Abstract
Background
Clonal hematopoiesis of indeterminate potential (CHIP), the age-related expansion of mutant hematopoietic stem cells, confers risk for multiple diseases of aging including hematologic cancer and cardiovascular disease. Whole-exome or genome sequencing can detect CHIP, but due to those assays’ high cost, most population studies have been cross-sectional, sequencing only a single timepoint per individual.
Results
We developed and validated a cost-effective single molecule molecular inversion probe sequencing (smMIPS) assay for detecting CHIP, targeting the 11 most frequently mutated genes in CHIP along with 4 recurrent mutational hotspots. We sequenced 548 multi-timepoint samples collected from 182 participants in the Women’s Health Initiative cohort, across a median span of 16 years. We detected 178 driver mutations reaching variant allele frequency ≥ 2% in at least one timepoint, many of which were detectable well below this threshold at earlier timepoints. The majority of clonal mutations (52.1%) expanded over time (with a median doubling period of 7.43 years), with the others remaining static or decreasing in size in the absence of any cytotoxic therapy.
Conclusions
Targeted smMIPS sequencing can sensitively measure clonal dynamics in CHIP. Mutations that reached the conventional threshold for CHIP (2% frequency) tended to continue growing, indicating that after CHIP is acquired, it is generally not lost. The ability to cost-effectively profile CHIP longitudinally will enable future studies to investigate why some CHIP clones expand, and how their dynamics relate to health outcomes at a biobank scale.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12979-022-00278-9.
Keywords: Clonal hematopoiesis, Somatic mutations, Longitudinal analysis, Aging
Background
Chronological age is the dominant risk factor for cancers and cardiovascular disease – the leading causes of death worldwide [1]. Aging is also associated with a higher prevalence of acquired somatic mutations, especially in frequently regenerating cells, such as hematopoietic stem cells (HSC). Clonal hematopoiesis of indeterminate potential (CHIP) is the age-related expansion (defined as variant allele fraction, VAF ≥2%) of cancer-associated somatic mutations (typically in DNMT3A, TET2, ASXL1, JAK2) in hematopoietic stem cells in the absence of unexplained cytopenia, dysplasia, or neoplasia [2]. Recent whole exome sequence (WES) and whole genome sequence (WGS) analyses of blood-derived DNA have shown that CHIP is increasingly common with advancing age (i.e., approximately 10% of asymptomatic adults older than 70 years of age) [3–6]. While CHIP is a risk factor for hematologic malignancy and all-cause mortality [3, 7, 8], a number of analyses have shown an association with atherosclerotic cardiovascular disease [4, 9, 10]. CHIP is also associated with heightened risk of therapy-related myeloid malignancies [11–14]. These studies underline the importance of CHIP as a novel biomarker for early detection and monitoring of multiple age-related diseases [15, 16]. However, further longitudinal studies are needed for a better understanding of the root causes of CHIP, surveillance strategies, and how CHIP dynamics influence the development of chronic diseases.
Sensitivity for the detection of driver mutations is highly dependent on sequencing depth. Both WGS and WES are suitable for the detection of larger clones (e.g., VAF > 5% in WGS [6, 7], and VAF > 3% in WES [3, 4]). By comparison, deeper coverage, error-corrected targeted sequencing techniques are capable of detecting very small clones [8, 15], which are nearly ubiquitous in healthy adults [17]. Additional studies of apparently healthy adults characterizing longitudinal changes in clone size over time may reveal genetic and environmental factors promoting clonal stability versus progression and yield new insights into mechanisms underlying somatic mutagenesis and aging as well as resultant disease pathogenesis and disease prediction.
Here we present a single-molecule molecular inversion probe sequencing (smMIPS) assay [18], that leverages a cost-effective, ultrasensitive, high-throughput targeted sequencing technique, for the detection of CHIP. We apply this assay to a set of longitudinal peripheral blood DNA samples obtained over a median range of 16 years from 182 post-menopausal women from the Women’s Health Initiative to compare to whole genome sequence analysis and evaluate clonal dynamics.
Results
CHIP panel design and assay validation
We designed a smMIPS capture panel tiling all coding exons (±5 bp) across the 11 most common CHIP genes, along with mutational hotspots in four other genes (Fig. 1A and Table 1). The final capture included 3526 probes, each containing a 9-mer unique molecular index (UMI) for duplicate read removal, spanning a total of 35.2 kb of genomic sequence in the target region. To validate this panel, we first re-sequenced five HapMap lymphoblastoid cell lines (LCLs) and successfully identified all variants defined by 1000G WGS datasets in the target region (n = 152), with no additional variants called. Focusing on positions invariant in these cell lines, we estimate a low sequencing error rate of 0.045% (~ 1/2200 bp; Supplementary Note). Next, to mimic driver mutations across a range of variant allele fractions (VAF) starting well below the conventional CHIP threshold of 2%, we mixed these cell lines’ genomic DNAs at known proportions, and sequenced the mixture. We detected all variants present in this mixture, at allelic fractions tightly correlated with those expected given the cell lines’ mixing proportions (Pearson’s r = 0.998; Fig. 2). Based on samples of known VAF sequenced in replicate, the between-variant reliability of VAF estimated as an intraclass correlation coefficient was 0.998 (95% confidence interval 0.998 to 0.999) (see Supplementary Note).
Fig. 1.
