Abstract
The initial step of organ infiltration of malignant cells is the interaction with host vascular endothelial cells, which is often mediated by specific combinations of cell adhesion molecules. Cell adhesion molecule 1 (CADM1) is overexpressed in adult T‐cell leukemia/lymphoma (ATL) and provides a cell‐surface diagnostic marker. CADM1 promotes the adhesion of ATL cells to vascular endothelial cells and multiple organ infiltration in mice. However, its binding partner on host cells has not yet been identified. In this study, we show that CADM1 promotes transendothelial migration of ATL cells in addition to the adhesion to vascular endothelial cells. Moreover, CADM1 enhances liver infiltration of mouse T‐cell lymphoma cells, EL4, after tail vein injection, whereas a CADM1 mutant lacking adhesive activity did not. Among the known CADM1‐binding proteins expressed in primary endothelial cells, only CADM1 and CADM4 could induce morphological extension of ATL cells when plated onto glass coated with these proteins. Furthermore, CADM1‐mediated liver infiltration of EL4 cells was canceled in conventional and vascular endothelium‐specific Cadm1 knockout mice, whereas it was not canceled in Cadm4 knockout mice. These results suggest that CADM1 on host vascular endothelial cells is required for organ infiltration of ATL and other T‐cell lymphomas expressing CADM1.
Keywords: ATL, CADM1, endothelial cell, lymphoma, migration
Trans‐homophilic interaction CADM1 between T‐lymphoma cells and host vascular endothelial cells is required for organ infiltration of lymphoma cells expressing CADM1.

Abbreviations
- ATL
adult T‐cell leukemia/lymphoma
- HTLV‐I
human T‐lymphotropic virus type I
- HUVEC
human umbilical vein endothelial cells
- SPRi
surface plasmon resonance imaging
1. INTRODUCTION
The CADM1 (Cell adhesion molecule 1) gene, which was originally identified as a tumor suppressor in non–small‐cell lung cancer, encodes a single‐pass transmembrane protein belonging to the immunoglobulin superfamily (IgSF). 1 CADM1 is highly expressed in the brain, testis, and lung in normal tissues, whereas loss of Cadm1 in mice causes autism spectrum disorder, male infertility, and increased tumorigenesis, indicating that CADM1, also known as TSLC1/Necl‐2/SynCAM1, plays diverse physiological roles by mediating cell–cell adhesion. 2 , 3 , 4 , 5 , 6 , 7
CADM1 was also identified as one of the markedly upregulated genes in ATL cells. 8 ATL is a malignancy of CD4+ T‐cells caused by human T‐lymphotropic virus type I (HTLV‐I) and develops in 3%–6% of HTLV‐I carriers with a long latency of about 60 years. 9 ATL is an aggressive form of leukemia with a poor prognosis. It is accompanied by the involvement of bone marrow, lymph nodes, spleen, and extranodal sites such as the skin, liver, central nervous system, and gastrointestinal tract. 10 CADM1 expression is observed in ATL cells and HTLV‐I‐infected T‐cells, but absent in normal and activated CD4+ T‐cells. Therefore, CADM1 is used as an established clinical diagnostic marker for ATL cells in combination with CD7, which is gradually downregulated according to the progression of ATL, in a flow cytometric analysis of peripheral blood mononuclear cells. 11 The ectopic expression of CADM1 is caused by constitutive activation of NF‐κB due to the autophagic degradation of p47, a negative regulator of NF‐κB, in ATL cells. 12
Previous studies have demonstrated that CADM1 promotes organ infiltration of ATL cells. Forced expression of CADM1 in ED‐40515(−), an ATL cell line without endogenous expression of CADM1, enhances its adhesion to HUVEC and liver infiltration in NOD‐SCID/γcnull (NOG) mice. 8 , 13 CADM1 induces the invasive phenotype of ATL cells by activating Rac through direct interaction with Tiam1, a Rac‐specific guanine nucleotide exchange factor. 14 Interestingly, CADM1 expression is also detected in cutaneous T‐cell lymphomas such as mycosis fungoides and Sézary syndrome and is associated with unfavorable outcomes, including skin infiltration. 15 , 16 , 17 These studies suggested that CADM1 is a potential therapeutic target for organ infiltration of ATL and other T‐cell lymphomas.
Although CADM1 is considered to enhance adhesion of T‐cell lymphoma cells to vascular endothelial cells, how CADM1 promotes organ infiltration, especially the expression of the homophilic or heterophilic binding partners for CADM1 in host cells, including vascular endothelial cells, is poorly understood. In this study, we focused on the tumor–host interaction mediated by CADM1 and demonstrated that CADM1 expression on host vascular endothelial cells is essential for CADM1‐induced organ infiltration of T‐cell lymphoma cells.
2. MATERIALS AND METHODS
2.1. Cells
Cells were obtained as listed in Table S1.
2.2. RNA interference
CADM1 was knocked down as previously described 18 (Document S1).
2.3. Expression vectors
Human CADM1 with an N‐terminal HA tag (HA‐CADM1) 19 and its F86S mutant were inserted into pLenti7.3‐V5/DEST (Thermo Fisher Scientific, Waltham, MA, USA). Jurkat and EL4 cells were introduced with the vectors, and stably expressing cells were obtained by sorting GFP‐positive cells using the FACSAria II system (BD Biosciences, San Jose, CA, USA).
2.4. Animals
NOD‐SCID/γcnull (NOG) mice and C57BL/6J mice were obtained from CLEA Japan (Tokyo, Japan). Cadm1‐deficient mice were generated as previously described. 5 Cadm4‐deficient mice were generated by inGenious Targeting Laboratory (Ronkonkoma, NY, USA) as described in Document S1 and Figure S1. Cadm1‐ and Cadm4‐deficient mice were backcrossed with C57BL/6J mice more than 10 times. Cadm1 [tm1a(EUCOMM)Wtsi] mice 20 were obtained from the European Conditional Mouse Mutagenesis Program and crossed with CAG‐FLPe transgenic mice (Jackson Laboratory, Bar Harbor, ME, USA) to obtain Cadm1‐flox mice (Figures S2 and S6B). Tie2‐Cre and CAG‐Cre 21 transgenic mice were obtained from the Jackson Laboratory and RIKEN BioResource Center (Tsukuba, Japan), respectively. All animal experiments were carried out under institutional guidelines. Details of the primers for genotyping are listed in Table S2.
