Abstract
Pancreatic adenocarcinoma (PAAD) remains an extremely fatal malignancy with a high mortality rate worldwide. This study focuses on the roles of ubiquitin‐specific peptidase 10 (USP10) and cysteine rich angiogenic inducer 61 (Cyr61) in macrophage polarization, immune escape, and metastasis of PAAD. USP10 showed a positive correlation with Yes1 associated transcriptional regulator (YAP1), which, according to the TCGA‐PAAD database, is highly expressed in PAAD and indicates poor patient prognosis. USP10 knockdown increased ubiquitination and degradation of YAP1, which further decreased the programmed cell death ligand 1 (PD‐L1) and Galectin‐9 expression, suppressed immune escape, and reduced the proliferation and metastasis of PAAD cells in vitro and in vivo. Cyr61, a downstream factor of YAP1, was overexpressed in PAAD cells after USP10 silencing for rescue experiments. Overexpression of Cyr61 restored the PD‐L1 and Galectin‐9 expression in cells and triggered M2 polarization of macrophages, which enhanced the immune escape and maintained the proliferation and metastasis ability of PAAD cells. In conclusion, this work demonstrates that USP10 inhibits YAP1 ubiquitination and degradation to promote Cyr61 expression, which induces immune escape and promotes growth and metastasis of PAAD.
Keywords: Cyr61, deubiquitinase USP10, Hippo signaling pathway, immune escape, macrophage polarization, pancreatic adenocarcinoma
This study confirmed that deubiquitinating enzyme USP10 suppresses ubiquitination and degradation of YAP1, which further elevates the expression of Cyr61 and enhances the immune escape and malignant behaviors of PAAD cells. USP10 and Cyr61 may serve as novel targets for overcoming the immunosuppression in PAAD cells and help improve the prognosis of patients.

Abbreviations
- ANOVA
analysis of variance
- CTG
CellTiter Glo
- Cyr61
cysteine rich angiogenic inducer 61
- FITC
fluorescein isothiocyanate
- GSEA
Gene Set Enrichment Analysis
- IHC
immunohistochemistry
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LDH
lactate dehydrogenase
- NK cells
natural killer cells
- OD
optical density
- PAAD
pancreatic adenocarcinoma
- PC
pancreatic cancer
- PFA
paraformaldehyde
- PI
propidium iodide
- RPMI
Roswell Park Memorial Institute
- TCGA
The Cancer Genome Atlas
- USP10
ubiquitin specific peptidase 10
- YAP1
Yes1 associated transcriptional regulator
- ZO‐1
tight junction protein 1
1. INTRODUCTION
Pancreatic cancer (PC) is an extremely malignant neoplastic disease, with approximately 459,000 new diagnoses and 432,000 deaths globally in 2018. 1 Pancreatic adenocarcinoma (PAAD) accounts for approximately 85% of all cases and represents the predominant type of PC, while pancreatic endocrine tumors account for no more than 5% of all cases. 2 Due to the lack of early signs, patients are often diagnosed at advanced stages. Consequently, the 5‐year survival rate has improved little and has remained at approximately 9% since the 1960s. 3 The combination chemotherapies had an unsatisfactory effect, only improving the median survival by 2–4 months and inducing toxic side effects. 4 , 5
Immunotherapy has shown considerable efficacy in several malignancies, 6 with promising therapeutic potential in PC as well. 7 The immune system exerts crucial functions in the pathogenesis of PAAD, which seems imbalanced in PC patients and spontaneously leads to cancer development. 8 Immune tolerance is an unavoidable problem contributing to the treatment failure of immunotherapy. The programmed cell death protein‐1 (PD‐1) and PD‐ligand‐1 (PD‐L1) axis represents a remarkable mechanism of adaptive immune escape for tumor cells by suppressing cytotoxic T lymphocytes and blocking T‐cell immune response. 9 Meanwhile, macrophages represent a major type of immune cell and crucial components in tumor microenvironment. They mainly polarize to the classical activated “M1” type that promotes anti‐tumor immune responses or to the alternatively activated “M2” phenotype that leads to immune suppression and tumor progression. 10 The PD‐1/PD‐L1 axis can trigger M2 skewing of macrophages, while the M2 macrophages can upregulate PD‑L1 expression on tumor cells. 11 Finding novel molecules involved in and developing more strategies to avoid escape from immune surveillance is an essential issue to improve the survival of patients.
Advanced bioinformatic analytical tools and systems such as The Cancer Genome Atlas (TCGA) dataset help greatly in the identification of key molecules involved in cancer initiation and development. 12 , 13 , 14 In the present study, a bioinformatic analysis using the TCGA‐PAAD database confirmed Yes1‐associated transcriptional regulator (YAP1) as a candidate oncogene in PAAD because it was upregulated and indicated poor prognosis in patients. YAP1 is a critical effector of the Hippo pathway and interacts with other cancer‐promoting pathways. It is frequently involved in tumorigenesis and immunosuppression. 15 The oncogenic effect of YAP1 has been reported in PC cells as well. 16 , 17 Intriguingly, PD‐1 was linked to YAP1 stabilization and the activation of its downstream molecules cysteine rich angiogenic inducer 61 (Cyr61) and cellular communication network factor 2 (CTGF) via the Hippo signaling pathway. In addition, the synergistic suppression of PD‐1 and Hippo signaling significantly suppressed pancreatic cancer cell growth in vitro. 18 Moreover, YAP1 has been correlated with PD‐L1 upregulation in the microenvironment of hepatocellular carcinoma. 19 This motivated us to explore the relevance of YAP1 to immune escape in PAAD. The subsequent bioinformatics analyses in this work suggested that ubiquitin‐specific peptidase 10 (USP10) had a strong positive correlation with YAP1 in PAAD. USP10 is a primary cytoplasmic deubiquitinase of the USPs and acts as either an oncogene or tumor suppressor depending on the different substrates. 20 Ubiquitination is one of the major posttranslational regulations of the genome that induces protein degradation through the proteasome pathway and the consequential protein relocation, and dysregulated ubiquitination may lead to tumorigenesis. 21 , 22 However, the roles of USP10 in PAAD development and immunosuppression are not clear yet. Here, the synergistical upregulation of USP10 and YAP1 in PAAD indicated that USP10 possibly deubiquitinates and stabilizes YAP1 to affect PAAD progression. Collectively, this study was performed to validate if there is an interaction between USP10 and YAP1 and to explore their functions in the development and immune regulation in PAAD.
2. MATERIALS AND METHODS
2.1. Bioinformatic analyses
The expression profiling and indicative value of YAP1 in PAAD were obtained from TCGA (https://www.cancer.gov/about‐nci/organization/ccg/research/structural‐genomics/tcga), in which the relevance of USP10 level to the prognosis of patients with PAAD was analyzed as well. The staining intensity of USP10 in PAAD tissues and normal pancreatic tissues was predicted via the Human Protein Atlas (HPA) system (https://www.proteinatlas.org/). Correlations between USP10, YAP1, Cyr61, and CTGF were analyzed using the Gene Expression Profiling Interactive Analysis (GEPIA) system (http://gepia.cancer‐pku.cn/index.html). Genes showing high correlation with YAP1 were analyzed using the correlation test (cor. test). A Gene Set Enrichment Analysis (GSEA) was conducted to examine the pathways enriched by the differentially expressed genes in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database.
