Abstract
Environmental and genetic factors play a critical role in the pathogenesis of pancreatic cancer, which is likely to follow a multistep process that includes intraductal papillary mucinous neoplasm. The pathogenesis of familial pancreatic cancer has been reported; however, epidemiological characteristics and causative genes remain unclear. This study aimed to determine the relationship between the family history of pancreatic cancer and tumor malignancy and identify novel susceptible germline variants of pancreatic cancer. We performed an epidemiologic study at our institute on a cohort of 668 patients with intraductal papillary mucinous neoplasm and 242 with pancreatic cancer but without associated intraductal papillary mucinous neoplasm stratified by family history of pancreatic cancer. Whole‐exome sequencing was conducted for 10 patients from seven families with familial pancreatic cancer and intraductal papillary mucinous neoplasm. We found that patients who had intraductal papillary mucinous neoplasm with positive family history of pancreatic cancer within first‐degree relatives were more likely to develop malignancy in a shorter period than those without family history. Duplicate frameshift variants in TET2 c.3180dupG (p.Pro1061fs) and ASXL1 c.1934dupG (p.Gly646fs) in one family and POLN c.1194dupT (p.Glu399fs) in another were identified as pathogenic truncating germline variants which were previously recognised susceptibility genes. Moreover, PDIA2 c.1403C>T (p.Pro468Leu) and DPYSL4 c.926C>A (p.Pro309Gln) were shared in four and two patients, respectively. In particular, PDIA2 was identified as a novel candidate for one of the deleterious variants of familial pancreatic cancer.
Keywords: cancer susceptibility genes, cross‐sectional study, familial pancreatic cancer, germline variant, intraductal papillary mucinous neoplasm
Patients with intraductal papillary mucinous neoplasm with a family history of pancreatic cancer are more likely to develop malignancy in a shorter period than those without a family history. Of 18 patients with a strong family history of pancreatic cancer, three previously known susceptibility genes and one novel candidate gene were identified.

Abbreviations
- EUS
endoscopic ultrasound
- IPMC
intraductal papillary mucinous carcinoma
- IPMN
intraductal papillary mucinous neoplasm
- MPD
main pancreatic duct
- MRCP
magnetic resonance cholangiopancreatography
1. INTRODUCTION
Pancreatic cancer is a malignant tumor with a poor prognosis, partly because of difficulty in detecting the cancer in early stages. Delineation of risk factors for pancreatic cancer could help initiate surveillance targeting a subpopulation at greater risk. Previous studies have shown that both environmental and genetic factors play important roles in the pathogenesis of pancreatic cancers. Known environmental risk factors include obesity, diabetes, smoking, and alcohol consumption. 1
Documentation of familial aggregation of pancreatic cancer in the literature points to the influence of genetic factors. A prospective epidemiological study of pancreatic cancer conducted by Klien et al. 2 in 2004 showed that the lifetime risk of pancreatic cancer was 6.4 times higher in family members with two first‐degree relatives with pancreatic cancer than in those without first‐degree relatives with pancreatic cancer. In addition, the risk was 32 times higher in family members with three first‐degree relatives with pancreatic cancer. Sequencing analysis of germline variants of families with pancreatic cancer unraveled the critical role of ATM, BRCA1, BRCA2, and CHEK2. 3 , 4 , 5 , 6 Besides studies on familial clusters, genome‐wide association studies demonstrated that the single‐nucleotide variants rs13303010 in NOC2L at 1p36.33 and rs78193826 in GP2 at 16p12.3 are associated with pancreatic cancer even among patients without apparent family histories. 7 , 8
Similar to other cancers, the pathogenesis of pancreatic cancers is likely a multistep process. The observation that familial pancreatic cancer was more likely to have earlier onset and mortality supports the multistep progression of pancreatic cancers. Intraductal papillary mucinous neoplasm (IPMN) has been regarded as the critical precancerous lesion of pancreatic cancers. A recent whole‐exome analysis of 350 patients with IPMN showed that germline variants in ATM and BRCA2 are associated with the progression of IPMN to pancreatic cancer. 9 However, whether genetic factors play a role in the development of IPMN remains unclear.
In this study, we performed epidemiologic and genetic studies on 668 patients with IPMN and 242 patients with pancreatic cancer but without associated IPMN. Among these two groups, we identified 18 patients with two or more affected family members, excluding the proband. Among these 18 patients, 10 patients from seven families underwent genomic studies. Identification of new risk genes for pancreatic cancer may facilitate genomic screening for early detection and treatment of pancreatic cancer.
