Abstract
The essential pre-mRNA splicing factor U2AF2 (also called U2AF65) identifies polypyrimidine (Py) tract signals of nascent transcripts, despite length and sequence variations. Previous studies have shown that the U2AF2 RNA recognition motifs (RRM1 and RRM2) preferentially bind uridine-rich RNAs. Nonetheless, the specificity of the RRM1/RRM2 interface for the central Py tract nucleotide has yet to be investigated. We addressed this question by determining crystal structures of U2AF2 bound to a cytidine, guanosine, or adenosine at the central position of the Py tract, and compared U2AF2-bound uridine structures. Local movements of the RNA site accommodated the different nucleotides, whereas the polypeptide backbone remained similar among the structures. Accordingly, molecular dynamics simulations revealed flexible conformations of the central, U2AF2-bound nucleotide. The RNA binding affinities and splicing efficiencies of structure-guided mutants demonstrated that U2AF2 tolerates nucleotide substitutions at the central position of the Py tract. Moreover, enhanced UV-crosslinking and immunoprecipitation of endogenous U2AF2 in human erythroleukemia cells showed uridine-sensitive binding sites, with lower sequence conservation at the central nucleotide positions of otherwise uridine-rich, U2AF2-bound splice sites. Altogether, these results highlight the importance of RNA flexibility for protein recognition and take a step towards relating splice site motifs to pre-mRNA splicing efficiencies.
Graphical Abstract
Graphical Abstract.
Splice site RNA sequence variants adapt to U2AF2 scaffold.
INTRODUCTION
The vast majority of human genes contain intervening introns that need to be spliced from the nascent transcript and the exons joined to form the mRNA before translation into a protein. Alternative splicing to join different subsets of exons expands the diversity of proteins encoded by a limited number of genes (1). The pre-mRNA splice sites are marked by relatively short, consensus motifs that can vary in length and sequence. Uridine (U)-rich polypyrimidine (Py) signals precede the major class of 3′ splice sites. Yet, purines often interrupt Py tract signals and can regulate alternative 3′ splice site selection in multicellular eukaryotes (2).
The essential pre-mRNA splicing factor U2AF2 (also called U2AF65) recognizes the Py tract signal to promote the earliest stage of pre-mRNA splicing. The U2AF2 protein forms a ternary complex with SF1 and U2AF1 (also called U2AF35), which ensures 3′ splice site fidelity by identifying the branchpoint and AG consensus sequences flanking the Py tract. In a series of ATP-dependent steps, the 5′ and 3′ splice sites ultimately are positioned for catalysis in the active spliceosome. Breakthrough cryo-electron microscopy structures have revealed the later stages of spliceosome assembly (reviewed in (3)), whereas piecewise X-ray crystallography and NMR structures provide snapshots of splicing factor domains during the transient, early stages of 3′ splice site recognition. The U2AF2 protein recognizes the Py tract via two tandem RNA recognition motifs (RRM1 and RRM2) and flanking α-helices (U2AF212L). In the absence of RNA or in the presence of degenerate Py tracts comprising less than four consecutive uridines, U2AF2 adopts a ‘closed’ conformation in which RRM1 is masked and only RRM2 is available for RNA binding (4–6). When bound to a longer uridine tract such as the 3′ splice site consensus, the U2AF2 RRMs have an ‘open’, side-by-side conformation with RRM1 and RRM2 contacting the respective 3′ and 5′ regions of the Py tract (4,7). Both RRMs prefer uridines (8,9), although the N-terminal RRM1 is more tolerant of cytidine and purine substitutions in the Py tract than is RRM2 (10,11). In particular, the uridine-specificity of a promiscuous RRM1 site can be enhanced by a structure-guided mutation (10). Yet, unlike the well-characterized RRM1 and RRM2 of U2AF2, the sequence specificity of the RRM1/RRM2 interface for the central nucleotide of the Py tract is unknown.
U2AF2 defects have been associated with a variety of human diseases. Acquired U2AF2 mutations recur among certain cancers (12–14), although with lower frequency than in the U2AF1 subunit (15). De novo mutations of U2AF2 are significantly associated with developmental delay and malformation (16). U2AF2 binding to RP2 and NF1 Py tracts is reduced by purine substitutions associated with retinitis pigmentosa and neurofibromatosis (10). U2AF2 has been shown to regulate splicing of an IL7R exon that is dysregulated in autoimmune disorders including multiple sclerosis (17). Moreover, disrupted association between U2AF2 and PTEN correlates with autism spectrum disorder (18). Structure/function studies of these disease-associated U2AF2 mutations highlight key interfaces for the normal functions of the protein and provide insight into mechanisms of disease progression. However, understanding the normal sequence specificity and adaptability of the protein is an important baseline for comparison with disease-associated mutants.
Here, we investigate the interactions and nucleotide sequence specificity of the U2AF2 RRM1/RRM2 interface. By X-ray crystallography and complementary molecular dynamics simulations, we find that a protein scaffold accommodates bulky purines at the RRM1/RRM2 interface by repositioning the central nucleotides of the bound Py tract. Structure-guided variants increased the ability of U2AF2 to distinguish purines from pyrimidines at the central Py tract position. In human cells, we found that the nucleotide consensus was more variable at the central positions of sequence logos for U2AF2 binding to sites that were otherwise uridine-rich. These results reveal that U2AF2, a key factor for early spliceosome assembly, adapts to natural splice site variations by offering alternative binding sites for different RNA conformations.
MATERIALS AND METHODS
Preparation of U2AF212L proteins and oligonucleotides
The wild-type and mutant U2AF212L proteins (residues 141–342 of NCBI RefSeq NP_009210) were expressed and purified as described (7,12). The final protein buffer was 100 mM NaCl, 15 mM HEPES pH 6.8, 0.2 mM TCEP following size exclusion chromatography. Purified, deprotected RNA oligonucleotides were purchased from Horizon Discovery Ltd.
Fluorescence anisotropy RNA binding assays
The RNA-binding experiments followed protocols described in (12,19). The 5′-fluorescein-labeled, RNA oligonucleotides were diluted >100-fold to 30 nM final concentration in a binding buffer comprising 100 mM NaCl, 15 mM HEPES at pH 6.8, 0.2 mM TCEP, 0.1 U ml−1 Superase-In™ (Invitrogen™). The changes in total volume following addition of the protein were <10% to minimize dilution effects. The fluorescence anisotropy changes during titration were measured using a FluoroMax-3 spectrophotometer, temperature-controlled at 23°C by a circulating water bath. Samples were excited at 490 nm and emission intensities recorded at 520 nm with slit widths of 5 nm. The fluorescence emission spectra also were monitored for similarity throughout the experiment. Each titration was fit with a nonlinear equation (12,19) to obtain the apparent equilibrium dissociation constant (KD). These fits and the P-values of a two-tailed unpaired t-test with Welch's correction were calculated using Prism v6.0 (GraphPad Software, Inc.). The apparent equilibrium affinities (KA) are the reciprocals of each KD. The average KD or KA values and standard deviations are given for three replicates of each experiment.
Crystallization, data collection and structure determination
Crystallization conditions were similar to those described (12). Following concentration to 20 mg ml−1, U2AF212L protein was mixed with 1.2-fold molar excess purified oligonucleotide variant (5′-phosphoryl-UU(dU)NU(5BrdU)CC-3′, where N is cytosine (C5), adenosine (A5), or guanosine (G5)). Crystals were obtained by hanging drop vapor diffusion experiments with precipitants composed either of 0.60 M succinic acid, 0.10 M HEPES pH 7.0, 2% PEG monomethyl ether 2000 (C5) or 0.24 M Na malonate, 26% PEG 3350 (G5, A5). Addition of 0.1 μl of 5% w/v LDAO detergent (Hampton Research) to the G5 or A5 drops and 10% sucrose to the A5 drops prior to incubation improved crystal quality. Crystals were flash-cooled in liquid nitrogen after coating with a mixture of 1:1 (v/v) paratone-N and silicone oil (G5), or sequential transfers to precipitant solutions containing either 21% glycerol (C5) or 28% sucrose/8% PEG 200 (A5). Crystallographic data sets were collected at 100 K by remotely using the Stanford Synchrotron Radiation Light (SSRL) source Beamline 12–2 (20) and processed using the SSRL AUTOXDS script (A. Gonzalez and Y. Tsai) implementation of XDS (21) and CCP4 packages (22). The structures were determined using the Fourier synthesis method starting from PDB code 6XLW. The models were adjusted using COOT (23) and refined using PHENIX (24). The crystallographic data and refinement statistics are given in Supplementary Table S1 and reduced-bias electron density maps (25) are shown in Supplementary Figure S3.
