Abstract
Cyclin‐dependent kinase 4/6 inhibitors (CDK4/6i) significantly improve progression‐free survival and have become the standard therapy for estrogen receptor‐positive/human epidermal growth factor receptor 2‐negative metastatic breast cancer patients. Treatment surveillance by radiological imaging has some limitations in detection and repeated biopsy genomic profiling is not clinically feasible. Serial circulating tumor DNA (ctDNA) analysis may provide insights into treatment response. Here we performed serial ctDNA analysis (n = 178) on 33 patients. Serial ctDNA analysis identified disease progression with sensitivity of 75% and specificity of 92%. In eight of 12 patients (61%) responding to CDK4/6i who eventually developed progressive disease, serial sampling every 3 or 6 months captured the initial rise of ctDNA with an average lead time of 3 months. In three of eight patients that did not respond to CDK4/6i (progressive disease at first radiological assessment, 3 months), biweekly sequencing within the first cycle of CDK4/6i treatment (1 month) detected sustained ctDNA levels (≥0.2% variant allele frequency), with lead time of 2 months. Serial ctDNA analysis tracked RECIST response, including clinically challenging scenarios (bone metastases or small‐sized target lesions), as well as detecting acquired genetic alterations linked to CDK4/6i resistance in the G1 to S transition phase. Circulating tumor DNA analysis was more sensitive than carcinoembryonic antigen or cancer antigen 15‐3 serum tumor markers at monitoring tumor response to CDK4/6i treatment. Our findings indicated the possible clinical utility of serial ctDNA analysis for earlier progressive disease detection and real‐time monitoring of CDK4/6i response.
Keywords: breast cancer, CDK4/6 inhibitor, circulating tumor DNA, liquid biopsy, targeted NGS
Cyclin‐dependent kinase 4/6 inhibitors (CDK4/6i) significantly improve progression‐free survival and have become the standard therapy for ER+/HER2− metastatic breast cancer patients. Treatment surveillance by radiological imaging has some limitations in detection and repeated biopsy genomic profiling is not clinically feasible. Our findings showed the possible clinical utility of serial circulating tumor DNA analysis for earlier progressive disease detection and real‐time monitoring of CDK4/6i response.

Abbreviations
- CA15‐3
Cancer antigen 15‐3
- CDK4/6i
Cyclin dependent kinase 4/6 inhibitor
- CEA
Carcinoembryonic antigen
- cfDNA
Cell‐free deoxyribonucleic acid
- cfTNA
Cell‐free total nucleic acid
- CNV
Copy number variant
- CR
Complete response
- ctDNA
Circulating tumor DNA
- ddPCR
Droplet digital polymerase chain reaction
- dUTP
Deoxyuridine triphosphate
- EDTA‐2Na
Ethylenediaminetetraacetic acid disodium
- ER+
Estrogen receptor positive
- FDA
U.S. Food and Drug Administration
- HER2−
Human epidermal growth factor receptor 2 negative
- HR
Hazards ratio
- IRB
Institutional review board
- MBC
Metastatic breast cancer
- NGS
Next generation sequencing
- NPA
Negative percent agreement
- PCR
Polymerase chain reaction
- PD
Progressive disease
- PFS
Progression free survival
- PR
Partial response
- RECIST
Response Evaluation Criteria in Solid Tumors
- SD
Stable disease
- SNV
Single nucleotide variant
- TMAP
Torrent mapping alignment program
- UMT
Unique molecular tags
- VAF
Variat allele frequency
- WBC
White blood cell
1. INTRODUCTION
Cyclin‐dependent kinase 4/6 inhibitors have become the standard treatment for ER+/HER2− advanced breast cancer patients. Preclinical studies have established the role of a CDK4/6–RB pathway in regulating the cell cycle through the G1 to S transition phase. 1 , 2 Multiple clinical trials have reported that patients receiving CDK4/6i showed significantly longer PFS in combination with endocrine therapy in ER+ breast cancer. 3 , 4 Despite improvements in clinical management, a subset of breast cancer patients will develop resistance to treatment.
Treatment response is mainly assessed by serial radiographic images. 5 Radiographic imaging provides assessment of the disease burden but can sometimes be hampered by suboptimal detection limit and inconsistencies in tumor size measurements. 6 , 7 Carcinoembryonic antigen and CA 15‐3 are serum biomarkers mainly used for treatment monitoring in patients with MBC, 8 with elevated levels often associated with poor prognosis. Although clinically useful in some breast cancer patients, CEA and CA 15‐3 have limited sensitivity of only 50% 9 and 60%–70%, 8 , 10 respectively. Tumor genome sequencing remains the gold standard for genomic profiling, providing insights into potential actionable targets for personalized treatment. 11 However, in a metastatic setting, repeated biopsy is not clinically feasible. Taken together, a more sensitive and less invasive approach is needed to provide an earlier measure of treatment response to inform individual treatment decisions.
Many cancers shed DNA into the bloodstream, referred to as ctDNA. Liquid biopsy through serial ctDNA analysis can complement radiographic imaging and CEA and CA15‐3 markers in assessing CDK4/6i responses. It is minimally invasive, feasible for repeated sampling, and is able to detect tumor‐specific alterations in circulation, reflecting disease burden in real time. 12 Previous studies have investigated the clinical utility of liquid biopsy in predicting response to CDK4/6i. 13 , 14 , 15 However, these studies were focused on ctDNA dynamics at early time points 14 , 15 or on the heterogeneity of the mutational landscapes between baseline and at disease progression. 13 None of the studies used monitoring through a granular time series to catch disease progression at an earlier time point.
In this single center, retrospective, exploratory biomarker study, we evaluated serial ctDNA analysis against radiographic imaging and CEA and CA 15‐3 markers as a biomarker for CDK4/6i response monitoring. We show that short interval serial ctDNA analysis was able to catch initial rise or reemergence of ctDNA, indicative of disease progression, well in advance of radiological assessment. This study also identified mutations potentially predictive of CDK4/6i response, tracked tumor response of RECIST‐defined nonmeasurable target lesions, and identified acquired genetic alterations linked to CDK4/6i resistance. Our results support the possible clinical utility of serial ctDNA analysis for earlier PD detection and real‐time monitoring of CDK4/6i response.
2. MATERIALS AND METHODS
2.1. Patient cohort
Thirty‐three ER+/HER2− female MBC patients scheduled to receive CDK4/6i and endocrine combination therapy either as a first line or subsequent line treatment were recruited in a consecutive, retrospective manner from the Cancer Institute Hospital of Japanese Foundation for Cancer Research from April 16, 2018 to September 30, 2020. Patient age varied from 35 to 73 years (median, 54 years) and were eligible if they had progressed on endocrine treatment for metastatic disease or during adjuvant therapy. Study has been approved by the ethical committee of Japanese Foundation for Cancer Research (Institutional Review Board No. 2015‐1056), with approval granted for collection and genomic profiling of patient samples. All 33 patients in the study provided written informed consent for cell‐free DNA and white blood cell sequencing, with the option to be excluded from the study at any time. No patient withdrew consent for the study. The majority of patients (30/33, 91%) received oral palbociclib 125 mg daily for 3 weeks on, and 1 week off protocol in combination with either fulvestrant (500 mg every 4 weeks), letrozole (2.5 mg daily), tamoxifen (20 mg daily), anastrazole (1 mg daily), or exemestane (25 mg daily). The remaining three patients received oral abemaciclib 300 mg daily in combination with fulvestrant (500 mg every 4 weeks) or anastrazole (1 mg daily). Premenopausal women additionally received goserelin.