2.5. Tumor formation assay
Intravenous injection was performed by injecting 5 × 104 of MT‐2 cells and 1 × 106 of EL4 cells into NOG mice and C57BL/6 background mice, respectively. The number of tumor nodules on the liver surface was counted 24 days after injection for MT‐2 cells or 14 days for EL4 cells. Subcutaneous injection was performed by injecting 1 × 106 of EL4 cells into C57BL/6J mice, and the tumor volumes were measured twice a week.
2.6. Extravasation assay
The extravasation assay was performed as described previously. 22 Briefly, NOG mice were injected with 2 × 106 of MT‐2 cells into the tail vein and sacrificed 24 h after the injection. The livers were fixed in PBS with 4% formaldehyde/0.3% Triton X‐100, washed in 0.3% Triton X‐100/PBS, permeabilized with acetone overnight at 4°C, washed again, and soaked in blocking buffer (10% normal goat serum, 1% Triton X‐100, 0.3 M glycine in PBS) overnight at 4°C. After washing, the livers were incubated with anti‐CD31 antibody (1:50; BD Pharmingen, San Diego, CA, USA) in blocking buffer without glycine overnight at 4°C, then washed and incubated with Alexa Fluor 568‐conjugated secondary antibody (Thermo Fisher Scientific) overnight at 4°C. The nuclei were stained using Hoechst 33342 (Nacalai Tesque). Fluorescent signals were obtained by confocal microscopy (Nikon A1+; Nikon, Tokyo, Japan).
2.7. Endothelium adhesion assay
HUVEC were seeded in a 6‐well plate precoated with 10 µg/ml fibronectin. Then, 1 × 106 of MT‐2 cells or 4 × 105 of ATN‐1 cells were overlaid onto the HUVEC monolayer, and unbound cells were washed off after 30‐min incubation for MT‐2 cells and 60 min for ATN‐1 cells at 37°C. The number of adhered cells to HUVEC was counted in 10 randomly selected microscopic fields.
2.8. Transendothelial migration assay
HUVEC were cultured on Corning Matrigel Invasion Chamber inserts (8.0‐µm pore, Corning, NY, USA), and 4 × 105 of ATN‐1 cells or 1 × 106 of MT‐2 cells were overlaid to HUVEC monolayer. A conditioned medium of NIH3T3 cells was added to the lower chamber as a chemoattractant. After incubation for 24 h at 37°C, the number of cells that migrated to the lower chamber was counted.
2.9. Production of proteins
The production of the ectodomain of human CADM1–4, NECTIN3, PVR, and CRTAM fused to the Fc region of human IgG2 has been described previously. 18 The production of CADM1 ectodomain with Cd4‐COMP‐FLAG‐His tag is described in Document S1.
2.10. Surface plasmon resonance imaging
Surface plasmon resonance imaging (SPRi) analyses were performed using an SPRi system OpenPlex (HORIBA France, Palaiseau, France) according to a previous report. 18 Briefly, 10 µM protein samples and normal human IgG (Sigma‐Aldrich) in PBS were spotted onto an SPRi‐Biochip (HORIBA France) at 10 nl/spot using a DNA Array Spotter (HORIBA Ltd., Kyoto, Japan). The reflectivity of each spot in response to 2.4 µM analyte samples diluted in running buffer (PBS with 0.2% BSA and 0.02% Tween20) was recorded, and the percentage change in reflectivity (%ΔR) was calculated by subtracting the background signal. Data were processed using ScrubberGen software (HORIBA France).
2.11. Flow cytometric analysis
For expression analysis, EL4 cells were stained with anti‐CADM1 antibody (3E1; Medical & Biological Laboratories, Nagoya, Japan) and APC anti‐chicken IgY antibody (Jackson Immuno Research Laboratories, West Grove, PA, USA). For binding analysis, EL4 cells were treated with CADM1‐Fc, CADM4‐Fc, or normal human IgG and APC anti‐Fc antibody (BioLegend, San Diego, CA, USA). The cells were analyzed using the FACSVerse system (BD Biosciences).
2.12. Chemical crosslinking
EL4 cells were incubated with PBS containing 3 mM nonpenetrating crosslinker BS 3 (FUJIFILM Wako Pure Chemical) for 30 min with rotation. Crosslinking was quenched by adding 20 mM Tris‐HCl (pH 7.4) and rotating the sample for 15 min. Then the cells were subjected to western blotting.
2.13. Cell adhesion assay to Fc‐fusion proteins
The coverslips were coated with 140 µg/ml of proteins, and 2 × 104 of Jurkat cells were plated onto coverslips in a 24‐well plate. After incubation for 2 h at 37°C, the cells were fixed with 4% paraformaldehyde and labeled with Alexa Fluor 488 Phalloidin (Thermo Fisher Scientific). The number of cells was counted in randomly selected 20 fields.
2.14. Cell spreading assay
Cell spreading assay was performed as previously described. 18 Briefly, coverslips were coated with 7 µg/ml of proteins, and 1 × 104 of ATN‐1 cells were plated onto the coverslips in a 24‐well plate and incubated for 60 min at 37°C. The cells were fixed in 4% paraformaldehyde and labeled with Alexa Fluor 488 Phalloidin (Thermo Fisher Scientific). The cell area was quantified using AxioVision software (Carl Zeiss, Oberkochen, Germany).