Molecular dynamics simulations and analysis
Molecular dynamics (MD) simulations were run using Amber 18 (26). The U2AF2-U5, U2AF2-C5, U2AF2-G5 and U2AF2-A5 crystal structures were solvated in a truncated octahedron of OPC water (27) with a 12 Å margin of the solute using Leap. The system was neutralized using eight Na+ atoms, and 20 Na+ and Cl− ions were added to model NaCl at a bulk concentration of 150 mM (28). The starting structures were energy-minimized using the steepest descent and then conjugate gradient methods, each for 500 steps. Subsequently, the systems were heated to 298 K in 200 ps with a timestep of 2 fs. These equilibrated structures were used to run the final production dynamics for 2 μs using Amber ff14SB (29) + RNA.OL3 (30–32) forcefields with periodic boundary conditions, using a 2 fs timestep and a direct space cutoff of 10 Å for non-bonded interactions. The structures were written to a trajectory file every 100 ps. Pressure was maintained at 1 atm using a Monte Carlo barostat and the temperature was maintained at 298 K using Langevin thermostat with a collision frequency of 1.0 ps−1. For the oligonucleotide-only simulations, the U2AF2 protein coordinates were removed to generate the starting structures, then the same steps used for the protein–RNA complex were followed.
For analysis of MD simulations, all the trajectories were merged, and the water and ions were removed using Ambertools 18 (33). The trajectories were aligned using the Cα of RRMs with the starting structures for U2AF2-RNA simulations and six-membered base rings for the simulations of the isolated oligonucleotide, using aligner in LOOS (34). Root mean square fluctuations were calculated for six-membered rings of RNA residues using rmsf in LOOS (34). Root mean squared deviations (RMSD) of the Cα were calculated using rmsd2ref tool in LOOS. Pairwise RMSD was calculated using custom python script, rmsds-align.py.
Enhanced UV-crosslinking and immunoprecipitation
U2AF2 eCLIP-seq experiments followed the protocol in (35) with modifications reported in (36). For consistency with eCLIP-seq of U2AF1 splicing factor complexes (36), we used a human erythroleukemia (HEL) cell line (ATCC, Cat #TIB-180) cultured in RPMI 1640 supplemented with 1% l-glutamine, 1% penicillin–streptomycin and 10% FBS (ThermoFisher Sci. Cat #’s 11875093, 25030081, 15140122 and Gemini Bio-Products Cat #’s 100–106). The HEL cells were subjected to UV-crosslinking and U2AF2–RNA complexes were immunoprecipitated with 8 μg anti-U2AF2 antibody (Sigma-Aldrich, Cat #U4758) and Dynabeads Protein G (ThermoFisher Sci., Cat #10004D). RNA was partially digested with RNase I (ThermoFisher Sci., Cat #AM2295) and P32-labeled (PerkinElmer, Cat #BLU002Z250UC), followed by RNA linker ligation. After SDS-PAGE and transfer to nitrocellulose membrane, a region between 65 – 110 kD was excised to obtain U2AF2-bound RNA complexes (Supplementary Figure S7). RNA was isolated using the RNA Clean & Concentrator-5 kit (Zymo Research, Cat #R1016) after treatment with proteinase K, then subjected to library preparation. Libraries were sequenced on Illumina NovaSeq 6000 system at the Yale Center for Genome Analysis (YCGA). The U2AF2 eCLIP-seq was performed in two replicates, compared with four replicates for the U2AF2 eCLIP-seq with U2AF1 overexpression (OE) (36). The U2AF2 eCLIP-seq reads were processed according to the pipeline reported in (36). After duplicate removal (FastUniq (37)) and adapter trimming (Cutadapt (38)), reads were aligned to the human genome (GRCh38.p10) with STAR (version 2.7.0f, GENCODE Release 27 for transcript annotation). The average alignment rates were 86.2% and 81.8% for libraries with endogenous (here) or OE U2AF1 (36). Crosslinked nucleotides were extracted from BAM files considering the genomic position right after the end of each sequenced read. Bound junctions were confidently identified considering a nucleotide region from –40 to +10 around the 3′ splice site in all the annotated splice junctions in the human genome and using a coverage threshold of at least 10 reads, resulting in 149 708 and 90 918 selected splice junctions, for samples with endogenous or OE U2AF1 (36). Binding metaprofiles were built after trimming outlier signals at each nucleotide position from –20 to +5 around the 3′ splice site.
RESULTS
U2AF2 has little sequence preference for the central Py tract nucleotide
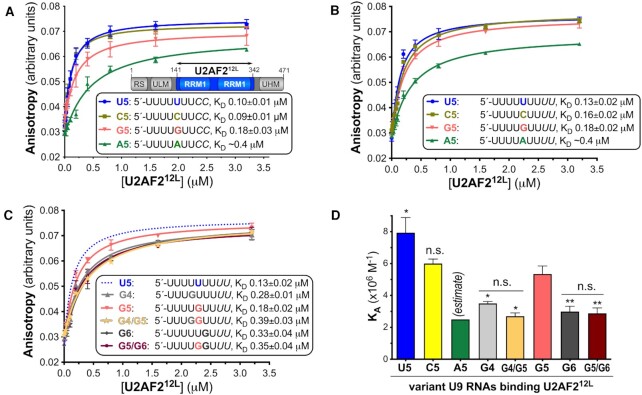
To fill a missing gap in previous studies of U2AF2–RNA sequence specificity (10,11), we investigated the preferences of U2AF2 for binding different nucleotides at the central position of the Py tract (Figure 1). Since nine nucleotide binding sites have been noted for the open conformation of U2AF212L (4,7), we compared the binding affinities of U2AF2 for nine-nucleotide RNAs substituted with U, C, G or A at the fifth nucleotide. We fit the fluorescence anisotropy changes of 5′-fluorescein-labeled oligonucleotides titrated with protein to obtain the apparent equilibrium dissociation constants (KD) using nonlinear regression as described (19). The KD’s of the A5-substituted RNAs are lower estimates, since the fluorescence anisotropies at the highest concentrations of U2AF212L in the titrations are less than the maxima of the fits. We first tested substitutions of a prototypical, strong Py tract from the adenovirus major late promoter transcript (AdML) (Figure 1A). The nine-nucleotide AdML Py tract bound U2AF212L with approximately three-fold lower affinity than a previously studied, 13-mer Py tract from the same intron (KD 100 nM versus 30 nM) (7). Substitution of a cytidine (C5) for the fifth uridine (U5), which is located between the RRM1 and RRM2 of the U2AF212L structure (4,7), does not significantly change the binding affinity. For purine substitutions, a guanosine (G5) incurred a subtle, approximately two-fold penalty, whereas an adenosine (A5) produced a more substantial decrease in affinity (at least 4-fold, equivalent to ∼1 kcal mol−1).
Figure 1.
The specificity of the U2AF2 RRM-containing region for centrally-substituted Py tract oligonucleotides. The boundary of the U2AF212L construct (blue) used for RNA binding and structure determination is inset in panel (A). Fluorescence anisotropy measurements of U2AF212L titrated into the given RNAs, including (A) an AdML Py tract (blue) and its central cytidine (mustard/yellow), guanosine (salmon), or adenosine (green) substitutions, (B) a nine-uridine tract (blue) and substituted with cytidine (mustard/yellow), adenosine (green), or guanosine at G5 (salmon), or (C) a nine-uridine tract (dashed blue line shown for reference) substituted with G4, (light gray), G5 (salmon), G4/G5 (orange-yellow), G6 (dark grey), or G5/G6 (maroon). The average data points and standard deviations of three experiments are overlaid with the fitted binding curves. The sequences of the 5′-fluorescein-labeled RNA oligonucleotides are inset, alongside average apparent equilibrium dissociation constants (KD) with standard deviations of three replicates. (D) Bar graph of U2AF212L binding affinities for the RNAs shown in B and C. The KD’s of U2AF212L for binding the A5 RNAs are estimates due to the very low affinities. The significance of the changes in the average apparent binding affinities compared to the G5-substituted oligonucleotide were calculated using two-tailed unpaired t-tests with Welch's correction in GraphPad Prism: P-values: n.s., not significant, *, <0.05; **, <0.005. The differences between the U2AF212L binding affinities for the G4 and G4/G5 RNAs, or between the G6 and G5/G6 RNAs, were not significant. The U2AF212L binding affinities for modified oligonucleotides used for co-crystallization are shown in Supplementary Figure 1.
We next introduced substitutions in the context of a consensus uridine tract (Figure 1B). The U2AF212L protein bound the uridine-tract with similar affinity as the AdML Py tract, consistent with a sequence difference of two terminal cytidines. As observed for the AdML Py tract, the effects of the nucleotide substitutions on U2AF212L binding ranged from no significant effect for C5, less than 2-fold for G5, to a more substantial estimated penalty for the A5 substitution. The greater discrimination of U2AF2 against adenosine could contribute to defining the AG-exclusion zone, a region devoid of AG-dinucleotides between the branchpoint and bona-fide AG at the 3′ splice site (39).