2.2. Cells
A human pancreatic duct epithelial cell line HPDE6 and PAAD cell lines (Capan‐2, Panc‐1, AsPC‐1, and BxPC‐1) were purchased from Genechem. Detailed descriptions of the methods are provided in Appendix S1.
2.3. Release of lactate dehydrogenase in cells
The Panc‐1 and BxPC‐1 cells were cultured in 96‐well plates (2 × 104 cells/well) as target cells. Detailed descriptions of the methods are provided in Appendix S1.
2.4. Cell viability examined by a CellTiter Glo kit
The Panc‐1 and BxPC‐1 cells were cultured in 96‐well plates at 5 × 103 cells/well. Each well was filled with 10 μL CellTiter Glo (CTG) solution (Promega) at the 0, 24, 48, and 72 h, followed by another 4 h of incubation. The optical density (OD) at 450 nm was read using a microplate reader.
2.5. EdU labeling assay
Proliferation of cells was determined using a 5‐ethynyl‐2′‐deoxyuridine (EdU) labeling kit (RiboBio). Detailed descriptions of the methods are provided in Appendix S1.
2.6. Flow cytometry for cell apoptosis
The Panc‐1 and BxPC‐1 cells were cultured in six‐well plates at 1.5 × 105 cells/well. The cells were resuspended in 1 × binding buffer. After that, 1 × 105 cells were added in 200‐μL tubes and mixed with 5 μL Annexin V‐fluorescein isothiocyanate (FITC) and 5 μL (PI) for 15 min of warm incubation at 25℃. Thereafter, each tube was filled with 400 μL 1 × binding buffer and examined on the flow cytometer (BD Biosciences) within 1 h. The green fluorescence of Annexin V‐FITC was examined at 530 nm, whereas the red fluorescence was detected at 585 nm. The Flow Jo software was used for result analysis.
2.7. Transwell assays
The migration and invasion of cells were determined using 24‐well (8 µm in diameter, Costar) Transwell chambers. Detailed descriptions of the methods are provided in Appendix S1.
2.8. Flow cytometry for cell identification
The PD‐L1‐APC antibody (BioLegend) and Galectin‐9 PE antibody (BD Biosciences) were used to examine the expression of surface biomarkers Galectin‐9 and PD‐L1 on the PAAD cells. The mean fluorescence intensity of each labeling was analyzed. After 48 h, the target cells were identified on a FACS AriaII flow cytometer (BD Biosciences). Detailed descriptions of the methods are provided in Appendix S1.
2.9. Ubiquitination examination
The YAP1‐FLAG were co‐transfected with HA‐Ub (all synthesized by GenePharma) and USP10 overexpressing vectors into HEK‐293T cells. The ubiquitinated YAP1 and USP10 proteins were incubated in 10 μM MG132 for 4 h and then precipitated by the anti‐FLAG (ab1162, Abcam) or anti‐MYC (ab32072, Abcam) affinity gels. After FLAG dilution and dialysis, the purified USP10 and YAP1 were obtained. The USP10 and YAP1 protein samples were further incubated in 30°C deubiquitination reaction buffer for 2 h and then quantified using western blot analysis.
2.10. Co‐IP
Before protein extraction, the cells were cultured in 10 μM MG132 for 4 h. The whole‐cell lysates were incubated with anti‐Flag M2 affinity gel for 12 h at 4°C. After the above endogenous Co‐IP determination, the total protein extracts were warmly hybridized with antibodies for 12 h at 4°C and conjugated with protein A/G agarose‐beads (#9863, Cell Signaling Technology). Next, the magnet beads were washed with RIPA cell lysis buffer, and the immunoprecipitated protein complexes were collected for western blot analysis.
2.11. Immunofluorescence staining
Cells were seeded on 18‐mm2 glass slides and fixed in acetic acid‐ethanol (1:3) solution at −20°C for 5 min. After blocking with 5% normal goat serum, the cell slides were co‐incubated with primary antibodies against ZO‐1 (1:1000, GTX108613, GeneTex), E‐cadherin (1:5000, ab40772, Abcam), Vimentin (1:2000, ab92547, Abcam) and N‐cadherin (1:2000, #MA1‐91128, ThermoFisher Scientific) at 4°C overnight and with luciferin‐labeled secondary antibody (1:50, ab150115 or ab150077, Abcam) at 37°C for 1 h. DAPI was used for nucleus staining. After that, the cell slides were washed twice in PBS, covered with DABCO (Sigma‐Aldrich Chemical Company), and then observed under a confocal laser scanning microscope (Olympus) at 568 nm.
2.12. RT‐qPCR
Total RNA from tissues and cells was isolated using a TRIzol Kit (15596‐026, Invitrogen, ThermoFisher Scientific). The RNA concentration and purity were determined using a nucleic acid and protein analyzer (Eppendorf) according to the ratio of OD at 260–280 nm. An OD260 nm/OD280 nm value of 1.8–2.0 indicated high RNA purity. The RNA was reverse‐transcribed into cDNA using a K1621 RT kit (Fermentas). The mRNA expression of genes was examined using a fluorescence quantitation PCR kit (Takara Biotechnology) and an ABI7500 PCR kit (ABI). The primers are presented in Table 1, where GAPDH is used as the internal control. Relative gene expression was determined using the 2−ΔΔCt method.
TABLE 1.
Primer sequences for RT‐qPCR
| Gene | Primer sequence (5′‐3′) |
|---|---|
| USP10 | F: GAGGTGGAAAGTTGTTTGCCTGG |
| R: TGCTCCATCTCTAAGAGGTGGC | |
| Cyr61 | F: GGAAAAGGCAGCTCACTGAAGC |
| R: GGAGATACCAGTTCCACAGGTC | |
| IL‐13 | F: CAGCCCTCAGCCATGAAATA |
| R: CTTGAGTGTGTAACAGGCCATTCT | |
| IL‐6 | F: GATAAGCTGGAGTCACAGAAGG |
| R: TTGCCGAGTAGATCTCAAAGTG | |
| IL‐1β | F: CCACAGACCTTCCAGGAGAATG |
| R: GTGCAGTTCAGTGATCGTACAGG | |
| iNOS | F: GCTCTACACCTCCAATGTGACC |
| R: CTGCCGAGATTTGAGCCTCATG | |
| Arg1 | F: TCATCTGGGTGGATGCTCACAC |
| R: GAGAATCCTGGCACATCGGGAA | |
| ZO‐1 | F: GTCCAGAATCTCGGAAAAGTGCC |
| R: CTTTCAGCGCACCATACCAACC | |
| E‐cadherin | F: GCCTCCTGAAAAGAGAGTGGAAG |
| R: TGGCAGTGTCTCTCCAAATCCG | |
| Vimentin | F: AGGCAAAGCAGGAGTCCACTGA |
| R: ATCTGGCGTTCCAGGGACTCAT | |
| N‐cadherin | F: CCTCCAGAGTTTACTGCCATGAC |
| R: GTAGGATCTCCGCCACTGATTC | |
| Galectin | F: ACACCCAGATCGACAACTCCTG |
| R: CAAACAGGTGCTGACCATCCAC | |
| GAPDH | F: CGGAGTCAACGGATTTGGTCGTAT |
| R: AGCCTTCTCCATCATTGTGGTGAAGAC |
Arg1, arginase 1; Cyr61, cysteine rich angiogenic inducer 61; F, forward; GAPDH, glyceraldehyde‐3‐phosphate dehydrogenase; IL, interleukin; iNOS, nitric oxide synthase 2; R, reverse; RT‐qPCR, reverse transcription quantitative polymerase chain reaction; USP10, ubiquitin specific peptidase 10; ZO‐1, tight junction protein 1.
