Abstract
The extremely thermophilic anaerobic archaeon strain B1001 was isolated from a hot-spring environment in Japan. The cells were irregular cocci, 0.5 to 1.0 μm in diameter. The new isolate grew at temperatures between 60 and 95°C (optimum, 85°C), from pH 5.0 to 9.0 (optimum, pH 7.0), and from 1.0 to 6.0% NaCl (optimum, 2.0%). The G+C content of the genomic DNA was 43.0 mol%. The 16S rRNA gene sequencing of strain B1001 indicated that it belongs to the genus Thermococcus. During growth on starch, the strain produced a thermostable cyclomaltodextrin glucanotransferase (CGTase). The enzyme was purified 1,750-fold, and the molecular mass was determined to be 83 kDa by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. Incubation at 120°C with SDS and 2-mercaptoethanol was required for complete unfolding. The optimum temperatures for starch-degrading activity and cyclodextrin synthesis activity were 110 and 90 to 100°C, respectively. The optimum pH for enzyme activity was pH 5.0 to 5.5. At pH 5.0, the half-life of the enzyme was 40 min at 110°C. The enzyme formed mainly α-cyclodextrin with small amounts of β- and γ-cyclodextrins from starch. This is the first report on the presence of the extremely thermostable CGTase from hyperthermophilic archaea.
Cyclomaltodextrin glucanotransferases (CGTase; EC 2.4.1.19) are able to convert starch into cyclodextrins (CDs), closed-ring structures in which six or more glucose units are joined by means of α-1,4 glucosidic bonds (15). Depending to the number of glucose units (six, seven, or eight), they are named α-, β-, or γ-CDs, respectively. They are able to form inclusion complexes with many organic and inorganic molecules, thereby changing the physical and chemical properties of the included compounds. Therefore, CGTase is an important enzyme for the food, cosmetic, and pharmaceutical industries. CGTase are known to catalyze four different transferase reactions: cyclization, coupling, disproportionation, and hydrolysis. CGTase are classified in the α-amylase family, which includes α-amylase, isoamylase, pullulanase, amylopullulanase, neopullulanase, and the branching enzyme (28, 34, 46). The α-amylase family enzymes include the four highly conserved amino acid sequences containing the substrate binding and active sites. The three-dimensional structure of CGTase closely resembles the (β/α)8-barrel structure of α-amylase (43).
CGTase is produced by many Bacillus strains, Klebsiella pneumoniae, Klebsiella oxytoca, a Brevibacterium sp., a Thermoanaerobacter sp., and Thermoanaerobacterium thermosulfurigenes (11, 21, 22, 26, 33, 38, 49). The thermophilic anaerobic bacteria Thermoanaerobacter sp. and T. thermosulfurigenes produce the thermostable CGTase. The optimum temperatures of these enzymes are 90 to 95°C and 80 to 90°C, respectively. Thermostable CGTase is very useful for industrial utilization. Liquefaction, which is the first step in the production of CDs from starch, is performed by jetcooking, where a starch slurry is treated with thermostable α-amylase at 105 to 110°C. Thermostable CGTase can be used in liquefaction and then in CD formation without being replaced (38).
Recently, hyperthermophilic archaea which can grow at temperatures higher than 90°C have been isolated, and their enzymes have been shown to be extremely thermostable. Extremely thermostable α-amylase family enzymes including α-amylase, pullulanase, and amylopullulanase have been characterized from the genera Thermococcus, Pyrococcus, Sulfolobus, Thermofilum, Desulfurococcus, and Staphylothermus (4–8, 12, 17, 24, 25, 29, 41, 45). However, CGTase has not been isolated from archaea. We have initiated a program to isolate hyperthermophiles which can secrete CGTases. In this paper, we report on the purification and characterization of an extremely thermostable CGTase from a newly isolated hyperthermophilic archaeon, Thermococcus sp. strain B-1001.
MATERIALS AND METHODS
Isolation of the hyperthermophile.
Hot sedimentary materials and venting water were collected at a thermal vents and hot spring environments in Japan. Samples were put into 50-ml screw cap bottles and brought back to the laboratory while being held at about 80°C for the transportation. The samples were inoculated immediately into anaerobic gas-flushed 15-ml test tubes containing 5 ml of medium. A mixture of anaerobic gases, N2-CO2-H2 (90:5:5 [vol/vol/vol]), was used. The tubes were closed tightly and incubated at 85°C until cell growth was observed. To obtain pure cultures, a dilution-to-extinction technique was used. After the cell density of an enrichment culture reached approximately 108 cells per ml, five serial transfers into same medium were carried out by performing 10 1:10 dilutions per transfer. Each dilution in the dilution-to-extinction series was incubated for at least 4 days. The culture in the tube showing growth at the highest dilution was chosen as the pure culture, which usually grew up to about 108 cells per ml in 2 days. Strain B1001 was isolated from the hot spring at Tottori prefecture, Japan.