2.15. Alpha technology
CADM1‐Fc, CADM1F86S‐Fc, and CADM4‐Fc were serially diluted from 400 nM in 0.2% BSA/PBS and added at a volume of 5 µl/well to 384‐well AlphaPlate (PerkinElmer, Waltham, MA, USA). CADM1 ectodomain with Cd4‐COMP‐FLAG‐His tag was added at 5 µl/well and incubated for 1 h. Alpha Screen Protein A Acceptor Beads (PerkinElmer) and Anti‐FLAG Alpha Donor Beads (PerkinElmer) were diluted to 80 µg/ml in 0.2% BSA/PBS, and 5 µl of each bead was added and incubated for 1 h in the dark. The interactions were detected by recording the emission at 520–620 nm using an EnSight Multimode Plate Reader (PerkinElmer).
3. RESULTS
3.1. Knockdown of CADM1 attenuates transendothelial migration of ATL cells and HTLV‐I‐transformed T‐cells
One of the most characteristic features of ATL is its infiltration into various organs, including the liver, spleen, and skin. To evaluate the role of CADM1 in the infiltration of ATL cells, we examined the involvement of CADM1 in the interaction with vascular endothelial cells and subsequent transendothelial migration in vitro. An ATL‐derived cell line, ATN1, and an HTLV‐I‐transformed T‐cell line, MT‐2, were transfected with two distinct siRNA against CADM1 (Figure 1A). CADM1 knockdown significantly decreased the adhesion of both cell lines to HUVEC by 20%–30% (Figure 1B), which is consistent with a previous study showing that forced expression of CADM1 in ATL cells promoted adhesion to HUVEC. 8 Moreover, CADM1 knockdown significantly suppressed the chemotactic migration of ATN1 and MT‐2 through the HUVEC monolayer (Figure 1C), whereas cells grown in vitro did not show any differences (Figure 1D). To further investigate the role of CADM1 in transendothelial migration in vivo, MT‐2 cells were introduced with shRNA against CADM1 (shCADM1‐5) (Figure 1E and Figure S6A) and injected intravenously into NOG mice. Infiltration of MT‐2 cells into the liver, lung, spleen, and bone was detected by microscopic analysis (Figure S3A), showing no discrepancy with the organotropism of ATL cells. Notably, large numbers of MT‐2 cell nodules were observed on the liver surface at autopsy 2 weeks after the injection. In contrast, the number of tumor nodules decreased after treating cells with shCADM1 (Figure 1F,G), suggesting that CADM1 is involved in the liver infiltration of MT‐2 cells. We further analyzed the positional relationship between MT‐2 cells and the liver blood vessels 24 h after the intravenous injection. When the immunofluorescence signal of an endothelial marker, CD31, was used to determine the localization of MT‐2 cells inside or outside of blood vessels in liver tissue (Figure 1H), knockdown of CADM1 decreased the number of extravascular cells, that is, infiltrated cells into the liver tissue (Figure 1I). Infiltration of MT‐2 cells in the lung and spleen was also attenuated significantly by knockdown of CADM1, and the bone infiltration appeared to be attenuated in the cells with shCADM1 (Figure S3B). These results suggested that CADM1 would promote the extravasation of ATL and HTLV‐I‐transformed T‐cells in multiple organ infiltration.
FIGURE 1.
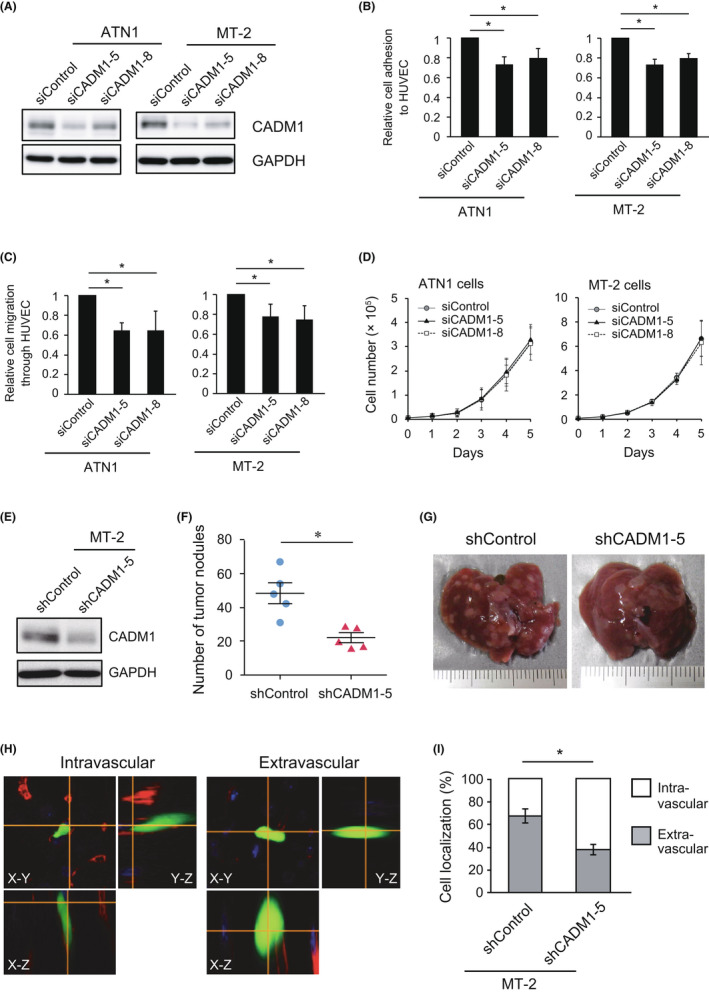
Knockdown of CADM1 in ATL and HTLV‐I‐transformed T‐cell reduced adhesion to HUVEC and transendothelial migration. (A) CADM1 was knocked down in ATN1 and MT‐2 cells by electroporation of two individual siRNA. (B) HUVEC adhesion assay. ATN1 and MT‐2 cells were plated onto a HUVEC monolayer, and the number of attached cells was counted. Means ± SD of the relative number of attached cells in three independent experiments were shown. *p < 0.05 using Student's t test. (C) Transendothelial migration assay. ATN1 and MT‐2 cells were plated onto a HUVEC monolayer, which was cultured on transwell culture inserts coated with Matrigel. The number of migrated cells to the lower chamber filled with NIH3T3‐conditioned medium was counted. Means ± SD of the relative number of migrated cells in three independent experiments are shown. *p < 0.05 using Student's t test. (D) In vitro cell growth of ATN1 and MT‐2 cells treated with siCADM1. The cells were seeded in a 48‐well plate at 1 × 104 cells/well, and the number of living cells was counted using a hemocytometer. Means ± SD of cell number in three independent experiments were shown. (E) CADM1 was stably knocked down in MT‐2 cells by introducing shRNA against CADM1. (F) The number of