2. MATERIALS AND METHODS
2.1. Study design and ethical approval
This retrospective and genome‐sequencing study was conducted according to the Declaration of Helsinki after approval from the Institutional Review Board of the Keio University School of Medicine (approval numbers: 20120443 and 20190042; date of approval: October 30, 2019).
2.2. Patients and data collection
We retrospectively reviewed the clinical characteristics and environmental factors of all patients who underwent endoscopic ultrasound (EUS) and endoscopic retrograde cholangiopancreatography between 2012 and 2020 and identified 668 patients with IPMN and 242 patients with non–IPMN‐associated pancreatic cancer (Figure S1). We defined non–IPMN‐associated pancreatic cancer as pancreatic cancer with no evidence of pancreatic cysts or IPMN based on magnetic resonance cholangiopancreatography (MRCP) or EUS scan interpreted by the radiologist and endoscopist.
The patients' clinical backgrounds were collected from medical records, and a questionnaire was provided during the first visit. Data collected included age, sex, body mass index (BMI), history of smoking and habitual alcohol consumption, medical history, family history of diabetes and cancer within first‐ or second‐degree relatives, and imaging findings of pancreatic lesions. Habitual alcohol consumption was defined as the consumption of more than 100 g of alcohol per week. History of diabetes was defined as receiving antidiabetic treatment before the diagnosis of pancreatic tumors. The clinical characteristics of all patients (668 IPMN and 242 non–IPMN‐associated pancreatic cancer) are shown in Table S1.
Patients with IPMN were classified as high risk or low risk according to the IPMN International Clinical Practice Guidelines 2017. 10 , 11 The high‐risk IPMN group included those who met the criteria for “worrisome features (imaging findings include cyst of ≥3 cm, enhancing mural nodule <5 cm, thickened enhanced cyst walls, main pancreatic duct [MPD] 5–9 mm, lymphadenopathy, an elevated serum level of carbohydrate antigen, and a rapid rate of cyst growth >5 mm per 2 years)” and “high‐risk stigma (obstructive jaundice, enhanced mural nodule ≥5 mm, MPD ≥10 mm).” Furthermore, those with intraductal papillary mucinous carcinoma (IPMC)‐, IPMN‐concomitant, and IPMN‐derived pancreatic cancer were included in the high‐risk IPMN group. The low‐risk IPMN group comprised patients with IPMN who did not meet the criteria defined above.
2.3. Whole‐exome sequencing of germline samples
After obtaining written informed consent, whole‐exome sequencing was performed on peripheral blood samples of 10 patients from seven families using the NovaSeq platform (Illumina) and Sure Select XT Human All Exon V6 (Agilent Technologies). Mapping of the sequenced reads to the reference human genome (GRCh37) and variant calling were performed according to the best practice guidelines of the Burrows‐Wheeler Aligner 12 and the Genome Analysis Tool Kit, 13 as packaged in the integrated analysis suite variant tools. 14 The variants were annotated with SnpEff. 15
2.4. Annotation of variants
To characterize potential functional significance of the variants revealed by whole‐exome analysis, the allele frequencies of the variants among Japanese patients were evaluated from epidemiological standpoints. Any variant for which the allele frequency among >7000 normal Japanese individuals was larger than 0.03 as per the ToMMo database was excluded from further consideration. 16 Thus, only variants for which the allele frequency was between 40% and 60% were retained. When two or more affected members including the proband were tested, only the shared variants were retained. Among the 10 patients from seven families, 4148 variants were retained according to these criteria.
2.4.1. Search for pathogenic variants in previously recognized susceptibility genes
We extracted frameshift, nonsense, and splicing variants and filtered them by previously reported pancreatic cancer–related genes (Table S2). Variants corresponding to a combined annotation‐dependent depletion (CADD) score of >20 were extracted. 17
2.4.2. Comparison with molecular epidemiological data from Japan
We further evaluated these filtered (nonsynonymous) variants based on our previous work on whole‐genome analysis of samples from Biobank Japan. We analyzed the table of variants and their allele count, allele number, and allele frequency of 6206 samples derived from patients with noncancer polygenic disorders and 1521 samples from patients with various kinds of cancer excluding pancreatic cancer. 18 , 19 Nonsynonymous variants with increased frequency among cancers were considered as candidate susceptibility variants. Filtered variants from familial pancreatic cancers and IPMN were identified from the table of candidate variants.
2.5. Statistical analyses
Categorical variables were compared between the two groups using the chi‐squared and Fisher's exact tests. The Mann‐Whitney U test was used to compare quantitative variables. Statistical analyses were performed using SPSS for Mac (version 25.0; IBM). Statistical significance was set at p < 0.05, and all tests were two‐sided. Kaplan‐Meier analysis was used to assess differences in survival between cohorts.
