ABSTRACT
Extreme Antarctic conditions provide one of the closest analogues of extraterrestrial environments. Since air and snow samples, especially from polar regions, yield DNA amounts in the lower picogram range, binning of prokaryotic genomes is challenging and renders studying the dispersal of biological entities across these environments difficult. Here, we hypothesized that dispersal of host-associated bacteriophages (adsorbed, replicating, or prophages) across the Antarctic continent can be tracked via their genetic signatures, aiding our understanding of virus and host dispersal across long distances. Phage genome fragments (PGFs) reconstructed from surface snow metagenomes of three Antarctic stations were assigned to four host genomes, mainly Betaproteobacteria, including Ralstonia spp. We reconstructed the complete genome of a temperate phage with nearly complete alignment to a prophage in the reference genome of Ralstonia pickettii 12D. PGFs from different stations were related to each other at the genus level and matched similar hosts. Metagenomic read mapping and nucleotide polymorphism analysis revealed a wide dispersal of highly identical PGFs, 13 of which were detected in seawater from the Western Antarctic Peninsula at a distance of 5,338 km from the snow sampling stations. Our results suggest that host-associated phages, especially of Ralstonia sp., disperse over long distances despite the harsh conditions of the Antarctic continent. Given that 14 phages associated with two R. pickettii draft genomes isolated from space equipment were identified, we conclude that Ralstonia phages are ideal mobile genetic elements to track dispersal and contamination in ecosystems relevant for astrobiology.
IMPORTANCE Host-associated phages of the bacterium Ralstonia identified in snow samples can be used to track microbial dispersal over thousands of kilometers across the Antarctic continent, which functions as an extraterrestrial analogue because of its harsh environmental conditions. Due to the presence of these bacteria carrying genome-integrated prophages on space-related equipment and the potential for dispersal of host-associated phages demonstrated here, our work has implications for planetary protection, a discipline in astrobiology interested in preventing contamination of celestial bodies with alien biomolecules or forms of life.
KEYWORDS: viruses, extreme environments, astrovirology, metagenomics, cryosphere, Ralstonia
INTRODUCTION
Due to harsh environmental conditions and isolation by the surrounding Southern Ocean’s circumpolar current, Antarctica is considered an analogue for multipurpose space exploration (1, 2). For example, its McMurdo Dry Valleys are regarded as a close terrestrial analogue to Mars (3). Astrobiology model organisms found on Antarctica are highly adapted to stressful conditions and comprise prokaryotes, such as spore-forming bacilli (4, 5), but also microfungi (3, 6). Understanding endurance and dispersal of microorganisms under conditions that mimic those on extraterrestrial planets, i.e., high ultraviolet (UV) radiation, low temperature, and low nutrient availability, has important implications for planetary protection, a discipline in astrobiology with the aim of preventing contamination of celestial bodies with foreign biomolecules or forms of life. For instance, the dispersal of microbes that hitchhike to a celestial body is currently not considered in planetary protection.
Among the potential candidates hitchhiking on spacecraft are Betaproteobacteria of the genus Ralstonia (order Burkholderiales). These bacteria are able to thrive under oligotrophic conditions (7) and were reported to be ubiquitous on space-related equipment, including water systems, of the International Space Station (ISS) (8, 9) and the Mir space station (10, 11). Likewise, they belonged to the microbial inventory of Mars Odyssey and Mars Phoenix lander facilities and can thus prevail under strict planetary protection regulations (12, 13). Ralstonia pickettii strains were found to thrive in simulated microgravity compared to normal gravity (11) and demonstrated high resistance against different metal ions and UV-C radiation (8).
Ralstonia spp. (mainly R. pickettii) were previously found in Antarctic soils (14), in Antarctic snow (15–17), in snow over Tibetan Plateau glaciers (18), and in the air of the Antarctic base Concordia (19). Interestingly, this genus has been regarded as an atmospheric traveler rather than being part of true snow microflora in Antarctica (15). Despite a report on bacterial activity at subzero temperatures in South Pole snow (20), Ralstonia was not among active bacterial communities inferred from cDNA-based 16S rRNA amplicon sequencing (15). This view was supported by a comprehensive Antarctic surface snow microbiome study that did not detect this bacterial genus (21) and by the fact that the spatial variability of snow microbiomes in Antarctica is high (22). Although aerial dispersal is probably the major contributor to (micro)biological input in remote regions (23), the role of bioaerosol transport in the microbial ecology of isolated systems such as the Antarctic continent is poorly understood (24). Applying high-throughput-sequencing approaches to study bacterial dispersal over the Antarctic continent remains a considerable challenge due to the low microbial biomass of atmosphere-derived samples regarding their DNA content (25) and resulting issues associated with recovering high-quality assemblies from metagenomic reads (26).
In addition to the limited knowledge about how transport via aerosols and snow across Antarctica shapes microbial dispersal patterns, another open question relates to the distances that microbes can cover within the atmosphere of extreme environments. A study on the dispersal of airborne fecal coliforms showed a distribution over about just 175 m from a sewage outfall at Rothera Research Station (Antarctic Peninsula), and thus, prolonged survival was considered unlikely (27). More stress-resistant microorganisms could, however, endure for much longer periods. Recently, Malard et al. (22) found high abundances of spore-forming bacilli and suggested that long-term dispersal may seed continental Antarctic snow ecosystems. However, to date, the role of aerial dispersal in shaping patterns of microbial biogeography is supported by little empirical evidence (23).
Here, we followed the hypothesis that geographically widespread (28) Ralstonia sp. prophages and/or replicating and adsorbed bacteriophages of this genus (all types referred to here as host-associated phages) can be used to study host bacterium dispersal across the Antarctic continent. Reconstructing prokaryotic genomes from samples containing small amounts of DNA, including air and precipitation, is challenging, as low-input libraries (~1 pg) can result in problems with genome binning (26). However, (pro)phage genomes and their fragments are much smaller than prokaryotic genomes and thus easier to identify, track, and compare. In this study, genome-resolved metagenomics was applied to demonstrate dispersal of host-associated phage genome fragments (PGFs) from surface snow across the Antarctic continent over hundreds to thousands of kilometers. We detected PGFs belonging to the orders Caudovirales and Tubulavirales in this extraterrestrial analogue and additionally show that similar genome-integrated phages are frequent colonizers of space equipment. Therefore, we suggest that host-associated PGFs represent a useful tool to study spatial dispersal of bacteria and their phages in extreme environmental settings and further envision implications for the dispersal of microbiological contaminations on spacecraft and celestial bodies that previously escaped planetary protection measures.
RESULTS
Reconstruction of low-coverage MAGs from Antarctic snow metagenomic data.
The microbial community composition of surface snow samples collected close to three Russian Antarctic stations, Druzhnaja, Mirnii, and Progress, based on 16S rRNA gene sequencing was described earlier (16) and showed that Ralstonia was the most dominant organism in snow collected at the Mirnii station, the second most dominant (after Janthinobacterium) at Druzhnaja, and the third most dominant (after Flavobacteria and Hydrogenophaga) around Progress station. We reconstructed four prokaryotic and one eukaryotic metagenome-assembled genomes (MAGs) from the three low-biomass snow metagenomes (Fig. 1A). Two MAGs related to Ralstonia sp. and R. pickettii were recovered from Druzhnaja and Mirnii samples and had 86%/10% and 55%/0% completeness/contamination scores, respectively. MAGs of Janthinobacterium lividum with scores of 92%/6% and of Flavobacterium micromati with scores of 96%/0% were recovered from Druzhnaja and Progress, respectively. We also identified a MAG of a diatom (likely Thalassiosira sp.) in the Mirnii metagenome, which we used for normalizing some of our viral analysis (see below) but which was not further characterized in this work. Read mapping revealed coverage scores of 5.9, 7.1, 6.7, and 14.6 for the MAGs of Ralstonia sp., R. pickettii, J. lividum, and F. micromati, respectively.
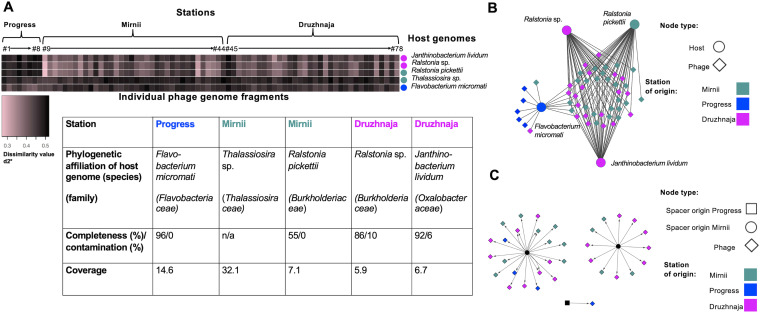
FIG 1.
Host-phage pairings based on shared k-mer frequency patterns. (A) Heat map representing dissimilarity values (d2*) for matches between five host MAGs and 78 PGFs derived from three Antarctic snow metagenomes from the stations Progress, Mirnii, and Druzhnaja. The PGF number corresponds to the number in the phage name; for instance, PGF#78 refers to Antarcphage78_Dr_3477 (Table S1). The table shows MAG characteristics (phylogenetic affiliation, completeness, contamination, and coverage). n/a, not available. (B) A network of phage-host interactions based on k-mer frequency pattern reveals a strong overlap between Ralstonia- and Janthinobacterium lividum-infecting PGFs. Here, only PGFs matching the host MAG below the defined threshold (see the text) are shown. (C) CRISPR spacer matches to PGFs. Different spacers matching the same PGFs are shown by multiple arrows. The two hosts of CRISPR arrays for Mirnii-derived spacers remain unidentified according to their direct repeat sequence, whereas the Progress spacer is derived from a Flavobacterium sp. (Table S2).
Prevalent absence of CRISPR-Cas systems suggest low adaptive immunity.