tumor nodules of MT‐2/shControl and MT‐2/shCADM1‐5 cells on the liver surface was counted. *p < 0.05 using Mann–Whitney U‐test. (G) Photographs of the livers 24 days after tail vein injection of MT‐2/shControl or MT‐2/shCADM1‐5 cells. (H) Representative images of MT‐2 cells in the liver. The blood vessels were stained using an anti‐CD31 antibody (red) to determine whether MT‐2 cells expressing EGFP were localized inside (intravascular, left) or outside blood vessels (extravascular, right) using confocal microscopy (magnification: ×600). (I) Percentage of intravascular and extravascular MT‐2 cells in the liver. Approximately 30 cells were analyzed in each sample. Means ± SD of three independent experiments were shown. *p < 0.05 using Student's t test
3.2. CADM1 promotes liver infiltration of T‐cell lymphoma cells through its binding activity to CADM family proteins
Although CADM1 on ATL and HTLV‐I‐transformed T‐cells is shown to be involved in adhesion to vascular endothelial cells, the binding partners for CADM1 on endothelial cells have not yet been identified. CADM1 forms trans‐interactions with several IgSF molecules, including all CADM family proteins (CADM1–4) and CRTAM, through its first Ig‐V loop. To generate a loss‐of‐function mutant of CADM1, we introduced an F86S mutation into the Ig‐V loop because a previous study had demonstrated that CADM3 with a substitution in the F82 residue, which corresponds to F86 of CADM1, completely lost its trans‐homophilic interaction activity. 23 We quantitatively analyzed the interactions of CADM1 or CADM1F86S with the CADM1‐binding proteins using surface plasmon resonance imaging (SPRi). We detected the interactions of CADM1‐Fc with CADM1‐Fc, CADM2‐Fc, CADM3‐Fc, CADM4‐Fc, and CRTAM‐Fc, which is consistent with our previous report. 18 In contrast, CADM1F86S‐Fc bound to CRTAM‐Fc with a lower affinity compared with CADM1‐Fc, whereas CADM1F86S‐Fc yielded no binding responses to CADM1–4 (Figure 2A). The same results were obtained using another quantitative interaction analysis and Alpha technology (Figure S4A–F). These results suggested that the F86 residue of CADM1 was critical for the interactions with the CADM family proteins and was partially involved in the interaction with CRTAM.
FIGURE 2.
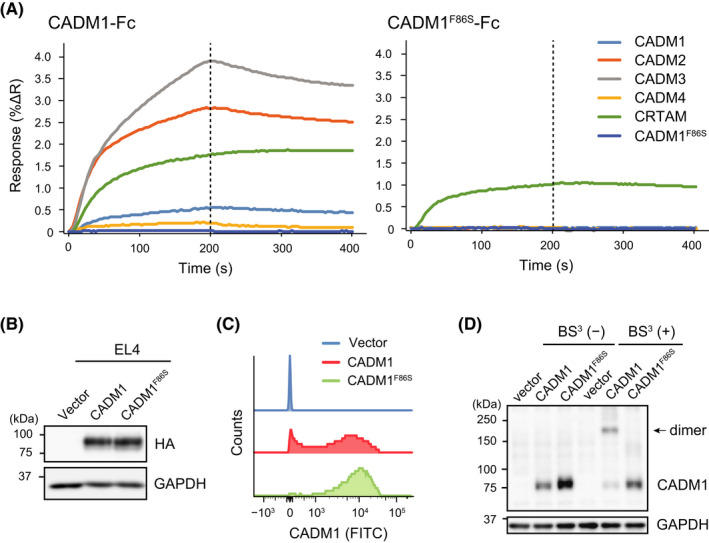
CADM1F86S is a loss‐of‐function mutant that lacks trans‐interaction activity with the CADM family proteins and homo‐cis‐dimer formation activity. (A) Quantitative analysis of the interactions of CADM1 with its binding proteins using surface plasmon resonance imaging (SPRi). Purified Fc‐fusion proteins of CADM1–4, CRTAM, and CADM1F86S were spotted in duplicate on a sensor chip, and the SPRi analysis was performed by injecting CADM1‐Fc (left) and CADM1F86S‐Fc (right) as analytes with an association phase of 200 s and dissociation phase of 200 s. The averaged response signals (percent change in reflectivity: %ΔR) of the two spots are shown. (B) Generation of EL4 cells stably expressing CADM1 or CADM1F86S. (C) A stacked histogram of cell‐surface expression of CADM1 in EL4/CADM1 and EL4/CADM1F86S cells analyzed by flow cytometry. (D) Crosslinking of CADM1 or CADM1F86S on the cell surface. A single‐cell suspension of EL4 cells was incubated in the presence or absence of BS, 3 and the cell lysates were subjected to western blotting. The arrow indicates homo‐cis‐dimers of CADM1
Next, CADM1F86S was introduced into mouse T‐cell lymphoma EL4 cells to characterize its physiological functions (Figure 2B). Although CADM1F86S was expressed on the cell surface (Figure 2C), the CADM family proteins could not bind to cell‐surface CADM1F86S (Figure S4G, H). CADM1F86S also lacked homo‐cis‐dimer formation (Figure 2D), indicating that CADM1F86S serves as a loss‐of‐function mutant lacking both cis‐dimer formation and trans‐interaction activity to the CADM family proteins. In addition, cell adhesion activity of the CADM1F86S mutant was examined using Jurkat T‐cell lymphoma cells (Figure 3A). CADM1 enhanced the adhesion of Jurkat cells to the culture dishes coated with the CADM family proteins and CRTAM, but CADM1F86S promoted adhesion only to CRTAM (Figure 3B). CADM1 also enhanced the cell aggregation activity of EL4 through trans‐homophilic interaction, whereas CADM1F86S did not (Figure 3C), showing that CADM1F86S lacked cell adhesion activity to the CADM family proteins.
