Abstract
Breast cancer is the most harmful malignancy in women worldwide. Therefore, in the current study, we investigated the combinatory effect of natural bioactive compounds, including curcumin (Cur) and thymoquinone (TQ), on MCF7 and MDA-MB-231 breast cancer cell lines’ progression. We investigated the Fa values and combination index of Cur and TQ in this context. Moreover, cytotoxicity percentages, annexin-V, proliferation, colony formation, and migration assays were used along with cell cycle analysis. In addition, caspase-3, phosphatidylinositol 3-kinase (PI3K), and protein kinase B (AKT) protein levels were determined by ELISA assessment. The results showed that Cur, TQ, and Cur + TQ induced apoptosis with cell cycle arrest and decreased cell proliferation, colony formation, and migration activities. Cur + TQ combination significantly increased caspase-3 and decreased PI3K and AKT protein levels. These results suggest the promising anticancer benefit of the Cur and TQ combination against breast cancer.
Keywords: natural bioactive compounds, curcumin; thymoquinone; combination; breast cancer; anticancer
Introduction
Breast cancer is the most common type of cancer found in women and today represents a significant challenge to public health. 1 Breast cancer has been classified by hormone receptors like estrogen or progesterone (some have both), called estrogen receptor-positive or progesterone receptor-positive breast cancer, respectively, and human epidermal growth factor receptor-2 positive expressed cancer.2,3 The triple-negative breast cancer treatment remains a challenge due to its aggressive characteristics and limited therapy. 4 Breast cancer therapies involve drugs that block cancer growth by interfering with the function of specific molecules responsible for tumor cell proliferation and survival. 5
A healthy diet reduced cancer incidence, leading many researchers to focus on natural products for the prevention of cancer6,7 through downregulating estrogen receptor-α expression and activity, inhibiting proliferation, migration, metastasis, angiogenesis, inducing apoptosis, cell cycle arrest, and increasing the sensitivity of breast tumor cells to radiotherapy and chemotherapy. 8 Among them, curcumin (Cur), the active ingredient of Curcuma longa, is the most studied compound described as a potential anticancer agent.9-11 Cur targets multiple signaling/molecular pathways including, Rb, p53, mitogen-activated protein kinase, phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT), and nuclear factor kappa B cells (NF-κB), pathways 10 in most cancer forms.9,12 Previous studies have demonstrated that Cur can inhibit cancer cell proliferation. However, the functional role and mechanism of Cur in breast cancer are still unclear and need more investigation. 13 Recently, Cur was reported to suppress breast cancer (MDA-MB-231) cell proliferation and migration through autophagy-dependent AKT degradation 14 and increased natural killer cells activity. 14
Thymoquinone (TQ), the bioactive constituent of Nigella sativa seeds, is a well-known natural bioactive compound used to manage several cancer types.9,15-19 The anticancer properties of TQ induced their anticancer potential through prevention of inflammation and oxidative stress, inhibition of angiogenesis and metastasis, induction of apoptosis, upregulation of specific tumor suppressor genes, and downregulation of tumor-promoting genes. 20 The anticancer effect of TQ is mainly due to the induction of apoptosis by activation of caspases, downregulation of oncogenes, inhibition of NF-κB, reactive oxygen species regulation, hypoxia, and anti-metastasis activity. 21 Furthermore, TQ inhibited various tumor cells’ proliferation via induction of cell cycle arrest, disruption of the microtubule organization, and downregulation of cell survival protein expression.20,22 Also, TQ potentiated the anticancer activity of many chemotherapeutic agents and sensitized cancer cells to radiotherapy making it a promising molecule for cancer therapy.16,17,19
In the current study, we investigated the combinatory effect of Cur and TQ on hormone receptor-positive (MCF7) and triple-negative (MDA-MB-231) breast cancer cells progression.
Materials and Methods
Cell Lines
Human breast cancer cell lines (MCF7 and MDA-MB-231) were purchased from ATCC (Manassas, VA, USA). MCF7 cells were grown in low-glucose Dulbecco’s Modified Eagle Medium (DMEM), while MDA-MB-231 cells were raised in a high-glucose DMEM medium. Both media were supplemented with 10% fetal bovine serum and protected with 1% penicillin/streptomycin solution.
MTT (3-[4,5-Dimethylthiazol-2-yl]-2,5-Diphenyltetrazolium Bromide) Assay and Combination Index
The half-maximal inhibitory concentration (IC50) of Cur and TQ were determined by seeding of MCF7 or MDA-MB-231 cells in 24-well plates (3 × 104 per well in 1.5 mL) and incubated for 48 hours at 37°C in a 5% CO2 incubator. Cells were treated with 1 mL of Cur (Sigma-Aldrich Co., MO, USA) in concentrations of 0, 5, 10, 25, 50, or 100 μM dissolved in DMSO (Sigma-Aldrich Co.) or TQ (Sigma-Aldrich Co.) in concentrations of 0, 10, 25, 50, 100, or 200 μM dissolved in DMSO, incubated for 24 hours, then treated with MTT reagent (1.25 mg/mL) and incubated for 2 hours. The resulting formazan crystals were dissolved in 1 mL DMSO, and the optical density was determined using a microplate reader at 570 nm. 9 The data of the MTT assay was analyzed by CompuSyn software (https://www.combosyn.com/) to determine the IC50 of Cur and TQ against MCF7 and MDA-MB-231 cells. These experiments were repeated 3 times.
Combination index (CI) of Cur and TQ was determined by CompuSyn software at Fa 75 (43.31 µM of Cur and 64.01 µM of TQ), Fa 90 (24.91 µM of Cur and 41.16 µM of TQ) for MCF7, Fa 75 (37.37 µM of Cur and 79.09 µM of TQ), and Fa 90 (24.50 µM of Cur and 51.76 µM of TQ) for MDA-MB-231. CI indicates the drug combination type: synergism (CI < 1), additive effect (CI = 1) or antagonism (CI > 1).
Cytotoxicity Assay
MCF7 or MDA-MB-231cells were seeded in 12-well plates (3 × 104 per well in 1.5 mL) and incubated for 48 hours at 37°C in a 5% CO2 incubator. After determination of Fa 75, 90, 95, and 97 of Cur and TQ by CompuSyn software against MCF7 and MDA-MB-231 cells, control, Cur, TQ, and Cur + TQ cell groups were treated with Fa 75 (43.31 µM of Cur and 64.01 µM of TQ), Fa 90 (24.91 µM of Cur and 41.16 µM of TQ), Fa 95 (17.10 µM of Cur and 30.48 µM of TQ), and Fa 97 (13.08 µM of Cur and 24.61 µM of TQ) for MCF7. The Fa 75 (37.37 µM of Cur and 79.09 µM of TQ), Fa 90 (24.50 µM of Cur and 51.76 µM of TQ), Fa 95 (18.38 µM of Cur and 38.79 µM of TQ), and Fa 97 (14.98 µM of Cur and 31.60 µM of TQ) were used for MDA-MB-231 cells treatment in 1 mL of the medium. After 24 hours, cells were treated with MTT and visualized as described in the MTT assay. These experiments were repeated 3 times.