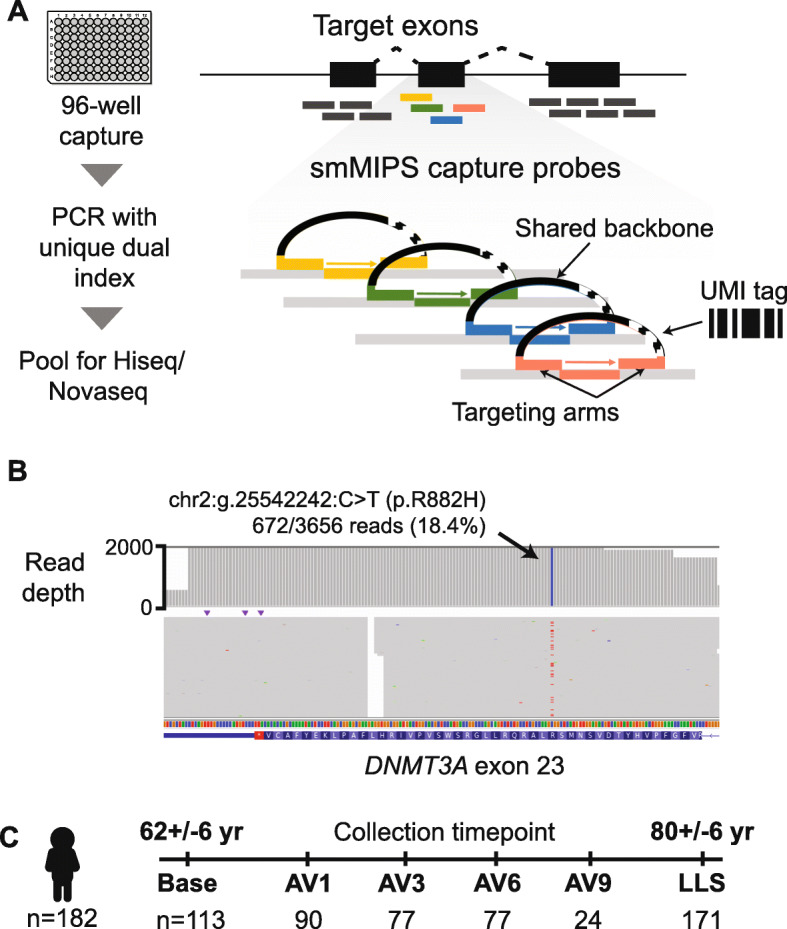
Study design and CHIP sequencing strategy. A Schematic of smMIPS assay design. B Somatic mutation identified as CHIP by smMIPS assay. C Schematic of study design, with sequencing of each subject (n = 182) using samples collected at up to six timepoints including a baseline visit, a series of annual visits (AV), and a final visit (LLS, Long Life Study)
Table 1.
Genes targeted on smMIPs assay and proportion of CHIP in population covered by these target regions
| Gene name | Target size (bp) | MIPS read depth | % of CHIP in Populationb | |
|---|---|---|---|---|
| Median | Mean | |||
| ASXL1 | 4687 | 2054 | 3891 | 7.41% |
| CBL | 2803 | 4880 | 6845 | 0.62% |
| DNMT3A | 3071 | 3163 | 4325 | 45.97% |
| GNB1 | 1023 | 4671 | 6139 | 0.97% |
| PPM1D | 1894 | 3889 | 4856 | 4.01% |
| SF3B1 | 4045 | 3936 | 5073 | 2.46% |
| TET2 | 6165 | 2033 | 3587 | 19.13% |
| TP53 | 1714 | 4063 | 5543 | 1.97% |
| U2AF1 | 880 | 4403 | 6126 | 0.23% |
| ZBTB33 | 2019 | 1983 | 2632 | 2.22% |
| ZNF318 | 6918 | 3240 | 4765 | 1.80% |
| SRSF2a | 1 | 289 | 512 | 1.88% |
| IDH1a | 1 | 1325 | 1797 | 0.09% |
| IDH2a | 1 | 3029 | 3909 | 0.30% |
| JAK2a | 1 | 3839 | 4944 | 1.65% |
| Overall | 35,223 bp | 2803 | 4480 | 88.83% |
aSRSF2, IDH1, IDH2, JAK2 target a single hotspot mutation
bPopulation frequencies of CHIP previously reported in NHLBI TOPMed cohorts [6]
Fig. 2.

Validation by sequencing defined sample mixtures. Observed (mean +/− s.e. across 27 replicates) vs expected variant allele frequency (VAF) for repeated smMIPS sequencing of a defined control mixture of gDNAs from five cell lines, across 152 polymorphic sites. Overall Pearson’s correlation r = 0.998, and r = 0.847 and r = 0.997 for variants with expected VAF ≤2 and > 2%, respectively
CHIP prevalence in WHI samples
We applied our new CHIP sequencing assay to samples collected longitudinally from 182 subjects in the Women’s Health Initiative (mean: 3.0, range: 1–6 samples per subject; summarized in Tables S1 and S2). We obtained an overall median sequencing depth of 2803 (Table 1). After filtering, we detected a total of 206 CHIP driver mutations (defined as VAF ≥2%; Table S3). In a subset of 97 individuals who previously underwent both whole-genome sequencing (to ~30X depth) and deep smMIPS targeted sequencing (>1000X depth) of the same blood sample, 75/81 (92.3%) of the driver mutations called by WGS (mean VAF = 13.2%) were also detected by the smMIPS capture panel. The six mutations found by WGS but missed by smMIPS were in genes not included in the panel (n = 2), in a low coverage target SRSF2 that was optimized in later captures (n = 2), or were long deletions that disrupted probe binding (n = 2 instances of a 23-bp ASXL1 deletion). Despite the difference in sequencing depth between the two methods, we observed a correlation of 0.79 between VAFs measured by WGS and smMIPS, among driver mutations reaching VAF ≥ 2% by both methods (Fig. S1). Due to the deeper sequencing coverage, smMIPS sequencing detected an additional 103 driver mutations that went undetected by WGS; as expected, these tended to be at lower VAFs (mean VAF = 2.6%) for which the read depth provided by WGS (~ 30X coverage) is insufficient.