Culture conditions.
Enrichments and isolation were carried out in a modified SME medium (2) containing the following components (per liter): 2 g of yeast extract (Difco), 2 g of NaCl, 3.5 g of MgSO4 · 2H2O, 2.75 g of MgCl2 · 6H2O, 1 g of CaCl2 · 2H2O, 0.5 g of KH2PO4, 0.325 g of KCl, 50 mg of NaBr, 15 mg of H3BO3, 7.5 mg of SrCl2 · 6H2O, 2 mg of (NH4)2Ni(SO4)2 · 2H2O, 2.5 μg of KI, 10 ml of trace minerals mixture, 10 g of elemental sulfur, 0.48 g of Na2S · 9H2O, and 1 mg of resazurine. The pH was adjusted to 7.0 with KOH. The trace minerals mixture (1) contained (per liter) 1.5 g of nitrilotriacetic acid, 3 g of MgSO4 · 7H2O, 0.5 g of MnCl2 · 4H2O, 1 g of NaCl, 0.1 g of FeSO4 · 7H2O, 0.1 g of CoSO4, 0.1 g of CaCl2 · 2H2O, 0.1 g of ZnSO4, 10 mg of CuSO4 · 5H2O, 10 mg of AlK(SO4)2, 10 mg of H3BO3, and 10 mg of Na2MoO4 · 2H2O.
Physiological characteristics.
The physiological characteristics of the isolated strain were investigated in anaerobic gas-flushed 15-ml test tubes containing 10 ml of medium supplemented with 0.2% peptone. To check the optimal growth conditions, the incubation temperature, pH, and NaCl concentration of the culture medium were varied over a wide range of conditions. The temperature was tested from 40 to 110°C. Temperatures were maintained with aluminum heating blocks in an anaerobic incubator (EAN-140; Tabai Espec Corp., Osaka, Japan). The pH of the medium was adjusted from 4 to 10 by the addition of H2SO4 or KOH. The NaCl concentration ranged from no NaCl added to 6% NaCl. Samples were collected every 2 h under anaerobic and high-temperature conditions, and the cells were counted. To determine substrate utilization, substrates were added at 0.2% (wt/vol) to yeast extract- and peptone-free medium.
Sensitivity to the antibiotics benzylpenicillin, vancomycin, streptomycin, chloramphenicol, and rifampin was examined by the addition of 100 μg of antibiotic per ml of medium under the same conditions as those reported for Thermococcus fumicolans (13) and Thermococcus celer (50).
H2S was qualitatively detected by adding 10 μl of a saturated lead acetate solution to 1 ml of culture sample based on the procedure of Williams (48). A blackish-brown precipitate indicated the presence of H2S.
Determination of cell numbers.
The number of cells in liquid cultures was determined by direct cell counting in a Thoma counting chamber (depth, 0.02 mm) under a phase-contrast microscope. Generation times were calculated by linear regression analysis from three points along the logarithmic part of the resulting growth curves.
Electron microscopy.
For scanning electron microscopy, the cells were fixed in 0.1 M phosphate-buffered glutaraldehyde (2.5% final concentration) at pH 7.0. The fixed samples were dehydrated through 25, 50, 75, and 100% ethanol and t-butanol, dried to the critical point, and sputter-coated with palladium-gold alloy. The samples were examined under an S-3200N scanning electron microscope (Hitachi, Tokyo, Japan).
DNA preparation and DNA base composition.
The genomic DNA was isolated with a GenomicPrep kit (Pharmacia, Uppsala, Sweden). The G+C content of the genomic DNA was determined by direct analysis of the mononucleoside content of the decomposed DNA (31) with a DNA-GC kit (Yamasa Shoyu, Tokyo, Japan). Calf thymus DNA (42 mol% G+C) (Sigma) was used as reference. The value used was the average of the results of three separate experiments.
Analysis of the 16S rRNA gene.
The 16S rRNA gene of strain B1001 was amplified by PCR with forward primer 5′-TTCCGGTTGATCCYGCCGGA-3′ and reverse primer 5′-GGTTACCTTGTTACGACTT-3′, which correspond to nucleotides 2 to 21 and 1510 to 1492 of the Escherichia coli 16S rRNA gene sequence, respectively (39). The amplified PCR product was cloned in pBluescript II SK(+), and both strands were sequenced by the dideoxy-chain termination method with fluorescent primer (A.L.F DNA sequencer; Pharmacia).
The 16S rRNA gene sequence was aligned with a representative group of archaeal sequences obtained from GenBank and EMBL. The phylogenetic tree was constructed based on the neighbor-joining (NJ) method (42). Alignment of sequences and estimations of the numbers of nucleotide substitutions were performed with the CLUSTAL W program (47).