We searched for clustered regularly interspaced short palindromic repeat (CRISPR) spacers in the snow metagenome reads to link them to potential protospacers on PGFs. CRISPR arrays and cas genes were absent from both Ralstonia and the J. lividum MAGs, as determined by CRISPRcasFinder (29). BLAST searching of direct repeat (DR) sequences against NCBI’s NR database also did not indicate Ralstonia or Janthinobacterium to be the host of the CRISPR array. Only the genome of F. micromati contained two CRISPR arrays with four spacers each (both evidence level 3, i.e., highly likely candidates). CRISPR arrays were not detected in R. pickettii draft genomes SSH4 and CW2, obtained from space equipment.
PGF-host analyses suggest shared hosts of Antarctic phages.
A total of 26 predicted PGFs, eight of them putative PGFs, were found in Antarctic snow metagenomic data (see Table S1 in the supplemental material). VIBRANT (30) additionally detected 52 PGFs, resulting in a total of 78 PGFs (Fig. 1A). PGFs with a minimum of 75% of the genome covered with reads had coverages ranging between 1.9 and 17.5. Most PGFs were partial sequences according to viralComplete (31) and CheckV (32); only Antarcphage10_Mi_4716 and Antarcphage49_Dr_7823_circ were estimated to be of full length (Table S1), although annotations of Antarcphage10_Mi_4716 in comparison to Ralstonia phage p12J revealed missing genes and caused us to question its completeness (Fig. 2). Of 78 PGFs, 77 were categorized as lytic by VirSorter (33) and 75 were found to be of viral or unclassified origin by CheckV (Table S1). Proportions of 43.6%, 46.1%, and 10.3% of PGFs identified originated from Druzhnaja, Mirnii, and Progress, respectively.
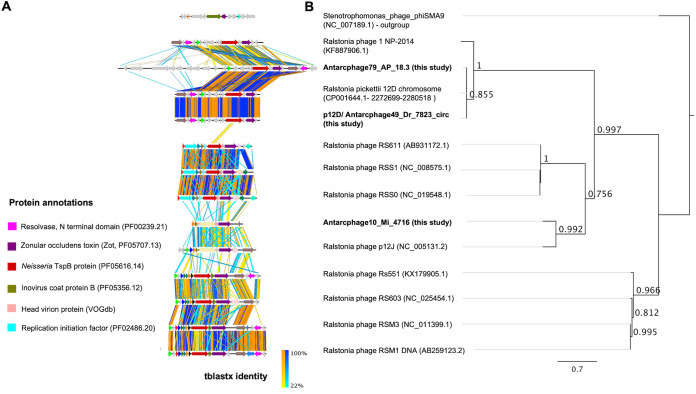
FIG 2.
Phage genome comparisons, functional annotations, and phylogenetic relationship based on zonula occludens toxin (Zot; Pfam ID PF05707). (A) Synteny of known Ralstonia sp. phages from NCBI and Antarctic PGFs from this study. Similar coding sequences (CDS) are in the same color if they occur on more than one phage genome. Functional annotations performed by DRAM-v (115) are given for all colored CDS where available. Vertical lines between sequences indicate regions of shared similarity shaded according to tBLASTx (orange gradient for matches in the same direction or blue gradient for inverted matches). The figure was created using Easyfig (119). (B) Phylogenetic tree built using FastTree 2.1.11 (122) in Geneious 11.1.5 (112) with default settings, showing four distinct clusters of Zot proteins based on their amino acid sequences (aligned with MUSCLE [121]).
All 78 PGFs were analyzed in conjunction with reconstructed MAGs using VirHostMatcher (34). We defined a dissimilarity threshold of a d2* value of 0.436, which corresponds to the lowest dissimilarity value (i.e., highest similarity) for PGFs matching the MAG of Thalassiosira sp. from the snow environment (Fig. 1A) by assuming that the eukaryotic MAG does not match any of our extracted prokaryote-infecting PGFs. In total, 50 of the 78 PGFs matched a host MAG below the defined dissimilarity threshold, based on their shared k-mer patterns. Most PGFs (i.e., 41) matched both MAGs of Ralstonia sp. and R. pickettii (Fig. 1A). In total, 38 and 14 PGFs matched J. lividum and F. micromati MAGs, respectively. Five of eight PGFs extracted from the Progress station metagenomic snow sequences matched the F. micromati MAG recovered from this station. Seven PGF matches were shared among all prokaryotic MAGs (Fig. S1). However, with 30 shared matches, the three MAGs belonging to the order Burkholderiales had the most overlap (Fig. 1B; Fig. S1).
Matching of CRISPR spacers derived from CRISPR loci of unknown hosts (reconstructed from DR sequences) revealed that spacers from Mirnii matched 19, 14, and three PGFs from Druzhnaja, Mirnii, and Progress, respectively, and one Flavobacterium sp. spacer from Progress matched a PGF from that station (Fig. 1C; Table S2) (≥80% similarity). Of these 37 spacer matches, three matched protospacers of PGFs of unknown hosts, and 26 were assigned to a Ralstonia host according to VirHostMatcher or other predictions (Table S1). Since p12D (see below) was among the PGF spacer targets and is a certain Ralstonia phage, this could indicate that an unknown host belonging to the order Burkholderiales uses adaptive immunity against viruses. We compiled evidence of host prediction for the 78 PGFs in Table S1.
The temperate phage p12D forms a distinct, monophyletic clade excluding most known Ralstonia phages.
Among the 78 PGFs, we detected a circular (and thus complete) 7.8-kb PGF termed Antarcphage49_Dr_7823_circ (Fig. 2A; Fig. S2). Since Antarcphage49_Dr_7823_circ PGF from the Druzhnaja station was found in the chromosome of a R. pickettii 12D strain isolated from copper-contaminated sediment from a lake in Michigan with 99.9% identity (Fig. 2A), we propose the name p12D, equivalent to Ralstonia p12J, a known phage infecting the R. pickettii 12J strain. Antarcphage49_Dr_7823_circ (referred to below as p12D) matched only the MAG of Ralstonia sp. and R. pickettii based on shared k-mer frequency patterns. Functional protein annotations provided evidence that p12D contained a gene for a resolvase domain containing protein/site-specific recombinase, likely used for integration into the host genome, as well as for the zonula occludens toxin (Zot; accession number PF05707) (Fig. 2A; Fig. S2; Table S3). The nontoxic component of Zot at the N-terminus represents a characteristic protein in filamentous phages that has been used for phage classification (35, 36). Reconstruction of phylogenetic relationships of Zot proteins from this study and respective references (Fig. 2B; Fig. S3) confirmed a close identity of p12D to Ralstonia phage 1 NP-2014 and Antarcphage79_WAP_18.3, all belonging to the same monophyletic clade. Zot of Antarcphage10_Mi_4716 was phylogenetically related to filamentous Ralstonia phage p12J. The tree shows four distinctive clusters for the Zot protein, reflecting the overall synteny of the gene order of the different Ralstonia phages very well.
Annotations against UniRef100 revealed that 68.8% of all annotated Antarctic PGF proteins remain hypothetical. The annotations taxonomically assigned 34.4% of all Antarctic PGF proteins to either Ralstonia or R. pickettii and 11.6% to phages of Janthinobacterium or J. lividum, potentially indicating lateral gene transfer between virus and host in their respective evolution (37, 38) or supporting the idea that these PGFs indeed represent prophages. Three of the Antarctic PGFs carried a site-specific integrase or resolvase domain, and 4.2% of genes were related to phage structural proteins, e.g., head, tail, or capsid proteins (Tables S4 and S5).
Cross-mapping and nucleotide variations reveal dispersal patterns of MAGs and PGFs across Antarctica.
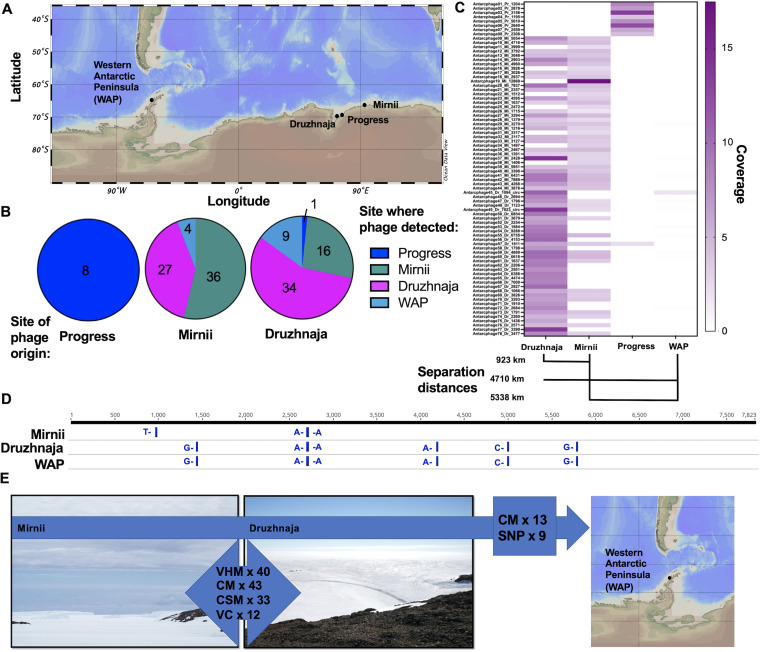
Mapping of 100% identical reads from the three snow samples and the Western Antarctic Peninsula (WAP) (Fig. 3A) seawater sample to the prokaryotic MAGs revealed that both Ralstonia MAGs were detected at all sites (89 to 100% genome coverage). J. lividum was considered absent from the Progress station (genome coverage of 62%) and detected at other stations (minimum of 95% covered genome). F. micromati was detected only in samples from Progress (100%) and Druzhnaja (96%) and was considered absent from Mirnii (58.7%) and the WAP (1.8%). Cross-mapping on the 78 PGFs demonstrated that PGFs derived from the Progress station could not be found in the other two snow metagenomes (Fig. 3B and C). In contrast, 16 different PGFs from Druzhnaja were detected at Mirnii, and 27 Mirnii PGFs were found at Druzhnaja (Fig. 3C). In addition, 4 and 9 PGFs from Mirnii and Druzhnaja, respectively, were detected in the WAP data set (Fig. 3B and C), all of them having a minimum of 97% of their lengths covered with reads from the snow metagenomes (Table S1). Of 43 PGFs that were found at both stations (Mirnii and Druzhnaja) based on read mapping, 26 PGFs shared a viral cluster (VC), and 34 matched prokaryotic hosts based on k-mer frequencies between stations (Table S1). Interestingly, nine of the 13 PGFs occurring in WAP and Mirnii/Druzhnaja samples contained identical single nucleotide polymorphisms (SNPs) (Fig. 3D and E; Table S6) or were missing common SNPs, pointing to a common phage population before dispersal led to separation.