Characteristics of CHIP mutations detected by smMIPS assay at initial WHI sampling
In the overall WHI sample (N = 182), at the initial timepoint sampled for each individual, 69/182 (38%) were CHIP-positive (carrying at least one driver mutation at VAF ≥2%), while 27/182 (15%) carried two or more such mutations (Fig. 3A). At the baseline timepoint, the most frequently mutated genes were DNMT3A (57% of driver mutations at VAF ≥2%), TET2 (19%), and ASXL1 (6%), consistent with prior WES or WGS reports [4, 6] (Fig. 3C). Among these were recurrent mutations at known hotspots including DNMT3A R882H/R882C (n = 8 individuals) and JAK2 V617F (n = 10 individuals). In aggregate, clone sizes estimated by VAF were not significantly different by the gene mutated (Fig. 3D).
Fig. 3.
CHIP at initial blood draw. A Number of CHIP clones (driver mutations with VAF ≥ 2%) identified per subject at initial draw. B Prevalence of CHIP at VAF ≥2% (blue) or ≥ 10% (orange) by age at initial draw. C Number of mutations (VAF ≥ 2%) per gene. D Driver mutation VAFs grouped by gene
As expected, the prevalence of CHIP (defined as VAF ≥2%) at the initial blood sampling increased with age (Fig. 3B), from 18% in individuals with initial samples taken at age 60 years or younger, compared to 84% among individuals with initial samples taken at 70 years or older, with the high prevalence reflecting in part the selection criteria for subjects previously known to be CHIP-positive at baseline. In cross-sectional analyses, we observed a significant association between baseline BMI and age-adjusted CHIP VAF (p-value = 0.0446), but no association with other available baseline participant characteristics (race/ethnicity, smoking status) in this sample (Table S4). The BMI association is consistent with results from a WGS-based CHIP analysis showing the association of CHIP with obesity in a larger WHI sample [19].
CHIP dynamics in longitudinal samples
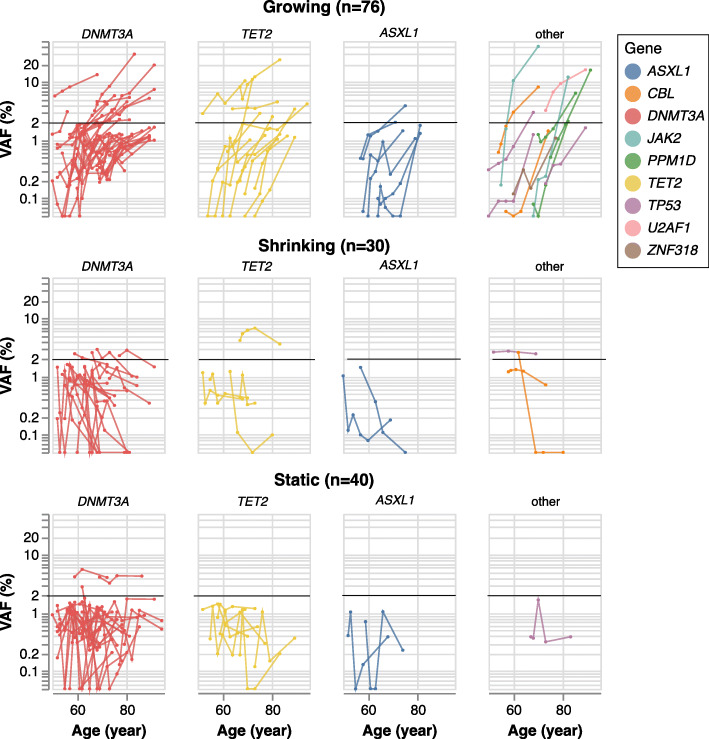
Of the 85 individuals with two or more blood draws for which no driver mutation at VAF ≥ 2% was detected in the initial sample, 49 (58%) developed at least one such driver mutation at the final sampling point an average of 13.9 years later. While these late-arising clones tended to remain small (only 11/49 reached VAF ≥ 10%), many were detectable above background at earlier time points even though they did not meet the working definition of CHIP. We classified the trajectories of each driver mutation, focusing on individuals (n = 65) for which there were three or more timepoints, with a VAF ≥ 1% clone in at least one of them. In these individuals, we identified 146 ‘trackable’ mutations, with 76 on growing trajectories, 30 shrinking, and 40 remaining static (Fig. 4). Among the mutations with a growing trajectory, the median rate of growth was 7.43 years (interquartile range: 4.48, 10.9 years) per doubling.
Fig. 4.
