Abstract
d-Pantothenate is synthesized via four enzymes from ketoisovalerate, which is an intermediate of branched-chain amino acid synthesis. We quantified three of these enzyme activities in Corynebacterium glutamicum and determined specific activities ranging from 0.00014 to 0.001 μmol/min mg (protein)−1. The genes encoding the ketopantoatehydroxymethyl transferase and the pantothenate synthetase were cloned, sequenced, and functionally characterized. These studies suggest that panBC constitutes an operon. By using panC, an assay system was developed to quantify d-pantothenate. The wild type of C. glutamicum was found to accumulate 9 μg of this vitamin per liter. A strain was constructed (i) to abolish l-isoleucine synthesis, (ii) to result in increased ketoisovalerate formation, and (iii) to enable its further conversion to d-pantothenate. The best resulting strain has ilvA deleted from its chromosome and has two plasmids to overexpress genes of ketoisovalerate (ilvBNCD) and d-pantothenate (panBC) synthesis. With this strain a d-pantothenate accumulation of up to 1 g/liter is achieved, which is a 105-fold increase in concentration compared to that of the original wild-type strain. From the series of strains analyzed it follows that an increased ketoisovalerate availability is mandatory to direct the metabolite flux into the d-pantothenate-specific part of the pathway and that the availability of β-alanine is essential for d-pantothenate formation.
d-Pantothenate is a water-soluble vitamin required as a pharmaceutical and a feed additive. About 4,000 tons of pantothenate are produced annually (48). The present method of production depends for the most part on chemical synthesis from bulk chemicals. However, this synthesis requires the optical resolution of racemic intermediates. Therefore, a variety of routes have been assayed to improve its synthesis, including enzyme conversions (41). One of the processes of d-pantothenate synthesis uses a lactonohydrolase activity of Fusarium oxysporum, which catalyzes the stereospecific hydrolysis of chemically made d,l-pantolactone to generate d-pantolactone as a chiral building block for its further chemical conversion to d-pantothenate (19). Therefore, there is still potential for further improving d-pantothenate production, for instance, by its direct microbial synthesis.
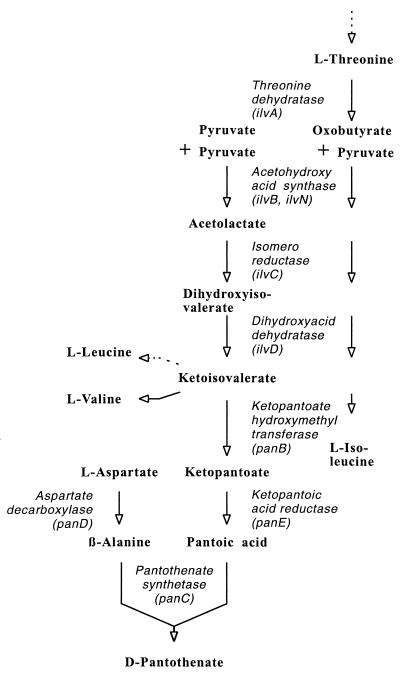
In Escherichia coli the specific biosynthesis pathway of this vitamin consists of only four steps (Fig. 1). The first reaction, catalyzed by the ketopantoatehydroxymethyl transferase, uses the l-valine intermediate 2-ketoisovalerate to generate ketopantoate, which is reduced to d-pantoic acid. An aspartate-α-decarboxylase activity generates β-alanine, which is ligated with pantoic acid to yield d-pantothenate. The respective enzymes of E. coli and Salmonella typhimurium have been characterized, and the corresponding genes have been identified (11, 15). Also for Bacillus subtilis, transferase and ketopantoate reductase activities have been demonstrated (1). In general, three different mechanisms of β-alanine formation are thought to be present in microorganisms (41).
FIG. 1.
The pathway of d-pantothenate biosynthesis and its integration into the synthesis of branched-chain amino acids.
We are interested in metabolite flux analysis in the gram-positive bacterium Corynebacterium glutamicum (25). This bacterium is used for the large-scale production of l-lysine and l-glutamate (22). It has a high capacity to supply precursor metabolites (26), and its molecular physiology of amino acid synthesis has been analyzed in detail (36). We have also developed strains producing l-isoleucine (7), the synthesis of which uses enzymes in part identical to those required for the synthesis of l-valine (Fig. 1). Due to the linkage of the branched-chain amino acid synthesis with the short reaction sequence of d-pantothenate synthesis, the analysis of d-pantothenate formation with C. glutamicum is an attractive target. Moreover, a closely related bacterium, Brevibacterium ammoniagenes, has already been reported to accumulate coenzyme A, which is synthesized from d-pantothenate (42). In the present work, we analyze enzymes and genes involved in d-pantothenate synthesis by C. glutamicum and study their use, together with genes of branched-chain amino acid synthesis, for the direct microbial synthesis of d-pantothenate.
MATERIALS AND METHODS
Strains, plasmids, and cultivations.