We further evaluated the consequence of a guanosine-substitution at the neighboring sites, G4 and G6, which are expected to bind RRM2 and RRM1 (Figure 1C, D). Although the G4- or G6-associated changes in U2AF212L binding affinities were moderate, the approximately three-fold decreases were comparable to the penalties for U2AF2 binding to disease-associated mutations in the RP2 and NF1 Py tracts (10). Addition of G5 to the G4 or G6 substitutions (G4/G5 or G5/G6) had no additional effect, again reflecting the promiscuity of the inter-RRM binding site at the fifth position of the oligonucleotide.
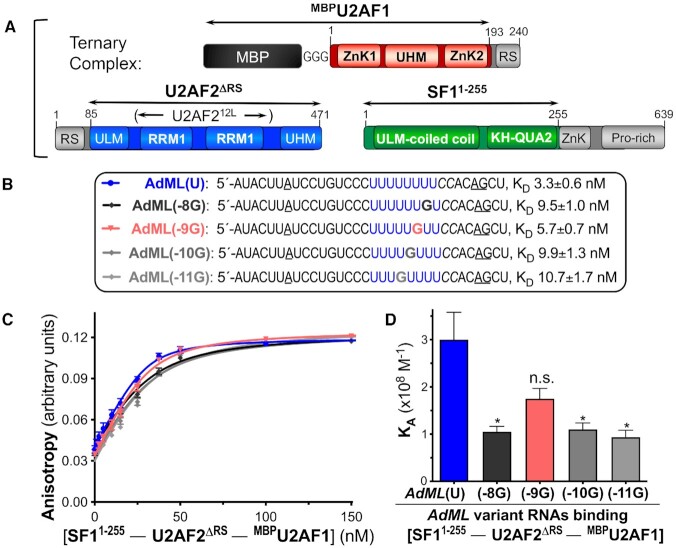
To relate U2AF212L’s subtle discrimination among different nucleotides at the center of the Py tract to intact 3′ splice site recognition, we compared the RNA affinities of a ternary complex among U2AF2, SF1 and U2AF1 subunits (Figure 2). The U2AF2 and U2AF1 constructs were nearly full length apart from RS domains that contact the branchpoint rather than the Py tract (40–42), and a zinc knuckle/proline-rich region of SF1 that have been implicated in protein-protein interactions (43–46). Although the U2AF1 subunit retained an MBP tag to enhance expression and solubility, this tag has no detectable effect on RNA affinity (6). We measured the binding affinities of the purified protein complex for AdML splice site RNAs spanning the branchpoint, Py tract, and 3′ splice site junction. We compared the effects of four guanosine substitutions at different positions of the Py tract. Similar to U2AF212L binding the G6-substituted Py tract, most guanosines reduced the RNA affinity of the ternary complex by approximately three-fold. Notably, a guanosine at the central position (–9G) had no significant effect on affinity for the protein complex, in agreement with the subtle effect of G5 on U2AF212L association with the isolated Py tract. This result supported the relevance of the nine nucleotide binding sites of U2AF212L to splice site recognition in the context of the ternary U2AF2–SF1–U2AF1 complex.
Figure 2.
The ternary complex of U2AF2 with SF1 and MBP-tagged U2AF1 has subtle preferences for G-substitutions in the Py tract of the AdML 3′ splice site sequence. (A) Domains of subunits in the ternary complex with construct boundaries indicated by double-headed arrows. (B) Sequences of 5′ fluorescein-labeled oligonucleotides used for RNA binding. (C) Fitted binding curves and (D) bar graph of average binding affinities and standard deviations, with the significance indicated as for Figure 1. The guanosine-substitutions of the wild-type AdML (blue) are numbered in reverse from the splice site junction following the AG consensus (underlined): –8G, black; –9G, salmon; –10G, dark grey; –11G, light grey.
Local shifts of the central nucleotides adapt to the U2AF212L structure
To view how U2AF2 adapts to different nucleotides at the RRM1/RRM2 interface, we determined three crystal structures of U2AF212L bound to Py tracts with various nucleotides at the central position (Figure 3, Supplementary Table S1). To promote crystallization and confirm the oligonucleotide binding register, we included 2′-deoxy-uridine (dU) and 5-bromo-dU modifications at the fourth and seventh positions of U2AF212L-oligonucleotide crystal structures as described (7,10,11,47). The U2AF212L protein binds the modified oligonucleotides with comparable affinity and specificity as the corresponding RNAs (KD 65 nM versus 100 nM for modified versus unmodified AdML oligonucleotides and approximately three-fold preference for U5 over A5; Supplementary Figure S1). Crystallization was facilitated further by using eight-mer oligonucleotides that omit the 5′-terminal uridine (7,12). Well-defined electron density for the eight nucleotides is observed in the documented nucleotide binding sites 2–9 of the open U2AF2 conformation (PDB ID 5EV4, PDB ID 2YH1). Electron density for the 5-bromo-modification, as well as distinct, atomic resolution shapes for the pyrimidine vs. purine bases, confirms the binding register for each complex (Supplementary Figure S3). To match PDB ID 5EV4, we numbered the eight bound nucleotides from 2–9 starting at U2 in the second documented nucleotide binding site of U2AF212L, as shown in Figure 3.
Figure 3.
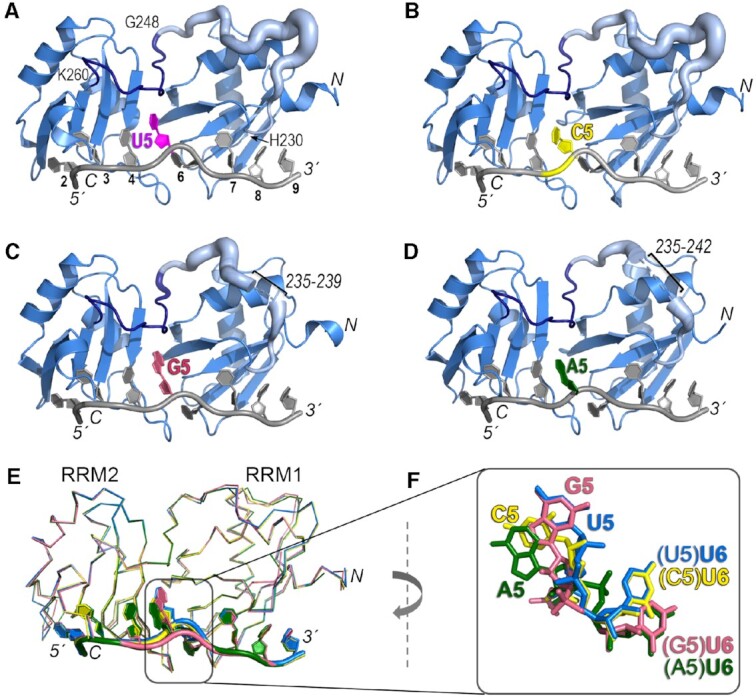
Crystal structures of U2AF212L recognizing AdML Py tract variants that differ in the identities of the central nucleotide. The amino (N)/carboxy (C)-termini of the polypeptide and 5′/3′ termini of the oligonucleotide are labeled in italics. The nucleotide positions are numbered on panel A. (A–D) Overall ribbon diagrams of the protein (blue) bound to oligonucleotides (grey) substituted with (A) uridine (U5, magenta, PDB ID 6XLW), (B) cytidine (C5, yellow, PDB ID 7S3A, this study), (C) guanosine (G5, salmon, PDB ID 7S3B, this study), or (D) adenosine (A5, green, PDB ID 7S3C, this study). On panels A–D, the temperature factors (mobility) of the inter-RRM linker (residues 230–260) are represented using cartoon putty in PyMOL, which scales the size of the coil proportionately to the temperature factors of the residues. Residues 230–247 are pale blue and residues 248–260 are dark blue (boundary residues labeled on panel A). Ranges of linker residues that unresolved in the G5 and A5 structures are labeled. (E) Superposition by matching Cα atoms in the four structures shows conformational changes at the fifth and sixth nucleotide positions (numbered according to the nine nucleotide-binding sites of PDB ID 5EV4). (F) Closer view of the fifth and sixth nucleotides shown in (E) following a 90° clockwise rotation about the y-axis relative to (E).
The overall conformations of the protein backbones remained similar (0.1–0.3 Å pairwise RMSD between matching Cα atoms of C5, A5 or G5-containing structures when compared to the U5 structure) (Figure 3E). In particular, the polypeptide backbones of an RRM2-proximal, nucleotide-bound region of the inter-RRM linker (residues 248–260), as well as of the modular RRM1 and RRM2 domains, were nearly identical among the structures. A distinct region of the linker (residues 230–247) near the alpha-helical surface of RRM1 was more divergent, consistent with its higher temperature factors and in some cases, missing residues (Figure 3A–D). Despite differences in the inter-RRM region, the nucleotides bound to the respective RRM2 and RRM1 also shared similar positions (0.2–0.4 Å pairwise RMSD between all atoms of nucleotides 2–4/7–9 of C5, A5 or G5-containing structures compared to the U5 structure). However, the central nucleotide substitutions dramatically shifted the local positions of the U2AF2-bound RNA (Figure 3F, Figure 4, Supplementary Movies S1-S3). A cytidine or adenosine (C5 or A5), for which the hydrogen bond groups differ from uridine, rotated ∼25° away from the U2AF2 inter-RRM linker relative to the U5 position. Notably, networks of ordered water molecules filled the resulting gaps and mediated contacts between the extruded cytosine or adenine bases and the protein backbone (Figure 4B, D and Supplementary Figure S3). The six-member ring of a guanine base at the central position (G5), on the other hand, superimposed with the uracil and equivalent atoms (U-O4/N3H and G-O6/N1H) maintained similar hydrogen bonds with the protein (Figure 3F, Figure 4A, C).