FIG 3.
Evidence for dispersal of PGFs across the Antarctic continent. (A) Map showing the different snow sampling stations in the East and the sampling station for seawater at the Western Antarctic Peninsula. The map was built using Ocean Data View (123). (B) Pie charts summarizing the number of PGFs considered present at each station based on cross mapping of reads and the site of PGF assembly (phage origin). (C) Heat map depicting the normalized coverages based on cross-mapping of reads against the 78 PGFs and separation distances between stations. White areas indicate PGFs that were not called present because they had less than 75% of scaffold length covered in mapping (with least 1-fold coverage). (D) Single-nucleotide polymorphism (SNP) analysis of p12D/Antarcphage49_Dr_7823_circ based on reads from Druzhnaja, Mirnii, and the WAP. This PGF was absent from the metagenome of the Progress station. Further SNP analysis data can be found in Table S6. (E) Summary of the major dispersal route and supporting evidence. The majority of PGFs disperse between Mirnii and Druzhnaja, and 13 PGFs additionally occurred at the WAP based on cross-mapping (CM) and SNP analysis. Values refer to the numbers of tested features, which include number of virus clusters (VC), number of shared viruses based on CM, virus-host matches based on k-mer links (VHM), and CRISPR-spacer matches (CSM). A VHM of 1 indicates that one PGF has infected host MAGs from two stations. No VC and only one CM were observed for Progress-Druzhnaja and Progress-Mirnii. Therefore, this station was excluded from the summary.
Clustering of PGFs in VICTOR (39) and vConTACT2 (40) revealed that groups of two to four PGFs could be assigned to 12 distinct viral genus-forming viral clusters (Fig. 3E; Fig. S4 and S5; Table S1). Members of the same cluster were often recovered from different stations, for example, Druzhnaja and Mirnii (Fig. 3B), which are located 923 km apart (Fig. 3C). The phylogenomic Genome-BLAST distance phylogeny (GBDP) tree yielded average support of 47% and 80% in the nucleic acid- and amino acid-based analysis, respectively. OPTSIL clustering yielded 78 and 64 species and genus clusters at the nucleic acid level and 78 and 70 at the amino acid level, respectively.
Many PGFs formed VCs with no genomic relatedness to any known phages from the ProkaryoticViralRefSeq v94 database. Others were, however, similar to known Ralstonia PGFs (current family Inoviridae, order Tubulavirales) and hence clustered with those, such as p12D with Ralstonia phage 1 NP-2014, and Antarcphage10_Mi_4716 with Ralstonia phage PE226 and Ralstonia phage p12J (Fig. 4; Table S1). Other Antarctic PGFs shared protein clusters with Ralstonia phages from space equipment based on vConTACT2 or were related to phages of the order Caudovirales from the RefSeq database (Fig. 4).
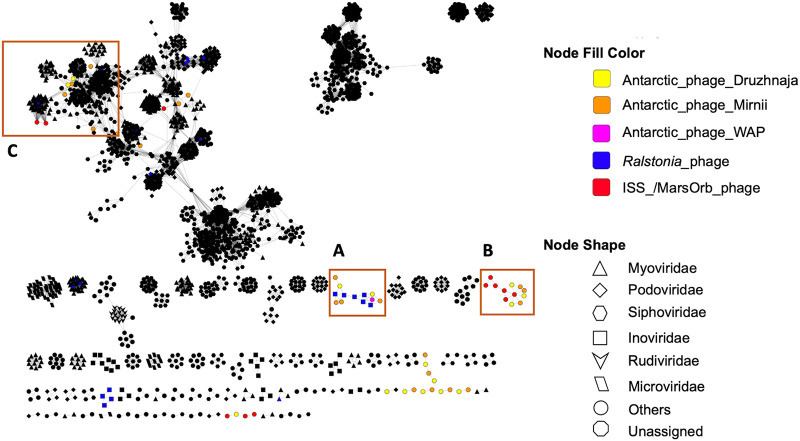
FIG 4.
Phage network of Antarctic PGFs derived from Druzhnaja, Mirnii, WAP, and space equipment (ISS/MarsOrb) clustered with viruses of the viral RefSeq database. Based on shared protein clusters, Antarctic PGFs from this study group with known Ralstonia phages of the family Inoviridae (A), with phages obtained from Ralstonia isolates from space equipment (B), or with known phages of the order Caudovirales (C). Black nodes refer prokaryotic viruses other than Ralstonia and PGFs from this study. Interactions show relatedness of genomes on viral genus or higher taxonomic level. “Others” refer to other known viral families not listed in the legend. Visualization was done using Cytoscape 3.8.2. (109). For details about viral clusters, see Table S1.
In summary, our results support the idea of dispersal of host-associated phages between widely separated stations because (i) cross-mapping revealed the presence of Mirnii PGFs at Druzhnaja and vice versa, the presence of a temperate phage from snow in the WAP seawater data set, and the presence of the hosts Ralstonia and J. lividum in metagenomes of most stations; (ii) PGFs in widely separated locations carry identical SNPs or lack nucleotide variations; (iii) some PGFs from Mirnii and Druzhnaja belong to the same genus/VC; and (iv) Mirnii and Druzhnaja PGFs share host MAGs according to their k-mer frequency patterns. These observations were mainly true for PGFs affiliated with Ralstonia and J. lividum MAGs and less so for F. micromati PGFs, which could indicate that certain bacterial species or their phages in Antarctic snow are more prone to atmospheric dispersal than others. Finally, we found CRISPR spacers from two unidentified hosts (represented by two distinct CRISPR DR sequences) at the Mirnii station to match PGFs from Druzhnaja, Mirnii, and Progress. Mapping revealed little or no abundance of Progress PGFs at other stations, and no F. micromati MAG was detected in Mirnii and WAP samples. This could be related to the low biomass and/or or insufficient sequencing coverage. However, spacers from Mirnii suggest a history of infection with PGF from Progress, thus reflecting (past) dispersal of host and/or phage.
Ralstonia phages occur across diverse ecosystems and on space equipment.
By BLAST searching all recovered PGFs against the IMG/VR viral database (41), we found that p12D shared high identity (85.9%) with a R. pickettii prophage (IMG/VR v.2 scaffold ID Ga0075447_10000781; genome ID 3300006191) from a seawater metagenome of the WAP (Fig. 2), which was further validated by cross-mapping of WAP reads to p12D delivering 100% scaffold coverage with 90% identical reads. The WAP is 4,710 km away from the Druzhnaja station (Fig. 3C). The PGF Antarcphage79_WAP_18.3 was obtained by BLAST searching the p12D scaffold against the WAP assembly but was assembled containing genomic regions extending the actual PGF. Based on vConTACT2, it was affiliated with the same viral genus as p12D. Other PGFs recovered from the Antarctic snow metagenome showed hits to phages deposited at IMG/VR. Hits were related to phages originating from freshwater, wastewater, groundwater, or phages that were associated with plant root microbial communities, which is in accordance with the fact that Ralstonia sp. is a frequent phytopathogen (42). Many of the matching entries in IMG/VR were identified as phages of Ralstonia due to matching with CRISPR spacers (mostly of Ralstonia solanacearum) (Table S7).
Altogether, 14 PGFs were found on space equipment, here referred to as ISSphage (number of phages [n] = 7) and MarsOrbphage (n = 7), for PGFs extracted from the R. pickettii strains CW2 and SSH4 draft genomes from the ISS cooling system and Mars Odyssey Orbiter, respectively. Seven of these PGFs were identified as lysogenic and according to protein annotations (Tables S4 and S5) rely on proteins of the integrase family for integration into the host chromosome. vConTACT2 revealed that ISSphage4 formed a common genus cluster with four Antarctic PGFs, MarsOrbphage5 shared protein clusters with ISSphage3, and MarsOrbphage7 formed a genus cluster with several Burkholderia phages, among other Caudovirales members (Fig. 4; Table S1).
DISCUSSION
CRISPR-Cas systems equip bacteria and archaea with a powerful defense system against invading mobile genetic elements, including plasmids, phages, and viruses. Only ca. 50% of bacteria rely on this adaptive immune system, and only 31% of genomes from public databases belonging to plant-pathogenic Ralstonia solanacearum contain CRISPR-Cas arrays (43). Upon exposure to a virulent phage under laboratory conditions, the CRISPR array of R. solanacearum strain CFBP2957 did not acquire new spacers from viral protospacers (43). Gonçalves et al. (28) reported that in the presence of CRISPR arrays, 27.9% of CRISPR spacers from Ralstonia genomes targeted prophage elements. In our study, two Ralstonia MAGs from a low-temperature environment were devoid of CRISPR systems, implying that representatives of this genus have alternative strategies for defense against mobile genetic elements (43). However, analyses of more Ralstonia genomes are necessary to corroborate this statement.