3. RESULTS
3.1. Clinical characteristics of patients with IPMN and pancreatic cancer with a strong family history of pancreatic cancer
Among the 668 patients with IPMN and the 242 with non–IPMN‐associated pancreatic cancer, we identified 15 patients with IPMN and three patients with non–IPMN‐associated pancreatic cancer who had two or more affected family members excluding the proband (i.e., two first‐degree relatives or one first‐degree and one second‐degree relative with pancreatic cancer; Figure S2). These patients were arbitrarily defined as “patients with a strong family history.”
The clinical characteristics of patients with IPMN and pancreatic cancer with a strong family history of pancreatic cancer (n = 18) are shown in Table 1. Seven patients with IPMN had branch‐type IPMN, two had high‐grade IPMN, three had IPMN‐derived pancreatic cancer, three had IPMN concomitant with pancreatic cancer, and three had non–IPMN‐related pancreatic cancer.
TABLE 1.
Characteristics of patients with a strong family history of pancreatic cancer (N = 18)
| Patient | Pedigree | Age | Sex | Disease | Family history of pancreatic cancer | Past cancer history | Genome sequencing | Outcome |
|---|---|---|---|---|---|---|---|---|
| #1 | #A | 41 | Female | Branch duct–type IPMN | Mother and grandmother | None | – | Alive |
| #2 | #B | 79 | Female | Branch duct–type IPMN | Older brother and younger sister | None | Whole‐exome sequencing | Alive |
| #3 | #B | 82 | Female | Branch duct–type IPMN | Younger brother and younger sister | None | Whole‐exome sequencing | Alive |
| #4 | #C | 52 | Female | Branch duct–type IPMN | Father, younger brother, and father's aunt | None | Gene panel testing | Alive |
| #5 | #D | 73 | Female | Branch duct–type IPMN | Older brother, mother, and mother's uncles | None | Whole‐exome sequencing | Alive |
| #6 | #E | 62 | Female | Branch duct–type IPMN | Father, father's aunt, and grand father | None | – | Alive |
| #7 | #F | 72 | Female | Branch duct–type IPMN | Father, older brother, and younger brother | Breast cancer | Whole‐exome sequencing | Alive |
| #8 | #G | 78 | Female | High‐grade IPMN | Younger sister and daughter | Cervical cancer | Whole genome sequencing | Alive |
| #9 | #G | 51 | Female | High‐grade IPMN | Mother and mother's aunt | None | Whole genome sequencing | Alive |
| #10 | #H | 54 | Female | IPMN‐derived pancreatic cancer | Father and father's grandmother | Breast cancer | – | Dead |
| #11 | #I | 75 | Male | IPMN‐derived pancreatic cancer | Older brothers, father, and mother | None | Whole‐exome sequencing | Alive |
| #12 | #J | 69 | Male | IPMN‐concomitant pancreatic cancer | Older sister and younger sister | Rectal cancer | – | Dead |
| #13 | #K | 72 | Male | IPMN‐concomitant pancreatic cancer | Older brother and younger brother | None | Whole‐exome sequencing | Alive |
| #14 | #L | 79 | Female | IPMN‐derived pancreatic cancer | Mother and older sister | Lung cancer | – | Dead |
| #15 | #M | 82 | Female | IPMN‐concomitant pancreatic cancer | Father and older sister | None | – | Dead |
| #16 | #G | 72 | Female | Non–IPMN‐related pancreatic cancer | Older sister and sister's daughter | None | Whole genome sequencing | Alive |
| #17 | #N | 71 | Male | Non–IPMN‐related pancreatic cancer | Father and father's uncle | Gastric cancer | Whole‐exome sequencing | Alive |
| #18 | #O | 53 | Male | Non–IPMN‐related pancreatic cancer | Father and father's aunt | None | – | Dead |
Abbreviation: IPMN, intraductal papillary mucinous neoplasm.
3.2. Malignant progression of IPMN with positive family history of pancreatic cancer
We compared the clinical backgrounds of the patients with IPMN and those with non–IPMN‐related pancreatic cancer by categorizing them into those with positive family history of pancreatic cancer (more than one first‐degree relative with pancreatic cancer) and those without (Table 2). Patients with IPMN‐related pancreatic cancer with positive family history of pancreatic cancer were more likely to be female and to have a personal history of cancer; however, this finding was not statistically significant (Table 2).
TABLE 2.