Figure 4.
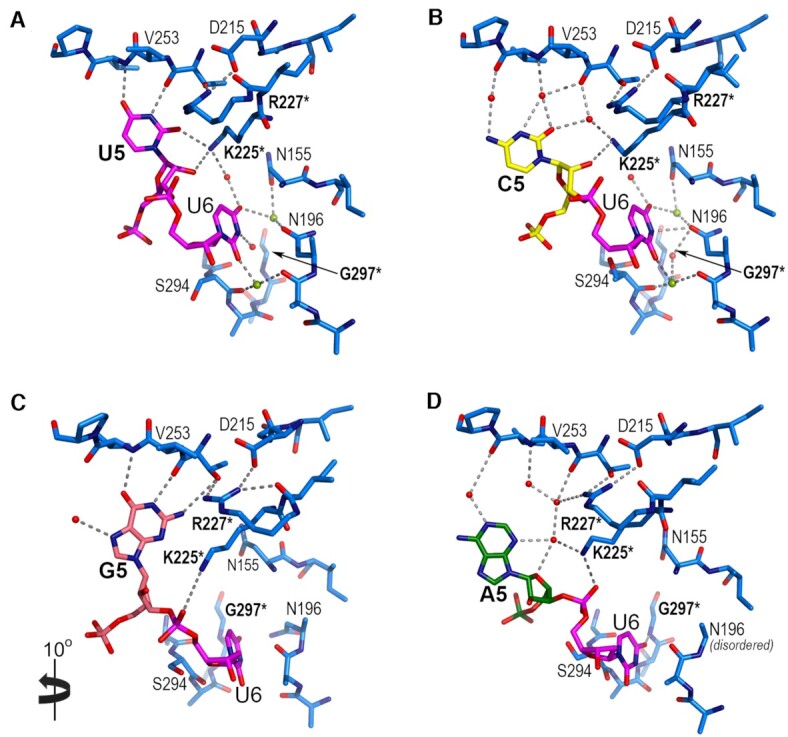
U2AF212L interactions with the fifth and sixth nucleotides of bound Py tracts (numbered with the convention of PDB ID 5EV4). Variants of the fifth nucleotide include (A) uridine (U5, magenta), (B) cytidine (C5, yellow), (C) guanosine (G5, salmon) or (D) adenosine (A5, green). Perspectives are similar to Figure 3F. Panel C is rotated 10° into the plane of the page for clarity of the interactions. Nitrogens (blue) and oxygens (red) are colored; interacting water molecules (red) and sodium ions (lime green) are indicated by spheres. Hydrogen bonds are indicated by dashed lines. Mutated residues are bold and marked by an asterisk. Representative electron density is shown for the nucleotides in Supplementary Figure S3.
Interestingly, the adjacent uridine on the 3′ side (U6) also shifted position when purine nucleotides were substituted at the fifth site (Figure 3F, Figure 4). In the U2AF2-bound, all-uridine oligonucleotide, RRM2 and RRM1 loops sandwiched the U6 base. In the presence of the bulky A5 or G5 purines, the downstream U6 rotated ∼25° away from the inter-RRM linker to settle in an alternative binding site, which also is located between the RRM1 and RRM2 loops. To achieve a comparable position of U6 despite the different locations of the A5 and G5 bases, the A5-linked U6 phosphate rotated over the ribose group (Figure 4D, Supplementary Movie S3). Although unique to the A5 nucleotide substitution, we cannot rule out that the neighboring 5-bromo-dU7 modification influenced this conformation of the A5-linked U6 phosphodiester group. Unlike the U5-linked U6 position, no direct or water-mediated U6 contacts with the protein were detected in either purine-containing structure. Instead, several ordered water molecules that mediated U6 contacts with U2AF2 in the U5/C5 structures appeared absent in the presence of the purine substitutions (Figure 4, Supplementary Figure S3). The purine-induced perturbations of the adjacent U6 site, coupled with the shifted position of A5, could account for the subtle differences in U2AF2 binding affinity (U5/C5 > G5 > A5) for the oligonucleotides (Figure 1).
U2AF212L-bound Py tract RNA is dynamic at the central nucleotides
To explore the conformations of the U2AF2–Py tract complex beyond the environment of the crystal structures, we performed all-atom molecular dynamics simulations using Amber (26). The simulations revealed differences in the conformational flexibility of the protein regions. The simulations also demonstrate that interaction with the protein reduced the intrinsic flexibility of the RNA.
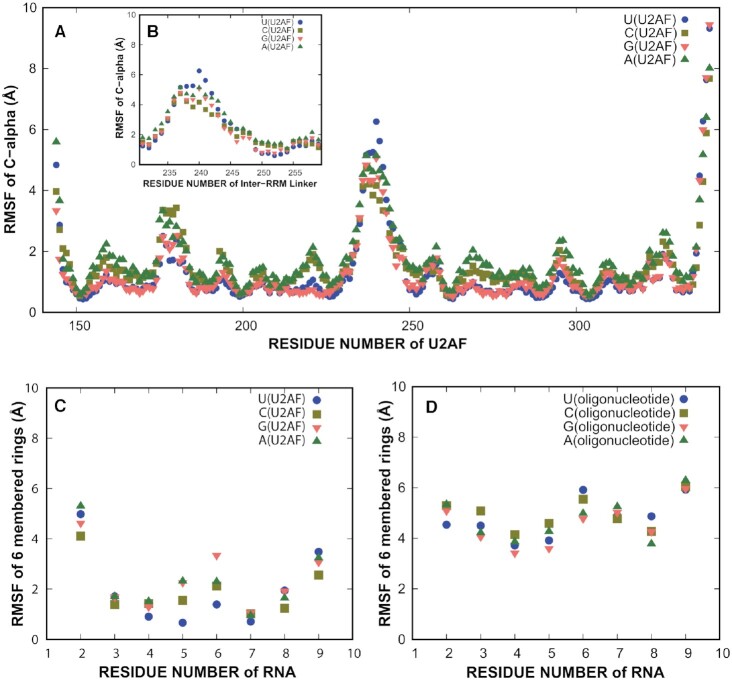
First, we ran 2 μs simulations of the U5, C5, G5 and A5 crystal structures, repeated five times each. Each protein–RNA structure was stable (Supplementary Figure S4), and pairwise RMSD plots (Supplementary Figure S5) demonstrated convergence. To quantify the dynamics of residues, we calculated the root mean squared fluctuation (RMSF) for each residue, which is the extent to which a residue fluctuates around the average structure during the simulation (Figure 5). The RRMs were found to be relatively static (Figure 5A). A portion of the linker region connecting the RRMs was flexible in the simulations (residues 236–242, Figure 5B). However, residues 250–255, the linker region bound to the central nucleotide of the Py tract, was static. The U2AF212L crystal structures are consistent with the results of the simulations, showing variability and sometimes disorder in residues 236–242 of the inter-RRM linker, whereas residues 250–255 and the RRMs remain similar among known structures (Figure 3A–D, Supplementary Figure S6) (7,12). When a purine was in the fifth position of the U2AF2-bound oligonucleotide, substantially more fluctuation was found in the fifth position than when a pyrimidine was in the fifth position (Figure 5C). The presence of a purine at the fifth position also increased the fluctuation of the nucleotide at the sixth position of the U2AF2-bound oligonucleotide.
Figure 5.
Molecular dynamics studies of conformational flexibility. (A) Root mean squared fluctuations (RMSF) of U2AF2 Cα by residue numbers. The RRM1 and RRM2 regions are relatively static. (B) Inset showing RMSF of the inter-RRM linker Cαs (residues 230–260). (C) RMSF values of six-membered rings of the RNA in the U2AF2-RNA complex simulation (2 μs). (D) RMSF values of six-membered rings of the RNA in the oligonucleotide simulations (1 μs). The oligonucleotide simulations have higher fluctuations than the U2AF2–RNA simulations. Supplementary Figures S4 and S5 show RMSDs to demonstrate stability of the protein during simulation and convergence of the simulation.
We also tested whether the conformation of the central nucleotide is related to an intrinsic property of the oligonucleotide. We ran five, 1 μs all-atom simulations of oligonucleotides (U5, C5, A5 and G5) in the absence of the protein. These simulations of the oligonucleotides exhibited substantial conformational fluctuations compared to the oligonucleotides bound to U2AF2 (Figure 5D). Specifically, the pairwise RMSD plots (Supplementary Figure S5) demonstrated no innate preferred conformation for the RNA. These plots compare the conformations sampled across trajectories, and are useful for comparing the consistency of the conformations across multiple simulations. These suggest that the RNA is flexible in nature, allowing the central nucleotide to adopt a conformation that accommodates protein binding.