Patient records were reviewed to obtain radiological images of computed tomography scans as well as tumor size measurements for the patients receiving CDK4/6i therapy in this study. Radiologic images were obtained at pretreatment and at all subsequent intervals until clinical progression, 24 months evaluation, or the most recent assessment by September 30, 2020 (Figure 1). Radiologic images of multiple target lesions were obtained when available and evaluated at every subsequent interval. Tumor response was assessed and defined by the study clinician according to RECIST version 1.1. 5 Patients who developed clinical progression at first radiographic assessment were defined as not responding to CDK4/6i and endocrine combination therapy. Tumor marker levels of CEA and CA15‐3 were obtained retrospectively from patient records at concurrent or nearest to the liquid biopsy sampling time points for comparison.
FIGURE 1.
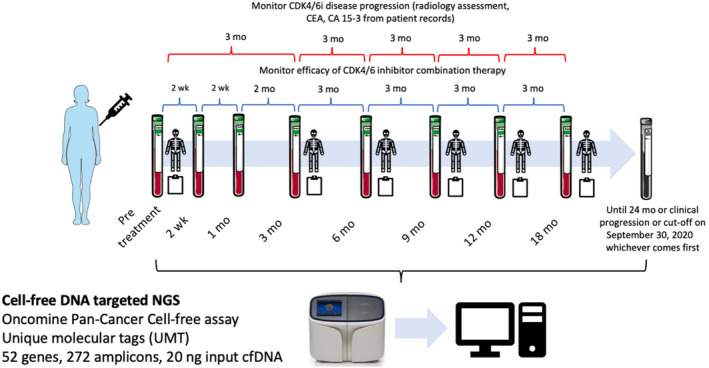
Sampling intervals for targeted next generation sequencing (NGS) circulating tumor DNA analysis in patients with metastatic breast cancer. Patients were followed until progressive disease, 24 months, or the cut‐off date (September 30, 2020), whichever comes first. Radiology assessment and tumor markers carcinoembryonic antigen (CEA), cancer antigen (CA) 15‐3 timeline might differ as decided by clinicians. Some figures in Figure 1 were adapted from Servier Medical Art (https://smart.servier.com)
2.2. Sample processing, library preparation, and sequencing
Blood samples were collected at pretreatment, 2 weeks, and 1, 3, 6, 9, 12, 18, and 24 months or until clinical progression, whichever comes first (Figure 1). For patients with no ctDNA mutations detected at the end of cycle 1 of CDK4/6i therapy (1M), the end of evaluation sample is sequenced to assess for detectable ctDNA mutations. If mutations are present, cfDNA from all sampling intervals are evaluated. Based on our data cut‐off on September 30, 2020, we collected 178 plasma samples from 33 patients. Collection and processing of whole blood has been described in previous publications. 16 , 17 , 18 Briefly, 14 mL whole blood was collected using EDTA‐2Na tubes (Terumo). Whole blood was centrifuged at 2000 g at 4°C for 10 minutes to separate plasma from white blood cells and red blood cells. The plasma layer was further centrifuged at 16 000 g at 4°C for 10 minutes to remove cell debris. Both plasma and white blood cells were stored at −80°C until nucleic acid extraction.
Plasma cfTNA was extracted using MagMAX Cell‐Free Total Nucleic Acid Isolation kit (Applied Biosystems) according to the manufacturer’s protocol. Genomic DNA from white blood cells was extracted using FlexiGene DNA Kit (Qiagen). Targeted NGS for cfTNA was carried out using Oncomine Pan‐Cancer Cell‐Free Assay following the manufacturer’s protocol (Ion Torrent). Oncomine Pan‐Cancer Cell‐Free Assay is an amplicon‐based ctDNA targeted assay, incorporating short oligonucleotides called UMTs to distinguish PCR/sequencing artifacts from actual variants (Figure 1). Library construction and subsequent NGS of cfTNA and genomic DNA were undertaken as previously described. 16 , 17 , 18 In most samples, 20 ng cfTNA was used for library construction. Sequencing of genomic DNA was carried out using the same methods using 30 ng genomic DNA for library construction.
2.3. Analysis of sequencing data
Sequencing reads were aligned to hg19 and variant calling was carried out using Torrent Suite 5.10.1 and Ion Reporter 5.10.3.0 software, respectively. Briefly, raw sequence files were aligned to hg19 using Torrent Mapping Alignment Program (TMAP) with default analysis parameters. Valid NGS runs have 90% or more of total reads mapped, alignment quality with read error rate of 2% or less (AQ17). The targeted minimum total coverage for each amplicon is at least 20,000 X. Library conversion rate was evaluated using the theoretical assumption that 10 ng cfDNA would be equivalent to approximately 3000 haploid genomes. Variant calling was undertaken using the Oncomine TagSeq Pan‐Cancer Liquid Biopsy w2.1 workflow with modifications. In order to reduce false positive detection in the plasma ctDNA assay, samples are defined as ctDNA‐positive if the following criteria are met: (a) a minimum of three reads with the same UMT is required to form a functional family; (b) SNVs and indels are detected with a minimum of two variants supporting functional families at VAF of 0.2% or more; (c) SNVs and indels are not detected with any counts in the corresponding white blood cells; and (d) minimum of 2.5‐fold change between CNV target amplicons and non‐CNV reference amplicons is required. Variants were annotated using Oncomine Pan‐Cancer Annotations version 1, a proprietary list of databases.
2.4. Digital droplet PCR analysis
For patient CDK28, plasma cfDNA was insufficient for targeted NGS in 1M, 3M, and 6M samples. Circulating tumor DNA mutations were evaluated focusing only on ESR1 Y537S detected by Oncomine Pan‐Cancer Cell‐Free Assay at disease progression. Digital droplet PCR was carried out for all seven time points for patient CDK28 from pretreatment to disease progression (10.5M). The assay used was LBx Probe Catalog no. A082 (65117) ESR1 multi probe targeting Y537N/S, D538G (Riken Genesis). All reactions were carried out on a QX200 ddPCR system (Bio‐Rad). A total of 10 ng cfDNA was used in each PCR reaction consisting of primers, probes, and ddPCR Supermix for probes (no dUTP). Reactions were divided into approximately 20,000 droplets per cell using the QX200 droplet generator. Emulsified PCR was carried out on a 96‐well thermal cycler using the following cycling conditions: (1 cycle of 95°C for 10 minutes; 40 cycles of 94°C for 30 seconds, 60°C for 1 minute, one cycle of 98°C for 10 minutes, and 4°C hold). Samples were analyzed using the QuantaSoft software to assess number of droplets harboring mutant ESR1 or WT ESR1.