Clinical backgrounds of patients with intraductal papillary mucinous neoplasm (IPMN) and pancreatic cancer stratified by family history of pancreatic cancer (N = 668)
| Characteristics | Positive family history of pancreatic cancer (n = 46) | No family history of pancreatic cancer (n = 622) | p |
|---|---|---|---|
| Age, years, median (range) | 68 (41–83) | 70 (28–90) | 0.384 |
| Age >70 years | 20 (43.5) | 312 (50.2) | 0.382 |
| Sex (male/female) | 18/28 | 316/306 | 0.127 |
| BMI (kg/m2) | 21.0 (16.7–31.6) | 21.8 (12.8–34.2) | 0.313 |
| BMI >25 | 7 (15.2) | 105 (16.9) | 0.704 |
| Smoking (Ex/Cur) | 15 [13/2] (32.6) | 216 [177/39] (34.7) | 0.765 |
| Alcohol consumption | 13 (28.3) | 119 (19.1) | 0.134 |
| History of diabetes | 5 (10.9) | 75 (12.1) | 0.889 |
| History of cancer | 7 (15.2) | 159 (25.6) | 0.117 |
Data are presented as n (%) unless otherwise indicated.
Abbreviations: BMI, body mass index; Cur, current smoker; Ex, ex‐smoker.
Progression to malignant pancreatic cancer or high‐risk IPMN (including high‐grade IPMN, IPMC, and IPMN‐related pancreatic cancer) was evaluated in these subgroups using Kaplan‐Meier analysis (Figure 1). We observed 242 patients with high‐risk IPMN during the study period, of which 23 had a positive family history of pancreatic cancer and 219 had no family history of pancreatic cancer. We found that patients with IPMN and positive family history were more likely to develop malignancy in a shorter period than those with no family history of pancreatic cancer (log‐rank test, p = 0.006).
FIGURE 1.
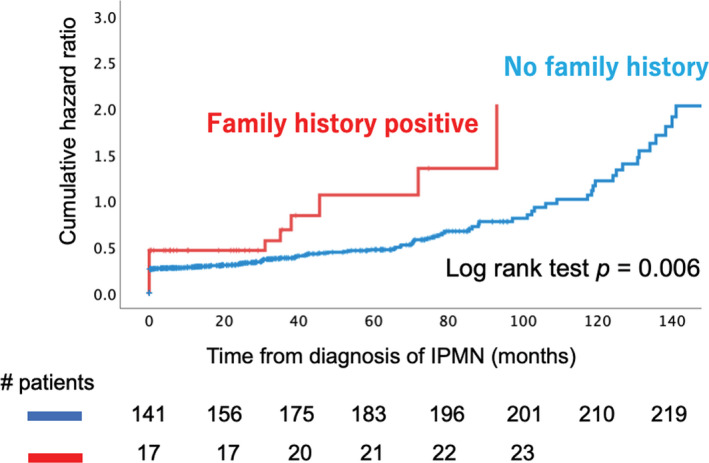
Cumulative risk of malignancy and overall survival of patients with intraductal papillary mucinous neoplasm (IPMN) stratified by family history of pancreatic cancer. The cumulative hazard ratio for malignant transformation in patients with IPMN with a family history of pancreatic cancer is shown. Malignant transformation includes intraductal papillary mucinous carcinoma and IPMN‐concomitant pancreatic cancer. The red line represents those with family history, whereas the blue line represents those without. p = 0.006 (analyzed by log rank test)
3.3. Families with truncating variants in known pancreatic cancer–related genes
Of the 18 patients with a strong family history, 11 patients underwent genomic analysis, five patients died, and two did not wish to undergo testing.
One patient (#7) underwent commercial testing for multiple cancer genes, and a heterozygous PALB2 pathogenic variant (c.1675_1676inv [p. Gln559*]) and a heterozygous NBN pathogenic variant (c.265C>T [p. Arg89*]) were identified that has been reported elsewhere. 20
Whole‐exome sequencing was performed for 10 patients from seven families. The summary of the whole‐exome sequencing results is shown in Table 3. Truncating variants of genes already implicated in the pathogenesis of pancreatic cancer (Table S1) were identified in two families. Heterozygous frameshift variants of NM_017628.4 (TET2; c.3180dupG, p. Pro1061fs) and NM_015338.6 (ASXL1; c.1934dupG, p. Gly646fs) were identified in patient #16 (Table 3a and Figure 2A), and a frameshift variant of NM_181808.4 (POLN; c.1194dupT, p. Glu399fs) was identified in patient #10 (Table 3b and Figure 2B).
TABLE 3.