Structure-guided mutations enhance U2AF212L specificity for a central uridine
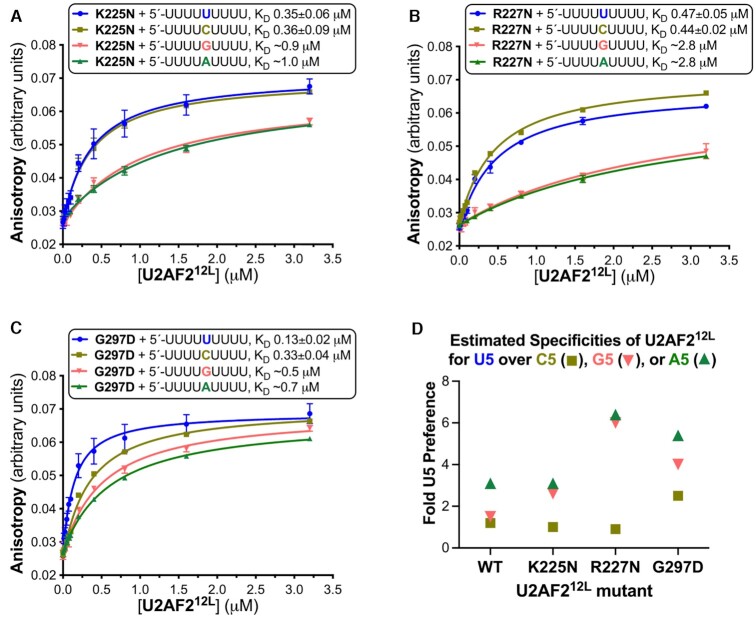
To test the U2AF2 interactions with central nucleotide viewed in the structures, we substituted either of the positively-charged K225 or R227 residues with negatively-charged glutamates (K225E and R227E) to nonspecifically reduce the RNA binding affinity. Compared to the wild-type protein, the K225E and R227E mutations reduced the U2AF212L affinities for the AdML Py tract and its G5 variant by approximately 20- and 80-fold (Supplementary Figure S2), most likely by general electrostatic repulsion of the phosphodiester backbone. This result supported the observed locations of K225 and R227 residues at the RNA interface of the open U2AF2 conformation.
We next considered whether the promiscuity of U2AF2 for various nucleotides at the central position of the Py tract could be altered by replacing key amino acids (Figure 6). Since the K225 side chain forms a salt bridge with a phosphoryl group of the A5/G5-containing RNAs, we reasoned that an asparagine at this position would penalize U2AF2 binding to purines at this position more than to pyrimidine-containing RNAs. Likewise, we conjectured that replacing R227 with the shorter side chain of asparagine would disrupt the direct and indirect networks of U2AF2 with G5 and A5 bases more than for U5 and C5. Third, we predicted that an aspartate substitution of G297 would repel the U6-O2 atom in the purine-bound conformation, thereby favoring U2AF2 binding to U5 and C5. Accordingly, the K225N and R227N variants significantly increased U2AF212L discrimination of U5/C5- from G5/A5-containing oligonucleotides (Figure 6A, B and D), by having substantially greater penalties for U2AF212L binding the purine-containing RNAs (at least five-fold penalties). The G297D replacement also increased the specificity of U2AF212L for binding to U5 > C5 > G5/A5 oligonucleotides (in order of preference, Figure 6C, D), by having no detectable effect on the all-uridine oligonucleotide and approximately two-fold penalties for binding the other nucleotide variants. These results demonstrated that single amino acid changes could increase the stringency of U2AF2 for distinguishing the identity of the central Py tract nucleotide.
Figure 6.
U2AF2 residues at the RNA interface influence its specificity for the central nucleotide. (A–C) Average fluorescence anisotropy data points and standard deviations from three replicates of the indicated U2AF212L mutants titrated into 5′-fluorescein-labeled RNA oligonucleotides. The fitted curves are overlaid. The RNA sequences comprising nine-uridines (blue) or its C5, G5 or A5 variants (mustard, salmon, or green), are inset alongside the apparent equilibrium dissociation constants (KD) and standard deviations. (D) Scatter graph of the ratios of the wild-type or mutant U2AF212L binding affinities for U5 to the affinities for the C5 (square), G5 (inverted triangle), or A5 (triangle) variants of the central nucleotide. The KD’s and specificities of the U2AF2 variants binding the G5 and A5 RNAs are estimates due to the very low affinities. Supplementary Figure S2 shows penalties of K225E or R227E mutations on U2AF212L–RNA binding.
U2AF2 interaction sites in human cells agree with U2AF212L–RNA binding specificity
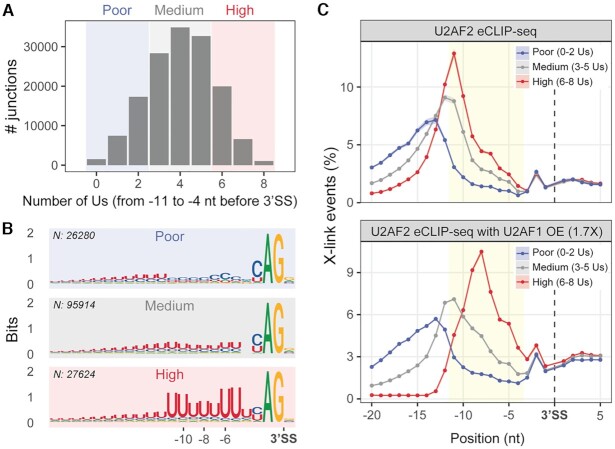
To further understand the organization of U2AF2 and the 3′ splice site, we used the enhanced UV crosslinking and immunoprecipitation (eCLIP) assay (35,36,48) to map the RNA interactome of U2AF2 in human erythroleukemia (HEL) cells. The HEL cell line represents a preclinical model for the study of myelodysplastic syndromes and acute myeloid leukemia, which are blood cancers frequently characterized by mutations in splicing factors such as U2AF1. Following U2AF2 immunoprecipitation and 32P labeling of the crosslinked RNA, the immunoprecipitated complexes were separated by denaturing gel electrophoresis (Supplementary Figure S7). We focused on analyzing the region with a molecular weight between 65 and 110 kD, corresponding to the expected size of U2AF2-RNA complexes. Overall, we could identify U2AF2-binding locations in 149 818 splice junctions across the human transcriptome.
As expected, significant peaks for U2AF2 interactions occurred in Py-rich regions upstream of 3′ splice site junctions (Figure 7). To specifically investigate the relationship between U2AF2 binding and the sequence-content of 3′ splice site signals, we divided the splice site junctions into three classes based on their uridine enrichment. These included splice sites with poor (0–2), medium (3–5), or high (6–8) numbers of uridines in the zone from –11 to –4 nucleotides upstream of the intron 3′ end (Figure 7A). Sequence logos were generated from splice junctions of the three classes (Figure 7B). Importantly, motif analysis of the high uridine-containing class showed two clusters of approximately two highly conserved uridines (–11, –10 and –6, –5), surrounding a core of less conserved uridines at the central positions (–9, –8, –7), in agreement with the RNA binding preferences of the U2AF212L protein and of the ternary SF1–U2AF2–U2AF1 complex (Figures 1 and 2). By comparing the U2AF2 binding signal in each class of splice junctions, we observed that the U2AF2 contacts with endogenous splice sites shifted position depending on the local uridine content. In particular, the interaction peak was broader and more distant from the intron 3′ end for the splice site junctions with few uridines, while the peak was narrowest, strongest and closest to the intron 3′ end for the high uridine class (Figure 7C, and for examples of U2AF2 binding on single junctions belonging to the three classes, Supplementary Figure S8). Furthermore, we observed that a modest increase of U2AF1 levels (OE, see Materials and Methods) specifically affected the contacts with the high uridine-containing class, shifting the maximum of the U2AF2 peak to position -8, thereby matching the core of less conserved uridines in positions –9, –8 and –7 (Figure 7C, bottom panel and Supplementary Figure S8A). The U2AF1-enhanced position of U2AF2 is consistent with U2AF1 stabilization of U2AF2 conformations (6) as well as U2AF1 recognition of the intron–exon junction (49–52). Collectively, these results demonstrated that the U2AF2 binding sites were responsive to the uridine contents and locations within the pre-mRNA splice site signals.
Figure 7.