Based on shared protein clusters, some Antarctic PGFs, such as Antarcphage10_Mi_4716, Antarcphage48_Dr_1123, Antarcphage17_Mi_3026, and p12D, clustered with known Ralstonia phages, including Ralstonia phage p12J (NC_005131.2), Ralstonia phage PE226 (NC_015297.1), and Ralstonia phage 1 NP-2014 (NC_023586.1). These are filamentous phages of the Inoviridae, a family for which reclassification to higher taxonomic ranks has been recently called for (36, 44). Inoviruses (order Tubulavirales) typically feature circular, single-stranded DNA genomes of ~5 to 15 kb, lead to chronic infections, and are globally abundant (36). As our protocol should have revealed only double-stranded DNA, detection of single-stranded DNA (ssDNA) inovirus sequences shows that these must be either replicating phages or genome-integrated prophages. Zot, which was detected on p12D, Antarcphage10_Mi_4716, and Antarcphage79_WAP_18.3, is a typical gene in filamentous Ralstonia phages (45, 46). This phage type places little burden on its host or can even serve it, e.g., by increasing its host’s virulence and evolutionary fitness and because virions can leave the host in a nondestructive way (35, 36). Ralstonia phages detected in Arctic viromes were shown to transduce genomic information of cold shock proteins to their hosts (47), a clear asset for microorganisms in polar environments. However, we did not find transduction of beneficial genes, which seems to be common in extreme environments (48).
Some Ralstonia phages occur as nonintegrative, episomal forms, e.g., RS603, a hybrid of RSM1/3 infecting the phytopathogen R. solanacearum (49), whose genome lacks a resolvase domain (Fig. 2A), but many mesophilic Ralstonia also occur as lysogens (50). Lysogeny, a lifestyle during which the phage genome becomes integrated into the host chromosome, is a widespread phenomenon in low-temperature environments (51–53) and likely attributed to prolonged starvation and low activity of host cells under harsh conditions, the latter being previously reported for Ralstonia (15). Since p12D and its counterpart from the WAP have a resolvase domain-containing protein likely functioning in integration/excision during lysogenization (54, 55), we conclude that they must be temperate phages of Ralstonia.
Many (i.e., 75) of the 78 Antarctic PGFs found in this study shared few protein clusters with known phages from public databases. This is certainly related to the often high diversity of viruses, limited accessibility to Antarctic environments as well as a stronger focus on sequencing metaviromes and viral isolates of direct human interest (56). The missing relatedness is known for ssDNA viruses originating from Antarctic cryoconite holes and was attributed to the isolation and extreme environmental conditions on the Antarctic continent (57). In total, 45 of the 78 PGFs could be assigned to a host, which in 71% of cases was Ralstonia (summarized in Table S1, column “host prediction”).
Aeolian transport of viruses over polar environments, especially attached to snowflakes, has barely been investigated to date. Former investigations, mainly conducted at low latitudes, demonstrated intercontinental transport of microbes by winds (58) and showed that highly identical phages can be found in distantly related areas and in various ecosystems around the globe (59–62). Bellas et al. (63) recently reported the presence of nearly identical phage genomes being spread up to 4,000 km in cryoconite holes of Svalbard, Greenland, and the Alps. Our data show that despite the isolation of the Antarctic continent, and under no consideration of anthropogenic dispersal (64), bacteria and phage distribution via snow over extensive distances across Antarctica is possible. Dispersal attributable to human activity seems unlikely for our samples, due to the relatedness of Antarctic phages to phages from environmental sources according to database hits. We cannot be certain about the nature of the PGFs (prophage or lytic phage) by metagenomic predictions alone. While the Inoviridae fraction likely occurs as prophages or episomal forms, detection of PGFs assigned to lytic categories by VIBRANT and VirSorter (categories 1 to 3) suggests that host-associated, lytic phages captured at the adsorption and infection stages were present as well. The degree of uncertainty about the category of a virus presumably results from many fragmented scaffolds with relatively low coverages, which were, however, sufficient to identify shared elements of viruses between stations. Conclusions about the presence/absence of hallmark genes should be drawn from more complete data sets based on greater sequencing depth of samples containing more biomass. We further recommend experiments that involve cultivation attempts and sequencing of metaviromes (free phages) to reveal the extent of lysogenic or chronic compared to lytic infection styles in Antarctic snow.
Snow PGFs carried SNPs identical to those from the seawater metagenome from the WAP, located 4,710 km and 5,338 km from Druzhnaja and Mirnii, respectively (Fig. 3C), implying long-distance transport. This is further supported by the detection of their hosts, Ralstonia and J. lividum, in the WAP. From seawater, a transmission route via sea spray aerosols to snow that is blown over ice surfaces (65) can be assumed. Aerosolization of bacteria from the sea surface is highly taxon-specific but seems to work well for R. pickettii (66) and also viruses (67). Alternatively, transport via aerosols to clouds and precipitating snow is possible. In the latter scenario, bioaerosols, including microbial cells, would act as ice nucleation particles (68), transferring microbes to ice clouds, where they might induce their own precipitation (69, 70). The abundance data support the isolation of Progress-derived PGFs and their potential host F. micromati and point toward a decreasing gradient of abundances from Druzhnaja to Mirnii to the WAP for many PGFs (Table S1). Thus, general dispersal patterns of microbes across Antarctica seem governed by westward drift and are probably mediated by the prevailing Southern Hemisphere westerly winds (71). On short spatial-temporal scales, dispersal seems more complex and is probably shaped by multifactorial dependencies, such as the different potential of a species to become airborne (66), meteorological conditions, or the local geography. For instance, while Druzhnaja is located ~50 km into the continent, the stations Mirnii and Progress are near the coast and more exposed to the sea in summer when ice breaks occur.
We assume that the transferability of the use of PGFs to study microbial dispersal in space analogues such as the Antarctic continent is likely applicable to other celestial bodies like Mars. Three celestial bodies in our solar system (Mars, Europa, and Enceladus) have environmental conditions that could favor microbial life (reviewed by Netea et al. [72]). Most microbial isolates (85 to 95%) obtained from spacecraft and assembly facilities are associated with humans (73). Since Ralstonia spp. can be human and plant pathogens (42, 74) and are able to thrive under harsh and oligotrophic conditions (7), they might contribute to the transmission of viruses to extraterrestrial environments, particularly via manned missions.
We found evidence for viral signatures associated with two R. pickettii strain draft genomes previously obtained from space equipment of a spacecraft assembly clean room and from water systems of the ISS. This result in conjunction with the result that temperate Ralstonia phages (and other viruses) can undergo long-range dispersal in association with their hosts across the extraterrestrial analogue Antarctica suggest that contaminations of space equipment with particularly persistent microbes such as Ralstonia should receive more focus during microbiological monitoring in the framework of planetary protection. However, the field of astrovirology has so far generally garnered little attention (75). The contribution of lysogenic and episomal phage (“hidden hitchhikers”) to overall viral loads on spacecraft and associated equipment has been overlooked despite early work reporting on alterations in prophage induction during spaceflight (76–78), the ability of tobacco mosaic virus to survive space flight-equivalent proton irradiation (79), the occurrence of phages and human-related circoviruses in clean rooms (80), and the presence of inoviruses on the ISS (81, 82).
Planetary protection aims to prevent the spread of biological contaminants (forward contamination) to space shuttles and stations as well as extraterrestrial environments of the solar system. However, this policy largely ignores the potential of escaped biological contaminants to heavily disperse on foreign celestial bodies once released, for instance after crash landings, as happened with the Schiaparelli module of the ExoMars program in 2016 (83). Our results not only show that host-associated PGFs are suitable indicators for tracking long-distance dispersal in space analogues but also demonstrate that the release of contaminants that previously escaped planetary protection measures could spread far across extraterrestrial ecosystems and, in the worst-case scenario, confound future life detection missions.
MATERIALS AND METHODS
Metagenomic and genomic data processing.
We analyzed publicly available metagenomic data sets, which correspond to Antarctic surface snow collected around three Russian stations (Druzhnaja, Mirnii, and Progress) and are deposited at NCBI’s Sequence Read Archive (SRA) as BioProject accession number PRJNA674475 (Table S8) and MG-RAST under project accession number mgp13052, including taxon abundance data. Snow sampling was conducted in December 2008 and 2009 as described previously (15); in brief, a sterile plastic scoop was used to sample ~10 kg corresponding to a 2- to 3-cm layer of surface snow across several 1-m2 areas. Snow was melted over a period of 12 h and concentrated using Pellicon tangential flow filters (Millipore, Burlington, MA, USA) to a final volume of ~10 mL. DNA extraction for these metagenomes was carried out with the DNA blood and tissue kit (Qiagen, Hilden, Germany), whose protocol results in removal of free viruses and resulted in DNA amounts of 170 to 490 ng. Sequencing libraries were prepared with a MiSeq reagent kit v.2 (Illumina, USA), targeting mainly double-stranded DNA (dsDNA) viruses. Metagenomic data of a seawater sample from the WAP was also obtained from SRA (accession number SRR5591034). Raw shotgun sequencing reads of the three snow metagenomes and the WAP data set were quality-trimmed using BBDuk (https://github.com/BioInfoTools/BBMap/blob/master/sh/bbduk.sh) from the BBTools package (84) and Sickle (85), resulting in read counts between 1.31 and 2.65 million for the three snow samples. Assembly of reads was done using MetaSPAdes version 3.13 (86), and scaffolds of <1 kbp were removed. Draft genome sequences of R. pickettii strains SSH4 and CW2 isolated from space equipment were obtained from GenBank, accession numbers JFZG00000000 and JFZH00000000. Strains SSH4 and CW2 were isolated preflight from the surface of the Mars Odyssey Orbiter during assembly and from a water sample taken in flight from the ISS cooling system, respectively (9).
Reconstruction of microbial genomes from metagenomes and CRISPR prediction.