Susceptible germline variants of pancreatic cancer and intraductal papillary mucinous neoplasm. (a) Susceptible frameshift variants of familial pancreatic cancer. (b) Susceptible single‐nucleotide variants of familial pancreatic cancer
| (a) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | NM | Patients | Codon change | Amino acid change | AF | CADD score | Category | |||
| TET2 | NM_017628.4 | #13 | c.3180dupG | p. Pro1061fs | – | 32.0 | Cancer driver gene | |||
| ASXL1 | NM_015338.6 | #13 | c.1934dupG | p. Gly646fs | 0.00084 | 34.0 | Cancer driver gene | |||
| POLN | NM_181808.4 | #5 | c.1194dupT | p. Glu399fs | 0.001 | 24.8 | DNA polymerase gene | |||
| (b) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | NM | Patients | Codon change | Amino acid change | AF of cancer | AF of non‐cancer | OR (95%CI) | p | PROVEAN score | SIFT score | Polyphen‐2 |
| PDIA2 | NM_006849.4 | #5, #8, #9, #16 | c.1403C>T | p. Pro468Leu | 0.0377 | 0.0265 | 1.48 | 0.005 | Deleterious (−8.50) | Damaging (0.003) | Possibly damaging |
| DPYSL4 | NM_006426.3 | #11, #17 | c.926C>A | p. Pro309Gln | 0.0301 | 0.0220 | 1.45 | 0.019 | Deleterious (−7.62) | Damaging (0.013) | Probably damaging |
Abbreviations: AF, allele frequency; CADD, combined annotation‐dependent depletion; CI, confidence interval; OR, odds ratio; PROVEAN, protein variation effect analyzer; SIFT, sorting intolerant from tolerant.
FIGURE 2.
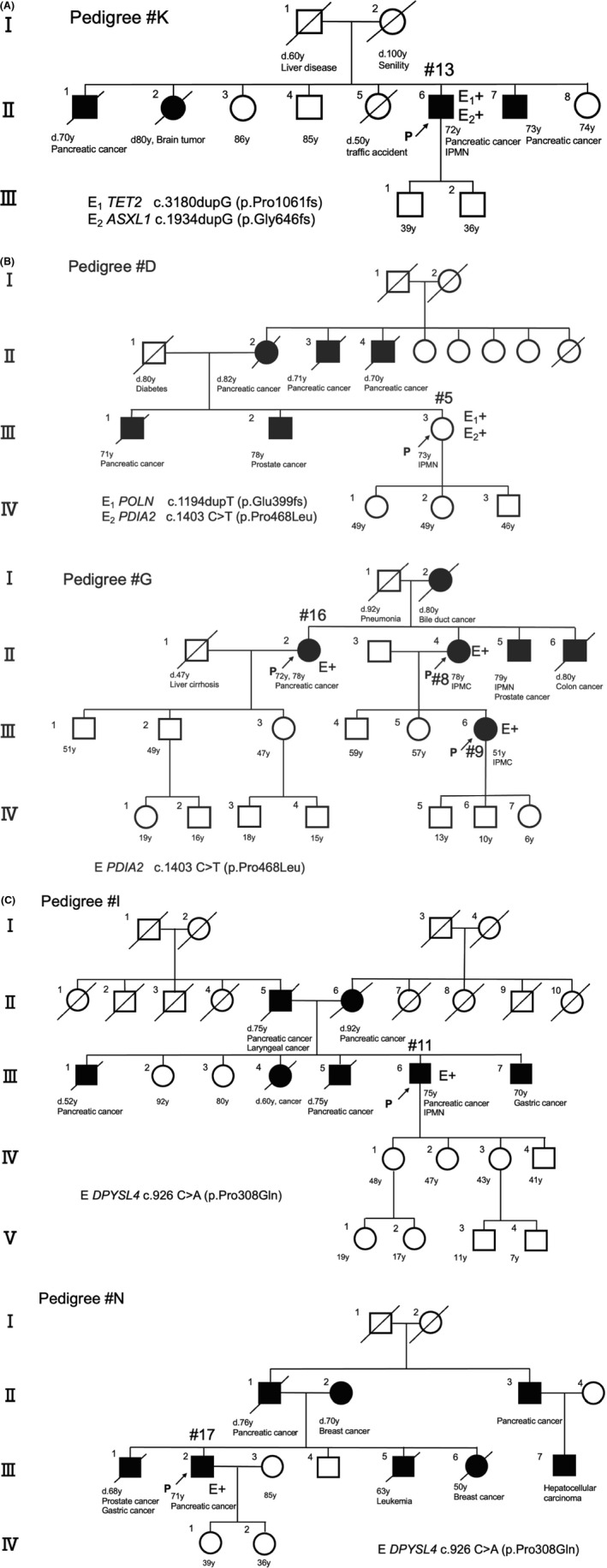
Family tree of patients who underwent whole‐exome sequencing analysis. A, Patient #13 (pedigree #K; pancreatic cancer) was identified to have TET2 and ASXL1 frameshift overlapped variants. B, Patient #5 (pedigree #D; branch duct–type intraductal papillary mucinous neoplasm [IPMN]) has a POLN frameshift variant and a PDIA2 missense variant, similar to pedigree #G of patients #16 (pancreatic cancer), #8 (high‐grade IPMN), and #9 (high‐grade IPMN). C, Patient #11 (pedigree #I; IPMN‐related pancreatic cancer) and patient #17 (pedigree #N; pancreatic cancer) have a common missense variant DPYSL4
3.4. Comparison with molecular epidemiological data from Japan
We then compared the 4148 variants extracted from the analyzed patients to the allele frequencies of the whole‐genome sequencing data from cancer (n = 1521) and noncancer patients (n = 6206). Of these variants, 172 variants with odds ratios >1.0 and P‐values <0.05 were extracted. We further tested whether any of the variants were recurrent among the 10 families in which the proband underwent whole‐exome analysis.