Annexin-V Assay
Apoptosis was analyzed by flow cytometry using annexin V-fluorescein isothiocyanate (annexin-FITC) and propidium iodide (PI) detection kit (BD Biosciences, San Jose, CA, USA). MCF7 and MDA-MB-231 cells treated with Cur (24.91 and 24.50 µM, respectively) and TQ (41.16 and 51.76 µM, respectively) and their corresponding control flasks were collected and centrifuged at 500 × g for 5 minutes at room temperature. The pellet was rinsed twice with PBS and then resuspended in a proper volume of binding buffer. After adding 10 μL of annexin V-FITC followed by gentle mixing, it was incubated for 15 minutes at room temperature in the dark and washed. The fluorescence intensity of FITC was carried on a FACSCalibur™ (Becton Dickinson) instrument, using Cell Quest software.9,16
Cell Cycle Analysis
Using a FACSCalibur™ flow cytometer (Becton Dickinson, San Jose, CA, USA) and Cell Quest software, the cell cycle status was analyzed in MCF7, and MDA-MB-231cells treated with Cur (24.91 and 24.50 µM, respectively) and TQ (41.16 and 51.76 µM, respectively).9,16,23
Cell Proliferation Assay
MCF7 or MDA-MB-231cells were cultured in 12-well plates (3 × 104 per well in 1.5 mL) and incubated for 48 hours at 37°C in a 5% CO2 incubator. Cells were treated with Fa 90 of 1 mL of Cur (24.91 and 24.50 µM) and TQ (41.16 and 51.76 µM) against MCF7 and MDA-MB-231 cells, respectively for 24 hours. The total viable cell number was counted using a hemocytometer using the dye exclusion method with 0.2% Trypan blue at room temperature (Thermo Fisher Scientific, Inc.) using an inverted light microscope (Primovert, Zeiss, Carl Zeiss Industrielle Messtechnik GmbH, Oberkochen, Germany) at magnification, ×4 objective. 24 These experiments were repeated 3 times.
Colony Formation Assay
About 2500 cells (MCF7 or MDA-MB-231) per well were plated in 12-well plates and were allowed to grow for about 4 to 5 days until small colonies could be seen. Cells were treated with Fa 90 of 1 mL of Cur (24.91 and 24.50 µM) and TQ (41.16 and 51.76 µM) against MCF7 and MDA-MB-231cells, respectively, for 24 hours. Cells were fixed with 4% formaldehyde in PBS and stained with 0.1% crystal violet diluted in water. 25 A camera took images for each well. These experiments were repeated 3 times.
Migration Assay
Cell migration was evaluated using the monolayer denudation assay as previously described. 24 Briefly, MCF7 or MDA-MB-231 cells were inoculated (5 × 104 per well in 1.5 mL) and were cultured to 100% confluence in a 12-well plate. Cells were then wounded by denuding a strip of the monolayer with a 200 μL pipette tip. Cells were washed twice with PBS and then incubated with 1 mL of Cur (24.91 and 24.50 µM) and TQ (41.16 and 51.76 µM) against MCF7 and MDA-MB-231 cells, respectively for 24 hours. The rate of wound closure was assessed in 4 separate fields of view using a light microscope (magnification, ×4 objective). These experiments were repeated 3 times.
Enzyme-Linked Immunosorbent (ELISA) Assay
MCF7 or MDA-MB-231cells at the density of 30 × 104 were cultured in a T25 flask for 48 hours. Seeded cells were treated with 10 mL of Cur (24.91 and 24.50 µM) and TQ (41.16 and 51.76 µM) against MCF7 and MDA-MB-231 cells, respectively for 24 hours. Cells in each flask were trypsinized and centrifuged to produce a clear cell pellet. Cells were homogenized in RIPA buffer using TissueLyser (Qiagen Co., Germantown, MD, USA). Caspase-3, PI3K, and AKT levels were determined by FineTest ELISA kit (Wuhan Fine Biotech Co., Wuhan, Hubei, China) according to the manufacturer’s instructions at 450 nm. Protein levels in all samples were determined by the Bradford 26 method.
Results
MTT and Fa Values of Cur and TQ Against MCF7 Cells
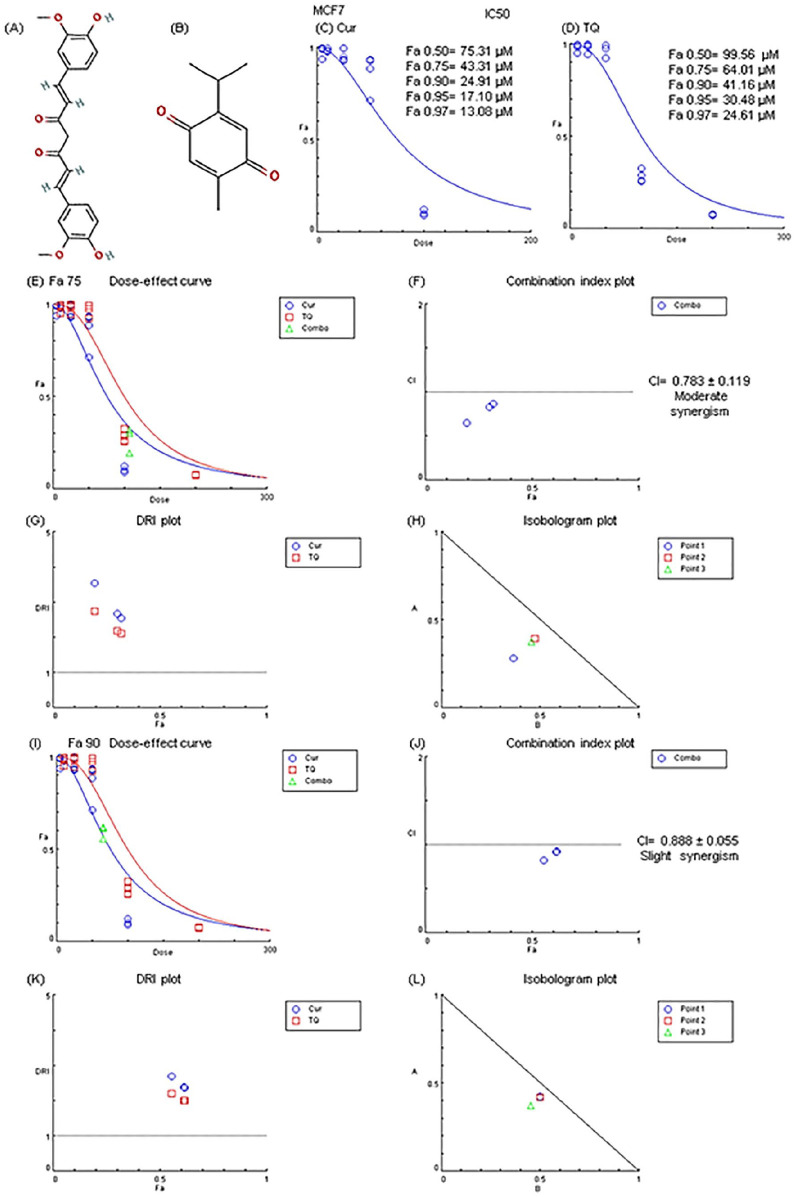
Here, we provide some experimental evaluation of the anticancer effect of Cur, TQ, and their combination against MCF7 cell lines. In this work, we first determined the Fa values of Cur and TQ against MCF7 (Figure 1C and D) using different serial concentrations of Cur and TQ.
Figure 1.