Binning of MAGs from the three stations Druzhnaja, Mirnii, and Progress was performed using emergent self-organizing maps (ESOMs) (87), ABAWACA (https://github.com/CK7/abawaca), and MaxBin 2.0 (88). Aggregation of bins was performed using DASTool (89), and curation of bins was done in uBin (90), also delivering contamination and completeness scores (91). Read coverage of recovered MAGs was obtained from read mapping using Bowtie2 (92) in sensitive mode to the individual bins, followed by mismatch filtering (2% mismatch allowance, depending on read length). To investigate the dispersal of MAGs, mismatch criteria were set to 0% mismatches (100% similarity) and calcopo.rb (https://github.com/ProbstLab/viromics/tree/master/calcopo) was used to calculate the coverage per nucleotide (breadth) and the percentage of positions in the genome covered by reads. Only genomes with a least 70% breadth were considered present in the respective metagenome.
CRISPR arrays, which represent prokaryotic adaptive immunity, were searched in host MAGs using CRISPRcasFinder and considering evidence level 3 or 4 (29). CRISPR loci consist of repeats interspaced with short DNA sequences (spacers) obtained from invading mobile genetic elements such as phages and thus provide a record of past infections. Since CRISPR arrays might get lost during the binning processes, e.g., due to fragmentation in assembly of strain variants, absence of CRISPR arrays in CRISPRcasFinder was further investigated by reconstructing CRISPR systems from raw reads using Crass (93) and BLAST searching the obtained direct repeat (DR) sequences, which are phylogenetically well conserved (94), against the NCBI nonredundant database (release 1 March 2021) using the BLASTn –short algorithm (95) with subsequent filtering at 80% similarity and an E value threshold of 10e−05. DR sequences were BLAST searched against the MAGs and used to extract CRISPR spacers from reads using MetaCRAST with the following settings: -d 3 -l 60 -c 0.99 -a 0.99 -r (96). Spacers were matched against PGFs as mentioned above for DRs. In most cases, BLAST searching the DR sequences against the NR database did not reveal the host’s identity. Nevertheless, spacers derived from unknown hosts were considered, as their matches show that targeted PGFs represent true mobile genetic elements, and matches can be used to infer infection patterns between stations.
PGF detection, host allocation, and viral clustering.
PGFs were identified from metagenome assemblies using a combination of bioinformatic tools, namely, VirSorter v1 (33), VirFinder (97), CircMG (98) (renamed VRCA) (https://github.com/alexcritschristoph/VRCA), VOGdb (version VOG93) (99), and Endmatcher (https://github.com/ProbstLab/viromics/tree/master/Endmatcher). The classification of predicted PGFs as “putative viruses” and “viruses” was done as described in supplementary Figure 3 of reference 100. VIBRANT v.1.2.1 (30) with default settings was used to find additional PGFs. No length cutoff was set (101), since many known Ralstonia phages and inoviruses have genome sizes of <10 kb (36, 50) and since, because of the low biomass of Antarctic snow samples, few PGFs compared to other ecosystems were expected. VirSorter and VIBRANT aided in (pro)phage detection in draft genomes of R. pickettii from space equipment.
CheckV (32) was used to determine the type of identified PGF and completeness, and viralComplete (31) was applied to predict closely related phages. PGFs associated with MAGs were identified by grepping the scaffold ID on the bins. Pairwise comparisons of Antarctic PGFs and clustering were done using nucleic acid- and amino acid-based VICTOR (39). The resulting intergenomic distances were used to infer a balanced minimum evolution tree with branch support via FASTME, including subtree pruning and regrafting (SPR) postprocessing (102) using the distance formula D0. Branch support was inferred from 100 pseudo-bootstrap replicates each. Trees were rooted at the midpoint (103) and visualized with FigTree v1.4.4 (104). Taxon boundaries at the species and genus levels were estimated with the OPTSIL program (105), using the recommended clustering thresholds (39) and an F value (fraction of links required for cluster fusion) of 0.5 (106).
Clustering of PGFs from Antarctic snow and space equipment was further substantiated via vConTACT2 v.0.9.19 (40, 107) in combination with the ProkaryoticViralRefSeq database (v94) (108) followed by visualization of viral clusters (VCs) in Cytoscape v. 3.8.2 (109). Virus-host matches were determined using the tool VirHostMatcher (34) with oligonucleotide frequency dissimilarity measures (d2*) for a k-mer length of 6. Viral genus clusters were determined using VIRIDIC (110). Venn diagrams were calculated using the VIB-ugent web tool (http://bioinformatics.psb.ugent.be/webtools/Venn/).
Mapping of reads to assembled PGFs and nucleotide polymorphism analysis.
We assumed that the presence of a PGF at two locations confirmed by read mapping represents dispersal. To determine if an assembled PGF from a single sample occurred in other metagenomes, even if not assembled, we performed read mapping following previously published guidelines (101): reads should map to a PGF with at least 90% identity (Bowtie2 settings as described in reference 111), and more than 75% of the scaffold should have a coverage of at least 1×. To detect the breadth of a PGF, we again used calcopo.rb (see above). Mean coverage of PGFs was calculated using calc_coverage_v3 (https://github.com/ProbstLab/uBin-helperscripts/blob/master/bin/04_01calc_coverage_v3.rb) and normalized to sequencing depth. Analysis of nucleotide polymorphisms was conducted for the 13 PGFs that underwent long-range dispersal, i.e., PGFs present in the WAP sample and at least one of the snow metagenomes based on read mapping (Table S1). Variant analysis was performed in Geneious 11.1.5 (112) by applying default settings to the read mappings generated as explained above.
Gene prediction and annotations.
Open reading frames in PGFs were detected using Prodigal in meta mode (113). Functional and taxonomic annotations of predicted proteins of PGFs were performed by DIAMOND searches with a E value of 10e−06 (114) against FunTaxDB (90) and by using DRAM-v (115). For the full-length genome of the PGF p12D, annotations were improved using HHpred against PDB, Pfam, UniProt-Swiss-Prot-viral and NCBI_Conserved Domains (116, 117) (https://toolkit.tuebingen.mpg.de/tools/hhpred) with a probability threshold of 70%. Sequences of PGFs were BLAST searched against IMG/VR 2.0 (118) with an E value cutoff of 1e-05 to find related phages from other metagenomic data sets.
Synteny of Ralstonia phage and phylogenetic comparison of the zot gene.
Synteny of known Ralstonia sp. phages from NCBI and all Antarctic PGFs that were identified herein and carried the zonula occludens toxin (zot; Pfam ID PF05707) was performed with tBLASTx comparisons using Easyfig v.2.2.5 (119) on .gbk files generated by Prokka (120). A phylogenetic tree for the MUSCLE-aligned (121) amino acid and nucleic acid sequences of zot was constructed using the FastTree (122) algorithm in Geneious 11.1.5 (112).
Data availability.
The sequence for the R. pickettii 12D strain isolated from copper-contaminated sediment from a lake in Michigan, in which the Antarcphage49_Dr_7823_circ PGF from the Druzhnaja station was found, is available from the corresponding author (Janina.rahlff@uol.de) on request.
ACKNOWLEDGMENTS
We acknowledge Ken Dreger for server administration and maintenance as well as Cristina Moraru for sharing insights on virus taxonomy. We thank the editor and three anonymous reviewers for their constructive comments that helped to improve the manuscript.
J.R. designed the study, wrote the manuscript, and carried out the analyses with input from T.L.V.B.; A.L. and K.S. generated the raw data and performed the sampling; A.J.P. conceptualized the project, provided supervision and resources, and was involved in data interpretation; all authors edited drafts of the manuscript.
For all authors, no competing financial interests exist.
We acknowledge funding from the German Aerospace Center (DLR) for the project DISPERS (50WB1922). J.R. was partially supported by the German Science Foundation for the project VIBOCAT (grant number DFG RA3432/1-1). A.J.P. and T.L.V.B. were supported by the Ministerium für Kultur und Wissenschaft des Landes Nordrhein-Westfalen (Nachwuchsgruppe Alexander Probst). K.S. lab was supported by the Ministry of Science and Higher Education of Russian Federation (agreement No. 075-10-2021-114).
Footnotes
Supplemental material is available online only.
Contributor Information
Janina Rahlff, Email: Janina.rahlff@uol.de.