Two were common among families and deleterious according to the PROVEAN, SIFT (http://provean.jcvi.org/protein_batch_submit.php?species=human), and PolyPhen‐2 scores (http://genetics.bwh.harvard.edu/pph2/): NM_006849.4 (PDIA2; c.1403C>T, p. Pro468Leu) was common to patients #1, #10, #11, and #12 (Table 3b and Figure 2B), and NM_006426.3 (DPYSL4; c.926C>A, p. Pro309Gln) was common to patients #2 and #14 (Table 3b and Figure 2C).
In most families, only the probands were tested. Intrafamilial segregation was confirmed in two families.
4. DISCUSSION
In this study, we found deleterious variants in cancer susceptibility genes, including TET2, ASXL1, and POLN, which are known susceptibility genes, and PDIA2 and DPSYL4, which are novel risk factors.
Premature truncating germline variants in TET2 and ASXL1 have been identified in patients with familial pancreatic cancer. 5 , 6 Tet methyl cytosine dioxygenase 2 (TET2) is involved in DNA demethylation, and its loss‐of‐function mutations result in hypermethylation. 21 Meanwhile, ASXL1 is a histone modifier, and gain‐of‐function mutations are associated with tumorigenesis. 22 Somatic variants in TET2 and ASXL1 represent poor prognostic indicators in hematological tumors. 23 It is notable that a patient with strong family history was a double heterozygote for truncating germline variants of TET2 and ASXL1. The two genes could have an additive effect in tumorigenesis.
POLN gene is a member of the DNA polymerase family and is responsible for repairing DNA damage. 24 Truncating variants of POLN have been previously reported as causative genes of germline variants in pancreatic cancer. To our knowledge, this is the first study to report that a truncating variant (i.e., a frameshift variant) of POLN could be associated with both IPMN and pancreatic cancer. 5 , 25
Two variants observed in patients with a strong family history were previously reported to have a significantly higher frequency in patients with various cancers than in the general population. First, the protein disulfide‐isomerase A2 (PDIA2), a protein expressed in the pancreas, is involved in the regulation of cellular levels and biological functions of estrogen and has been identified as an autoantibody for autoimmune pancreatitis in vivo. 26 , 27 Although the association between a pathogenic PDIA2 variant and pancreatic cancer has not yet been reported, all four patients in the two families with this variant were females with a strong family history, which could lead to pancreatic cancer or cystic changes. Second, DPYSL4 is an intracellular metabolic regulator induced by p53, a known tumor suppressor, and is found in mitochondria of mast cells. 28 However, its relatively high allele frequency among Southeast Asians (0.098) makes the candidacy of variants less likely.
This study has some limitations. First, protein functions by candidate gene variants obtained from our analysis have only been investigated using in silico analysis. According to the American College of Medical Genetics and Genomics/the Association for Molecular Pathology (ACMG/AMP) guideline 2015, NM_006849.4 (PDIA2: c.1403C>T, p. Pro468Leu) and NM_015338.6 (DPYSL4: c.926C>A, p. Pro309Gln) are classified as variants of uncertain significance, suggesting the need for functional and segregation analysis. 29 Second, only subsets of the affected family members and obligate carriers underwent genomic analysis. The segregation analysis is incomplete because of the limited availability of samples from family members.