2.5. Statistical analysis
The primary outcome was to assess whether granular sampling time series ctDNA analysis could indicate earlier treatment response compared to RECIST‐based clinical diagnosis. Secondary outcomes include identifying ctDNA biomarkers predictive of CDK4/6i response, evaluating ctDNA surveillance of clinical responses to CDK4/6i compared to radiological images, CEA and CA15‐3 markers, and identifying acquired ctDNA alterations linked to treatment resistance. Circulating tumor DNA monitoring markers are defined as follows: (a) ctDNA detected within cycle 1 of CDK4/6i treatment – within 1 month; (b) ctDNA of 0.2% or more for SNVs and at 2.5‐fold or more for CNVs at least once throughout the monitoring period; and (c) ctDNA alteration was not detected with any counts in the corresponding WBC sequencing. Acquired ctDNA alterations are defined as follows: (a) ctDNA undetected within the first cycle of CDK4/6i treatment (SNV count = 0 or CNV = 2.0‐fold); or (b) at disease progression, SNV of 0.2% or more or CNV of at least 2.5‐fold. For sensitivity and specificity of ctDNA‐detected disease progression: (a) ctDNA levels at point of progression shows a minimum of 0.2% for SNVs and at least 2.5‐fold for CNVs. Sensitivity is defined as (true positives)/(true positives + false negatives) and expresses the fraction of ctDNA positive patients at point of progression made using plasma targeted NGS. Specificity, or negative percent agreement is defined as (true negatives)/(true negatives + false positives), and expresses the ratio of patients expected to be ctDNA negative at the end of evaluation by plasma targeted NGS. To define lead time for ctDNA analysis, we set the following criteria: (a) SNVs of 0.2% or more or CNVs of 2.5‐fold or more in consecutive intervals from initial rise until progressive disease; (b) lead time calculated from 1 month onwards (end of cycle 1 treatment); and (c) lead time is calculated from ctDNA initial rise to nearest sampling interval to clinical progression. Clinical implications of acquired ctDNA alterations were annotated by OncoKB. 19 Association of pretreatment markers with CDK4/6i response was evaluated using Kaplan‐Meier and Cox regression analyses, with response time measured from time of CDK4/6i treatment initiation to RECIST‐defined clinical progression or tumor response by cut‐off date of September 30, 2020. Multivariable Cox regression analysis was used to test the independent prognostic value of pretreatment markers, adjusted for age, presence of visceral metastases, prior lines of endocrine therapy, and prior lines of metastatic therapy. These variables were preselected as known prognostic factors. Kaplan‐Meier and Cox regression analyses were undertaken using the “survival” and “survminer” packages in R version 4.0.3. Randomization and blinding were not applicable to our study; as this was a biomarker observational and comparison study, patients were recruited irrespective of the CDK4/6i or combination endocrine therapy they received. Comparison of ctDNA and detection rates of tumor markers CEA and CA 15‐3 were evaluated using Fisher’s exact test. All statistical analyses were carried out in R version 4.0.3 using the default statistical package. All statistical tests were two‐sided, with P‐values less than .05 considered statistically significant.
3. RESULTS
3.1. Patient characteristics
By the data cut‐off date of September 30, 2020, 178 plasma samples from 33 patients (average, 54 years [35–73 years]) had been recruited with a median follow‐up period of 12 months (1–24 months; Table 1). Complete response was observed in one patient and partial response in nine patients, giving an objective response rate of 30% (10 of 33 patients). Sixteen patients had stable disease (Table 1). Eight patients (24%) had no clinical benefit (PD at first radiological assessment) while 12 patients developed disease progression during the follow‐up period (Figure 2A). The majority of patients received palbociclib in combination with endocrine therapy (91%; 30/33). At pretreatment, 17 patients (52%) had visceral metastasis, 16 patients (48%) had nonvisceral metastasis with eight of the 16 patients presenting only bone metastasis. All 33 patients were evaluable according to RECIST 1.1 with 13 patients (39%) having measurable disease and 20 (61%) having nonmeasurable disease.
TABLE 1.
Clinical characteristics of 33 metastatic breast cancer patients at pretreatment
| Description | Clinical and pathological features |
|---|---|
| Number of patients | 33 |
| Median age, y (range) | 54 (35–73) |
| Pathology (n = 33) | |
| Invasive ductal carcinoma | 32 (97) |
| Others | 1 (3) |
| Receptor status (n = 33) | |
| ER+PR+ HER2− | 30 (91) |
| ER+PR− HER2− | 3 (9) |
| Tumor response (n = 33) | |
| Complete response | 1 (4) |
| Partial response | 9 (27) |
| Stable disease | 15 (45) |
| Progressive disease | 8 (24) |
| Menopausal status (n = 33) | |
| Premenopausal | 12 (36) |
| Postmenopausal | 21 (64) |
| CDK4/6i (n = 33) | |
| Palbociclib + endocrine therapy | 30 (91) |
| Abemaciclib + endocrine therapy | 3 (9) |
| Visceral metastases (n = 33) | |
| Yes | 17 (52) |
| No | 16 (48) |
| RECIST criteria (n = 33) | |
| Measurable | 13 (39) |
| Nonmeasurable | 20 (61) |
| Prior lines of endocrine therapy (n = 33) a | |
| 0 line | 5 (15) |
| 1 line | 13 (39) |
| 2 lines | 5 (15) |
| 3+ lines | 10 (31) |
| Prior lines of metastatic therapy (n = 33) | |
| 0 line | 12 (36) |
| 1 line | 10 (30) |
| 2 lines | 4 (12) |
| 3+ lines | 7 (22) |
| ctDNA median follow‐up, mo (range) | |
| All patients (n = 33) | 12 (1–24) |
| Progressive disease (n = 20) | 6 (1–24) |
| Responders (n = 13) | 18 (6–24) |
Data are shown as n (%) unless otherwise noted.
Abbreviations: CDK4/6i, cyclin‐dependent kinase 4/6 inhibitor; ctDNA, circulating tumor DNA; ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; PR, progesterone receptor.
Prior lines of endocrine therapy include adjuvant and metastatic regimens prior to CDK4/6i treatment.
FIGURE 2.