2.13. Western blot analysis
Detailed descriptions of the methods are provided in Appendix S1.
2.14. Tumorigenesis of pancreatic adenocarcinoma in vivo
All animal procedures were approved by the Animal Ethical Committee of Shengjing Hospital of China Medical University and adhered to the Guide for the Care and Use of Laboratory Animals (NIH, Bethesda, Maryland, USA). Detailed descriptions of the methods are provided in Appendix S1.
2.15. Metastasis of tumors in vivo
Tumor metastasis was determined as well. Another batch of nude mice were injected with 4 × 106 cells through tail veins. These mice were killed on the day 45 after cell injection. After that, the mouse lung and liver tissues were harvested for HE staining to examine the tumor metastasis.
2.16. HE staining
The paraffin‐embedded sections were dewaxed, hydrated, and stained with hematoxylin for 5 min. Next, the section slides were immersed in 1% acid alcohol (1% HCl in 70% alcohol) for 3 s and then stained with eosin (Sangon Biotech) for 3 min. The slides were dehydrated in ethanol, cleared in xylene, and observed and imaged under the microscope.
2.17. Immunohistochemistry
Detailed descriptions of the methods are provided in Appendix S1.
2.18. Statistical analysis
Data were analyzed using the SPSS22.0 (IBM). Three independent experiments were performed. Measurement data are exhibited as the mean ± SD. Differences were determined by t test or one‐ or two‐way analysis of variance (ANOVA) followed by Tukey’s post‐hoc test. Spearman’s rank correlation coefficient was used to analyze correlations between variables. Fisher’s exact test was used for gene enrichment analysis. The log‐rank test was used for post‐statistical analysis. Statistical significance was set at P < 0.05.
3. RESULTS
3.1. USP10 is abundantly expressed in pancreatic adenocarcinoma tissues and cells and positively correlated with YAP1
We first examined the YAP1 expression and its correlation with the prognosis of 178 PAAD patients in TCGA‐PAAD. It was suggested that YAP1 was highly expressed in PAAD tissues compared to the normal tissues. Meanwhile, high YAP1 level was correlated with poor patient survival. Moreover, the GSEA suggested that high expression of YAP1 was correlated with increased hydrolysis of ubiquitination‐related proteins, and the hydrolysis‐related signaling pathway was enriched on the positive side of the YAP1 (Figure 1A), indicating that the aberrantly high expression of YAP1 is caused by dysregulated ubiquitination. Thereafter, the genes showed close correlation with YAP1 were analyzed using a cor. test. We focused on the deubiquitinases, among which USP10 showed a strong positive correlation with YAP1 (R = 0.68) (Figure 1B). In addition, USP10 was suggested to have significant differential expression between PAAD and normal tissues in TCGA‐PAAD and GTEx databases (Figure 1C), and high USP10 expression was correlated with poor prognosis of PAAD patients as well (Figure 1D). Moreover, data on the HPA website suggested that staining intensity of USP10 in the PAAD tissues was notably stronger than that in the normal pancreatic tissues (Figure 1E). We further explored the expression profile of the downstream genes of YAP1 (Cyr61 and CTGF) in TCGA‐PAAD. Importantly, both Cyr61 and CTGF were predicted to be abundantly expressed in the PAAD tissues (Figure 1F,G) and had a positive association with USP10 (Figure 1H,I). We further examined the USP10 expression in the acquired PAAD cell lines Capan‐2, AsPC‐1, Panc‐1, and BxPC‐1 and the normal HPDE6 cells. Consequently, elevated USP10 expression was detected in all PAAD cell lines compared to the HPDE6 cells (Figure 1J).
FIGURE 1.
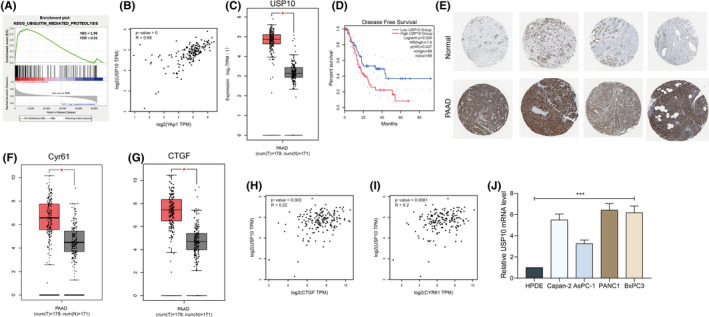
Ubiquitin‐specific peptidase 10 (USP10) is abundantly expressed in pancreatic adenocarcinoma (PAAD) tissues signaling pathway enriched on the positive side of the YAP1 according to Gene Set Enrichment Analysis (GSEA and cells and positively correlated with YAP1. (A) Hydrolysis‐related); (B) genes show high correlation with YAP1 analyzed using a correlation test (cor. Test); (C) USP10 expression in TCGA‐PAAD and GTEx databases; (D) association between USP10 level and the prognosis of patients analyzed in TCGA‐PAAD; (E) staining intensity of USP10 in the PAAD tissues and normal pancreatic tissues in the HPA system; (F and G) expression of Cyr61 (F) and CTGF (G) in TCGA‐PAAD; (H and I) positive correlations between CTGF (H) and Cyr61 (I), and USP10 examined by Spearman's rank correlation coefficient; (J) expression of USP10 in PAAD cell lines (Capan‐2, AsPC‐1, Panc‐1, and BxPC‐1), and in the normal HPDE6 cells determined by RT‐qPCR. Differences were analyzed by one‐way ANOVA (J), ***P < 0.001
3.2. Knockdown of USP10 increases ubiquitination and degradation of YAP1 in pancreatic adenocarcinoma cells
To examine the correlation between USP10 and YAP1, the Panc‐1 and BxPC‐1 cells were transfected with shRNA of USP10 (shUSP10). It was found that USP10 knockdown in Panc‐1 and BxPC‐1 cells led to a notable decline in the expression of YAP1 (Figure 2A). We then proposed that USP10, as a deubiquitinase, possibly regulates deubiquitination and maintains the stability of YAP1, while knockdown of USP10 might increase ubiquitination and degradation of YAP1. To examine this, we then validated the binding relationship between USP10 and YAP1 in cells using the Co‐IP assay. Importantly, an abundancy of YAP1 was found in the complexes precipitated by anti‐USP10, whereas an enrichment of USP10 fragments was found in the complexes precipitated by anti‐YAP1 (Figure 2B). These findings indicated that YAP1 can bind to USP10. We then examined the sub‐cellular localization of USP10 and YAP1 in Panc‐1 and BxPC‐1 cells using dual immunofluorescence staining. It was found that the green fluorescence (USP10) was mainly distributed in cytoplasm, while the red fluorescence (YAP1) was distributed both in cytoplasm and in the nucleus, and the yellow fluorescence (merge) was distributed in cytoplasm as well (Figure 2C). To further determine if USP10 can affect the ubiquitination of YAP1, overexpression of HA‐ubiquitin, Flag‐YAP1, or USP10 was introduced in 293T cells. It was observed that the ubiquitination level of YAP1 was significantly decreased (Figure 2D). These results indicated that USP10 mainly binds to YAP1 in cytoplasm. Moreover, we further observed that the ubiquitination level of YAP in Panc‐1 and BxPC‐3 cells was significantly elevated after USP10 knockdown (Figure 2E). However, treatment of MG132 restored the protein level of YAP1 in Panc‐1 and BxPC‐3 cells (Figure 2F). Collectively, these results suggested that USP10 can bind to YAP1 and suppress ubiquitination and maintain the stability of YAP1 in PAAD cells.