Nevertheless, even among families who were not shown to have relevant variants in known cancer susceptibility genes, IPMN and strong family history were demonstrated as significant factors predictive of progression to IPMC or pancreatic cancer. Furthermore, patients with IPMN and strong family history tended to be negative for known environmental risk factors for pancreatic cancer. Patients with IPMN and a strong family history of pancreatic cancer could benefit from surveillance using EUS and/or MRCP regardless of the known environmental risk factors. 30 , 31
The identification of novel pancreatic cancer or IPMN susceptibility genes, including PDIA2, could advance the genomic screening of patients who would benefit from regular imaging studies for IPMN and pancreatic cancers and provide a better understanding of the multistep pathogenic progression of pancreatic cancer. 32
In conclusion, in our cohort of 668 patients with IPMN and 244 with pancreatic cancer, we identified 18 patients with a strong family history who are at high risk of progression to pancreatic cancer. Genomic analysis of these patients identified three previously recognized susceptibility genes and a novel potential candidate, PDIA2.
DISCLOSURE
Yuko Kitagawa received grants from CHUGAI PHARMACEUTICAL CO., LTD.; TAIHO PHARMACEUTICAL CO., LTD; Yakult Honsha Co. Ltd.; ASAHI KASEI PHARMA CORPORATION EA Pharma Co., Ltd.; Astellas Pharma Inc.; Toyama Chemical Co., Ltd.; MEDICON INC.; KAKEN PHARMACEUTICAL CO. LTD.; Eisai Co., Ltd.; Otsuka Pharmaceutical Factory Inc.; TEIJIN PHARMA LIMITED; NIHON PHARMACEUTICAL CO., LTD.; Nippon Covidien Inc.; SHIONOGI & CO., LTD.; Olympus Corporation AstraZeneca K.K.; Ethicon, Inc.; MSD K.K.; Otsuka Pharmaceutical Co., Ltd.; Takeda Pharmaceutical Co., Ltd.; ONO PHARMACEUTICAL CO., LTD.; TSUMURA & CO.; Kyouwa Hakkou Kirin Co., Ltd.; DAINIPPON SUMITOMO PHARMA Co., Ltd.; Smith&Nephew KK; and Bristol‐Myers Squibb K.K.
Supporting information
Supplementary Material
ACKNOWLEDGEMENTS
This study was supported by the Kenko Kagaku Zaidan (grant number KenkoKagakuZaidan201902). We would like to thank Kazumasa Fukuda, a staff member of the Department of Surgery of the Keio University School of Medicine, for his help in preparing this manuscript. We would also like to thank Editage (www.editage.com) for English language editing.
Abe K, Kitago M, Kosaki K, et al. Genomic analysis of familial pancreatic cancers and intraductal papillary mucinous neoplasms: A cross‐sectional study. Cancer Sci. 2022;113:1821–1829. doi: 10.1111/cas.15316
Funding information
Kenko Kagaku Zaidan (Grant/Award Number: ‘KenkoKagakuZaidan201902’)
REFERENCES
- 1. Midha S, Chawla S, Garg PK. Modifiable and non‐modifiable risk factors for pancreatic cancer: a review. Cancer Lett. 2016;381:269‐277. [DOI] [PubMed] [Google Scholar]
- 2. Klein AP, Brune KA, Petersen G, et al. Prospective risk of pancreatic cancer in familial pancreatic cancer kindreds. Cancer Res. 2004;64:2634‐2638. [DOI] [PubMed] [Google Scholar]
- 3. Hu C, Hart S, Polly E, et al. Association between inherited germline mutations in cancer predisposition genes and risk of pancreatic cancer. JAMA. 2018;319:2401‐2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mizukami K, Iwasaki Y, Kawakami E, et al. Genetic characterization of pancreatic cancer patients and prediction of carrier status of germline pathogenic variants in cancer‐predisposing genes. EBioMedicine. 2020;60: 103033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roberts NJ, Norris AL, Petersen GM, et al. Whole genome sequencing defines the genetic heterogeneity of familial pancreatic cancer. Cancer Discov. 2016;6:166‐175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Takai E, Nakamura H, Chiku S, et al. Whole‐exome sequencing reveals new potential susceptibility genes for Japanese familial pancreatic cancer. Ann Surg. 2020. 10.1097/SLA.0000000000004213 [DOI] [PubMed] [Google Scholar]
- 7. Klien A, Wolpin B, Risch H, et al. Genome‐wide meta‐analysis identifies five new susceptibility loci for pancreatic cancer. Nat Commun. 2018;9:556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lin Y, Nakatochi M, Hosono Y, et al. Genome‐wide association meta‐analysis identifies GP2 gene risk variants for pancreatic cancer. Nat Commun. 2020;11:3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Skaro M, Nanda N, Gauthier C, et al. Prevalence of germline mutations associated with cancer risk in patients with intraductal papillary mucinous neoplasms. Gastroenterology. 2019;156:1905‐1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tanaka M, Castillo C, Adsay V, et al. International consensus guidelines 2012 for the management of IPMN and MCN of the pancreas. Pancreatology. 2012;12:183‐197. [DOI] [PubMed] [Google Scholar]