Chemical structure, Fa values, and combination index of curcumin (Cur) and thymoquinone (TQ) against MCF7. (A) Chemical structure of Cur. (B) Chemical structure of TQ. (C) Fa values of Cur against MCF7. (D) Fa values of TQ against MCF7. (E) Dose-effect curve of Fa 75 of Cur (43.31 µM) and TQ (64.01 µM). (F) Combination index plot of Fa 75 of Cur and TQ. (G) DRI plot of Fa 75 of Cur and TQ. (H) Isobologram plot of Fa 75 of Cur and TQ. (I) Dose-effect curve of Fa 90 of Cur (24.91 µM) and TQ (41.16 µM). (J) Combination index plot of Fa 90 of Cur and TQ. (K) DRI plot of Fa 90 of Cur and TQ. (L) Isobologram plot of Fa 90 of Cur and TQ. The CI values represent the mean of 4 experiments. CI > 1.3: antagonism; CI (1.1-1.3): moderate antagonism; CI (0.9-1.1): additive effect; CI (0.8-0.9): slight synergism; CI (0.6-0.8): moderate synergism; CI (0.4-0.6): synergism; CI (0.2-0.4): strong synergism.
After rigorous determination of combination assessment, it was discovered that Cur and TQ cotreatment exhibited moderate synergistic effect against MCF7 using Fa 75 and slight synergism at Fa 90 concentrations of Cur and TQ (Figure 1E-L).
Cytotoxicity Percentages of Cur and TQ Against MCF7 Cells
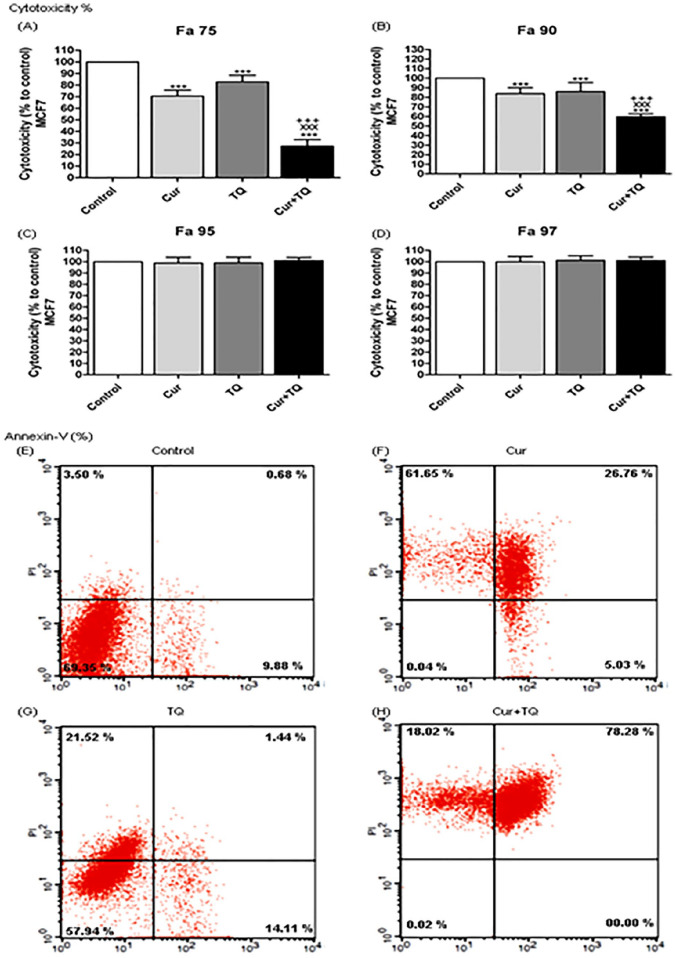
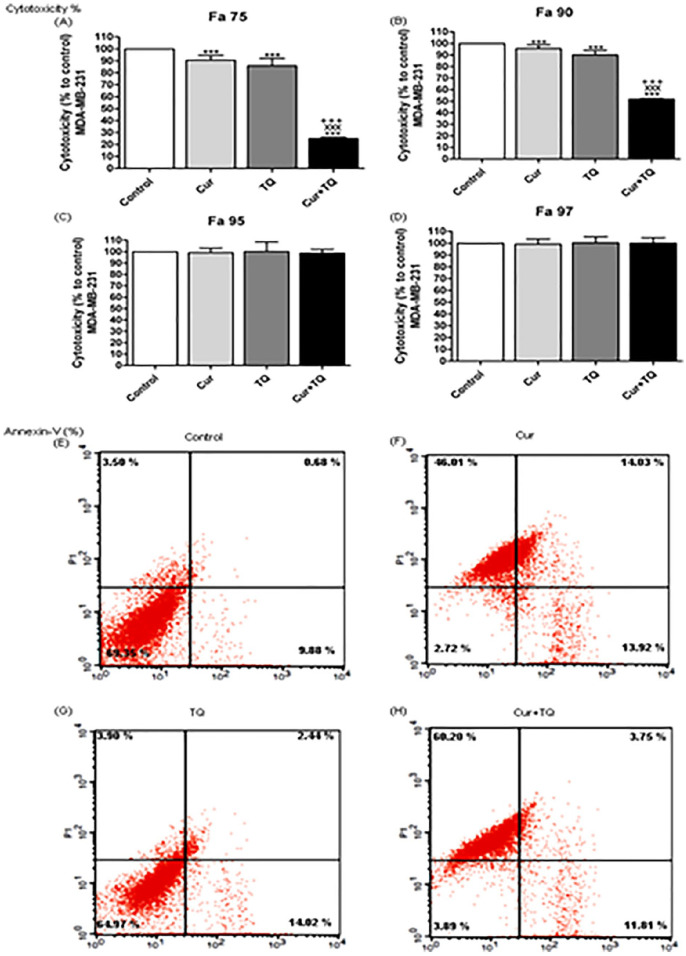
After determination of Fa 50, Fa 75, Fa 90, Fa 95, and Fa 97 values of Cur (75.31, 43.31, 24.91, 17.10, and 13.08 µM, respectively) and TQ (99.56, 64.01, 41.16, 30.48, and 24.61 µM, respectively) the cytotoxicity percentages of these concentrations were evaluated against MCF7 using MTT assay (Figure 2A-D). Fa 95 and Fa 97 concentrations of Cur and TQ induced nonsignificant changes, while both significantly (P < .001) induced cytotoxicity against MCF7 either alone or in combination using Fa 75 (Figure 2A) and Fa 90 (Figure 2B) concentrations. Consequently, the low concentrations (Fa 90) of Cur and TQ were used in annexin-V, cell cycle analysis, proliferation, colony formation, migration, and ELISA assays.
Figure 2.
Cytotoxicity and annexin-V positive cell percentages. Cytotoxicity percentages of (A) Fa 75, (B) Fa 90, (C) Fa 95, and (D) Fa 97 of curcumin (Cur), thymoquinone (TQ), and their combination against MCF7. Annexin-V positive cell percentages of MCF7 (E) control, (F) Cur (Fa 90 = 24.91 µM), (G) TQ (Fa 90 = 41.16 µM), and (H) their combination (Cur + TQ) groups. The data were analyzed with one-way ANOVA followed by Tukey’s multiple comparison test. Error bars represent mean ± SD. ***P < .001 versus control. xxxP < .001 versus Cur. +++P < .001 versus TQ.
Annexin-V Positive Cell Percentages of MCF7 Cells Treated With Cur, TQ, and Their Combination
As seen in Figure 2E to H, MCF7 cells had more annexin-V positive cells in Cur (93.44%), TQ (37.07%), and Cur + TQ (96.3%) compared with the control (14.06%) group.
