Abstract
Background
Tick hemolymph bathes internal organs, acts as an exchange medium for nutrients and cellular metabolites, and offers protection against pathogens. Hemolymph is abundant in proteins. However, there has been limited integrated protein analysis in tick hemolymph thus far. Moreover, there are difficulties in differentiating tick-derived proteins from the host source. The aim of this study was to profile the tick/host protein components in the hemolymph of Haemaphysalis flava.
Methods
Hemolymph from adult engorged H. flava females was collected by leg amputation from the Erinaceus europaeus host. Hemolymph proteins were extracted by a filter-aided sample preparation protocol, digested by trypsin, and assayed by liquid chromatography–tandem mass spectrometry (LC–MS/MS). MS raw data were searched against the UniProt Erinaceidae database and H. flava protein database for host- and tick-derived protein identification. Protein abundance was further quantified by intensity-based absolute quantification (iBAQ).
Results
Proteins extracted from hemolymph unevenly varied in size with intense bands between 100 and 130 kDa. In total, 312 proteins were identified in the present study. Therein 40 proteins were identified to be host-derived proteins, of which 18 were high-confidence proteins. Top 10 abundant host-derived proteins included hemoglobin subunit-α and subunit-β, albumin, serotransferrin-like, ubiquitin-like, haptoglobin, α-1-antitrypsin-like protein, histone H2B, apolipoprotein A-I, and C3-β. In contrast, 169 were high-confidence tick-derived proteins. These proteins were classified into six categories based on reported functions in ticks, i.e., enzymes, enzyme inhibitors, transporters, immune-related proteins, muscle proteins, and heat shock proteins. The abundance of Vg, microplusin and α-2-macroglobulin was the highest among tick-derived proteins as indicated by iBAQ.
Conclusions
Numerous tick- and host-derived proteins were identified in hemolymph. The protein profile of H. flava hemolymph revealed a sophisticated protein system in the physiological processes of anticoagulation, digestion of blood meal, and innate immunity. More investigations are needed to characterize tick-derived proteins in hemolymph.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s13071-022-05287-7.
Keywords: Tick, Hemolymph, Enzyme, Enzyme inhibitors, Vitellogenin, Serpin, Cystatin, Microplusin
Background
Tick hemolymph is a circulating fluid that fills the body cavity and bathes the inner organs. It serves as an exchange medium for the transport of nutrients, hormones, and products of cellular metabolism, and offers protection against pathogens to which ticks are exposed [1]. Hemolymph consists of hemocytes and plasma. Plasma predominates in tick hemolymph, representing approximately 90% by weight.
In tick plasma, proteins are the most soluble components (11.5–14.3% by weight) [2]. Thus far, a few proteins have been isolated, identified, and partially characterized in tick hemolymph, including vitellogenins (Vgs) [3, 4], macroglobulins [5], antimicrobial peptides [6], defensins [7], lectins [8], carrier proteins [9, 10], myeloid differentiation-2-related lipid-recognition domain [11], serine proteinase inhibitor (serpin) [12], and protein disulfide isomerases [13]. However, owing to the tiny volume of hemolymph from ticks, it is difficult to characterize proteins in hemolymph using traditional protein chemistry methods. The limited and unsystematic information obstructs our understanding of the roles of hemolymph in tick development, reproduction, endocrine function, and defense against pathogens; it also hinders the discovery of new targets against ticks [14].
Proteomic approaches are efficient tools for mapping protein profiles in ticks. Madden et al. initially reported saliva protein profiles of two related tick species, Amblyomma americanum and Amblyomma maculatum, by matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry (MALDI-TOF MS) [15]. Since then, proteomic investigations have been performed in Ixodes scapularis (saliva) [16], Ornithodoros moubata and Ornithodoros erraticus (salivary proteins) [17], Rhipicephalus sanguineus (saliva) [18], Haemaphysalis flava (fecal proteins and midgut contents) [19, 20], and Rhipicephalus microplus (saliva) [21]. Nevertheless, thus far there have been only two reports describing the protein profile in tick hemolymph. Stopforth et al. conducted a proteomic study to identify proteins secreted in the hemolymph of Ornithodoros savignyi ticks following immune challenge with yeasts [22]. Aguilar-Díaz et al. compared hemolymph proteomes of two R. microplus strains with different degrees of resistance to ixodicides [23]. Because of the lack of a transcriptome library of the tested ticks at the time, the number of hemolymph proteins identified in both studies was quite low.
In this study, hemolymph was collected from adult H. flava females. Proteins contained in the hemolymph were analyzed by liquid chromatography–tandem MS (LC–MS/MS) in combination with a search against the UniProt database and self-constructed H. flava transcriptome library, aiming to provide the most comprehensive data to data regarding host- and tick-derived proteins in tick hemolymph.
Methods
Collection of tick hemolymph
All experimental procedures were approved and overseen by the Institutional Animal Care and Use Committee at Hunan Agricultural University, with approval no. 2021085. Fully engorged H. flava ticks were picked from naturally infected hedgehogs in our experimental and observation station located in Xinyang City, Henan Province, China (31°44′N, 114°10′E). Hedgehogs, which are common hosts of H. flava ticks [24], were housed with no recent exposure to any chemical acaricides. Hemolymph from the ticks was collected according to a previous study [25]. Briefly, 45 engorged adult H. flava females were randomly selected, rinsed with water, and sterilized with 70% ethanol. Ticks were immobilized on Petri dishes with their ventral sides up using double-sided tape. The legs were cut off with ophthalmic scissors. Then, gentle pressure was applied to the tick body, and hemolymph was collected using a glass-capillary tube with 2 μl of protease inhibitor cocktail [26]. Hemolymph from 15 ticks was pooled to ensure adequate size for further analysis. Thus, these 45 ticks represent three replicates. The pooled hemolymph sample was transferred into a clean tube and centrifuged at 14,000×g for 10 min at 4 °C. Supernatant was collected and quantified with a Bradford Protein Assay Kit (Beyotime Biotechnology, Shanghai, China).
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis
Ten microliters of supernatant was mixed with SDT buffer (30 μl, 4% sodium dodecyl sulfate, 100 mM dithiothreitol, and 150 mM Tris–HCl pH 8.0). The mixture was subjected to a boiling-water bath for 5 min. After cooling to room temperature and centrifuging at 14,000×g for 10 min at 4 °C, samples were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) with omniPAGE™ precast gels (LK204, 4–15%, Epizyme Biomedical Technology, Shanghai, China).
Protein digestion by filter-aided sample preparation
We followed a filter-aided sample preparation protocol before LC–MS/MS analysis [27]. An aliquot of 20 μl supernatant was added to 5 μl 200 mM dithiothreitol, boiled in water for 5 min, and cooled to room temperature. Next, 200 μl 8 M urea buffer was introduced and mixed well. The mixture was transferred into an ultrafiltration tube fitted with a 10 kDa centrifugal filter unit, and centrifuged at 14,000×g for 15 min. Proteins retained on the filter were washed several times with 8 M urea buffer to ensure maximal removal of impurities. They were then mixed with 100 μl iodoacetamide solution, shaken at 600 rpm for 1 min, kept away from light at room temperature for 30 min, and then centrifuged at 14,000×g for 10 min. Proteins on the filter were rinsed twice with 8 M urea buffer and then incubated with 40 μl trypsin solution (3 μg trypsin in 40 μl 25 mM NH4HCO3, Sigma-Aldrich, MO, USA) at 37 °C for 16–18 h. Then, the centrifugal filter unit with digests on it was inserted into a new collection tube and centrifuged at 14,000×g for 10 min. Filtrates were collected and submitted to a C18 cartridge (Empore™ solid-phase extraction (SPE) C18 cartridges, bed I.D. 7 mm, volume 3 ml; Sigma-Aldrich, St. Louis, MO, USA) for desalination. Then they were concentrated by vacuum centrifugation and reconstituted in 40 µl of 0.1% (v/v) trifluoroacetic acid.
Analysis by LC–MS/MS
LC–MS/MS analyses were performed on a Q Exactive mass spectrometer coupled to an EASY-nLC system (Thermo Fisher Scientific, Waltham, MA, USA). A total of 5 μg of peptides was injected. Peptides were passed through a C18 reversed-phase column (Thermo Scientific EASY-Column, 10 cm, 75 μm inner diameter, 3 μm resin) in buffer A (2% acetonitrile and 0.1% formic acid) and separated with a linear gradient of buffer B (80% acetonitrile and 0.1% formic acid) at a flow rate of 250 nl/min controlled by IntelliFlow technology over 60 min.
MS data were acquired using a data-dependent top 10 method dynamically choosing the most abundant precursor ions from the survey scan (300–1800 m/z) for higher-energy collisional dissociation (HCD) fragmentation. The target value was determined based on predictive automatic gain control (gAGC). The dynamic exclusion duration was set at 25 s. Survey scans were acquired at a resolution of 70,000 at m/z 200. The resolution for HCD spectra was set to 17,500 at m/z 200. The normalized collision energy was 30 eV. The underfill ratio was defined as 0.1%.