- 11. Tanaka M, Castillo C, Kamisawa T, et al. Revisions of international consensus Fukuoka guidelines for the management of IPMN of the pancreas. Pancreatology. 2017;17:738‐753. [DOI] [PubMed] [Google Scholar]
- 12. McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: a mapreduce framework for analyzing next‐generation DNA sequencing data. Genome Res. 2010;20:1297‐1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li H, Durbin R. Fast and accurate short read alignment with Burrows‐Wheeler transform. Bioinformatics. 2009;25:1754‐1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lucas F, Wang G, Sheet P, Peng B. Integrated annotation and analysis of genetic variants from next‐generation sequencing studies with variant tools. Bioinformatics. 2012;28:421‐422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cingaloni P, Platts A, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: NPs in the genome of Drosophila melanogaster strain w1118; iso‐2; iso‐3. Fly (Austin). 2012;6:80‐92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yamaguchi‐Kabata Y, Nariai N, Kawai Y, et al. iJGVD: an integrative Japanese genome variation database based on whole‐genome sequencing. Hum Genome Var. 2015;2:15050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kircher M, Witten D, Jain P, O’Roak B, Cooper G, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310‐315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Koyama S, Ito K, Terao C, et al. Population‐specific and trans‐ancestry genome‐wide analyses identify distinct and shared genetic risk loci for coronary artery disease. Nat Genet. 2020;52:1169‐1177. [DOI] [PubMed] [Google Scholar]
- 19. Liu X, Takata S, Ashikawa K, et al. Prevalence and spectrum of pathogenic germline variants in japanese patients with early‐onset colorectal, breast, and prostate cancer. JCO Precis Oncol. 2020;4:183‐191. [DOI] [PubMed] [Google Scholar]
- 20. Abe K, Ueki A, Urakawa Y, et al. Familial pancreatic cancer with PALB2 and NBN pathogenic variants: a case report. Hered Cancer Clin Pract. 2021;19:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ferrone C, Blydt‐Hansen M, Rauh M. Age‐associated TET2 mutations: common drivers of myeloid dysfunction, cancer and cardiovascular disease. Int J Mol Sci. 2020;21:626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fujino T, Goyama S, Sugiura Y, et al. Mutant ASXL1 induces age‐related expansion of phenotypic hematopoietic stem cells through activation of Akt/mTOR pathway. Nat Commun. 2021;12:1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Patnaik M, Lasho T, Vijayvargiya P, et al. Prognostic interaction between ASXL1 and TET2 mutations in chronic myelomonocytic leukemia. Blood Cancer J. 2016;6:e385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marini F, Kim N, Schuffert A, Wood R. POLN, a nuclear PolA family DNA polymerase homologous to the DNA cross‐link sensitivity protein Mus308. J Biol Chem. 2003;278:32014‐32019. [DOI] [PubMed] [Google Scholar]
- 25. Grant R, Denroche R, Borgida A, et al. Exome‐wide association study of pancreatic cancer risk. Gastroenterology. 2018;154:719‐722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Klappa P, Stromer T, Zimmermann R, Ruddock L, Freedman R. A pancreas‐specific glycosylated protein disulphide‐isomerase binds to misfolded proteins and peptides with an interaction inhibited by oestrogens. Eur J Biochem. 1998;254:63‐69. [DOI] [PubMed] [Google Scholar]
- 27. Kurisaki H, Nagao Y, Nagafuchi S, Mitsuyama M. Autoimmune gastro‐pancreatitis with anti‐protein disulfide isomerase‐associated 2 autoantibody in aire‐deficient BALB/cAnN mice. PLoS One. 2013;8:e73862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kimura J, Kudoh T, Miki Y, Yoshida K. Identification of dihydropyrimidinase‐related protein 4 as a novel target of the p53 tumor suppressor in the apoptotic response to DNA damage. Int J Cancer. 2011;128:1524‐1531. [DOI] [PubMed] [Google Scholar]
- 29. Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet Med. 2015;17:405‐424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Canto MI, Almario JA, Schulick RD, et al. Risk of neoplastic progression in individuals at high risk for pancreatic cancer undergoing long‐term surveillance. Gastroenterology. 2018;155:740‐751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Canto MI, Harinck F, Hruban RH, et al. International cancer of the pancreas screening (CAPS) consortium summit on the management of patients with increased risk for familial pancreatic cancer. Gut. 2013;62:339‐347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Abe K, Kitago M, Kitagawa Y, Hirasawa A. Hereditary pancreatic cancer. Int J Clin Oncol. 2021;26(10):1784‐1792. 10.1007/s10147-021-02015-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