FIGURE 3.
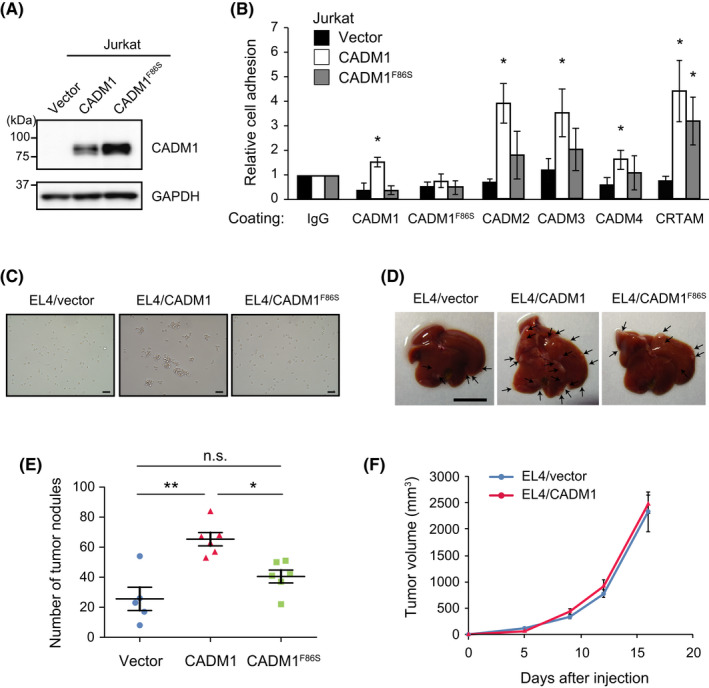
The adhesive activity of CADM1 is essential for promoting liver infiltration of T‐cell lymphoma cells. (A) Generation of Jurkat cells stably expressing CADM1 or CADM1F86S. (B) Adhesion assay on recombinant CADM1‐binding proteins. Jurkat cells with vector, CADM1, and CADM1F86S were plated onto coverslips coated with the indicated proteins, and the number of attached cells was counted. Means ± SD of relative cell number in three experiments are shown. *p < 0.05 using one‐sample t test (versus IgG). (C) Cell aggregation activity of CADM1 and CADM1F86S was examined by incubating for 24 h after plating 1 × 104 single cells in a 24‐well plate. Bars, 50 µm. (D) Representative photographs of the livers after the tail vein injection of EL4 cells expressing CADM1. Bar, 1 cm. (E) The number of tumor nodules on the liver surface. Means ± SD and each value of six experiments are shown. *p < 0.05; **p < 0.01 using Mann–Whitney U‐test. (F) Subcutaneous tumor formation of EL4/vector and EL4/CADM1 cells. Means ± SD of tumor volume in five mice are shown
Then, we injected EL4 cells expressing CADM1 or CADM1F86S into the tail vein of C57BL/6 mice, and tumor nodules on the liver surface were examined 2 weeks after the injection. CADM1 promoted liver nodule formation of EL4 cells, whereas CADM1F86S did not (Figure 3D,E). Because CADM1 did not affect subcutaneous tumor growth of EL4 cells (Figure 3F), CADM1 appeared to promote the infiltration step, not the growing step in liver nodule formation after intravenous injection. Therefore, the trans‐interaction activity of CADM1 is essential for liver infiltration, and the CADM1 family proteins on vascular endothelial cells may mediate the interaction with CADM1 on T‐cell lymphoma cells.
3.3. CADM1 on T‐cell lymphoma cells promote liver infiltration through interacting with CADM1 on vascular endothelial cells
To identify the binding partner of CADM1 on vascular endothelial cells, we examined the expression of all seven CADM1‐interacting molecules: CADM1–4, NECTIN3, PVR, and CRTAM. Among them, the expression of CADM1, CADM4, NECTIN3, and PVR was detected in HUVEC by RT‐PCR, but that of CADM2, CADM3, and CRTAM was not (Figure 4A). Next, we examined the cell spreading of ATN1 cells on the recombinant proteins of the molecules expressed in HUVEC because T‐cell lymphoma cells, including ATL cells, show a characteristic extended morphology in a CADM1‐dependent manner when attached to fibroblast or endothelial cells. 14 Quantification of cell area revealed that ATN1 cells showed significant cell extension on CADM1‐Fc and CADM4‐Fc, but not on NECTIN3‐Fc or PVR‐Fc (Figure 4B,C). Thus, we focused on CADM1 and CADM4 as promising candidates for the binding partner of CADM1 on vascular endothelial cells. Because ATN1 cells with shCADM1 did not exhibit cell spreading on CADM1‐Fc or CADM4‐Fc (Figures S5A,B and S6C), the extension of ATN1 cells on CADM1‐Fc or CADM4‐Fc is dependent on the CADM1 expression on the cell surface. Moreover, forced expression of CADM1 in Jurkat cells enhanced adhesion activity to the culture dishes coated with CADM1‐Fc or CADM4‐Fc, whereas CADM1F86S did not (Figure 3B). These results suggest that CADM1 on ATL and T‐cell lymphoma cells induces cell adhesion and extension through the interaction of CADM1 and CADM4.
FIGURE 4.