FIGURE 2.
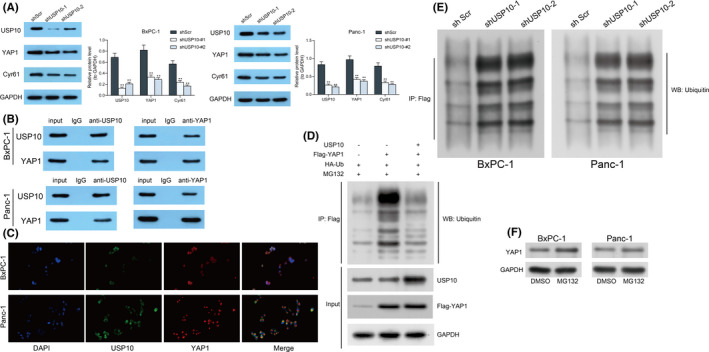
Ubiquitin‐specific peptidase 10 (USP10) knockdown increases ubiquitination and degradation of YAP1 in pancreatic adenocarcinoma (PAAD) cells. (A) Protein levels of YAP1, Cyr61, and USP10 in Panc‐1 and BxPC‐1 cells after shUSP10 transfection examined by western blot analysis; (B) binding relationship between USP10 and YAP1 in Panc‐1 and BxPC‐1 cells examined by the Co‐IP assay; (C) sub‐cellular distribution of USP10 and YAP1 in Panc‐1 and BxPC‐1 cells examined using dual immunofluorescence staining; (D) ubiquitination level of YAP1 in 293T cells after HA‐ubiquitin, Flag‐YAP1, or USP10 overexpression; (E) ubiquitination level of YAP1 in Panc‐1 and BxPC3 cells after USP10 knockdown examined by Co‐IP; and (F) protein level of YAP1 in Panc‐1 and BxPC3 cells after 10 μM MG132 treatment determined by western blot analysis. Differences were analyzed by two‐way ANOVA (A), **P < 0.01
3.3. Knockdown of USP10 reduces the immune escape of pancreatic adenocarcinoma cells
As mentioned above, synergistic suppression of PD‐1 and Hippo signaling has been found to significantly suppress the growth of pancreatic cancer cell in vitro. 18 Of note, YAP1 has been correlated with PD‐L1 upregulation in the microenvironment of hepatocellular carcinoma. 19 Whether the USP10/YAP1 axis regulates the immune escape of PAAD cells aroused our interest. Therefore, the levels of PD‐L1 and Galectin‐9, two important immune checkpoint molecules, were determined in the Panc‐1 and BxPC‐1 cells after shUSP10 transfection. It was found that downregulation of USP10 decreased the PD‐L1 and Galectin‐9 levels in cells (Figure 3A,B). The flow cytometry also suggested that the number of PD‐L1‐ and Galectin‐9‐positive cells was significantly reduced as well upon USP10 downregulation (Figure 3C). Next, the shUSP10‐transfected Panc‐1 and BxPC‐1 cells were co‐incubated with the PMA‐treated THP‐1 cells (macrophages). The polarization of the macrophages was examined by flow cytometry and RT‐qPCR. It was found that after co‐incubation with the cells with stable USP10 downregulation, the portion of M1‐polarized macrophages was increased and, accordingly, the portion of M2‐polarized macrophages was reduced (Figure 3D,E). In addition, the cells stably transfected with shUSP10 were co‐cultured with natural killer (NK) cells, and the degree of cell lysis was determined by an LDH kit. It was found that knockdown of USP10 increased the LDH release in the cells; namely, it increased the sensitivity of PAAD cells to NK cells (Figure 3F). Collectively, this ample of evidence suggests that USP10 knockdown‐mediated YAP1 suppression potentially leads to increased expression of immunosuppressive molecules, such as PD‐L1, Galectin‐9, and M2 macrophages.
FIGURE 3.
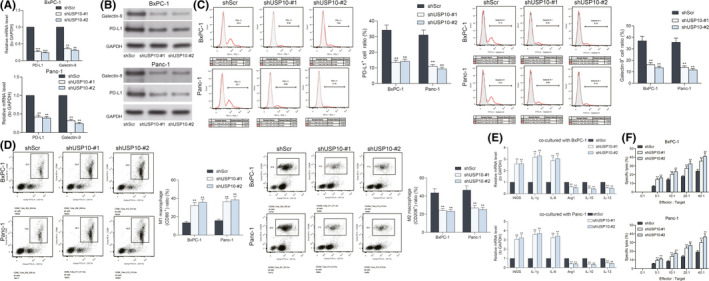
Ubiquitin‐specific peptidase 10 (USP10) knockdown reduces the immune escape of pancreatic adenocarcinoma (PAAD) cells. (A and B) expression of PD‐L1 and Galectin‐9 in Panc‐1 and BxPC‐1 cells detected by RT‐qPCR and western blot analysis; (C) portion of programmed death ligand‐1 (PD‐L1)‐ and Galectin‐9‐positive Panc‐1 and BxPC‐1 cells examined by flow cytometry; (D) portion of CD86‐positive (M1‐polarized) and CD206‐positive (M2‐polarized) macrophages after co‐incubation with Panc‐1 and BxPC‐1 cells evaluated by flow cytometry; (E) expression of the M1‐macrophage markers (iNOS, IL‐1β, and IL‐6) and M2‐macrophage markers (Arg1, IL‐10, and IL‐13) in cells examined by RT‐qPCR; (F) the degree of Panc‐1 and BxPC‐1 cells lysed by natural killer (NK) cells examined using an LDH kit. Differences were analyzed by two‐way ANOVA (A–E), **P < 0.01; ***P < 0.001
3.4. Knockdown of USP10 reduces growth and invasiveness of pancreatic adenocarcinoma cells in vitro
The exact roles of USP10 in the behaviors of PAAD cells were further examined. First, the viability of cancer cells was evaluated with a CTG kit. It was found that USP10 knockdown significantly suppressed the viability of Panc‐1 and BxPC‐1 cells (Figure 4A). Likewise, the EdU labeling assay showed that shUSP10 also reduced the EdU‐positive rate in both cells (Figure 4B). After that, the flow cytometry concerning cell apoptosis indicated that USP10 downregulation significantly increased the number of apoptotic Panc‐1 and BxPC‐1 cells (Figure 4C). Moreover, the levels of epithelial‐mesenchymal transition (EMT)‐related factors ZO‐1, E‐cadherin, Vimentin, and N‐cadherin in Panc‐1 and BxPC‐1 cells were examined by RT‐qPCR, western blot analysis, and immunofluorescence staining. After USP10 knockdown, the ZO‐1 and E‐cadherin levels in cells were increased, whereas the Vimentin and N‐cadherin levels in cells were reduced (Figure 4D–F). The Transwell assays showed that the number of migratory or invasive cells on the lower membrane was significantly reduced upon USP10 knockdown (Figure 4G,H). This body of evidence indicated that USP10 knockdown can limit proliferation and dissemination of PAAD cells in vitro.
