Abstract
Perioperative systemic chemotherapy improves the prognosis of upper tract urothelial carcinoma (UTUC). The first objective of this study was to verify whether perioperative circulating tumor DNA (ctDNA) analysis using a pan‐cancer gene panel and next‐generation sequencing could identify patients with poor prognosis who require perioperative chemotherapy. Second, we investigated whether ctDNA is useful for minimal residual disease (MRD) detection and treatment monitoring in UTUC. This study included 50 patients with untreated UTUC, including 43 cases of localized UTUC. We performed targeted ultradeep sequencing of plasma cell‐free DNA (cfDNA) and buffy coat DNA and whole‐exome sequencing of cancer tissues, allowing exclusion of possible false positives. We attempted to stratify the prognosis according to the perioperative ctDNA levels in patients with localized UTUC. In patients with metastatic UTUC, ctDNA was evaluated before, during, and after systemic treatment. In total, 23 (46%) of 50 patients with untreated UTUC were ctDNA positive, and 17 (40%) of 43 patients with localized UTUC were ctDNA positive. Of the detected TP53 mutations, 19% were false positives due to clonal hematopoiesis of indeterminate potential. Among preoperative risk factors, only the preoperative ctDNA fraction>2% was a significant and independent risk factor associated with worse recurrence‐free survival (RFS). Furthermore, the existence of ctDNA early points after the operation was significantly associated with worse RFS, suggesting the presence of MRD. ctDNA also showed a potential as a real‐time marker for systemic therapy in patients with metastatic UTUC. Detection of ctDNA may indicate potential metastasis and guide decisions on perioperative chemotherapy.
Keywords: cell‐free DNA, circulating tumor DNA, next‐generation sequencing, upper tract urothelial carcinoma
In this study, we performed ultradeep sequencing of cell‐free DNA (cfDNA) in the plasma of 50 patients with untreated upper tract urothelial carcinoma (UTUC), of whom 23 (46%) were circulating tumor DNA (ctDNA) positive. Among the preoperative risk factors, only a preoperative ctDNA proportion >2% was found to be a significant and independent risk factor associated with worse recurrence‐free survival (RFS). Furthermore, the presence of ctDNA in the early postoperative period was significantly associated with worse RFS, suggesting the presence of minimal residual disease.

Abbreviations
- CHIP
clonal hematopoiesis of indeterminate potential
- CNVs
copy‐number variants
- CR
complete response
- ctDNA
circulating tumor DNA
- HR
hazard ratio
- ICI
immune checkpoint inhibitor
- MAF
mutant allele frequency
- MRD
minimal residual disease
- OS
overall survival
- RFS
recurrence‐free survival
- SD
stable disease
- SNVs
single nucleotide variants
- UTUC
upper tract urothelial carcinoma
1. INTRODUCTION
Upper tract urothelial carcinoma is uncommon cancer that accounts for ~5%–10% of all urothelial carcinomas. 1 , 2 In total, ~60% of UTUCs are invasive at diagnosis (compared with 15%–25% of bladder cancers), 3 while ~7% are metastases. 4 Standard treatment for patients with nonmetastatic UTUC is radical nephroureterectomy, while systemic treatment with platinum‐based chemotherapy or immune checkpoint inhibitors is administered for metastatic UTUC. Patients with UTUC are often diagnosed at a relatively advanced stage and showed a poor prognosis. Multidisciplinary treatment with perioperative chemotherapy is used to improve the prognosis in locally advanced UTUC. 5 The randomized phase III POUT trial of platinum‐based adjuvant chemotherapy in patients with high‐risk UTUC (pT2‐4 or N+) showed that adjuvant chemotherapy significantly improved disease‐free survival, demonstrating the benefit of perioperative chemotherapy. 6 However, questions remain regarding the optimal timing of perioperative chemotherapy in patients with UTUC. Neoadjuvant chemotherapy before radical cystectomy has been shown to improve survival in patients with muscle‐invasive bladder cancer. 7 Although neoadjuvant chemotherapy is generally recommended as perioperative chemotherapy for advanced UTUCs—especially as patients are ineligible for chemotherapy due to postoperative renal dysfunction and complications—inaccurate clinical staging might lead to overtreatment. 8 Therefore, a biomarker is needed that can preoperatively identify patients with poor prognosis UTUC who will benefit from perioperative chemotherapy.
Radiological imaging studies, including computed tomography and magnetic resonance imaging, cannot accurately predict pathological depth or detect postoperative MRD. Circulating tumor DNA is a promising biomarker for noninvasive molecular profiling, monitoring, and predicting the response to systemic therapy. 9 , 10 , 11 , 12 , 13 , 14 Studies have shown that ctDNA is a reliable biomarker for detecting MRD in various cancers. 15 , 16 , 17 , 18 , 19 , 20 , 21 , 22 In one study on muscle‐invasive bladder cancer, researchers performed next‐generation sequencing on patients’ primary tumors, then used the data to develop a customized, patient‐specific gene panel for ctDNA detection, which could then be used to test for metastatic recurrence after total cystectomy. 23 It has also been shown that the ctDNA fraction >2% is associated with worse OS in metastatic urothelial carcinoma. 24 Because preoperative tissue collection is more challenging for UTUC than bladder cancer, a plasma‐based deep sequencing approach may be preferable for UTUC. However, such sequencing approaches are subject to false positives due to white blood cell‐derived variants of cfDNA associated with CHIP. 25 , 26 , 27 Previous studies on ctDNA in UTUC have been in the setting of metastatic disease and have not excluded such false positives. 28
We hypothesized that a platform to detect ctDNA while excluding false positives would enable clinicians to monitor the response to systemic therapy and detect MRD.
The first objective of this study was to verify whether perioperative ctDNA analysis using a pan‐cancer gene panel and next‐generation sequencing could identify patients with poor prognosis who required perioperative chemotherapy. Second, we investigated whether ctDNA is useful for MRD detection and treatment monitoring in UTUC.
We performed a multiomics analysis using whole‐exome sequencing and transcriptome analysis of cancer tissues and targeted sequencing of plasma cfDNA and buffy coat DNA.
2. MATERIALS AND METHODS
2.1. Study design
Between January 2016 and January 2020, 50 patients with UTUC were enrolled in this study; no patients had developed other signs of active cancer at the time of enrollment. The study was approved by the Institutional Review Board of Osaka University Hospital (#13397‐14). Blood sampling was performed before and after treatments for all patients with UTUC. In some patients, multiple blood draws were performed over time. All patients were pathologically diagnosed by surgically resected specimen or needle biopsy specimen. Histological diagnosis was determined based on standard H&E‐stained sections. Two or more experienced senior pathologists evaluated the pathological diagnosis according to the 7th American Joint Committee on Cancer TNM staging system (AJCC 2010). RFS and OS were calculated using the Kaplan–Meier method from the date of surgery. Treatment response was evaluated according to RECIST criteria by radiological imaging.