Longitudinal measurement of CHIP dynamics. Trajectories are shown, grouped by direction (rows) and gene (columns). Each trajectory corresponds to a single driver mutation in one subject, shaded by gene; black horizontal represents the VAF = 2% threshold
Once mutations reached appreciable frequency, they tended to continue growing, indicating that after CHIP is acquired, it is generally not lost. Among the 76 growing trajectories, 34 (44.7%) reached the CHIP threshold of VAF ≥ 2% and 16 of these (21.1%) reached a VAF of ≥5%. By contrast, among the shrinking or static trajectories, only 10 and 2 mutations reached these respective VAF thresholds at any single timepoint. Of the 10 non-growing clones that reached VAF ≥ 2% at any timepoint, half involved another, growing trajectory detected in the same individual, and likely reflect competition from separate, fitter clones (Fig. 5).
Fig. 5.
Competition among multiple different driver mutations in individual subjects. Each panel represents a single subject, and each driver mutation is a single line shaded by trajectory (red: growing, gray: static, black: shrinking)
Growth rate varies by driver gene
We next examined driver mutation growth rate by CHIP gene and participant characteristics, as driver mutations in different CHIP genes may confer differential fitness advantages [20–23]. To examine this, we selected the dominant (largest VAF) trajectory from each individual, removing individuals whose dominant clone trajectories are shrinking or static, leading to 43 independent trajectories from 8 CH driver genes (Table 2). Due to the small number of clones for all but the 3 major CHIP driver genes (DNMT3A, TET2, ASXL1; n = 35 trajectories), we grouped trajectories for the other genes into a single “Other” category (n = 8). The rate of growth was higher among the “other” group, which included CBL, JAK2, TP53, and U2AF1, compared to the three major CHIP genes (P = 0.0013, Mann-Whitney U test). In addition, DNMT3A mutant clones were less likely to be in growing trajectories compared with other driver genes (OR = 0.52, P = 0.0085). Similar trends held among participants with only two timepoints, in which CHIP clones were classified as growing or non-growing. No association was observed between age-adjusted change in VAF (after log10 transformation) of dominant clones and any baseline participant characteristics (race/ethnicity, smoking status and BMI) in this sample (Table S5).
Table 2.
Number of trajectories for each driver gene
| Gene | DNMT3A | TET2 | ASXL1 | CBL | JAK2 | PPM1D | TP53 | U2AF1 | Total |
|---|---|---|---|---|---|---|---|---|---|
| Growing, dominant | 23 | 9 | 3 | 2 | 2 | 2 | 1 | 1 | 43 |
| Growing, non-dominant | 20 | 6 | 3 | 0 | 0 | 1 | 2 | 0 | 33 |
| Shrinking | 20 | 5 | 2 | 2 | 0 | 0 | 1 | 0 | 30 |
| Static | 30 | 7 | 2 | 0 | 0 | 0 | 1 | 0 | 40 |
Discussion
Here we describe a rapid and cost-effective smMIPS-based assay that enables detection of CHIP in large scale longitudinal populations. Applying this assay to multi-time point samples from WHI participants demonstrates robust real-world performance in a large collection of longitudinal samples and reveals novel insights on clonal dynamics in a population without hematologic malignancy.
Most studies of CHIP to date rely upon either WGS or WES, or commercial capture kits which have high sequencing or library preparation costs, respectively, ranging from $150–$1000 per sample. The smMIPS approach offers a sensitive alternative at much lower per-sample cost ($30 per sample). Previous work using smMIPS for CHIP detection has been focused on individual hotspots [24] with full gene tiling of only DNMT3A [22]. Our results demonstrate that smMIPS can scale to fully tile gene sets which cumulatively account for nearly 90% of CHIP as determined by WGS.
Our application of the smMIPS assay to WHI reveals several important insights. We observe a significant burden of driver mutations below the conventional CHIP definition (VAF ≥ 2%) [4], enabled by deep sequencing coverage this assay provides. Indeed, for 97 individuals sequenced by both smMIPS and WGS, smMIPS detected 75 of the 81 driver mutations found by WGS, and an even greater number (n = 103) of driver mutations missed by WGS. Although these smMIPS-only clones tended to be less abundant, as expected, a subset nevertheless exceeded the working VAF ≥ 2% definition of CHIP. While these lower-VAF driver mutations may be less likely to have a clinical impact, in another WHI study, somatic mutations with any detectable VAF > 1% were associated with increased risk of acute myeloid leukemia [8]. Likewise, driver mutations in DNMT3A and TET2 at frequencies as low as 1% have been associated with poor prognosis in chronic ischemic heart failure [25]. Thus, the clinical implications of these small clones (VAF range 0.1–2%) remain to be determined in future work, enabled through cost-efficient sequencing via assays like the one described here.
Our results add to recent observations regarding the longitudinal dynamics of clonal hematopoiesis suggesting driver gene-specific differences in clonal fitness. We find that CHIP clones detected among individuals without cancer do not inexorably grow: just over half of those observed did expand, with the remaining, mostly low-frequency clones divided roughly evenly between static and shrinking trajectories. Once mutations reached appreciable frequency, they tended to continue growing. Our results showing that DNMT3A mutant clones are less likely to be in growing trajectories are consistent with those of Fabre et al. [21] who found that clonal growth rate varies according to both age and driver gene mutation, with DNMT3A having a comparatively slow clonal growth rate in older aged adults. Similarly, using longitudinal targeted error-corrected sequence analysis in the Lothian Birth Cohorts, Robertson et al. [23] showed that clonal growth and fitness can differ substantially by gene, with splicing genes (such as SF3B1) having higher growth rates and clonal fitness compared to mutations in common genes such as DNMT3A, TET2 or ASXL1.