FIGURE 4.
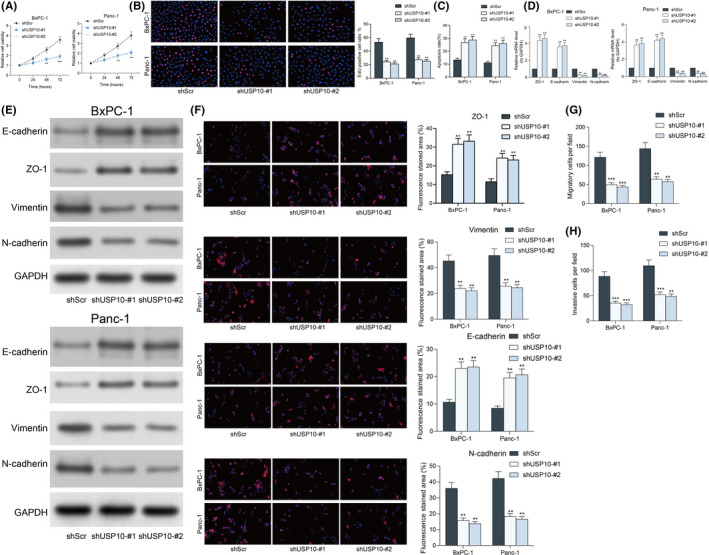
Ubiquitin‐specific peptidase 10 (USP10) knockdown reduces growth and invasiveness of pancreatic adenocarcinoma (PAAD) cells in vitro. (A) viability of Panc‐1 and BxPC‐1 cells examined using a CTG kit; (B) proliferation rate of the Panc‐1 and BxPC‐1 cells using the EdU labeling kit; (C) apoptosis of Panc‐1 and BxPC‐1 cells examined using flow cytometry; (D and E) levels of EMT‐related biomarkers ZO‐1, E‐cadherin, Vimentin, and N‐cadherin in Panc‐1 and BxPC‐1 cells determined by RT‐qPCR (D), western blot analysis (E), and immunofluorescence staining (F), respectively; (G and H) migration (G) and invasion (H) ability of Panc‐1 and BxPC‐1 cells determined by Transwell assays. Differences were analyzed by two‐way ANOVA (A–E), **P < 0.01; ***P < 0.001
3.5. Knockdown of USP10 reduces pancreatic adenocarcinoma tumorigenesis in mice
We further examined the function of USP10 knockdown on cell growth in vivo. Panc‐1 and BxPC‐1 cells stably transfected with shUSP10 were subcutaneously injected into NOD/SCOD mice. The volume of xenograft tumors was examined every 7 days. Importantly, knockdown of USP10 blocked the growth rate of tumors in mice (Figure 5A). Likewise, the weight of tumors on the 35th day was reduced as well in the setting of USP10 inhibition (Figure 5B). The tumor tissues were further collected, and the IHC staining intensity of KI67 and PCNA in tissues was significantly reduced after USP10 knockdown (Figure 5C,D). In addition, USP10 inhibition decreased the staining intensity of YAP1, Cyr61 and CTGF in the tissues of xenograft tumors (Figure 5E–G). Moreover, for metastasis measurement, the Panc‐1 and BxPC‐1 cells stably transfected with shUSP10 were also injected into mice through tail veins. After 45 days, the animals were killed to collect the lung tissues for HE staining. Importantly, USP10 knockdown reduced the tumor dissemination in lung and liver tissues (Figure 5H,I). Namely, USP10 inhibition reduced the metastasis of PAAD cells in vivo as well.
FIGURE 5.
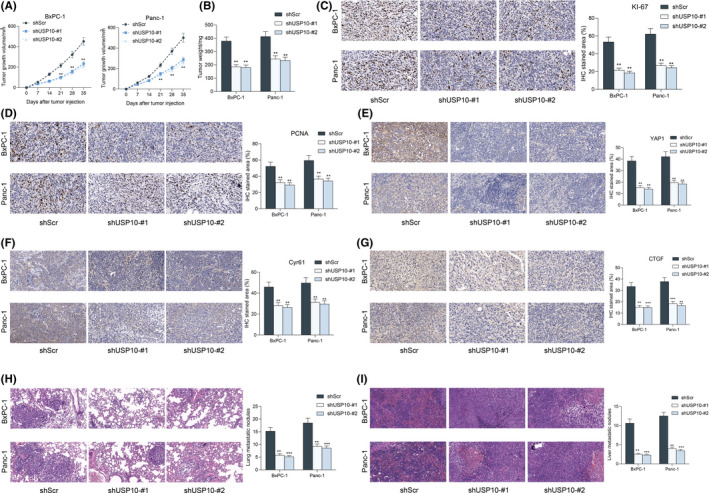
Ubiquitin‐specific peptidase 10 (USP10) knockdown suppresses growth and metastasis of pancreatic adenocarcinoma (PAAD) cells in vivo. (A) Growth rate of the xenograft tumors; (B) weight of the xenograft tumors on the 35th day; (C–G) staining intensity of KI67 (C), PCNA (D), YAP1 (E), Cyr61 (F), and CTGF (G) in the tissues of xenograft tumors detected by IHC staining; and (H and I) tumor metastasis in murine lung (H) and liver (I) tissues on the 45th day examined by HE staining. In each group, n = 6. Differences were analyzed by two‐way ANOVA (A–E), **P < 0.01; ***P < 0.001
3.6. Overexpression of Cyr61 enhances immune tolerance of pancreatic adenocarcinoma cells
As aforementioned, USP10 might maintain the stability of YAP1 and promote the expression of downstream Cyr61. The relevance of Cyr61 to the immune escape of PAAD cells remains unknown. Therefore, the Panc‐1 and BxPC‐1 cells transfected with shUSP10 were further transfected with oe‐Cyr61. Successful upregulation of Cyr61 was confirmed by western blot analysis (Figure 6A). After that, we examined the PD‐L1 and Galectin‐9 levels in cells. Importantly, the PD‐L1 and Galectin‐9 expression in Panc‐1 and BxPC‐1 that was initially decreased after USP10 knockdown was restored after further Cyr61 overexpression (Figure 6B). In concert with this, the flow cytometry results suggested that the number of PD‐L1‐ and Galectin‐9‐positive cells was increased upon Cyr61 upregulation (Figure 6C). These cells were co‐incubated with the PMA‐treated THP‐1 cells. Importantly, the flow cytometry and RT‐qPCR results showed that the portion of M2‐polarized macrophages was significantly increased after Cyr61 overexpression (Figure 6D,E). It was also noteworthy that upregulation of Cyr61 increased the resistance of Panc‐1 and BxPC‐1 cells to NK cells since the LDH release in cell lysates was notably reduced (Figure 6F).