The strains and plasmids used are shown in Table 1. C. glutamicum was grown on brain heart infusion medium or minimal medium CGXII (20). E. coli was grown in Luria broth or minimal medium M9 (45). The cultivations of strains containing the pEKx2 plasmids were done in the presence of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) added 5 h after inoculation.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| E. coli | ||
| S17-1 | Mobilizing strain | 43 |
| SJ2 | panB mutant | 5 |
| DV39 | panC mutant | 47 |
| C. glutamicum | ||
| ATCC 13032 | Wild type | |
| ATCC 13032 ΔilvA | ilvA deletion mutant | This work |
| R127 | Restriction negative | 23 |
| R127::orf1 | ORF 1 integration mutant | This work |
| R127::panC | panC integration mutant | This work |
| L. plantarum | ||
| ATCC 8014 | Pantothenate auxotroph | |
| Plasmids | ||
| pJC1 | E. coli-C. glutamicum shuttle vector, Kanr | 3 |
| pZ1 | E. coli-C. glutamicum shuttle vector, Kanr | 27 |
| pUR1 | pBR322 with 9.3-kb chromosomal SauIIIA fragment encompassing panBC and xylB | This work |
| pUR1.1 | pUC19 with 2.4-kb SspI/PvuII fragment encompassing panB | This work |
| pUR1.2 | pUC18 with 3.9-kb SspI/SalI fragment encompassing panBC | This work |
| pEC7panD | pEC7 with 900-bp fragment encompassing panD of E. coli | This work |
| pEC7 | E. coli-C. glutamicum shuttle vector, Cmr | 10 |
| pJC1ilvBNCD | pKK5 with 2.6-kb XbaI fragment encompassing ilvD | This work |
| pJC1ilvBNC | pJC1 with 5.7-kb HindIII/EcoRI fragment encompassing ilvBNC | This work |
| pKK5 | pJC4 with 5.7-kb HindIII/EcoRI fragment encompassing ilvBNC | 2 |
| pECM3ilvBNCD | pECM3 with 5.7-kb XbaI fragment encompassing ilvBNC and 3.1-kb XbaI fragment encompassing ilvD | This work |
| pECM3ilvBNC | pECM3 with 5.7-HindIII/EcoRI fragment encompassing ilvBNC | This work |
| pECM3 | Shuttle vector, derived from pECM2, Cmr | 16 |
| pZ1panBC | pZ1 with 3.5-kb ScaI/SalI fragment encompassing panBC | This work |
| pZ1panC | pZ1 with 2.4-kb BstEII/SalI fragment encompassing panC | This work |
| pEKEx2 | Expression vector, Kanr Ptac | 9 |
Metabolite quantifications.
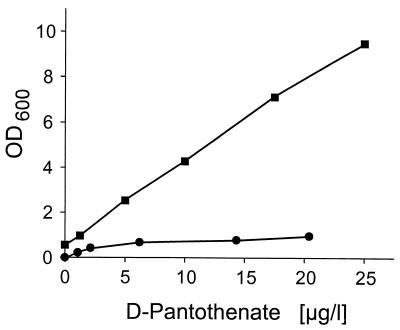
d-Pantothenate was quantified in a microbiological assay with C. glutamicum R127::panC (this work). For this purpose, cells of this strain were precultivated overnight on brain heart infusion medium (with 25 μg of kanamycin [Difco]), washed twice with 9 g of NaCl per liter, and inoculated into minimal medium CGXII (with 25 μg of kanamycin) to obtain an initial optical density at 600 nm (OD600) of 0.5. This served to deprive the cells of d-pantothenate. Although d-pantothenate had not been supplied, growth of the cells was possible up to an OD600 of about 20 (the control strain, C. glutamicum R127, reaches an OD600 of about 40). One milliliter of the pantothenate-deprived culture (taken after 30 h) was mixed with 700 μl of glycerol and stored at −70°C. Sixty-microliter aliquots of these stocks were used to inoculate assay tubes. These assay tubes (Falcon 2057; Becton and Dickinson) contained 3 ml of four-thirds concentrated CGXII medium (with 25 μg of kanamycin), 1 ml of sterile filtered d-pantothenate sample, and C. glutamicum R127::panC (60 μl). Tubes were cultivated for 40 h at 30°C with shaking, and the OD600 was determined. On the basis of results from this procedure, growth is linearly dependent on the concentration of d-pantothenate over a broad concentration range (Fig. 2), which is a clear advantage compared to the standard d-pantothenate determination with Lactobacillus plantarum ATCC 8014 according to the U.S. Pharmacopoeial Convention. When assays for one sample were repeated, the standard deviation for 10 ng of d-pantothenate per ml was ± 0.9 ng. The assay is linear in the range from 0 to 100 ng of d-pantothenate per assay (4 ml). In the case of nonlinear growth obtained with new glycerol stock cultures of the panC mutant, the inoculum varied between 60 and 100 μl.
FIG. 2.
d-Pantothenate quantification for C. glutamicum::panC (■) and L. plantarum (●). The d-pantothenate concentration given is the final concentration in the assay.
l-Valine and β-alanine were quantified by automated precolumn derivatization with ortho-phthaldialdehyde (24), followed by reversed-phase chromatography with fluorometric detection (model HP LC1090; Hewlett Packard). α-Ketoisovalerate was derivatized with diaminomethoxybenzole (12) and again quantified by reversed-phase chromatography and fluorometric detection.
Enzyme activity determinations.
A crude extract of cells taken from the late-exponential phase was prepared by sonication. The level of ketopantoatehydroxymethyl transferase activity was determined by quantifying the ketoisovalerate formation from ketopantoate. The assay mixture consisted of 71 mM potassium phosphate (pH 6.8), 1 mM MgSO4, 3.6 mM ketopantoate, and 0.71 mM tetrahydrofolate. The reaction was started by the addition of the crude extract, which was equilibrated prior to use on PD10 columns (LKB-Pharmacia) with 100 mM potassium phosphate (pH 6.8), and was run at 37°C for 60 min.
The pantothenate synthetase activity was assayed, with minor modifications, as described previously (29). The assay mixture consisted of 100 mM Tris-HCl (pH 10), 10 mM MgSO4, 5 mM d,l-pantoate, 5 mM β-alanine, and 10 mM ATP. After the addition of the crude extract the assay mixture was incubated at 30°C for 40 min, and the assay was then terminated by the addition of 5 volumes of isatoic acid anhydride (to a concentration of 3.2 mM in dimethylformamide). This enabled the quantification of pantothenate by reversed-phase chromatography as described by Julliard (18).