Sequence database search and data processing
MS data were processed by MaxQuant software (version 1.6.1.0., https://maxquant.net/maxquant/). The MS/MS raw files were searched against the UniProt Erinaceidae database (28,253 entries, downloaded on 03/01/2021) for the identification of host proteins, and then against a H. flava protein database constructed in parallel with the transcriptome (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA756707/) for identification of tick proteins, which contained 57,024 clusters and 10,859 predicted proteins. An initial search was set at a precursor mass window of 6 parts per million (ppm). The search followed an enzymatic cleavage rule of trypsin/P, and allowed a maximum of two missed cleavage sites and a mass tolerance of 20 ppm for fragment ions. Carbamidomethylation of cysteines was defined as fixed modification, while protein N-terminal acetylation and methionine oxidation were defined as variable modification. The cutoff for the global false discovery rate for peptide and protein identification was set to 0.01. Intensity-based absolute quantification (iBAQ) was carried out in MaxQuant.
Results and discussion
SDS-PAGE for total proteins in tick hemolymph
The concentration of protein in tick hemolymph was determined to be 5.03 ± 0.19 μg/μl. Figure 1 presents an SDS-PAGE image of total hemolymph proteins. The electrophoretogram indicated that proteins in H. flava hemolymph varied greatly in size, with intense bands at 100–130 kDa.
Fig. 1.
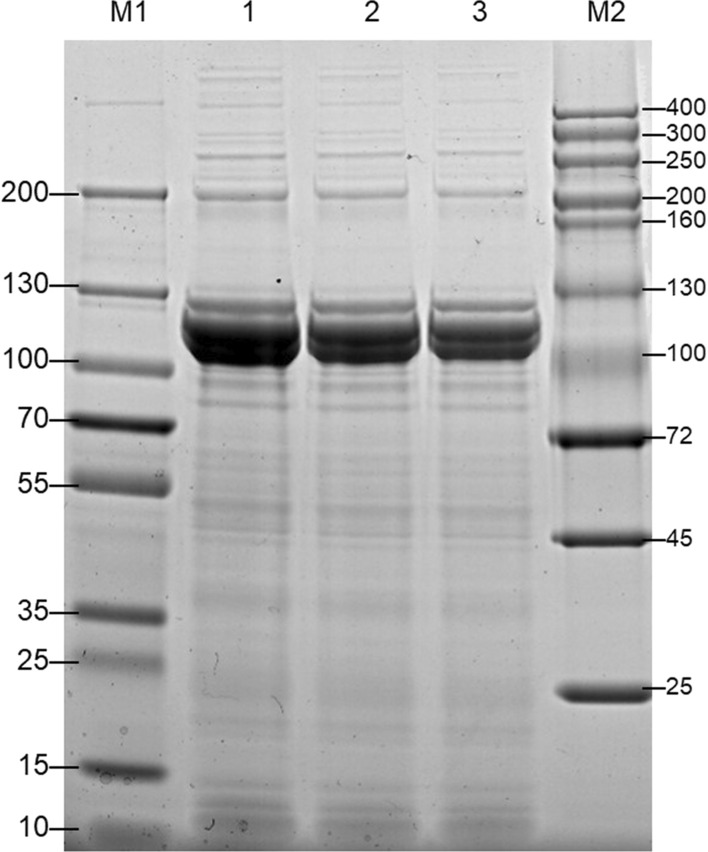
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis analysis of proteins from Haemaphysalis flava hemolymph. M1, M2 = markers (kDa); 1, 2, 3 = three replicates of proteins from the H. flava hemolymph
Tatchell et al. firstly reported that hemolymph proteins from female R. microplus immediately after engorgement revealed 15 bands by SDS-PAGE, but 14 bands in the case of ovipositing females [28]. Thereafter, investigations of hemolymph proteins were undertaken in O. moubata [4], Ornithodoros parkeri [29], Dermacentor variabilis [30–32], and Haemaphysalis longicornis [33] using SDS-PAGE and native PAGE.
Protein components in hemolymph vary with tick species, and also display dynamic changes in various physiological processes. Stopforth et al. showed the size of hemolymph proteins of O. savignyi in the range of 14–200 kDa [22], but Boldbaatar et al. demonstrated that some hemolymph proteins in H. longicornis could be as large as 669 kDa [33]. Protein concentration and composition changed greatly in the hemolymph of female O. parkeri during blood-feeding [29]. Hefnawy revealed that the total content of hemolymph varied according to life stage and engorgement level [34].
Host proteins in tick hemolymph
A search of the UniProt Erinaceidae database identified a total of 40 host proteins. Among these, 18 belonged to high-confidence proteins (unique peptides ≥ 2, Table 1). Of the 18 high-confidence proteins, 12 were components from host plasma, namely, albumin, ubiquitin-like, serotransferrin-like, α-1-antitrypsin-like protein, α-2-macroglobulin-like, fibrinogen α/β/γ chain, haptoglobin, C3-β, hemopexin, and apolipoprotein A-I. The other six proteins were from host blood cells, including hemoglobin (Hb) subunit-α/β, tubulin, histone H2B, carbonic anhydrase, and HSP90α.
Table 1.
High-confidence proteins from the host (Erinaceus europaeus) identified in hemolymph of Haemaphysalis flava ticks
| Protein overview | No. of unique peptides | Coverage (%) | Identity (%) | iBAQ (× 106) |
|---|---|---|---|---|
| P01949, hemoglobin subunit α, Erinaceus europaeus | 8 | 64.5 | 64.5 | 271.90 ± 59.13 |
| A0A1S3WPY1, hemoglobin subunit β, E. europaeus | 11 | 74.8 | 74.8 | 257.22 ± 55.55 |
| A0A1S2ZRW6, serum albumin, E. europaeus | 38 | 65.8 | 65.8 | 112.09 ± 27.87 |
| A0A1S3W2S7, serotransferrin-like, E. europaeus | 33 | 59.3 | 59.3 | 17.93 ± 4.42 |
| A0A1S3W634, ubiquitin-like, E. europaeus | 2 | 32.5 | 32.5 | 17.77 ± 4.71 |
| A0A1S3WFY0, haptoglobin, E. europaeus | 9 | 29.5 | 29.5 | 8.28 ± 2.12 |
| A0A1S3WJ22, α-1-antitrypsin-like protein, E. europaeus | 2 | 20.4 | 7 | 3.86 ± 0.87 |
| A0A1S3WR78, histone H2B, E. europaeus | 2 | 20.8 | 20.8 | 2.52 ± 0.63 |
| Q9TS49, apolipoprotein A-I, E. europaeus | 3 | 13.7 | 13.7 | 2.21 ± 0.68 |
| A0A1S3WDL1, C3-β, E. europaeus | 6 | 14.7 | 5.8 | 1.84 ± 0.51 |
| A0A1S3W909, tubulin β chain, E. europaeus | 5 | 15.9 | 15.9 | 1.77 ± 0.37 |
| A0A1S3WJA6, fibrinogen γ chain, E. europaeus | 4 | 14.5 | 14.5 | 1.46 ± 0.38 |
| A0A1S3W2L2, α-2-macroglobulin-like, E. europaeus | 13 | 12.4 | 12.4 | 1.34 ± 0.38 |
| A0A1S3AIG2, carbonic anhydrase, E. europaeus | 3 | 11.5 | 11.5 | 1.21 ± 0.30 |
| A0A1S3A1I2, hemopexin, E. europaeus | 3 | 8.4 | 8.4 | 0.86 ± 0.11 |
| A0A1S2ZF33, HSP90-α, E. europaeus | 4 | 7.1 | 7.1 | 0.80 ± 0.12 |
| A0A1S3A559, fibrinogen α chain, E. europaeus | 4 | 6.5 | 6.5 | 0.74 ± 0.21 |
| A0A1S3WJK5, fibrinogen β chain, E. europaeus | 4 | 8.6 | 8.6 | 0.71 ± 0.37 |
Host serum constituents have been detected in tick hemolymph, including Hb hydrolyzed fragments, immunoglobulin G (IgG), transferrin, and albumin [35]. However, the full spectrum of host proteins that could be transferred to tick hemolymph remained unknown [36]. Our data demonstrated that at least these 40 host plasma proteins could be transferred into tick hemolymph.
Mammalian fibrinogen is composed of two identical subunits, each subunit containing one α, β, and γ chain. Our data indicated the presence of a considerable number of host fibrinogen α, β, and γ chains in hemolymph, but did not detect any fibrinogen of tick origin. This observation implies that host fibrinogen was transferred intact from the midgut to the hemolymph. It is possible that the molecules and mechanisms involved in coagulation in tick hemolymph are the same as those in the host blood. In other words, ticks may share the same coagulation machinery as the hosts. Consistent with this assumption, anticoagulants used during the collection of tick hemolymph were protease inhibitor cocktail and ethylenediamine tetraacetic acid (EDTA) [26, 37]. The former inhibited serine protease, cysteine protease, aspartic protease, metalloprotease, and aminopeptidase, whereas the latter prevented blood from clotting by Ca2+ chelation.
Tick-derived proteins in hemolymph
In total, 1196 unique peptides and 312 deduced protein sequences were identified by searching the H. flava transcriptome database (Additional file 1: Table S1). Among these tick sequences, 175 were high-confidence-deducing sequences (unique peptides ≥ 2) and belonged to 169 proteins, as several peptides were from the same protein. For instance, α2-macroglobulin-like protein covered peptides Cl-k.18726, Cl-k.14664, Cl-k.14873 and Cl-k.18058. No albumin of tick origin was detected.