2.2. Extraction of plasma cfDNA and buffy coat DNA from blood samples
Whole blood (2.0–7.0 ml) was collected directly into EDTA tubes. Within 3 h of collection, all blood samples were centrifuged sequentially at 900 and 20,000 g for 10 min each, and supernatants were stored at −80°C as plasma. cfDNA was collected from 0.6–2.8 ml plasma samples using the QIAamp Circulating Nucleic Acid Kit® (QIAGEN) according to the manufacturer's protocol. Buffy coat DNA was isolated from blood lymphocytes using the QIAamp DNA Blood Mini kit® (QIAGEN, Hilden, Germany) according to the manufacturer's protocol.
2.3. Library preparation and targeted next‐generation sequencing
Library preparation was performed using the amplicon‐based Oncomine Pan‐Cancer Cell‐Free Assay® according to the manufacturer's protocol (Thermo Fisher Scientific) with an input of 1.7–20 ng cfDNA, as described previously. 29 The panel comprised 272 multiplex PCR primers that covered 969 hotspot SNVs and 12 copy‐number variations across 50 genes. Samples were barcoded for multiplexing using the Tag Sequencing Barcode system (Thermo Fisher Scientific), then purified with AMPure XP beads® (Beckman Coulter) and quantified using the Ion TaqMan Quantification Kit® (Thermo Fisher Scientific). Libraries were multiplexed for templating on the Ion Chef Instrument and sequenced on the Ion Gene Studio S5 Prime System® using the Ion 540 Chip Kit®. Buffy coat DNA was mechanically sheared to 150 bp to mimic the average DNA length of cfDNA before library construction. A similar methodology for cfDNA was applied to DNA extracted from the buffy coat with an input of 40 ng.
2.4. Targeted sequencing data analysis and statistical analysis
Alignment of sequencing data was performed using the torrent suite Software version 5.8.0 (Thermo Fisher Scientific) using the default parameters of the torrent mapping alignment program (TMAP). The BAM files generated were analyzed and were then further analyzed using Oncomine TagSeq Pan‐Cancer Liquid Biopsy w2.4 version 5.10 workflow for variant calling. A minimum of three reads with the same barcode was required to form a functional family, and a minimum of two variants supporting functional families was required to make SNP, MNP, and indel calls.
2.5. Estimation of ctDNA fraction
ctDNA fractionation was estimated based on allelic fractionation of somatic mutations in autosomal bodies, as described previously. 30
Because MAF in diploid chromosomes is highest when mutations, except somatic mutations in genes with detectable amplification, are combined with LOH, we assumed that all mutations could be associated with LOH. In this situation, the MAF and ctDNA fractions are related as MAF = (ctDNA ×1)/[(1 − ctDNA) ×2 + ctDNA ×1], so ctDNA =2/((1/MAF) +1).
2.6. Preparation of genomic DNA and total RNA from cancer tissue
Upper tract urothelial carcinoma tissues with surgical resection or needle biopsy were frozen and preserved at −80°C. Genomic DNA was isolated from UTUC tissue using the QIAamp DNA Mini kit® (QIAGEN) according to the manufacturer's protocol. Tissue samples were also taken to soak in RNAlater® (Thermo Fisher Scientific) and stored at −20°C for stable storage. Total RNA was isolated from UTUC tissue using the RNeasy mini kit® (QIAGEN) according to the manufacturer's protocol.
2.7. Whole‐exome sequencing
The whole‐exome sequencing of genomic DNA from UTUC tissue as well as matched germline samples were performed using target capture with Agilent SureSelect XT Human All Exon V6 (Agilent Technologies). The raw sequence data were generated using the Illumina NovaSeq6000 platform (Illumina) with a standard 150‐bp paired‐end read protocol at Macrogen Japan.
2.8. Analysis of somatic mutations
For our sequencing data, FASTQ files were generated by bcl2fastq2. Candidate somatic mutations were identified using the Genomon pipeline (https://github.com/Genomon‐Project/genomon‐docs/tree/v2.0). The human reference file that was used was GRCh37/hg19. The candidate mutations in a tumor sample were identified using the following criteria: (i) Fisher's exact p ≤ 0.1; (ii) ≥5 variant reads in the tumor sample; (iii) variant allele frequency (VAF) in the tumor sample ≥0.08; and (iv) VAF of the matched normal sample <0.07, with the exclusion of synonymous SNVs and known variants listed in NCBI dbSNP build 131.
2.9. Analysis of somatic copy‐number alterations
Copy‐number aberrations were quantified and reported for each bed size as the segmented, normalized, log2‐transformed exon coverage ratio between each tumor sample and its matched normal sample.
2.10. RNA sequencing
RNA integrity was verified using an Agilent 2100 Bioanalyzer with RNA Nano reagents (Agilent Technologies). High‐quality RNA from 27 UTUC tissue samples was subjected to polyA+ selection and chemical fragmentation, and the 100‐bp RNA fraction was used to construct cDNA libraries using the TruSeq Stranded mRNA Prep kit (Illumina) according to the manufacturer's protocol. These paired‐end libraries were sequenced using the Illumina NovaSeq6000 platform with a standard 100‐bp paired‐end read protocol at Macrogen Japan.
2.11. Analysis of gene expression
Gene expression values were estimated from the RNA‐seq data from the tumor samples using the Genomon pipeline (https://github.com/Genomon‐Project/genomon‐docs/tree/v2.0). The alignment was performed with the STAR aligner (v.2.5.2a) against the hg19 human genome. BAM files named Aligned.sortedByCoord.out.bam, which was generated by the STAR, were used to quantify the expression data using GenomonExpression.
2.12. Hierarchical clustering to infer UTUC subtypes
The UTUC cohort was classified using previously published subtype‐specific gene signatures. For this purpose, normalized expression values corresponding to BASE47 signature genes were extracted from RNA‐seq results. Heatmaps were generated using R Studio's ComplexHeatmap v2.6.0 package, and all clustering was performed using average linkage clustering with the central correlation similarity metric.
2.13. Statistical analysis
Patient and cfDNA characteristics were presented as median + range, and correlation analysis was performed using the Wilcoxon test. RFS and OS were calculated using the Kaplan–Meier method. Differences among the two groups were assessed by log‐rank test and considered statistically significant when the p‐value was <0.05. Univariate and multivariate Cox proportional hazard analysis was performed to assess the relative contributions of perioperative ctDNA status and clinicopathological factors for RFS. All statistical analysis was performed in JMP Pro v.15.0.0 (SAS Institute Inc.).
