Abstract
Background
Chronic myelomonocytic leukemia (CMML) is a rare and heterogeneous hematological malignancy. It has been shown that the molecular abnormalities such as ASXL1, TET2, SETBP1, and SRSF2 mutations are common in Caucasian population.
Methods
We retrospectively analyzed 178 Chinese CMML patients. The targeted next generation sequencing (NGS) was used to evaluate 114 gene variations, and the prognostic factors for OS were determined by COX regression analysis.
Results
The CMML patients showed a unique mutational spectrum, including TET2 (36.5%), NRAS (31.5%), ASXL1 (28.7%), SRSF2 (24.7%), and RUNX1 (21.9%). Of the 102 patients with clonal analysis, the ancestral events preferentially occurred in TET2 (18.5%), splicing factors (16.5%), RAS (14.0%), and ASXL1 (7.8%), and the subclonal genes were mainly ASXL1, TET2, and RAS. In addition, the secondary acute myeloid leukemia (sAML) transformed from CMML often had mutations in DNMT3A, ETV6, FLT3, and NPM1, while the primary AML (pAML) demonstrated more mutations in CEBPA, DNMT3A, FLT3, IDH1/2, NPM1, and WT1. It was of note that a series of clones were emerged during the progression from CMML to AML, including DNMT3A, FLT3, and NPM1. By univariate analysis, ASXL1 mutation, intermediate- and high-risk cytogenetic abnormality, CMML-specific prognostic scoring system (CPSS) stratifications (intermediate-2 and high group), and treatment options (best supportive care) predicted for worse OS. Multivariate analysis revealed a similar outcome.
Conclusions
The common mutations in Chinese CMML patients included epigenetic modifiers (TET2 and ASXL1), signaling transduction pathway components (NRAS), and splicing factor (SRSF2). The CMML patients with DNMT3A, ETV6, FLT3, and NPM1 mutations tended to progress to sAML. ASXL1 mutation and therapeutic modalities were independent prognostic factors for CMML.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40164-022-00284-z.
Keywords: CMML, Gene mutation, Clonal evolution, Secondary AML (sAML), Overall survival (OS)
Background
Chronic myelomonocytic leukemia (CMML) is a clonal and heterogeneous hematological neoplasm, characterized by persistent monocytosis and hematopoietic dysplasia. Approximately 15–30% of CMML patients progress to secondary acute myeloid leukemia (sAML) [1–5]. The median age of CMML is 65–75 year-old with a disposition to male (1.5–3:1) [6, 7]. According to the WHO classification, CMML is classified into three subtypes: CMML-0 (< 2% in peripheral blood [PB] and < 5% in bone marrow [BM]), CMML-1 (2–4% in PB and 5–9% in BM), and CMML-2 (5–19% in PB and 10–19% in BM). Based on white blood cell (WBC) count, CMML is further defined as myeloproliferative CMML (MP-CMML) or myelodysplastic CMML (MD-CMML) [8].
Recent studies have demonstrated that CMML carried multiple mutations, involving epigenetic regulation, such as DNA methylation (TET2, DNMT3A, IDH1/2) and histone transcription (RUNX1, EZH2, ASXL1), splicing factors (SF3B1, U2AF1, ZRSR2, SRSF2), and signaling transduction pathway (JAK2, KRAS, NRAS, CBL, FLT3) [7, 9–12]. Among them, mutations in TET2, SRSF2, and ASXL1 tended to occurred in MD-CMML, whereas mutations in RAS signaling transduction pathway were more prevalent in MP-CMML [13, 14]. The prognostic system for CMML generally included clinical features and lab findings, such as blood/bone marrow biopsies, as well as mutations in ASXL1, NRAS, RUNX1, and SETBP1 et al. [15–20].
To reveal the molecular landscape, we retrospectively analyzed 178 Chinese CMML patients and investigated their mutational spectrum. The clonal dynamics upon disease progression from CMML to sAML was studied, and the prognostic factors for the overall survival (OS) of CMML patients were evaluated.
Methods
Patients
A total of 411patients from June 2015 to January 2021 were collected and sorted in this study, including 178 CMML, 13 sAML (3 transformed from the CMML cases), and 223 primary AML (AML-M4/M5). The risk stratification was as follows [1, 2]: low-risk, a diploid karyotype or sole -Y; high-risk, trisomy 8, alterations of chromosome 7, or complex karyotype; intermediate-risk, all other karyotypes. In addition, 92 CMML patients with available clinical information were evaluated for OS in our cohort. The median follow-up time was 32 months. The OS was calculated from the day of diagnosis to the day of death regardless of cause or last contact. This study was approved by the IRB of Sino-US Diagnostics.
Next-generation sequencing (NGS)
A total of 114 genes associated with hematological disorders were analyzed using NGS (Additional file 1: Table S1). Genomic DNAs were extracted from the BM mononuclear cells according to the manufacturer’s instructions (TIANGEN, China). The genes were amplified in the 275-bp libraries by customized primers (Life Technologies, USA), and the primers covered the entire coding regions and canonical splice sites. The amplified products were sequenced on Ion Torrent platform (Life Technologies, USA). The average sequencing depth was 1000× per patient and mutations with VAF (variant allele frequency) > 2% were considered positive.
The variants were annotated by ANNOVAR using following resources: RefGene, public population databases, 1000 Genomes Project Phase 3 database, protein function prediction databases [21, 22], and in-house database. These databases were listed in Additional file 2: Table S2. In addition, some variants, such as frameshift mutations or total reads less than 30, were manually reviewed using the Integrative Genomics Viewer (Broad Institute, bam) [23].
Ancestral vs. sub-clonal variants
To distinguish the ancestral/founder from the secondary/subclonal variants, the VAF was used to estimate the clonal hierarchy of each sample and the following criteria were formulated: (a) a cut-off value of at least 5% difference between VAFs was used to define an ancestral mutation. If the difference was less than 5%, a co-dominant was called [24–26]; and (b) to analyze the transformed cases, mutations appearing at progression to CMML-2/AML but not present initially were deemed subclones [27]. The ambiguous data were disregarded.
Statistical analysis
The Chi-square test was used to compare the enumeration data between different groups, and the student t-test or analysis of variance was used to compare the measurement data. All statistical analyses were performed using SPSS version 24.0 (IBM, Armonk, NY, USA) and R software. Correlations between mutations were conducted by Pearson coefficients. The forest plots were performed by the GraphPad Prism 7.0. The univariate analysis was performed to reveal the OS related factors, and the multivariate Cox proportional hazards regression analysis was used to further evaluate the factors with p < 0.05 from the univariate analysis. The OS curve was plotted by the Kaplan-Meier method and tested by the log-rank. All p-values were two-tailed, and p < 0.05 was considered to be statistically significant.