U2AF2–RNA binding adapts to the local uridine content of the 3′ splice site in vivo. (A) Classification of U2AF2-bound splice site junctions according to the number of uridines (Us) in the –11 to –4 region of the 3′ splice site. Shadowed areas distinguish the three classes of junctions: blue, poor uridine content (0–2 Us); grey, medium uridine content (3–5 Us); red, high uridine content (6–8 Us). This analysis is focused on internal and last exons of all spliced transcripts. (B) 3′ splice site sequence logos for each class. N, number of junctions per class. (C) Binding metaprofiles (y-axis, mean ± SEM of the percentage of crosslinking events) for each class of uridine-containing junctions in U2AF2 eCLIP-seq (top panel, n = 2) and in U2AF2 eCLIP-seq in the presence of modestly overexpressed (OE) U2AF1, at 1.7X in comparison to endogenous levels (36) (bottom panel, n = 4). Yellow area: –11 to –4 region. Supplementary Figure S7 shows steps and results of the U2AF2 eCLIP-seq sample preparation. Supplementary Figure S8 shows binding profiles of representative junctions from each class of uridine content.
DISCUSSION
Here, we expand our view of U2AF2 – splice site recognition by demonstrating that a relatively static region of the inter-RRM linker contributes to versatile U2AF2–RNA associations through inherent flexibility of the RNA site itself. Local rearrangements of the bound RNA, rather than protein backbone, contributed to an innate ability of U2AF2 to accommodate different nucleotides at the center of the Py tract (Figure 3E–F, Figure 4, Movies S1–S3). Bulky purines fit the central U2AF2 binding site through adjustments of the oligonucleotide backbone, which in turn shifted the adjacent, 3′ uridine (U6) into a distinct binding site. Cytidine or adenosine have rotated away from the protein at this inter-RRM site, and instead, intermediary water molecules glued the mismatch nucleobases to the inter-RRM surface. Otherwise, the U2AF2 RRMs maintained unperturbed contacts with the surrounding pyrimidines. Prior studies of U2AF2 RRM1/RRM2 bound to noncognate RNAs reveal a variety of changes, ranging from subtle shifts of the side chains and protein backbone to nucleotide rotations and syn/anti-conformer flips (10,11). In particular, we had observed flexible nucleotide conformations facilitating U2AF2 promiscuity at one other site (position 8 bound to RRM1). At this site, a guanosine binds the U2AF2 RRM1 in an unusual syn-conformer (10) or a cytosine shifts to optimize hydrogen bonds with the U2AF2 backbone and side chains (11). A distinct, previously-established means for U2AF2 to fulfill its multifaceted role in 3′ splice site recognition is to rely on its modular architecture of tandem RRMs, which differ in uridine-specificity and switch between ‘open’ and ‘closed’ conformations in response to the RNA sequence (4,6,11). Consistent with the sequence-sensitivity of U2AF2 conformations, the uridine contents of the splice sites modulate the U2AF2–3′ splice site binding registers (Figure 7 and (36)).
These expanding views of U2AF2 complexes with different oligonucleotides reinforce an emerging theme among ribonucleoprotein structures, which is that the RNA conformation frequently adapts to fit (or is conformationally selected by) the surface of the protein binding site. Beyond U2AF2, syn/anti base flipping enables SRSF2 to recognize either tandem cytosines or guanosines with similar affinities (53). In the structures of Csr/Rsm with various noncoding RNA substrates, rearrangements of bound nucleotides facilitate recognition of the different RNA sequences (54). In another well-studied example, one mechanism for PUF family repeat proteins to bind a large set of degenerate RNA sequences is to eject noncognate nucleotides from the modular RNA binding surface (55). Altogether, these findings highlight the importance of RNA flexibility for proteins to associate with appropriate sites amidst the milieu of cellular RNAs.
Molecular dynamics simulations, starting from the U2AF2–RNA crystal structures, revealed that the oligonucleotides were inherently flexible in the absence of protein, and that the central nucleotides (positions 5 and 6) remain flexible in the U2AF2-bound complex (Figure 5). Although more studies of ribonucleoproteins have focused on the dynamics of the protein than on the RNA components, RNA flexibility clearly is an important contributor to versatile RNA–protein recognition. Several proteins have been shown to select an RNA structure with optimal intermolecular contacts among multiple conformations sampled by the protein-free RNA site (56–58). Indeed, a survey of RNA-binding proteins in the bound and free states implies that nucleic acid movements are a key aspect of protein-RNA recognition (59). In some cases, nucleotides making important contacts increase (rather than diminish) dynamics in the protein complex compared to the free state (57,58). Here, molecular dynamics simulations demonstrated that the Py tract RNAs likewise possessed a conformational repertoire in the absence of protein cofactors. Accordingly, polyuridine lacks a uniform structure in solution and shows the least base-stacking among the nucleotide polymers (60,61). From the ensemble of Py tract RNA conformations, we propose that U2AF2 selects a particular RNA conformation, thereby optimizing the intermolecular contacts with the altered central nucleotide and adjacent uridines. The molecular dynamics simulations further suggest that the central nucleotides remain flexible in the U2AF2–RNA complex, such as observed for other RRM-bound RNAs (57,58), and this facilitates recognition of alternative nucleotides in the fifth position.
The ability to structurally adapt to diverse splice sites is likely to represent a key functional characteristic of metazoan U2AF2. The transcriptome of human cells offers a vast number of sequence combinations, from which U2AF2 must select the bona fide splice sites during the initial stages of spliceosome assembly. Indeed, transcriptome-wide mapping of U2AF2 binding sites in cells (Figure 7 and (36,62)) demonstrates widespread association of U2AF2 with a plethora of RNA sites comprising various sequences. We have established that structure-guided mutations, including R227N, K225N and G297D at the central site (Figure 6) and D231V at position 8 (10), could artificially increase the uridine-specificity of human U2AF2. These results suggest that the subtle RNA sequence preferences of human U2AF2 have evolved to support the broad identification of a wide range of 3′ splice sites. Yet, accurate identification of the 3′ splice site signals is critical for the fidelity of gene expression. Even the relatively ‘small’, 2–4-fold changes in binding affinities, such as observed here for U2AF2 binding to the Py tract variants, can evoke relevant changes in gene expression in certain contexts. Specific Py tract mutations that penalize U2AF2 binding by a few fold, have been associated with specific diseases, including retinitis pigmentosa and cystic fibrosis (10). Likewise, cancer-associated mutations of U2AF2 that modulate its RNA binding affinities have significant consequences for splicing of pre-mRNA transcripts (12,14). Moreover, a cancer-associated S34F mutation of U2AF1, which affects association with 3′ splice sites to a similar extent as the nucleotide substitutions studied here, in turn alters splicing, 3′ end processing, and translation of transcripts in cells (63–67). Altogether, these studies support that U2AF2 transcends a traditional classification of either a ‘specific’ or ‘nonspecific’ RNA binding protein, and has critical functional requirements to adapt to a variety of splice sites while serving as a sensitive rheostat for splicing.
We note that many factors, beyond the scope of the studies in this work, contribute to the physiological RNA binding preferences of U2AF2 in cells. Multiple partners work to enhance and regulate U2AF2 conformations and RNA interactions, including U2AF1, SF1, SF3B1 and PUF60/RBM39, among others. Already, the distribution of U2AF2 binding sites observed in CLIP experiments reflects the ensemble of all spliceosome assembly states. Accordingly, when U2AF1 levels increase, the conglomerate of U2AF2 binding sites shift closer to the junctions for 3′ splice sites with high uridine content (Figure 7). This U2AF1-enhanced position is consistent with the RNA binding preferences of the ternary SF1–U2AF2–U2AF1 complex (Figure 2), conformational stabilization of U2AF2 by the U2AF1 heterodimer (6), and the function of the U2AF1 subunit to direct the ternary complex to the 3′ splice site junction (49–52). Cancer-associated mutations of U2AF1 also influence the binding register of U2AF2-containing splicing complexes relative to 3′ splice site junctions (6,36)). Moreover, perturbation of U2AF1, and by extension U2AF2, affects transcription rates and coupled splicing events (68,69). Altogether, these diverse factors in the context of coupled gene expression processes converge to modulate the pre-mRNA sites associated with U2AF2. Resolving how RNA sequence contexts, spliceosome components, cancer-associated mutations, transcription rates, and coupled pre-mRNA processing events influence the U2AF2–RNA conformation for 3′ splice site recognition remain important directions for future studies.
DATA AVAILABILITY
Data deposition: The coordinates for the U2AF structures have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 7S3A, 7S3B, 7S3C for C5, G5 and A5 structures). The U2AF2 eCLIP-seq files have been deposited in the GEO database, https://www.ncbi.nlm.nih.gov/geo/ (GSE195669). The eCLIP-seq files for U2AF2 with OE U2AF1 are available with GEO accession GSE195620 (36).
Supplementary Material
ACKNOWLEDGEMENTS
We thank M.J. Pulvino for insightful discussions and S. Henderson for initial refinement of the C5 and G5 structures.
Contributor Information
Eliezra Glasser, Department of Biochemistry and Biophysics, and the Center for RNA Biology, University of Rochester School of Medicine and Dentistry, Rochester, NY 14642, USA.
Debanjana Maji, Department of Biochemistry and Biophysics, and the Center for RNA Biology, University of Rochester School of Medicine and Dentistry, Rochester, NY 14642, USA.