Assay of CGTase activity.
Potato starch (1 g) was suspended in 20 ml of distilled water and then dissolved by adding 5 ml of 2 N NaOH in boiling water. Then 2 N CH3COOH (5 ml) and distilled water (25 ml) were added to neutralize the solution. The solution was adjusted to the appropriate pH with 0.1 N CH3COOH at 80°C and made up to 100 ml with distilled water as the substrate.
Two assay methods for determining CGTase activity were used. The starch-degrading activity assay was based on the decrease in absorbance (blue value) of the iodine-amylose complex as a result of the starch degradation. The enzyme solution (6 μl) was incubated at 90°C for 10 min with 60 μl of substrate (1% starch), and the reaction was terminated by cooling the solution in ice-water. The reaction mixture (10 μl) was removed and mixed with 100 μl of 0.1 N HCl. This solution (100 μl) was mixed with 2 ml of an iodine solution (0.005% I2, 0.05% KI), and the absorbance of the mixture at 660 nm was measured. One unit of starch-degrading activity was defined as the amount of enzyme which can decrease the absorbance at 660 nm of the amylose-iodine complex by 1% per min.
The CD synthesis activity was examined by measurement of liberated α-CD by the methyl orange method (30) with slight modifications. The reaction mixture (50 μl) was mixed with 6 N HCl (7.5 μl) and 52.5 μM methyl orange solution (100 μl), and then incubated at 25°C for 30 min. The absorbance of the mixture at 505 nm was measured. One unit of CD synthesis activity was defined as the amount of enzyme which released 1 μmol of α-CD per min.
The amount of liberated reducing sugar was determined by a dinitrosalicyclic acid method (3) with slight modifications (on a 1/10 scale). The protein concentration was measured with a NanoOrange protein quantitation kit (Molecular Probes, Eugene, Oreg.).
Purification of CGTase.
Culture broth (70 liters) of strain B1001 was filtered through Toyo no. 2 filter paper (Toyo Roshi Co., Tokyo, Japan) to remove elemental sulfur and then centrifuged (8,000 × g for 10 min) to obtain the supernatant. The supernatant was concentrated to 500 ml by an ultrafiltration system (AIP-1010; Asahikasei Co., Tokyo, Japan), brought to 80% ammonium sulfate saturation, and kept at 4°C overnight. The precipitate was collected by centrifugation (27,000 × g for 30 min), dissolved in 25 mM sodium acetate buffer (pH 5.0), and dialyzed overnight against the same buffer. The dialysate was applied to a Resource Q column (Pharmacia) equipped with a fast protein liquid chromatography system (Pharmacia), equilibrated with 25 mM sodium acetate buffer (pH 5.0), and eluted with a linear gradient of NaCl (0 to 1.0 M) at a flow rate of 1 ml/min. The eluted fractions containing CGTase activity were pooled and then brought to 80% ammonium sulfate saturation. The precipitate was collected by centrifugation (27,000 × g for 30 min), dissolved in 25 mM Tris-HCl buffer (pH 7.0) containing 12% ammonium sulfate saturation, and applied to a phenyl-Superose HR 5/5 column (Pharmacia) previously equilibrated with the same buffer. The column was washed with the same buffer and eluted with 25 mM Tris-HCl buffer (pH 7.0) at a flow rate of 0.3 ml/min. The eluted fractions containing CGTase activity were pooled and applied to an α-CD-(epoxy)–Sepharose 6B affinity column (Pharmacia) previously equilibrated with 25 mM Tris-HCl buffer (pH 7.0) containing 100 mM NaCl. The column was washed with 25 mM Tris-HCl buffer (pH 7.0) containing 1.0 M NaCl, and the CGTase was eluted at a flow rate of 0.2 ml/min with 25 mM Tris-HCl buffer (pH 7.0) supplemented with 1% α-CD. The eluted fractions containing CGTase activity were pooled and dialyzed against distilled water for 3 days. All enzyme purification procedures were performed at 4°C.
Gel electrophoresis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and native PAGE were carried out with the Phastsystem (Pharmacia) by the procedures recommended in the Phastsystem manual. Protein bands were detected by staining with 0.1% Coomassie brilliant blue R-250. For amylase activity staining, after gel electrophoresis the gel was incubated at 85°C for 1 h in 100 mM sodium acetate buffer (pH 5.0) containing 5% soluble starch. After being washed with distilled water, the gel was subjected to staining with 50% methanol solution containing 1% iodine and 10% potassium iodide.
N-terminal analysis.
The NH2-terminal amino acid sequence was determined by using the automated Edman degradation system of ABI protein sequencer 473A (Applied Biosystems Japan, Tokyo, Japan).