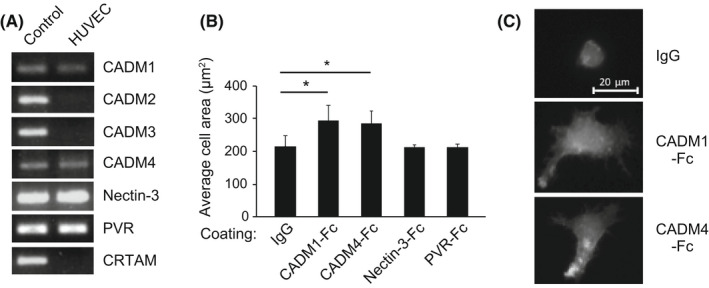
ATL cells show extended morphology by interacting with CADM1 and CADM4. (A) The expression of CADM1‐binding molecules in HUVEC was examined using RT‐PCR. The primers used for RT‐PCR are listed in Table S3. Positive controls are shown in the left lane (A549 cells for CADM1, NECTIN3, and PVR; SH‐SY5Y cells for CADM2, CADM3, and CADM4; U937 cells for CRTAM). (B) Spreading assay of ATN1 cells on CADM1‐binding proteins. ATN1 cells were incubated on coverslips coated with the indicated proteins, and cell spreading was quantified by measuring the area of 100 cells in an assay. Means ± SD of three independent experiments was shown. *p < 0.05 using Student's t test. (C) Representative images of the spread morphology of ATN1 cells incubated on CADM1‐Fc‐ or CADM4‐Fc‐coated glasses. Scale bar, 20 µm
Then, to clarify possible involvement of CADM1 and CADM4 on host cells in the infiltration of T‐cell lymphoma cells expressing CADM1, we performed tail vein injection of EL4 cells into Cadm1‐ or Cadm4‐knockout mice of C57BL/6 background. Interestingly, forced expression of CADM1 into EL4 promoted liver nodule formation in wild‐type mice, whereas this promoting activity was canceled when injected into Cadm1 −/− mice, suggesting that trans‐homophilic interaction of CADM1 is involved in liver nodule formation (Figure 5A). In sharp contrast, the promoting activity of CADM1 on EL4 cells was not canceled when injected into Cadm4 −/− mice, showing that heterophilic interaction of CADM1 and CADM4 is not implemented in liver nodule formation (Figure 5B). Furthermore, to confirm the involvement of endothelial cells in the interaction with leukemic cells, we generated a conditional knockout mouse of Cadm1 in vascular endothelial cells by crossing Tie2‐Cre mice with Cadm1‐flox mice. Targeted deletion of Cadm1 in vascular endothelial cells canceled liver infiltration of EL4 cells induced by forced expression of CADM1 (Figure 5C). These results strongly suggest that CADM1 expression on host vascular endothelial cells is essential for CADM1‐induced liver infiltration of T‐cell lymphoma cells.
FIGURE 5.
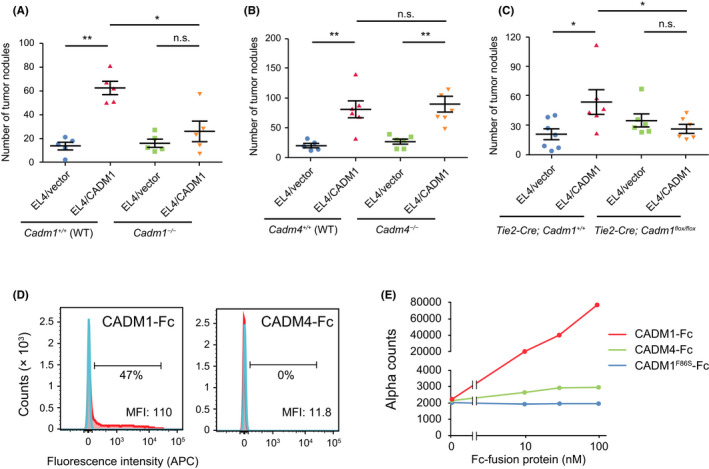
Liver nodule formation of EL4 mouse T‐cell lymphoma cells in Cadm1‐ or Cadm4‐knockout mice. (A, B) The number of tumor nodules on the liver was counted after the tail vein injection of EL4/vector and EL4/CADM1 cells into Cadm1 −/− (A) or Cadm4 −/− mice (B) and their littermate wild‐type mice. *p < 0.05; **p < 0.01 using Mann–Whitney U‐test. (C) The number of tumor nodules on the liver was counted after the tail vein injection of EL4/vector and EL4/CADM1 cells into Cadm1 conditional knockout mice in vascular endothelial cells (Tie2‐Cre; Cadm1flox / flox ) and their littermate controls (Tie2‐Cre; Cadm1 +/+). *p < 0.05 using Mann–Whitney U‐test. (D) Flow cytometry‐based binding analysis of CADM1‐Fc or CADM4‐Fc to the cell‐surface CADM1. Histograms of cell counts treated with IgG (blue) or Fc‐fusion protein (red) are shown. The percentage of cell population bound to the Fc‐fusion protein was indicated. MFI: median fluorescence intensity. (E) The binding of FLAG‐tagged CADM1 with CADM1‐Fc or CADM4‐Fc was examined by Alpha technology. CADM1F86S‐Fc was used as a negative control
In fact, we have previously reported that CADM1–CADM1 interaction showed higher cell aggregation activity than did the CADM1–CADM4 interaction. 24 Finally, to confirm the difference in the affinity of these interactions, we performed flow cytometry‐based binding analysis and showed that CADM1‐Fc, but not CADM4‐Fc, bound considerable portions of EL4 cells expressing CADM1 (Figure 5D). Furthermore, we examined quantitative analysis of molecular interaction between CADM1‐FLAG and CADM1‐Fc or CADM4‐Fc using Alpha technology. The binding signal in a unit of alpha counts was much stronger in the CADM1–CADM1 interaction than in the CADM1–CADM4 interaction (Figure 5E). These findings would provide the molecular basis of the different activity of endothelial CADM1 and CADM4 in organ infiltration of T‐cell lymphoma cells (Table S4).