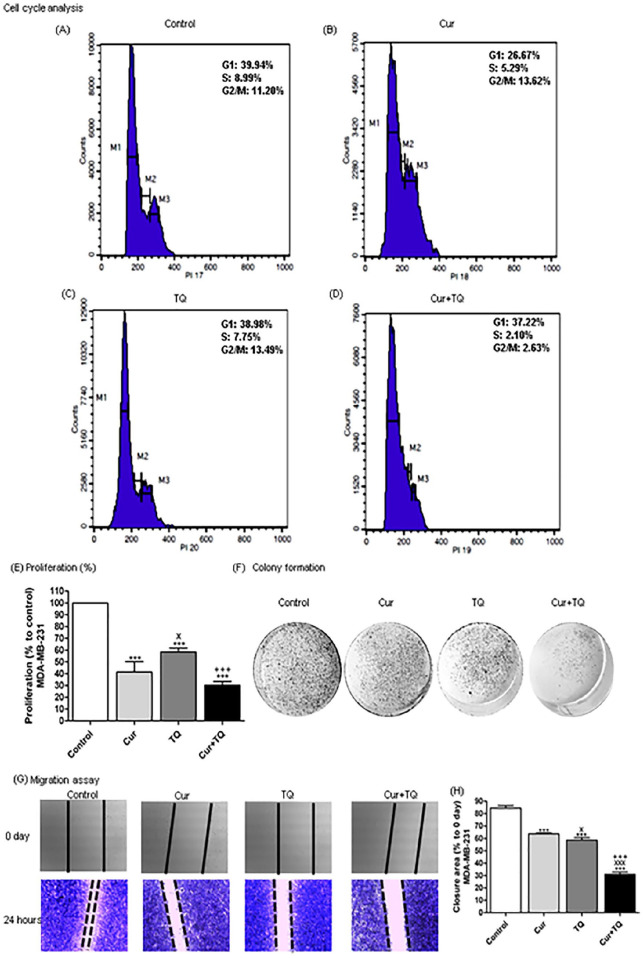
Cell Cycle Analyses of MCF7 Cells Treated With Cur, TQ, and Their Combination
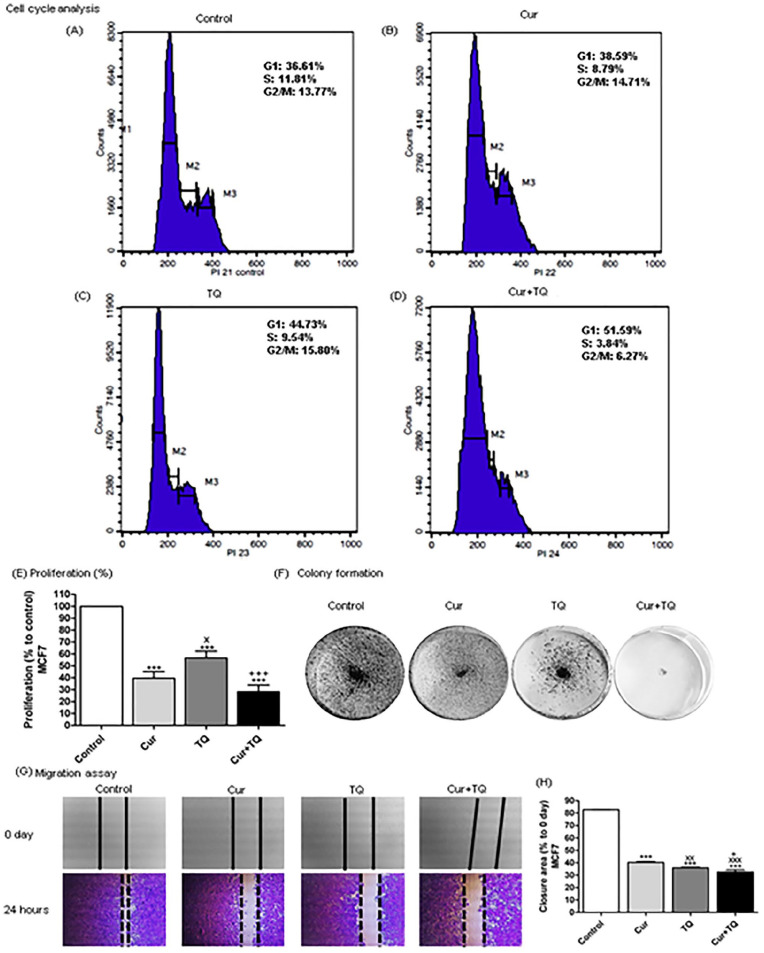
Cell cycle analysis of MCF7 in Cur, TQ, and Cur + TQ showed decreases in S phase values compared with the control group (Figure 3A-D).
Figure 3.
Cell cycle analysis, proliferation percentage, colony formation, and migration assays for MCF7. Cell cycle analysis of MCF7 (A) control, (B) Cur (Fa 90 = 24.91 µM), (C) TQ (Fa 90 = 41.16 µM), and (D) their combination (Cur + TQ) groups. (E) Proliferation percentage of MCF7 control, Cur (Fa 90 = 24.91 µM), TQ (Fa 90 = 41.16 µM), and their combination (Cur + TQ) groups. (F) Colony formation assay of MCF7 control, Cur (Fa 90 = 24.91 µM), TQ (Fa 90 = 41.16 µM), and their combination (Cur + TQ) groups. (G, H) Migration assay of MCF7 control, Cur (Fa 90 = 24.91 µM), TQ (Fa 90 = 41.16 µM), and their combination (Cur + TQ) groups at 0 and 24 hours. The data were analyzed with one-way ANOVA followed by Tukey’s multiple comparison test. Error bars represent mean ± SD. ***P < .001 versus control. xP < .05, xxP < .01, and xxxP < .001 versus Cur. +P < .05 and +++P < .001 versus TQ.
Proliferation, Colony Formation, and Migration Assays of MCF7 Cells Treated With Cur, TQ, and Their Combination
The results of proliferation percentages of MCF7 treated with Cur, TQ, and Cur + TQ are set out in Figure 3E. It is apparent from this data that Cur, TQ, and Cur + TQ significantly (P < .001) decreased MCF7 proliferation compared with control. In contrast, Cur and TQ combination exhibited higher decreases in proliferation percentages. Similarly, Cur, TQ, and Cur + TQ reduced colony formation of MCF7 in comparison with control (Figure 3F).
In Figure 3G and H, there is a clear trend of decreasing the extent of closure area in Cur, TQ, and Cur + TQ treated MCF7 compared with control cells assessed with migration assay.
Caspase-3, PI3K, and AKT Protein Levels in MCF7 Cells Treated With Cur, TQ, and Their Combination
The results, as shown in Figure 4A, indicate that treatment of MCF7 with Cur, TQ, and Cur + TQ significantly (P < .01) increased caspase-3 levels compared with control. PI3K (Figure 4B) and AKT (Figure 4C) protein levels were significantly decreased in Cur (P < .01 and P < .001, respectively), TQ (P < .05 and P < .01, respectively), and Cur + TQ (P < .01 and P < .001, respectively) in comparison with the control group at 24 hours.
Figure 4.