In a longitudinal study of ultra-sensitive smMIP-based targeted gene sequencing of obese individuals, Van Deuren et al. [22] reported that metabolic factors such as insulin resistance and high density lipoprotein cholesterol may accelerate expansion of CHIP clones. While we did not detect any association of other baseline participant characteristics such as race/ethnicity, BMI, or smoking on clonal growth, larger sample sizes with serial sampling will be required to identify the genetic and environmental factors contributing to the differing outcomes of clonal competition and growth. This area of investigation has important clinical implications because mutations driving faster clonal growth, as reflected by a more rapid rise in VAF, carry a higher risk of malignant progression [21] and shorter time to development of AML [8].
Our study has several limitations. First, our assay robustly targets genes which account for ~ 90% of CHIP present in the population, so we may be misclassifying ~ 10% of CHIP-positive individuals due to omission of minor CHIP genes from the sequencing panel. This tradeoff was required to make the platform highly cost effective. However, a key benefit of the assay is that it is simple to extend to cover new targets, or to optimize coverage at existing ones, by spiking in new probes. In the present study, we leveraged this capability to add additional probes targeting a highly G + C-rich mutational hotspot in SRSF2, which increased its mean coverage from < 1 to 508. A second limitation of our study is that the availability of multi-time point samples was not uniform due to differences in the WHI study protocol. Third, there are other kinds of clonal hematopoiesis, such as mosaic chromosomal abnormalities (e.g. structural variants) that are not detected with our CHIP assay. These limitations are balanced by the significant strengths of the novel CHIP detection assay applied to one of the largest sample sizes studied to date.
Conclusions
Our development of a novel smMIPS assay for CHIP detection enables scalable and cost-effective identification of CHIP in longitudinal multi-timepoint samples from WHI. This data enabled new observations on the spectrum of clonal hematopoiesis and clonal dynamics. Future investigations using this assay at scale may enable understanding of causes of these clonal dynamic phenomena and how changes in CHIP dynamics relate to diseases of aging associated with clonal hematopoiesis.
Methods
Samples
The Women’s Health Initiative (WHI) is a multicenter prospective study of risk factors for CVD, cancer, osteoporotic fractures, and other causes of morbidity and mortality among postmenopausal women [26]. Between 1993 and 1998, women aged 50–79 years from forty WHI clinical centers throughout the United States (US) were enrolled. All WHI participants completed a baseline screening visit at the time of enrollment which included blood sample collection. WHI participants have been followed prospectively for over 25 years. A subset of participants had blood collected at annual visits (AV) occurring at one, three, six, and 9 years after enrollment (AV1, AV3, AV6, AV9). An additional visit occurred between 2012 and 2013 (mean 15.4 years; range from 14 to 19 years after enrollment) as part of the WHI Long Life Study (LLS), which recruited a subset of 7875 surviving women ranging in age from 63 to 99 years at the time of LLS recruitment [27]. At each visit (baseline, AV1, AV3, AV6, AV9, LLS) genomic DNA was extracted from peripheral blood leukocytes using the 5 Prime DNA extraction kit.
A total of 182 WHI participants (without known prevalent hematological malignancy) were included in the current smMIPS-based sequencing study. These 182 individuals were selected either on the basis of either (a) having previously undergone WGS-based or targeted sequencing -based CHIP determination through the NHLBI TOPMed project (sample set A; N = 100) or having DNA samples at 3 or more time points (sample set B; N = 86). Sample set A was used to compare the detection of driver mutations between WGS and our new smMIPS capture panel. Therefore, we intentionally over-sampled WHI TOPMed participants who were previously determined to have CHIP (driver mutations at VAF ≥ 2% based on WGS or targeted sequencing) in order to directly compare intra-subject CHIP detection and VAF as determined by different assays using the same blood sample at the same time point. Sample set B was primarily to maximize our ability to assess longitudinal CHIP trajectories over time, and therefore includes mainly individuals who had DNA samples available at 4, 5 or 6 different time points. A detailed breakdown of number of samples at each time point (baseline, AV1, AV3, AV6, AV9, LLS) is provided in Table S1. Median age of participants was 62 years at baseline (range: 50–78 years) and 81 years (range: 66–95 years) at the LLS visit, respectively.
Single molecule molecular inversion probe sequencing (smMIPS) assay
A smMIPS capture panel was designed to tile coding exons (+/− 5 bp) of the 11 most common CHIP genes [9] and recurrent mutational hotspots in four others (Table 1). Probe sequences were selected as previously described [28], with adjustments to eliminate the need for custom sequencing primers. Briefly, probe libraries were synthesized as a 12 k oligo pool by CustomArray (Bothell, WA) Inc., and subjected to bulk PCR amplification using flanking primers jklab0255_2019mipsPrep1f (GAGATCGGCGCGTTAGAAGAC) and jklab0256_2019mipsPrep1r (TGCAGGATCTAGGGCGAAGAC). PCR product was cleaned with 2.5X SPRI beads and eluted in 1X NEB cut smart buffer. To generate capture-ready probe pools, flanking adaptors were removed by BbsI-HF (#R3539L, NEB; Ipswitch, MA) digestion, overnight at 37 °C. Digested probes were cleaned by incubating with 1x volume SPRI beads (supplemented with 5 volumes isopropanol for 20 minutes), followed by washes in 70% ethanol and elution in Tris-EDTA pH 8. Poorly captured regions were tiled with additional probes (N = 112), synthesized as an oPool library by Integrated DNA Technologies (Coralville, IA) lacking flanking amplification adaptors and with 5′ phosphates. Original and make-up probes were combined into a single pool before use.