The aspartate α-decarboxylase activity was quantified by β-alanine formation in a reaction mixture containing 100 mM potassium phosphate buffer (pH 7.5), 5 mM EDTA (pH 7.5), and 5 mM l-aspartate. The reaction was started by the addition of the crude extract to the mixture and was run for 60 min at 37°C.
Gene bank and sequence analysis.
The gene bank used was as described previously (32). The sequence for both strands of the 2,164-bp fragment was determined by the dideoxy chain termination method on subclones derived from exonuclease treatment of pUR1.1 and pUR1.2. Additional sequence information covering xylB was obtained by primer walking.
Plasmid constructions.
All plasmid constructions were done in E. coli DH5αmcr. Plasmid pJC1ilvBNCD was obtained by ligating a 2.6-kb XbaI fragment containing ilvD into the BamHI site of pKK5 (2). To obtain pECM3ilvBNC a 5.7-kb fragment of pKK5 encompassing ilvBNC was cloned into the EcoRV site of pECM3. Additionally, a 5.7-kb XbaI fragment (ilvBNC) of pKK5 and a 3.1-kb XbaI fragment (ilvD) were ligated with EcoRV-digested pECM3 to yield pECM3ilvBNCD. Plasmid pEC7panD was constructed by ligating a 900-bp PvuII fragment of pDKS1, containing panD of E. coli (35), with SmaI-digested pEC7. To construct pEKEx2panBC, the 5′ region of panB was amplified with the primers 5′GATCGTCGACCATCACATCTATACTCATGCCC and 5′ACCCGATGTGGCCGACAACC. The resulting PCR fragment was treated with SalI and EcoRI and ligated with the identically treated pEKEx2. The plasmid obtained was cleaved with EcoRI and ligated with the 1.8-kb EcoRI fragment of pUR1, containing the 3′ end of panB and panC.
Strain constructions.
To construct the ilvA deletion mutant of C. glutamicum ATCC 13032, the 242-bp BglII fragment of ilvA was deleted in pBM21 (30). Subsequently, the fragment with the deletion was excised as a 1.3-kb EcoRI fragment, which was ligated with pK19mobsacB (39). The resulting mobilizable E. coli vector enabled the transfer of the deletion into the chromosome of C. glutamicum by two rounds of positive selection. The deletion was confirmed by PCR.
To construct the panC insertion mutant of C. glutamicum R127, an internal 168-bp panC fragment was amplified with the primers 5′GTTCGCACCCGATGTGGAGG and 5′ATGCACGATCAGGGCGCACC. The fragment was cloned into the SmaI site of pUC18 with the SureClone ligation kit (Amersham), subsequently excised as an EcoRI/SalI fragment, and finally ligated with EcoRI/SalI-treated pK18mob (39). The resulting vector was transferred to C. glutamicum via conjugation (38), and kanamycin-resistant transconjugants were obtained. One strain selected was termed C. glutamicum R127::panC. Its d-pantothenate auxotrophy was verified, as well as the vector integration into the chromosome.
To construct the open reading frame (ORF) 1 insertion mutant of C. glutamicum R127, an internal 202-bp ORF 1 fragment was amplified with the primers 5′GATCGAATTCCCGATTAAATCGCGGAGACGG and 5′GATCGTCGACCTTTGCTGCCGATTCAAGTG. The fragment was digested with EcoRI and SalI and ligated with the EcoRI/SalI-treated pK19mobsacB. This vector was used to construct C. glutamicum R127::orf1, whose correct integration of the vector into ORF 1 was verified via PCR.
Nucleotide sequence accession number.
The sequence for both strands of the 2,164-bp fragment was deposited in the EMBL and GenBank databases under accession no. X96580.
RESULTS
Cloning and sequence analysis of panBC.
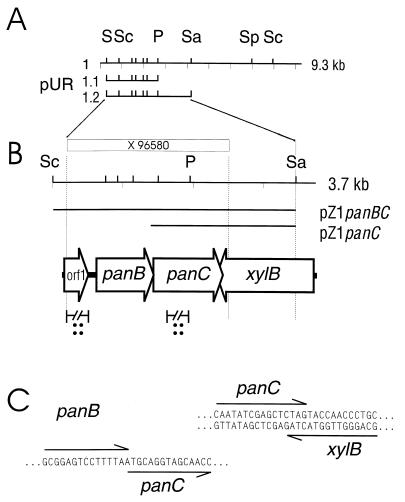
The E. coli panB mutant SJ2 (4) was transformed to ampicillin resistance with genomic DNA of C. glutamicum ATCC 13032 ligated with pBR322. This yielded eight plasmids able to restore the growth of SJ2 on minimal medium plates. They were found to contain three different inserts of 9.3, 2.1, and 1.8 kb, respectively. The insert in the largest plasmid, named pUR1 (Fig. 3), was assayed by a Southern blot analysis of ScaI-digested chromosomal DNA (data not shown) with the 1.5-kb PvuII/SalI fragment as a probe. This confirmed the origin and structural identity of the large fragment cloned. The isolated plasmids were used to assay for an additional complementation of the panC mutation of E. coli DV39 (47). Plasmid pUR1 complemented this mutation, whereas the two smaller plasmids failed to do so. To further confine the complementing functions, several subclones were made. Whereas pUR1.1 only complemented the mutation in E. coli SJ2, pUR1.2 complemented the mutations of both E. coli strains. A nucleotide sequence of 2.2 kb from the insert of pUR1 was determined on both strands, whereas the sequence for an adjacent 1.5-kb part was established on one strand only (Fig. 3).
FIG. 3.
Overview of the cloned and subcloned chromosomal fragments (A), the sequenced part and organization of the genes (B), and the overlaps of panB with panC and those of panC with xylB (C). S, SalI; Sc, ScaI; Sa, SalI; P, PstI; Sp, SphI.