Results
Clinical characteristics
The study included 178 CMML patients with 65 CMML-0, 58 CMML-1, and 55 CMML-2. The median age was 62 year-old (17–90), and 66.3% of CMML patients were male. Most of MD-CMML patients (66/83; 79.5%) presented in the low-risk group, more than MP-CMML patients (63/95; 66.3%). The MP-CMML patients preferentially exhibited splenomegaly than the MD-CMML patients (p = 0.036). Moreover, the MP-CMML patients showed significantly higher levels of absolute neutrophil count (ANC), absolute monocyte count (AMC), and lactate dehydrogenase (LDH) than the MD-CMML patients (p = 0.000, p = 0.002, p = 0.008, respectively) (Table 1). The CMML-0 had more low-risk patients (55/65; 84.6%) in contrast to the CMML-1 (38/58, 65.5%; p = 0.014) and CMML-2 (36/55, 65.5%; p = 0.015), whereas the CMML-1 (13/58, 22.4%; p = 0.021) and CMML-2 (13/55; 23.6%; p = 0.015) tended to be in the high-risk group compared with the CMML-0 (5/65, 7.7%). Up to 28.4% of CMML patients exhibited splenomegaly. The platelet (PLT) count increased along with the higher disease stage (p = 0.015), whereas the WBC count showed a reverse trend (p = 0.007). Most CMML-1/2 patients received hypomethylating agents (HMAs) ± chemotherapy/allogeneic hematopoietic stem cell transplantation (allo-HSCT) (Table 2).
Table 1.
The clinical information of CMML patients classified as MD/MP subtypes
| Variables | Total CMML (n = 178) |
MD-CMML (n = 83) |
MP-CMML (n = 95) |
P value |
|---|---|---|---|---|
| Age in years; median (range) | 62 (17–90) | 63 (24–90) | 60 (17–85) | 0.362 |
| Sex (male); n (%) | 118 (66.3%) | 54 (65.1%) | 64 (67.4%) | 0.745 |
| Hemoglobin g/L; median (range) | 85 (38–173) | 85 (38–140) | 87 (40–173) | 0.410 |
| ANC ×109/L; median (range) | 4.75 (0.45–29.97) | 2.17 (0.45–6.15) | 10.9 (1.14–29.97) | 0.000 |
| AMC ×109/L; median (range) | 2.43 (1.01–16.32) | 1.8 (1.01–5.69) | 3.23 (1.11–16.32) | 0.002 |
| Platelets×109/L; median (range) | 76 (3–977) | 69 (11–900) | 85 (3–977) | 0.828 |
| LDH IU/ml; median (range) | 279.1 (120–1403) | 224.6 (120–977.1) | 333.5 (152.4–1403) | 0.008 |
| Cytogenetic risk groupa n (%) | 0.032 | |||
| Low-risk | 129 (72.5%) | 66 (79.5%) | 63 (66.3%) | 0.049 |
| Intermediate-risk | 18 (10.1%) | 9 (10.8%) | 9 (9.5%) | 0.762 |
| High-risk | 31 (17.4%) | 8 (9.7%) | 23 (24.2%) | 0.011 |
| Splenomegaly ; n (%) | 42 (28.4%) | 13 (19.6%) | 29 (35.3%) | 0.036 |
| Treatment strategy; n (%) | 0.821 | |||
| Best supportive care | 28 (30.4%) | 12 (28.6%) | 16 (32.0%) | |
| HMAs ± chemotherapy/allo-HSCT | 64 (69.6%) | 30 (71.4%) | 34 (68.0%) | |
FAB classification MD-CMML: WBC < 13 × 109/L, (109: 9 needs to be superscripted) MP-CMML: WBC ≥ 13 × 109/L (109: 9 needs to be superscripted)
LDH: Lactate dehydrogenase
aCytogenetic risk groups [1, 2]: low-risk: a diploid karyotype or -Y; high-risk: trisomy 8, alterations of chromosome 7, as well as complex karyotype; intermediate-risk: all other karyotypes
Table 2.
The clinical information of 178 CMML patients classified as CMML-0, -1, and -2
| Variables | CMML-0 (n = 65) |
CMML-1 (n = 58) |
CMML-2 (n = 55) |
P value | 0 vs. 1 | 0 vs. 2 | 1 vs. 2 |
|---|---|---|---|---|---|---|---|
| Age in years; median (range) | 65 (24–86) | 62 (23–82) | 59 (17–90) | 0.085 | – | – | – |
| Sex (male); n (%) | 41 (63.1%) | 43 (73.1%) | 34 (61.8%) | 0.383 | – | – | – |
| Hemoglobin (g/L); median (range) | 89 (53–173) | 82 (38–153) | 84.0 (39–131) | 0.049 | 0.267 | 0.058 | 1.000 |
| WBC (×109/L); median (range) | 9.32 (1.38–158.9) | 12.79 (3.15–177.7) | 23.7 (1.22-323.66) | 0.007 | 1.000 | 0.007 | 0.068 |
| ANC (×109/L); median (range) | 4.66 (0.51–32) | 3.92 (0.45–20.62) | 6.35 (1.06–29.97) | 0.606 | – | – | – |
| AMC (×109/L); median (range) | 2.4 (1.11–6.5) | 2.43 (1.01–6.45) | 2.85 (1.01–16.32) | 0.600 | – | – | – |
| Platelets(×109/L); median (range) | 101 (11–977) | 66 (6-900) | 62 (3-610) | 0.015 | 0.046 | 0.037 | 1.000 |
| LDH (IU/ml); median (range) | 230.5 (120–1403) | 266 (152.4–623) | 352 (121.6–1298) | 0.046 | 1.000 | 0.097 | 0.136 |
| Cytogenetic risk groupa; n (%) | |||||||
| Low-risk | 55 (84.6%) | 38 (65.5%) | 36 (65.5%) | 0.023 | 0.014 | 0.015 | 0.994 |
| Intermediate-risk | 5 (7.7%) | 7 (12.1%) | 6 (10.9%) | 0.704 | – | – | – |
| High-risk | 5 (7.7%) | 13 (22.4%) | 13 (23.6%) | 0.034 | 0.021 | 0.015 | 0.877 |
| Splenomegaly; n (%) | 16 (28.6%) | 12 (26.1%) | 14 (30.4%) | 0.898 | – | – | – |
| Treatment strategy; n (%) | |||||||
| Best supportive care | 17 (51.5%) | 6 (23.1%) | 5 (15.2%) | 0.004 | 0.034 | 0.004 | 0.512 |
| HMAs ± chemotherapy/allo-HSCT | 16 (48.5%) | 20 (76.9%) | 28 (84.8%) | ||||
The spectrum of gene mutations
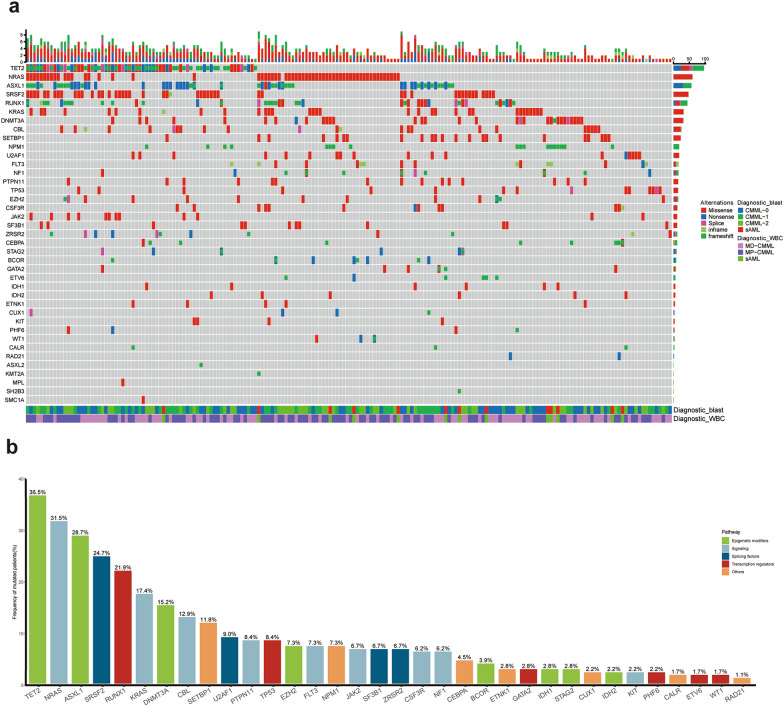
The targeted panel NGS was performed in 178 CMML, 13 sAML (3 transformed from CMML), and 223 pAML. As shown in Fig. 1a, the CMML patients had a unique mutational spectrum, and 83.1% (148/178) carried at least two oncogenic mutations. The common mutations included TET2 (36.5%), NRAS (31.5%), ASXL1 (28.7%), SRSF2 (24.7%), and RUNX1 (21.9%) (Fig. 1b). Among those mutations, TET2 and RUNX1 had multiple mutation forms, including frameshift/inframe, nonsense, splicing, and missense mutations. ASXL1 preferentially presented with nonsense or frameshift mutations, whereas N/KRAS, SRSF2, SETBP1, DNMT3A, and CBL were mainly missense mutations.