The sequences of other CGTases were obtained from the NBRF database (accession numbers are as follows: Bacillus macerans, S31281; B. licheniformis, S15920; Bacillus sp., S26399; B. stearothermophilus, S26588) and the EMBL database (accession numbers are as follows: Thermoanaerobacterium thermosulfurigenes, M57580; Thermoanaerobacter sp., Z35484).
Determination of the amounts of α-, β-, and γ-CD.
Reaction mixtures (100 μl) containing 2.5% (wt/vol) soluble starch and 50 mM sodium acetate buffer (pH 5.0) were incubated with the purified enzyme (8 mU as CD synthesis activity) at 90°C. Samples were taken at convenient time intervals, and the reaction was terminated by cooling the mixtures on ice. The reaction mixture (50 μl) was removed and mixed with 1 μl (2 U) of glucoamylase (Toyobo Co., Osaka, Japan), 9 μl of 2 M sodium acetate buffer (pH 5.0), and 40 μl of distilled water. The mixture was incubated at 40°C for 60 min, mixed with 150 μl of acetonitrile, and then filtered through a membrane filter (pore size, 0.45 μm). The filtered sample (10 μl) was analyzed by high-performance liquid chromatography (HPLC) with a TSKgel Amide-80 column (TOSOH, Tokyo, Japan) and eluted with acetonitrile-water (60:40 [vol/vol]) at 1 ml/min. The flow cell was set at 30°C, and products were detected with a refractive index detector.
The amount of total sugar in the reaction mixtures was assayed by the phenol-sulfuric acid method (9) with glucose as a standard.
Nucleotide sequence accession number.
The nucleotide sequence data of the gene for 16S rRNA reported in this paper has been submitted to the DDBJ, EMBL, and GenBank DNA databases under accession no. AB016298.
RESULTS
Isolation of the CGTase-producing hyperthermophile.
To isolate the CGTase-producing hyperthermophile, various hyperthermophilic strains were isolated by enrichment of the water and sediment samples of hot spring environments and serial dilutions of positive enrichments. These strains were incubated in medium containing 0.2% soluble starch, and the extracellular supernatants were prepared. After incubation of starch with the supernatant, the degradation of starch (the decrease in the blue value) and the liberated reducing power were measured. When the starch is incubated with CGTase, which produces mainly CD (no reducing sugar), the blue value decreases rapidly and the reducing power increases slightly. One strain, named B1001, for which the blue value decreased but the reducing power increased only slightly, was selected. The α-, β-, and γ-CD-specific synthesis activity measurement (see Materials and Methods) (18, 19) and the HPLC of reaction products suggested the formation of CDs (data not shown).
Characterization of isolate B1001.
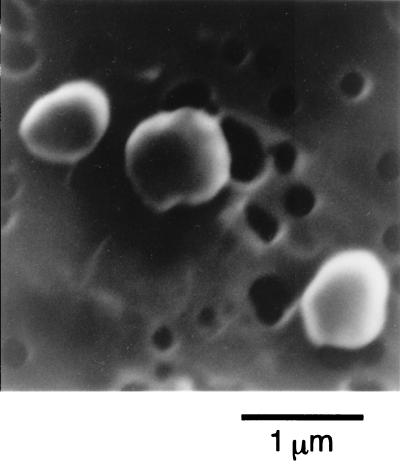
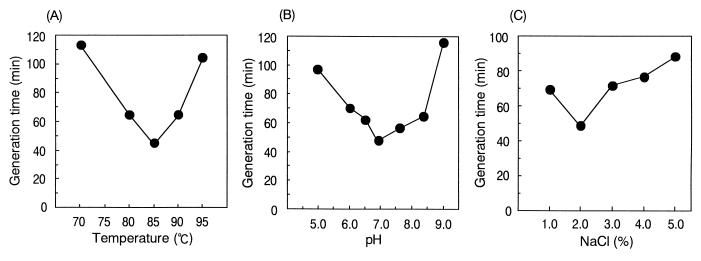
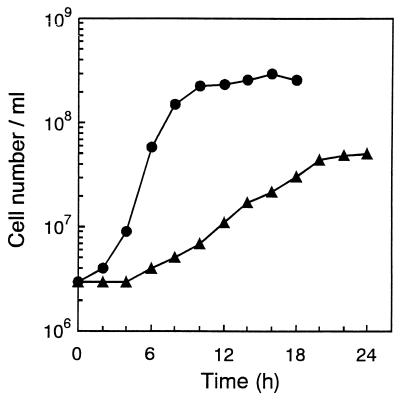
Cells of B1001 were irregular cocci of 0.5 to 1.0 μm in diameter (Fig. 1) and grew between 60 and 95°C; the optimum growth temperature was 85°C (Fig. 2A). The pH range for growth was 5.0 to 9.0, and the optimum pH was around 7.0 (Fig. 2B). Growth was observed in medium containing 1 to 6% NaCl, with the optimum at 2% (Fig. 2C). The presence of elemental sulfur stimulated growth, which was accompanied by H2S production. The generation time for strain B1001 was 44 min under the optimal cultivation conditions, and the cell density reached a maximum of 3 × 108 cells/ml after 16 h of incubation (Fig. 3). Cells were also grown in the absence of elemental sulfur; however, growth was very slow. The cell density in medium without elemental sulfur reached a maximum of 5 × 107 cells/ml after 24 h of incubation (Fig. 3). Cell growth was not observed in the absence of Na2S (oxidized conditions).