The sequence analyses identified four ORFs. ORF 1 exhibits no identities with known sequences. Inactivation of ORF 1 in C. glutamicum R127::orf1 resulted in a decreased growth rate (μ = 0.31 h−1; μ = 0.38 h−1 [for C. glutamicum R127]), which could not be restored by the addition of d-pantothenate (data not shown). Interestingly, amino acid residues 64 to 89 encoded by ORF 1 fit exactly to the consensus sequence of the helix-turn-helix motif of LysR-type regulators (40). Therefore, it is proposed that ORF 1 encodes a transcriptional regulator which is functionally not related to d-pantothenate synthesis in C. glutamicum. The deduced amino acid sequence encoded by the second ORF (nucleotides 351 to 1166) exhibits a high identity with PanB, as does that encoded by the third ORF (nucleotides 1166 to 2005) with PanC polypeptides. The highest identities are shared with PanB of Mycobacterium tuberculosis (52%) and PanC of Schizosaccharomyces pombe (45%), respectively. The fourth ORF is located on the strand opposite to that of the pan genes. Its deduced polypeptide shows significant homology to xylulokinases (encoded by xylB).
Enzyme activity determinations.
To functionally characterize the genes, enzyme activity determinations in the homologous background were performed. For this purpose, the ScaI/ SalI fragment of pUR1.2 was ligated with the E. coli-C. glutamicum shuttle vector pZ1 (27) to yield pZ1panBC and with the BstEII/SalI fragment to yield pZ1panC (Fig. 3). With these plasmids the wild type of C. glutamicum was transformed. The resulting recombinant strains were grown on minimal medium, and cells were harvested for activity determinations.
The ketopantoatehydroxymethyl transferase activity (panB) was determined in a novel assay based on the quantification of ketoisovalerate formed from ketopantoate (see Materials and Methods). With C. glutamicum/pZ1panBC a specific activity of 1.9 nmol/min/mg of protein was obtained, whereas the control yielded an activity of 0.14 nmol/min/mg of protein (Table 2). This ∼13-fold increase in synthesis confirms the identity of panB. It is reported that d-pantothenate and d-pantoate inhibit the transferase activity in E. coli (33) and salicylate the enzyme in S. typhimurium (34). Therefore, these compounds were included individually at 10 mM concentrations in the enzyme assay with the extract of C. glutamicum. Only a marginal effect with d-pantothenate was detected, but d,l-pantoate reduced the transferase activity to 20% and salicylate to 22%.
TABLE 2.
Enzyme activities of d-pantothenate synthesis in C. glutamicum, and E. colia
| Enzyme and strain | Sp act (nmol/min/mg of protein) |
|---|---|
| Ketopantoate hydroxymethyltransferase | |
| C. glutamicum | 0.14 |
| C. glutamicum pZ1panBC | 1.9 |
| C. glutamicum pEKEx2panBC | 1.9 |
| E. coli | 3–7 |
| Pantothenate synthetase | |
| C. glutamicum | 1 |
| C. glutamicum pZ1panBC | 12 |
| C. glutamicum pZ1panC | 1 |
| E. coli | 4.2 |
| Aspartate decarboxylase | |
| C. glutamicum | 0.11 |
| E. coli | 0.004–0.17 |
The activity of the pantothenate synthetase (encoded by panC) was determined in C. glutamicum/pZ1panBC. It is 12 nmol/min/mg of protein, opposed to 1 nmol/min/mg of protein in the control (Table 2). However, with C. glutamicum/pZ1panC, no increased specific activity was detected. This suggests the organization of panBC as an operon, as indicated from the sequence of the cluster.
In addition to the quantification of the transferase and synthase activities, we also assayed C. glutamicum for aspartate decarboxylase activity with a novel assay by quantification of β-alanine via high-pressure liquid chromatography. As shown in Table 2, this enzyme has a specific activity of 0.11 nmol/min/mg of protein. For comparison Table 2 also includes decarboxylase, transferase, and synthetase activities for E. coli. It can be seen that the enzyme activities in E. coli are in the same order of magnitude, except that of the transferase, which is at least 1 order of magnitude higher.
d-Pantothenate formation by the wild type.
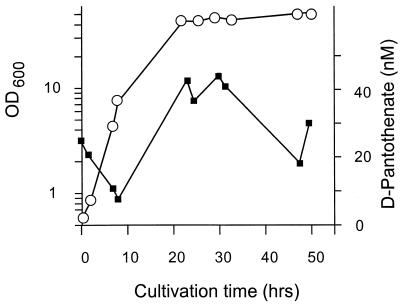
To assay for d-pantothenate accumulation by C. glutamicum, the wild type was grown in minimal medium. In samples of sterile filtered culture supernatants, d-pantothenate was quantified in the assay developed (see Materials and Methods). As can be seen in Fig. 4, there is only a very weak accumulation of maximal 42 nM d-pantothenate in culture supernatants, which is in accord with the low activities and/or a tight control of d-pantothenate synthesis. The unexpected time course of the d-pantothenate concentrations shown in Fig. 4 was verified in a separate experiment. With the wild type of E. coli a d-pantothenate accumulation of 3 mg/liter has been reported (14). In an additional experiment a recombinant C. glutamicum strain was made and assayed for d-pantothenate formation. This strain was C. glutamicum/pZ1panBC, which additionally contained the plasmid-encoded l-aspartate decarboxylase (panD) of E. coli (see Materials and Methods). This strain overexpressing three of the d-pantothenate biosynthesis genes again exhibited a time course of d-pantothenate accumulation almost identical to that of the wild type and also not exceeding 42 nM as the highest concentration. Further engineering was therefore required.
FIG. 4.
Time course of d-pantothenate accumulation (■) and growth (○).
Increased d-pantothenate formation by ilvA deletion.
