Abstract
The relative importance of viral lysis and bacterivory as causes of bacterial mortality were estimated. A laboratory experiment was carried out to check the kind of control that viruses could exert over the bacterial assemblage in a non-steady-state situation. Virus-like particles (VLP) were determined by using three methods of counting (DAPI [4′,6-diamidino-2-phenylindole] staining, YOPRO staining, and transmission electron microscopy). Virus counts increased from the beginning until the end of the experiment. However, different methods produced significantly different results. DAPI-stained VLP yielded the lowest numbers, while YOPRO-stained VLP yielded the highest numbers. Bacteria reached the maximal abundance at 122 h (3 × 107 bacteria ml−1), after the peak of chlorophyll a (80 μg liter−1). Phototrophic nanoflagellates followed the same pattern as for chlorophyll a. Heterotrophic nanoflagellates showed oscillations in abundance throughout the experiment. The specific bacterial growth rate increased until 168 h (2.6 day−1). The bacterivory rate reached the maximal value at 96 hours (0.9 day−1). Bacterial mortality due to viral infection was measured by using two approaches: measuring the percentage of visibly infected bacteria (%VIB) and measuring the viral decay rates (VDR), which were estimated with cyanide. The %VIB was always lower than 1% during the experiment. VDR were used to estimate viral production. Viral production increased 1 order of magnitude during the experiment (from 106 to 107 VLP ml−1 h−1). The percentage of heterotrophic bacterial production consumed by bacterivores was higher than 60% during the first 4 days of the experiment; afterwards, this percentage was lower than 10%. The percentage of heterotrophic bacterial production lysed by viruses as assessed by the VDR reached the highest values at the beginning (100%) and at the end (50%) of the experiment. Comparing both sources of mortality at each stage of the bloom, bacterivory was found to be higher than viral lysis at days 2 and 4, and viral lysis was higher than bacterivory at days 7 and 9. A balance between bacterial losses and bacterial production was calculated for each sampling interval. At intervals of 0 to 2 and 2 to 4 days, viral lysis and bacterivory accounted for all the bacterial losses. At intervals of 4 to 7 and 7 to 9 days, bacterial losses were not balanced by the sources of mortality measured. At these time points, bacterial abundance was about 20 times higher than the expected value if viral lysis and bacterivory had been the only factors causing bacterial mortality. In conclusion, mortality caused by viruses can be more important than bacterivory under non-steady-state conditions.
A few years ago bacterivory was considered to be the most important bacterial-loss factor in aquatic environments (e.g., reference 22). More recently, however, viral infection has been found to account for a significant proportion of bacterial mortality in some aquatic environments (8, 34, 39, 42).
The first attempt to incorporate viruses into the budget of microbial carbon transfer was done by Bratbak et al. (5). These authors measured viral lysis and bacterivory simultaneously in a mesocosm experiment. However, they could not balance bacterial losses with both loss factors. Viral lysis exceeded bacterial heterotrophic production (BHP) by a factor of 6, while bacterivory exceeded it by a factor of 2. Later, two studies, which measured viral lysis and bacterivory simultaneously, found that both factors accounted for the same proportion of bacterial mortality (8, 34). However, another study showed a small contribution of viral lysis to bacterial mortality compared to bacterivory (12). Recently, in a similar study from a eutrophic lake, grazers regulated bacterial production in the epilimnion, whereas in the anoxic hypolimnion was regulated by viral lysis (39). Thus, it is not clear in what situations viral lysis could prevail over bacterivory in controlling bacterial abundance. Weinbauer and Peduzzi (42) concluded, by evaluating the relationship between viral and bacterial abundance, that viral infection could prevail over bacterivory at a high bacterial concentration. These results were in agreement with the uncoupling found between bacteria and heterotrophic nanoflagellates at a high bacterial abundance (9).
All of these studies were done in a steady-state situation in which cells and viruses showed small temporal fluctuations. Large variations in the abundance of organisms in short periods of time occur during phytoplankton blooms. Bratbak et al. (4) have studied the fluctuations in the abundance of virus-like particles (VLP) during a phytoplankton bloom. In their study they found a rapid increase in VLP abundance after the maximal bacterial abundance had been reached. These authors suggested that part of the bacterial population had been lysed by viruses. However, they did not quantify the proportion of bacterial mortality attributable to viral lysis during the bloom. During a phytoplankton bloom, a non-steady-state situation is established and the biological diversity of the environment decreases. Due to the specificity of viral attack, viruses could be a significant source of bacterial mortality in these conditions.
The objective of the present study was to investigate the different proportions of bacterial mortality attributable to viral lysis or bacterivory at different stages of a phytoplankton bloom. In order to avoid the difficulties associated with the drifting of water masses, a situation inherent to marine environments, we carried out a microcosm experiment. The appearance of a phytoplankton bloom was stimulated by adding nutrients to the natural-water sample. Changes in chlorophyll a, flagellate, bacterial, and VLP abundance were monitored over time by different counting methods. Bacterial heterotrophic production, bacterivory, and viral lysis were also measured. The proportion of bacterial mortality due to viruses and to bacterivory was then estimated. In this work, we could assess whether one factor prevailed over the other or whether both acted simultaneously at each stage of the bloom.
MATERIALS AND METHODS
Experimental design.
A microcosm experiment to evaluate the microbial populations during a phytoplankton bloom was performed in November 1995 with water from Masnou harbor, located on the Mediterranean coast (20 km north of Barcelona). The same input of inorganic nutrients was added to two polypropylene bottles with 20 liters of the sampled water filtered throughout 150-μm-pore-size nylon mesh. The inorganic nutrient concentrations at the beginning of the experiment were 46 μM nitrate, 7 μM phosphate, and 60 μM silicate. Both replicates of the enriched cultures were incubated for 12 days at a temperature similar to the original sample (16°C). Cultures were incubated with periods of 12 h of light and 12 h of dark. Light intensity was about 100 to 120 microeinsteins m−2 s−2 during the light period. Samples for chlorophyll a concentration, bacterial, flagellate, and VLP abundance were taken daily from both cultures. Bacterial heterotrophic production, bacterivory, and the percentage of visibly infected bacteria were also measured at different times during the experiment (for the initial sample and after 2, 4, 7, and 9 days). At the same sampling days, viral-decay experiments were performed with water samples from the cultures.
Chlorophyll a determination and direct counts of microorganisms.
The chlorophyll a concentration was determined fluorometrically in 100-ml samples that were filtered through GF/F glass fiber filters and then frozen. The filters were extracted overnight in 90% acetone at 4°C, and the fluorescence of the extract measured with a Turner Designs fluorometer (45).
Samples for bacterial, flagellate, and VLP abundance studies were fixed with glutaraldehyde (1% final concentration) in polypropylene bottles.