FIGURE 6.
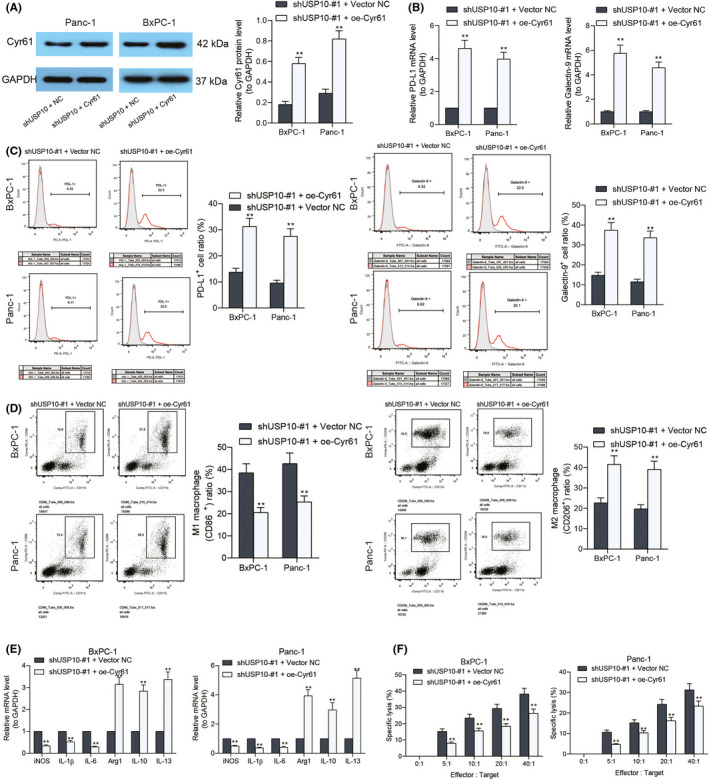
Overexpression of Cyr61 restores immune tolerance of pancreatic adenocarcinoma (PAAD) cells. (A) Protein level of Cyr61 in Panc‐1 and BxPC‐1 cells after oe‐Cyr61 transfection examined by western blot analysis; (B) PD‐L1 and Galectin‐9 expression in Panc‐1 and BxPC‐1 cells examined by RT‐qPCR; (C) portion of PD‐L1‐ and Galectin‐9‐positive cells evaluated by the flow cytometry; (D) portion of CD86‐positive (M1‐polarized) and CD206‐positive (M2‐polarized) macrophages after co‐incubation with Panc‐1 and BxPC‐1 cells detected by flow cytometry; (E) expression of the M1‐macrophage markers (iNOS, IL‐1β, and IL‐6) and M2‐macrophage markers (Arg1, IL‐10, and IL‐13) in cells examined by RT‐qPCR; (F) the degree of Panc‐1 and BxPC‐1 cells lysed by natural killer (NK) cells examined using an LDH kit. Differences were analyzed by two‐way ANOVA (A–E), **P < 0.01
3.7. Overexpression of Cyr61 restores pancreatic adenocarcinoma cell proliferation in vitro
The focus of the study then shifted to the functions of Cyr61 on the activity of PAAD cells in vitro. First, by using the CTG kit, we found that the viability of cells, which was suppressed after USP inhibition, was restored after Cyr61 overexpression (Figure 7A). In agreement with this, the EdU labeling assay indicated that the number of proliferative cells was increased by oe‐Cyr61 as well (Figure 7B). In addition, the apoptosis rate in Panc‐1 and BxPC‐1, which was elevated by shUSP10, was reduced after further Cyr61 overexpression (Figure 7C). The expression of ZO‐1, E‐cadherin, Vimentin, and N‐cadherin in cells was detected as well. Importantly, in the setting of Cyr61 upregulation, the expression of ZO‐1 and E‐cadherin was reduced, whereas the expression of N‐cadherin was significantly increased (Figure 7D–F). Moreover, the Transwell assay results also showed that upregulation of Cyr61 led to an increase in the number of Panc‐1 and BxPC‐1 migrated or invaded to the lower membranes (Figure 7G,H).
FIGURE 7.
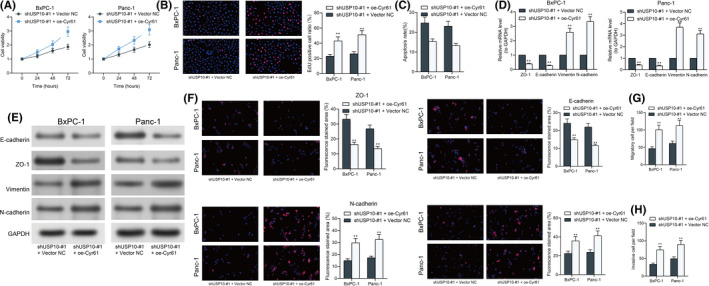
Overexpression of Cyr61 restores pancreatic adenocarcinoma (PAAD) cell proliferation in vitro. (A) Viability of Panc‐1 and BxPC‐1 cells examined using a CellTiter Glo (CTG) kit; (B) proliferation rate of the Panc‐1 and BxPC‐1 cells using the EdU labeling kit; (C) apoptosis of Panc‐1 and BxPC‐1 cells examined using flow cytometry; (D–F) levels of EMT‐related biomarkers ZO‐1, E‐cadherin, Vimentin, and N‐cadherin in Panc‐1 and BxPC‐1 cells determined by RT‐qPCR (D), western blot analysis (E), and immunofluorescence staining (F), respectively; and (G and H) migration (G) and invasion (H) ability of Panc‐1 and BxPC‐1 cells determined by Transwell assays. Differences were analyzed by two‐way ANOVA (A–G), **P < 0.01
4. DISCUSSION
Pancreatic cancer has some special characteristics, including an enrichment of immunosuppressive mediators in the microenvironment and a dense stroma, which serves as a physical barrier to drug delivery and a dynamic entity implicated in immune system regulation. Therefore, the immune system exerts a crucial function in PC development and effective management approaches for innate or adaptive immunity are being explored. 23 In this work, we confirmed that the deubiquitinate USP10 suppressed ubiquitination and degradation of YAP1, which further elevated the expression of Cyr61 and enhanced the immune escape and malignant development of PAAD.