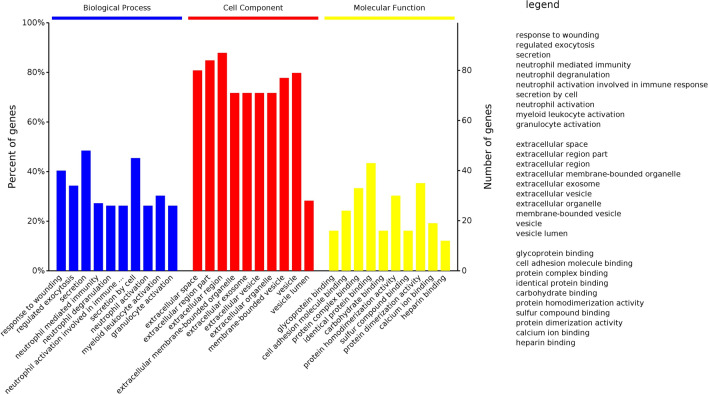
Gene Ontology (GO) analysis using OmicsBean (http://www.omicsbean.cn/) revealed that these 169 proteins were mainly enriched in biological processes of neutrophil, leukocyte, and granulocyte activation, and were significantly located in the extracellular space. Their molecular functions mainly included binding of proteins, carbohydrates, and other molecules such as sulfur compounds and calcium. The top ten GO terms of each category are displayed in Fig. 2.
Fig. 2.
GO analysis of tick-derived proteins in hemolymph. The top ten GO terms of each category are as indicated
We searched the literature in PubMed and the Chinese National Knowledge Infrastructure (CNKI), and found that 76 homologues of 169 high-confidence proteins were studied in the literature. Based on the conclusions of studies, these homologues were classified into six categories, including enzymes, enzyme inhibitors, transporters, immune-related proteins, muscle proteins, and others. Among them, enzymes were the most abundant. Their substrates included proteins, lipids, carbohydrates, and chitins. In addition, there were many types of serine proteases and their inhibitors (serpins).
Though some tick-derived proteins in hemolymph and other tissues have been characterized, the number of characterized proteins is relatively low compared to the total number of proteins detected in hemolymph. Hence, the functions of the majority of tick-derived proteins in hemolymph are as yet unknown, making it impossible to classify all of them based on function. The major tick-derived proteins in hemolymph with known functions will be discussed below.
Enzymes in tick hemolymph
Although just a portion of enzymes are listed in Table 2, it is clear that the enzyme composition in tick hemolymph was diverse and complex. These enzymes were mainly involved in anticoagulation, digestion of blood meal, and innate immunity. They also participated in substance metabolism and even molting.
Table 2.
High-confidence tick proteins with reported functions in the hemolymph of H. flava ticks
| No. | Protein | Alignment | iBAQ (× 106) | ||||
|---|---|---|---|---|---|---|---|
| ID | Length (amino acids) | Entry and overview | E value | Score | Identity (%) | ||
| I. Enzymes | |||||||
| 1 | Cl-k.18156 ① | 376 | L7M876, tick serine protease, Rhipicephalus pulchellus | 0 | 1897 | 84.6 | 5.96 ± 1.32 |
| 2 | Cl-k.17502 ① | 484 | A0A131Z7A0, tick serine protease, Rhipicephalus appendiculatus | 0 | 1451 | 67.8 | 0.39 ± 0.04 |
| 3 | Cl-k.19217 ① | 437 | A0A1E1X8K7, serine protease, Amblyomma aureolatum | 0 | 1907 | 85.1 | 2.13 ± 0.47 |
| 4 | Cl-k.19341 | 348 | A1IHG0, longipain, H. longicornis | 0 | 1814 | 93.5 | 7.34 ± 1.74 |
| 5 | Cl-k.18108 | 443 | A0A097CK68, enolase, H. flava | 0 | 2233 | 99.5 | 1.16 ± 0.35 |
| 6 | Cl-k.14381 | 507 | A0A6M2D6D9, serine carboxypeptidase, R. microplus | 0 | 2115 | 83.6 | 0.49 ± 0.15 |
| 7 | Cl-k.18093 | 164 | A0A131XJQ8, metalloproteinase, Hyalomma excavatum | 1.7E−91 | 686 | 75.0 | (7) 221.26 ± 58.79 |
| 8 | Cl-k.18993 | 398 | Q2WFX6, aspartic protease, H. longicornis | 0 | 1990 | 95.9 | 1.47 ± 0.34 |
| 9 | Cl-k.7217 | 397 | A0A034WWI5, heme-binding aspartic peptidase, R. microplus | 0 | 1343 | 68.0 | 3.03 ± 0.93 |
| 10 | Cl-k.14313 | 561 | A0A1E1XAU4, cysteine proteinase, A. aureolatum | 0 | 2602 | 84.4 | 5.24 ± 1.17 |
| 11 | Cl-k.18480 | 326 | A0A023FWK4, cathepsin L, Amblyomma parvum | 0 | 1604 | 88.7 | 99.93 ± 26.58 |
| 12 | Cl-k.24797 | 110 | A0A023GJU1, cathepsin C, Amblyomma triste | 2.6E−70 | 563 | 92.7 | 2.08 ± 0.83 |
| 13 | Cl-k.18626 | 416 | L7M0J1, phospholipase a2, R. pulchellus | 0 | 1704 | 80.1 | 25.19 ± 5.41 |
| 14 | Cl-k.30316 ② | 566 | Q9U6M8, carboxylic ester hydrolase, R. microplus | 5.1E−171 | 1290 | 47.3 | 10.59 ± 2.67 |
| 15 | Cl-k.24717 ② | 564 | A0A6M2CHI4, carboxylic ester hydrolase, R. microplus | 0 | 1366 | 49.7 | 0.25 ± 0.05 |
| 16 | Cl-k.18635 | 521 | A5LHV9, disulfide isomerase, H. longicornis | 0 | 2514 | 96.2 | 1.74 ± 0.43 |
| 17 | Cl-k.17461 | 163 | F2Z7L0, lysozyme, H. longicornis | 3.1E−70 | 544 | 87.1 | 2.44 ± 0.86 |
| 18 | Cl-k.17638 | 397 | G8C7A0, lysosomal acid phosphatase, H. longicornis | 0 | 1878 | 93.5 | 75.50 ± 18.32 |
| 19 | Cl-k.18136 | 233 | Q6JVN0, glutathione S-transferase, H. longicornis | 3.5E−159 | 1144 | 93.7 | 1.74 ± 0.50 |
| 20 | Cl-k.18835 | 526 | A0A131YK94, superoxide dismutase (Cu–Zn), R. appendiculatus | 0 | 1475 | 61.8 | 18.83 ± 4.63 |
| 21 | Cl-k.19315 ③ | 316 | A0A131YMH9, chitinase, R. appendiculatus | 0 | 1777 | 85.6 | 1.46 ± 0.44 |
| 22 | Cl-k.18448 ③ | 536 | A0A023FND8, chitinase, A. cajennense | 0 | 1919 | 80.3 | 21.40 ± 4.31 |
| 23 | Cl-k.17863 | 367 | A0A286R6W4, fructose-bisphosphate aldolase, Dermacentor silvarum | 0 | 1796 | 94.2 | 2.26 ± 0.57 |
| 24 | Cl-k.35642 | 331 | A0A1E1X9Y4, alpha-L-fucosidase, A. aureolatum | 0 | 1414 | 81.7 | 1.87 ± 0.38 |
| 25 | Cl-k.18180 | 430 | A0A2P1DPZ4, glyceraldehyde-3-phosphate dehydrogenase, H. flava | 0 | 1730 | 100 | 2.37 ± 0.62 |
| 26 | Cl-k.17933 | 549 | A0A6M2CTD4, ATP synthase subunit beta, R. microplus | 0 | 2628 | 96.3 | 0.39 ± 0.02 |
| II. Enzyme inhibitors | |||||||
| 27 | Cl-k.20245 ④ | 256 | A0A5P8H6S1, serpin-a, H. longicornis | 9.3E−97 | 753 | 61.9 | 55.88 ± 13.09 |
| 28 | Cl-k.17714 ④ | 415 | A0A5P8H6S1, serpin-a, H. longicornis | 6.9E−169 | 1246 | 63.4 | 0.25 ± 0.24 |
| 29 | Cl-k.16905 ④ | 143 | A0A5P8H6S1, serpin-a, H. longicornis | 6.4E−49 | 422 | 61.8 | 32.01 ± 6.67 |
| 30 | Cl-k.18212 ④ | 427 | A0A5P8H6S1, serpin-a, H. longicornis | 0 | 1727 | 87.5 | 45.04 ± 10.59 |
| 31 | Cl-k.19946 ④ | 406 | Q75Q63, serpin-2 , H. longicornis | 0 | 1686 | 82.4 | 11.48 ± 2.66 |
| 32 | Cl-k.16646 ④ | 201 | A0A6M2E637, tick serpins 8, A. tuberculatum | 7.5E−68 | 555 | 63.0 | ⑻ 178.59 ± 44.58 |
| 33 | Cl-k.22217 ④ | 398 | A0A023GN51, tick serpins 13, A. triste | 9.5E−165 | 1216 | 61.8 | 38.49 ± 10.90 |
| 34 | Cl-k.14429 | 421 | A0A023G8Z1, serine proteinase inhibitor, A. triste | 0 | 1409 | 65.5 | 71.47 ± 17.26 |
| 35 | Cl-k.18644 ⑤ | 229 | A0A224YJB7, α2-macroglobulin splicing variant, Rhipicephalus zambeziensis | 3.7E−141 | 1119 | 91.7 | ⑸ 435.08 ± 100.49 |
| 36 | Cl-k.18677 ⑤ | 1142 | A0A1E1XEL3, α-macroglobulin, A. aureolatum | 0 | 5401 | 89.6 | ⑹ 243.02 ± 53.71 |