Bacteria were stained with DAPI (4′,6-diamidino-2-phenylindole; 1 μg ml−1, final concentration), filtered onto black 0.2-μm-pore-size polycarbonate filters (25), mounted on microscope slides, and then frozen. The bacterial abundance was determined with a Nikon epifluorescence microscope at a magnification of ×1,250. About 200 to 300 bacteria were counted per sample. Flagellates were stained with DAPI (1 μg ml−1, final concentration) and filtered onto black 0.6-μm-pore-size polycarbonate filters mounted on microscope slides and frozen. Flagellate abundance was determined with a Nikon epifluorescence microscope at a magnification of ×1,250. About 200 to 300 flagellates were counted per sample. Phototrophic nanoflagellates (PNFs) were distinguished from heterotrophic nanoflagellates (HNFs) based on the fluorescence of chlorophyll a.
Viral direct counts.
VLP abundance was determined by three different methods: YOPRO staining (Molecular Probes YO-PRO 1) and epifluorescence microscopy, DAPI staining and epifluorescence microscopy, and transmission electron microscopy (TEM). Unfixed samples for VLP counting with YOPRO were immediately filtered (14). Samples of 100 μl were diluted with 700 μl of Mili-Q water filtered through a 0.02-μm-pore-size filter (Anodisc). Each diluted sample was gently filtered through a 0.02-μm-pore-size Anodisc 25 filter. The Anodisc filters with the filtered sample were placed on 80 μl of the staining solution (YO-PRO 1; 50 μM final concentration) in a petri dish and incubated in the dark for 2 days at room temperature. The filters were then washed twice by filtering 800 μl of Mili-Q water through the membrane. Filters were transferred to glass slides and immediately covered with a drop of spectrophotometric-grade glycerol and a coverslip. Filters were stored at −20°C until counted.
VLP abundance was determined also by using DAPI staining and counting the particles under the epifluorescence microscope (35). VLP were stained with DAPI (1 μg ml−1, final concentration) overnight and filtered onto 0.02-μm-pore-size filters (Anodisc). Filters were mounted on microscope slides with nonfluorescent oil (R. P. Cargille Laboratories, Inc.) and frozen. Both YOPRO- and DAPI-stained samples were counted with a Nikon epifluorescence microscope at a magnification of ×1,250. Ca. 200 to 300 VLP were counted per sample.
In samples examined by TEM, viruses were harvested onto the grids (400-mesh Ni electron microscope grids with carbon-coated Formvar film) by using a Beckman SW41 swing-out rotor run at 100,000 × g for 30 min at 20°C (3, 35). For each sample, duplicate grids were stained for 1 min with uranyl acetate (2% [wt/wt]). VLP were enumerated and sized in a Hitachi 600 transmission electron microscope operated at 80 kV and at a magnification of ×100,000. Fields were randomly selected and counted until the total counts exceeded 200 VLP. Because of the high acceleration voltage (80 kV) used in this study, we were able to identify cells containing mature phages in the same grids (41). A cell was considered infected when the phage inside could be clearly recognized on the basis of shape and size (5, 41). The minimal number of phages found in an infected cell was six. At least 500 cells were inspected at a ×20,000 magnification for potential infection in each sample.
Bacterial heterotrophic production and cell volume.
Bacterial heterotrophic production was determined by measuring the incorporation of [3H]leucine into the cells (15). Two replicates and a formaldehyde (4% final concentration)-killed control were incubated with [3H]leucine (40 nM final concentration) at the same temperature as the original cultures. Incubations were terminated after 1 h and 45 min by the addition of formaldehyde (4% final concentration). The samples were then filtered through 0.22-μm-pore-size cellulose acetate filters and then rinsed twice in 5% ice-cold trichloroacetic acid and three times with 80% ethanol. The filters were dissolved with 0.5 ml of ethyl acetate, and 4.5 ml of Optiphase Hisafe II scintillation cocktail was added before counting was done with a Beckman scintillation counter. The amount of [3H]leucine incorporated was converted to the amount of carbon produced by using an empirical conversion factor estimated for coastal Mediterranean waters (24). The carbon-produced value was converted to the number of cells produced by dividing by the carbon content per cell. This was calculated with the equation reported by Norland (20): picograms of carbon cell−1 = 0.09 × (μm3)0.9.
Cell volumes were determined with an image analysis system that measured at least 200 cells per sample. A Hamamatsu C2400-08 video camera was used to examine microscopic preparations. Objects occupying less than 7 pixels (equivalent to a sphere with a diameter less than 0.2 μm) were discarded. The remaining objects were measured, and the volume was calculated from the area and perimeter measurements with the formula of Fry (6). The system was calibrated with fluorescent latex beads and with natural bacterioplankton samples measured simultaneously by phase-contrast microscopy and epifluorescence (16).
Bacterivory by protists.
This parameter was measured with fluorescently labeled bacteria (FLB [31]) by using the FLB disappearance method (30). FLB were prepared from a heterotrophic bacterium isolated from the Mediterranean coast. One-liter samples were incubated at the same temperature as the original cultures in polycarbonate bottles in the dark. Incubations lasted 48 h and were stopped by fixing subsamples with glutaraldehyde (final concentration, 2%). One experiment for each culture and a control killed with formaldehyde (final concentration, 4%) were done at the times indicated above.
Viral-decay experiments.
Incubations for VLP decay experiments were carried out in 1.5-liter polyethylene bottles. One experiment was carried out for each culture at the indicated times. Experiments were incubated at the same temperature as the original cultures for 48 h. VLP decay was recorded after inhibiting the production of new viruses by adding KCN to a final concentration of 2 mM (13). Samples for [3H]leucine incorporation were taken at the beginning and at the end of each experiment in order to make sure that the microbial activity was stopped by KCN. The viral-decay rate (VDR) was calculated from a log-linear part of the decay curves by using linear regression (13, 17). Samples for counting VLP were taken at intervals of 1 to 2 h for the first 9 to 10 h of the experiment. After this time, samples were taken less frequently until the end of the experiments. The changes in the number of VLP in the microcosms without added KCN were used as controls.
RESULTS
VLP abundance.
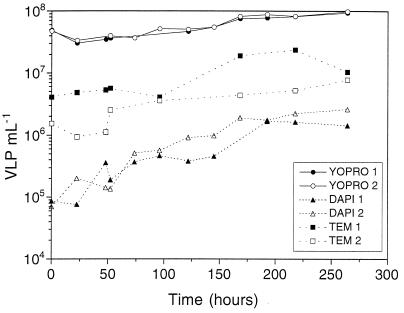
VLP abundance counted with YOPRO staining doubled from the beginning (5 × 107 VLP ml−1) to the end (1 × 108 VLP ml−1) of the experiment (Fig. 1). Both replicate cultures showed similar numbers. The average coefficient of variation (CV) for YOPRO counts was 12%. With the TEM method, VLP counts increased from 3 × 106 VLP ml−1 at the beginning to 5 × 106 VLP ml−1 at the end of the experiment (Fig. 1). The average CV for TEM counts was 20%. VLP abundance determined with DAPI staining gave the lowest counts (Fig. 1). With this method, however, viruses increased 1 order of magnitude (from 105 VLP ml−1 to 106 VLP ml−1). The average CV for DAPI counts was 10%. A linear function could be fitted to the log-transformed VLP abundance with time for each method. Analysis of covariance was used to test for significant differences among the three methods of viral counting. The three methods gave significantly different results (P < 0.001, n = 74).