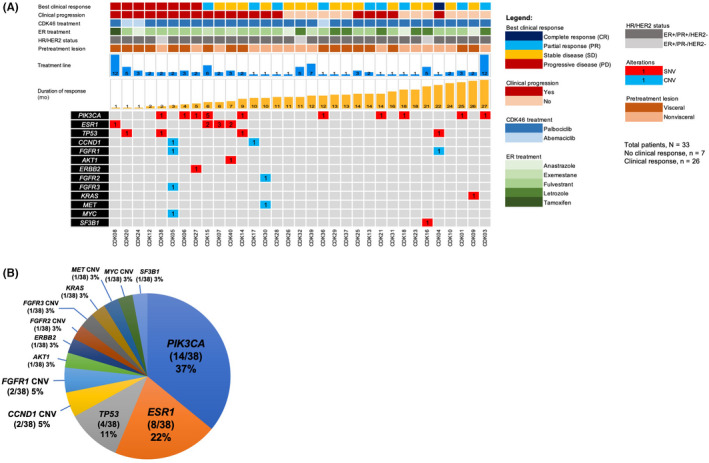
Genomic landscape of circulating tumor DNA (ctDNA) monitoring markers (excluding clonal hematopoiesis (CH)‐related mutations) for 33 estrogen receptor (ER)+/human epidermal growth factor receptor 2 (HER2)− metastatic breast cancer patients. (A) Clinicopathological characteristics and mutation matrix of ctDNA alterations. Patients showing no response to cyclin‐dependent kinase (CDK)4/6 inhibitor shown with progressive disease as best clinical response and are grouped to the left. (B) The distribution of alterations according to the respective genes. Percentages calculated using total number of ctDNA alterations detected from 33 patients. CNV, copy number variant; HR, hormone receptor; PR, progesterone receptor; SNV, single nucleotide variant
3.2. Cell‐free DNA isolation and ctDNA NGS analysis
Cell‐free DNA was successfully extracted from all 178 samples of 33 patients (average, 49.8 ng per 14 mL blood) with 175 of 178 samples successfully sequenced using targeted NGS covering 52 genes (average total coverage, 58,616× [27,488×–90,339×]; average molecular coverage, 3997× [1670×–7283×]; Table S1). Pretreatment WBC sequencing achieved similar coverage to the plasma cfDNA sequencing (average total coverage, 60,439× [49,930×–80,008×]; average molecular coverage, 5381× [2554×–12,856×]; Table S1). The average library conversion rate was 76% (47%–100%; Table S1). In three samples (P290, P328, and P385) of patient CDK28, ctDNA was evaluated using ddPCR due to insufficient cfDNA. White blood cell sequencing identified possible clonal hematopoiesis (CH) mutations in TP53, FB×W7, and IDH2 (Data S1). After excluding clonal hematopoiesis‐related mutations, 38 ctDNA alterations remained as ctDNA monitoring markers (61% detection; 20/33 patients), mostly in PIK3CA (37%), ESR1 (22%), and TP53 (11%; Figure 2, Data S1 and S2).
3.3. Prognostic impact of pretreatment ESR1 ctDNA mutations
Kaplan‐Meier analysis indicated shorter PFS for patients carrying ESR1 mutations at pretreatment compared to noncarriers (median, 5.7 and 22.2 months, respectively; HR = 5.25, 95% CI, 1.52–18.11; P = 9.0 × 10−3; Figure S1), whereas no difference was observed for patients with or without PIK3CA mutations (median, 9.4 and 16 months, respectively; HR = 1.16, 95% CI, 0.41–3.8; P = .78; Figure S1). The association of pretreatment ESR1 mutations remained statistically significant after multivariable adjustment (HR = 6.01, 95% CI, 1.50–24.53; P = 1.16 × 10−2; Table S2).
3.4. Circulating tumor DNA analysis for earlier detection of PD
Serial ctDNA analysis accurately identified disease progression in most patients, achieving analytical sensitivity of 75% and specificity of 92%. In 20 patients who eventually developed PD, 15 patients (75%) were ctDNA positive at or near point of clinical progression with SNV VAF of 0.2% or more or CNV of at least 2.5‐fold (Figure 3). In contrast, only 1 of 13 patients (8%) that were still responding to CDK4/6i by the end of the monitoring period were ctDNA positive. Our study adopted a granular time series for early disease progression detection. Serial sampling every 3 or 6 months post cycle 1 of CDK4/6i treatment was able to capture the initial rise of ctDNA earlier than clinical progression. In 8 of 12 patients responding to CDK4/6i (≥6 months) but eventually developing PD, ctDNA analysis detected earlier treatment response, with initial rise or reemergence of ctDNA observed prior to RECIST‐defined disease progression (average lead time, 3 months; range, 1.5–6 months; Figures 3 and S2). In three of eight patients that did not respond to CDK4/6i (PD at first radiologic assessment ~3 months), biweekly sequencing within the first cycle of CDK4/6i treatment (1 month) detected sustained ctDNA levels (≥0.2% VAF), with an average lead time of 2 months (Figures 3 and S2). Based on our findings, ctDNA analysis, through granular sampling intervals, detected tumor‐derived mutations indicative of disease progression well in advance of radiologic assessments.
FIGURE 3.
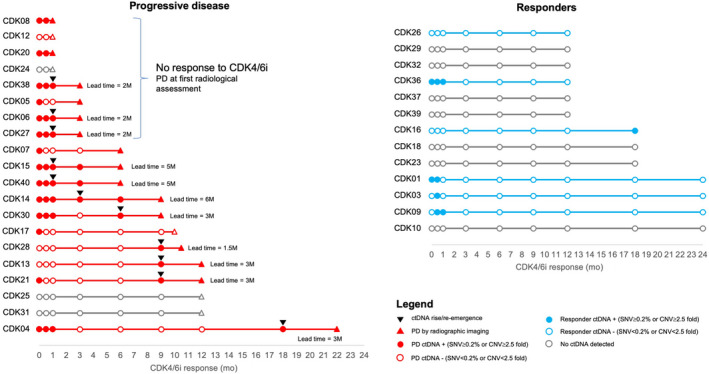
Earlier disease progression detected by circulating tumor DNA (ctDNA). Earlier disease progression detected by ctDNA is denoted with black triangles, and lead time is stated. CDK, cyclin‐dependent kinase; CDK4/6i, cyclin‐dependent kinase 4/6 inhibitor; CNV, copy number variant; PD, progressive disease; SNV, single nucleotide variant
Carcinoembryonic antigen and CA 15‐3 are the most widely used serum markers in patients with MBC. From our findings, ctDNA analyses were more sensitive and reflective of tumor response compared to CEA or CA15‐3. More patients (20/33 patients, 61%) had detectable ctDNA alterations in at least one time point compared to elevated CEA (>5 ng/mL: 12/33 patients, 36%) or CA15‐3 levels (>30 U/ml: 15/33 patients, 45%; Figures S2 and S3), although this trend is suggestive and not statistically significant (P Fisher > .05).
3.5. Circulating tumor DNA analysis to monitor small‐sized tumors, bone metastasis, and heterogenous tumors
Consistent with previous findings, ctDNA analysis concordantly tracked tumor response of RECIST‐measurable disease (Figure 4). However, for more challenging scenarios, such as patients showing only bone metastasis or small‐sized tumors below RECIST thresholds, the comparisons are less clear. In our study, 11 patients were defined as having nonmeasurable disease at pretreatment due to small tumor sizes that were below RECIST criteria (Table S3). Eight of the 11 patients had detectable ctDNA alterations. In patient CDK05 with tumor sizes below RECIST criteria, ctDNA analysis detected multigene genomic amplifications in FGFR3, FGFR1, MYC, and CCND1 at pretreatment. These CNVs reduced to basal levels during treatment, and reemerging during clinical progression at levels similar to pretreatment (Figure 5A). The ctDNA profiles in patient CDK05 were consistent with presence of liver target lesions observed at both pretreatment and disease progression. Similarly, in patient CDK14, driver mutations PIK3CA H1047R and TP53 Y220C detected at pretreatment increased in tandem with the lung target lesion until clinical progression (Figure 5B). Circulating tumor DNA analysis could be useful considering challenging and potentially inconclusive tumor response interpretation for patients who were not assessable using RECIST criteria.