| 37 | Cl-k.19779 ⑤ | 1820 | A0A1E1XL07, α-macroglobulin, A. sculptum | 0 | 6650 | 76.1 | 1.10 ± 0.27 |
| 38 | Cl-k.18726 et al. | 1915 | A0A023FNM2, α2-macroglobulin splicing variant, Amblyomma cajennense | 0 | 5819 | 78.9 | 35.72 ± 8.99 |
| 39 | Cl-k.18944 | 269 | A0A023GP16, Kazal-type serine protease inhibitor, A. triste | 5.8E−155 | 1123 | 84.0 | 8.02 ± 1.79 |
| 40 | Cl-k.12087 ⑥ | 183 | A0A6B9DA14, cystatin, H. flava | 0 | 757 | 100 | 15.11 ± 4.89 |
| 41 | Cl-k.17388 ⑥ | 185 | A0A3G6VF56, cystatin, H. flava | 1.9E−100 | 745 | 99.3 | 65.43 ± 15.54 |
| 42 | Cl-k.20981 ⑥ | 164 | A0A6M3YRY3, cystatin, H. flava | 4.2E−89 | 667 | 100 | 15.73 ± 4.93 |
| 43 | Cl-k.21288 | 228 | A0A023GEH6, thyropin, A. triste | 1.5E−94 | 723 | 60.0 | 22.19 ± 7.67 |
| 44 | Cl-k.23450 | 229 | A0A023GAB0, thyropin, A. triste | 2.2E−89 | 689 | 59.4 | 35.86 ± 8.77 |
| III. Immune-related proteins | |||||||
| 45 | Cl-k.18200 ⑦ | 84 | A0A6G5A751, microplusin, R. microplus | 9.5E−31 | 260 | 54.8 | ⑴ 6514.56 ± 803.43 |
| 46 | Cl-k.18906 ⑦ | 134 | A0A6G5A751, microplusin, R. microplus | 8.0E−34 | 295 | 52.5 | ⑷ 699.92 ± 23.27 |
| 47 | Cl-k.20235 ⑦ | 87 | A0A6G5A751, microplusin, R. microplus | 2.1E−29 | 266 | 46.2 | 61.60 ± 17.16 |
| 48 | Cl-k.3924 | 78 | A0A2D1CLH7, defensin DFS2, H. longicornis | 2.1E−48 | 361 | 83.6 | 11.98 ± 3.53 |
| 49 | Cl-k.23590 ⑧ | 1612 | A0A131ZDX3, TIL domain-containing protein, R. appendiculatus | 0 | 7053 | 77.0 | 11.48 ± 2.56 |
| 50 | Cl-k.13586 ⑧ | 2610 | A0A131Z678, TIL domain-containing protein, R. appendiculatus | 0 | 8535 | 77.2 | 9.72 ± 2.21 |
| 51 | Cl-k.18775 ⑧ | 2252 | A0A131YJS1, TIL domain-containing protein, R. appendiculatus | 0 | 8966 | 68.9 | 1.42 ± 0.30 |
| 52 | Cl-k.25067 ⑧ | 109 | A0A6M2CNI0,TIL domain-containing protein, R. microplus | 4.1E−39 | 329 | 69.9 | 26.91 ± 4.60 |
| 53 | Cl-k.18521 ⑨ | 235 | F0J8I6, ixoderin, A. variegatum | 5.4E−93 | 707 | 66.3 | 6.23 ± 1.40 |
| 54 | Cl-k.17959 ⑨ | 313 | A0A1E1XEF5, ixoderin, A. aureolatum | 5.7E−153 | 1118 | 73.8 | 6.92 ± 1.81 |
| 55 | Cl-k.19166 | 177 | A0A6M2D1K3, C2b, R. microplus | 6.9E−55 | 488 | 62.3 | 2.75 ± 0.88 |
| 56 | Cl-k.21838 | 428 | A0A7L9DI94, C3, Ixodes ricinus | 1.0E−134 | 1104 | 50.9 | 10.48 ± 2.55 |
| 57 | Cl-k.14141 | 152 | A0A224Z7V2, serum amyloid A protein, R. zambeziensis | 1.4E−77 | 592 | 72.7 | 28.19 ± 6.81 |
| 58 | Cl-k.17842 | 332 | A0A0S3Q1T5, leucine-rich repeat containing protein, H. longicornis | 0 | 1464 | 90.1 | 19.96 ± 4.17 |
| 59 | Cl-k.18342 | 233 | A0A0M3TC17, AV422, H. flava | 8.4E−171 | 1221 | 100 | 12.15 ± 2.84 |
| 60 | Cl-k.18520 ⑩ | 181 | Q08G07, Hq05, Haemaphysalis qinghaiensis | 1.1E−129 | 940 | 97.2 | 6.53 ± 1.43 |
| 61 | Cl-k.18575 ⑩ | 180 | G3BJU6, immunogenic protein, H. longicornis | 4.5E−120 | 877 | 93.9 | 16.62 ± 4.03 |
| IV. Transporters | |||||||
| 62 | Cl-k.25224 ⑪ | 1538 | Q5EG54, Vg, D. variabilis | 0 | 5360 | 67.5 | 0.21 ± 0.10 |
| 63 | Cl-k.16576 ⑪ | 686 | E1CAX9, vitellogenin-1, H. longicornis | 0 | 1248 | 98.5 | 126.50 ± 31.13 |
| 64 | Cl-k.19213-k18886 ⑪ | 351 | B1B544, vitellogenin-2, H. longicornis | 8.7E−86 | 732 | 98.3 | ⑶ 1606.10 ± 295.28 |
| 65 | Cl-k.18067 ⑪ | 463 | B1B544, vitellogenin-2, H. longicornis | 0 | 2072 | 85.1 | ⑵ 1829.13 ± 312.65 |
| 66 | Cl-k.19483 ⑪ | 1386 | B1B544, vitellogenin-2, H. longicornis | 0 | 4657 | 62.4 | ⑽ 138.31 ± 29.98 |
| 67 | Cl-k.16789 ⑪ | 114 | E1CAY0, vitellogenin-3, H. longicornis | 3.1E−62 | 535 | 85.7 | 95.40 ± 25.38 |
| 68 | Cl-k.18851-18114 ⑪ | 2279 | G9M4L6, vitellogenin B, H. longicornis | 0 | 13,028 | 85.9 | 63.60 ± 12.52 |
| 69 | Cl-k.21299 | 207 | M5AYG7, ferritin 2, H. longicornis | 9.9E−133 | 965 | 93.5 | 22.51 ± 4.62 |
| 70 | Cl-k.19103 | 152 | A0A023G718, fatty acid-binding protein, A. triste | 4.6E−83 | 628 | 85.4 | 15.26 ± 2.76 |
| V. Muscle proteins | |||||||
| 71 | Cl-k.18720 | 877 | J7LVN2, paramyosin, H. longicornis | 0 | 4196 | 98.2 | 0.53 ± 0.13 |
| 72 | Cl-k.18460 | 424 | A8E4J9, calreticulin, H. qinghaiensis | 0 | 2247 | 99.5 | 2.42 ± 0.76 |
| 73 | Cl-k.18394 | 52 | A0A131ZAE8, tropomyosin, R. appendiculatus | 0 | 1423 | 93.2 | 3.76 ± 1.07 |
| 74 | Cl-k.18452 | 83 | A0A0N6X2B1, muscle LIM protein, H. longicornis | 7.8E−57 | 417 | 95.8 | 4.98 ± 1.14 |
| VI. Heat shock proteins | |||||||
| 75 | Cl-k.18505 ⑫ | 655 | A0A097A1J8, heat shock 70 kDa protein 8, H. flava | 0 | 3290 | 100 | 2.09 ± 0.55 |
| 76 | Cl-k.18161 ⑫ | 683 | E4W3Z2, heat shock 70 kDa protein 5, H. longicornis | 0 | 3317 | 99.1 | 0.45 ± 0.22 |
| Other | |||||||
| Cl-k.18334 | 255 | A0A023FPM9, glycine-rich secreted cement protein, A. cajennense | 1.1E−118 | 884 | 74.0 | ⑼ 170.44 ± 38.51 | |
Proteins sharing the same number in the Protein ID column belong to the same family. Length indicates the number of amino acid residues of protein fractions detected by MS. The number in the iBAQ column indicates the top 10 most abundant peptides detected by MS
There were many enzymes with anticoagulation activity in tick hemolymph. Among them, serine protease was the most abundant. Three serine proteinase genes from H. longicornis (Hl-Sp1, Hl-Sp2, and Hl-Sp3) were cloned, and their recombinant enzymes efficiently hydrolyzed substrates specific for serine proteinases [38]. RNA interference (RNAi) of Hl-Sp1, Hl-Sp2, and Hl-Sp3 genes synchronously caused a decrease in the body weight of engorged ticks, suggesting their synergistic roles in blood-feeding and digestion [38]. Longistatin is an unconventional serine protease that has been shown to hydrolyze fibrinogen and efficiently induce high titers of protective IgG antibodies against ticks [39–41]. Metalloproteinases in tick saliva were found to be essential for blood-feeding [42, 43]. During the initial feeding stage, metalloproteinases suppressed blood clotting and degraded extracellular matrix proteins, which is critical for the preparation of the feeding site. As these enzymes also demonstrated anti-angiogenic activity, they were of importance in the late feeding stage by inhibiting tissue repair in the host. Rhipicephalus microplus secreted carboxylic ester hydrolase in the skin of calves, immediately adjacent to mouthparts, or in the attachment cone [44]. This constitutes an enzyme system against coagulation together with serine protease and metalloproteinases, among others.