Capture reactions were assembled in a 96-well format, in 20 ul volume containing: probes (150:1 M excess to genomic DNA targets), 1X Ampligase buffer, 1 U Ampligase (Lucigen; Madison, WI), dNTPs at 0.4 uM each and 0.32 ul Hemo KlenTaq polymerase (NEB). Plates were incubated in a thermocycler at 95 °C for 10 minutes, 95 °C → 60 °C at − 0.1 °C/sec, followed by a hold at 60 °C for 18–24 hours. Exonuclease treatment was continued immediately after capture by adding 2 ul of mix containing 1X Ampligase buffer, 5 U Exonuclease I (NEB), and 25 U Exonuclease III (NEB) to each sample. Reactions were incubated at 37 °C for 45 minutes and 95 °C for 2 minutes. Dual indexed sequencing libraries were constructed by PCR amplification using indexing primers directed against common sequences on the probe backbone. Libraries were pooled at equal volumes, purified by 0.9X SPRI beads, and sequenced in batches of 196 on Hiseq 4000 or Novaseq instruments with paired-end 150-bp reads. Reagent, consumable, and sequencing costs total approximately $30 USD/sample.
Sequencing reads were aligned to the human reference genome (build 37) with bwa mem [29], and a custom sequencing pipeline (https://github.com/kitzmanlab/mimips) was used for post-alignment processing to remove probe arm sequences from each alignment and filter reads with duplicate unique molecular identifiers (UMIs).
smMIPS assay validation and reliability
To validate the clone size detection limit of the smMIPS method, we prepared mixtures of gDNAs from five lymphoblastoid cell lines (GM06994, GM12878, GM20847, GM12877 and GM18507) with known genotypes, combined at 78.8, 16, 4, 1, 0.25%. Within the target region, these cell lines have 152 known variants as defined by the 1000 Genome Project (1000G) WGS genotypes and by detecting germline variants by sequencing cell lines individually. In the resulting mixture, their expected VAFs range from 0.125 to 100%. These variants constituted the true positive variant set. We also defined as ‘true negative’ sites 13 common polymorphism SNVs absent from all of the five cell lines, and those sites (+/− 50 bp) were defined as true negative variants. The positive control mixture was included with each sequencing batch for a total of 27 replicates. The between-variant reliability of VAF estimated as an intraclass correlation coefficient was 0.998 (95% confidence interval 0.998 to 0.999) (see Supplementary Note).
Variant calling
Somatic SNPs and indels were called using LoFreq 2.1.3.1 [30], requiring minimum coverage 40, with ≥5 reads supporting the alternate allele and a variant allele frequency (VAF) ≥ 0.1%. Variants present in ≥5% of samples at a VAF of 1–10% were discarded as likely recurrent artifacts.
CHIP calling
Variants were annotated using ANNOVAR software [31]. Variant calls processed using an existing filtering pipeline based upon gene name, variant functional class, and populational allele frequency [6]; workflow is available at available at https://app.terra.bio/#workspaces/terra-outreach/CHIP-Detection-Mutect2/notebooks. For ZBTB33 and ZNF318, two genes not listed in [6], we included variants annotated as frameshift/splice-site/nonsense or nonsynonymous [32]. The full list of specific mutations queried is presented in Table S5. We manually reviewed alignments for selected CHIP variant calls using Integrative Genomics Viewer (IGV) [33].
CHIP clone trajectories
To characterize the longitudinal trajectory of each CHIP clone over time, we restricted our analysis to individuals who (a) underwent smMIPS sequencing at least 3 time points and (b) had at least one driver mutation detectable at VAF > 1% at any of the timepoints. We excluded any variants with alternate read count < 2 or total read depth < 200. For each driver mutation meeting these criteria, we modeled the trajectory by fitting a linear regression: log10(VAF) = C + β * age; VAFs of zero were set to a minimum of 10− 4 (reflecting a conservative limit of detection for smMIPS), and each observation was weighted by the square root of the read depth. To further characterize clonal dynamics, we classified each trajectory based on linear trajectory, as (a) growing (β > 0, P < 0.5), (b) shrinking (β ≤ 0, P < 0.5), or (c) static (P ≥ 0.5). For trajectory analysis, we excluded CHIP clones with starting VAF > 10%, for which an exponential growth assumption may not fit.
Association between participant characteristics and CHIP VAF and growth rate
For cross-sectional analyses at a single time point, we fit linear and logistic regression models to assess the relationship of either CHIP prevalence (total clones or large clones only) or log-transformed VAF to age at blood draw, race/ethnicity, smoking status, or BMI. We used the first visit time point for each subject. To assess the relationship of these same participant characteristics, to clone growth over time, we utilized longitudinal data from all individuals with sequencing data from at least two time points with positive VAF observations (N = 148 individuals). For each participant, we first selected a single driver clone, prioritizing as the predominant clone those with the highest VAF at any follow-up timepoint. We used a linear regression to determine the effect of age, race/ethnicity, smoking, or BMI on the difference (after log10 transformation) between the first non-zero VAF value and the last non-zero VAF. All statistical analyses were adjusted for the first visit time of year and performed in R version 4.2 (R Core Team, URL https://www.R-project.org/).