We first assayed the consequences of the deletion of the threonine dehydratase gene ilvA on a flux increase towards d-pantothenate. This was based on the idea that due to the prevention of l-isoleucine synthesis an increased pyruvate availability could result in increased ketoisovalerate accumulation with further conversion to d-pantothenate (Fig. 1). By the application of two rounds of positive selection for the presence and absence of vector sequences, respectively (39), a C. glutamicum wild-type derivative was constructed with the internal 242-bp BglII fragment of ilvA deleted from the chromosome. The d-pantothenate concentration after 24 h of cultivation in minimal medium by the C. glutamicum ilvA deletion mutant obtained was 236 nM, which is about a fivefold increase compared to that of the wild type (see above).
l-Valine accumulation by overexpressing ilv genes.
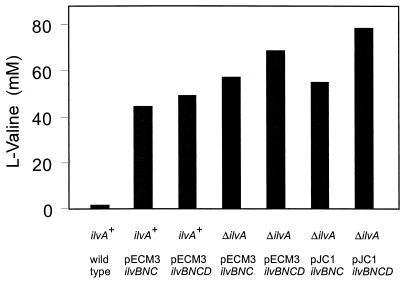
Based on the increased d-pantothenate accumulation as a consequence of the ilvA deletion, a further flux increase was attempted by overexpressing the common genes required for l-valine and l-isoleucine synthesis (Fig. 1). For this purpose pKK5 encoding ilvBNC (2) was used, thus resulting in high-level acetohydroxy acid synthase and isomeroreductase activities. In addition the recently cloned ilvD gene (30a) was used. This gene was ligated with pKK5 to yield pJC1ilvBNCD. As a further construct pECM3ilvBNCD was made, which confers chloramphenicol resistance in contrast to pJC1ilvBNCD. The plasmids were used to transform C. glutamicum and its ilvA deletion mutant. The strains constructed were cultivated in minimal medium, and l-valine accumulations were determined after 48 h of cultivation when glucose was consumed. The highest l-valine concentration obtained was 79 mM, whereas the wild type accumulated only 1 mM (Fig. 5). The strains with the ilvA deletions accumulated higher l-valine concentrations than the ilvA+ strains. Furthermore, ilvD overexpression is necessary to obtain the maximal l-valine accumulation. As a third outcome, it is evident that the basis vector itself influences l-valine accumulations, although both vectors use the same C. glutamicum replicon.
FIG. 5.
l-Valine accumulation with isogenic C. glutamicum strains. Below the columns the genotype of each strain is given, which is either ilvA+ or ΔilvA. The strains additionally carry the plasmid pECM3 or pJC1 carrying ilvBNC or ilvBNCD, respectively.
d-Pantothenate accumulation by combined overexpression of ilv and pan genes.
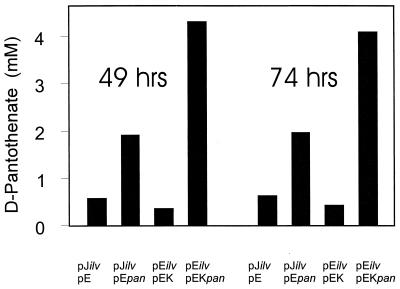
To exploit the increased capability of l-valine formation for increased d-pantothenate accumulation, the four ilv genes were overexpressed together with panBC. To enable their common overexpression, compatible plasmids were required. For this purpose, the chromosomal 2.2-kb fragment encompassing panBC was engineered, as outlined in Materials and Methods, to be cloned into the expression vector pEKEx2, which carries the pBL1 replicon (37) and confers kanamycin resistance, to yield pEKEx2panBC. Furthermore, panBC was ligated with pEC7 by using the same replicon (but conferring chloramphenicol resistance) to yield pEC7panBC. Starting from the C. glutamicum ilvA deletion mutant carrying pJC1ilvBNCD, a C. glutamicum ilvA deletion mutant carrying pJC1ilvBNCD and pEC7panBC was made, and starting from the C. glutamicum ilvA deletion mutant carrying pECM3ilvBNCD, a C. glutamicum ilvA deletion mutant carrying pECM3ilvBNCD and pEKEx2panBC was made. The d-pantothenate accumulations obtained with these strains in the standard minimal medium containing 20 mM β-alanine are shown in Fig. 6. First of all, the l-valine-producing C. glutamicum ilvA deletion mutant carrying pJC1ilvBNCD and pEC7 (control [without panBC overexpressed]) already accumulated up to 0.53 mM d-pantothenate after 49 h (Fig. 6). Without β-alanine addition the accumulation was only 0.87 μM, thus showing the absolute requirement of this amino acid derivative for increased d-pantothenate formation (data not shown). When additionally panBC was overexpressed (the ilvA deletion mutant carrying pJC1ilvBNCD and pEC7panBC) the d-pantothenate was accumulated to a concentration of 2.1 mM. This strong effect of panBC overexpression is also apparent with the second gene combination. Whereas the ilvA deletion mutant carrying pECM3ilvBNCD and pEKEx2 accumulated 0.43 mM d-pantothenate, the ilvA deletion mutant carrying pEKEx2panBC accumulated as much as a 4.2 mM concentration of the vitamin, which is a 105-fold-higher concentration than that obtained with the wild type. After 74 h the pantothenate accumulations quantified were almost the same as those at the earlier point in time.
FIG. 6.
d-Pantothenate accumulation with plasmid-carrying strains derived from the C. glutamicum ilvA deletion mutant. pJilv, pJC1ilvBNCD; pEilv, pECM3ilvBNCD; pEpan, pEC7panBC; pEKpan, pEKEx2panBC; pE, pEC7.