3. RESULTS
3.1. Overall approach
We enrolled 50 patients with UTUC; clinicopathological information for these patients is shown in Table 1. In total, 43 patients with nonmetastatic cancer underwent radical surgery (nephroureterectomy); seven patients with metastatic disease were treated with systemic therapy (platinum‐based chemotherapy or ICI). The workflow of blood and cancer tissue sampling in the 50 patients is shown in Figure 1. Before the initial treatment, blood samples were collected to obtain plasma cfDNA and buffy coat DNA. Cancer tissue samples were collected at the time of radical surgery or needle biopsy. For patients with nonmetastatic cancer, blood samples were collected before postoperative adjuvant chemotherapy and within 3 months after surgery. In our institution, pT3‐4 or N+ stage patients are eligible for adjuvant chemotherapy, but 10 of 21 patients (48%) could not be treated due to postoperative renal dysfunction or complications. For patients with metastatic cancer, blood samples were collected before, during, and after systemic treatment, and during the follow‐up period as appropriate. Plasma cfDNA and buffy coat DNA were extracted from blood samples and subjected to targeted sequencing. We performed whole‐exome sequencing and transcriptome analysis of DNA and RNA recovered from cancer tissues.
TABLE 1.
Patient characteristics
| Nonmetastatic cancer (n = 43) | Metastatic cancer (n = 7) | |
|---|---|---|
| Age (years) [median (range)] | 76 (58–88) | 73 (62–80) |
| Gender, n (%) | ||
| Male | 36 (84) | 7 (100) |
| Female | 7 (16) | 0 (0) |
| BMI (kg/m2) [median (range)] | 22.9 (16.8–33.5) | 21.6 (18.4–27.4) |
| Smoking history, n (%) | ||
| Yes | 33 (77) | 5 (71) |
| No | 10 (23) | 2 (29) |
| Tumor location, n (%) | ||
| Renal pelvis | 17 (40) | 6 (86) |
| Ureter | 25 (58) | 0 (0) |
| Both | 1 (2) | 1 (14) |
| Urine cytology, n (%) | ||
| 1–3 | 24 (56) | 3 (43) |
| 4–5 | 19 (44) | 4 (57) |
| Hydronephrosis, n (%) | ||
| Yes | 18 (42) | 5 (71) |
| No | 25 (58) | 2 (29) |
| UBC history, n (%) | ||
| Former | 12 (28) | 2 (29) |
| Never | 31 (72) | 5 (71) |
| NLR [median (range)] | 2.6 (1.3–9.4) | 6.9 (2.6–14.0) |
| Clinical T stage, n (%) | ||
| cTa,is,1 | 14 (33) | 0 (0) |
| cT2 | 22 (51) | 2 (29) |
| cT3 | 7 (16) | 2 (29) |
| cT4 | 0 (0) | 3 (42) |
| Clinical N stage, n (%) | ||
| cN0 | 42 (98) | 1 (14) |
| cN1 | 1 (2) | 2 (29) |
| cN2 | 0 (0) | 4 (57) |
| Clinical M stage, n (%) | ||
| cM0 | 43 (100) | 3 (43) |
| cM1 | 0 (0) | 4 (57) |
| Pathological T stage, n (%) | ||
| pTa,is,1 | 18 (42) | |
| pT2 | 7 (16) | |
| pT3 | 17 (40) | |
| pT4 | 1 (2) | |
| Pathological N stage, n (%) | ||
| pN0 | 29 (67) | |
| pN1 | 5 (12) | |
| pN2 | 2 (5) | |
| pNx | 7 (16) | |
| CIS, n (%) | ||
| Negative | 38 (88) | |
| Positive | 5 (12) | |
| Lymphovascular invasion, n (%) | ||
| Negative | 23 (53) | |
| Positive | 20 (47) | |
| Adjuvant chemotherapy, n (%) | ||
| Yes | 11 (26) | |
| No | 32 (74) | |
Abbreviations: CIS, concomitant carcinoma in situ; NLR, neutrophil‐to‐lymphocyte ratio; UBC, urothelial bladder cancer.
FIGURE 1.
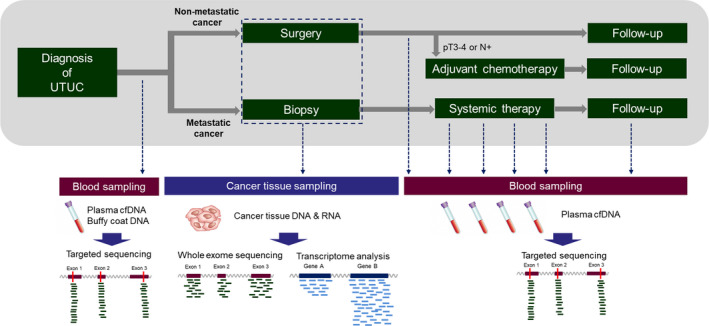
Study design. To render longitudinal plasma analysis comparable, sample collection was done before and within 3 months after surgery in patients with nonmetastatic upper tract urothelial carcinoma (UTUC). In patients with metastatic UTUC, sample collection was done before and during systemic chemotherapy or immunotherapy. Plasma cfDNA and buffy coat DNA were subjected to targeted sequencing with next‐generation sequencing. Cancer tissues were harvested by surgery or biopsy. Genomic DNA and total RNA were extracted for whole‐exome sequencing and transcriptome analysis
3.2. Identification of circulating tumor DNA (ctDNA) variants in plasma cfDNA
Targeted sequencing of plasma cfDNA in the 50 UTUC patients at untreated time points detected 47 genetic alterations in 25 patients (50%) (Table S1). The median sequence depth of ctDNA was 1516 (478–4661), and the median distribution of the ctDNA fraction was 3.1 (0.14%–71.5%). Detected genetic alterations included TP53 (53%), PIK3CA (4%), FGFR3 (4%), KRAS (4%), MAP2K1 (4%), and ERBB2 (4%) (Figure 2A). When these genetic alterations were classified by oncogenic signaling pathway, most were associated with the p53 and RTK/RAS pathways. (Figure 2B). To exclude false positives due to genetic alterations derived from normal cells, such as CHIP, we compared the sequencing of plasma cfDNA, buffy coat DNA, and cancer tissue DNA. According to our flowchart, we determined positive, negative, and false‐positive results for circulating tumor DNA (ctDNA) (Figure S1). Whole‐exome sequencing we performed on cancer tissue DNA and matching buffy coat DNA did not allow us to evaluate CHIP due to insufficient depth. The pan‐cancer gene panel used in this study sequenced limited regions, including hotspot mutations, for genes other than TP53. Therefore, as described in Figure S1, the genes other than TP53 can be considered as ctDNA when SNVs, indels, or CNVs are detected, and not as false positives by CHIP. Conversely, as TP53 does not have hotspots, we sequenced all exomes for TP53 only. For these reasons, we could not exclude the possibility of CHIP for TP53 so, in cases in which TP53 SNVs, indels, or CNVs were detected, we re‐sequenced buffy coat DNA using the same assay as ctDNA. Of all the TP53 mutations detected at untreated time points, 52% were detected only in plasma, and 29% were matched in plasma and cancer tissue. The remaining 19% were detected in plasma and buffy coat DNA, suggesting the false positives derived from CHIP (Figure 2C and Table S2). After excluding false positives for TP53 mutations, we found that 23 of 50 (46%) patients with UTUC were ctDNA positive. Of all cases, 8 (16%) had ctDNA fraction ≤2% and 15 (30%) were higher than 2% (Figure 2D). Further analysis indicated that the ctDNA positivity rate and ctDNA fraction increased with the progression of histopathological depth (Figure 2E). The clinicopathological information and the landscape of genetic alterations in the ctDNA‐positive group are shown in Figure 2F. Clinical stage Ⅳ means patients with metastasis. Of 18 patients with a postoperative diagnosis of pathological T3‐4, only five (28%) had a preoperative diagnosis of T3‐4. Among seven patients with metastatic cancer, six (86%) were ctDNA positive, while ctDNA were detected in 17 patients (40%) of 43 patients with nonmetastatic cancer, suggesting that the detection rate of ctDNA depended on the clinical stage of UTUC. We classified 16 cases into luminal type and 11 cases into basal type based on mRNA expression obtained from transcriptome analysis. There was no significant difference in the detection rate of ctDNA for each subtype (Figures 2F, and S2).