Enzymes in tick hemolymph also take part in nutrient metabolism. Aspartic and cysteine proteinases and exopeptidases were shown to catalyze the decomposition of Vg and Hb [45, 46]. Cathepsin L-like cysteine endopeptidase was reported to hydrolyze synthetic substrates and protein substrates including Hb [47, 48], and serine carboxypeptidase and cathepsin C broke down small peptides, releasing free amino acids [46, 49]. Glutathione S-transferases facilitated the excretion of physiological and xenobiotic substances, protecting cells against chemical toxicity and stress [50]. Although specific roles of glyceraldehyde-3-phosphate dehydrogenase and fructose-1,6-bisphosphate aldolase have not been verified in ticks, they are key enzymes in carbohydrate metabolism.
Some enzymes have appeared in hemolymph with innate immune activity. Liao et al. cloned genes encoding putative protein disulfide isomerase (Hl-PDI1, Hl-PDI2, Hl-PDI3), lysozyme (Hl-lysozyme), and lysosomal acid phosphatase (HL-3) in H. longicornis ticks. Hl-PDI1/2/3 were expressed in all developmental stages and in organs including the midgut, salivary gland, ovary, hemolymph, and fat body of adult females, and Hl-PDI1/3 was possibly involved in Babesia infection [13]. Increased gene expression of Hl-Lysozyme was observed in female ticks challenged with bacteria, implying a possible role in the innate immunity of ticks against microorganisms [51]. HL-3 transcripts were significantly induced by blood-feeding, and were involved in tick innate immunity [52]. Superoxide dismutase (SOD) was reported as a key enzyme in detoxification of reactive oxygen species, and silencing of Cu/Zn-SOD decreased the colonization of O parkeri in A. maculatum ticks [53].
We also detected chitinase in H. flava hemolymph. Chitinase was induced by ecdysteroids to degrade older chitin at the time of molting, and recombinant chitinase from H. longicornis was capable of chitin degradation [54, 55].
Protease inhibitors in tick hemolymph
Numerous protease inhibitors have been detected in tick hemolymph, including serine protease inhibitors, tight-binding inhibitors, cystatins, and thyropins.
α-2-macroglobulin, serpins, and Kunitz/Kazal domain-containing proteins were found to belong to the serine protease inhibitors [56]. Serpins were the most abundant protease inhibitors in H. flava hemolymph. In total, about 120 serpins were recorded in ticks [57–59]; 20 of them in different tick genera have been functionally characterized [60]. A serpin from Rhipicephalus haemaphysaloides was shown to participate in vitellogenesis [61]. Additionally, serpin-2 and other serpins were directly related to tick blood-feeding, facilitating successful acquisition of a blood meal [62–64].
Cystatins and thyropins were found to be inhibitors of cysteine peptidases. Tick cystatins either regulated cathepsin-mediated Hb digestion [46] or played a role in tick embryogenesis [65]. In addition to these functions, a type-2 cystatin in the hemocytes of R. microplus was related to tick immunity [66].
Immune-related proteins in tick hemolymph
Three microplusins were detected. They were 103 amino acids in length; all contained signal peptides. They displayed similarity of 46.2–52.5% compared with a microplusin from R. microplus [67]. Microplusin was shown to have bacteriostasis activity (gram-positive bacterium) and to offer protection against Rickettsia rickettsii infection [67, 68]. Microplusin gene expression was verified in several organs, including fat body, hemocyte, ovary, and midgut [67, 68]. A microplusin-like peptide was identified in A. hebraeum hemolymph [69].
Transporters in tick hemolymph
There were three types of transporters in tick hemolymph, i.e., Vg, ferritin, and fatty acid-binding protein. The Vg family contained the most members. Vgs have been investigated and partially characterized in O. moubata [4, 70], O. parkeri [29], D. variabilis [31, 71], I. scapularis [3], H. longicornis [33], and R. microplus [72], among others. Vg is synthesized in fat bodies, gut cells, and, to a lesser extent, ovaries of tick females after mating and feeding [2]. Based on the unique peptides obtained, a minimum of seven types of Vgs were retrieved in H. flava. Cl-k.16576, Cl-k.19213, and Cl-k.16789 displayed a notable similarity with Hl-Vg1, Hl-Vg2, and Hl-Vg3, respectively. Hl-Vgs consisted of four major polypeptides [33]. Hl-Vg RNAi-challenged ticks displayed lower body weight and egg weight and higher mortality in engorged females [33]. Cl-k.25224 was structurally similar to Vg in D. variabilis (UniProt accession number: Q5EG54, GenBank accession number: AY885250). Vg mRNA was detected in replete (mated) pre-ovipositional female D. variabilis, increased to a more notable level in ovipositing females, and was absent after completion of egg-laying [31].
Muscle proteins and heat shock proteins in tick hemolymph
There were four muscle proteins detected in tick hemolymph, including paramyosin, calreticulin, tropomyosin, and muscle LIM protein. Aside from muscle composition, they demonstrated other special functions. For example, the recombinant B. microplus paramyosin was able to bind both IgG and collagen [73], while calreticulin from A. americanum was found to bind to C1q [74]. Silencing of H. longicornis tropomyosin (HL-Tm) led to a reduction in tick engorgement and oviposition [75].
Two heat shock proteins 70 (HSP70) were found in tick hemolymph, and their expression was significantly upregulated upon blood-feeding [76, 77]. HSP70-8 and HSC70 were shown to exert an anticoagulation effect in vitro [78].
Quantitative analysis of proteins in tick hemolymph
The iBAQ of high-confidence host proteins in H. flava hemolymph is listed in Table 1. The top 10 abundant host-derived proteins included Hb subunit-α and subunit-β, albumin, serotransferrin-like, ubiquitin-like, haptoglobin, α-1-antitrypsin-like protein, histone H2B, apolipoprotein A-I, and C3-β.
Vitellogenin, microplusin and α-macroglobulin were the top three abundant tick proteins (Table 2). The abundance of Vg1, Vg2 and Vg3 was extremely high in the hemolymph of H. longicornis [33]. Our unpublished data indicate that the egg protoplasm did not contain large quantities of these Vgs, implying that the main role of these Vgs might not be as nutrients. The high abundance of Vg and microplusin indicated that the major function of tick hemolymph was the transport of substances and participation in the immune responses.
Of note, Cl-k.18334, which was annotated as a glycine-rich secreted cement protein (A0A023FPM9), was ranked as the ninth most abundant tick-derived protein in hemolymph. Thus far, there have been no reports on its function in ticks.
Protein families in tick hemolymph
Twelve protein families were identified in this study: serine protease, carboxylic ester hydrolase, chitinase, serpin, α-macroglobulin, cystatin, microplusin, TIL domain-containing protein, ixoderin, immunogenic protein, Vg, and heat shock protein. However, only four families, namely, serpin, Vg, cystatin, and microplusin, have been investigated extensively in ticks.
Sequence analysis revealed that some families had extremely similar sequences among members. For instance, three microplusins shared up to 86.22% sequence similarity (Additional file 2: Fig. S1). All had an N-terminal sequence MKA, six C residues, and signal peptides.
Proteins in some families, such as cystatin and serpin, shared remarkable similarity in structure, although their amino acid sequences were quite different. Cl-k.17388, Cl-k.20981, and Cl-k.12087 were all cystatins; the similarity between them was 35.92%. However, both had conserved a GG at the N-terminal, and a QXVXG motif of cystatin2 and a typical C-PW-C motif at the C-terminal (Additional file 3: Fig. S2).
Seven serpins all contained serpin consensus amino acid motif N-[AT]-[VIM]-[YLH]-F-[KRT]-[GS], [DERQ]-[VL]-[NDS]-E-[EVDKQ]-G, and serpin signature PS00284 ([LIVMFY]-[G]-[LIVMFYAC]-[DNQ]-[rkHQs]-[PST]-F-[LIVMFY]-[LIVMFYC]-X-[LIVMFAH]).