Supplementary Information
Authors’ contributions
A.G.B., A.P.R., P.N. and J.O.K. conceived and designed the study. B.B. performed DNA library preparation and sequencing. M.M.U., Y.Z., A.G.B., and J.O.K. analyzed the data. A.G.B., P.N., A.P.R., M.M.U., Y.Z., and J.O.K. wrote the manuscript. All authors received and approved the manuscript.
Funding
This work was supported National Institutes of Health grant R01HL148565 (A.P.R., E.W., P.N., S.J., J.O.K., C.K., P.D., J.M.), Burroughs Wellcome Fund Career Award for Medical Scientists (S.J), ASH Scholar Award (S.J.), Fondation Leducq grant TNE-18CVD04 (S.J.), Support from the Ludwig Center for Cancer Stem Cell Research at Stanford University (S.J.), National Institutes of Health grant DP2-HL157540 (S.J.), National Institutes of Helath grant R01HL146500 (A.P.R.), National Institutes of Health grant DP5OD029586 (A.G.B.), a Burroughs Wellcome Fund Career Award for Medical Scientists (A.G.B), and funding to A.G.B. from the E.P. Evans Foundation and the RUNX1 Research Program, the American Heart Association grant 940136 (M.C.H.), National Institutes of Health R01HL34594 and R01HL145386 (J.M.). The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contracts 75N92021D00001, 75N92021D00002, 75N92021D00003, 75N92021D00004, 75N92021D00005.
Availability of data and materials
Sequencing data underlying this study is deposited in dbGaP (phs000200.v12.p3).
Declarations
Ethics approval and consent to participate
Human samples were obtained with informed consent, and our study was reviewed and approved by the Fred Hutchinson Cancer Center Institutional Review Board (IRB #10186).
Consent for publication
Not applicable.
Competing interests
All unrelated to the present work: S.J. is a consultant to Novartis, Roche Genentech, AVRO Bio, and Foresite Labs, and on the scientific advisory board for Bitterroot Bio; S.J., P.N., and A.G.B. are founders, equity holders, and/or scientific advisory board members of TenSixteen Bio; J.O.K is an advisor to MyOme Inc. P.N. reports grant support from Amgen, Apple, AstraZeneca, Boston Scientific, and Novartis, spousal employment and equity at Vertex, consulting income from Apple, AstraZeneca, Novartis, Genentech / Roche, Blackstone Life Sciences, and Foresite Labs, and is a scientific advisor board member of geneXwell, all unrelated to this work. M.C.H. reports consulting fees from CRISPR Therapeutics. All other authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Md Mesbah Uddin, Ying Zhou, Alexander G. Bick, Bala Bharathi Burugula are contributed equally to this work.
Pradeep Natarajan, Alexander P. Reiner and Jacob O. Kitzman jointly supervised this work.
References
- 1.GBD 2016 Causes of Death Collaborators Global, regional, and national age-sex specific mortality for 264 causes of death, 1980–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390(10100):1151–1210. doi: 10.1016/S0140-6736(17)32152-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126(1):9–16. doi: 10.1182/blood-2015-03-631747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Genovese G, Kahler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20(12):1472–1478. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bick AG, Weinstock JS, Nandakumar SK, Fulco CP, Bao EL, Zekavat SM, et al. Inherited causes of clonal haematopoiesis in 97,691 whole genomes. Nature. 2020;586(7831):763–768. doi: 10.1038/s41586-020-2819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zink F, Stacey SN, Norddahl GL, Frigge ML, Magnusson OT, Jonsdottir I, et al. Clonal hematopoiesis, with and without candidate driver mutations, is common in the elderly. Blood. 2017;130(6):742–752. doi: 10.1182/blood-2017-02-769869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desai P, Mencia-Trinchant N, Savenkov O, Simon MS, Cheang G, Lee S, et al. Somatic mutations precede acute myeloid leukemia years before diagnosis. Nat Med. 2018;24(7):1015–1023. doi: 10.1038/s41591-018-0081-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaiswal S, Natarajan P, Silver AJ, Gibson CJ, Bick AG, Shvartz E, et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med. 2017;377(2):111–121. doi: 10.1056/NEJMoa1701719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bick AG, Pirruccello JP, Griffin GK, Gupta N, Gabriel S, Saleheen D, et al. Genetic interleukin 6 signaling deficiency attenuates cardiovascular risk in clonal hematopoiesis. Circulation. 2020;141(2):124–131. doi: 10.1161/CIRCULATIONAHA.119.044362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillis NK, Ball M, Zhang Q, Ma Z, Zhao Y, Yoder SJ, et al. Clonal haemopoiesis and therapy-related myeloid malignancies in elderly patients: a proof-of-concept, case-control study. Lancet Oncol. 2017;18(1):112–121. doi: 10.1016/S1470-2045(16)30627-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu JI, Dayaram T, Tovy A, De Braekeleer E, Jeong M, Wang F, et al. PPM1D mutations drive clonal hematopoiesis in response to cytotoxic chemotherapy. Cell Stem Cell. 2018;23(5):700–13 e6. doi: 10.1016/j.stem.2018.