FIG. 1.
VLP abundance during the experiment as determined by the different counting methods.
Nutrients and chlorophyll a.
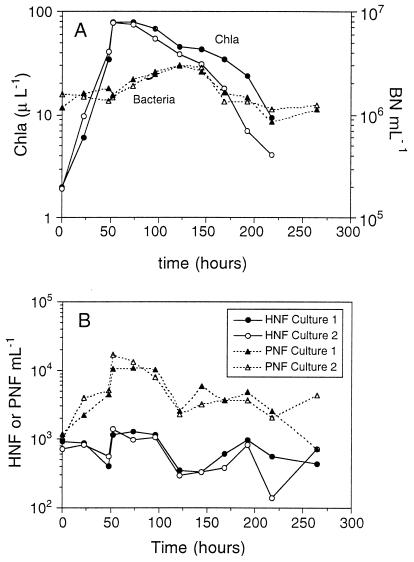
Nutrient concentrations showed a depletion of nitrate and silicate after 48 h. Phosphate levels decreased slowly during the experiment (data not shown). The chlorophyll a concentration showed a rapid increase (>10-fold) after nutrient addition for up to the 48 h after initiation of the experiment. The chlorophyll a concentration then decreased until the end of the experiment (Fig. 2A).
FIG. 2.
(A) Chlorophyll a (Chla) and bacterial abundance (BN) throughout the experiment. (B) HNF and PNF abundance throughout the experiment.
Abundance of bacteria and flagellates.
Bacterial abundance reached the maximal concentration at 122 h. Bacteria started to increase in number after the peak concentration of chlorophyll a had been reached. Bacteria increased their initial abundance by a factor of 3. After 122 h, bacteria decreased to levels similar to the initial value (Fig. 2A).
PNFs showed the same pattern as chlorophyll a. Their concentration from the beginning of the experiment to 52 h increased 10-fold. After this time PNF abundance started to decrease until 122 h and remained constant thereafter (Fig. 2B). HNFs showed oscillations with minimal values after 48 and 122 h and higher values for the period in between these two time points (Fig. 2B).
Bacterial production, cell volume, and growth rate.
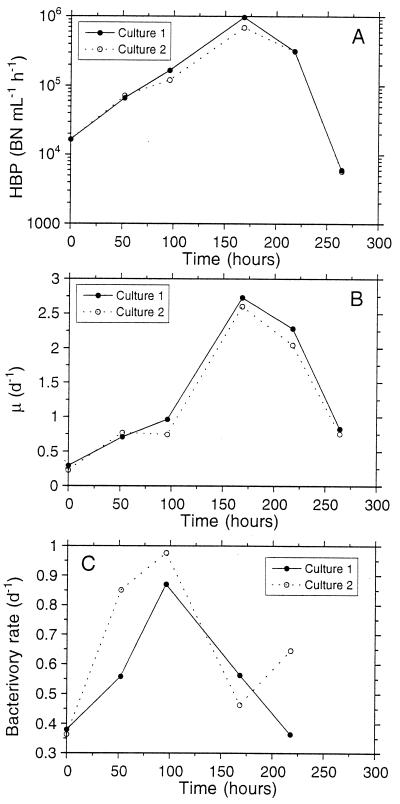
From the beginning of the experiment until 168 h, BHP increased 100 fold (Fig. 3A). After 218 h, BHP showed a rapid decrease until the end of the experiment. Bacterial cell volumes during the experiment are shown in Table 1. The volumes measured were used to convert BHP from carbon units to cell units (see Materials and Methods). The specific bacterial growth rate (μ) showed a similar pattern to that of BHP. Values of μ at 168 h were 5 times higher than those at the beginning of the experiment (Fig. 3B). The bacterivory rate increased about threefold from the beginning point to the 96-h point. The maximal bacterivory rate appeared 3 days before the maximal specific bacterial growth rate. Between 96 and 168 h, the bacterivory rate rapidly decreased (Fig. 3C).
FIG. 3.
(A) BHP during the experiment. Bars indicate the standard errors based on two replicates for each culture. When no bars are visible, the errors were smaller than the marker points. (B) Specific growth rate (μ) of the bacteria during the experiment. (C) Bacterivory rate during the experiment. d, day.
TABLE 1.
Bacterial volumes at different times during the experiment for both cultures
| Time (days) | Culture | Mean vol (μm3) ± SE |
|---|---|---|
| 0 | 1 | 0.03 ± 0.01 |
| 2 | 0.04 ± 0.02 | |
| 1 | 1 | 0.04 ± 0.02 |
| 2 | 0.04 ± 0.02 | |
| 2 | 1 | 0.04 ± 0.02 |
| 2 | 0.04 ± 0.02 | |
| 4 | 1 | 0.03 ± 0.01 |
| 2 | 0.04 ± 0.01 | |
| 7 | 2 | 0.05 ± 0.02 |
| 9 | 1 | 0.06 ± 0.03 |
| 2 | 0.05 ± 0.03 |
Viral decay and visibly infected bacteria.