DISCUSSION
In the present work the genes panB and panC of C. glutamicum were cloned and were found to be clustered. In B. subtilis and E. coli an identical organization of the two genes is present (28, 44). For the latter organism a transcriptional analysis has revealed a significantly larger transcript of panB than expected from the size of that gene, suggesting the cotranscription of a second gene (17). According to recent genome information this could well be panC. There is evidence that in C. glutamicum panB and panC constitute an operon. The sequence shows that both genes overlap by one nucleotide (Fig. 3), which has been demonstrated for amino acid biosynthetic genes to be evidence of a close translational coupling (31). In addition to this structural feature, the functional characterization of pantothenate synthetase activities supports the conclusion that panBC in C. glutamicum forms an operon. Whereas with a panBC-containing fragment an increased synthetase activity was the result, this was not the case with a fragment containing panC, which included a significant chromosomal part of the 5′ region of the gene.
The three enzymes of the d-pantothenate synthesis quantified have specific activities of around 1 nmol/min/mg of protein. This is extremely low compared to the specific activities of enzymes of amino acid synthesis, which are about 2 orders of magnitude higher, or that of enzymes of the central metabolism, whose specific activities are increased by as many as 3 orders of magnitude. This may be due to different enzyme amounts and consequently different expression levels. As the expression levels of genes of amino acid synthesis and of the central metabolism are shown to be directly related to the degree of codon bias in C. glutamicum (10), as is the case for other organisms too, it was interesting to inspect the codon usage of the cloned pan genes. This was done together with the biotin biosynthesis genes (bioABD) of C. glutamicum (13). This analysis revealed that in fact the codon usage of the vitamin biosynthesis genes of C. glutamicum is less biased than that of the high and moderately expressed genes. As a consequence, the preferred codon for vitamin biosynthesis genes is, in 6 of 19 cases, different from that of the high and moderately expressed genes, for which almost exclusively the same codon is used (data not shown). Since the d-pantothenate accumulation is in part dependent on the vector used (Fig. 6), which might reflect different expression levels, the design of pan genes by the use of appropriate codons is an option to obtain optimal expression levels for increased product accumulation.
From the enzyme activity determinations it is furthermore evident that the pathway of d-pantothenate synthesis in the gram-positive bacterium C. glutamicum is identical to that of the gram-negative bacterium E. coli, where β-alanine is not uracil derived as in plants, for instance. Also the feedback inhibition of the ketopantoatehydroxymethyl transferase by pantoate is comparable in both organisms (33). The inhibition of the transferase activity of C. glutamicum by the false feedback inhibitor salicylate reflects the situation described for S. typhimurium (34). An important difference is the inhibition of the transferase in E. coli by d-pantothenate. In a concentration of 2.5 mM this effector reduces the enzyme activity of E. coli by about 50% (33), whereas the enzyme of C. glutamicum is almost unaffected by 10 mM d-pantothenate.
The increased d-pantothenate accumulation by C. glutamicum required a concerted engineering of the metabolite flux similar to that experienced during the construction of l-isoleucine-producing strains (7). One important feature in obtaining a d-pantothenate accumulation is the deletion of ilvA, which encodes the key enzyme of isoleucine synthesis (30). There are three possibilities of explaining this effect. The first is that the catalytic activity of the single acetohydroxy acid synthase present in C. glutamicum (20) is, after the deletion of ilvA, exclusively available for ketoisovalerate synthesis. The second is that l-isoleucine no longer exerts its inhibitory effect by an allosteric interaction with the acetohydroxy acid synthase (6). The third is that due to the introduced growth limitations, increased precursor metabolite concentrations are available to enter the biosynthesis pathway. This is known from several examples. For instance, a molecularly introduced growth limitation results in an increased l-lysine accumulation by C. glutamicum (8, 32), and a growth limitation obtained by an appropriate process management results in an increased l-phenylalanine accumulation by E. coli (21).
The successful use of ilvBNCD overexpression to obtain an increased d-pantothenate accumulation is due to the increased ketoisovalerate availability. Only then does the panBC overexpression result in a substantial accumulation of d-pantothenate. It therefore follows that an increased ketoisovalerate availability is mandatory to direct the metabolite flux into the d-pantothenate-specific part of the pathway with its low specific activities. Furthermore, the availability of β-alanine is essential, since without its addition no substantial amounts of d-pantothenate accumulate with the strain constructed. By using the appropriate tools and procedures developed in this study the low concentration of 10 μg of d-pantothenate per liter accumulated by the wild type of C. glutamicum was increased to the high concentration of about 1 g of the vitamin per liter. A further improvement of C. glutamicum appears possible to reach concentrations which are in the range of those obtained for the amino acids produced with this organism.
ACKNOWLEDGMENTS
We thank S. Jackowski for the E. coli strains, A. Ondrejková for the use of ilvD from C. glutamicum, K. Krumbach for help during the work, and Degussa AG for the synthesis of enzyme substrates.