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolton KL, Ptashkin RN, Gao T, Braunstein L, Devlin SM, Kelly D, et al. Cancer therapy shapes the fitness landscape of clonal hematopoiesis. Nat Genet. 2020;52(11):1219–1226. doi: 10.1038/s41588-020-00710-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coombs CC, Zehir A, Devlin SM, Kishtagari A, Syed A, Jonsson P, et al. Therapy-related clonal hematopoiesis in patients with non-hematologic cancers is common and associated with adverse clinical outcomes. Cell Stem Cell. 2017;21(3):374–82 e4. doi: 10.1016/j.stem.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abelson S, Collord G, Ng SWK, Weissbrod O, Mendelson Cohen N, Niemeyer E, et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature. 2018;559(7714):400–404. doi: 10.1038/s41586-018-0317-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolton KL, Zehir A, Ptashkin RN, Patel M, Gupta D, Sidlow R, et al. The clinical Management of Clonal Hematopoiesis: creation of a clonal hematopoiesis clinic. Hematol Oncol Clin North Am. 2020;34(2):357–367. doi: 10.1016/j.hoc.2019.11.006. [DOI] [PubMed] [Google Scholar]
- 17.Young AL, Challen GA, Birmann BM, Druley TE. Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat Commun. 2016;7:12484. doi: 10.1038/ncomms12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hiatt JB, Pritchard CC, Salipante SJ, O'Roak BJ, Shendure J. Single molecule molecular inversion probes for targeted, high-accuracy detection of low-frequency variation. Genome Res. 2013;23(5):843–854. doi: 10.1101/gr.147686.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haring B, Reiner AP, Liu J, Tobias DK, Whitsel E, Berger JS, et al. Healthy lifestyle and clonal hematopoiesis of indeterminate potential: results from the Women's Health Initiative. J Am Heart Assoc. 2021;10(5):e018789. doi: 10.1161/JAHA.120.018789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watson CJ, Papula AL, Poon GYP, Wong WH, Young AL, Druley TE, et al. The evolutionary dynamics and fitness landscape of clonal hematopoiesis. Science. 2020;367(6485):1449–1454. doi: 10.1126/science.aay9333. [DOI] [PubMed] [Google Scholar]
- 21.Fabre MA, JGd A, Fiorillo E, Mitchell E, Damaskou A, Rak J, et al. The longitudinal dynamics and natural history of clonal haematopoiesis. bioRxiv. 2021; 2021.08.12.455048. [DOI] [PMC free article] [PubMed]
- 22.van Deuren RC, Andersson-Assarsson JC, Kristensson FM, Steehouwer M, Sjöholm K, Svensson P-A, et al. Expansion of mutation-driven haematopoietic clones is associated with insulin resistance and low HDL-cholesterol in individuals with obesity. bioRxiv. 2021; 2021.05.12.443095.
- 23.Robertson NA, Latorre-Crespo E, Terradas-Terradas M, Purcell AC, Livesey BJ, Marsh JA, et al. Longitudinal dynamics of clonal hematopoiesis identifies gene-specific fitness effects. bioRxiv. 2021; 2021.05.27.446006. [DOI] [PMC free article] [PubMed]
- 24.Acuna-Hidalgo R, Sengul H, Steehouwer M, van de Vorst M, Vermeulen SH, Kiemeney L, et al. Ultra-sensitive sequencing identifies high prevalence of clonal hematopoiesis-associated mutations throughout adult life. Am J Hum Genet. 2017;101(1):50–64. doi: 10.1016/j.ajhg.2017.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Assmus B, Cremer S, Kirschbaum K, Culmann D, Kiefer K, Dorsheimer L, et al. Clonal haematopoiesis in chronic ischaemic heart failure: prognostic role of clone size for DNMT3A- and TET2-driver gene mutations. Eur Heart J. 2021;42(3):257–265. doi: 10.1093/eurheartj/ehaa845. [DOI] [PubMed] [Google Scholar]
- 26.The Women's Health Initiative Study Group Design of the Women's health initiative clinical trial and observational study. Control Clin Trials. 1998;19(1):61–109. doi: 10.1016/S0197-2456(97)00078-0. [DOI] [PubMed] [Google Scholar]
- 27.The Women's Health Initiative Study Group . Long Life Study (W64) 2013. [Google Scholar]
- 28.Pérez Millán MI, Vishnopolska SA, Daly AZ, Bustamante JP, Seilicovich A, Bergadá I, Braslavsky D, Keselman AC, Lemons RM, Mortensen AH, Marti MA, Camper SA, Kitzman JO. Next generation sequencing panel based on single molecule molecular inversion probes for detecting genetic variants in children with hypopituitarism. Mol Genet Genomic Med. 2018;6(4):514–25. https://doi.org/10.1002/mgg3.395. Epub ahead of print. PMID: 29739035; PMCID: PMC6081231. [DOI] [PMC free article] [PubMed]
- 29.Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilm A, Aw PP, Bertrand D, Yeo GH, Ong SH, Wong CH, et al. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012;40(22):11189–11201. doi: 10.1093/nar/gks918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beauchamp EM, Leventhal M, Bernard E, Hoppe ER, Todisco G, Creignou M, et al. ZBTB33 is mutated in clonal hematopoiesis and myelodysplastic syndromes and impacts RNA splicing. Blood Cancer Discov. 2021;2(5):500–517. doi: 10.1158/2643-3230.BCD-20-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson JT, Thorvaldsdottir H, Wenger AM, Zehir A, Mesirov JP. Variant review with the integrative genomics viewer. Cancer Res. 2017;77(21):e31–ee4. doi: 10.1158/0008-5472.CAN-17-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data underlying this study is deposited in dbGaP (phs000200.v12.p3).