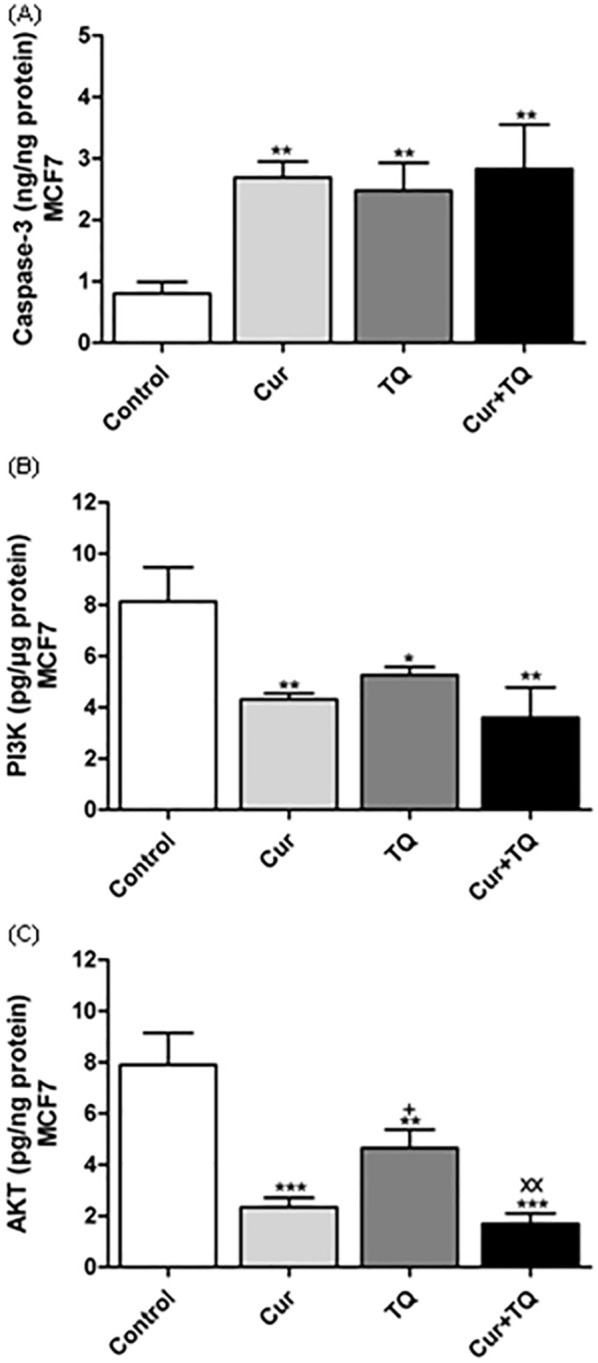
Enzyme-linked immunosorbent (ELISA) assay. (A) Caspase-3 (ng/ng protein), (B) Phosphatidylinositol 3-kinase (PI3K, pg/µg protein), and (C) Protein kinase B (AKT, pg/ng protein) levels in MCF7 control, Cur (Fa 90 = 24.91 µM), TQ (Fa 90 = 41.16 µM), and their combination (Cur + TQ) groups. The data were analyzed with one-way ANOVA followed by Tukey’s multiple comparison test. Error bars represent mean ± SD. *P < .05, **P < .01, and ***P < .001 versus control. xxP < .01 versus Cur. +P < .05 versus TQ.
MTT and Fa Values of Cur and TQ Against MDA-MB-231 Cells
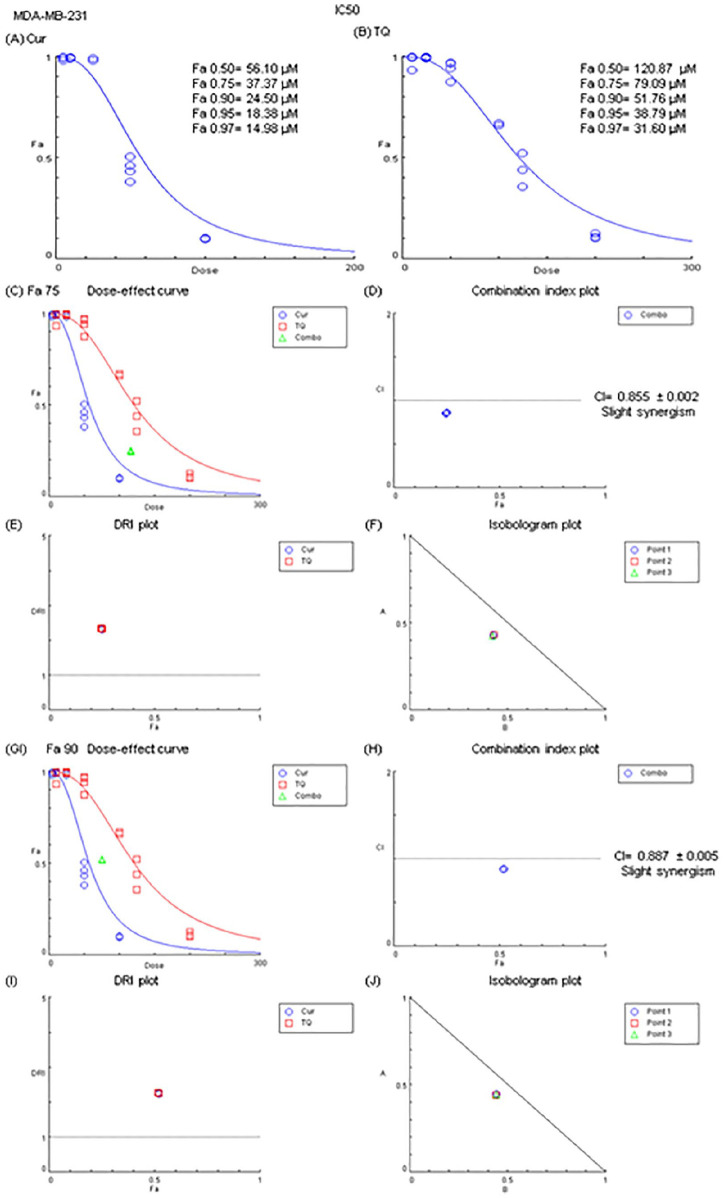
Figure 5 presents the results of the MTT assay and Fa values of Cur and TQ against MDA-MB-231 cells. Cur exhibited Fa 0.50, Fa 0.75, Fa 0.90, Fa 0.95, and Fa 0.97 of 56.10, 37.37, 24.50, 18.38, and 14.98 µM, respectively (Figure 5A). By the same manner, TQ had Fa 0.50, Fa 0.75, Fa 0.90, Fa 0.95, and Fa 0.97 of 120.87, 79.09, 51.76, 38.79, and 31.60 µM, respectively (Figure 5B).
Figure 5.
Fa values and combination index of curcumin (Cur) and thymoquinone (TQ) against MDA-MB-231. (A) Fa values of Cur against MDA-MB-231. (B) Fa values of TQ against MDA-MB-231. (C) Dose-effect curve of Fa 75 of Cur (37.37 µM) and TQ (79.09 µM). (D) Combination index plot of Fa 75 of Cur and TQ. (E) DRI plot of Fa 75 of Cur and TQ. (F) Isobologram plot of Fa 75 of Cur and TQ. (G) Dose-effect curve of Fa 90 of Cur (24.50 µM) and TQ (51.76 µM). (H) Combination index plot of Fa 90 of Cur and TQ. (I) DRI plot of Fa 90 of Cur and TQ. (J) Isobologram plot of Fa 90 of Cur and TQ. The CI values represent the mean of 4 experiments. CI > 1.3: antagonism; CI (1.1-1.3): moderate antagonism; CI (0.9-1.1): additive effect; CI (0.8-0.9): slight synergism; CI (0.6-0.8): moderate synergism; CI (0.4-0.6): synergism; CI (0.2-0.4): strong synergism.
The combination between Cur and TQ at Fa 0.75 (37.37 and 79.09, respectively) and Fa 0.90 (24.50 and 51.76, respectively) concentrations of each revealed slight synergistic effects (Figure 5C-J).