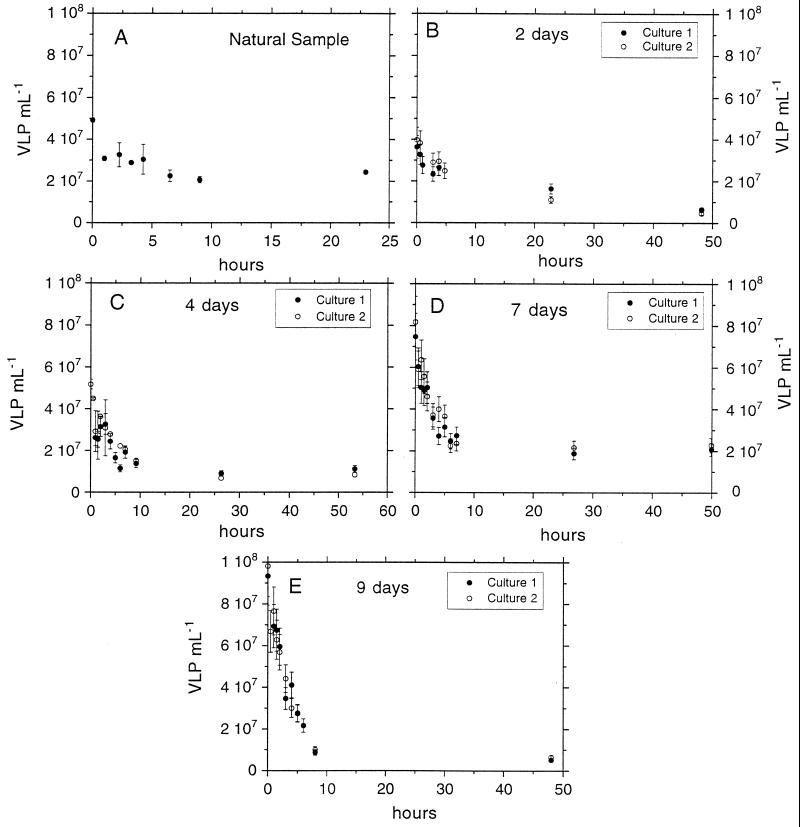
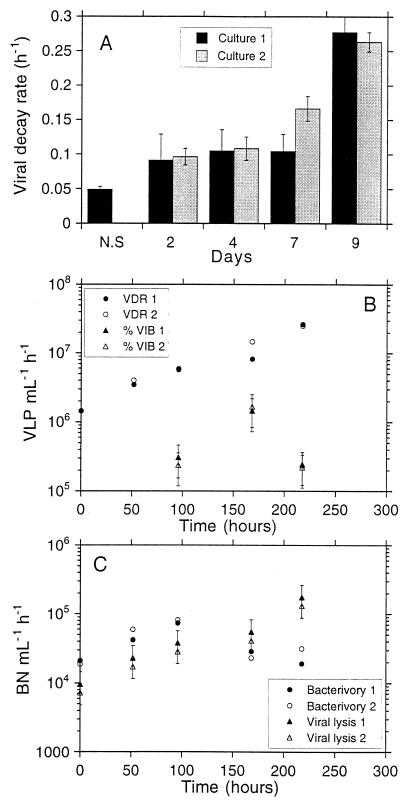
Viral decay experiments showed a similar pattern of VLP abundance over time (Fig. 4). VLP abundance decreased rapidly at first (<10 h) and afterwards either decreased more slowly (see experiment carried out on day 2 [Fig. 4B]) or remained constant (see remaining experiments). In the original sample, VLP abundance rapidly decreased over the first 30 min and decreased more slowly from this point to 10 h (Fig. 4A). The VDRs are shown in Table 2 and Fig. 5A. The VDR for the original water showed the lowest value. The VDR at 9 days showed the maximal values. The VDRs for both replicate cultures were similar (Fig. 5A). The changes in VLP levels in microcosms without KCN that were used as controls were always very small for the few hours of the decay experiments and thus insignificant compared to the fast decay seen in the sample bottles with KCN.
FIG. 4.
Viral-decay experiments performed after 0 (A), 2 (B), 4 (C), 7 (D), and 9 (E) days. At day 0, the viral-decay experiment was performed with the natural sample. Bars indicate the standard errors based on two replicates for each experiment. When no bars are visible, the errors were smaller than the marker points.
TABLE 2.
Statistical parameters of the slopes (VDR) obtained from the decay experiments performed on different days and for both cultures 1 and 2
| Time (days) | Culture | VDR (h−1) | n | P | r2 | Time (h) |
|---|---|---|---|---|---|---|
| 0 | 1 and 2 | 0.049 | 6 | 0.002 | 0.975 | 0–9 |
| 2 | 1 | 0.091 | 5 | 0.045 | 0.675 | 0–5 |
| 2 | 0.096 | 5 | 0.004 | 0.955 | 0–5 | |
| 4 | 1 | 0.104 | 9 | 0.012 | 0.611 | 0–10 |
| 2 | 0.108 | 10 | <0.001 | 0.830 | 0–10 | |
| 7 | 1 | 0.104 | 9 | 0.003 | 0.722 | 0–9 |
| 2 | 0.166 | 10 | <0.001 | 0.917 | 0–9 | |
| 9 | 1 | 0.277 | 9 | <0.001 | 0.955 | 0–9 |
| 2 | 0.263 | 10 | <0.001 | 0.980 | 0–9 |
FIG. 5.
(A) VDR calculated as the slope of the log-linear part of each decay experiment. Bars indicate the standard error for each slope. Significance and r2 values for these slopes and the interval of hours used to calculate them are shown in Table 2. (B) VLP produced per milliliter per hour, calculated according to the VDR and according to the percentage of infected cells (%VIB). In the latter case we have assumed a burst size of between 100 to 300 VLP per cell. Error bars correspond to the standard error of the estimated values with this range of burst sizes. (C) Bacterial cells lost per hour and per milliliter due to viral lysis and due to bacterivory. The bacterial mortality due to viral lysis corresponds to the values calculated from the VDR. Error bars correspond to the standard error of the estimated values with a range of burst sizes of between 100 and 300 VLP per cell.
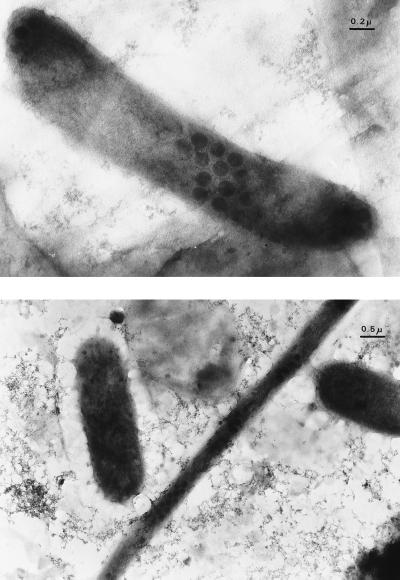
The percentage of visibly infected bacteria (%VIB; Fig. 6) was always lower than 1% (Table 3). We converted VIB to total infected bacteria with the factors reported by Proctor et al. (29). The resulting percentage was still lower than 5%. Initially and at day 2, no infected cells could be detected. The maximal VIB value was found at day 7.
FIG. 6.
Visibly infected bacteria at day 7 of the experiment.
TABLE 3.
%VIB for the different sampling days and for both cultures
| Time (days) | Culture | %VIBa |
|---|---|---|
| 0 | 1 and 2 | ND |
| 1 | 1 and 2 | ND |
| 2 | 1 and 2 | ND |
| 4 | 1 | 0.2 |
| 2 | 0.2 | |
| 7 | 1 | 0.5 |
| 2 | 0.6 | |
| 9 | 1 | 0.4 |
| 2 | 0.4 |
Infected cells were not detected (ND) during the first days of the experiment.