FIGURE 4.
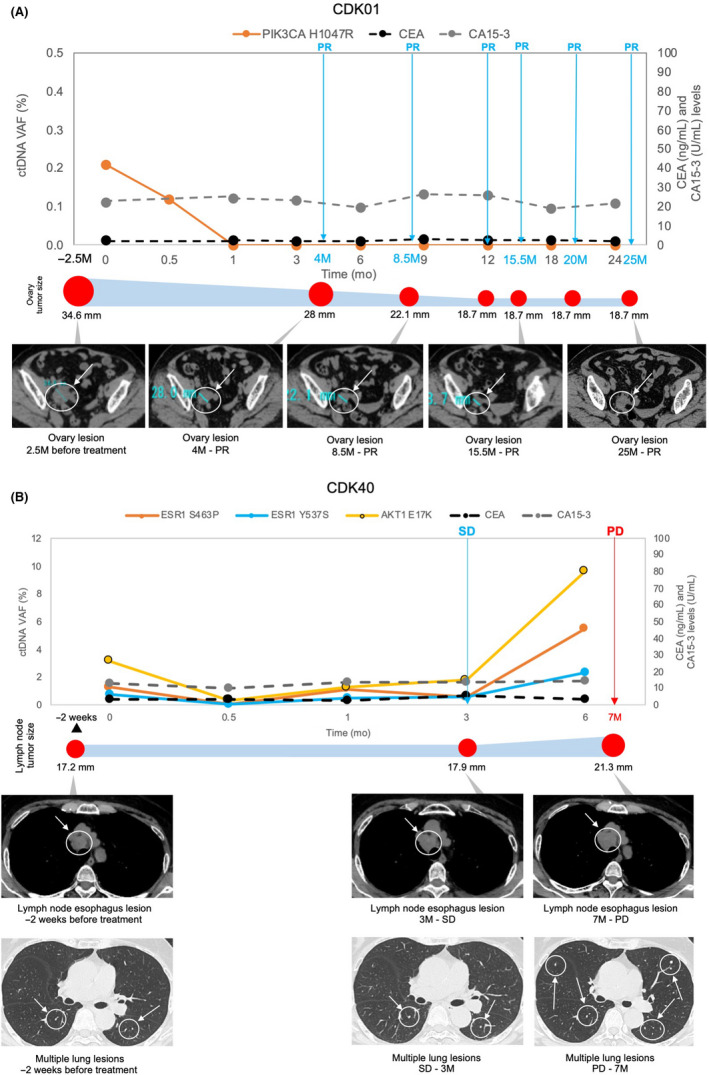
Circulating tumor DNA (ctDNA) analysis and radiographic images of RECIST‐measurable disease. (A) CDK01, (B) CDK40 ctDNA levels compared with tumor size changes. Target lesion sizes are shown as graphical representations corresponding to the radiographic assessment intervals and are denoted with arrows on the radiographic images. CA 15‐3, cancer antigen 15‐3; CEA, carcinoembryonic antigen; PD, progressive disease; PR, partial response; SD, stable disease; VAF, variant allele frequency
FIGURE 5.
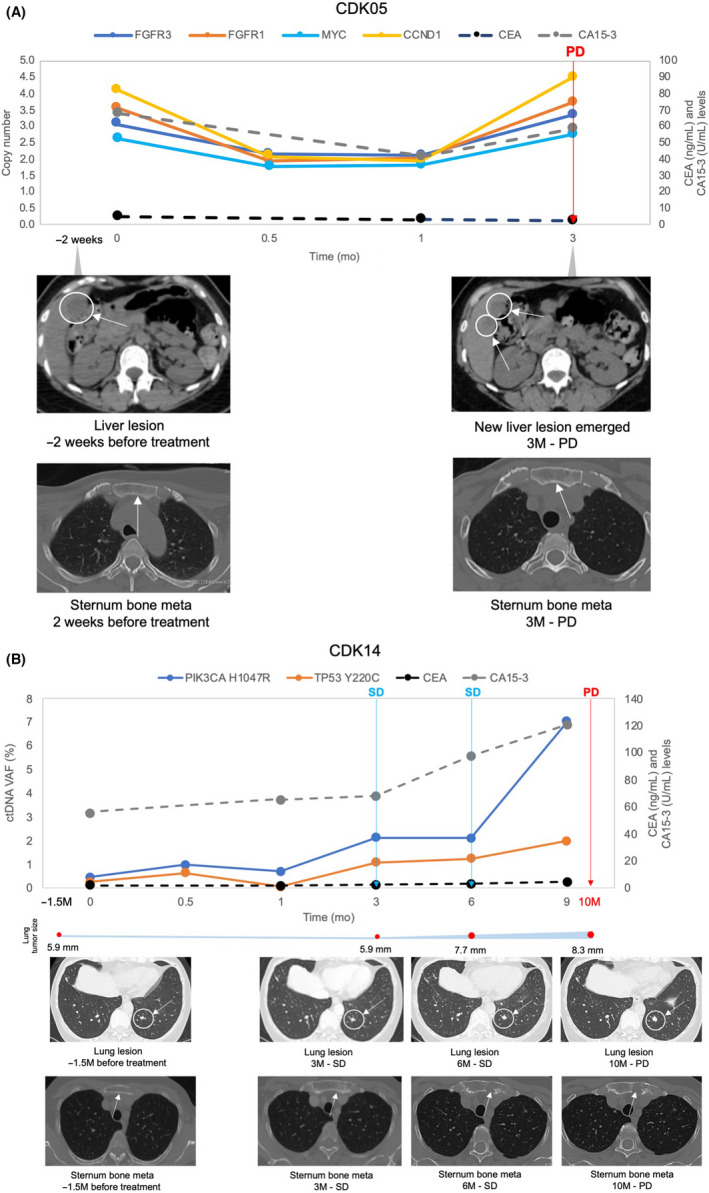
Circulating tumor DNA (ctDNA) analysis and radiographic images for tumor sizes below RECIST. (A) CDK05. (B) CDK14. RECIST tumor response is shown in colored arrows in the ctDNA profile. Target lesion size is shown as a graphical representation corresponding to the radiographic assessment intervals. CA 15‐3, cancer antigen 15‐3; CEA, carcinoembryonic antigen; CNV, copy number variant; CR, complete response; PD, progressive disease; SD, stable disease
Monitoring bone metastasis is challenging due to difficulty in interpreting radiologic images or in obtaining tissue biopsies. In the nine patients presenting with only bone metastasis at pretreatment, ctDNA analysis detected breast cancer‐related alterations in 67% (6 of 9) of patients (Table S3). In 4 of 6 bone metastasis patients (CDK04, CDK07, CDK16, and CDK21) with detected ctDNA alterations, ctDNA analysis detected rapid increase of pretreatment alterations prior to or at clinical progression, implying detection of possible tumor‐specific alterations reflecting resistance towards CDK4/6i treatment (Figure 6). Particularly for patients CDK04, CDK07, and CDK21, increase in pretreatment ctDNA alterations at clinical progression coincided with acquisition of new metastatic lesions in the liver and lung (Figure 6A,B,D). As for patient CDK16 (Figure 6C), pretreatment ctDNA mutation SF3B1 K700E reemerged and has shown rapid increase in ctDNA levels while tumor response continues to be defined as stable disease based on bone metastasis target lesion. Our findings highlight the potential clinical utility of liquid biopsy in complementing radiologic assessment of tumor response when interpretation is challenging or inconclusive.