FIGURE 2.
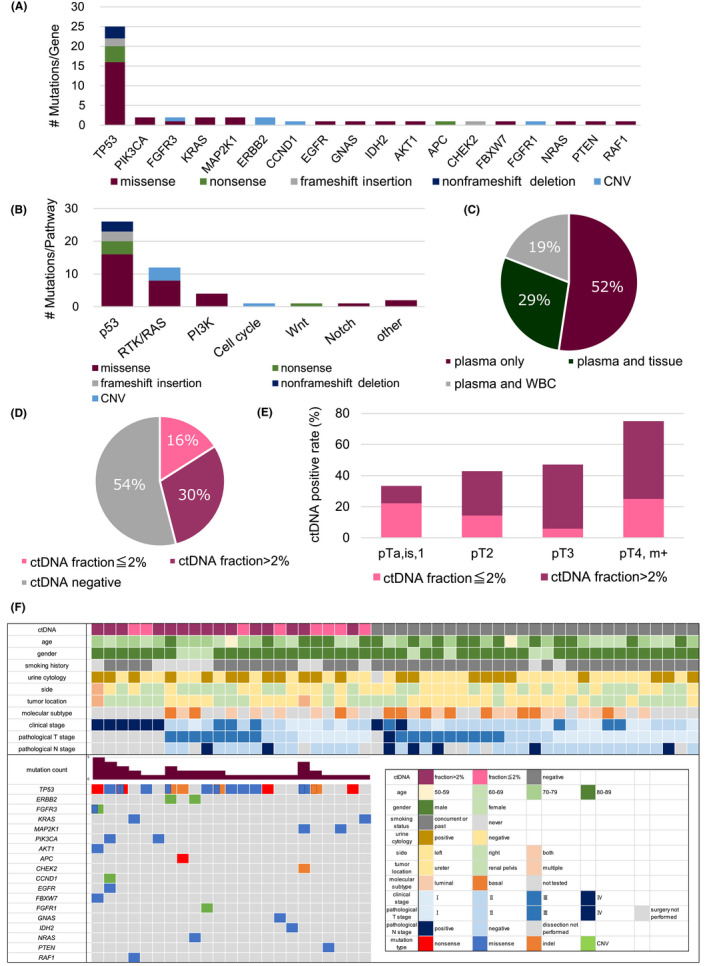
Identification of circulating tumor DNA (ctDNA) variants in plasma cell‐free DNA (cfDNA). Distribution graphs depict genomic alterations (A) and classification of oncogenic signaling pathways (B) of plasma cfDNA variants detected in 25 patients with upper tract urothelial carcinoma. (C). Pie chart showing the rate of concordance between TP53 mutations in plasma cfDNA and tumor DNA or WBC DNA. (D). Pie chart showing the positivity rate of ctDNA in 50 patients with UTUC after exclusion of false positives. (E). Bar chart showing ctDNA positivity rate at different histopathological stages. Patients with metastatic cancer (m+) were classified into the pT4 group because their pathological stage was not obtained and they had a poor prognosis. (F). Clinicopathological demographics of cases, colored according to their categorization (bottom right) for ctDNA‐positive (top left) and ctDNA‐negative groups (top right), respectively. The mutational landscape of each patient in the ctDNA‐positive group is also shown (bottom left). Clinical stage Ⅳ means cancers with metastasis
3.3. Association of preoperative ctDNA status and prognosis in localized UTUC
We investigated the association of ctDNA status at the preoperative time points with RFS and OS in 43 patients with localized UTUC. At baseline, the ctDNA fraction >2% group had significantly worse RFS (p < 0.0001) and OS (p = 0.0027) compared with the ctDNA fraction ≤2% or negative group (Figure 3A,B). There was no statistically significant difference in the RFS between ctDNA‐negative patients and ≤2% patients. As shown in Figure 3A, only four of the 32 patients who were ctDNA negative or ≤2% had postoperative recurrences, which meant these patients had a good prognosis. Multivariate Cox proportional hazard analysis revealed that only the ctDNA fraction >2% was significantly and independently associated with worse RFS (HR =4.565, 95% CI =1.433–14.539, p = 0.0102) (Figure 3C).
FIGURE 3.