Among the four TIL domain-containing proteins, Cl-k.25067 shared similarity with ixodidin, an antimicrobial peptide from hemocytes of R. microplus with inhibitory activity against serine proteinases [79]; the other three TIL domain-containing proteins (Cl-k.23590, Cl-k.13586, and Cl-k.18775) all included a trypsin inhibitor-like, cysteine-rich domain and a von Willebrand factor type domain in their structures, and might play a role in hemolymph anticoagulation [80].
Importantly, the present study only provided a protein profile in the hemolymph of fully engorged ticks at a single time point in blood-feeding. Further studies will address the importance of hemolymph proteins during feeding, and will include the unfed tick stage and different time points.
Conclusion
Based on a search against the UniProt Erinaceidae database and H. flava proteome library, we identified 18 host-derived high-confidence proteins and 169 tick-derived high-confidence proteins, providing the most comprehensive protein composition in tick hemolymph thus far. The protein profile of the H. flava hemolymph mirrored a sophisticated protein system in the physiological processes of anticoagulation, blood meal digestion, and innate immunity. As the bulk of proteins detected in hemolymph have not been functionally characterized in ticks, further investigations are needed to decipher their roles in tick biology.
Supplementary Information
Additional file 1: Table S1. Unique peptides and deduced protein sequences identified by searching against the H. flava transcriptome database.
Additional file 2: Figure S1. Amino acid sequences of three microplusins in H. flava hemolymph.
Additional file 3: Figure S2. Amino acid sequences and domains of two cystatins in H. flava hemolymph.
Acknowledgements
Not applicable.
Abbreviations
- Hb
Hemoglobin
- iBAQ
Intensity-based absolute quantification
- LC–MS/MS
Liquid chromatography–tandem mass spectrometry
- MS
Mass spectrometry
- SDS-PAGE
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- Serine proteinase inhibitor
Serpin
- Vg
Vitellogenin
Author contributions
TC and LL designed the experiments. ZL and ZW collected the tick hemolymph and prepared the protein extracts for high-performance LC–MS/MS. TC, LL, FY, and DD analyzed the data and prepared the tables and figures. TC, LL, and FY collaborated in writing and editing the manuscript. All authors read and approved the final manuscript.
Funding
This research was supported by a Grant from the Science Fund for Distinguished Young Scholars of Changsha City, China (Foundation for Distinguished Young Talents in Higher Education of Guangdong) (Changsha, China, kq2009055), the National Science Foundation for Young Scientists of China (Beijing, China, No. 31902294), the Hunan Province Technology Breakthrough Project of 2021 for the open competition mechanism to select the best candidates, and the Double first-class construction project of Hunan Agricultural University (SYL201802016).
Availability of data and materials
The data supporting the conclusions of this article are available in the iProX repository, https://www.iprox.cn/page/DSV021.html;?url=1642501651308hONl, with the key 3ABj.
Declarations
Ethics approval and consent to participate
All procedures involving animals in the present study were approved and overseen by the Hunan Agricultural University Institutional Animal Care and Use Committee (No. 2021085).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Lei Liu, Email: ll434@hunau.edu.cn.
Fen Yan, Email: 631109101@qq.com.
Lu Zhang, Email: 1197192694@qq.com.
Zhi-feng Wu, Email: 158530545@qq.com.
De-yong Duan, Email: duandy@hunau.edu.cn.
Tian-yin Cheng, Email: hn5368@163.com.
References
- 1.Sonenshine DE. Biology of ticks. Oxford: Oxford University; 1992. [Google Scholar]
- 2.Libor G, Nataliia R, Marie V, Maryna G, Jan S. Circulatory system and hemolymph. In: Sonenshine DE, Roe R, editors. Biology of ticks. Oxford: Oxford University; 2013. p. 263. [Google Scholar]
- 3.James AM, Oliver JH. Purification and partial characterization of vitellin from the black-legged tick, Ixodes scapularis. Insect Biochem Mol Biol. 1997;27:639–649. doi: 10.1016/s0965-1748(97)00038-6. [DOI] [PubMed] [Google Scholar]
- 4.Chinzei Y, Chino H, Takahashi K. Purification and properties of vitellogenin and vitellin from a tick, Ornithodoros moubata. J Comp Physiol. 1983;152:13–21. [Google Scholar]
- 5.Kopacek P, Weise C, Saravanan T, Vitova K, Grubhoffer L. Characterization of an αmacroglobulin-like glycoprotein isolated from the plasma of the soft tick Ornithodoros moubata. Eur J Biochem. 2000;267:465–475. doi: 10.1046/j.1432-1327.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 6.Fogaça A, da Silva PJ, Miranda M, Bianchi A, Miranda A, Ribolla P, et al. Antimicrobial activity of a bovine hemoglobin fragment in the tick Boophilus microplus. J Biol Chem. 1999;274:25330–25334. doi: 10.1074/jbc.274.36.25330. [DOI] [PubMed] [Google Scholar]
- 7.Johns R, Sonenshine DE, Hynes WL. Identification of a defensin from the hemolymph of the American dog tick, Dermacentor variabilis. Insect Biochem Mol Biol. 2001;31:857–865. doi: 10.1016/s0965-1748(01)00031-5. [DOI] [PubMed] [Google Scholar]
- 8.Sterba J, Dupejova J, Fiser M, Vancova M, Grubhoffer L. Fibrinogen-related proteins in ixodid ticks. Parasit Vectors. 2011;4:127. doi: 10.1186/1756-3305-4-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dupejova J, Sterba J, Vancova M, Grubhoffer L. Hemelipoglycoprotein from the ornate sheep tick, Dermacentor marginatus: structural and functional characterization. Parasit Vectors. 2011;4:4. doi: 10.1186/1756-3305-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kluck GEG, Silva Cardoso L, De Cicco NNT, Lima MS, Folly E, Atella GC. A new lipid carrier protein in the cattle tick Rhipicephalus microplus. Ticks Tick Borne Dis. 2018;9:850–859. doi: 10.1016/j.ttbdis.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 11.Horáčková J, Rudenko N, Golovchenko M, Havlíková S, Grubhoffer L. IrML–a gene encoding a new member of the ML protein family from the hard tick, Ixodes ricinus. J Vector Ecol. 2010;35:410–418. doi: 10.1111/j.1948-7134.2010.00100.x. [DOI] [PubMed] [Google Scholar]
- 12.Du W, Gao Z, Wang K, Zhao Y, Zheng P, Yu Z, et al. Expression and function assessment of two serpin-type serine protease inhibitors from Haemaphysalis doenitzi. Res Vet Sci. 2020;132:1–9. doi: 10.1016/j.rvsc.2020.05.015. [DOI] [PubMed] [Google Scholar]
- 13.Liao M, Hatta T, Umemiya R, Huang P, Jia H, Gong H, et al. Identification of three protein disulfide isomerase members from Haemaphysalis longicornis tick. Insect Biochem Mol Biol. 2007;37:641–654. doi: 10.1016/j.ibmb.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 14.Gudderra NP, Sonenshine DE, Apperson CS, Roe RM. Hemolymph proteins in ticks. J Insect Physiol. 2002;48:269–278. doi: 10.1016/s0022-1910(02)00050-1. [DOI] [PubMed] [Google Scholar]
- 15.Madden RD, Sauer JR, Dillwith JW. A proteomics approach to characterizing tick salivary secretions. Exp Appl Acarol. 2002;28:77–87. [PubMed] [Google Scholar]
- 16.Tirloni L, Kim TK, Pinto AFM, Yates JR, da Silva VI, Mulenga A. Tick-host range adaptation: changes in protein profiles in unfed adult Ixodes scapularis and Amblyomma americanum saliva stimulated to feed on different hosts. Front Cell Infect Microbiol. 2017;7:517. doi: 10.3389/fcimb.2017.00517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oleaga A, Escudero-Población A, Camafeita E, Pérez-Sánchez R. A proteomic approach to the identification of salivary proteins from the argasid ticks Ornithodoros moubata and Ornithodoros erraticus. Insect Biochem Mol Biol. 2007;37:1149–1159. doi: 10.1016/j.ibmb.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 18.Tirloni L, Lu S, Calvo E, Sabadin G, Di Maggio LS, Suzuki M, et al. Integrated analysis of sialotranscriptome and sialoproteome of the brown dog tick Rhipicephalus sanguineus (s.l.): insights into gene expression during blood feeding. J Proteomics. 2020;229:103899. doi: 10.1016/j.jprot.2020.103899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu L, Cheng TY, He XM. Proteomic profiling of the midgut contents of Haemaphysalis flava. Ticks Tick Borne Dis. 2018;9:490–495. doi: 10.1016/j.ttbdis.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 20.Liu L, Liu YS, Liu GH, Cheng TY. Proteomics analysis of faecal proteins in the tick Haemaphysalis flava. Parasit Vectors. 2018;11:89. doi: 10.1186/s13071-018-2673-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tirloni L, Reck J, Terra RMS, Martins JR, Mulenga A, Sherman NE, et al. Proteomic analysis of cattle tick Rhipicephalus (Boophilus) microplus saliva: a comparison between partially and fully engorged females. PLoS ONE. 2014;9:1–19. doi: 10.1371/journal.pone.0094831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stopforth E, Neitz AWH, Gaspar ARM. A proteomics approach for the analysis of hemolymph proteins involved in the immediate defense response of the soft tick, Ornithodoros savignyi, when challenged with Candida albicans. Exp Appl Acarol. 2010;51:309–325. doi: 10.1007/s10493-010-9338-z. [DOI] [PubMed] [Google Scholar]