Fig. 1.
a The distribution of somatic mutations in CMML. Each column represents a patient and each row corresponds to a gene. The color of each rectangle represents the type of gene mutation, the diagnosis, and the karyotype of each patient. The bar graph indicates the mutation frequency, the mutation type, and the cytogenetics of each patient; b The overview of pathways involving in the identified mutations. The histogram represents the frequency of common gene mutations, and the mutation frequency is expressed as a percentage. FLT3 mutations consist of FLT3-ITD and FLT3-TKD
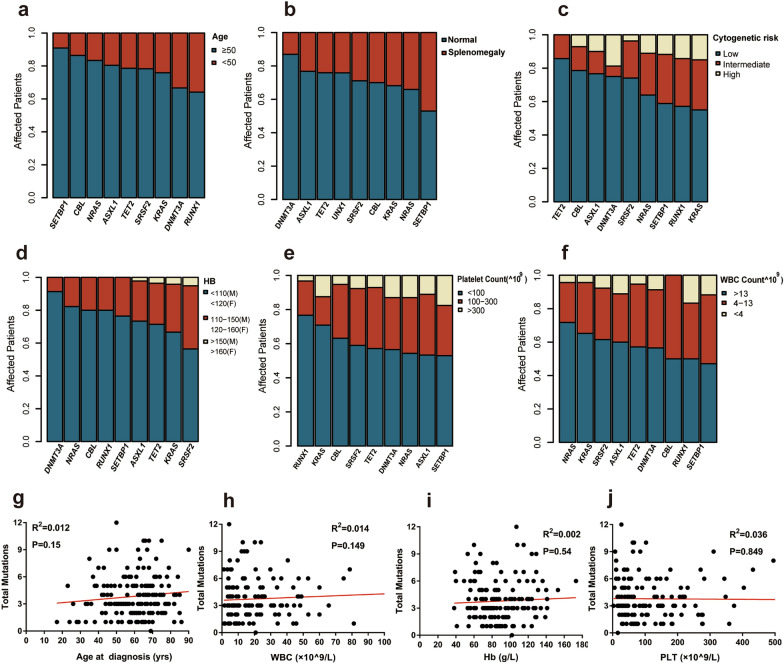
Further analysis showed that RUNX1 mutation occurred preferentially in CMML patients less than 50 year-old (p < 0.05). Additionally, patients with SRSF2 mutation were prone to have normal hemoglobin (Hb) level (p < 0.05), whereas patients with RUNX1 mutation tended to have lower PLT count (p = 0.05). Notably, NRAS mutation was correlated with leukocytosis (p < 0.05) (Fig. 2a–f). No significant correlation was found between the total number of mutations and age, Hb, PLT count, or WBC count (Fig. 2g–j).
Fig. 2.
The proportions of patients with various mutations based on age (a), splenomegaly (b), cytogenetic risk (c), HB (d), PLT (e), and WBC (f). The correlation of total CMML mutations versus age at diagnosis (g), WBC (h), Hb (i), and PLT level (j)
The correlation between genetic mutations
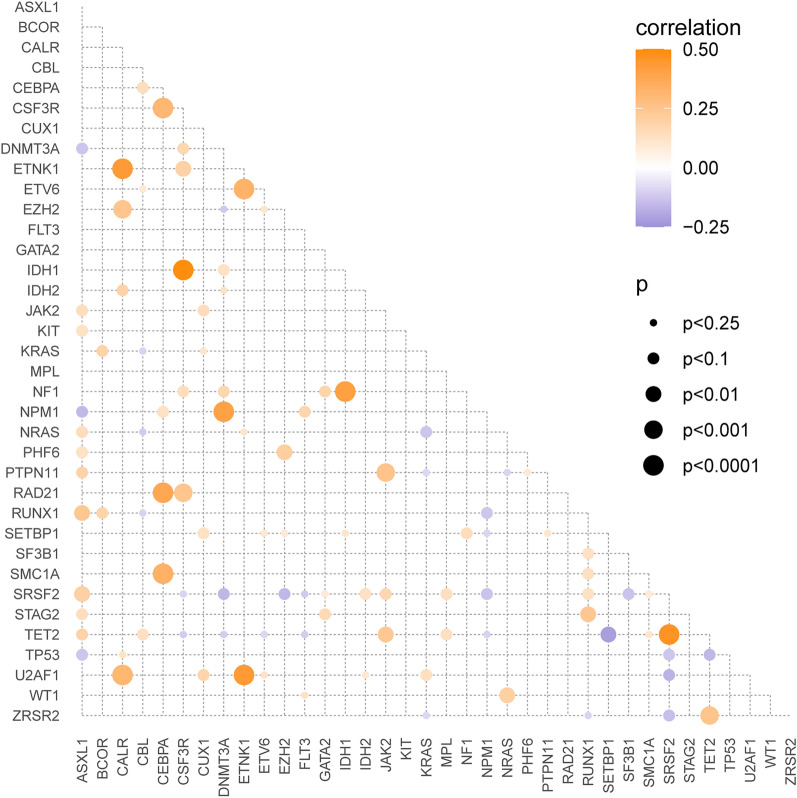
As shown in Fig. 3, a co-mutation was detected between IDH1 and CSF3R or NF1 (r = 0.490, p < 0.0001; r = 0.414, p < 0.0001, respectively). Similarly, SRSF2 mutation co-presented with TET2 mutation (r = 0.465, p < 0.0001), so did ETNK1 and U2AF1, CALR, or ETV6 mutations (r = 0.439, p < 0.0001; r = 0.435, p < 0.0001; r = 0.331, p < 0.001, respectively), as well as DNMT3A and NPM1 mutations (r = 0.404, p < 0.0001). By contrast, TET2 and SETBP1 mutations were mutually exclusive (r=-0.219, p < 0.01).
Fig. 3.