FIGURE 6.
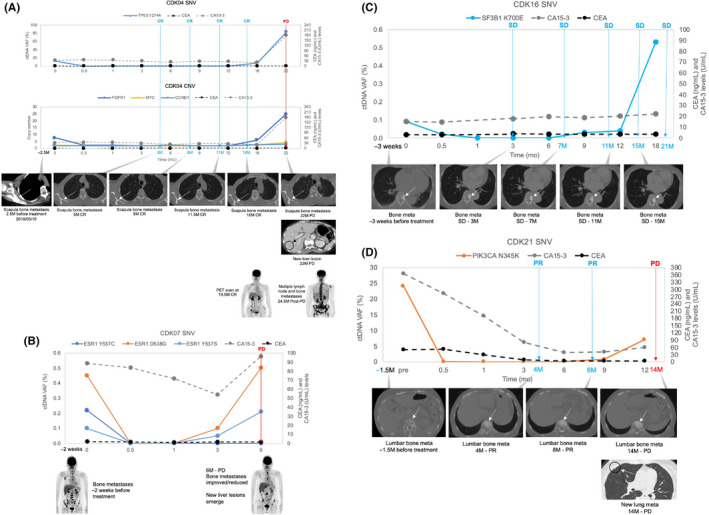
Circulating tumor DNA (ctDNA) analysis and radiographic images for bone metastasis target lesions. (A). CDK04, (B). CDK07, (C). CDK16, (D). CDK21. RECIST tumor response is shown in colored arrows in the ctDNA profile. CA 15‐3, cancer antigen 15‐3; CEA, carcinoembryonic antigen; CNV, copy number variant; CR, complete response; PD, progressive disease; PR, partial response; SD, stable disease; SNV, single nucleotide variant; VAF, variant allele frequency
Circulating tumor DNA analysis has the advantage of capturing genomic heterogeneity of multiple tumor lesions. In patient CDK15, a total of eight ctDNA mutations were detected within cycle 1 of treatment for serial monitoring, comprising of six PIK3CA and two ESR1 mutations (Figure S4). Diverse tumor responses were observed; ctDNA profiles 1, 2, and 4 show constant residual amounts of ctDNA variants throughout CDK4/6i treatment, followed by subsequent increase of ctDNA levels prior to clinical progression. The ctDNA analysis suggests partial inhibition of these tumors, eventually leading to resistance and clinical progression. Circulating tumor DNA profile 5 shows acquisition of AKT1 and PIK3CA mutations, previously undetected at pretreatment, near to or at clinical progression, possibly reflecting acquisition of the new liver lesion at disease progression.
3.6. Circulating tumor DNA analysis of acquired mutations associated with resistance
In some patients, acquired mutations might emerge due to treatment pressure. Evaluation focused on 12 of 20 patients who initially responded to therapy but later developed disease progression. Serial ctDNA analysis detected seven acquired genetic alterations from five patients in genes from several signaling pathways related to cell cycle checkpoint. Figure 7 shows acquired variants ERBB2 (HER2 tyrosine kinase pathway), ESR1 (estrogen receptor signaling) as well as in downstream genes such as PIK3CA (PI3K‐AKT signaling), MYC, and cyclin D1 (cell cycle checkpoint). These genes are known to play critical roles in the G1 to S transition in the cell cycle. We define “acquired variants” as ctDNA that was undetected within first cycle of CDK4/6i treatment (SNV count = 0 or CNV = 2.0‐fold) and detected at disease progression (SNV 0.2% or CNV 2.5‐fold). The VAF or copy numbers of paired pretreatment and PD time points are listed in the colored boxes directly below the detected variants. Five of the seven acquired alterations have clinical relevance as potential actionable targets for molecular‐based therapeutics, with ERBB2 amplification and PIK3CA N345K being FDA recognized biomarkers predictive of response to the corresponding therapy while AKT1 E17K, ESR1 D538G, and ESR1 Y537S show compelling clinical evidence predicting response to the targeted therapy (Table S4). We show some key examples. In patients CDK13 and CDK28, acquired ESR1 mutations, undetected in pretreatment samples, appeared prior to disease progression (Figure S5, Data S1 and S3). For patient CDK13, ESR1 mutation emerged when pretreatment liver target lesion increased in size leading up to disease progression (Figure S5). As for patient CDK28, ESR1 mutation was detected when a new liver lesion, previously undetected at pretreatment, was observed (Figure S5).
FIGURE 7.
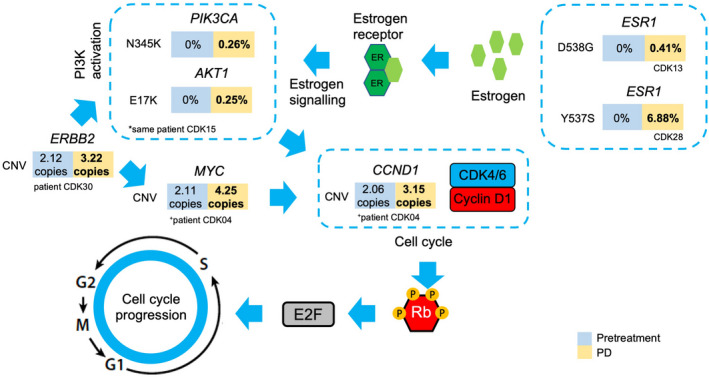
Newly detected circulating tumor DNA (ctDNA) alterations at disease progression linked to resistance. Arrows indicate the direction of pathway cascades leading to cell cycle checkpoint regulation. “Acquired variants” were ctDNA that were undetected within the first cycle of cyclin‐dependent kinase 4/6 (CDK4/6) inhibitor treatment (single nucleotide variant [SNV] count = 0 or copy number variant [CNV] = 2.0‐fold) and detected at disease progression (SNV ≥0.2% or CNV ≥2.5‐fold). Variant allele frequencies (VAF) or copy numbers of paired pretreatment and progressive disease (PD) time points are listed in the colored boxes directly below the detected variants
4. DISCUSSION
In this retrospective, longitudinal monitoring study of CDK4/6i therapeutic response, we show serial ctDNA analysis to be a highly informative biomarker that accurately captures the clonal dynamics of MBC tumors. This study presents comprehensive evaluation of MBC response to CDK4/6i through short interval serial ctDNA analysis, using targeted NGS in every interval (175 samples for 33 patients). Both DNA and RNA extraction was carried out as the ctDNA assay can detect somatic alterations as well as gene fusions from plasma. This approach did not impact recovery of cfDNA, achieving an average library conversion rate of 76%.