Nicole R. Buan, University of Nebraska-Lincoln
REFERENCES
- 1.Pyne SJ. 2007. The extraterrestrial Earth: Antarctica as analogue for space exploration. Space Policy 23:147–149. 10.1016/j.spacepol.2007.06.006. [DOI] [Google Scholar]
- 2.Lugg D, Shepanek M. 1999. Space analogue studies in Antarctica. Acta Astronaut 44:693–699. 10.1016/s0094-5765(99)00068-5. [DOI] [PubMed] [Google Scholar]
- 3.Onofri S, Selbmann L, Zucconi L, Pagano S. 2004. Antarctic microfungi as models for exobiology. Planet Space Sci 52:229–237. 10.1016/j.pss.2003.08.019. [DOI] [Google Scholar]
- 4.Puskeppeleit M, Quintern LE, El Naggar S, Schott JU, Eschweiler U, Horneck G, Bucker H. 1992. Long-term dosimetry of solar UV radiation in Antarctica with spores of Bacillus subtilis. Appl Environ Microbiol 58:2355–2359. 10.1128/aem.58.8.2355-2359.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicholson WL, Munakata N, Horneck G, Melosh HJ, Setlow P. 2000. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol Mol Biol Rev 64:548–572. 10.1128/MMBR.64.3.548-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Onofri S, Barreca D, Selbmann L, Isola D, Rabbow E, Horneck G, De Vera J, Hatton J, Zucconi L. 2008. Resistance of Antarctic black fungi and cryptoendolithic communities to simulated space and Martian conditions. Stud Mycol 61:99–109. 10.3114/sim.2008.61.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McAlister MB, Kulakov LA, O'Hanlon JF, Larkin MJ, Ogden KL. 2002. Survival and nutritional requirements of three bacteria isolated from ultrapure water. J Ind Microbiol Biotechnol 29:75–82. 10.1038/sj.jim.7000273. [DOI] [PubMed] [Google Scholar]
- 8.Mijnendonckx K, Provoost A, Ott CM, Venkateswaran K, Mahillon J, Leys N, Van Houdt R. 2013. Characterization of the survival ability of Cupriavidus metallidurans and Ralstonia pickettii from space-related environments. Microb Ecol 65:347–360. 10.1007/s00248-012-0139-2. [DOI] [PubMed] [Google Scholar]
- 9.Monsieurs P, Mijnendonckx K, Provoost A, Venkateswaran K, Ott CM, Leys N, Van Houdt R. 2014. Draft genome sequences of Ralstonia pickettii strains SSH4 and CW2, isolated from space equipment. Genome Announc 2:e00887-14. 10.1128/genomeA.00887-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ott CM, Bruce RJ, Pierson DL. 2004. Microbial characterization of free floating condensate aboard the Mir space station. Microb Ecol 47:133–136. 10.1007/s00248-003-1038-3. [DOI] [PubMed] [Google Scholar]
- 11.Baker PW, Leff L. 2004. The effect of simulated microgravity on bacteria from the Mir space station. Microgravity Sci Technol 15:35–41. 10.1007/BF02870950. [DOI] [PubMed] [Google Scholar]
- 12.Vaishampayan P, Osman S, Andersen G, Venkateswaran K. 2010. High-density 16S microarray and clone library-based microbial community composition of the Phoenix spacecraft assembly clean room. Astrobiology 10:499–508. 10.1089/ast.2009.0443. [DOI] [PubMed] [Google Scholar]
- 13.La Duc MT, Nicholson W, Kern R, Venkateswaran K. 2003. Microbial characterization of the Mars Odyssey spacecraft and its encapsulation facility. Environ Microbiol 5:977–985. 10.1046/j.1462-2920.2003.00496.x. [DOI] [PubMed] [Google Scholar]
- 14.Lysak V, Maksimova IA, Nikitin DA, Ivanova AE, Kudinova AG, Soina VS, Marfenina OE. 2018. Soil microbial communities of Eastern Antarctica. Moscow Univ Biolsci Bull 73:104–112. 10.3103/S0096392518030124. [DOI] [Google Scholar]
- 15.Lopatina A, Krylenkov V, Severinov K. 2013. Activity and bacterial diversity of snow around Russian Antarctic stations. Res Microbiol 164:949–958. 10.1016/j.resmic.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 16.Lopatina A, Medvedeva S, Shmakov S, Logacheva MD, Krylenkov V, Severinov K. 2016. Metagenomic analysis of bacterial communities of Antarctic surface snow. Front Microbiol 7:398. 10.3389/fmicb.2016.00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Antony R, Mahalinganathan K, Krishnan KP, Thamban M. 2012. Microbial preference for different size classes of organic carbon: a study from Antarctic snow. Environ Monit Assess 184:5929–5943. 10.1007/s10661-011-2391-1. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y, Yao T, Jiao N, Kang S, Xu B, Zeng Y, Huang S, Liu X. 2009. Bacterial diversity in the snow over Tibetan Plateau Glaciers. Extremophiles 13:411–423. 10.1007/s00792-009-0227-5. [DOI] [PubMed] [Google Scholar]
- 19.Van Houdt R, De Boever P, Coninx I, Le Calvez C, Dicasillati R, Mahillon J, Mergeay M, Leys N. 2009. Evaluation of the airborne bacterial population in the periodically confined Antarctic base Concordia. Microb Ecol 57:640–648. 10.1007/s00248-008-9462-z. [DOI] [PubMed] [Google Scholar]
- 20.Carpenter EJ, Lin S, Capone DG. 2000. Bacterial activity in South Pole snow. Appl Environ Microbiol 66:4514–4517. 10.1128/AEM.66.10.4514-4517.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michaud L, Lo Giudice A, Mysara M, Monsieurs P, Raffa C, Leys N, Amalfitano S, Van Houdt R. 2014. Snow surface microbiome on the High Antarctic Plateau (DOME C). PLoS One 9:e104505. 10.1371/journal.pone.0104505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malard LA, Sabacka M, Magiopoulos I, Mowlem M, Hodson A, Tranter M, Siegert MJ, Pearce DA. 2019. Spatial variability of Antarctic surface snow bacterial communities. Front Microbiol 10:461. 10.3389/fmicb.2019.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pearce DA, Alekhina IA, Terauds A, Wilmotte A, Quesada A, Edwards A, Dommergue A, Sattler B, Adams BJ, Magalhaes C, Chu WL, Lau MC, Cary C, Smith DJ, Wall DH, Eguren G, Matcher G, Bradley JA, de Vera JP, Elster J, Hughes KA, Cuthbertson L, Benning LG, Gunde-Cimerman N, Convey P, Hong SG, Pointing SB, Pellizari VH, Vincent WF. 2016. Aerobiology over Antarctica—a new initiative for atmospheric ecology. Front Microbiol 7:16. 10.3389/fmicb.2016.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bottos EM, Woo AC, Zawar-Reza P, Pointing SB, Cary SC. 2014. Airborne bacterial populations above desert soils of the McMurdo Dry Valleys, Antarctica. Microb Ecol 67:120–128. 10.1007/s00248-013-0296-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Behzad H, Gojobori T, Mineta K. 2015. Challenges and opportunities of airborne metagenomics. Genome Biol Evol 7:1216–1226. 10.1093/gbe/evv064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bowers RM, Clum A, Tice H, Lim J, Singh K, Ciobanu D, Ngan CY, Cheng JF, Tringe SG, Woyke T. 2015. Impact of library preparation protocols and template quantity on the metagenomic reconstruction of a mock microbial community. BMC Genomics 16:856. 10.1186/s12864-015-2063-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hughes KA. 2003. Aerial dispersal and survival of sewage-derived faecal coliforms in Antarctica. Atmos Environ 37:3147–3155. 10.1016/S1352-2310(03)00207-3. [DOI] [Google Scholar]
- 28.Gonçalves OS, de Oliveira Souza F, Bruckner FP, Santana MF, Alfenas-Zerbini P. 2021. Widespread distribution of prophages signaling the potential for adaptability and pathogenicity evolution of Ralstonia solanacearum species complex. Genomics 113:992–1000. 10.1016/j.ygeno.2021.02.011. [DOI] [PubMed] [Google Scholar]
- 29.Couvin D, Bernheim A, Toffano-Nioche C, Touchon M, Michalik J, Neron B, Rocha EPC, Vergnaud G, Gautheret D, Pourcel C. 2018. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res 46:W246–W251. 10.1093/nar/gky425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kieft K, Zhou Z, Anantharaman K. 2020. VIBRANT: automated recovery, annotation and curation of microbial viruses, and evaluation of viral community function from genomic sequences. Microbiome 8:90. 10.1186/s40168-020-00867-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antipov D, Raiko M, Lapidus A, Pevzner PA. 2020. Metaviral SPAdes: assembly of viruses from metagenomic data. Bioinformatics 36:4126–4129. 10.1093/bioinformatics/btaa490. [DOI] [PubMed] [Google Scholar]
- 32.Nayfach S, Camargo AP, Schulz F, Eloe-Fadrosh E, Roux S, Kyrpides NC. 2021. CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat Biotechnol 39:578–585. 10.1038/s41587-020-00774-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roux S, Enault F, Hurwitz BL, Sullivan MB. 2015. VirSorter: mining viral signal from microbial genomic data. PeerJ 3:e985. 10.7717/peerj.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahlgren NA, Ren J, Lu YY, Fuhrman JA, Sun F. 2017. Alignment-free d2* oligonucleotide frequency dissimilarity measure improves prediction of hosts from metagenomically-derived viral sequences. Nucleic Acids Res 45:39–53. 10.1093/nar/gkw1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hay ID, Lithgow T. 2019. Filamentous phages: masters of a microbial sharing economy. EMBO Rep 20:e47427. 10.15252/embr.201847427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roux S, Krupovic M, Daly RA, Borges AL, Nayfach S, Schulz F, Sharrar A, Matheus Carnevali PB, Cheng JF, Ivanova NN, Bondy-Denomy J, Wrighton KC, Woyke T, Visel A, Kyrpides NC, Eloe-Fadrosh EA. 2019. Cryptic inoviruses revealed as pervasive in bacteria and archaea across Earth's biomes. Nat Microbiol 4:1895–1906. 10.1038/s41564-019-0510-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Filée J, Forterre P, Laurent J. 2003. The role played by viruses in the evolution of their hosts: a view based on informational protein phylogenies. Res Microbiol 154:237–243. 10.1016/S0923-2508(03)00066-4. [DOI] [PubMed] [Google Scholar]
- 38.Moreira D. 2000. Multiple independent horizontal transfers of informational genes from bacteria to plasmids and phages: implications for the origin of bacterial replication machinery. Mol Microbiol 35:1–5. 10.1046/j.1365-2958.2000.01692.x. [DOI] [PubMed] [Google Scholar]
- 39.Meier-Kolthoff JP, Göker M. 2017. VICTOR: genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics 33:3396–3404. 10.1093/bioinformatics/btx440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolduc B, Jang HB, Doulcier G, You ZQ, Roux S, Sullivan MB. 2017. vConTACT: an iVirus tool to classify double-stranded DNA viruses that infect Archaea and Bacteria. PeerJ 5:e3243. 10.7717/peerj.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prospero JM, Blades E, Mathison G, Naidu R. 2005. Interhemispheric transport of viable fungi and bacteria from Africa to the Caribbean with soil dust. Aerobiologia 21:1–19. 10.1007/s10453-004-5872-7. [DOI] [Google Scholar]