Data from the TCGA‐PAAD dataset suggested that YAP1 was abundantly expressed in PAAD patients and indicated poor prognosis. YAP1 is a key effector of the Hippo pathway, whose dysfunction is frequently involved in many human diseases, including cancer. 24 Phosphorylated YAP1 is prohibited from nuclear accumulation and degraded in cytoplasm, whereas the non‐phosphorylated YAP1 trans‐locates into the nucleus and induces cell proliferation and development by activating the most reliable downstream targets CTGF and Cyr61. 25 , 26 Likewise, activation of YAP1 was found to promote malignant behaviors of PC cells. 17 , 27 Our subsequent integrated bioinformatic analyses suggested that USP10 showed a strong positive correlation with YAP1, and its aberrant high expression indicated poor prognosis in PAAD patients. This trend was validated in the acquired cell lines, where we found increased expression of USP10 in the PAAD cells compared to the HPDE cells. USP10 has shown oncogenic roles in several human malignancies and indicated dismal prognosis of patients. 28 , 29 As a deubiquitinating enzyme, USP10 has been found to maintain the stability of EMT‐transcription factor Slug/SNAI2. 30 Similarly, USP10 was reported to deubiquitinate and stabilize Smad4 protein to augment metastasis of hepatocellular carcinoma. 31 Interestingly, USP10 has been reported to stabilize YAP and its paralog TAZ to promote proliferation of hepatocellular carcinoma. 32 Given that USP10 was predicted to be positively linked to YAP1 expression, we therefore surmised that there might be a similar correlation between USP10 and YAP1 in PAAD. The subsequent Co‐IP, ubiquitination measurement and double‐labeled immunofluorescence assays confirmed that USP10 prevents YAP from ubiquitination and degradation and maintains its stability in PAAD cells.
USP10 has been found to mediate deubiquitination and upregulation of the NLR family pyrin domain containing 7 to promote M2 polarization of macrophage and tumor progression in colorectal cancer. 33 In relation to the role of the USP10/YAP1 axis in immune escape and malignant development in PAAD, we first examined the PD‐L1 and galectin‐9 levels in cells. As aforementioned, PD‐L1 plays a critical role in the potential of cancer cells to evade the host’s immune system. 6 , 9 The anti‐PD‐1 and anti‐PD‐L1 drugs have been approved by the Food and Drug Administration. 34 Galectin‐9 represents another immune checkpoint molecule that may block the anti‐tumor activity of cytotoxic lymphoid cells, such as NK cells. 35 Moreover, galectin‐9 has been reported to be correlated with the development of regulatory T (Treg) cells, 36 which is closely linked to immune tolerance as well. 37 Importantly, we found that the PD‐L1 and Galectin‐9 levels in PAAD cells were significantly reduced, whereas the release of LDH in cells after NK cell treatment was increased in the setting of USP10 knockdown. Moreover, the portion of M1 macrophages was increased after USP10 knockdown, indicating that USP10 and YAP1 were positively correlated with the immune escape of PAAD cells. Consequently, the proliferation and metastasis of PAAD cells, both in vitro and in vivo, were reduced after USP10 downregulation. Similarly, YAP1 has been correlated with elevated PD‐L1 expression in hepatocellular carcinoma. 19 Likewise, phosphorylation and restricted nuclear translocation of YAP1 has been correlated with reduced PD‐L1 levels in colorectal cancer. 38 Feng et al. (2021) reported that YAP1 can bind to the enhancer region of PD‐L1 to induce its transcriptional activation. 39 In addition, YAP1 upregulation has been correlated with immunosuppression via upregulation of PD‐1 and M2 macrophage levels. 25
To further validate the interactions among USP10, YAP1, and Cyr61, rescue experiments were performed through further upregulation of Cyr61 in cells with stable transfection of shUSP10. Consequently, it was found that the PD‐L1 and Galectin‐9 levels, the resistance of cells to NK cells, the portion of M2 macrophages, as well as the malignant growth and invasiveness of PAAD cells were restored after Cyr61 overexpression. This body of evidence proves that the USP10/YAP1 axis is linked to the increase of immunosuppression‐related factors and triggers immune escape and tumor development.
Collectively, this study demonstrates that deubiquitinating enzyme USP10 suppresses ubiquitination and degradation of YAP1, which further elevates the expression of Cyr61 and subsequently elevates PD‐L1 and Galectin‐9 levels as well as the portion of M2 macrophages in the tumor microenvironment, leading to immune escape and malignant development of PAAD cells. USP10 and Cyr61 may serve as novel targets for overcoming the immunosuppression in PAAD cells and help improve the prognosis of patients.
DISCLOSURE
The authors have no conflicts of interest to declare.
Supporting information
Appendix S1
ACKNOWLEDGMENTS
None.