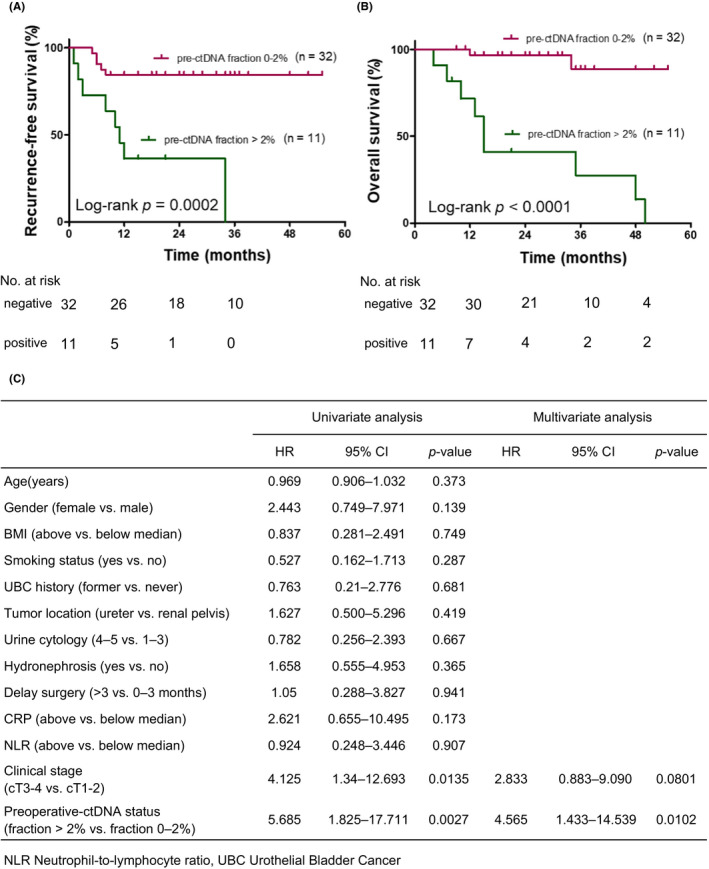
Preoperative ctDNA and recurrence‐free survival (RFS)/overall survival (OS) using Kaplan–Meier survival estimates. RFS (HR =13.75, 95% CI =3.50–54.03, p = 0.0002; (A)) and OS (HR =32.93, 95% CI =7.87–137.8, p < 0.0001; (B)])in preoperative ctDNA fraction >2% (n = 11) versus preoperative ctDNA fraction ≤2% or negative (n = 32). (C). Multivariate analysis results are shown
3.4. Postoperative ctDNA detects clinically significant MRD
We investigated whether postoperative ctDNA could detect postoperative MRD in patients with nonmetastatic cancer who had undergone radical surgery. Among 43 such patients in our cohort, we followed the clinical course of all 17 patients who were ctDNA positive at the preoperative stage (Figure 4A). Of 17 patients with preoperative ctDNA‐positive results, seven patients had postoperative ctDNA negative results. Patient UTUC138 was initially ctDNA negative after surgery, but ctDNA was detected at 30 months after surgery. At 34 months after the surgery, lymph node metastasis was observed by radiological imaging, indicating that cfDNA analysis detected the metastasis 4 months earlier than radiological imaging. For patient UTUC101, the ctDNA was sustained after the operation, and this patient was confirmed to have muscle‐invasive bladder cancer at 8 months after radical surgery. After radical cystectomy for muscle‐invasive bladder cancer, ctDNA disappeared and no further evidence of cancer was detected. Among 17 preoperative ctDNA‐positive cases, those with residual ctDNA after surgery had significantly poorer RFS (p = 0.0125, HR =6.259, 95% CI =1.485–26.38) and OS (p = 0.0065, HR =9.867, 95% CI =1.899–51.28) compared with those who were ctDNA negative (Figure 4B,C). We concluded that ctDNA is informative for the detection of MRD after radical surgery in patients with UTUC. Multivariate Cox proportional hazard analysis revealed that the postoperative ctDNA and pathological T stage were significantly and independently associated with worse RFS (HR = 4.565, 95% CI = 1.433–14.539, p = 0.0102; Figure 4D). Based on these results, we concluded that ctDNA is informative for the detection of MRD after radical surgery in patients with UTUC.
FIGURE 4.
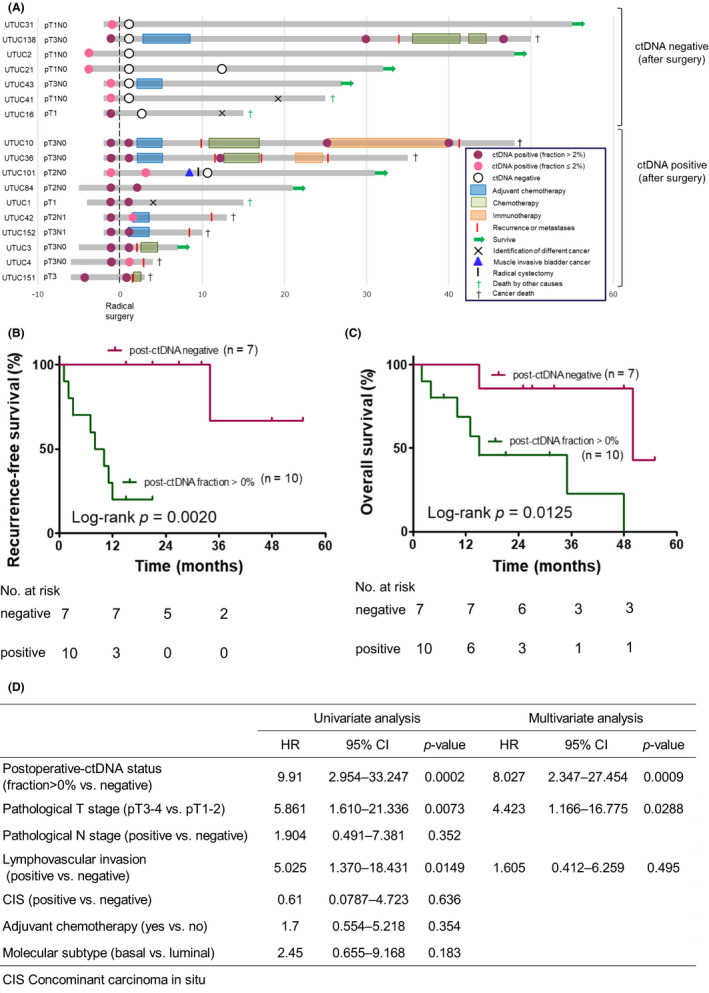
(A) Longitudinal monitoring of ctDNA levels during clinical disease course in patients with nonmetastatic UTUC. Individual swimmer plots for each patient showing the duration of treatment after surgery for the ctDNA‐negative cohort (upper panel, n = 7) and the ctDNA‐positive cohort (lower panel, n = 10). Horizontal lines represent the clinical course of each patient. Circles represent ctDNA status (red and white circles mean positive and negative, respectively). Red bars indicate recurrence or metastasis as evaluated by radiological images. Patients UTUC41, UTUC16, and UTUC1 developed cholangiocarcinoma, acute lymphocytic leukemia (ALL), and esophageal cancer, respectively. Postoperative ctDNA and recurrence‐free survival (RFS)/overall survival (OS) using Kaplan–Meier survival estimates. RFS (HR =9.26, 95% CI =2.25–38.10, p = 0.0020; (B)) and OS (HR =6.26, 95% CI =1.49–26.38, p = 0.013; (C)) in postoperative ctDNA detectable (n = 10) versus undetectable (n = 7). (D) Multivariate analysis results are shown
3.5. Longitudinal measurement of ctDNA in patients with metastatic cancer
Next, to verify whether ctDNA reflected the treatment effects, we performed sequential measurements of ctDNA in patients with metastatic cancer who received systemic therapy. The clinical course of all seven patients with metastatic cancer is shown in Figure S3. Patients with metastatic cancer, except for one case, were found to be ctDNA positive at more than one time point (Figure S3). Patient UTUC23 was a 73‐year‐old man with left renal pelvic cancer (cT4N2M1). Single nucleotide variants of FGFR3, FBXW7, AKT1, and TP53 and CNVs of FGFR3 were detected in plasma cfDNA at the untreated time points. We collected tissue from the primary lesion and a pelvic lymph node metastasis by needle biopsy. Whole‐exome sequencing of these tissues revealed SNV consistent with ctDNA of FGFR3, AKT1, and TP53 in the primary lesion, but SNV of FBXW7 was not detected. In contrast, SNVs consistent with all ctDNAs, including FBXW7, were detected in pelvic lymph node metastases. This result supports the existing paradigm that ctDNA reflected systemic tumor information, a phenomenon that can be explained by tumor heterogeneity. We performed cisplatin‐based anticancer chemotherapy as the initial treatment for patient UTUC23. The response to anticancer chemotherapy was SD on radiological imaging following the RECIST 1.1 criteria. Subsequent