Our study brings additional insights regarding the biology of CDK4/6i response. Through granular time series ctDNA analysis, we were able to capture the initial rise of ctDNA levels, indicative of disease progression, well in advance of radiological assessment, in patients responding to CDK4/6i treatment. In addition, for patients who did not respond to CDK4/6i, sustained ctDNA detection in consecutive intervals within cycle 1 of CDK4/6i treatment enabled possible earlier detection of disease progression. The potential of ctDNA to detect early disease progression is crucial as it could provide an opportunity for earlier termination of CDK4/6i treatment, avoiding unnecessary CDK4/6i‐related side effects. The clinical implications of earlier detection of disease progression remains a contentious point for discussion, with very little data to support claims of improved clinical outcome. However, early data from the PADA‐1 study lends support to this claim. PADA‐1 is an ongoing ctDNA‐based interventional clinical trial, evaluating the efficacy of switching therapy upon detection of plasma ESR1 mutations in ER+HER2− MBC. 20 Patients were initially treated with combination of aromatase inhibitors and palbociclib as line 1 treatment. In a subcohort of patients with detected “rising” ESR1 mutations, patients were randomized (1:1) into two separate treatment arms: (a) aromatase inhibitors + palbociclib; and (b) fulvestrant + palbociclib. After further follow‐up of 34.5 months, the median PFS of patients who switched to fulvestrant was double compared to those who remained on aromatase inhibitor, 11.9 months vs 5.7 months. Although results are preliminary, data from the PADA‐1 study has indicated that early detection of plasma ESR1 mutations could guide treatment switch to another ER targeted combination with palbociclib, giving significant gain in PFS. 20
Our findings also build on previous studies, 13 , 14 , 15 , 21 providing a more detailed assessment of breast cancer clonal evolution during response to CDK4/6i. Other MBC liquid biopsy monitoring studies applied targeted NGS at pretreatment/baseline followed by personalized ddPCR of NGS alterations in subsequent surveillance 22 , 23 , 24 or serial targeted NGS focused on key intervals. 25 Our approach has the added advantage of detecting newly acquired alterations that emerge during CDK4/6i endocrine combination therapy, and precisely documenting the initial rise of ctDNA levels.
The prognostic role of ESR1 mutations is widely recognized as a resistance factor for endocrine therapy, 26 , 27 although not observed in the palbociclib and fulvestrant treatment arm of the phase III PALOMA clinical trial. 21 In the PALOMA‐3 study, when evaluating the predictive ability of baseline plasma ESR1 mutations on control arm (fulvestrant and placebo), results show shorter response for mutant ESR1 carriers (ESR1 mutants median PFS, 3.6 months [95% CI, 2‐5.4 months]; ESR1 WT median PFS, 5.5 months [95% CI, 3.5‐7.4 months]; P = .04). 14 In our study, we did not include a control arm (patients receiving only endocrine therapy) to evaluate predictive ability of baseline ESR1 mutations towards treatment efficacy. However, results from the PALOMA‐3 trial indicate that patients receiving only endocrine therapy, carrying ESR1 mutations at baseline, will not benefit from treatment, as shown by the shorter median PFS. In our study, patients carrying ESR1 mutations prior to treatment were unlikely to benefit from the CDK4/6i endocrine combination therapy, with the trend remaining statistically significant even after adjusting with known prognostic factors. Although potentially interesting, we acknowledge that the prognostic significance of ESR1 is suggestive and lacks external validation in a larger independent dataset.
Currently, clinical response of solid tumors is assessed by RECIST 1.1 criteria with target lesions selected based on size and suitability for repeated evaluations. Our results imply that the dynamic range of serial ctDNA analysis are concordant with radiologic assessments, supporting the adoption of ctDNA as a monitoring biomarker. When RECIST criteria is not applicable (bone lesions or target lesions below RECIST size), the mutation profiles detected from serial ctDNA analysis could complement radiographic imaging in providing a more decisive assessment of tumor response. In our analysis, we observed that liquid biopsy provided an earlier measure of treatment response, with increasing ctDNA levels detected before radiographic assessment of disease progression or changes in CEA and CA 15‐3 levels (Figures S2 and S3). This observation has been supported by other recent studies. 22 , 24
In this study, ctDNA analysis detected acquisition of new alterations in ERBB2, PIK3CA, AKT1, MYC, CCND1, and ESR1. These alterations might have emerged due to prolonged exposure to the CDK4/6i and endocrine combination therapy. These mutated genes are involved in dysregulation of key pathways upstream of the CDK4/6‐RB1, possibly rendering cancer cells resistant to CDK4/6 inhibition, in line with previous findings. 13 , 28 ESR1 mutations not only have significant clinical implications at the pretreatment stage, but also at disease progression as a possible mechanism in evading CDK4/6 and endocrine therapy inhibition. Some acquired alterations are clinically relevant as potential actionable targets for molecular based therapeutics, possibly aiding treatment selection after CDK4/6i treatment resistance.
Our study has its limitations. This study was designed to evaluate the clinical validity of liquid biopsy to monitor response to CDK4/6i. A larger population cohort is needed to confidently establish the association of ESR1 mutations towards CDK4/6i response. Further refining the granular time series and a larger cohort will help establish the clinical significance of ctDNA‐detected earlier treatment response and the key landmark intervals predictive of CDK4/6i response. The ctDNA alterations detected were largely known breast cancer associated markers. This is due to the ctDNA assay, with targeted alterations largely limited to known mutations in frequently occurring cancers to maximize assay sensitivity. This probably explains the lack of ctDNA detection in some patients, particularly CDK12 and CDK24 who rapidly developed PD. With genes that do not harbor hotspot mutations (eg, RB1), using hybrid‐capture assays covering more target regions at landmark intervals to guide ctDNA monitoring might improve detection sensitivity. Nevertheless, our findings have indicated possible clinical utility of serial ctDNA analysis for early detection of disease progression as well as real‐time monitoring of CDK4/6i response. Serial monitoring using targeted NGS liquid biopsy reveal a multifaceted view of MBC clonal evolution during CDK4/6i treatment. The findings from this study are crucial to advancing disease management of advanced breast cancer patients, in particular patients receiving CDK4/6i treatment.
DISCLOSURE
Y.M.C. reported employment with Cancer Precision Medicine, Inc. Japan during the submitted work. Y.N. reported receiving advisory fees and stocks from OncoTherapy Science, Inc. Japan outside the submitted work. S.‐K.L. reported receiving advisory fees from Cancer Precision Medicine, Inc. Japan outside the submitted work. The remaining authors declare no potential conflicts of interest.
Supporting information
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by the Council for Science, Technology and Innovation (CSTI), cross‐ministerial Strategic Innovation Promotion Program (SIP), “Innovative AI Hospital System”; Funding Agency: National Institute of Biomedical Innovation, Health and Nutrition (NIBIOHN); Grant SIPAIH18C03. We thank all contributing patients and their families as well as staff of the Breast Oncology Center, Cancer Institute Hospital, Japanese Foundation for Cancer Research.