REFERENCES
- 1.Baigori M, Grau R, Morbidoni H R, de Mendoza D. Isolation and characterization of Bacillus subtilis mutants blocked in the synthesis of pantothenic acid. J Bacteriol. 1991;173:4240–4242. doi: 10.1128/jb.173.13.4240-4242.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cordes C, Möckel B, Eggeling L, Sahm H. Cloning and functional analysis of ilvA, ilvB and ilvC genes from Corynebacterium glutamicum. Gene. 1992;112:113–116. doi: 10.1016/0378-1119(92)90311-c. [DOI] [PubMed] [Google Scholar]
- 3.Cremer J, Eggeling L, Sahm H. Cloning of the dapA dapB cluster of Corynebacterium glutamicum. Mol Gen Genet. 1990;220:478–480. [Google Scholar]
- 4.Cronan J E., Jr β-Alanine synthesis in Escherichia coli. J Bacteriol. 1980;141:1291–1297. doi: 10.1128/jb.141.3.1291-1297.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cronan J E, Jr, Littel K J, Jackowski S. Genetic and biochemical analyses of pantothenate biosynthesis in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1982;149:916–922. doi: 10.1128/jb.149.3.916-922.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eggeling I, Cordes C, Eggeling L, Sahm H. Regulation of acetohydroxy acid synthase in Corynebacterium glutamicum during fermentation of α-ketobutyrate to l-isoleucine. Appl Microbiol Biotechnol. 1987;25:346–351. [Google Scholar]
- 7.Eggeling L, Morbach S, Sahm H. The fruits of molecular physiology: engineering the l-isoleucine biosynthesis pathway in Corynebacterium glutamicum. J Biotechnol. 1997;56:167–182. [Google Scholar]
- 8.Eggeling L, Oberle S, Sahm H. Improved l-lysine yield with Corynebacterium glutamicum: use of dapA resulting in increased flux combined with growth limitation. Appl Microbiol Biotechnol. 1997;49:24–30. doi: 10.1007/s002530051132. [DOI] [PubMed] [Google Scholar]
- 9.Eikmanns B J, Kleinertz E, Liebl W, Sahm H. A family of Corynebacterium glutamicum/Escherichia coli shuttle vectors for cloning, controlled gene expression, and promoter probing. Gene. 1991;102:93–98. doi: 10.1016/0378-1119(91)90545-m. [DOI] [PubMed] [Google Scholar]
- 10.Eikmanns B J. Identification, sequence analysis, and expression of a Corynebacterium glutamicum gene cluster encoding the three glycolytic enzymes glyceraldehyde-3-phosphate dehydrogenase, 3-phosphoglycerate kinase, and triosephosphate isomerase. J Bacteriol. 1992;174:6076–6086. doi: 10.1128/jb.174.19.6076-6086.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frodyma M E, Downs D. AbpA, the ketopantoate reductase enzyme of Salmonella typhimurium is required for the synthesis of thiamine via the alternative pyrimidine biosynthetic pathway. J Biol Chem. 1998;273:5572–5576. doi: 10.1074/jbc.273.10.5572. [DOI] [PubMed] [Google Scholar]
- 12.Hara S, Takemori Y, Iwata T, Yamaguchi M, Nakamura M. Fluorimetric determination of α-keto acids with 4,5-dimethoxy-1,2-diamino-benzene and its application to high-performance liquid chromatography. Anal Chim Acta. 1985;172:167–173. [Google Scholar]
- 13.Hatakeyama K, Kohama K, Vertès A A, Kobayashi M, Kurusu Y, Yukawa H. Genomic organization of the biotin biosynthetic genes of coryneform bacteria: cloning and sequencing of the bioA-bioD genes from Brevibacterium flavum. DNA Sequence. 1993;4:177–184. doi: 10.3109/10425179309015630. [DOI] [PubMed] [Google Scholar]
- 14.Jackowski S, Rock C O. Regulation of coenzyme A biosynthesis. J Bacteriol. 1981;146:926–932. doi: 10.1128/jb.148.3.926-932.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackowski S. Biosynthesis of pantothenic acid and coenzyme A. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 687–694. [Google Scholar]
- 16.Jäger W, Schäfer A, Pühler A, Labes G, Wohlleben W. Expression of the Bacillus subtilis sacB gene leads to sucrose sensitivity in the gram-positive bacterium Corynebacterium glutamicum but not Streptomyces lividans. J Bacteriol. 1992;174:5462–5465. doi: 10.1128/jb.174.16.5462-5465.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones C E, Brook J M, Buck D, Abell C, Smith A G. Cloning and sequencing of the Escherichia coli panB gene, which encodes ketopantoate hydroxymethyltransferase, and overexpression of the enzyme. J Bacteriol. 1993;175:2125–2130. doi: 10.1128/jb.175.7.2125-2130.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Julliard J H. Purification and characterization of oxopantoyl lactone reductase from higher plants: role in pantothenate biosynthesis. Bot Acta. 1994;107:191–200. [Google Scholar]
- 19.Kataoka M, Shimizu K, Sakamoto K, Yamada H, Shimizu S. Lactonohydrolase-catalyzed optical resolution of pantoyl lactone: selection of a potent enzyme producer and optimization of culture and reaction conditions for practical resolution. Appl Microbiol Biotechnol. 1995;44:333–338. [Google Scholar]
- 20.Keilhauer C, Eggeling L, Sahm H. Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J Bacteriol. 1993;175:5595–5603. doi: 10.1128/jb.175.17.5595-5603.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Konstantinov K B, Nishino N, Seki T, Yoshida T. Physiologically motivated strategies for control of the fed-batch cultivation of recombinant Escherichia coli for phenylalanine production. J Ferment Bioeng. 1991;71:350–355. [Google Scholar]
- 22.Leuchtenberger W. Amino acids—technical production and use. In: Rehm H J, Reed G, editors. Biotechnology. 6. Products of primary metabolism. Weinheim, Germany: VCH Verlagsgesellschaft; 1996. pp. 455–502. [Google Scholar]
- 23.Liebl W, Bayerl A, Schein B, Stillner U, Schleifer K H. High efficiency electroporation of intact Corynebacterium glutamicum cells. FEMS Microbiol Lett. 1989;65:299–394. doi: 10.1016/0378-1097(89)90234-6. [DOI] [PubMed] [Google Scholar]
- 24.Lindroth P, Mopper K. High performance liquid chromatographic determination of subpicomole amounts of amino acids by precolumn fluorescence derivatization with o-phthaldialdehyde. Anal Chem. 1979;51:1167–1174. [Google Scholar]
- 25.Marx A, de Graaf A A, Wiechert W, Eggeling L, Sahm H. Determination of the fluxes in the central metabolism of Corynebacterium glutamicum by NMR spectroscopy combined with metabolite balancing. Biotechnol Bioeng. 1996;49:111–129. doi: 10.1002/(SICI)1097-0290(19960120)49:2<111::AID-BIT1>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 26.Marx A, Striegel K, de Graaf A A, Sahm H, Eggeling L. Response of the central metabolism of Corynebacterium glutamicum to different flux burdens. Biotechnol Bioeng. 1997;56:168–180. doi: 10.1002/(SICI)1097-0290(19971020)56:2<168::AID-BIT6>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 27.Menkel E, Thierbach G, Eggeling L, Sahm H. Influence of increased aspartate availability on lysine formation by a recombinant strain of Corynebacterium glutamicum and utilization of fumarate. Appl Environ Microbiol. 1989;55:684–688. doi: 10.1128/aem.55.3.684-688.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merkel W K, Nichols B. Characterization and sequence of the Escherichia coli panBCD gene cluster. FEMS Microbiol Lett. 1996;143:247–252. doi: 10.1111/j.1574-6968.1996.tb08488.x. [DOI] [PubMed] [Google Scholar]
- 29.Miyatake K, Nakano Y, Kitaoka S. Pantothenate synthetase from Escherichia coli. J Biochem. 1976;79:673–678. doi: 10.1093/oxfordjournals.jbchem.a131112. [DOI] [PubMed] [Google Scholar]
- 30.Möckel B, Eggeling L, Sahm H. Threonine dehydratases of Corynebacterium glutamicum with altered allosteric control: their generation and biochemical and structural analysis. Mol Microbiol. 1994;13:833–842. doi: 10.1111/j.1365-2958.1994.tb00475.x. [DOI] [PubMed] [Google Scholar]
- 30a.Ondrejková, A. Unpublished data.