- 42.Askora A, Kawasaki T, Usami S, Fujie M, Yamada T. 2009. Host recognition and integration of filamentous phage phiRSM in the phytopathogen, Ralstonia solanacearum. Virology 384:69–76. 10.1016/j.virol.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 43.da Silva Xavier A, de Almeida JCF, de Melo AG, Rousseau GM, Tremblay DM, de Rezende RR, Moineau S, Alfenas-Zerbini P. 2019. Characterization of CRISPR-Cas systems in the Ralstonia solanacearum species complex. Mol Plant Pathol 20:223–239. 10.1111/mpp.12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koonin EV, Dolja VV, Krupovic M, Varsani A, Wolf YI, Yutin N, Zerbini FM, Kuhn JH. 2020. Global organization and proposed megataxonomy of the virus world. Microbiol Mol Biol Rev 84:e00061-19. 10.1128/MMBR.00061-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murugaiyan S, Bae JY, Wu J, Lee SD, Um HY, Choi HK, Chung E, Lee JH, Lee SW. 2011. Characterization of filamentous bacteriophage PE226 infecting Ralstonia solanacearum strains. J Appl Microbiol 110:296–303. 10.1111/j.1365-2672.2010.04882.x. [DOI] [PubMed] [Google Scholar]
- 46.Mai-Prochnow A, Hui JG, Kjelleberg S, Rakonjac J, McDougald D, Rice SA. 2015. Big things in small packages: the genetics of filamentous phage and effects on fitness of their host. FEMS Microbiol Rev 39:465–487. 10.1093/femsre/fuu007. [DOI] [PubMed] [Google Scholar]
- 47.Sanguino L, Franqueville L, Vogel TM, Larose C. 2015. Linking environmental prokaryotic viruses and their host through CRISPRs. FEMS Microbiol Ecol 91:fiv046. 10.1093/femsec/fiv046. [DOI] [PubMed] [Google Scholar]
- 48.Hwang Y, Rahlff J, Schulze-Makuch D, Schloter M, Probst AJ. 2021. Diverse viruses carrying genes for microbial extremotolerance in the Atacama Desert hyperarid soil. mSystems 6:e00385-21. 10.1128/mSystems.00385-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van TTB, Yoshida S, Miki K, Kondo A, Kamei K. 2014. Genomic characterization of ϕRS603, a filamentous bacteriophage that is infectious to the phytopathogen Ralstonia solanacearum. Microbiol Immunol 58:697–700. 10.1111/1348-0421.12203. [DOI] [PubMed] [Google Scholar]
- 50.Askora A, Yamada T. 2015. Two different evolutionary lines of filamentous phages in Ralstonia solanacearum: their effects on bacterial virulence. Front Genet 6:217. 10.3389/fgene.2015.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Filippova SN, Surgucheva NA, Sorokin VV, Akimov VN, Karnysheva EA, Brushkov AV, Andersen D, Gal’chenko VF. 2016. Bacteriophages in Arctic and Antarctic low-temperature systems. Microbiology 85:359–366. 10.1134/S0026261716030048. [DOI] [Google Scholar]
- 52.Dziewit L, Radlinska M. 2016. Two inducible prophages of an Antarctic Pseudomonas sp. ANT_H14 use the same capsid for packaging their genomes—characterization of a novel phage helper-satellite system. PLoS One 11:e0158889. 10.1371/journal.pone.0158889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Filippova SN, Surgucheva NA, Kulikov EE, Sorokin VV, Akimov VN, Bej AK, McKay C, Andersen D, Galchenko VF. 2013. Detection of phage infection in the bacterial population of Lake Untersee (Antarctica). Microbiology 82:383–386. 10.1134/S0026261713030041. [DOI] [PubMed] [Google Scholar]
- 54.Askora A, Kawasaki T, Fujie M, Yamada T. 2011. Resolvase-like serine recombinase mediates integration/excision in the bacteriophage phiRSM. J Biosci Bioeng 111:109–116. 10.1016/j.jbiosc.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 55.Ahmad AA, Stulberg MJ, Mershon JP, Mollov DS, Huang Q. 2017. Molecular and biological characterization of varphiRs551, a filamentous bacteriophage isolated from a race 3 biovar 2 strain of Ralstonia solanacearum. PLoS One 12:e0185034. 10.1371/journal.pone.0185034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodrigues RAL, Andrade A, Boratto PVM, Trindade GS, Kroon EG, Abrahao JS. 2017. An anthropocentric view of the virosphere-host relationship. Front Microbiol 8:1673. 10.3389/fmicb.2017.01673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sommers P, Fontenele RS, Kringen T, Kraberger S, Porazinska DL, Darcy JL, Schmidt SK, Varsani A. 2019. Single-stranded DNA viruses in Antarctic cryoconite holes. Viruses 11:1022. 10.3390/v11111022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith DJ, Timonen HJ, Jaffe DA, Griffin DW, Birmele MN, Perry KD, Ward PD, Roberts MS. 2013. Intercontinental dispersal of bacteria and archaea by transpacific winds. Appl Environ Microbiol 79:1134–1139. 10.1128/AEM.03029-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Short CM, Suttle CA. 2005. Nearly identical bacteriophage structural gene sequences are widely distributed in both marine and freshwater environments. Appl Environ Microbiol 71:480–486. 10.1128/AEM.71.1.480-486.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Breitbart M, Rohwer F. 2005. Here a virus, there a virus, everywhere the same virus? Trends Microbiol 13:278–284. 10.1016/j.tim.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 61.Breitbart M, Miyake JH, Rohwer F. 2004. Global distribution of nearly identical phage-encoded DNA sequences. FEMS Microbiol Lett 236:249–256. 10.1016/j.femsle.2004.05.042. [DOI] [PubMed] [Google Scholar]
- 62.Li Y, Endo H, Gotoh Y, Watai H, Ogawa N, Blanc-Mathieu R, Yoshida T, Ogata H. 2019. The Earth is small for “leviathans”: long distance dispersal of giant viruses across aquatic environments. Microbes Environ 34:334–339. 10.1264/jsme2.ME19037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bellas CM, Schroeder DC, Edwards A, Barker G, Anesio AM. 2020. Flexible genes establish widespread bacteriophage pan-genomes in cryoconite hole ecosystems. Nat Commun 11:4403. 10.1038/s41467-020-18236-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hughes KA, Convey P, Pertierra LR, Vega GC, Aragon P, Olalla-Tarraga MA. 2019. Human-mediated dispersal of terrestrial species between Antarctic biogeographic regions: a preliminary risk assessment. J Environ Manage 232:73–89. 10.1016/j.jenvman.2018.10.095. [DOI] [PubMed] [Google Scholar]
- 65.Benninghoff W, Benninghoff A. 1985. Wind transport of electrostatically charged particles and minute organisms in Antarctica, p 592–596. In Siegfried WR, Condy PR, Laws RM (ed), Antarctic nutrient cycles and food webs. Springer, Berlin, Germany. 10.1007/978-3-642-82275-9_80. [DOI] [Google Scholar]
- 66.Michaud JM, Thompson LR, Kaul D, Espinoza JL, Richter RA, Xu ZZ, Lee C, Pham KM, Beall CM, Malfatti F, Azam F, Knight R, Burkart MD, Dupont CL, Prather KA. 2018. Taxon-specific aerosolization of bacteria and viruses in an experimental ocean-atmosphere mesocosm. Nat Commun 9:2017. 10.1038/s41467-018-04409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aller JY, Kuznetsova MR, Jahns CJ, Kemp PF. 2005. The sea surface microlayer as a source of viral and bacterial enrichment in marine aerosols. J Aerosol Sci 36:801–812. 10.1016/j.jaerosci.2004.10.012. [DOI] [Google Scholar]
- 68.Wilbourn EK, Thornton DCO, Ott C, Graff J, Quinn PK, Bates TS, Betha R, Russell LM, Behrenfeld MJ, Brooks SD. 2020. Ice nucleation by marine aerosols over the North Atlantic Ocean in late spring. J Geophys Res Atmos 125:e2019JD030913. 10.1029/2019JD030913. [DOI] [Google Scholar]
- 69.Amato P. 2012. Clouds provide atmospheric oases for microbes. Microbe 7:119–123. 10.1128/microbe.7.119.1. [DOI] [Google Scholar]
- 70.Christner BC, Morris CE, Foreman CM, Cai R, Sands DC. 2008. Ubiquity of biological ice nucleators in snowfall. Science 319:1214. 10.1126/science.1149757. [DOI] [PubMed] [Google Scholar]
- 71.Strother SL, Salzmann U, Roberts SJ, Hodgson DA, Woodward J, Van Nieuwenhuyze W, Verleyen E, Vyverman W, Moreton SG. 2015. Changes in Holocene climate and the intensity of Southern Hemisphere westerly winds based on a high-resolution palynological record from sub-Antarctic south Georgia. Holocene 25:263–279. 10.1177/0959683614557576. [DOI] [Google Scholar]
- 72.Netea MG, Dominguez-Andres J, Eleveld M, Op den Camp HJM, van der Meer JWM, Gow NAR, de Jonge MI. 2020. Immune recognition of putative alien microbial structures: host-pathogen interactions in the age of space travel. PLoS Pathog 16:e1008153. 10.1371/journal.ppat.1008153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nicholson WL, Schuerger AC, Race MS. 2009. Migrating microbes and planetary protection. Trends Microbiol 17:389–392. 10.1016/j.tim.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 74.Coenye T, Vandamme P, LiPuma JJ. 2002. Infection by Ralstonia species in cystic fibrosis patients: identification of R. pickettii and R. mannitolilytica by polymerase chain reaction. Emerg Infect Dis 8:692–696. 10.3201/eid0807.010472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Berliner AJ, Mochizuki T, Stedman KM. 2018. Astrovirology: viruses at large in the universe. Astrobiology 18:207–223. 10.1089/ast.2017.1649. [DOI] [PubMed] [Google Scholar]
- 76.Mattoni RHT, Keller EC, Jr., Ebersold WT, Eiserling FA, Romig WR. 1971. Induction of lysogenic bacteria in the space environment, p 309–324. In Saunders JF (ed), The experiments of Biosatellite II. NASA, Washington, DC. [Google Scholar]
- 77.Mattoni RHT. 1968. Influence of spaceflight and radiation on induction of prophage P-22 in Salmonella typhimurium. Jpn J Genet 43:465–465. 10.1266/jjg.43.465. [DOI] [Google Scholar]
- 78.Mattoni RHT. 1968. Space-flight effects and gamma radiation interaction on growth and induction of lysogenic bacteria, a preliminary report. Bioscience 18:602–608. 10.2307/1294308. [DOI] [Google Scholar]
- 79.Koike J. 1991. Fundamental questions concerning the contamination of other planets with terrestrial microorganisms carried by space-probes. J Space Technol Sci 7:9–14. 10.11230/jsts.7.2_9. [DOI] [Google Scholar]
- 80.Weinmaier T, Probst AJ, La Duc MT, Ciobanu D, Cheng JF, Ivanova N, Rattei T, Vaishampayan P. 2015. A viability-linked metagenomic analysis of cleanroom environments: eukarya, prokaryotes, and viruses. Microbiome 3:62. 10.1186/s40168-015-0129-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mora M, Wink L, Kogler I, Mahnert A, Rettberg P, Schwendner P, Demets R, Cockell C, Alekhova T, Klingl A, Krause R, Zolotariof A, Alexandrova A, Moissl-Eichinger C. 2019. Space Station conditions are selective but do not alter microbial characteristics relevant to human health. Nat Commun 10:3990. 10.1038/s41467-019-11682-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pavletić B, Runzheimer K, Siems K, Koch S, Cortesão M, Ramos-Nascimento A, Moeller R. 2022. Spaceflight virology: what do we know about viral threats in the spaceflight environment? Astrobiology 22:210–224. 10.1089/ast.2021.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Aboudan A, Colombatti G, Bettanini C, Ferri F, Lewis S, Van Hove B, Karatekin O, Debei S. 2018. ExoMars 2016 Schiaparelli module trajectory and atmospheric profiles reconstruction. Space Sci Rev 214:1–31. 10.1007/s11214-018-0532-3. [DOI] [Google Scholar]
- 84.Bushnell B. 2014. BBTools software package. http://sourceforge.net/projects/bbmap.