treatment with an ICI was successful, leading to a CR. Sequencing of plasma cfDNA after cisplatin‐based chemotherapy showed a slight decrease in total MAF from the baseline; FBXW7 SNVs disappeared, but FGFR3, AKT1, and TP53 SNVs remained. No genetic alterations were detected in plasma cfDNA at the time of confirmation of CR by radiological imaging during ICI therapy (Figure 5A). Although patient UTUC23 developed colorectal cancer at 28 months, this patient was found in a different analysis to have Lynch syndrome, and it is not uncommon for patients with Lynch syndrome to have UTUC and colorectal cancer. In a second case, patient UTUC62 represented a 77‐year‐old man with left renal pelvic cancer (cT3N2M0) who was initially treated with cisplatin‐based chemotherapy, resulting in 28% tumor shrinkage and judged as SD by radiological imaging. Subsequently, the patient was treated with ICI therapy, but liver metastasis appeared 3 months after beginning treatment. Platinum‐based chemotherapy was readministered, but the disease progressed. Finally, he died of cancer 18 months after diagnosis. We found that SNVs of RAF1 and KRAS as ctDNA observed at baseline increased or decreased reflecting the disease status (Figure 5B). In the third case, patient UTUC25 represented a 67‐year‐old male patient with left renal pelvic cancer (cT2N0M1) who had responded well to cisplatin‐based chemotherapy as initial therapy. At 18 months after diagnosis, the primary lesion increased in size and muscle‐invasive bladder cancer was detected, and treatment with ICI was initiated. The patient did not respond to ICI treatment and died of cancer 22 months after initial diagnosis. The SNVs of PIK3CA, TP53, and EGFR as ctDNA at baseline were eliminated during anticancer chemotherapy, but the SNVs of PIK3CA and TP53 were re‐detected during immunotherapy after progression (Figure 5C). In patient UTUC25, the TP53 V173L mutation was also detected in buffy coat DNA and determined to be CHIP. TP53 V173L was detected in all three‐time points, regardless of treatment or cancer status.
FIGURE 5.
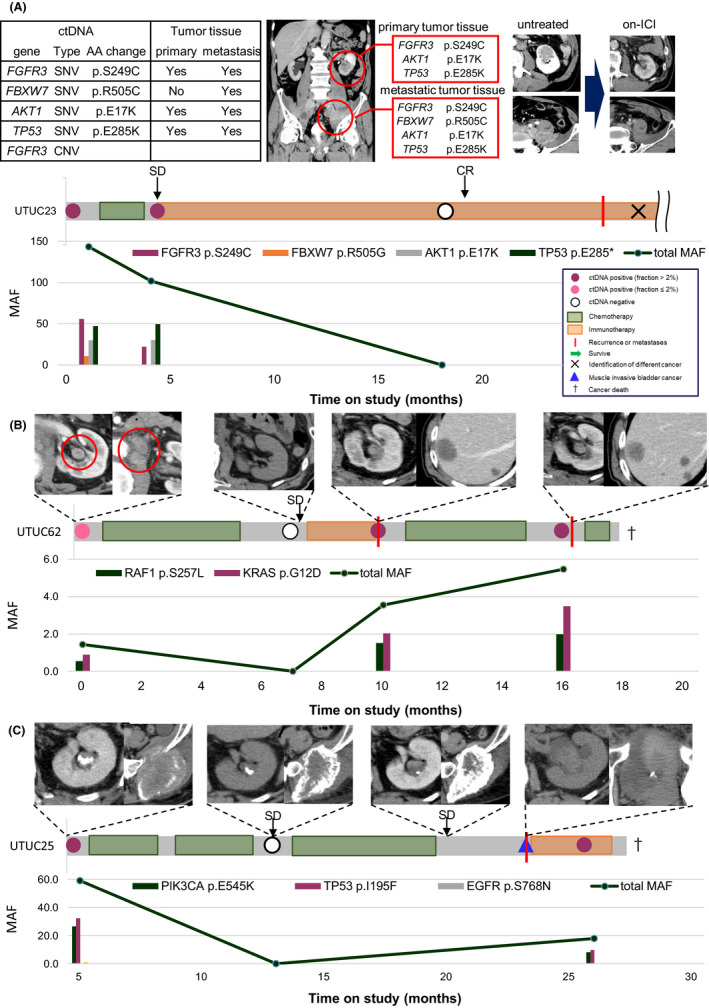
(A–C) Longitudinal monitoring of ctDNA levels during the clinical disease course in patients with metastases. The horizontal bar chart represents the clinical course and indicates the events of blood collection, treatment, and so on, as explained in the associated inlet. Zero indicates the first day to draw plasma in patients with UTUC. The black arrows show the evaluation of the target lesions after treatment according to the RECIST criteria by radiological imaging. The vertical bar chart shows the mutant allele frequency (MAF) of each detected ctDNA, and the line graph shows the total MAF of detected ctDNAs
4. DISCUSSION
Agarwal et al. 28 revealed the genetic profile of plasma cfDNA in metastatic UTUC. However, the clinical usefulness of ctDNA in UTUC, especially localized cancers, has not been fully validated. In our study, ctDNA was detected in 46% of cases by genetic profiling of plasma cfDNA from patients with untreated UTUC. We observed that, among the detected genetic aberrations, those related to TP53 were the most common, in agreement with another study on metastatic UTUC. 28 Christensen et al. 23 reported that preoperative and postoperative ctDNA are markers for early detection of recurrence and metastasis in muscle‐invasive bladder cancer. Their study detected ctDNA in 30 of 67 patients (44%) by creating a patient‐specific gene panel based on the results of whole‐exome sequencing of cancer tissue DNA and performing ultradeep sequencing. As this method searches plasma ctDNA only for genetic alterations detected in cancer tissue, it is unnecessary to exclude false positives such as CHIP. In contrast, our study compared mutations detected with a pan‐cancer gene panel for multiple cancer types with the results of whole‐exome sequencing of cancer tissue DNA. Our study excluded CHIP false positives and detected multiple ctDNAs found only in plasma. We also observed a case in which ctDNA‐matched mutations were detected only in metastases, but not in the primary tumor, indicating that ctDNA reflected the systemic tumor information. We believe that the use of a pan‐cancer panel is advantageous in that it is inexpensive, simple, and allows easy comparison between patients. Using such a panel also obviates the need to perform cancer tissue sequencing and design custom panels for each patient, enabling preoperative evaluation of ctDNA. Additionally, ctDNA may be detected from metastasis that have new mutations not present in the primary tumor, and so the selection of target genes based on the primary tumor may not be advantageous.