The relationships among the driver mutations in CMML patients. The red and blue circles represent co-occurrence and mutually exclusive changes respectively. The circle size indicates the size of the effect, and the p value is expressed by colored gradient
Ancestral and sub-clonal events
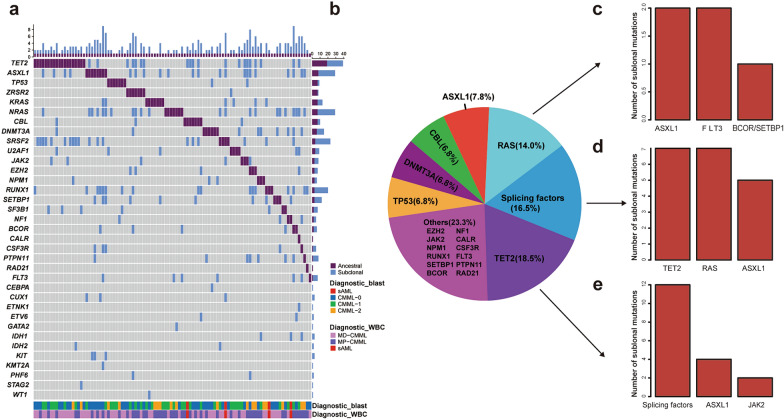
To analyze the ancestral and subclonal events in the 178 CMML patients, mutations with highest VAF were defined as the ancestral/dominant mutation, and others with similar VAFs were defined as co-dominant. As shown in Figs. 4a and 57.3% (102/178) of the patients had a single unique dominant event. Of the 102 patients, the ancestral events preferentially occurred in the following genes: TET2 (18.5%), splicing factors (16.5%), RAS (14.0%), ASXL1(7.8%), CBL (6.8%), DNMT3A (6.8%), and TP53 (6.8%); the subclonal genes were mainly ASXL1, TET2, and NRAS. (Fig. 4a, b). The ancestral events with TET2 mutations were concomitantly accompanied by splicing factors, ASXL1, and JAK2 subclones, whereas the ancestral events with splicing factors mutations were accompanied by TET2, RAS, and ASXL1 subclones. The ancestral events with RAS signaling mutations were usually accompanied by ASXL1 and FLT3 subclones (Fig. 4c–e).
Fig. 4.
The ancestral and subclonal events in CMML patients. a The distribution of the ancestral and subclonal mutations. Each column represents a patient and each row corresponds to a gene. The bar graph on the right represents the number of primary clones or subclones of each gene. A purple square represents one ancestral clonal event, and a blue square represents one subclonal event. b The pie chart lists the distribution of more than 6% of ancestral genes. c–e In CMML, the bar graphs represent the most common subclonal events for the first three ancestral events
Driver mutation enrichment in CMML and sAML
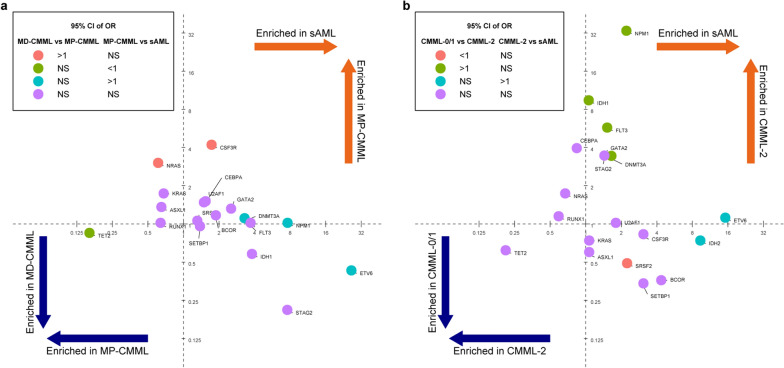
The enrichment of driver mutations in various malignancies has been observed. As seen in Fig. 5a, mutations in NPM1, ETV6, FLT3, and DNMT3A were preferentially enriched in sAML, TET2 mutation was enriched in MD-CMML, and NRAS mutation was enriched in MP-CMML. It was also found that IDH1, FLT3, NPM1, DNMT3A, and ETV6 mutations were enriched in CMML-2 and/or sAML, and TET2 mutation was enriched in CMML-0/1 using univariate comparison (Fig. 5b). Those results indicated that ETV6, FLT3, DNMT3A, and NPM1 mutations may play a role in the transformation from CMML to sAML.
Fig. 5.
The driver mutations from CMML subtypes and sAML. a The enrichment shows the odds ratio (OR) of mutation rates in sAML (n = 13) vs. MP-CMML (n = 95) and MP-CMML (n = 95) vs. MD-CMML (n = 83) on the x-axis and y-axis, respectively. b The enrichment shows the OR of mutation rates in sAML (n = 13) vs. CMML-2 (n = 55) and CMML-2 (n = 55) vs. CMML-0/1 (n = 123) on the x-axis and y-axis, respectively The enrichments from the comparisons are indicated by colors according to odds ratios (OR) and 95% confidence intervals (CI) limits being above (if OR > 1) or below (if OR < 1)
Prevalent gene mutations in CMML and pAML
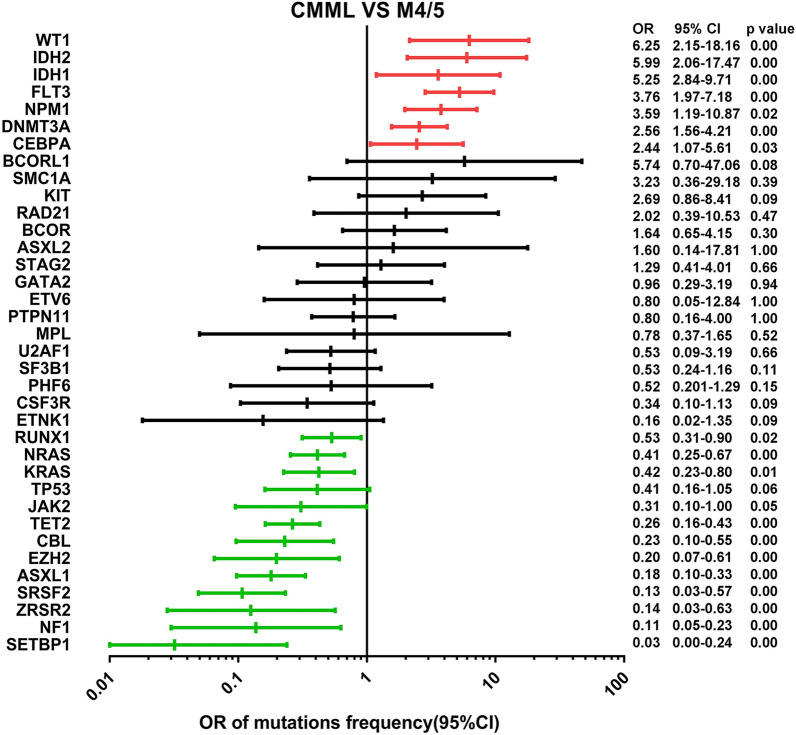
Two different mutational spectrums were found when CMML and pAML were co-evaluated. As shown in Fig. 6, seven mutations including WT1 (OR 6.25; p = 0.00), IDH2 (OR 5.99; p = 0.00), IDH1 (OR 5.25; p = 0.00), FLT3 (OR 3.76; p = 0.00), NPM1 (OR 3.59; p = 0.02), DNMT3A (OR 2.56; p = 0.00), and CEBPA (OR 2.44; p = 0.03) were enriched in pAML, whereas twelve mutations from RUNX1 (OR 0.53; p = 0.02), NRAS (OR 0.41; p = 0.00), KRAS (OR 0.42; p = 0.01), JAK2 (OR 0.31; p = 0.05), TET2 (OR 0.26; p = 0.00), CBL (OR 0.23; p = 0.00), EZH2 (OR 0.20; p = 0.00), ASXL1 (OR 0.18; p = 0.00), SRSF2 (OR 0.13; p = 0.00), ZRSR2 (OR 0.14; p = 0.00), NF1 (OR 0.11; p = 0.00), and SETBP1 (OR 0.03; p = 0.00) were enriched in CMML.