Giulia Biancon, Section of Hematology, Department of Internal Medicine and Yale Cancer Center, Yale University School of Medicine, New Haven, CT 06520, USA.
Anees Mohammed Keedakkatt Puthenpeedikakkal, Department of Biochemistry and Biophysics, and the Center for RNA Biology, University of Rochester School of Medicine and Dentistry, Rochester, NY 14642, USA.
Chapin E Cavender, Department of Biochemistry and Biophysics, and the Center for RNA Biology, University of Rochester School of Medicine and Dentistry, Rochester, NY 14642, USA.
Toma Tebaldi, Section of Hematology, Department of Internal Medicine and Yale Cancer Center, Yale University School of Medicine, New Haven, CT 06520, USA; Department of Cellular, Computational and Integrative Biology (CIBIO), University of Trento, Trento, Italy.
Jermaine L Jenkins, Department of Biochemistry and Biophysics, and the Center for RNA Biology, University of Rochester School of Medicine and Dentistry, Rochester, NY 14642, USA.
David H Mathews, Department of Biochemistry and Biophysics, and the Center for RNA Biology, University of Rochester School of Medicine and Dentistry, Rochester, NY 14642, USA.
Stephanie Halene, Section of Hematology, Department of Internal Medicine and Yale Cancer Center, Yale University School of Medicine, New Haven, CT 06520, USA; Yale Center for RNA Science and Medicine, Yale University School of Medicine, New Haven, CT 06520, USA; Department of Pathology, Yale University School of Medicine, New Haven, CT 06520, USA.
Clara L Kielkopf, Department of Biochemistry and Biophysics, and the Center for RNA Biology, University of Rochester School of Medicine and Dentistry, Rochester, NY 14642, USA; Wilmot Cancer Institute, University of Rochester School of Medicine and Dentistry, Rochester, NY 14642, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01 GM070503 to C.L.K., R01 GM132185 to D.H.M., R01 DK102792 to S.H.]; Yale Cooperative Center of Excellence in Hematology (YCCEH) [NIH U54 DK106857 to G.B. and T.T.]; AIRC [MFAG 2020 (ID 24883 project) to T.T.]; the Edward P. Evans Foundation supported work in the labs of C.L.K. and S.H. Funding for open access charge: NIH [R01 GM070503].
Conflict of interest statement. None declared
REFERENCES
- 1.Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B.. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008; 456:470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nguyen H., Xie J.. Widespread separation of the polypyrimidine tract from 3' AG by g tracts in association with alternative exons in metazoa and plants. Front Genet. 2018; 9:741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wan R., Bai R., Shi Y.. Molecular choreography of pre-mRNA splicing by the spliceosome. Curr. Opin. Struct. Biol. 2019; 59:124–133. [DOI] [PubMed] [Google Scholar]
- 4.Mackereth C.D., Madl T., Bonnal S., Simon B., Zanier K., Gasch A., Rybin V., Valcarcel J., Sattler M.. Multi-domain conformational selection underlies pre-mRNA splicing regulation by U2AF. Nature. 2011; 475:408–411. [DOI] [PubMed] [Google Scholar]
- 5.Voith von Voithenberg L., Sanchez-Rico C., Kang H.S., Madl T., Zanier K., Barth A., Warner L.R., Sattler M., Lamb D.C.. Recognition of the 3' splice site RNA by the U2AF heterodimer involves a dynamic population shift. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E7169–E7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warnasooriya C., Feeney C.F., Laird K.M., Ermolenko D.N., Kielkopf C.L.. A splice site-sensing conformational switch in U2AF2 is modulated by U2AF1 and its recurrent myelodysplasia-associated mutation. Nucleic Acids Res. 2020; 48:5695–5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agrawal A.A., Salsi E., Chatrikhi R., Henderson S., Jenkins J.L., Green M.R., Ermolenko D.N., Kielkopf C.L.. An extended U2AF65-RNA-binding domain recognizes the 3' splice site signal. Nat. Commun. 2016; 7:10950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh R., Valcarcel J., Green M.R.. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995; 268:1173–1176. [DOI] [PubMed] [Google Scholar]
- 9.Singh R., Banerjee H., Green M.R.. Differential recognition of the polypyrimidine-tract by the general splicing factor U2AF65 and the splicing repressor Sex-lethal. RNA. 2000; 6:901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agrawal A.A., McLaughlin K.J., Jenkins J.L., Kielkopf C.L.. Structure-guided U2AF65 variant improves recognition and splicing of a defective pre-mRNA. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:17420–17425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jenkins J.L., Agrawal A.A., Gupta A., Green M.R., Kielkopf C.L.. U2af65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs. Nucleic Acids Res. 2013; 41:3859–3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maji D., Glasser E., Henderson S., Galardi J., Pulvino M.J., Jenkins J.L., Kielkopf C.L.. Representative cancer-associated U2AF2 mutations alter RNA interactions and splicing. J. Biol. Chem. 2020; 295:17148–17157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glasser E., Agrawal A.A., Jenkins J.L., Kielkopf C.L.. Cancer-associated mutations mapped on high-resolution structures of the U2AF2 RNA recognition motifs. Biochemistry. 2017; 56:4757–4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kralovicova J., Borovska I., Kubickova M., Lukavsky P.J., Vorechovsky I.. Cancer-Associated substitutions in RNA recognition motifs of PUF60 and U2AF65 reveal residues required for correct folding and 3' splice-site selection. Cancers (Basel). 2020; 12:1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kielkopf C.L. Insights from structures of cancer-relevant pre-mRNA splicing factors. Curr. Opin. Genet. Dev. 2017; 48:57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiraide T., Tanaka T., Masunaga Y., Ohkubo Y., Nakashima M., Fukuda T., Ogata T., Saitsu H.. Global developmental delay, systemic dysmorphism and epilepsy in a patient with a de novo U2AF2 variant. J. Hum. Genet. 2021; 66:1185–1187. [DOI] [PubMed] [Google Scholar]
- 17.Schott G., Galarza-Munoz G., Trevino N., Chen X., Weirauch M., Gregory S.G., Bradrick S.S., Garcia-Blanco M.A.. U2AF2 binds IL7R exon 6 ectopically and represses its inclusion. RNA. 2021; 27:571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thacker S., Sefyi M., Eng C.. Alternative splicing landscape of the neural transcriptome in a cytoplasmic-predominant pten expression murine model of autism-like behavior. Transl. Psychiatry. 2020; 10:380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenkins J.L., Shen H., Green M.R., Kielkopf C.L.. Solution conformation and thermodynamic characteristics of RNA binding by the splicing factor U2AF65. J. Biol. Chem. 2008; 283:33641–33649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soltis S.M., Cohen A.E., Deacon A., Eriksson T., Gonzalez A., McPhillips S., Chui H., Dunten P., Hollenbeck M., Mathews I.et al.. New paradigm for macromolecular crystallography experiments at SSRL: automated crystal screening and remote data collection. Acta. Crystallogr. D Biol. Crystallogr. 2008; 64:1210–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta. Crystallogr. D Biol. Crystallogr. 2010; 66:133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A.et al.. Overview of the CCP4 suite and current developments. Acta. Crystallogr. D Biol. Crystallogr. 2011; 67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of coot. Acta. Crystallogr. D Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W.et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Afonine P.V., Moriarty N.W., Mustyakimov M., Sobolev O.V., Terwilliger T.C., Turk D., Urzhumtsev A., Adams P.D.. FEM: feature-enhanced map. Acta Crystallogr. D Biol. Crystallogr. 2015; 71:646–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gotz A.W., Williamson M.J., Xu D., Poole D., Le Grand S., Walker R.C.. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized born. J. Chem. Theory Comput. 2012; 8:1542–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Izadi S., Anandakrishnan R., Onufriev A.V.. Building water models: a different approach. J. Phys. Chem. Lett. 2014; 5:3863–3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmit J.D., Kariyawasam N.L., Needham V., Smith P.E.. SLTCAP: a simple method for calculating the number of ions needed for MD simulation. J. Chem. Theory Comput. 2018; 14:1823–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C.. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015; 11:3696–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perez A., Marchan I., Svozil D., Sponer J., Cheatham T.E. III, Laughton C.A., Orozco M.. Refinement of the AMBER force field for nucleic acids: improving the description of alpha/gamma conformers. Biophys. J. 2007; 92:3817–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zgarbova M., Otyepka M., Sponer J., Mladek A., Banas P., Cheatham T.E. III, Jurecka P.. Refinement of the Cornell et al. Nucleic acids force field based on reference quantum chemical calculations of glycosidic torsion profiles. J. Chem. Theory Comput. 2011; 7:2886–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J., Cieplak P., Kollman P.A.. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules?. J. Comput. Chem. 2000; 21:1049–1074. [Google Scholar]