Viral production rates calculated from the VDR or from the %VIB are shown in Fig. 5B. To convert the %VIB to viral production, we assumed that the viral latent period was approximately equal to the host generation time (12, 29), and we used a range of burst sizes between 100 and 300 viruses released per lysed cell. Viral production calculated by using the VDR increased exponentially from the beginning to 218 h (Fig. 5B). Viral production at this point was about 1 order of magnitude higher than that at the beginning of the experiment. Viral production calculated from the %VIB showed a different pattern, with the maximal production occurring at 168 h (Fig. 5B). By using this approach the viral production measured was at least ten times lower than when measured with the VDR.
Bacterial losses due to bacterivory or to viral lysis.
In order to calculate the number of bacteria lysed per milliliter and per hour from the VDR, we used the same range of burst sizes as described above. The number of bacteria ingested by bacterivores peaked at 96 h, while the number of bacteria lysed by viruses increased exponentially from the beginning to the end of the experiment (Fig. 5C).
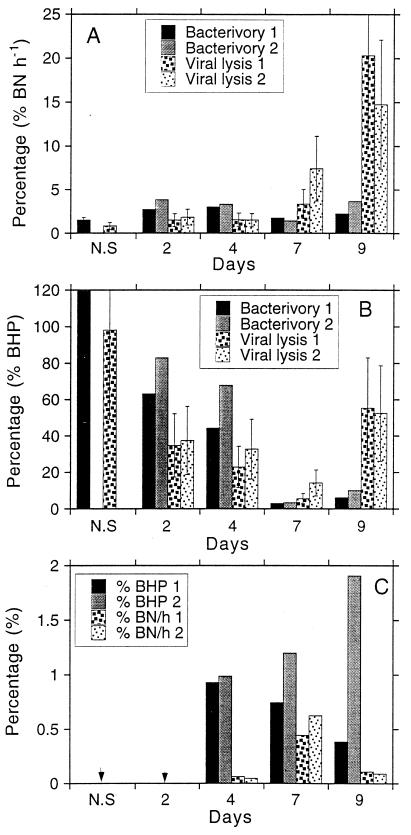
The percentages of bacterial abundance and production lost due to viral lysis or to bacterivory are shown in Fig. 7. The percentage of bacteria ingested by bacterivores per hour was always lower than 5% (Fig. 7A). Maximal values corresponded to days 2 and 4. The percentage of bacteria lysed by viruses per hour (calculated from the VDR) was lower than that for bacteria ingested by bacterivory during the first few sampling days (Fig. 7A). At days 7 and 9, however, the percentage of bacterial mortality due to viral infection was higher than that due to bacterivory.
FIG. 7.
(A) Bacteria ingested by bacterivores or lysed by viruses as percentages of bacterial abundance (% BN) per hour at different days of the experiment. The values of viral lysis were calculated from the VDR. N.S., natural sample. Error bars indicate the lowest and the highest numbers of bacteria lysed by viruses calculated by assuming a range of burst sizes of between 100 and 300 VLP released per cell. (B) Bacteria ingested by bacterivores or lysed by viruses as percentages of BHP at different days of the experiment for both cultures. The values of viral lysis were calculated from the VDR. N.S., natural sample. Error bars indicate the lowest and highest values of bacteria lysed by viruses calculated by assuming a range of burst sizes of between 100 and 300 VLP released per cell. (C) Bacteria ingested by bacterivores or lysed by viruses as percentages of BHP and abundance (BN) per hour at different days of the experiment. Numbers of bacteria lysed by viruses have been calculated from the percentage of infected cells. The two arrowheads indicate samples where infected cells could not be detected.
The percentage of BHP ingested by bacterivores was higher than 100% at time of the original sampling (Fig. 7B). At days 2 and 4, it was between 40 and 80%. At days 7 and 9, the percentage of BHP ingested by bacterivores was lower than 10%. The percentage of BHP lysed by viruses (calculated by using the VDR) was also higher at the time of the original sampling (100%). At days 2 and 4 this percentage was about half the percentage of BHP ingested by bacterivores. At day 7, viral lysis and bacterivory accounted for a similar percentage of BHP, although viral lysis accounted for a slightly higher percentage than bacterivory. At day 9 viral lysis accounted for a much larger fraction of BHP than bacterivory (Fig. 7B).
The percentage of bacterial losses due to viral infection determined by using the second approach (i.e., %VIB) is shown in Fig. 7C. This percentage was always lower than 5% of BHP. Bacterial abundance lost per hour was always lower than 1%. At day 9 the percentage of mortality due to viral infection in one replicate was four times higher than in the other replicate.
DISCUSSION
Methods of viral counting.
VLP abundance was found to vary by an order of magnitude or more depending on the analytic method used. The slopes of the increase in abundance over time found with each method were significantly different. However, using the data presented here plus other data from different environments, we have found a significant linear relationship between DAPI and TEM counts and between YOPRO and TEM counts (10, 11). Thus, for a broad range of environments it seems safe to assume that VLP abundance shows a proportional difference among the methods used (14, 43). However, the differences found in the present study suggest that in highly productive environments the methods of counting VLP could be influenced to a different extent by the particulate material in the sample, since the slopes found here were not equal.
DAPI counts seem to underestimate the number of VLP (35), mostly because the small DAPI-stained VLP are not visible under the epifluorescence microscope with this dye. It has been also reported that TEM counts underestimate VLP abundance to a degree depending on the amount of organic matter present in the sample (14). Moreover, this method presents the highest CV (20% in this study) compared to DAPI counts (CV = 10%) and YOPRO counts (CV = 12%). The YOPRO counting method has some advantages over TEM (14). Unfortunately, there is no test that can absolutely eliminate the possibility that virus-size particles other than viruses are stained by YOPRO. Hennes and Suttle (14) investigated this possibility and concluded that the discrepancy between TEM and YOPRO estimates of viral abundance results from the TEM protocol underestimating the virus concentration. Recently, Noble and Fuhrman (19) also estimated VLP abundance with another fluorescent dye (SYBR green I) and the counts were higher than the TEM counts. Thus, for the rest of the discussion we will refer to the VLP abundance obtained with the YOPRO method alone.
Limitations of the measurement of bacterial mortality due to viral lysis.
We used two approaches to estimate bacterial mortality due to viral infection: determining the %VIB and measuring the VDR in cyanide-amended cultures. The two methods showed different results.
The direct count of infected cells presents several problems (29, 34, 41). First, viruses are only visible in infected bacteria during a part of their latent period. To be able to convert the VIB to total infected cells, Proctor et al. (29) calculated a conversion factor derived for some specific host-virus systems. The factor, however, was calculated for the conversion of %VIB in thin sections and not in whole cells, as we determined the %VIB. Second, to convert total infected cells to bacterial mortality per time period, the length of the viral latent period must be known. At the moment there is no way to measure this period in natural samples. However, for some host-virus systems investigated in cultures it has been reported that the latent period is similar to the host generation time (29).