Fig. 6.
The forest plot with the OR and 95% CI of rates from the common driver mutations between CMML and pAML. The significant discrimination is shown in red and green lines
Clonal dynamics
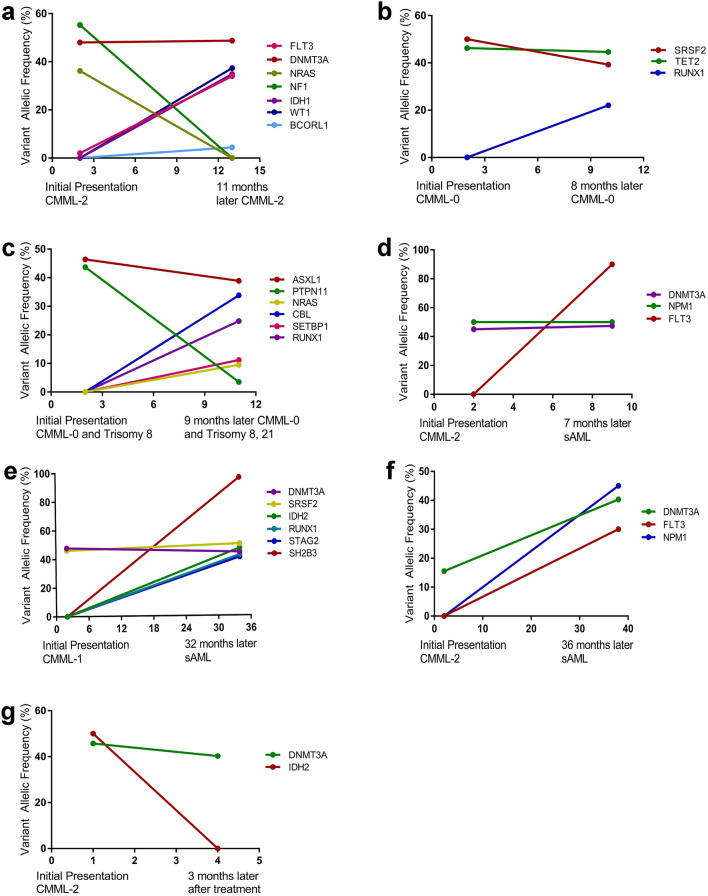
Clonal evolution was a common phenomenon in cancers, especially during disease progression, so as to CMML. The clonal dynamics from seven patients were evaluated with available molecular data, including 2 cases of CMML-0, 2 cases of CMML-2, and 3 cases of sAML transformed from CMML. The secondary clones were divided into emerging and vanishing clones. As demonstrated in Fig. 7, the emerging/increasing clones were FLT3/WT1 (Fig. 7a), RUNX1 (Fig. 7b), CBL/ RUNX1/SETBP1/NRAS (Fig. 7c), FLT3 (Fig. 7d), SH2B3/IDH2/STAG2/RUNX1 (Fig. 7e), and DNMT3A/FLT3/NPM1 (Fig. 7f). The vanishing/decreasing clones included NF1/NRAS (Fig. 7a), SRSF2 (Fig. 7b), PTPN11/ASXL1 (Fig. 7c), and DNMT3A/IDH2 (Fig. 7g). An illustrative example was shown in Fig. 7d–f, including three sAML cases transformed from CMML. Among them one acquired an emerging FLT3 mutation, another one acquired three emerging mutations (FLT3, DNMT3A, and NPM1), and the third one acquired an emerging SH2B3 mutation.
Fig. 7.
The clonal evolution in CMML. The clonal dynamics from seven patients with the VAF of numerous mutations (a–g) using different line colors, followed by the acquisition of new mutations (a–f), cytogenetic abnormalities (c), and progression (d–f)
Survival analysis
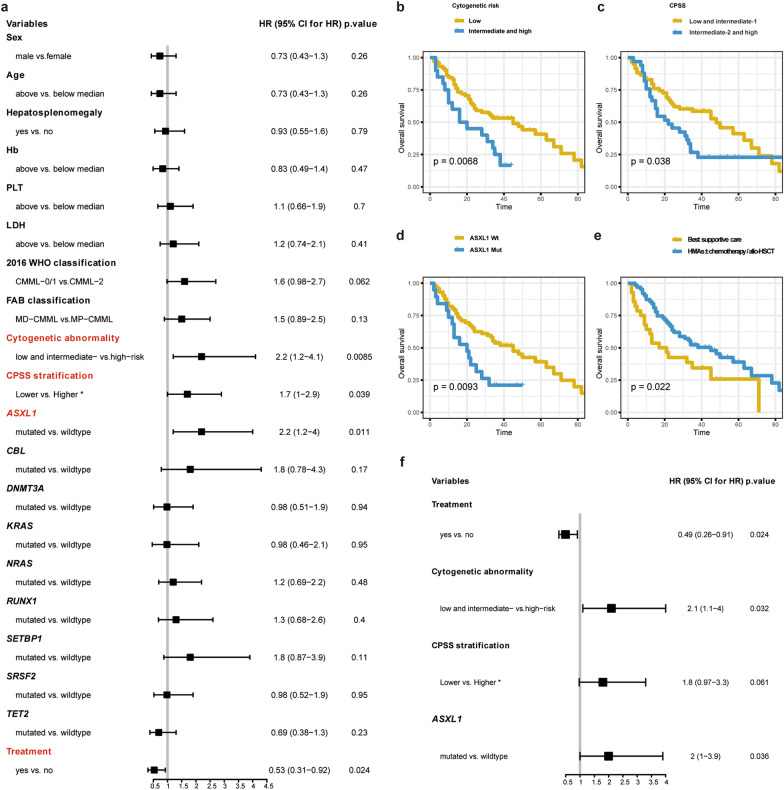
The univariate analysis included gender, adjusted age, splenomegaly, Hb, PLT, LDH, 2016 WHO classification, FAB subtypes, cytogenetic abnormality, CPSS risk stratifications, gene mutations and total number, and treatment options. Among them, cytogenetic abnormality [intermediate- and high-risk, hazard ratio (HR): 2.2, 95% confidence intervals (CI) 1.2–4.1; p = 0.009], and CPSS risk stratifications (intermediate-2 and high group, HR: 1.7, 95% CI 1.0-2.9; p = 0.039), and ASXL1 mutation [HR: 2.2, 95% CI 1.2-4.0; p = 0.011] indicated a shorter OS, and treatment modalities (HMAs ± chemotherapy/allo-SCT) (HR: 0.53, 95% CI 0.31–0.92; p = 0.024) predicted a better OS (Fig. 8a–e). The multivariate analysis revealed a similar result, except that CPSS risk stratifications (intermediate-2 and high group, HR: 1.8, 95% CI 0.97–3.3; p = 0.061) was not associated with the prognosis (Fig. 8f).
Fig. 8.