FIG. 1.
Scanning electron micrograph of Thermococcus sp. strain B1001.
FIG. 2.
Influence of temperature (A), pH (B), and NaCl concentration (C) on the growth of Thermococcus sp. strain B1001. The generation times were calculated from the slopes of the growth curves. All values are the average of two separate experiments.
FIG. 3.
Growth of strain B1001. Cultivations were carried out under optimal conditions (●) and in medium lacking elemental sulfur (▴).
Strain B1001 was capable of growing on proteinaceous complex substrates such as yeast extract, peptone, and tryptone, but it did not grow on Casamino Acids. No growth was observed on glucose, maltose, fructose, sucrose, lactose, soluble starch, α-CD, ethanol, or methanol as the sole carbon source. Growth of strain B1001 was not influenced by benzylpenicillin, vancomycin, or streptomycin. It was partly inhibited by chloramphenicol and completely inhibited by rifampin.
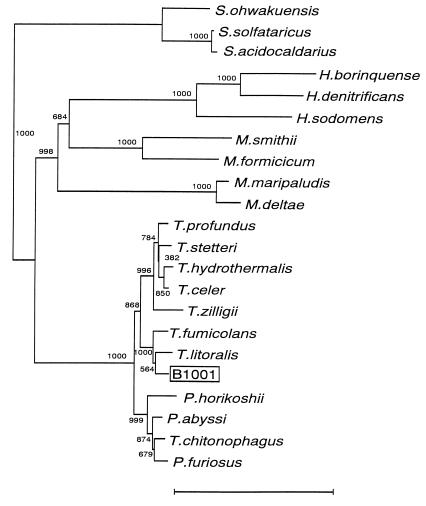
The genomic DNA of strain B1001 had a G+C content of 43.0 mol% as calculated by direct analysis of the nucleotides. The 16S rRNA gene sequence (1,416 bases) of strain B1001 was determined. The sequence of B1001 was aligned and compared to the 16S rRNA gene sequences of various species of the domain Archaea. It showed high sequence similarity to the 16S rRNA from Thermococcus litoralis (97.7%), T. fumicolans (97.0%), and T. celer (96.6%). For a more precise classification, the phylogenetic tree was inferred by the NJ method (Fig. 4). The result indicates that strain B1001 belongs to the genus Thermococcus and is most closely related to Thermococcus litoralis.
FIG. 4.
Phylogenetic tree based on 16S rRNA gene sequences. The tree was constructed by the NJ method. Numbers indicate bootstrap values of 1,000 times trial. The scale bar represents 10 nucleotide substitutions per 100 nucleotides. The accession numbers of 16S rRNA sequences used for the unrooted tree are as follows: Sulfurishaera ohwakuensis, D85507; Sulfolobus solfataricus, X03235; Sulfolobus acidocaldarius, D14053; Halogeometricum borinquense, AF002984; Haloferax denitrificans, D14128; Halobacterium sodomense, D13379; Methanobrevibacter smithii, U55234; Methanobacterium formicicum, AF028689; Methanococcus maripaludis, AF005049; Methanococcus deltae, U38485; T. profundus, Z75233; T. stetteri, Z75240; T. hydrothermalis, Z70244; T. celer, M21529; T. zilligii, U76534; T. fumicolans, Z70250; T. litoralis, Z70252; Pyrococcus horikoshii, D45214; Pyrococcus abyssi, Z70246; T. chitonophagus, X99570; and Pyrococcus furiosus, U20163.
Purification of the CGTase.
Strain B1001 culture broth (70 liters) was concentrated to a final volume of 500 ml. The purification steps are summarized in Table 1. By applying anion-exchange chromatography, hydrophobic interaction chromatography, and affinity chromatography, CGTase was purified 1,750-fold with a yield of 10%. The purified enzyme exhibited a specific CD synthesis activity of 1,400 U/mg of protein.
TABLE 1.
Summary of CGTase purification steps
| Fraction | Activitya (U) | Amt of protein (mg) | Sp act (U/mg) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|
| Crude sample | 95.8 | 123.5 | 0.8 | 1.0 | 100 |
| Resource Q | 59.5 | 12.8 | 4.6 | 5.8 | 62 |
| Phenyl-Superose | 15.8 | 1.680 | 9.4 | 11.8 | 16 |
| α-CD affinity | 9.8 | 0.007 | 1,400 | 1,750 | 10 |
CD synthesis activity was measured at 90°C.