The most important problems that we found in the present study in quantifying the %VIB were (i) the possible rupture of some infected cells by use of a high centrifugal speed (100,000 × g [39, 40]) and (ii) the fact that a high percentage of bacteria were opaque under TEM despite the use of a high acceleration voltage (80 kV). Thus, our estimates of bacterial mortality from the %VIB are clearly underestimates. Therefore, we will not refer to them for the rest of this discussion.
Calculations of the VDR inhibiting viral production with KCN (13) also present problems (7, 13). VDR values obtained by this method represent a minimal estimation of viral decay because only abiotic loss factors are considered. However, VDR found in some studies by this method exceeded the BHP severalfold and thus the bacterial assemblage would have to disappear in a short period of time (13). This is unrealistic and, therefore, it seems that the results obtained by this method have to be examined with caution. In the present study, viral production calculated by this method showed results consistent with the parallel estimations of bacterial production and bacterivory. Likewise, in another study (17), a good agreement was found between this method and the %VIB. The main problem of the cyanide method is the difficult interpretation of viral-decay experiments. Curves obtained from these experiments showed a rapid decrease at the several first hours of the experiment and a slow rate thereafter (>10 h), as has been found in other studies (13, 17). We have calculated our VDR with data from the initial 10-h period of the experiments, assuming that the later rate might be an artifact of the incubation in a small volume for a longer period of time. Thus, even given that this method presents some uncertainties, we still consider the results obtained here as reasonable.
In order to convert the VDR to yield bacterial mortality, the burst size must be known. The method of counting VLP inside the VIB has been used in some studies to estimate the burst size in natural environments (8, 12, 39, 40, 41). However, this method could underestimate the burst size because the stage of the latent period to which the VLP observed in the cells correspond is unknown. Also, phages lying on top of each other may be counted as one phage (41). Different studies have used a range of burst sizes between 10 and 300 phages released per lysed bacterium (13) to calculate the viral impact on the bacterial assemblage from the viral production rates. Other studies have used an average burst size of 50 to calculate the viral production from the VIB (37). Burst size has been shown to be dependent on the bacterial growth rate (26) and on the bacterial cell volume (12, 41). This implies that nutrient supply may indirectly determine the total number of phage set free per bacterium (41). Børsheim (2) calculated an average burst size of 185 phages in cultured marine bacteria. Given the conditions of our experiment (high nutrient supply in enclosure cultures), we used the upper range of the burst sizes reported in the literature (i.e., 100 to 300) to convert our estimated VDR to bacterial mortality values.
Viral lysis and bacterivory as factors of bacterial mortality during a phytoplankton bloom.
Given the enriched conditions of the experiment, bacteria were expected to grow rapidly, and in order to control bacterial abundance and growth the loss process should be significant and therefore measurable. The bacterivory rate was maximal before reaching the peaks of bacterial heterotrophic production and bacterial specific growth rate. By this time the phytoplankton bloom was declining (96 h). Immediately after the maximum bacterivory peak, HNF levels decreased and increased only slightly after 3 days. PNF levels followed the pattern of chlorophyll a for up to 96 h and increased slightly after the peak of bacterivory. This could indicate that part of the phototrophic nanoflagellate assemblage was mixotrophic. After a diatom bloom, when mineral nutrients are depleted, the diatoms will sink out, mainly as cysts. During these periods mixotrophy has been shown to be a successful strategy for the small algae to retain a C/N/P ratios close to the Redfield ratio (21).
The fact that heterotrophic nanoflagellates increased only slightly after the bacterivory peak could suggest that not all the bacterivory was due to the HNF assemblage. Mixotrophy is one possibility, but ciliates could also be responsible for a portion of the bacterivory during the experiment. Ciliates would pass through the 150-μm-pore-size filters used to set up the cultures. In spite of their expected low initial concentration (38), the ciliates might have grown during the experiment and become important bacterivores in an advanced stage of the bloom. At the same time, ciliates that ingest flagellates could have been also responsible for the slight fluctuations in flagellate abundance during the experiment.
VLP abundance increased from the first day to the end of the experiment. This was a consequence of an increase in viral production. Maximal viral production appeared 2 days after the maximal bacterial heterotrophic production point was reached. This could indicate that part of the actively growing bacterial assemblage was susceptible to viral attack.
Part of the viral assemblage could be infecting phytoplankton. However, our results showed a clear decrease in chlorophyll a when the nutrient concentration was depleted, while the number of VLP increased until the end of the experiment. This suggests that most of the VLP were not produced by phytoplankton.
Bacterivory and viral lysis did not present their maximal values at the same stage of the bloom. While bacterivory was maximal immediately after the bloom when bacterial abundance was maximal, viral production showed maximal values when the phytoplankton bloom had declined. This could be a consequence of the different strategies of both groups of organisms: the host-selective predators (viruses) determine the abundance of bacteria in each host-virus system, while the nonselective predators (HNFs) determine the size of the total bacterial assemblage (37).
Balance between bacterial losses and bacterial production.
Bacterivory plus viral lysis balanced all the BHP before, during, and immediately after the bloom period. From the beginning to day 4, bacterivory accounted for a higher percentage of the bacterial production and abundance per hour than did viral lysis. From day 7 onward, bacterial heterotrophic production could not be balanced by viral lysis plus bacterivory. In the postbloom period, bacterivory accounted for a lower percentage of BHP and bacterial abundance per hour than did viral lysis.
In this experiment bacterial cells did not show a constant abundance over time. Thus, for each period of time the net changes in bacterial abundance should be balanced by the heterotrophic bacterial production minus the bacterial losses (if these were the only factors responsible for bacterial mortality). Therefore, bacterial abundance (BN) plus BHP measured for a fixed day (di), minus the losses due to bacterivory (BTV) and minus the losses due to viral lysis (VL) measured during the same day, would be equivalent to the bacterial abundance observed for the next sampling time (df) as follows: BNdf = BNdi + (BHP − BTV − VL)di, where df = di + 2 days.
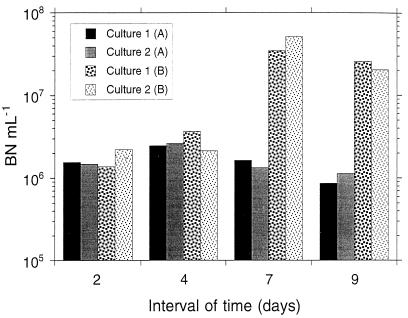
Because the interval of sampling activities (BHP, BTV, and VL) was 2 days (except from days 4 to 7), we calculated the balance for this time interval. In doing this, however, we are probably introducing an error, because BHP and viral lysis were measured over a period of a few hours, whereas bacterivory was measured for a 2-day period. This error could be especially important in a system such as this, which is changing continuously. Thus, in order to be able to compare these processes, we used the averaged BHP and the averaged viral lysis measured at the beginning and at the end of this 2-day period. The results, presented in Fig. 8, therefore correspond to the following equation: BNdi + 2 = BNdi + average BHPdi, di + 2 − average VLdi, di + 2 − BTVdi, where di = 0, 2, 4, and 7 days.