The univariate and multivariate Cox regression analysis of clinical and biological factors influencing OS. a The univariate analysis defines these variables by the hazard ratio (HR) with 95% CI, and gene mutations are included in the analysis if presenting in at least 10% patients. b Survival curves are charted for variables affecting the OS. c The multivariate analysis contains the index if presenting in the univariate analysis with p-value < 0.05 for OS
Discussion
In this study 178 CMML cases were analyzed including demographic, clinical, and laboratory features. This has been the largest cohort of CMML in Chinese population so far. Our study revealed multiple mutations in CMML and investigated their correlation during the clonal evolution. The paired data of gene mutations in CMML between Chinese and Caucasian patients were as follows: TET2 (36.5% vs. 29–61%), ASXL1 (28.7% vs. 30–50%), SRSF2 (24.7% vs. 29–52%), KRAS (17.4% vs. 7–16%), CBL (12.9% vs. 8–22%), RUNX1 (21.9% vs. 8–25%), and SETBP1 (11.8% vs. 4–18%). Nevertheless, Chinese patients gained higher mutation rates in NRAS (31.5% vs. 4–25%), DNMT3A (15.2% vs. 2–12%), PTPN11 (8.4% vs. < 5%), TP53 (8.4% vs. 1–3%), FLT3 (7.3% vs. 1–3%), and NPM1 (7.3% vs. 1–3%) [9, 14]. It has been reported that mutations in RAS/MAPK signaling genes such as NRAS and PTPN11 were associated with CMML-MP [1, 3, 28]. Both NRAS and DNMT3A mutations played a role in CMML transformation to AML [29, 30]. NRAS mutations were usually obtained in the later stage of clonal hematopoiesis, and transplanted CMML patients with NRAS mutation had an unfavorable prognosis [28]. Azacitidine and trametinib delivered a synergistic effect in NRAS-mutated CMML [31]. Several studies demonstrated that DNMT3A mutation was an independent adverse prognostic factor for OS and LFS [12, 32, 33]. CMML patients with NPM1 and FLT3 mutations tended to have a rapid progress to AML [3, 9, 28, 34], so did TP53 mutation [3, 35, 36]. The similar findings in our cohort suggested that those genes support a clonal expansion in CMML, as well as leading to a poor prognosis.
The order of acquired molecular abnormalities is particularly important. The ancestral/primary clonal lesions usually present the main clinical phenotypes, and the secondary or tertiary mutations likely predict disease progression. Several studies identified that TET2 and ASXL1 were the most predominant primary/ancestral events in CMML patients [24, 27, 37]. Unlike the reported studies, our investigation demonstrated that splicing factors (SRSF2, U2AF1, SF3B1, and ZRSR2) and RAS related genes (NRAS and KRAS) were common in primary/ancestral events. ASXL1 and FLT3 mutations were considered to be the adverse prognostic factors in CPSS-mol and MMM prognostic stratification systems [6, 9, 20]. In addition, the ancestral and subclonal events are exchangeable. The driver genes such as IDH1/CSF3R, IDH1/NF1, TET2/SRSF2 [2, 10, 28, 38, 39] and ETNK1/U2AF1 could mutate simultaneously, resulting in an instability of the genome and promoting the acquisition of additional mutations. On the other side, mutations in TET2/SETBP1 was observed in a mutually exclusive manner [33, 40].
During the transformation of CMML to sAML, one distinct feature was the emergence of new gene mutations or increase of VAF [41]. It was of note that a series of clones were emerged or increased from the seven transformed CMML cases in our study, including FLT3, NPM1, IDH2, DNMT3A et al. Some newly acquiring mutations, the so-called type 1 mutations FLT3, NPM1, and IDH2, seemed to predict a much shorter time for disease progression from MDS to sAML than type 2 mutations that were present in MDS [41–44]. The mutational tendency was evident in our study, and mutations in WT1, IDH2, IDH1, FLT3, NPM1, DNMT3A, and CEBPA were more common in pAML than in CMML. In addition, sAML and pAML overlapped with FLT3, NPM1, DNMT3A mutations, while WT1 and IDH1/2 mutations typically occurred in pAML. Therefore, closely monitoring of the emergence of DNMT3A, ETV6, FLT3, and NPM1 mutations would be an efficient way to predict the early transformation of CMML.
ASXL1 mutation (only nonsense and frameshift mutations) was an independent prognostic factor in our study by univariate and multivariate analyses, which was consistent with Mayo molecular model (MMM) and CPSS molecular model (CPSS-mol) [1, 6, 45]. The treatment options for CMML were various given the fact of disease heterogeneity. The strategies included: best supportive treatment, HMAs, chemotherapy, and allo-HSCT [46, 47]. In our study, the Cox model showed that treatment modalities (HMAs ± chemotherapy/allo-HSCT) was closely correlated with a better OS. A retrospective study showed that HMAs monotherapy group had a prolonged OS and achieved high response, but could not significantly modify the disease process [48]. In addition, the universal applicability of this treatment option was limited by the complication of allo-HSCT, such as non-recurrent mortality (NRM) and acute and chronic graft-versus-host disease (GVHD) [5, 47]. An open-label, non-randomized phase 2 clinical trial is undergoing to evaluate the efficacy of cobimetinib, a selective a reversible ATP-noncompetitive MEK inhibitor, in CMML patients with activated RAS pathway (mutations in NRAS, KRAS, PTPN11, FLT3,CBL, JAK2, BRAF, and NF1 at VAF ≥ 5%) (NCT04409639) [49]. The gain of function mutation in SRSF2 resulted in transcriptome-wide mis-splicing, leaving the mutated cells more susceptible to splicing inhibitory molecules than their counterpart wild-type cells [50, 51]. H3B-8800, a splicing modulator, is currently being investigated in patients with MDS, AML, and CMML (NCT02841540). These data suggested that specific molecular targets for gene mutations be an alternative and effective choice for the treatment of CMML.
Conclusions
The molecular profile of Chinese CMML patients displayed a wide range of mutations, and the emerging genes such as DNMT3A, ETV6, FLT3, and NPM1 usually indicated a dismal outcome of disease progression. In addition to the cytogenetic abnormality and treatment options (best supportive care), ASXL1 mutation also negatively affected the OS. Therefore, those molecules provided some potential biomarkers for diagnostic and prognostic prediction.
Supplementary Information
Additional file 1: Table S1. Thelist of 114 genes by NGS.
Additional file 2: Table S2. Reference database website.
Acknowledgements
Not applicable.
Abbreviations
- CMML
Chronic myelomonocytic leukemia
- sAML
Secondary acute myeloid leukemia
- pAML
Primary AML
- MDS
Myelodysplastic syndrome
- PB
Peripheral blood
- BM
Bone marrow
- WHO
World Health Organization
- MP-CMML
Myeloproliferative CMML
- MD-CMML
Myelodysplastic CMML
- WBC
White blood cell
- Hb
Hemoglobin
- PLT
Platelet
- WES
Whole-exome sequencing
- VAF
Variant allelic frequency
- NGS
Next-generation sequencing
- ExAc
Exome aggregation consortium
- ESP6500
Exome sequencing project 6500
- OR
Odds ratio
- HR
Hazard ratio
- CI
Confidence intervals
- OS
Overall survival
Author contributions
YL and KR provide research design and ideas, as well as modification and approval of the literature. YN, LS, and HZ are responsible for sorting out patients’ data and writing the paper. HZ and CKH are responsible for statistical processing and drawing charts; HJ, HL, JZ, LC, and HT are responsible for case collection and trial operation. All authors contributed to the review of research data and manuscripts. All authors read and approved the final manuscript.
Funding
This work was supported by the Major Special Project of Tianjin Science and Technology Service (No. 17ZXFWGX00110) to KR.
Availability of data and materials
The data generated and analyzed in the current study are not public, but it can be obtained from the authors upon request.