One challenge in ctDNA analysis is excluding false positives. Several studies have confirmed the presence of hematopoietic cells in healthy individuals, reaching 10% in those over age 70. 25 The most frequently mutated genes in hematopoietic cells of nonhematologic cancer patients are DNMT3A, TET2, PPM1D, ASXL1, ATM, and TP53. 27 In our study, we detected hematocyte‐derived TP53 mutations in four (8%) out of 50 patients. We also detected CHIP‐related mutations in serial plasma samples regardless of treatment response, reinforcing the importance of using buffy coat DNA sequencing to exclude CHIP‐derived mutations. With the accumulation of CHIP data and the creation of a CHIP database in the future, CHIP would be bioinformatically excluded and buffy coat DNA sequencing may become unnecessary.
In muscle‐invasive bladder cancer, neoadjuvant chemotherapy before radical cystectomy has been shown to improve survival, and the use of neoadjuvant chemotherapy for patients with cT2‐4N0M0 bladder cancer in the USA increased from 22.9% in 2011 to 32.3% in 2015. 31 For UTUC, the phase III POUT trial showed a 2‐year disease‐free survival benefit of postoperative adjuvant chemotherapy in high‐risk patients (pT2‐4 or N+). 6 The optimal timing of perioperative chemotherapy in patients with UTUC remains controversial. Because there are many cases in which patients are ineligible for adjuvant chemotherapy based on renal dysfunction or postoperative complications, current studies are evaluating the efficacy of neoadjuvant chemotherapy for high‐grade UTUC. 8 Several studies have shown the efficacy of neoadjuvant chemotherapy before radical surgery for UTUC. 32 , 33 However, neoadjuvant chemotherapy tends to be avoided because of the difficulty in predicting the depth of disease preoperatively by radiological imaging and due to concerns about overtreatment. Currently, adjuvant chemotherapy based on the pathological stage of the resected tissue is the common strategy.
Our data indicated a discrepancy between preoperative depth diagnosis by radiological imaging and the pathological depth of the resected tissue. Among the preoperative clinicopathological factors, only the ctDNA fraction >2% was associated with significant independent poor RFS. In this study, as patients who were ctDNA negative or ≤2% had a good prognosis regardless of pT or pN stage, we considered that neoadjuvant chemotherapy can be omitted in patients with less than 2% ctDNA. These facts indicate that preoperative ctDNA may be an important indicator for neoadjuvant chemotherapy. Therefore, it is important to exclude false positives; the absence of which may lead to overtreatment.
In this study, we evaluated ctDNA for detection of MRD and long‐term disease monitoring in patients with nonmetastatic UTUC undergoing curative radical surgery. We found that ctDNA positivity immediately after surgery was significantly associated with worse RFS and OS, suggesting the presence of MRD. Among the postoperative clinicopathological factors, postoperative ctDNA and pathological T stage were significant independent factors associated with poor RFS. One patient was ctDNA negative immediately after surgery, but experienced recurrence after 34 months; a liquid biopsy obtained 30 months postoperatively indicated he was ctDNA positive again, indicating that ctDNA can reflect disease status before radiological imaging. We also demonstrated the potential of ctDNA as a real‐time marker of efficacy for systemic treatment of patients with metastatic UTUC, in agreement with previous studies analyzing multiple cancer types. 11 , 12 , 13 , 16 , 17 , 18 , 19 , 20 , 21 , 22 Detection of ctDNA may indicate potential metastasis and guide decisions regarding neoadjuvant therapy for preoperative cases and adjuvant therapy for postoperative cases. Limitations of this study include the relatively small cohort size.
ctDNA analysis of patients with UTUC can identify patients with localized UTUC who are at significantly higher risk of recurrence and death and may be a useful biomarker for detection of postoperative MRD and response to systemic therapy. Prospective studies are required to validate our findings and test the utility of liquid biopsies to guide decisions regarding perioperative chemotherapy in localized UTUC.
DISCLOSURE
Dr. Norio Nonomura is an Editorial Board Member of Cancer Science. All other authors have no conflicts of interest.
Supporting information
Fig S1
Fig S2
Fig S3
Table S1
Table S2
ACKNOWLEDGMENTS
This study was supported by a JSPS KAKENHI grant (19K18558). We thank Mutsumi Tuchiya and Atsuko Yasumoto for their excellent technical support.