FIG. 8.
Bacterial abundance (BN ml−1) as determined by epifluorescence microscopy at the end of each time interval BNdi + 2 (A) and bacterial abundance calculated as follows: BNdi + average HBPdi, di + 2 + average VLdi, di + 2 + BVdi (B), where i is 0, 2, 4, and 7 days. Values were calculated for each culture separately.
In the two first intervals of the experiment (0 to 2 and 2 to 4 days), viral lysis and bacterivory balanced bacterial losses with the net changes observed in bacterial abundance. During this period of time, although bacterivory and viral production increased, BHP increased faster. From days 4 to 7, bacterivory and viral lysis did not balance the zero increase in bacterial abundance. After the losses due to bacterivory and viral lysis from the BHP are subtracted, the bacterial abundance should have been about 30 times higher than the observed abundance. Between days 7 and 9, we did not find a balance: bacterial abundance was about 20 times lower than the expected value if viral lysis and bacterivory were the only factors causing bacterial mortality.
A factor of 20 to 30 is difficult to reconcile unless at this time other factors were responsible for bacterial mortality. The attachment to algae could be an important factor impacting bacterial losses during this period. It has been reported that bacterial cells attach mainly to moribund algae (1). During this period (days 7 to 8) mineral nutrients were depleted and diatoms could sink out. If bacteria attached to them, they would be removed from the water column and they would not be counted. At the same time, it has been reported that attached bacteria show higher values of production than free-living bacteria (23). In the method to measure BHP we would not distinguish between production corresponding to attached or free-living bacteria. Thus, in this case BHP could be overestimated with respect to bacterial abundance and bacterivory measured by microscopy. The percentage of VIB has also been reported to be higher in attached bacteria than in free-living bacteria (28). By the viral decay method to measure viral production, we could only detect the decay of free VLP. Thus, viral lysis might be also underestimated here in relation to the BHP.
The experiment cannot be directly extrapolated to nature because the food web was simplified by eliminating organisms larger than 150 μm. However, it gives an indication of the kind of control that viruses could exert over the bacterial assemblage in a non-steady-state situation such as a phytoplankton bloom. As has been pointed out before (4), viruses show intense activity in environments that have received a load of nutrients. In such systems, bacteria will be able to grow rapidly because of the increased amount of carbon provided by the phytoplankton. Bacterivores will respond to the increase in bacterial biomass. They will exert nonspecific control on bacterial abundance and, at the same time, they will stimulate growth of part of the bacterial assemblage able to take the organic carbon released by their activity. At this time viruses will have their maximal impact, thus preventing any particular species from becoming too dominant (37).
ACKNOWLEDGMENTS
We thank Ricardo Guerrero for making possible the use of the electron microscope and Rosina Gironés, Montse Puig, and Sonia Pina for providing the ultracentrifuge.
This work was supported by PB95-0222-C02-01. N.G.-B. was supported by an FI scholarship from the “Generalitat de Catalunya,” and K.L. was supported by an Erasmus grant from the European Union.
REFERENCES
- 1.Azam F, Fenchel T, Field J G, Gray J S, Mayer-Reil L A, Thingstad F. The ecological role of water-column microbes in the sea. Mar Ecol Prog Ser. 1983;10:257–263. [Google Scholar]
- 2.Børsheim K I. Native marine bacteriophages. FEMS Microbiol Ecol. 1993;102:141–159. [Google Scholar]
- 3.Bratbak G, Heldal M. Total count of viruses in aquatic environment. In: Kemp P F, Sherr B F, Sherr E B, Cole J J, editors. Handbook of methods in aquatic microbial ecology. Boca Raton, Fla: Lewis Publishers; 1993. pp. 135–138. [Google Scholar]
- 4.Bratbak G, Heldal M, Norland S, Thingstad T F. Viruses as partners in spring bloom microbial trophodynamics. Appl Environ Microbiol. 1990;56:1400–1405. doi: 10.1128/aem.56.5.1400-1405.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bratbak G, Heldal M, Thingstad T F, Riemann B, Haslund O H. Incorporation of viruses into the budget of microbial C-transfer. A first approach. Mar Ecol Prog Ser. 1992;83:273–280. [Google Scholar]
- 6.Fry J C. Direct methods and biomass estimation. Methods Microbiol. 1990;22:41–85. [Google Scholar]
- 7.Fuhrman J A, Suttle C A. Viruses in marine planktonic systems. Oceanography. 1993;6:51–63. [Google Scholar]
- 8.Fuhrman J A, Noble R T. Viruses and protists cause similar bacterial mortality in coastal seawater. Limnol Oceanogr. 1995;40:1236–1242. [Google Scholar]
- 9.Gasol J M, Vaqué D. Lack of coupling between heterotrophic nanoflagellates and bacteria: a general phenomenon across aquatic systems? Limnol Oceanogr. 1993;38:657–665. [Google Scholar]
- 10.Guixa-Boixereu, N., J. I. Calderón-Paz, J. M. Gasol, and C. Pedrós-Alió. Low viral impact on bacteria in an oligotrophic marine system: the northwestern Mediterranean Sea. Aquat. Microb. Ecol., in press.
- 11.Guixa-Boixereu, N., C. Pedrós-Alió, J. I. Calderón-Paz, and J. M. Gasol. Relationships between viral abundance and environmental factors across diverse aquatic environments. Submitted for publication.