Declarations
Ethics approval and consent to participate
This study was approved by the Institutional Review Board of Sino-US diagnostics.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yanbo Nie, Liang Shao and Hong Zhang contributed equally to this work
Contributor Information
Yani Lin, Email: yanilin@sino-us-diagnostics.com.
Kun Ru, Email: kunru@yahoo.com.
References
- 1.Lee SC, Dvinge H, Kim E, Cho H, Micol JB, Chung YR, Elena C, Gallì A, Such E, Meggendorfer M, Germing U, Rizzo E, et al. Integrating clinical features and genetic lesions in the risk assessment of patients with chronic myelomonocytic leukemia. Blood. 2016;128(10):1408–17. doi: 10.1182/blood-2016-05-714030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loghavi S, Khoury JD. Recent updates on chronic myelomonocytic leukemia. Curr Hematol Malig Rep. 2018;13(6):446–54. doi: 10.1007/s11899-018-0475-5. [DOI] [PubMed] [Google Scholar]
- 3.Arber DA, Orazi A. Update on the pathologic diagnosis of chronic myelomonocytic leukemia. Mod Pathol. 2019;32(6):732–40. doi: 10.1038/s41379-019-0215-y. [DOI] [PubMed] [Google Scholar]
- 4.Keykhaei M, Masinaei M, Mohammadi E, Azadnajafabad S, Rezaei N, Saeedi Moghaddam S, et al. A global, regional, and national survey on burden and quality of care index (QCI) of hematologic malignancies; global burden of disease systematic analysis 1990–2017. Exp Hematol Oncol. 2021;10(1):11. doi: 10.1186/s40164-021-00198-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong Y, Shi O, Zeng Q, Lu X, Wang W, Li Y, et al. Leukemia incidence trends at the global, regional, and national level between 1990 and 2017. Exp Hematol Oncol. 2020;19:14. doi: 10.1186/s40164-020-00170-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Itzykson R, Kosmider O, Renneville A, Gelsi-Boyer V, Meggendorfer M, Morabito M, et al. Prognostic score including gene mutations in chronic myelomonocytic leukemia. J Clin Oncol. 2013;31(19):2428–36. doi: 10.1200/JCO.2012.47.3314. [DOI] [PubMed] [Google Scholar]
- 7.Patnaik MM, Tefferi A. Chronic myelomonocytic leukemia: 2020 update on diagnosis, risk stratification and management. Am J Hematol. 2020;95(1):97–115. doi: 10.1002/ajh.25684. [DOI] [PubMed] [Google Scholar]
- 8.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 9.Itzykson R, Fenaux P, Bowen D, Cross NCP, Cortes J, De Witte T, et al. Diagnosis and treatment of chronic myelomonocytic leukemias in adults: recommendations from the European Hematology Association and the European LeukemiaNet. Hemasphere. 2018;2(6):e150. doi: 10.1097/HS9.0000000000000150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Itzykson R, Duchmann M, Lucas N, Solary E. CMML: clinical and molecular aspects. Int J Hematol. 2017;105(6):711–19. doi: 10.1007/s12185-017-2243-z. [DOI] [PubMed] [Google Scholar]
- 11.Jian J, Qiao Y, Li Y, Guo Y, Ma H, Liu B. Mutations in chronic myelomonocytic leukemia and their prognostic relevance. Clin Transl Oncol. 2021;23(9):1731–42. doi: 10.1007/s12094-021-02585-x. [DOI] [PubMed] [Google Scholar]
- 12.Coltro G, Patnaik MM. Chronic myelomonocytic leukemia: insights into biology, prognostic factors, and treatment. Curr Oncol Rep. 2019;21(11):101. doi: 10.1007/s11912-019-0855-6. [DOI] [PubMed] [Google Scholar]
- 13.Carr RM, Vorobyev D, Lasho T, Marks DL, Tolosa EJ, Vedder A, et al. RAS mutations drive proliferative chronic myelomonocytic leukemia via a KMT2A-PLK1 axis. Nat Commun. 2021;12(1):2901. doi: 10.1038/s41467-021-23186-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel AB, Deininger MW. Genetic complexity of chronic myelomonocytic leukemia. Leuk Lymphoma. 2021;62(5):1031–45. doi: 10.1080/10428194.2020.1856837. [DOI] [PubMed] [Google Scholar]
- 15.Nazha A, Patnaik MM. Making sense of prognostic models in chronic myelomonocytic leukemia. Curr Hematol Malig Rep. 2018;13(5):341–7. doi: 10.1007/s11899-018-0465-7. [DOI] [PubMed] [Google Scholar]
- 16.Onida F, Kantarjian HM, Smith TL, Ball G, Keating MJ, Estey EH, et al. Prognostic factors and scoring systems in chronic myelomonocytic leukemia: a retrospective analysis of 213 patients. Blood. 2002;99(3):840–9. doi: 10.1182/blood.V99.3.840. [DOI] [PubMed] [Google Scholar]
- 17.Beran M, Wen S, Shen Y, Onida F, Jelinek J, Cortes J, et al. Prognostic factors and risk assessment in chronic myelomonocytic leukemia: validation study of the M.D. Anderson prognostic scoring system. Leuk Lymphoma. 2007;48(6):1150–60. doi: 10.1080/10428190701216386. [DOI] [PubMed] [Google Scholar]
- 18.Tang G, Zhang L, Fu B, Hu J, Lu X, Hu S, et al. Cytogenetic risk stratification of 417 patients with chronic myelomonocytic leukemia from a single institution. Am J Hematol. 2014;89(8):813–8. doi: 10.1002/ajh.23751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Such E, Germing U, Malcovati L, Cervera J, Kuendgen A, Della Porta MG, et al. Development and validation of a prognostic scoring system for patients with chronic myelomonocytic leukemia. Blood. 2013;121(15):3005–15. doi: 10.1182/blood-2012-08-452938. [DOI] [PubMed] [Google Scholar]
- 20.Patnaik MM, Padron E, LaBorde RR, Lasho TL, Finke CM, Hanson CA, et al. Mayo prognostic model for WHO-defined chronic myelomonocytic leukemia: ASXL1 and spliceosome component mutations and outcomes. Leukemia. 2013;27(7):1504–10. doi: 10.1038/leu.2013.88. [DOI] [PubMed] [Google Scholar]
- 21.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Q, Wang K, InterVar Clinical interpretation of genetic variants by the 2015 ACMG-AMP guidelines. Am J Hum Genet. 2017;100(2):267–80. doi: 10.1016/j.ajhg.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palomo L, Meggendorfer M, Hutter S, Twardziok S, Ademà V, Fuhrmann I, et al. Molecular landscape and clonal architecture of adult myelodysplastic/myeloproliferative neoplasms. Blood. 2020;136(16):1851–62. doi: 10.1182/blood.2019004229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Awada H, Nagata Y, Goyal A, Asad MF, Patel B, Hirsch CM, et al. Invariant phenotype and molecular association of biallelic TET2 mutant myeloid neoplasia. Blood Adv. 2019;3(3):339–49. doi: 10.1182/bloodadvances.2018024216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirsch CM, Nazha A, Kneen K, Abazeed ME, Meggendorfer M, Przychodzen BP, et al. Consequences of mutant TET2 on clonality and subclonal hierarchy. Leukemia. 2018;32(8):1751–61. doi: 10.1038/s41375-018-0150-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel BJ, Przychodzen B, Thota S, Radivoyevitch T, Visconte V, Kuzmanovic T, et al. Genomic determinants of chronic myelomonocytic leukemia. Leukemia. 2017;31(12):2815–23. doi: 10.1038/leu.2017.