Liu X, Chen B, Chen J, Su Z, Sun S. Deubiquitinase ubiquitin‐specific peptidase 10 maintains cysteine rich angiogenic inducer 61 expression via Yes1 associated transcriptional regulator to augment immune escape and metastasis of pancreatic adenocarcinoma. Cancer Sci. 2022;113:1868–1879. doi: 10.1111/cas.15326
DATA AVAILABILITY STATEMENT
Data are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394‐424. [DOI] [PubMed] [Google Scholar]
- 2. Ilic M, Ilic I. Epidemiology of pancreatic cancer. World J Gastroenterol. 2016;22:9694‐9705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ansari D, Tingstedt B, Andersson B, et al. Pancreatic cancer: yesterday, today and tomorrow. Future Oncol. 2016;12:1929‐1946. [DOI] [PubMed] [Google Scholar]
- 4. Conroy T, Desseigne F, Ychou M, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;364:1817‐1825. [DOI] [PubMed] [Google Scholar]
- 5. Von Hoff DD, Ervin T, Arena FP, et al. Increased survival in pancreatic cancer with nab‐paclitaxel plus gemcitabine. N Engl J Med. 2013;369(18):1691‐1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brahmer JR, Tykodi SS, Chow LQM, et al. Safety and activity of anti‐PD‐L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455‐2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Morrison AH, Byrne KT, Vonderheide RH. Immunotherapy and prevention of pancreatic cancer. Trends Cancer. 2018;4:418‐428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wormann SM, Diakopoulos KN, Lesina M, Algul H. The immune network in pancreatic cancer development and progression. Oncogene. 2014;33:2956‐2967. [DOI] [PubMed] [Google Scholar]
- 9. Kim DH, Kim HR, Choi YJ, et al. Exosomal PD‐L1 promotes tumor growth through immune escape in non‐small cell lung cancer. Exp Mol Med. 2019;51:1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pan Y, Yu Y, Wang X, Zhang T. Tumor‐associated macrophages in tumor immunity. Front Immunol. 2020;11:583084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cai J, Qi QI, Qian X, et al. The role of PD‐1/PD‐L1 axis and macrophage in the progression and treatment of cancer. J Cancer Res Clin Oncol. 2019;145:1377‐1385. [DOI] [PubMed] [Google Scholar]
- 12. Borgmastars E, de Weerd HA, Lubovac‐Pilav Z, Sund M. miRFA: an automated pipeline for microRNA functional analysis with correlation support from TCGA and TCPA expression data in pancreatic cancer. BMC Bioinform. 2019;20:393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ho J, Li X, Zhang L, et al. Translational genomics in pancreatic ductal adenocarcinoma: a review with re‐analysis of TCGA dataset. Semin Cancer Biol. 2019;55:70‐77. [DOI] [PubMed] [Google Scholar]
- 14. Xu X, Yu Y, Zong K, Lv P, Gu Y. Up‐regulation of IGF2BP2 by multiple mechanisms in pancreatic cancer promotes cancer proliferation by activating the PI3K/Akt signaling pathway. J Exp Clin Cancer Res. 2019;38:497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shibata M, Ham K, Hoque MO. A time for YAP1: tumorigenesis, immunosuppression and targeted therapy. Int J Cancer. 2018;143:2133‐2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gruber R, Panayiotou R, Nye E, Spencer‐Dene B, Stamp G, Behrens A. YAP1 and TAZ control pancreatic cancer initiation in mice by direct up‐regulation of JAK‐STAT3 signaling. Gastroenterology. 2016;151:526‐539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xia P, Liu P, Fu Q, et al. Long noncoding RNA EPIC1 interacts with YAP1 to regulate the cell cycle and promote the growth of pancreatic cancer cells. Biochem Biophys Res Commun. 2020;522:978‐985. [DOI] [PubMed] [Google Scholar]
- 18. Pu N, Gao S, Yin H, et al. Cell‐intrinsic PD‐1 promotes proliferation in pancreatic cancer by targeting CYR61/CTGF via the hippo pathway. Cancer Lett. 2019;460:42‐53. [DOI] [PubMed] [Google Scholar]
- 19. Li S, Ji J, Zhang Z, et al. Cisplatin promotes the expression level of PD‐L1 in the microenvironment of hepatocellular carcinoma through YAP1. Mol Cell Biochem. 2020;475:79‐91. [DOI] [PubMed] [Google Scholar]
- 20. Kim K, Huh T, Park Y, et al. Prognostic significance of USP10 and p14ARF expression in patients with colorectal cancer. Pathol Res Pract. 2020;216:152988. [DOI] [PubMed] [Google Scholar]
- 21. Jeusset LM, McManus KJ. Developing targeted therapies that exploit aberrant histone ubiquitination in cancer. Cells. 2019;8:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Debelouchina GT, Gerecht K, Muir TW. Ubiquitin utilizes an acidic surface patch to alter chromatin structure. Nat Chem Biol. 2017;13:105‐110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Aroldi F, Zaniboni A. Immunotherapy for pancreatic cancer: present and future. Immunotherapy. 2017;9:607‐616. [DOI] [PubMed] [Google Scholar]
- 24. Zheng Y, Pan D. The Hippo signaling pathway in development and disease. Dev Cell. 2019;50:264‐282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim EH, Sohn BH, Eun Y‐G, et al. Silence of Hippo pathway associates with pro‐tumoral immunosuppression: potential therapeutic target of glioblastomas. Cells. 2020;9:1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wennmann DO, Schmitz J, Wehr MC, et al. Evolutionary and molecular facts link the WWC protein family to Hippo signaling. Mol Biol Evol. 2014;31:1710‐1723. [DOI] [PubMed] [Google Scholar]
- 27. Huang H, Xiong G, Shen P, et al. MicroRNA‐1285 inhibits malignant biological behaviors of human pancreatic cancer cells by negative regulation of YAP1. Neoplasma. 2017;64:358‐366. [DOI] [PubMed] [Google Scholar]
- 28. Hu C, Zhang MU, Moses N, et al. The USP10‐HDAC6 axis confers cisplatin resistance in non‐small cell lung cancer lacking wild‐type p53. Cell Death Dis. 2020;11:328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Takayama KI, Suzuki T, Fujimura T, Takahashi S, Inoue S. Association of USP10 with G3BP2 inhibits p53 signaling and contributes to poor outcome in prostate cancer. Mol Cancer Res. 2018;16:846‐856. [DOI] [PubMed] [Google Scholar]
- 30. Ouchida AT, Kacal M, Zheng A, et al. USP10 regulates the stability of the EMT‐transcription factor Slug/SNAI2. Biochem Biophys Res Commun. 2018;502:429‐434. [DOI] [PubMed] [Google Scholar]
- 31. Yuan T, Chen Z, Yan F, et al. Deubiquitinating enzyme USP10 promotes hepatocellular carcinoma metastasis through deubiquitinating and stabilizing Smad4 protein. Mol Oncol. 2020;14:197‐210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhu H, Yan F, Yuan T, et al. USP10 promotes proliferation of hepatocellular carcinoma by deubiquitinating and stabilizing YAP/TAZ. Cancer Res. 2020;80:2204‐2216. [DOI] [PubMed] [Google Scholar]
- 33. Li B, Qi Z‐P, He D‐L, et al. NLRP7 deubiquitination by USP10 promotes tumor progression and tumor‐associated macrophage polarization in colorectal cancer. J Exp Clin Cancer Res. 2021;40:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Martin‐Liberal J, Ochoa de Olza M, Hierro C, Gros A, Rodon J, Tabernero J. The expanding role of immunotherapy. Cancer Treat Rev. 2017;54:74‐86. [DOI] [PubMed] [Google Scholar]
- 35. Gonçalves Silva I, Yasinska IM, Sakhnevych SS, et al. The Tim‐3‐galectin‐9 secretory pathway is involved in the immune escape of human acute myeloid leukemia cells. EBioMedicine. 2017;22:44‐57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen H, Huang NA, Tian H, et al. Splenectomy provides protective effects against CLP‐induced sepsis by reducing TRegs and PD‐1/PD‐L1 expression. Int J Biochem Cell Biol. 2021;136:105970. [DOI] [PubMed] [Google Scholar]
- 37. Walker LS. Treg and CTLA‐4: two intertwining pathways to immune tolerance. J Autoimmun. 2013;45:49‐57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang JJ, Zhang QS, Li ZQ, Zhou JW, Du J. Metformin attenuates PD‐L1 expression through activating Hippo signaling pathway in colorectal cancer cells. Am J Transl Res. 2019;11:6965‐6976. [PMC free article] [PubMed] [Google Scholar]
- 39. Feng S, Sun H, Zhu W. MiR‐92 overexpression suppresses immune cell function in ovarian cancer via LATS2/YAP1/PD‐L1 pathway. Clin Transl Oncol. 2021;23:450‐458. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1
Data Availability Statement
Data are available from the corresponding author upon reasonable request.