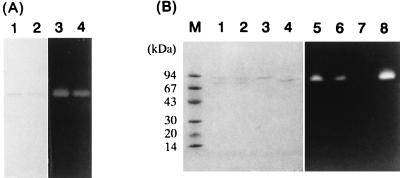
The purified enzyme was analyzed by native PAGE and SDS-PAGE. After native PAGE, the mobility of the active band as determined by starch-degrading activity staining coincided with that of the single protein band stained with Coomassie brilliant blue (Fig. 5A). It indicated that the purification of B1001 CGTase had been accomplished. Furthermore, after heating at 100°C for 40 min, the same result was observed, suggesting that the enzyme was highly stable to heating. However, unusual mobility was observed when the purified CGTase was subjected to SDS-PAGE (Fig. 5B). When the enzyme was incubated in denaturing buffer (2.3% SDS, 5% 2-mercaptoethanol) at 60°C for 5 min, a single protein band with a molecular mass of 72 kDa was observed, and this protein band was visualized by starch-degrading activity staining. The 72-kDa band was considered to be detected due to incomplete denaturation. Incubation at 100 or 110°C for 5 min caused the appearance of another protein band, with a molecular mass of 83 kDa, which was not detected by activity staining. Furthermore, the 72-kDa band visible with activity staining became weaker with increasing temperature during heating treatment. After incubation at 120°C for 5 min, the 72-kDa band completely disappeared and only the 83-kDa band was observed. The CGTase was considered to be switched from the active form of 72 kDa to the unfolded, denatured form of 83 kDa in SDS-PAGE. Hence, the size of B1001 CGTase was estimated to be approximately 83 kDa by SDS-PAGE.
FIG. 5.
Behavior of CGTase in native PAGE (A) and SDS-PAGE (B). (A) Enzyme samples were visualized by Coomassie brilliant blue staining (lanes 1 and 2) and activity staining (lanes 3 and 4). The purified samples (lanes 1 and 3) were treated in 0.1 M sodium acetate buffer (pH 5.0) at 100°C for 40 min (lanes 2 and 4). (B) The purified samples were visualized by Coomassie brilliant blue staining (lanes 1 to 4) and activity staining (lanes 5 to 8). The samples were treated at 60°C (lanes 4 and 8), 100°C (lanes 1 and 5), 110°C (lanes 2 and 6), or 120°C (lanes 3 and 7) for 5 min in denaturing buffer containing 2.3% SDS and 5% 2-mercaptoethanol. Protein molecular mass standards are shown in lane M and are listed on the left.
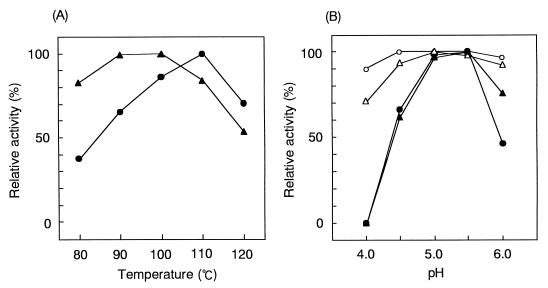
Effects of temperature and pH on activity, and thermal stability of the enzyme.
The optimum starch-degrading activity of CGTase was observed at 110°C, and the optimum CD synthesis activity was noted at 90 to 100°C. Almost 70% starch-degrading activity and 50% (CD synthesis) of the maximum enzyme activity were detected at 120°C (Fig. 6A). The pH optimum of CGTase was 5.0 to 5.5 at 110°C for both the starch-degrading activity and the CD synthesis activity. At 90°C, the optimum pH curve changed to a broad curve in the range of pH 4.5 to 6.0 (Fig. 6B). The stability of CGTase at various temperatures (at pH 5.0) was examined. Significant loss of CGTase activity was not observed after heat treatment at 90°C for 40 min. About 5% of the initial activity was lost after boiling (100°C) for 40 min. Further treatment at 110°C for 40 min induced activity loss, resulting in 50% of the maximal activity. These results indicate that CGTase from strain B1001 is extremely thermostable in comparison with other known bacterial CGTases.
FIG. 6.
Effects of temperature (A) and pH (B) on enzyme activity. (A) Starch-degrading activity (●) and CD synthesis activity (▴) were measured at pH 5.0 and various temperatures. (B) Starch-degrading activity (●, ○) and CD synthesis activity (▴, ▵) were measured at various pHs and 90°C (open symbols) or 110°C (solid symbols). The values are shown as relative activity. The activity under the optimized condition is indicated as 100%. Each experiment was performed in triplicate.
CD production.