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patnaik MM, Lasho TL. Genomics of myelodysplastic syndrome/myeloproliferative neoplasm overlap syndromes. Hematol Am Soc Hematol Educ Program. 2020;2020(1):450–9. doi: 10.1182/hematology.2020000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patnaik MM, Lasho T. Myelodysplastic syndrome/myeloproliferative neoplasm overlap syndromes: a focused review. Hematol Am Soc Hematol Educ Program. 2020;2020(1):460–4. doi: 10.1182/hematology.2020000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palomo L, Ibáñez M, Abáigar M, Vázquez I, Álvarez S, Cabezón M, et al. Spanish Guidelines for the use of targeted deep sequencing in myelodysplastic syndromes and chronic myelomonocytic leukaemia. Br J Haematol. 2020;188(5):605–22. doi: 10.1111/bjh.16175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kloos A, Mintzas K, Winckler L, Gabdoulline R, Alwie Y, Jyotsana N, et al. Effective drug treatment identified by in vivo screening in a transplantable patient-derived xenograft model of chronic myelomonocytic leukemia. Leukemia. 2020;34(11):2951–63. doi: 10.1038/s41375-020-0929-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patnaik MM, Barraco D, Lasho TL, Finke CM, Hanson CA, Ketterling RP, et al. DNMT3A mutations are associated with inferior overall and leukemia-free survival in chronic myelomonocytic leukemia. Am J Hematol. 2017;92(1):56–61. doi: 10.1002/ajh.24581. [DOI] [PubMed] [Google Scholar]
- 33.Kar SA, Jankowska A, Makishima H, Visconte V, Jerez A, Sugimoto Y, et al. Spliceosomal gene mutations are frequent events in the diverse mutational spectrum of chronic myelomonocytic leukemia but largely absent in juvenile myelomonocytic leukemia. Haematologica. 2013;98(1):107–13. doi: 10.3324/haematol.2012.064048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guan W, Zhou L, Li Y, Yang E, Liu Y, Lv N, et al. Profiling of somatic mutations and fusion genes in acute myeloid leukemia patients with FLT3-ITD or FLT3-TKD mutation at diagnosis reveals distinct evolutionary patterns. Exp Hematol Oncol. 2021;10(1):27. doi: 10.1186/s40164-021-00207-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patnaik MM, Vallapureddy R, Lasho TL, Hoversten KP, Finke CM, Ketterling R, et al. EZH2 mutations in chronic myelomonocytic leukemia cluster with ASXL1 mutations and their co-occurrence is prognostically detrimental. Blood Cancer J. 2018;8(1):12. doi: 10.1038/s41408-017-0045-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang W, Routbort MJ, Loghavi S, Tang Z, Medeiros LJ, Wang SA. Characterization of chronic myelomonocytic leukemia with TP53 mutations. Leuk Res. 2018;70:97–9. doi: 10.1016/j.leukres.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 37.Solary E, Wagner-Ballon O, Selimoglu-Buet D. Incorporating flow cytometry and next-generation sequencing in the diagnosis of CMML. Are we ready for prime? Best Pract Res Clin Haematol. 2020;33(2):101134. doi: 10.1016/j.beha.2019.101134. [DOI] [PubMed] [Google Scholar]
- 38.Carr RM, Patnaik MM. Genetic and epigenetic factors interacting with clonal hematopoiesis resulting in chronic myelomonocytic leukemia. Curr Opin Hematol. 2020;27(1):2–10. doi: 10.1097/MOH.0000000000000553. [DOI] [PubMed] [Google Scholar]
- 39.Thomopoulos TP, Bouhla A, Papageorgiou SG, Pappa V. Chronic myelomonocytic leukemia—a review. Expert Rev Hematol. 2021;14(1):59–77. doi: 10.1080/17474086.2021.1860004. [DOI] [PubMed] [Google Scholar]
- 40.Meggendorfer M, Roller A, Haferlach T, Eder C, Dicker F, Grossmann V, et al. SRSF2 mutations in 275 cases with chronic myelomonocytic leukemia (CMML) Blood. 2012;120(15):3080–8. doi: 10.1182/blood-2012-01-404863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogawa S. Genetics of MDS. Blood. 2019;133(10):1049–59. doi: 10.1182/blood-2018-10-844621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu J, Li Y, Zhang D, Wan D, Jiang Z. Clinical implications of recurrent gene mutations in acute myeloid leukemia. Exp Hematol Oncol. 2020;27:4. doi: 10.1186/s40164-020-00161-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu J, Li Y, Li T, Li Y, Xing H, Sun H, et al. Gene mutational analysis by NGS and its clinical significance in patients with myelodysplastic syndrome and acute myeloid leukemia. Exp Hematol Oncol. 2020;9:2. doi: 10.1186/s40164-019-0158-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Makishima H, Yoshizato T, Yoshida K, Sekeres MA, Radivoyevitch T, Suzuki H, et al. Dynamics of clonal evolution in myelodysplastic syndromes. Nat Genet. 2017;49(2):204–12. doi: 10.1038/ng.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patnaik MM, Itzykson R, Lasho TL, Kosmider O, Finke CM, Hanson CA, et al. ASXL1 and SETBP1 mutations and their prognostic contribution in chronic myelomonocytic leukemia: a two-center study of 466 patients. Leukemia. 2014;28(11):2206–12. doi: 10.1038/leu.2014.125. [DOI] [PubMed] [Google Scholar]
- 46.Kongtim P, Popat U, Jimenez A, Gaballa S, El Fakih R, Rondon G, et al. Treatment with hypomethylating agents before allogeneic stem cell transplant improves progression-free survival for patients with chronic myelomonocytic leukemia. Biol Blood Marrow Transplant. 2016;22(1):47–53. doi: 10.1016/j.bbmt.2015.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun YQ, Zhao C, Wang Y, Yan CH, Zhang XH, Xu LP, et al. Haploidentical stem cell transplantation in patients with chronic myelomonocytic leukemia. Sci China Life Sci. 2020;63(8):1261–4. doi: 10.1007/s11427-019-1606-3. [DOI] [PubMed] [Google Scholar]
- 48.Ma L, Jiang L, Yang W, Luo Y, Mei C, Zhou X, Tong H, et al. Real-world data of chronic myelomonocytic leukemia: a chinese single-center retrospective study. Cancer Med. 2021;10(5):1715–25. doi: 10.1002/cam4.3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramos Perez J, Montalban-Bravo G. Emerging drugs for the treatment of chronic myelomonocytic leukemia. Expert Opin Emerg Drugs. 2020;25(4):515–29. doi: 10.1080/14728214.2020.1854224. [DOI] [PubMed] [Google Scholar]
- 50.Kim E, Ilagan JO, Liang Y, Daubner GM, Lee SC, Ramakrishnan A, et al. SRSF2 mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer Cell. 2015;27(5):617–30. doi: 10.1016/j.ccell.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee SC, Dvinge H, Kim E, Cho H, Micol JB, Chung YR, et al. Modulation of splicing catalysis for therapeutic targeting of leukemia with mutations in genes encoding spliceosomal proteins. Nat Med. 2016;22(6):672–8. doi: 10.1038/nm.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Thelist of 114 genes by NGS.
Additional file 2: Table S2. Reference database website.
Data Availability Statement
The data generated and analyzed in the current study are not public, but it can be obtained from the authors upon request.