Cytotoxicity Percentages of Cur and TQ Against MDA-MB-231 Cells
Cur, TQ, and their combinations by Fa 0.75 and Fa 90 concentrations significantly (P < .001) increased cytotoxicity percentages of MDA-MB-231 cells compared with control cells. In the current study, we used the lower concentrations (Fa 90) in the following assays of Cur and TQ against MDA-MB-231 cells (Figure 6).
Figure 6.
Cytotoxicity and annexin-V positive cell percentages of MDA-MB-231. Cytotoxicity percentages of (A) Fa 75, (B) Fa 90, (C) Fa 95, and (D) Fa 97 of curcumin (Cur), thymoquinone (TQ), and their combination against MDA-MB-231. Annexin-V positive cell percentages of MDA-MB-231 (E) control, (F) Cur (Fa 90 = 24.50 µM), (G) TQ (Fa 90 = 51.76 µM), and (H) their combination (Cur + TQ) groups. The data were analyzed with one-way ANOVA followed by Tukey’s multiple comparison test. Error bars represent mean ± SD. ***P < .001 versus control. xxxP < .001 versus Cur. +++P < .001 versus TQ.
Annexin-V Positive Cell Percentages of MDA-MB-231 Cells Treated With Cur, TQ, and Their Combination
The annexin-V positive cells shown in Figure 6E-H revealed that Cur induced apoptosis of MDA-MB-231 cells in the percentage of 73.96. Also, TQ-treated cells exhibited apoptosis by the percentage of 20.36, while Cur + TQ-treated MDA-MB-231 cells had 75.76% of apoptosis.
Cell Cycle Analyses of MDA-MB-231 Cells Treated With Cur, TQ, and Their Combination
Data represented in Figure 7A-D showed decreases in S phase values of MDA-MB-231cell cycle analysis in Cur, TQ, and Cur + TQ-treated cells compared with the control group.
Figure 7.
Cell cycle analysis, proliferation percentage, colony formation, and migration assays of MDA-MB-231. Cell cycle analysis of MDA-MB-231 (A) control, (B) Cur (Fa 90 = 24.50 µM), (C) TQ (Fa 90 = 51.76 µM), and (D) their combination (Cur + TQ) groups. (E) Proliferation percentage of MDA-MB-231 control, Cur (Fa 90 = 24.50 µM), TQ (Fa 90 = 51.76 µM), and their combination (Cur + TQ) groups. (F) Colony formation assay of MDA-MB-231 control, Cur (Fa 90 = 24.50 µM), TQ (Fa 90 = 51.76 µM), and their combination (Cur + TQ) groups. (G, H) Migration assay of MDA-MB-231 control, Cur (Fa 90 = 24.50 µM), TQ (Fa 90 = 51.76 µM), and their combination (Cur + TQ) groups at 0 and 24 hours. The data were analyzed with one-way ANOVA followed by Tukey’s multiple comparison test. Error bars represent mean ± SD. ***P < .001 versus control. xP < .05 and xxxP < .001 versus Cur. +++P < .001 versus TQ.
Proliferation, Colony Formation, and Migration Assays of MDA-MB-231 Cells Treated With Cur, TQ, and Their Combination
MDA-MB-231 cells treated with Cur, TQ, and Cur + TQ revealed significant decreases in cell proliferation percentages (Figure 7E), colony formation (Figure 7F), and area of closure of migration assay (Figure 7G and H) compared with control untreated cells.
Caspase-3, PI3K, and AKT Protein Levels in MDA-MB-231 Cells Treated With Cur, TQ, and Their Combination
Caspase-3 protein levels were significantly increased in Cur and Cur + TQ-treated MDA-MB-231 cells compared with control cells (Figure 8A). On the contrary, PI3K (Figure 8B) and AKT (Figure 8C) levels were significantly decreased in Cur (P < .01 and P < .001, respectively), TQ (P < .05), and Cur + TQ (P < .001)-treated MDA-MB-231 cells compared with control cells.
Figure 8.
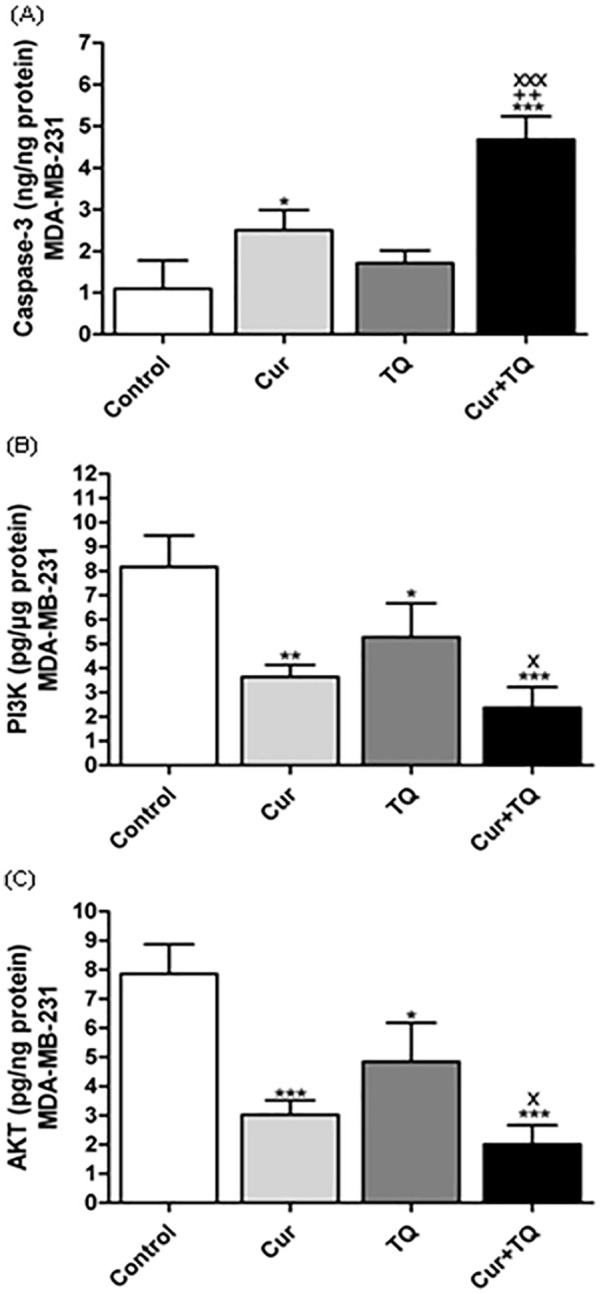
Enzyme-linked immunosorbent (ELISA) assay. (A) Caspase-3 (ng/ng protein), (B) Phosphatidylinositol 3-kinase (PI3K, pg/µg protein), and (C) Protein kinase B (AKT, pg/ng protein) levels in MDA-MB-231 control, Cur (Fa 90 = 24.50 µM), TQ (Fa 90 = 51.76 µM), and their combination (Cur + TQ) groups. The data were analyzed with one-way ANOVA followed by Tukey’s multiple comparison test. Error bars represent mean ± SD. *P < .05, **P < .01, and ***P < .001 versus control. xP < .05 and xxxP < .001 versus Cur. ++P < .01 versus TQ.