- 12.Guixa-Boixereu N, Calderón-Paz J I, Heldal M, Bratbak G, Pedrós-Alió C. Viral lysis and bacterivory as prokaryotic loss factors along a salinity gradient. Aquat Microb Ecol. 1996;11:215–227. [Google Scholar]
- 13.Heldal M, Bratbak G. Production and decay of viruses in aquatic environments. Mar Ecol Prog Ser. 1991;72:205–212. [Google Scholar]
- 14.Hennes K P, Suttle C A. Direct counts of viruses in natural waters and laboratory cultures by epifluorescence microscopy. Limnol Oceanogr. 1995;40:1050–1055. [Google Scholar]
- 15.Kirchman D L. Leucine incorporation as a measure of biomass production by heterotrophic bacteria. In: Kemp P F, Sherr B F, Sherr E B, Cole J J, editors. Handbook of methods in aquatic microbial ecology. Boca Raton, Fla: Lewis Publishers; 1993. pp. 509–512. [Google Scholar]
- 16.Massana R, Gasol J M, Bjørnsen P K, Blackburn N, Hangström A, Hietanen S, Hygum J, Kuparinen J, Pedrós-Alió C. Measurement of bacterial size via image analysis of epifluorescence preparations: description of an inexpensive system and solutions to some of the most common problems. Sci Mar. 1997;61:397–407. [Google Scholar]
- 17.Mathias C B, Kirschner A K T, Velimirov B. Seasonal variations of virus abundance and viral control of the bacterial population in backwater system of the Danube River. Appl Environ Microbiol. 1995;61:3734–3740. doi: 10.1128/aem.61.10.3734-3740.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagata T, Kirchman D L. Release of dissolved organic matter by heterotrophic protozoa: implications for microbial food webs. Arch Hydrobiol. 1992;35:99–109. [Google Scholar]
- 19.Noble R T, Fuhrman J A. Use of SYBR green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat Microb Ecol. 1998;14:113–118. [Google Scholar]
- 20.Norland S. The relationship between biomass and volume of bacteria. In: Kemp P F, Sherr B F, Sherr E B, Cole J J, editors. Handbook of methods in aquatic microbial ecology. Boca Raton, Fla: Lewis Publishers; 1993. pp. 303–307. [Google Scholar]
- 21.Nygaard K, Tobiesen A. Bacterivory in algae: a survival strategy during nutrient limitation. Limnol Oceanogr. 1993;38:273–279. [Google Scholar]
- 22.Pace M L. Bacterial mortality and the fate of bacterial production. Hydrobiology. 1988;159:41–50. [Google Scholar]
- 23.Pedrós-Alió C, Brock T D. The importance of attachment to particles for planktonic bacteria. Arch Hydrobiol. 1983;98:354–379. [Google Scholar]
- 24.Pedrós-Alió, C., J. I. Calderón-Paz, N. Guixa-Boixereu, M. Estrada, and J. M. Gasol. Relationships between bacterioplankton and phytoplankton biomass and production in an oligotrophic marine environment during summer stratification. Deep-Sea Res., in press.
- 25.Porter K G, Feig Y S. The use of DAPI for identification and enumeration of bacteria and blue-green algae. Limnol Oceanogr. 1980;25:943–948. [Google Scholar]
- 26.Probst-Ricciuti C. Host virus interactions in Escherichia coli: effect of stationary phase on release from MS2-infected bacteria. J Virol. 1972;10:162–165. doi: 10.1128/jvi.10.1.162-165.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Proctor L M, Fuhrman J A. Viral mortality of marine bacteria and cyanobacteria. Nature (London) 1990;343:60–62. [Google Scholar]
- 28.Proctor L M, Fuhrman J A. Roles of marine infection in organic particle flux. Mar Ecol Prog Ser. 1991;69:133–142. [Google Scholar]
- 29.Proctor L M, Okubo A, Fuhrman J A. Calibrating estimates of phage-induced mortality in marine bacteria: ultrastructural studies of marine bacteriophage development from one-step growth experiments. Microb Ecol. 1993;25:161–182. doi: 10.1007/BF00177193. [DOI] [PubMed] [Google Scholar]
- 30.Salat J, Marrasé C. Exponential and linear estimation of grazing on bacteria: effects of changes in the proportion of marked cells. Mar Ecol Prog Ser. 1994;104:205–209. [Google Scholar]
- 31.Sherr B F, Sherr E B, Fallon R D. Use of monodispersed, fluorescently labeled bacteria to estimate in situ protozoan bacterivory. Appl Environ Microbiol. 1987;53:958–965. doi: 10.1128/aem.53.5.958-965.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sherr B F, Sherr E B, Pedrós-Alió C. Simultaneous measurement of bacterioplankton production and protozoan bacterivory in estuarine water. Mar Ecol Prog Ser. 1989;54:209–219. [Google Scholar]
- 33.Steward G F, Wikner J, Cochlan W P, Smith D C, Azam F. Estimation of virus production in the sea: II. Field results. Mar Microb Food Webs. 1992;6:79–90. [Google Scholar]
- 34.Steward G F, Smith D C, Azam F. Abundance and production of bacteria and viruses in the Bering and Chukchi Seas. Mar Ecol Prog Ser. 1996;131:287–300. [Google Scholar]
- 35.Suttle C A. Enumeration and isolation of viruses. In: Kemp P F, Sherr B F, Sherr E B, Cole J J, editors. Handbook of methods in aquatic microbial ecology. Boca Raton, Fla: Lewis Publishers; 1993. pp. 121–133. [Google Scholar]
- 36.Taylor G T, Iturriaga R, Sullivan C W. Interactions of bacterivores grazers and heterotrophic bacteria with dissolved organic matter. Mar Ecol Prog Ser. 1985;23:129–141. [Google Scholar]
- 37.Thingstad T F, Bratbak G, Heldal M, Dundas I. Proceedings of the International Symposium in Microbial Ecology (ISME-7), 27.08-01.09.1995. 1998. Trophic interactions controlling the diversity in pelagic microbial food webs. ISME, Santos, Brazil. [Google Scholar]
- 38.Vaqué D, Blough H A, Duarte C M. Dynamic of ciliate abundance, biomass and community composition in an oligotrophic coastal environment (NW Mediterranean) Aquat Microb Ecol. 1997;12:71–83. [Google Scholar]
- 39.Weinbauer M G, Höfle M G. Significance of viral lysis and flagellate grazing as factors controlling bacterioplankton production in a eutrophic lake. Appl Environ Microbiol. 1998;64:431–438. doi: 10.1128/aem.64.2.431-438.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weinbauer M G, Höfle M G. Size-specific mortality of lake bacterioplankton by natural virus communities. Aquat Microb Ecol. 1998;15:103–113. [Google Scholar]
- 41.Weinbauer M G, Peduzzi P. Frequency, size and distribution of bacteriophages in different marine bacterial morphotypes. Mar Ecol Prog Ser. 1994;108:11–20. [Google Scholar]
- 42.Weinbauer M G, Peduzzi P. Significance of viruses versus heterotrophic nanoflagellates for controlling bacterial abundance in the Northern Adriatic sea. J Plankton Res. 1995;17:1851–1856. [Google Scholar]
- 43.Weinbauer M G, Suttle C A. Comparison of epifluorescence and transmission electron microscope for counting viruses in natural marine waters. Aquat Microb Ecol. 1997;13:225–232. [Google Scholar]
- 44.Weinbauer M G, Fuks D, Peduzzi P. Distribution of viruses and dissolved DNA along a coastal trophic gradient in the Northern Adriatic Sea. Appl Environ Microb. 1993;59:4074–4082. doi: 10.1128/aem.59.12.4074-4082.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yentsch C S, Menzel D W. A method for determination of phytoplankton chlorophyll and phaeophytin by fluorescence. Deep-Sea Res. 1963;10:221–231. [Google Scholar]