Nakano K, Koh Y, Yamamichi G, et al. Perioperative circulating tumor DNA enables the identification of patients with poor prognosis in upper tract urothelial carcinoma. Cancer Sci. 2022;113:1830–1842. doi: 10.1111/cas.15334
REFERENCES
- 1. Rouprêt M, Babjuk M, Burger M, et al. European association of urology guidelines on upper urinary tract urothelial carcinoma: 2020 update. Eur Urol. 2021;79(1):62‐79. [DOI] [PubMed] [Google Scholar]
- 2. Nakano K, Yamamoto Y, Yamamichi G, et al. Fragmentation of cell‐free DNA is induced by upper‐tract urothelial carcinoma‐associated systemic inflammation. Cancer Sci. 2021;112(1):168‐177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Margulis V, Shariat SF, Matin SF, et al. Outcomes of radical nephroureterectomy: a series from the upper tract urothelial carcinoma collaboration. Cancer. 2009;115(6):1224‐1233. [DOI] [PubMed] [Google Scholar]
- 4. Soria F, Shariat SF, Lerner SP, et al. Epidemiology, diagnosis, preoperative evaluation and prognostic assessment of upper‐tract urothelial carcinoma (UTUC). World J Urol. 2017;35(3):379‐387. [DOI] [PubMed] [Google Scholar]
- 5. Leow JJ, Chong YL, Chang SL, et al. Neoadjuvant and adjuvant chemotherapy for upper tract urothelial carcinoma: a 2020 systematic review and meta‐analysis, and future perspectives on systemic therapy. Eur Urol. 2021;79(5):635‐654. [DOI] [PubMed] [Google Scholar]
- 6. Birtle A, Johnson M, Chester J, et al. Adjuvant chemotherapy in upper tract urothelial carcinoma (the POUT trial): a phase 3, open‐label, randomised controlled trial. Lancet. 2020;395(10232):1268‐1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. International Collaboration of Trialists . International phase III trial assessing neoadjuvant cisplatin, methotrexate, and vinblastine chemotherapy for muscle‐invasive bladder cancer: long‐term results of the BA06 30894 trial. J Clin Oncol 2011;29(16):217‐218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Margulis V, Puligandla M, Trabulsi EJ, et al. Phase II trial of neoadjuvant systemic chemotherapy followed by extirpative surgery in patients with high grade upper tract urothelial carcinoma. J Urol 2020;203(4):690‐698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Parikh AR, Mojtahed A, Schneider JL, et al. Serial ctDNA monitoring to predict response to systemic therapy in metastatic gastrointestinal cancers. Clin Cancer Res. 2020;26(8):1877‐1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vidal J, Muinelo L, Dalmases A, et al. Plasma ctDNA RAS mutation analysis for the diagnosis and treatment monitoring of metastatic colorectal cancer patients. Ann Oncol. 2017;28(6):1325‐1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tsui DWY, Murtaza M, Wong ASC, et al. Dynamics of multiple resistance mechanisms in plasma DNA during EGFR‐targeted therapies in non‐small cell lung cancer. EMBO Mol Med. 2018;10(6):e7945 10.15252/emmm.201707945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nong J, Gong Y, Guan Y, et al. Circulating tumor DNA analysis depicts subclonal architecture and genomic evolution of small cell lung cancer. Nat Commun. 2018;9(1):3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yamamoto Y, Uemura M, Fujita M, et al. Clinical significance of the mutational landscape and fragmentation of circulating tumor DNA in renal cell carcinoma. Cancer Sci. 2019;110(2):617‐628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Powles T, Assaf ZJ, Davarpanah N, et al. ctDNA guiding adjuvant immunotherapy in urothelial carcinoma. Nature. 2021;595(7867):432‐437. [DOI] [PubMed] [Google Scholar]
- 15. Yamamoto Y, Uemura M, Nakano K, et al. Increased level and fragmentation of plasma circulating cell‐free DNA are diagnostic and prognostic markers for renal cell carcinoma. Oncotarget. 2018;9(29):20467‐20475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Garcia‐Murillas I, Schiavon G, Weigelt B, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med. 2015;7(302):302ra133. [DOI] [PubMed] [Google Scholar]
- 17. Tie J, Cohen JD, Wang Y, et al. Circulating tumor DNA analyses as markers of recurrence risk and benefit of adjuvant therapy for stage III colon cancer. JAMA Oncol. 2021;5(12):1710‐1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tarazona N, Gimeno‐Valiente F, Gambardella V, et al. Targeted next‐generation sequencing of circulating‐tumor DNA for tracking minimal residual disease in localized colon cancer. Ann Oncol. 2019;30(11):1804‐1812. [DOI] [PubMed] [Google Scholar]
- 19. Chaudhuri AA, Chabon JJ, Lovejoy AF, et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA Profiling. Cancer Discov. 2017;7(12):1394‐1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Leal A, van Grieken NCT, Palsgrove DN, et al. White blood cell and cell‐free DNA analyses for detection of residual disease in gastric cancer. Nat Commun. 2020;11(1):525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang J, Gong Y, Lam VK, et al. Deep sequencing of circulating tumor DNA detects molecular residual disease and predicts recurrence in gastric cancer. Cell Death Dis. 2020;11(5):346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Azad TD, Chaudhuri AA, Fang P, et al. Circulating tumor DNA analysis for detection of minimal residual disease after chemoradiotherapy for localized esophageal cancer. Gastroenterology. 2020;158(3):494‐505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Christensen E, Birkenkamp‐Demtröder K, Sethi H, et al. Early detection of metastatic relapse and monitoring of therapeutic efficacy by ultra‐deep sequencing of plasma cell‐free dna in patients with urothelial bladder carcinoma. J Clin Oncol. 2019;37(18):1547‐1557. 10.1200/JCO.18.02052 [DOI] [PubMed] [Google Scholar]
- 24. Chalfin HJ, Glavaris SA, Gorin MA, et al. Circulating tumor cell and circulating tumor DNA assays reveal complementary information for patients with metastatic urothelial cancer. Eur Urol Oncol. 2021;4:310‐314. [DOI] [PubMed] [Google Scholar]
- 25. Xie M, Lu C, Wang J, et al. Age‐related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20(12):1472‐1478. 10.1038/nm.3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Razavi P, Li BT, Brown DN, et al. High‐intensity sequencing reveals the sources of plasma circulating cell‐free DNA variants. Nat Med. 2019;25(12):1928‐1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Coombs CC, Zehir A, Devlin SM, et al. Therapy‐related clonal hematopoiesis in patients with non‐hematologic cancers is common and associated with adverse clinical outcomes. Cell Stem Cell. 2017;21(3):374‐382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Agarwal N, Pal SK, Hahn AW, et al. Characterization of metastatic urothelial carcinoma via comprehensive genomic profiling of circulating tumor DNA. Cancer. 2018;124(10):2115‐2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chan HT, Nagayama S, Chin YM, et al. Clinical significance of clonal hematopoiesis in the interpretation of blood liquid biopsy. Mol Oncol. 2020;14(8):1719‐1730. 10.1002/1878-0261.12727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vandekerkhove G, Todenhöfer T, Annala M, et al. Circulating tumor DNA reveals clinically actionable somatic genome of metastatic bladder cancer. Clin Cancer Res. 2017;23(21):6487‐6497. 10.1158/1078-0432.CCR-17-1140 [DOI] [PubMed] [Google Scholar]
- 31. McFerrin C, Davaro F, May A, et al. Trends in utilization of neoadjuvant and adjuvant chemotherapy for muscle invasive bladder cancer. Invest Clin Urol. 2020;61(6):565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Porten S, Siefker‐Radtke AO, Xiao L, et al. Neoadjuvant chemotherapy improves survival of patients with upper tract urothelial carcinoma. Cancer. 2014;120(12):1794‐1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hosogoe S, Hatakeyama S, Kusaka A, et al. Platinum‐based neoadjuvant chemotherapy improves oncological outcomes in patients with locally advanced upper tract urothelial carcinoma. Eur Urol Focus. 2018;4(6):946‐953. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Fig S3
Table S1
Table S2