- 85.Joshi N, Fass J. 2011. Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ files (version 1.33).
- 86.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res 27:824–834. 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dick GJ, Andersson AF, Baker BJ, Simmons SL, Thomas BC, Yelton AP, Banfield JF. 2009. Community-wide analysis of microbial genome sequence signatures. Genome Biol 10:R85. 10.1186/gb-2009-10-8-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu YW, Simmons BA, Singer SW. 2016. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32:605–607. 10.1093/bioinformatics/btv638. [DOI] [PubMed] [Google Scholar]
- 89.Sieber CMK, Probst AJ, Sharrar A, Thomas BC, Hess M, Tringe SG, Banfield JF. 2018. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat Microbiol 3:836–843. 10.1038/s41564-018-0171-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bornemann TLV, Esser SP, Stach L, Burg T, Probst AJ. 2020. uBin—a manual refining tool for metagenomic bins designed for educational purposes. bioRxiv 10.1101/2020.07.15.204776. [DOI]
- 91.Probst AJ, Castelle CJ, Singh A, Brown CT, Anantharaman K, Sharon I, Hug LA, Burstein D, Emerson JB, Thomas BC, Banfield JF. 2017. Genomic resolution of a cold subsurface aquifer community provides metabolic insights for novel microbes adapted to high CO2 concentrations. Environ Microbiol 19:459–474. 10.1111/1462-2920.13362. [DOI] [PubMed] [Google Scholar]
- 92.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Skennerton CT, Imelfort M, Tyson GW. 2013. Crass: identification and reconstruction of CRISPR from unassembled metagenomic data. Nucleic Acids Res 41:e105. 10.1093/nar/gkt183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mojica FJ, Diez-Villasenor C, Soria E, Juez G. 2000. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol Microbiol 36:244–246. 10.1046/j.1365-2958.2000.01838.x. [DOI] [PubMed] [Google Scholar]
- 95.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. Journal of Molecular Biology 215(3):403–410. 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 96.Moller AG, Liang C. 2017. MetaCRAST: reference-guided extraction of CRISPR spacers from unassembled metagenomes. PeerJ 5:e3788. 10.7717/peerj.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ren J, Ahlgren NA, Lu YY, Fuhrman JA, Sun F. 2017. VirFinder: a novel k-mer based tool for identifying viral sequences from assembled metagenomic data. Microbiome 5:69. 10.1186/s40168-017-0283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Crits-Christoph A, Gelsinger DR, Ma B, Wierzchos J, Ravel J, Davila A, Casero MC, DiRuggiero J. 2016. Functional interactions of archaea, bacteria and viruses in a hypersaline endolithic community. Environ Microbiol 18:2064–2077. 10.1111/1462-2920.13259. [DOI] [PubMed] [Google Scholar]
- 99.Marz M, Beerenwinkel N, Drosten C, Fricke M, Frishman D, Hofacker IL, Hoffmann D, Middendorf M, Rattei T, Stadler PF, Topfer A. 2014. Challenges in RNA virus bioinformatics. Bioinformatics 30:1793–1799. 10.1093/bioinformatics/btu105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rahlff J, Turzynski V, Esser SP, Monsees I, Bornemann TLV, Figueroa-Gonzalez PA, Schulz F, Woyke T, Klingl A, Moraru C, Probst AJ. 2021. Lytic archaeal viruses infect abundant primary producers in Earth's crust. Nat Commun 12:4642. 10.1038/s41467-021-24803-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Roux S, Emerson JB, Eloe-Fadrosh EA, Sullivan MB. 2017. Benchmarking viromics: an in silico evaluation of metagenome-enabled estimates of viral community composition and diversity. PeerJ 5:e3817. 10.7717/peerj.3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lefort V, Desper R, Gascuel O. 2015. FastME 2.0: a comprehensive, accurate, and fast distance-based phylogeny inference program. Mol Biol Evol 32:2798–2800. 10.1093/molbev/msv150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Farris JS. 1972. Estimating phylogenetic trees from distance matrices. Am Nat 106:645–668. 10.1086/282802. [DOI] [Google Scholar]
- 104.Rambaut A. 2006. FigTree, a graphical viewer of phylogenetic trees and as a program for producing publication-ready figures.
- 105.Göker M, Garcia-Blazquez G, Voglmayr H, Telleria MT, Martin MP. 2009. Molecular taxonomy of phytopathogenic fungi: a case study in Peronospora. PLoS One 4:e6319. 10.1371/journal.pone.0006319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Meier-Kolthoff JP, Hahnke RL, Petersen J, Scheuner C, Michael V, Fiebig A, Rohde C, Rohde M, Fartmann B, Goodwin LA, Chertkov O, Reddy T, Pati A, Ivanova NN, Markowitz V, Kyrpides NC, Woyke T, Göker M, Klenk HP. 2014. Complete genome sequence of DSM 30083(T), the type strain (U5/41(T)) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand Genomic Sci 9:2. 10.1186/1944-3277-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bin Jang H, Bolduc B, Zablocki O, Kuhn JH, Roux S, Adriaenssens EM, Brister JR, Kropinski AM, Krupovic M, Lavigne R, Turner D, Sullivan MB. 2019. Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat Biotechnol 37:632–639. 10.1038/s41587-019-0100-8. [DOI] [PubMed] [Google Scholar]
- 108.Brister JR, Ako-Adjei D, Bao Y, Blinkova O. 2015. NCBI viral genomes resource. Nucleic Acids Res 43:D571–D577. 10.1093/nar/gku1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Moraru C, Varsani A, Kropinski AM. 2020. VIRIDIC—a novel tool to calculate the intergenomic similarities of prokaryote-infecting viruses. Viruses 12:1268. 10.3390/v12111268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nilsson E, Bayfield OW, Lundin D, Antson AA, Holmfeldt K. 2020. Diversity and host interactions among virulent and temperate Baltic Sea Flavobacterium phages. Viruses 12:158. 10.3390/v12020158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 115.Shaffer M, Borton MA, McGivern BB, Zayed AA, La Rosa SL, Solden LM, Liu P, Narrowe AB, Rodriguez-Ramos J, Bolduc B, Gazitua MC, Daly RA, Smith GJ, Vik DR, Pope PB, Sullivan MB, Roux S, Wrighton KC. 2020. DRAM for distilling microbial metabolism to automate the curation of microbiome function. Nucleic Acids Res 48:8883–8900. 10.1093/nar/gkaa621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zimmermann L, Stephens A, Nam SZ, Rau D, Kubler J, Lozajic M, Gabler F, Soding J, Lupas AN, Alva V. 2018. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J Mol Biol 430:2237–2243. 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 117.Söding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33:W244–W248. 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Paez-Espino D, Roux S, Chen IA, Palaniappan K, Ratner A, Chu K, Huntemann M, Reddy TBK, Pons JC, Llabres M, Eloe-Fadrosh EA, Ivanova NN, Kyrpides NC. 2019. IMG/VR v.2.0: an integrated data management and analysis system for cultivated and environmental viral genomes. Nucleic Acids Res 47:D678–D686. 10.1093/nar/gky1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 121.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schlitzer R. 2015. Ocean Data View. https://odv.awi.de/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S5. Download aem.00315-22-s0001.pdf, PDF file, 1.6 MB (1.6MB, pdf)
Tables S1 to S8. Download aem.00315-22-s0002.xlsx, XLSX file, 0.4 MB (433.8KB, xlsx)
Data Availability Statement
The sequence for the R. pickettii 12D strain isolated from copper-contaminated sediment from a lake in Michigan, in which the Antarcphage49_Dr_7823_circ PGF from the Druzhnaja station was found, is available from the corresponding author (Janina.rahlff@uol.de) on request.