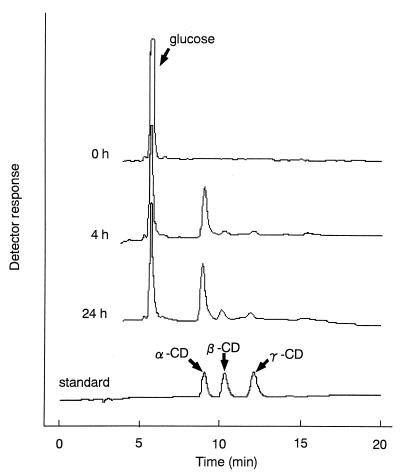
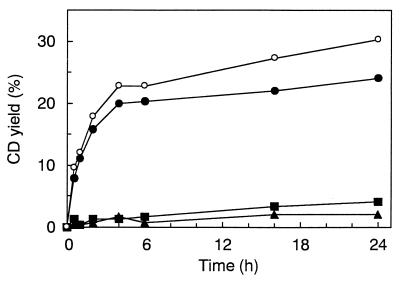
Production of CDs was analyzed by HPLC (Fig. 7). α-, β-, and γ-CDs produced from starch by purified enzyme are clearly indicated. The profiles of CD production with time were analyzed (Fig. 8). B1001 CGTase produced α-CD as the major compound. The yield of 12% conversion of soluble starch into CDs could be obtained after 1 h; the production ratio at that point was 92% α-CD, 4% β-CD, and 4% γ-CD. After 24 h, the conversion yield was 30% and the production profile was still biased toward α-CD formation (79% α-CD, 14% β-CD, and 7% γ-CD).
FIG. 7.
HPLC analysis of the formation of CDs. The reaction mixture containing 2.5% soluble starch was incubated with 8 mU of the enzyme (as CD synthesis activity). Samples were taken at various times during incubation (90°C) after addition of the purified enzyme. The amounts of CDs were determined by HPLC as described in Materials and Methods. The CD standard is a mixture of α-, β-, and γ-CDs.
FIG. 8.
Time course of CD production by purified CGTase. CDs produced at the indicated reaction times were separated by HPLC as described in the text. All values are the average of two separate experiments. Symbols: ●, α-CD; ■, β-CD; ▴, γ-CD; ○, total of α-, β-, and γ-CDs.
Under industrial CD production conditions, CGTase is incubated with high concentrations of starch. Thus, B1001 CGTase was incubated at 96°C for 24 h with 30% (wt/vol) cornstarch (with no pH adjustment; the pH was 4.5). The enzyme converted 34.4% of the starch into CDs, and the production ratios were 69% α-CD, 20% β-CD, and 11% γ-CD (data not shown).
N-terminal sequence.
The N-terminal amino acid sequence of the purified B1001 CGTase was determined. The sequence is shown in comparison with the N-terminal sequences of CGTase from various sources in Fig. 9. Interestingly, the sequence (positions 13 to 20) of B1001 CGTase shows significant similarity to those of known CGTases, suggesting that archaeal CGTase has an evolutionary relationship to bacterial CGTase.
FIG. 9.
Comparison of the N-terminal sequences between B1001 CGTase and other CGTases. The highly homologous regions are shaded. Abbreviations: B. ma, Bacillus macerans; B. li, B. licheniformis; B. sp, Bacillus sp.; B. st, B. stearothermophilus; T. th, Thermoanaerobacterium thermosulfurigenes; T. sp, Thermoanaerobacter sp.; B1001, Thermococcus sp. strain B1001.
DISCUSSION
On the basis of its G+C content and 16S rRNA sequence, strain B1001 clearly belongs to the genus Thermococcus. This classification is consistent with the morphological and physiological characteristics of strain B1001, which are similar to the characteristics of this genus. The genus Thermococcus includes nine described species: T. celer (50), T. litoralis (37), T. stetteri (32), T. profundus (23), T. peptonophilus (14), T. chitonophagus (16), T. fumicolans (13), T. alcaliphilus (20), and T. zilligii (40). All species and strain B1001 are cocci or irregular cocci, 0.5 to 2 μm in diameter, and are anaerobes that grow preferentially on peptides and yeast extract. Elemental sulfur has always been found to considerably stimulate growth, with the concomitant formation of H2S. Typical optimal temperatures are 80 to 88°C (75°C for T. stetteri and T. chitonophagus), and the G+C contents are approximately 40 to 50 mol%, except for those of T. celer (57 mol%) and T. litoralis (38 mol%). Thermococcus strains show a unique bias for antibiotic sensitivity. Strain B1001 resembles T. litoralis, T. stetteri, T. profundus, T. peptonophilus, and T. fumicolans in its susceptibility to 100 μg of rifampin per ml (T. celer and T. zilligii have no sensitivity).