4. DISCUSSION
CADM1 has been shown to enhance adhesion to vascular endothelial cells and liver infiltration of ATL cells, 8 , 13 and promote ATL cell extension through Tiam1‐Rac signaling. 14 This study demonstrated that CADM1 enhances organ infiltration of ATL and HTLV‐1‐transformed T‐cells by promoting adhesion to vascular endothelial cells and transendothelial migration (Figure 1). Because a loss‐of‐function mutant in adhesion activity of CADM1, CADM1F86S did not promote liver infiltration of EL4 T‐cell lymphoma cells (Figure 3), CADM1‐mediated cell adhesion would trigger subsequent cell migration. Therefore, CADM1 may function to transduce cell–cell contact signals bidirectionally into T‐cell lymphoma cells and vascular endothelial cells and promote the migration of T‐cell lymphoma cells.
This study showed that CADM1 on vascular endothelial cells facilitates liver infiltration of T‐cell lymphoma cells expressing CADM1 by promoting cell–cell adhesion through homophilic interaction. In contrast, CADM1–CADM4 interaction was not essential for organ infiltration, although CADM4 is expressed in vascular endothelial cells and could promote adhesion and spreading of T‐cell lymphoma cells in vitro. To understand the difference in the activity and affinity between CADM1–CADM1 and CADM1–CADM4 interactions, we summarized the results obtained from a series of interaction analyses at the molecular, cellular, and tissue levels in Table S4. At the molecular level, Alpha technology and SPRi analyses demonstrated that the CADM1–CADM1 interaction had higher affinity than the CADM1–CADM4 interaction (Figures 2A and 5E). Flow cytometry‐based binding assay also showed that CADM1‐Fc protein bound to the surface of EL4 cells expressing CADM1, but CADM4‐Fc protein did not (Figure 5D). Conversely, the adhesion and spreading assays of T‐cell lymphoma cells expressing CADM1 on the coverslips coated with CADM1‐Fc or CADM4‐Fc failed to show a significant difference between CADM1–CADM1 and CADM1–CADM4 interactions (Figures 3B and 4B), probably because these assays are rather qualitative due to a very high protein concentration. In a physiological condition of cell–cell adhesion activity, we had examined cell aggregation assay in a previous study 24 and shown that the CADM1–CADM1 interaction had a higher cell adhesion activity than the CADM1–CADM4 interaction. Based on these results, we conclude that the CADM1–CADM1 interaction shows higher affinity and biological activity than the CADM1–CADM4 interaction.
In this study, due to the technical limitation of using knockout mice, syngeneic T‐lymphoma cells, EL4, were used instead of human ATL or other T‐cell lymphoma cells, which are only transplantable to immunocompromised mice. We believe, however, that this finding would be explored in human T‐cell lymphomas, providing a new paradigm for understanding the infiltration of T‐cell lymphomas. CADM1 is reported to regulate endothelial progenitor cell migration under the control of the TNFα–NF‐κB pathway 25 and may be involved in the vascular endothelial cell barrier. 26 More importantly, blocking the trans‐homophilic interaction of CADM1 between T‐cell lymphoma cells and vascular endothelial cells could provide a therapeutic target for the organ infiltration of T‐cell lymphoma. In fact, a previous study has shown that an anti‐CADM1 neutralizing antibody suppressed the adhesion of ATL cells to HUVEC and liver nodule formation of EL4 cells expressing CADM1. 27 Although ATL cells frequently infiltrate into the skin rather than the liver, CADM1 expression on vascular endothelial cells could be a risk factor for the infiltration of T‐cell lymphoma cells; therefore, polymorphic variants of the CADM1 gene in host individuals might be associated with the malignant progression of T‐cell lymphomas expressing CADM1. Further studies on CADM1 expression in vascular endothelial cells would lead to a better understanding of the infiltration of ATL and other cutaneous lymphomas such as mycosis fungoides and Sézary syndrome.
DISCLOSURE
YM is a current Associate Editor of Cancer Science. YM obtained a joint research fund and party participated in a social cooperation research program with the NTT Corporation, Japan. The other authors have no conflict of interest.
AUTHOR CONTRIBUTIONS
YK, SPG, TF, YOK, MT, SJS, and DS performed molecular and cellular analyses and animal experiments. DM performed pathological analysis. TS, MSY, TI, and YM designed the experiments. YK, TI, and YM wrote the manuscript.
Supporting information
Fig S1‐S6
Table S1‐S4
Document S1
ACKNOWLEDGMENTS
The authors thank Drs. Kaoru Uchimaru and Satoru Nagatoishi, the University of Tokyo, for fruitful discussion. This work was supported by JSPS KAKENHI grant numbers 24790310 to MSY, 19K16708 and 20H05028 to TI, and 20H03525 and 20K21539 to YM.