Discussion
Several reports have shown that nutraceutical application in cancer therapy has been introduced for investigation as an alternative or combinatory for chemotherapeutics resulting in a promising era for cancer therapy, especially natural bioactive compounds.9,15,16,19,27 In the current study, we studied the anticancer effect of Cur, TQ, and their combination against MCF7 and MDA-MB-231 cell lines. The most interesting finding was that Cur, TQ, and their combination induced significant apoptosis of both cells and hindered their progression. Several reports have shown the anticancer effect of either Cur or TQ against MCF7 and MDA-MB-231 cell lines.9,16,28 Still, no literature considers the combinatory benefits of Cur and TQ against both cell lines.
One interesting finding is Cur, TQ, and their combination had synergistic effects against MCF7 and MDA-MB-231 cell lines at Fa 75 and Fa 90 concentrations. Therefore, we chose the low concentrations (Fa 90) in the remaining assays. Fa 90 concentrations of Cur, TQ, and Cur + TQ induced apoptosis, increased cytotoxicity, induced cell cycle arrest, and decreased proliferation, colony formation, and migration of MCF7 and MDA-MB-231 cells. El-Far et al9,16 stated that TQ induced apoptosis of proliferative and senescent MCF7. Also, TQ induced apoptosis and inhibited metastasis of MDA-MB-231 cells. 29 Hu et al 28 stated that Cur decreased proliferation and colony formation activities in MCF7 and MDA-MB-231 cell lines.
Mechanistically, the anticancer effect of drugs or natural bioactive compounds might contribute to several pathway involvements, including caspase-3 or PI3k/AKT. Caspase-3, a key executioner in apoptosis, is involved in cancer growth and progression. 30 In this study, Cur, TQ, and Cur + TQ were found to cause a significant increases in caspase-3 levels in treated MCF7 and MDA-MB-231 cell lines, enhancing their apoptosis. This study supports evidence from previous observations of Effenberger-Neidnicht and Schobert, 31 Attoub et al, 32 Masuelli et al, 33 Zhou et al, 34 and El-Far et al 16 who reported significant increases in caspase-3 expression, protein levels, or activities in either MCF7 or MDA-MB-231 cell lines treated with Cur or TQ separately. We did not recognize previous studies concerning Cur and TQ combination against MCF7 and MDA-MB-231 cell lines. In the current study Cur and TQ combination increased caspase-3 more than Cur or TQ alone. Caspase-3 induced apoptosis of cancer cells and regulated migration, invasion, and metastasis of colon 35 and melanoma 36 cancer cells.
PI3k/AKT is considered an anti-apoptotic pathway that hinders cancer cell apoptosis leading to cancer cell proliferation and is important for regulating cell cycle progression. 37 PI3K phosphorylated AKT, which promoted cell growth, proliferation, and motility by activating many downstream kinases, including the mammalian target of the rapamycin complex. 38 Also, the activation of PI3K/AKT and their downstream factors, NF-κB, c-Jun, and c-Fos, increased the matrix metalloproteinases expression and promoted cancer invasion and migration.39,40 Therefore, PI3k/AKT is an interesting target for cancer therapy. 41 Another important finding of the current study was that Cur, TQ, and Cur + TQ significantly decreased the PI3k and AKT protein levels in MCF7 and MDA-MB-231 cell lines. Jia et al 42 and Guan et al 43 recognized that Cur suppressed proliferation and migration of MCF7 and MDA-MB-231, interfering with PI3K/AKT signaling. Similarly, TQ induced cell cycle arrest and apoptosis of MCF7 and MDA-MB-231 through downregulation of PI3K/AKT signaling.44,45
Conclusion
The purpose of the current study was to determine the combinatory benefits of Cur and TQ to control MCF7 and MDA-MB-231 cells’ progression. The most obvious finding from this study is Cur + TQ combination induction of apoptosis and hindering proliferation of MCF7 and MDA-MB-231 cells. This effect is attributed to the increased caspase-3 and decreased PI3K and AKT protein levels. Further research might explore the impact of Cur + TQ combination on other apoptotic and anti-apoptotic pathways in breast cancer.
Footnotes
Author Contributions: Conceptualization, A.H.E.-F., A.A.S., S.A.M., O.A.A., S.A.M.; formal analysis, A.H.E.-F., S.A.M., O.A.A.; investigation, A.H.E.-F., A.A.S., S.A.M. O.A.A.; project administration, A.H.E.-F., A.A.S., O.A.A., and S.A.M.; software, A.H.E.-F., S.A.M., and S.A.M.; validation, A.H.E.-F.,, A.A.S., and S.A.M.; visualization, A.H.E.-F.; writing—original draft, A.H.E.-F.; writing—review and editing, A.H.E.-F., A.A.S., S.A.M., O.A.A., and S.A.M. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement: Data are contained within the article.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The authors extend their appreciation to the Deputyship for Research and Innovation, Ministry of Education in Saudi Arabia, to fund this research work through the project number MoE-IF-G-20-02.
ORCID iD: Ali H. El-Far  https://orcid.org/0000-0001-9721-4360
https://orcid.org/0000-0001-9721-4360
References
- 1. Masoud V, Pagès G. Targeted therapies in breast cancer: new challenges to fight against resistance. World J Clin Oncol. 2017;8:120-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gluz O, Liedtke C, Gottschalk N, Pusztai L, Nitz U, Harbeck N. Triple-negative breast cancer—current status and future directions. Ann Oncol. 2009;20:1913-1927. [DOI] [PubMed] [Google Scholar]
- 3. Murphy CG, Dickler MN. Endocrine resistance in hormone-responsive breast cancer: mechanisms and therapeutic strategies. Endocr Relat Cancer. 2016;23:R337-R352. [DOI] [PubMed] [Google Scholar]
- 4. Wang C, Kar S, Lai X, et al. Triple negative breast cancer in Asia: an insider’s view. Cancer Treat Rev. 2018;62:29-38. [DOI] [PubMed] [Google Scholar]
- 5. Rugo HS. Dosing and safety implications for oncologists when administering everolimus to patients with hormone receptor-positive breast cancer. Clin Breast Cancer. 2016;16:18-22. [DOI] [PubMed] [Google Scholar]
- 6. den Hollander P, Savage MI, Brown PH. Targeted therapy for breast cancer prevention. Front Oncol. 2013;3:250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mohsen E, El-Far AH, Godugu K, Elsayed F, Mousa SA, Younis IY. SPME and solvent-based GC–MS metabolite profiling of Egyptian marketed Saussurea costus (Falc.) Lipsch. concerning its anticancer activity. Phytomed Plus. 2022;2:100209. [Google Scholar]
- 8. Li Y, Li S, Meng X, Gan R-Y, Zhang JJ, Li HB. Dietary natural products for prevention and treatment of breast cancer. Nutrients. 2017;9:728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. El-Far AH, Darwish NHE, Mousa SA. Senescent colon and breast cancer cells induced by doxorubicin exhibit enhanced sensitivity to curcumin, caffeine, and thymoquinone. Integr Cancer Ther. 2020;19:1534735419901160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wong SC, Kamarudin MNA, Naidu R. Anticancer mechanism of curcumin on human glioblastoma. Nutrients. 2021;13:950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Farghadani R, Naidu R. Curcumin as an enhancer of therapeutic efficiency of chemotherapy drugs in breast cancer. Int J Mol Sci. 2022;23:2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Joshi P, Joshi S, Semwal D, et al. Curcumin: an insight into molecular pathways involved in anticancer activity. Mini Rev Med Chem. 2021;21:2420-2457. [DOI] [PubMed] [Google Scholar]