Chin YM, Shibayama T, Chan HT, et al. Serial circulating tumor DNA monitoring of CDK4/6 inhibitors response in metastatic breast cancer. Cancer Sci. 2022;113:1808–1820. doi: 10.1111/cas.15304
Funding information
National Institute of Biomedical Innovation, Grant/Award Number: SIPAIH18C03; JSPS KAKENHI, Grant/Award Number: 20K07709
DATA AVAILABILITY STATEMENT
All data supporting the findings of this study are available within the article and its supplementary information/data files, and from the corresponding author upon reasonable request.
REFERENCES
- 1. Yu Q, Geng Y, Sicinski P. Specific protection against breast cancers by cyclin D1 ablation. Nature. 2001;411:1017‐1021. [DOI] [PubMed] [Google Scholar]
- 2. Yu Q, Sicinska E, Geng Y, et al. Requirement for CDK4 kinase function in breast cancer. Cancer Cell. 2006;9:23‐32. [DOI] [PubMed] [Google Scholar]
- 3. Cristofanilli M, Turner NC, Bondarenko I, et al. Fulvestrant plus palbociclib versus fulvestrant plus placebo for treatment of hormone‐receptor‐positive, HER2‐negative metastatic breast cancer that progressed on previous endocrine therapy (PALOMA‐3): final analysis of the multicentre, double‐blind, phase 3 randomised controlled trial. Lancet Oncol. 2016;17:425‐439. [DOI] [PubMed] [Google Scholar]
- 4. Johnston S, Martin M, Di Leo A, et al. MONARCH 3 final PFS: a randomized study of abemaciclib as initial therapy for advanced breast cancer. NPJ Breast Cancer. 2019;5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45:228‐247. [DOI] [PubMed] [Google Scholar]
- 6. Erdi YE. Limits of tumor detectability in nuclear medicine and PET. Mol Imaging Radionucl Ther. 2012;21:23‐28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Oxnard GR, Zhao B, Sima CS, et al. Variability of lung tumor measurements on repeat computed tomography scans taken within 15 minutes. J Clin Oncol. 2011;29:3114‐3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Duffy MJ, Evoy D, McDermott EW. CA 15–3: uses and limitation as a biomarker for breast cancer. Clin Chim Acta. 2010;411:1869‐1874. [DOI] [PubMed] [Google Scholar]
- 9. Laessig D, Nagel D, Heinemann V, et al. Importance of CEA and CA 15–3 during disease progression in metastatic breast cancer patients. Anticancer Res. 2007;27:1963‐1968. [PubMed] [Google Scholar]
- 10. Lauro S, Trasatti L, Bordin F, et al. Comparison of CEA, MCA, CA 15–3 and CA 27–29 in follow‐up and monitoring therapeutic response in breast cancer patients. Anticancer Res. 1999;19:3511‐3515. [PubMed] [Google Scholar]
- 11. Carr TH, McEwen R, Dougherty B, et al. Defining actionable mutations for oncology therapeutic development. Nat Rev Cancer. 2016;16:319‐329. [DOI] [PubMed] [Google Scholar]
- 12. Lo YM, Zhang J, Leung TN, Lau TK, Chang AM, Hjelm NM. Rapid clearance of fetal DNA from maternal plasma. Am J Hum Genet. 1999;64:218‐224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. O'Leary B, Cutts RJ, Liu Y, et al. The genetic landscape and clonal evolution of breast cancer resistance to palbociclib plus fulvestrant in the PALOMA‐3 trial. Cancer Discov. 2018;8:1390‐1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. O'Leary B, Hrebien S, Morden JP, et al. Early circulating tumor DNA dynamics and clonal selection with palbociclib and fulvestrant for breast cancer. Nat Commun. 2018;9:896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martinez‐Saez O, Pascual T, Braso‐Maristany F, et al. Circulating tumor DNA dynamics in advanced breast cancer treated with CDK4/6 inhibition and endocrine therapy. NPJ Breast Cancer. 2021;7:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chin YM, Takahashi Y, Chan HT, et al. Ultradeep targeted sequencing of circulating tumor DNA in plasma of early and advanced breast cancer. Cancer Sci. 2021;112(1):454‐464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pittella‐Silva F, Chin YM, Chan HT, et al. Plasma or serum: which is preferable for mutation detection in liquid biopsy? Clin Chem. 2020;66:946‐957. [DOI] [PubMed] [Google Scholar]
- 18. Chan HT, Nagayama S, Chin YM, et al. Clinical significance of clonal hematopoiesis in the interpretation of blood liquid biopsy. Mol Oncol. 2020;14:1719‐1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chakravarty D, Gao J, Phillips SM, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017;(1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bidard FC, Hardy‐Bessard A‐C, Bachelot T, et al. Fulvestrant‐palbociclib vs continuing aromatase inhibitor‐palbociclib upon detection of circulating ESR1 mutation in HR+ HER2‐ metastatic breast cancer patients: Results of PADA‐1, a UCBG‐GINECO randomized phase 3 trial. Paper presented at: San Antonio Breast Cancer Symposium; 2021. [Google Scholar]
- 21. O'Leary B, Cutts RJ, Huang X, et al. Circulating tumor DNA markers for early progression on fulvestrant with or without palbociclib in ER+ advanced breast cancer. J Natl Cancer Inst. 2021;113(3):309‐317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dawson SJ, Tsui DW, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368:1199‐1209. [DOI] [PubMed] [Google Scholar]
- 23. Zivanovic Bujak A, Weng CF, Silva MJ, et al. Circulating tumour DNA in metastatic breast cancer to guide clinical trial enrolment and precision oncology: a cohort study. PLoS Medicine. 2020;17:e1003363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hrebien S, Citi V, Garcia‐Murillas I, et al. Early ctDNA dynamics as a surrogate for progression‐free survival in advanced breast cancer in the BEECH trial. Ann Oncol. 2019;30:945‐952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gerratana L, Davis AA, Zhang Q, et al. Longitudinal dynamics of circulating tumor cells and circulating tumor DNA for treatment monitoring in metastatic breast cancer. JCO Precis Oncol. 2021;5:943‐952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chandarlapaty S, Chen D, He W, et al. Prevalence of ESR1 mutations in cell‐free DNA and outcomes in metastatic breast cancer: a secondary analysis of the BOLERO‐2 clinical trial. JAMA Oncol. 2016;2:1310‐1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schiavon G, Hrebien S, Garcia‐Murillas I, et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci Transl Med. 2015;7:313ra182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wander SA, Cohen O, Gong X, et al. The genomic landscape of intrinsic and acquired resistance to cyclin‐dependent kinase 4/6 inhibitors in patients with hormone receptor‐positive metastatic breast cancer. Cancer Discov. 2020;10:1174‐1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Data Availability Statement
All data supporting the findings of this study are available within the article and its supplementary information/data files, and from the corresponding author upon reasonable request.


