC, Karydas AM, Goldman JS. Genetic counseling for FTD/ALS caused by the C9ORF72 hexanucleotide expansion. Alzheimers Res Ther. 2012;4(4):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Akimoto C, Volk AE, van Blitterswijk M, et al. A blinded international study on the reliability of genetic testing for GGGGCC-repeat expansions in C9orf72 reveals marked differences in results among 14 laboratories. J Med Genet. 2014;51(6):419-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nordin A, Akimoto C, Wuolikainen A, et al. Sequence variations in C9orf72 downstream of the hexanucleotide repeat region and its effect on repeat-primed PCR interpretation: a large multinational screening study. Amyotroph Lateral Scler Frontotemporal Degener. 2017;18(3-4):256-264. [DOI] [PubMed] [Google Scholar]
- 56.Warner JP, Barron LH, Goudie D, et al. A general method for the detection of large CAG repeat expansions by fluorescent PCR. J Med Genet. 1996;33(12):1022-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roggenbuck J. C9orf72 and the care of the patient with ALS or FTD: progress and recommendations after 10 years. Neurol Genet. 2021;7(1):e542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klepek H, Goutman SA, Quick A, Kolb SJ, Roggenbuck J. Variable reporting of C9orf72 and a high rate of uncertain results in ALS genetic testing. Neurol Genet. 2019;5(1):e301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murphy NA, Arthur KC, Tienari PJ, Houlden H, Chiò A, Traynor BJ. Age-related penetrance of the C9orf72 repeat expansion. Sci Rep. 2017;7(1):2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Turner MR, Al-Chalabi A, Chio A, et al. Genetic screening in sporadic ALS and FTD. J Neurol Neurosurg Psychiatry. 2017;88(12):1042-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- eReferences e1-e48 available at: links.lww.com/NXG/A520.