- 23.Aguilar-Díaz H, Esquivel-Velázquez M, Quiroz-Castañeda RE, Miranda-Miranda E, Conde-Baeye RJP, Cobaxín-Cárdenas M, et al. Comparative hemolymph proteomic and enzymatic analyses of two strains of Rhipicephalus (Boophilus) microplus ticks resistant and susceptible to ixodicides. Biomed Res Int. 2018;2018:9451547. doi: 10.1155/2018/9451547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng WQ, Xuan XN, Fu RL, Tao HY, Liu YQ, Liu XQ, et al. Tick-borne pathogens in ixodid ticks from Poyang lake region, southeastern China. Korean J Parasitol. 2018;56:589–596. doi: 10.3347/kjp.2018.56.6.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Au-Fiorotti J, Au-Gôlo PS, Au-Marciano AF, Au-Camargo MG, Au-Angelo IC, Au-Bittencourt VREP. Disclosing hemolymph collection and inoculation of Metarhizium Blastospores into Rhipicephalus Microplus ticks towards invertebrate pathology studies. JoVE. 2019;148:e59899. doi: 10.3791/59899. [DOI] [PubMed] [Google Scholar]
- 26.Golo PS, dos Santos ASDO, Monteiro CMO, Perinotto WMDS, Quinelato S, Camargo MG, et al. Lab-on-a-chip and SDS-PAGE analysis of hemolymph protein profile from Rhipicephalus microplus (Acari: Ixodidae) infected with entomopathogenic nematode and fungus. Parasitol Res. 2016;115:3459–68. doi: 10.1007/s00436-016-5109-z. [DOI] [PubMed] [Google Scholar]
- 27.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 28.Tatchell RJ. Electrophoretic studies on the proteins of the haemolymph, saliva, and eggs of the cattle tick, Boophilus microplus. Insect Biochem. 1971;1:47–55. [Google Scholar]
- 29.Taylor D, Chinzei Y, Miura K, Ando K. Vitellogenin synthesis, processing and hormonal regulation in the tick, Ornithodoros parkeri (Acari:Argasidae) Insect Biochem. 1991;21:723–733. [Google Scholar]
- 30.Rosell-Davis R, Coons LB. Relationship between feeding, mating, vitellogenin production and vitellogenesis in the tick Dermacentor variabilis. Exp Appl Acarol. 1989;7:95–105. doi: 10.1007/BF01200456. [DOI] [PubMed] [Google Scholar]
- 31.Thompson DM, Khalil SMS, Jeffers LA, Sonenshine DE, Mitchell RD, Osgood CJ, et al. Sequence and the developmental and tissue-specific regulation of the first complete vitellogenin messenger RNA from ticks responsible for heme sequestration. Insect Biochem Mol Biol. 2007;37:363–374. doi: 10.1016/j.ibmb.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 32.Donohue KV, Khalil SMS, Mitchell RD, Sonenshine DE, Michael RR. Molecular characterization of the major hemelipoglycoprotein in ixodid ticks. Insect Mol Biol. 2008;17:197–208. doi: 10.1111/j.1365-2583.2008.00794.x. [DOI] [PubMed] [Google Scholar]
- 33.Boldbaatar D, Umemiya-Shirafuji R, Liao M, Tanaka T, Xuan X, Fujisaki K. Multiple vitellogenins from the Haemaphysalis longicornis tick are crucial for ovarian development. J Insect Physiol. 2010;56:1587–1598. doi: 10.1016/j.jinsphys.2010.05.019. [DOI] [PubMed] [Google Scholar]
- 34.Hefnawy T. Biochemical and physiological studies of certain ticks (Ixodoidea). Hemolymph volume determined by isotope and dye dilution during the gonotrophic cycle of Argas (Persicargas) persicus (Oken) and A. (P.) arboreus Kaiser, Hoogstraal, and Kohls (Argasidae) J Parasitol. 1972;58:358–64. [PubMed] [Google Scholar]
- 35.Ackerman S, Clare FB, McGill TW, Sonenshine DE. Passage of host serum components, including antibody, across the digestive tract of Dermacentor variabilis (Say) J Parasitol. 1981;67:737–740. [PubMed] [Google Scholar]
- 36.Jeffers LA, Michael RR. The movement of proteins across the insect and tick digestive system. J Insect Physiol. 2008;54:319–332. doi: 10.1016/j.jinsphys.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 37.Maya-Monteiro CM, Daffre S, Logullo C, Lara FA, Alves EW, Capurro ML, et al. HeLp, a heme lipoprotein from the hemolymph of the cattle tick, Boophilus microplus. J Biol Chem. 2000;275:36584–36589. doi: 10.1074/jbc.M007344200. [DOI] [PubMed] [Google Scholar]
- 38.Miyoshi T, Tsuji N, Islam MK, Alim MA, Hatta T, Huang X, et al. A set of serine proteinase paralogs are required for blood-digestion in the ixodid tick Haemaphysalis longicornis. Parasitol Int. 2008;57:499–505. doi: 10.1016/j.parint.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 39.Anisuzzaman, Islam MK, Miyoshi T, Alim MA, Hatta T, Yamaji K, et al. Longistatin, a novel EF-hand protein from the ixodid tick Haemaphysalis longicornis, is required for acquisition of host blood-meals. Int J Parasitol. 2010;40:721–9. doi: 10.1016/j.ijpara.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 40.Anisuzzaman, Islam MK, Alim MA, Miyoshi T, Hatta T, Yamaji K, et al. Longistatin, a plasminogen activator, is key to the availability of blood-meals for ixodid ticks. PLoS Pathog. 2011;7:1–13. doi: 10.1371/journal.ppat.1001312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anisuzzaman, Islam MK, Alim MA, Miyoshi T, Hatta T, Yamaji K, et al. Longistatin is an unconventional serine protease and induces protective immunity against tick infestation. Mol Biochem Parasitol. 2012;182:45–53. doi: 10.1016/j.molbiopara.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 42.Francischetti IMB, Mather TN, Ribeiro JMC. Cloning of a salivary gland metalloprotease and characterization of gelatinase and fibrin(ogen)lytic activities in the saliva of the Lyme disease tick vector Ixodes scapularis. Biochem Biophys Res Commun. 2003;305:869–875. doi: 10.1016/s0006-291x(03)00857-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Francischetti IMB, Mather TN, Ribeiro JMC. Tick saliva is a potent inhibitor of endothelial cell proliferation and angiogenesis. Thromb Haemost. 2005;94:167–174. doi: 10.1267/THRO05010167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schleger AV, Lincoln DT. Boophilus microplus: characterization of enzymes introduced into the host. Aust J Biol Sci. 1976;29:487–498. doi: 10.1071/bi9760487. [DOI] [PubMed] [Google Scholar]
- 45.Cruz CE, Fogaca AC, Nakayasu ES, Angeli CB, Belmonte R, Almeida IC, et al. Characterization of proteinases from the midgut of Rhipicephalus (Boophilus) microplus involved in the generation of antimicrobial peptides. Parasit Vectors. 2010;3:63. doi: 10.1186/1756-3305-3-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horn M, Nussbaumerova M, Sanda M, Kovarova Z, Srba J, Franta Z, et al. Hemoglobin digestion in blood-feeding ticks: mapping a multipeptidase pathway by functional proteomics. Chem Biol. 2009;16:1053–1063. doi: 10.1016/j.chembiol.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seixas A, Dos Santos PC, Velloso FF, Da Silva VI, Masuda A, Horn F, et al. A Boophilus microplus vitellin-degrading cysteine endopeptidase. Parasitology. 2003;126:155–163. doi: 10.1017/s0031182002002731. [DOI] [PubMed] [Google Scholar]
- 48.Renard G, Garcia JF, Cardoso FC, Richter MF, Sakanari JA, Ozaki LS, et al. Cloning and functional expression of a Boophilus microplus cathepsin L-like enzyme. Insect Biochem Mol Biol. 2000;30:1017–1026. doi: 10.1016/s0965-1748(00)00070-9. [DOI] [PubMed] [Google Scholar]
- 49.Motobu M, Tsuji N, Miyoshi T, Huang X, Islam MK, Alim MA, et al. Molecular characterization of a blood-induced serine carboxypeptidase from the ixodid tick Haemaphysalis longicornis. FEBS J. 2007;274:3299–3312. doi: 10.1111/j.1742-4658.2007.05852.x. [DOI] [PubMed] [Google Scholar]
- 50.Freitas DRJ, Rosa RM, Moraes J, Campos E, Logullo C, Da Silva VI, et al. Relationship between glutathione S-transferase, catalase, oxygen consumption, lipid peroxidation and oxidative stress in eggs and larvae of Boophilus microplus (Acarina: Ixodidae) Comp Biochem Physiol A Mol Integr Physiol. 2007;146:688–694. doi: 10.1016/j.cbpa.2006.04.032. [DOI] [PubMed] [Google Scholar]