- 13. Cao X, Li Y, Wang Y, et al. Curcumin suppresses tumorigenesis by ferroptosis in breast cancer. PLoS One. 2022;17:e0261370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee HH, Cho H. Improved anti-cancer effect of curcumin on breast cancer cells by increasing the activity of natural killer cells. J Microbiol Biotechnol. 2018;28:874-882. [DOI] [PubMed] [Google Scholar]
- 15. El-Far AH, Tantawy MA, Al Jaouni SK, Mousa SA. Thymoquinone-chemotherapeutic combinations: new regimen to combat cancer and cancer stem cells. Naunyn Schmiedebergs Arch Pharmacol. 2020;393:1581-1598. [DOI] [PubMed] [Google Scholar]
- 16. El-Far AH, Godugu K, Noreldin AE, et al. Thymoquinone and costunolide induce apoptosis of both proliferative and doxorubicin-induced-senescent colon and breast cancer cells. Integr Cancer Ther. 2021;20:15347354211035450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. El-Far AH, Al Jaouni SK, Li W, Mousa SA. Protective roles of thymoquinone nanoformulations: potential nanonutraceuticals in human diseases. Nutrients. 2018;10:1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. El-Far AH. Thymoquinone anticancer discovery: possible mechanisms. Curr Drug Discov Technol. 2015;12:80-89. [DOI] [PubMed] [Google Scholar]
- 19. El-Far AH, Salaheldin TA, Godugu K, Darwish NH, Mousa SA. Thymoquinone and its nanoformulation attenuate colorectal and breast cancers and alleviate doxorubicin-induced cardiotoxicity. Nanomed. 2021;16:1457-1469. [DOI] [PubMed] [Google Scholar]
- 20. Alhmied F, Alammar A, Alsultan B, Alshehri M, Pottoo FH. Molecular mechanisms of thymoquinone as anticancer agent. Comb Chem High Throughput Screen. 2021;24:1644-1653. [DOI] [PubMed] [Google Scholar]
- 21. Phua CYH, Teoh ZL, Goh BH, Yap WH, Tang YQ. Triangulating the pharmacological properties of thymoquinone in regulating reactive oxygen species, inflammation, and cancer: therapeutic applications and mechanistic pathways. Life Sci. 2021;287:120120. [DOI] [PubMed] [Google Scholar]
- 22. Almajali B, Al-Jamal HAN, Taib WRW, et al. Thymoquinone, as a novel therapeutic candidate of cancers. Pharmaceuticals. 2021;14:369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sliwinska MA, Mosieniak G, Wolanin K, et al. Induction of senescence with doxorubicin leads to increased genomic instability of HCT116 cells. Mech Ageing Dev. 2009;130:24-32. [DOI] [PubMed] [Google Scholar]
- 24. El-Far AHAM, Munesue S, Harashima A, et al. In vitro anticancer effects of a RAGE inhibitor discovered using a structure-based drug design system. Oncol Lett. 2018;15:4627-4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guzmán C, Bagga M, Kaur A, Westermarck J, Abankwa D. Colony area: an ImageJ plugin to automatically quantify colony formation in clonogenic assays. PLoS One. 2014;9:e92444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248-254. [DOI] [PubMed] [Google Scholar]
- 27. Calvani M, Pasha A, Favre C. Nutraceutical boom in cancer: inside the labyrinth of reactive oxygen species. Int J Mol Sci. 2020;21:1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hu C, Li M, Guo T, et al. Anti-metastasis activity of curcumin against breast cancer via the inhibition of stem cell-like properties and EMT. Phytomedicine. 2019;58:152740. [DOI] [PubMed] [Google Scholar]
- 29. Shanmugam MK, Ahn KS, Hsu A, et al. Thymoquinone inhibits bone metastasis of breast cancer cells through abrogation of the CXCR4 signaling axis. Front Pharmacol. 2018;9:1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang Q, Li F, Liu X, et al. Caspase 3-mediated stimulation of tumor cell repopulation during cancer radiotherapy. Nat Med. 2011;17:860-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Effenberger-Neidnicht K, Schobert R. Combinatorial effects of thymoquinone on the anti-cancer activity of doxorubicin. Cancer Chemother Pharmacol. 2011;67:867-874. [DOI] [PubMed] [Google Scholar]
- 32. Attoub S, Sperandio O, Raza H, et al. Thymoquinone as an anticancer agent: evidence from inhibition of cancer cells viability and invasion in vitro and tumor growth in vivo. Fundam Clin Pharmacol. 2013;27:557-569. [DOI] [PubMed] [Google Scholar]
- 33. Masuelli L, Benvenuto M, Fantini M, et al. Curcumin induces apoptosis in breast cancer cell lines and delays the growth of mammary tumors in neu transgenic mice. J Biol Regul Homeost Agents. 2013;27:105-119. [PubMed] [Google Scholar]
- 34. Zhou Q-M, Sun Y, Lu YY, Zhang H, Chen Q-L, Su SB. Curcumin reduces mitomycin C resistance in breast cancer stem cells by regulating Bcl-2 family-mediated apoptosis. Cancer Cell Int. 2017;17:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhou M, Liu X, Li Z, Huang Q, Li F, Li CY. Caspase-3 regulates the migration, invasion and metastasis of colon cancer cells. Int J Cancer. 2018;143:921-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu YR, Sun B, Zhao XL, et al. Basal caspase-3 activity promotes migration, invasion, and vasculogenic mimicry formation of melanoma cells. Melanoma Res. 2013;23:243-253. [DOI] [PubMed] [Google Scholar]
- 37. Chang F, Lee JT, Navolanic PM, et al. Involvement of PI3K/Akt pathway in cell cycle progression, apoptosis, and neoplastic transformation: a target for cancer chemotherapy. Leukemia. 2003;17:590-603. [DOI] [PubMed] [Google Scholar]
- 38. Bilanges B, Posor Y, Vanhaesebroeck B. PI3K isoforms in cell signalling and vesicle trafficking. Nat Rev Mol Cell Biol. 2019;20:515-534. [DOI] [PubMed] [Google Scholar]
- 39. Fan L, Zhang Y, Zhou Q, et al. Casticin inhibits breast cancer cell migration and invasion by down-regulation of PI3K/Akt signaling pathway. Biosci Rep. 2018;38:BSR20180738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Giehl K. Oncogenic ras in tumour progression and metastasis. Biol Chem. 2005;386:193-205. [DOI] [PubMed] [Google Scholar]
- 41. Porta C, Paglino C, Mosca A. Targeting PI3K/Akt/mTOR signaling in cancer. Front Oncol. 2014;4:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jia T, Zhang L, Duan Y, et al. The differential susceptibilities of MCF-7 and MDA-MB-231 cells to the cytotoxic effects of curcumin are associated with the PI3K/Akt-SKP2-Cip/Kips pathway. Cancer Cell Int. 2014;14:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Guan F, Ding Y, Zhang Y, Zhou Y, Li M, Wang C. Curcumin suppresses proliferation and migration of MDA-MB-231 breast cancer cells through autophagy-dependent Akt degradation. PLoS One. 2016;11:e0146553. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44. Haiaty S, Rashidi M-R, Akbarzadeh M, et al. Thymoquinone inhibited vasculogenic capacity and promoted mesenchymal-epithelial transition of human breast cancer stem cells. BMC Complement Med Ther. 2021;21:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rajput S, Kumar BN, Dey KK, Pal I, Parekh A, Mandal M. Molecular targeting of Akt by thymoquinone promotes G1 arrest through translation inhibition of cyclin D1 and induces apoptosis in breast cancer cells. Life Sci. 2013;93:783-790. [DOI] [PubMed] [Google Scholar]