Kasai Y, Gan SP, Funaki T, et al. Trans‐homophilic interaction of CADM1 promotes organ infiltration of T‐cell lymphoma by adhesion to vascular endothelium. Cancer Sci. 2022;113:1669–1678. doi: 10.1111/cas.15307
Funding information
Japan Society for the Promotion of Science (grant/award numbers: “19K16708,” “20H05028,” “20K21539,” and “24790310”)
REFERENCES
- 1. Kuramochi M, Fukuhara H, Nobukuni T, et al. TSLC1 is a tumor‐suppressor gene in human non‐small‐cell lung cancer. Nat Genet. 2001;27:427‐430. [DOI] [PubMed] [Google Scholar]
- 2. Biederer T, Sara Y, Mozhayeva M, et al. SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science. 2002;297:1525‐1531. [DOI] [PubMed] [Google Scholar]
- 3. Shingai T, Ikeda W, Kakunaga S, et al. Implications of nectin‐like molecule‐2/IGSF4/RA175/SgIGSF/TSLC1/SynCAM1 in cell‐cell adhesion and transmembrane protein localization in epithelial cells. J Biol Chem. 2003;278:35421‐35427. [DOI] [PubMed] [Google Scholar]
- 4. Murakami Y. Involvement of a cell adhesion molecule, TSLC1/IGSF4, in human oncogenesis. Cancer Sci. 2005;96:543‐552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yamada D, Yoshida M, Williams YN, et al. Disruption of spermatogenic cell adhesion and male infertility in mice lacking TSLC1/IGSF4, an immunoglobulin superfamily cell adhesion molecule. Mol Cell Biol. 2006;26:3610‐3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Takayanagi Y, Fujita E, Yu Z, et al. Impairment of social and emotional behaviors in Cadm1‐knockout mice. Biochem Biophys Res Comm. 2010;396:703‐708. [DOI] [PubMed] [Google Scholar]
- 7. van der Weyden L, Arends MJ, Rust AG, Poulogiannis G, McIntyre RE, Adams DJ. Increased tumorigenesis associated with loss of the tumor suppressor gene Cadm1. Mol Cancer. 2012;11:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sasaki H, Nishikata I, Shiraga T, et al. Overexpression of a cell adhesion molecule, TSLC1, as a possible molecular marker for acute‐type adult T‐cell leukemia. Blood. 2005;105:1204‐1213. [DOI] [PubMed] [Google Scholar]
- 9. Yasunaga J, Matsuoka M. Leukaemogenic mechanism of human T‐cell leukaemia virus type I. Rev Med Virol. 2007;17:301‐311. [DOI] [PubMed] [Google Scholar]
- 10. Watanabe T. Adult T‐cell leukemia: molecular basis for clonal expansion and transformation of HTLV‐1‐infected T cells. Blood. 2017;129:1071‐1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kobayashi S, Nakano K, Watanabe E, et al. CADM1 expression and stepwise downregulation of CD7 are closely associated with clonal expansion of HTLV‐I‐infected cells in adult T‐cell leukemia/lymphoma. Clin Cancer Res. 2014;20:2851‐2861. [DOI] [PubMed] [Google Scholar]
- 12. Sarkar B, Nishikata I, Nakahata S, et al. Degradation of p47 by autophagy contributes to CADM1 overexpression in ATLL cells through the activation of NF‐κB. Sci Rep. 2019;9:3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dewan MZ, Takamatsu N, Hidaka T, et al. Critical role for TSLC1 expression in the growth and organ infiltration of adult T‐cell leukemia cells in vivo. J Virol. 2008;82:11958‐11963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Masuda M, Maruyama T, Ohta T, et al. CADM1 interacts with Tiam1 and promotes invasive phenotype of human T‐cell leukemia virus type I‐transformed cells and adult T‐cell leukemia cells. J Biol Chem. 2010;285:15511‐15522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mashima E, Sawada Y, Yamaguchi T, et al. A high expression of cell adhesion molecule 1 (CADM1) is an unfavorable prognostic factor in mycosis fungoides. Clin Immunol. 2018;193:121‐122. [DOI] [PubMed] [Google Scholar]
- 16. Yuki A, Shinkuma S, Hayashi R, et al. CADM1 is a diagnostic marker in early‐stage mycosis fungoides: multicenter study of 58 cases. J Am Acad Dermatol. 2018;79:1039‐1046. [DOI] [PubMed] [Google Scholar]
- 17. Yamaguchi M, Morizane S, Hamada T, et al. The expression of cell adhesion molecule 1 and its splicing variants in Sezary cells and cell lines from cutaneous T‐cell lymphoma. J Dermatol. 2019;46:967‐977. [DOI] [PubMed] [Google Scholar]
- 18. Ito T, Kasai Y, Kumagai Y, et al. Quantitative analysis of interaction between CADM1 and its binding cell‐surface proteins using surface plasmon resonance imaging. Front Cell Dev Biol. 2018;6:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ito T, Nakamura A, Tanaka I, et al. CADM1 associates with Hippo pathway core kinases; membranous co‐expression of CADM1 and LATS2 in lung tumors predicts good prognosis. Cancer Sci. 2019;110(7):2284–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. White JK, Gerdin AK, Karp NA, et al. Genome‐wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell. 2013;154:452‐464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sakai K, Miyazaki J. A transgenic mouse line that retains Cre recombinase activity in mature oocytes irrespective of the cre transgene transmission. Biochem Biophys Res Comm. 1997;237:318‐324. [DOI] [PubMed] [Google Scholar]
- 22. Hara T, Nakaoka HJ, Hayashi T, et al. Control of metastatic niche formation by targeting APBA3/Mint3 in inflammatory monocytes. Proc Natl Acad Sci USA. 2017;114:E4416‐e4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dong X, Xu F, Gong Y, et al. Crystal structure of the V domain of human Nectin‐like molecule‐1/Syncam3/Tsll1/Igsf4b, a neural tissue‐specific immunoglobulin‐like cell‐cell adhesion molecule. J Biol Chem. 2006;281:10610‐10617. [DOI] [PubMed] [Google Scholar]
- 24. Williams YN, Masuda M, Sakurai‐Yageta M, Maruyama T, Shibuya M, Murakami Y. Cell adhesion and prostate tumor‐suppressor activity of TSLL2/IGSF4C, an immunoglobulin superfamily molecule homologous to TSLC1/IGSF4. Oncogene. 2006;25:1446‐1453. [DOI] [PubMed] [Google Scholar]
- 25. Prisco AR, Hoffmann BR, Kaczorowski CC, et al. Tumor necrosis factor α regulates endothelial progenitor cell migration via CADM1 and NF‐kB. Stem Cells. 2016;34:1922‐1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hasstedt SJ, Bezemer ID, Callas PW, et al. Cell adhesion molecule 1: a novel risk factor for venous thrombosis. Blood. 2009;114:3084‐3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chilmi S, Nakahata S, Fauzi YR, et al. Development of anti‐human CADM1 monoclonal antibodies as a potential therapy for adult T‐cell leukemia/lymphoma. Int J Hematol. 2020;112:496‐503. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1‐S6
Table S1‐S4
Document S1