- 51.Tanaka T, Kawano S, Nakao S, Umemiya-Shirafuji R, Rahman MM, Boldbaatar D, et al. The identification and characterization of lysozyme from the hard tick Haemaphysalis longicornis. Ticks Tick Borne Dis. 2010;1:178–185. doi: 10.1016/j.ttbdis.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 52.Zhang P, Tian Z, Liu G, Xie J, Luo J, Zhang L, et al. Characterization of acid phosphatase from the tick Haemaphysalis longicornis. Vet Parasitol. 2011;182:287–296. doi: 10.1016/j.vetpar.2011.05.053. [DOI] [PubMed] [Google Scholar]
- 53.Crispell G, Budachetri K, Karim S. Rickettsia parkeri colonization in Amblyomma maculatum: the role of superoxide dismutases. Parasit Vectors. 2016;9:291. doi: 10.1186/s13071-016-1579-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.You M, Xuan X, Tsuji N, Kamio T, Taylor D, Suzuki N, et al. Identification and molecular characterization of a chitinase from the hard tick Haemaphysalis longicornis. J Biol Chem. 2003;278:8556–8563. doi: 10.1074/jbc.M206831200. [DOI] [PubMed] [Google Scholar]
- 55.You M, Fujisaki K. Vaccination effects of recombinant chitinase protein from the hard tick Haemaphysalis longicornis (Acari: Ixodidae) J Vet Med Sci. 2009;71:709–712. doi: 10.1292/jvms.71.709. [DOI] [PubMed] [Google Scholar]
- 56.Blisnick AA, Foulon T, Bonnet SI. Serine protease inhibitors in ticks: an overview of their role in tick biology and tick-borne pathogen transmission. Front Cell Infect Microbiol. 2017;7:1–24. doi: 10.3389/fcimb.2017.00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karim S, Ribeiro JMC. An insight into the sialome of the lone star tick, Amblyomma americanum, with a glimpse on its time dependent gene expression. PLoS ONE. 2015;10:1–17. doi: 10.1371/journal.pone.0131292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Porter L, Radulović Ž, Kim T, Braz GRC, Da Silva VI, Mulenga A. Bioinformatic analyses of male and female Amblyomma americanum tick expressed serine protease inhibitors (serpins) Ticks Tick Borne Dis. 2015;6:16–30. doi: 10.1016/j.ttbdis.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Porter LM, Radulović ŽM, Mulenga A. A repertoire of protease inhibitor families in Amblyomma americanum and other tick species: inter-species comparative analyses. Parasit Vectors. 2017;10:152. doi: 10.1186/s13071-017-2080-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chmelař J, Kotál J, Langhansová H, Kotsyfakis M. Protease inhibitors in tick saliva: the role of serpins and cystatins in tick-host-pathogen interaction. Front Cell Infect Microbiol. 2017;7:216. doi: 10.3389/fcimb.2017.00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu Z, Yan Y, Zhang H, Cao J, Zhou Y, Xu Q, et al. A serpin from the tick Rhipicephalus haemaphysaloides: involvement in vitellogenesis. Vet Parasitol. 2020;279:109064. doi: 10.1016/j.vetpar.2020.109064. [DOI] [PubMed] [Google Scholar]
- 62.Imamura S, Junior IDSV, Sugino M, Ohashi K, Onuma M. A serine protease inhibitor (serpin) from Haemaphysalis longicornis as an anti-tick vaccine. Vaccine. 2005;23:1301–11. doi: 10.1016/j.vaccine.2004.08.041. [DOI] [PubMed] [Google Scholar]
- 63.Bakshi M, Kim TK, Mulenga A. Disruption of blood meal-responsive serpins prevents Ixodes scapularis from feeding to repletion. Ticks Tick Borne Dis. 2018;9:506–518. doi: 10.1016/j.ttbdis.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ibelli AMG, Kim TK, Hill CC, Lewis LA, Bakshi M, Miller S, et al. A blood meal-induced Ixodes scapularis tick saliva serpin inhibits trypsin and thrombin, and interferes with platelet aggregation and blood clotting. Int J Parasitol. 2014;44:369–379. doi: 10.1016/j.ijpara.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lima CA, Sasaki SD, Tanaka AS. Bmcystatin, a cysteine proteinase inhibitor characterized from the tick Boophilus microplus. Biochem Biophys Res Commun. 2006;347:44–50. doi: 10.1016/j.bbrc.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 66.Lu S, Soares TS, Vaz Junior IS, Lovato DV, Tanaka AS. Rmcystatin3, a cysteine protease inhibitor from Rhipicephalus microplus hemocytes involved in immune response. Biochimie. 2014;106:17–23. doi: 10.1016/j.biochi.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 67.Fogaça AC, Lorenzini DM, Kaku LM, Esteves E, Bulet P, Daffre S. Cysteine-rich antimicrobial peptides of the cattle tick Boophilus microplus: isolation, structural characterization and tissue expression profile. Dev Comp Immunol. 2004;28:191–200. doi: 10.1016/j.dci.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 68.Martins LA, Malossi CD, Galletti M, Ribeiro JM, Fujita A, Esteves E, et al. The transcriptome of the salivary glands of Amblyomma aureolatum reveals the antimicrobial peptide microplusin as an important factor for the tick protection against Rickettsia rickettsii infection. Front Physiol. 2019;10:529. doi: 10.3389/fphys.2019.00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lai R, Takeuchi H, Lomas LO, Jonczy J, Rigden DJ, Rees HH, et al. A new type of antimicrobial protein with multiple histidines from the hard tick, Amblyomma hebraeum. FASEB J. 2004;18:1447–1449. doi: 10.1096/fj.03-1154fje. [DOI] [PubMed] [Google Scholar]
- 70.Chinzei Y, Yano I. Vitellin is the nutrient reserve during starvation in the nymphal stage of a tick. Experientia. 1985;41:948–950. [Google Scholar]
- 71.Khalil SMS, Donohue KV, Thompson DM, Jeffers LA, Ananthapadmanaban U, Sonenshine DE, et al. Full-length sequence, regulation and developmental studies of a second vitellogenin gene from the American dog tick, Dermacentor variabilis. J Insect Physiol. 2011;57:400–408. doi: 10.1016/j.jinsphys.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 72.Granjeno-Colin G, Hernandez-Ortiz R, Mosqueda J, Estrada-Mondaca S, Figueroa JV, Garcia-Vazquez Z. Characterization of a vitellogenin gene fragment in Boophilus microplus ticks. Ann NY Acad Sci. 2008;1149:58–61. doi: 10.1196/annals.1428.034. [DOI] [PubMed] [Google Scholar]
- 73.Ferreira CAS, Barbosa MC, Silveira TCL, Valenzuela JG, Da Silva Vaz Jr I, Masuda A. cDNA cloning, expression and characterization of a Boophilus microplus paramyosin. Parasitology. 2002;125:265–74. doi: 10.1017/s0031182002002019. [DOI] [PubMed] [Google Scholar]
- 74.Kim TK, Ibelli AMG, Mulenga A. Amblyomma americanum tick calreticulin binds C1q but does not inhibit activation of the classical complement cascade. Ticks Tick Borne Dis. 2015;6:91–101. doi: 10.1016/j.ttbdis.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tian M, Tian Z, Luo J, Xie J, Yin H, Zeng Q, et al. Identification of the tropomyosin (HL-Tm) in Haemaphysalis longicornis. Vet Parasitol. 2015;207:318–323. doi: 10.1016/j.vetpar.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 76.Tian Z, Liu G, Zhang L, Yin H, Wang H, Xie J, et al. Identification of the heat shock protein 70 (HLHsp70) in Haemaphysalis longicornis. Vet Parasitol. 2011;181:282–290. doi: 10.1016/j.vetpar.2011.04.026. [DOI] [PubMed] [Google Scholar]
- 77.Liu L, Cheng TY, Yang Y. Cloning and expression pattern of a heat shock cognate protein 70 gene in ticks (Haemaphysalis flava) Parasitol Res. 2017;116:1695–1703. doi: 10.1007/s00436-017-5444-8. [DOI] [PubMed] [Google Scholar]
- 78.He XM, Liu L, Cheng TY. HSC70 from Haemaphysalis flava (Acari: Ixodidae) exerts anticoagulation activity in vitro. Ticks Tick Borne Dis. 2019;10:170–175. doi: 10.1016/j.ttbdis.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 79.Fogaça AC, Almeida IC, Eberlin MN, Tanaka AS, Bulet P, Daffre S. Ixodidin, a novel antimicrobial peptide from the hemocytes of the cattle tick Boophilus microplus with inhibitory activity against serine proteinases. Peptides. 2006;27:667–674. doi: 10.1016/j.peptides.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 80.Kotani E, Yamakawa M, Iwamoto S, Tashiro M, Mori H, Sumida M, et al. Cloning and expression of the gene of hemocytin, an insect humoral lectin which is homologous with the mammalian von Willebrand factor. Biochim Biophys Acta. 1995;1260:245–258. doi: 10.1016/0167-4781(94)00202-e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Unique peptides and deduced protein sequences identified by searching against the H. flava transcriptome database.
Additional file 2: Figure S1. Amino acid sequences of three microplusins in H. flava hemolymph.
Additional file 3: Figure S2. Amino acid sequences and domains of two cystatins in H. flava hemolymph.
Data Availability Statement
The data supporting the conclusions of this article are available in the iProX repository, https://www.iprox.cn/page/DSV021.html;?url=1642501651308hONl, with the key 3ABj.