- 31.Oppenheim D S, Yanofski C. Translational coupling during expression of the tryptophan operon of Escherichia coli. Genetics. 1980;95:785–795. doi: 10.1093/genetics/95.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pátek M, Krumbach K, Eggeling L, Sahm H. Leucine synthesis in Corynebacterium glutamicum: enzyme activities, structure of leuA, and effect of leuA inactivation on lysine synthesis. Appl Environ Microbiol. 1994;60:133–140. doi: 10.1128/aem.60.1.133-140.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Powers S G, Snell E E. Ketopantoate hydroxymethyltransferase. J Biol Chem. 1976;251:3786–3793. [PubMed] [Google Scholar]
- 34.Primerano D A, Burns R O. Metabolic basis for the isoleucine, pantothenate or methionine requirement of ilvG strains of Salmonella typhimurium. J Bacteriol. 1982;150:1202–1211. doi: 10.1128/jb.150.3.1202-1211.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramjee M K, Genschel U, Abell C, Smith A G. Escherichia colil-aspartate-α-decarboxylase: preprotein processing and observation of reaction intermediates by electrospray mass spectrometry. Biochem J. 1997;323:661–669. doi: 10.1042/bj3230661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sahm H, Eggeling L, Eikmanns B J, Krämer R. Metabolic design in amino acid producing bacterium Corynebacterium glutamicum. FEMS Microbiol Rev. 1995;16:243–252. [Google Scholar]
- 37.Santamaria R I, Gil J A, Martin J F. High-frequency transformation of Brevibacterium lactofermentum protoplasts by plasmid DNA. J Bacteriol. 1985;162:463–467. doi: 10.1128/jb.162.1.463-467.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schäfer A, Kalinowski J, Simon R, Seep-Feldhaus A-H, Pühler A. High-frequency conjugal plasmid transfer from gram-negative Escherichia coli to various gram-positive coryneform bacteria. J Bacteriol. 1990;172:1663–1666. doi: 10.1128/jb.172.3.1663-1666.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 40.Schell M A. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 41.Shimizu S, Yamada H. Enzymatic synthesis of chiral intermediates for d-pantothenate synthesis. In: Heinemann R, Wolnak B, editors. Opportunities with industrial enzymes. Chicago, Ill: Bernard and Associates, Inc.; 1992. pp. 227–241. [Google Scholar]
- 42.Shimizu S, Esumi A, Komaki R, Yamada H. Production of coenzyme A by a mutant of Brevibacterium ammoniagenes resistant to oxypantetheine. Appl Environ Microbiol. 1984;48:1118–1122. doi: 10.1128/aem.48.6.1118-1122.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 44.Sorokin A, Azevedo V, Zumstein E, Galleron N, Ehrlich S D, Serror P. Sequence analysis of the Bacillus subtilis chromosome region between the serA and kdg loci cloned in a yeast artificial chromosome. Microbiology. 1996;142:2005–2016. doi: 10.1099/13500872-142-8-2005. [DOI] [PubMed] [Google Scholar]
- 45.Tanaka S, Lerner S A, Lin E C C. Replacement of a phosphoenolpyruvate-dependent phosphotransferase by a nicotinamide adenine dinucleotide-linked dehydrogenase for the utilization of mannitol. J Bacteriol. 1967;93:642–648. doi: 10.1128/jb.93.2.642-648.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teller J H, Powers S G, Snell E E. Ketopantoate hydroxymethyltransferase. Purification and role in pantothenate biosynthesis. J Biol Chem. 1976;251:3780–3785. [PubMed] [Google Scholar]
- 47.Vallari D S, Rock C O. Pantothenate transport in Escherichia coli. J Bacteriol. 1985;162:1156–1161. doi: 10.1128/jb.162.3.1156-1161.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vandamme E J. Production of vitamins, coenzymes and related biochemicals by biotechnological processes. J Chem Technol Biotechnol. 1992;53:313–327. doi: 10.1002/jctb.280530402. [DOI] [PubMed] [Google Scholar]
- 49.Williamson J M, Brown G M. Purification and properties of l-aspartate α-decarboxylase, an enzyme that catalyses the formation of β-alanine in Escherichia coli. J Biol Chem. 1979;254:8074–8082. [PubMed] [Google Scholar]