- 33.Case D.A., Ben-Shalom I.Y., Brozell S.R., Cerutti D.S., Cheatham T.E., Cruzeiro V.W.D., Darden T.A., Duke R.E., Ghoreishi D., Gilson M.K.et al.. 2018; San Francisco: University of California. [Google Scholar]
- 34.Romo T.D., Leioatts N., Grossfield A.. Lightweight object oriented structure analysis: tools for building tools to analyze molecular dynamics simulations. J. Comput. Chem. 2014; 35:2305–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Nostrand E.L., Pratt G.A., Shishkin A.A., Gelboin-Burkhart C., Fang M.Y., Sundararaman B., Blue S.M., Nguyen T.B., Surka C., Elkins K.et al.. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods. 2016; 13:508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Biancon G., Joshi P., Zimmer J.T., Hunck T., Gao Y., Lessard M.D., Courchaine E., Barentine A.E.S., Machyna M., Botti V.et al.. Precision analysis of mutant U2AF1 activity reveals deployment of stress granules in myeloid malignancies. Mol. Cell. 2022; 82:1107–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu H., Luo X., Qian J., Pang X., Song J., Qian G., Chen J., Chen S.. FastUniq: a fast de novo duplicates removal tool for paired short reads. PLoS One. 2012; 7:e52249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet j. 2011; 17:10. [Google Scholar]
- 39.Gooding C., Clark F., Wollerton M.C., Grellscheid S.N., Groom H., Smith C.W.. A class of human exons with predicted distant branch points revealed by analysis of AG dinucleotide exclusion zones. Genome Biol. 2006; 7:R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee C.G., Zamore P.D., Green M.R., Hurwitz J.. RNA annealing activity is intrinsically associated with U2AF. J. Biol. Chem. 1993; 268:13472–13478. [PubMed] [Google Scholar]
- 41.Gaur R.K., Valcarcel J., Green M.R.. Sequential recognition of the pre-mRNA branch point by U2AF65 and a novel spliceosome-associated 28-kDa protein. RNA. 1995; 1:407–417. [PMC free article] [PubMed] [Google Scholar]
- 42.Valcarcel J., Gaur R.K., Singh R., Green M.R.. Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA. Science. 1996; 273:1706–1709. [DOI] [PubMed] [Google Scholar]
- 43.Crisci A., Raleff F., Bagdiul I., Raabe M., Urlaub H., Rain J.C., Kramer A.. Mammalian splicing factor SF1 interacts with SURP domains of U2 snRNP-associated proteins. Nucleic Acids Res. 2015; 43:10456–10473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wiesner S., Stier G., Sattler M., Macias M.J.. Solution structure and ligand recognition of the WW domain pair of the yeast splicing factor prp40. J. Mol. Biol. 2002; 324:807–822. [DOI] [PubMed] [Google Scholar]
- 45.Bedford M.T., Reed R., Leder P.. WW domain-mediated interactions reveal a spliceosome-associated protein that binds a third class of proline-rich motif: the proline glycine and methionine-rich motif. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:10602–10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abovich N., Rosbash M.. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997; 89:403–412. [DOI] [PubMed] [Google Scholar]
- 47.Sickmier E.A., Frato K.E., Shen H., Paranawithana S.R., Green M.R., Kielkopf C.L.. Structural basis of polypyrimidine tract recognition by the essential pre-mRNA splicing factor, U2AF65. Mol. Cell. 2006; 23:49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Nostrand E.L., Pratt G.A., Yee B.A., Wheeler E.C., Blue S.M., Mueller J., Park S.S., Garcia K.E., Gelboin-Burkhart C., Nguyen T.B.et al.. Principles of RNA processing from analysis of enhanced CLIP maps for 150 RNA binding proteins. Genome Biol. 2020; 21:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zorio D.A., Blumenthal T.. Both subunits of U2AF recognize the 3' splice site in Caenorhabditiselegans. Nature. 1999; 402:835–838. [DOI] [PubMed] [Google Scholar]
- 50.Wu S., Romfo C.M., Nilsen T.W., Green M.R.. Functional recognition of the 3' splice site AG by the splicing factor U2AF35. Nature. 1999; 402:832–835. [DOI] [PubMed] [Google Scholar]
- 51.Merendino L., Guth S., Bilbao D., Martinez C., Valcarcel J.. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3' splice site aG. Nature. 1999; 402:838–841. [DOI] [PubMed] [Google Scholar]
- 52.Guth S., Martinez C., Gaur R.K., Valcarcel J.. Evidence for substrate-specific requirement of the splicing factor U2AF35 and for its function after polypyrimidine tract recognition by U2AF65. Mol. Cell. Biol. 1999; 19:8263–8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Daubner G.M., Clery A., Jayne S., Stevenin J., Allain F.H.. A syn-anti conformational difference allows SRSF2 to recognize guanines and cytosines equally well. EMBO J. 2011; 31:162–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Duss O., Michel E., Diarra dit Konte N., Schubert M., Allain F.H.. Molecular basis for the wide range of affinity found in csr/rsm protein-RNA recognition. Nucleic Acids Res. 2014; 42:5332–5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hall T.M. De-coding and re-coding RNA recognition by PUF and PPR repeat proteins. Curr. Opin. Struct. Biol. 2016; 36:116–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leulliot N., Varani G.. Current topics in RNA-protein recognition: control of specificity and biological function through induced fit and conformational capture. Biochemistry. 2001; 40:7947–7956. [DOI] [PubMed] [Google Scholar]
- 57.Shajani Z., Drobny G., Varani G.. Binding of U1A protein changes RNA dynamics as observed by 13C NMR relaxation studies. Biochemistry. 2007; 46:5875–5883. [DOI] [PubMed] [Google Scholar]
- 58.Oberstrass F.C., Allain F.H., Ravindranathan S.. Changes in dynamics of SRE-RNA on binding to the VTS1p-SAM domain studied by 13C NMR relaxation. J. Am. Chem. Soc. 2008; 130:12007–12020. [DOI] [PubMed] [Google Scholar]
- 59.Ellis J.J., Jones S.. Evaluating conformational changes in protein structures binding RNA. Proteins. 2008; 70:1518–1526. [DOI] [PubMed] [Google Scholar]
- 60.Inners L.D., Felsenfeld G.. Conformation of polyribouridylic acid in solution. J. Mol. Biol. 1970; 50:373–389. [DOI] [PubMed] [Google Scholar]
- 61.Norberg J., Nilsson L.. Stacking free energy profiles for all 16 natural ribodinucleoside monophosphates in aqueous solution. J. Am. Chem. Soc. 1995; 117:10832–10840. [Google Scholar]
- 62.Shao C., Yang B., Wu T., Huang J., Tang P., Zhou Y., Zhou J., Qiu J., Jiang L., Li H.et al.. Mechanisms for U2AF to define 3' splice sites and regulate alternative splicing in the human genome. Nat. Struct. Mol. Biol. 2014; 21:997–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Okeyo-Owuor T., White B.S., Chatrikhi R., Mohan D.R., Kim S., Griffith M., Ding L., Ketkar-Kulkarni S., Hundal J., Laird K.M.et al.. U2AF1 mutations alter sequence specificity of pre-mRNA binding and splicing. Leukemia. 2015; 29:909–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fei D.L., Motowski H., Chatrikhi R., Prasad S., Yu J., Gao S., Kielkopf C.L., Bradley R.K., Varmus H.. Wild-type U2AF1 antagonizes the splicing program characteristic of U2AF1-mutant tumors and is required for cell survival. PLoS Genet. 2016; 12:e1006384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Park S.M., Ou J., Chamberlain L., Simone T.M., Yang H., Virbasius C.M., Ali A.M., Zhu L.J., Mukherjee S., Raza A.et al.. U2AF35(S34F) promotes transformation by directing aberrant ATG7 Pre-mRNA 3' end formation. Mol. Cell. 2016; 62:479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Akef A., McGraw K., Cappell S.D., Larson D.R.. Ribosome biogenesis is a downstream effector of the oncogenic U2AF1-S34F mutation. PLoS Biol. 2020; 18:e3000920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Palangat M., Anastasakis D.G., Fei D.L., Lindblad K.E., Bradley R., Hourigan C.S., Hafner M., Larson D.R.. The splicing factor U2AF1 contributes to cancer progression through a noncanonical role in translation regulation. Genes Dev. 2019; 33:482–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Coulon A., Ferguson M.L., de Turris V., Palangat M., Chow C.C., Larson D.R.. Kinetic competition during the transcription cycle results in stochastic RNA processing. Elife. 2014; 3:e03939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wan Y., Anastasakis D.G., Rodriguez J., Palangat M., Gudla P., Zaki G., Tandon M., Pegoraro G., Chow C.C., Hafner M.et al.. Dynamic imaging of nascent RNA reveals general principles of transcription dynamics and stochastic splice site selection. Cell. 2021; 184:2878–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data deposition: The coordinates for the U2AF structures have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 7S3A, 7S3B, 7S3C for C5, G5 and A5 structures). The U2AF2 eCLIP-seq files have been deposited in the GEO database, https://www.ncbi.nlm.nih.gov/geo/ (GSE195669). The eCLIP-seq files for U2AF2 with OE U2AF1 are available with GEO accession GSE195620 (36).