The effects of pH on the starch-degrading activity and the CD synthesis activity were virtually identical, but the effects of temperature on these two activities were not. To explain this different behavior, temperature, the reaction mechanism of CGTase has to be taken into account. Since the catalytic residues of CGTase are proposed to be equivalent to those of α-amylases, CGTase will cleave the α-1,4-glucosidic bond of amylose in the same way as α-amylases do. The transglycosylation reaction of CGTase is operated by a “ping-pong” mechanism (35). In this mechanism, the transglycosylation occurs after the reducing side of the cleaved amylose is released from the enzyme. Then the enzyme transfers the newly formed reducing end of the substrate either to the nonreducing end of a separate linear acceptor molecule or glucose (the disproportionation reaction) or to its own nonreducing end (the cyclization reaction or CD synthesis reaction). The hydrolysis reaction (the starch-degrading reaction) will occur when this intermediate is attacked nucleophilically by a water molecule. For preferential CD synthesis, the efficient formation of the helical structure of amylose in the active-site cleft of enzyme is required (10, 36). In a crystal structure, amylose can occur as a single helix with six to eight glucose molecules in one helical turn (27). The most widely accepted hypothesis describes amylose in solution having an interrupted coil-like structure composed of helical and nonhelical segments (44). Therefore, the formation of CD by CGTase can be explained as a consequence of a preferential helical structure of the amylose in solution. However, the high temperature will destabilize the helical structure of the amylose, resulting in a shift to random structure. Accordingly, it is considered that the reaction at high temperature by B1001 CGTase resulted in a shift toward the starch-degrading reaction.
There are several processing difficulties in the present CD production system. The first step in the industrial production of CDs from starch is liquefaction. This is carried out by a bacterial thermostable α-amylase in a jetcooking process at 105 to 110°C and pH 6.0 to 6.5. However, further α-amylase treatment reduces the CD yield, because the maltodextrins, oligosaccharides, and glucose formed during this process act as acceptors, so that the coupling reaction (the degradation of CD) is accelerated. Another problem relating to the pH of liquefaction is the need to raise the pH of the 30 to 35% (wt/vol) starch slurry (the pH in general is 4.5 to 5.0) to pH 6.0 to 6.5. This pH adjustment requires the costly addition of acid-neutralizing chemicals. Moreover, in the next CD formation step, the pH must be adjusted to the optimum for CGTase. To overcome these two complications, thermostable CGTase of Thermoanaerobacter sp. as the liquefying enzyme can be applied applicable (38). In addition, B1001 CGTase is the most thermoactive and thermostable enzyme in reported CGTases and can react at acidic pH ranges (pH 4 to 5). Therefore, B1001 CGTase is more applicable than that of Thermoanaerobacter sp. for industrial application as a liquefying and CD-forming enzyme without pH adjustment prior to reaction by jetcooking.
Depending on the most abundant type of CD produced, CGTase is sometimes classified into three types (α-, β-, and γ-CGTase). The B1001 enzyme is the α-CGTase, which was reported from B. macerans, B. stearothermophilus, and K. pneumoniae (22). In general, α-CD formed mainly at early stage of the reaction is decomposed and α-, β-, and γ-CD are produced from decomposition products. Interestingly, B1001 CGTase produced predominantly α-CD in the later reaction as well as in the initial reaction. B1001-derived CGTase, which produced mainly α-CD with a small amount of β-CD and γ-CD from starch, can provide advantages in α-CD manufacturing.
We are currently working on isolation of the CGTase gene by using an oligonucleotide mixture as a probe that corresponds to the N-terminal sequence of the CGTase.
REFERENCES
- 1.Balch W E, Fox G E, Magrum L J, Woese C R, Wolfe R S. Methanogens: reevaluation of a unique biological group. Microbiol Rev. 1979;43:260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balch W E, Wolfe R S. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanessulfonic acid (HS-CoM)-dependent growth of Methanobactiumm ruminantium in a pressurized atmosphere. Appl Environ Microbiol. 1976;32:781–791. doi: 10.1128/aem.32.6.781-791.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernfeld P. Amylases α- and β- Methods Enzymol. 1955;1:149–158. [Google Scholar]
- 4.Bragger J M, Daniel R M, Coolbear T, Morgan H W. Very stable enzymes from extremely thermophilic archaebacteria and eubacteria. Appl Microbiol Biotechnol. 1989;31:556–561. [Google Scholar]
- 5.Brown S H, Kelly R M. Characterization of amylolytic enzymes, having both α-1,4 and α-1,6 hydrolytic activity, from the thermophilic archaea Pyrococcus furiosus and Thermococcus litoralis. Appl Environ Microbiol. 1993;59:2614–2621. doi: 10.1128/aem.59.8.2614-2621.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canganella F, Andrade C M, Antranikian G. Characterization of amylolytic and pullulytic enzymes from thermophilic archaea and from a new Fervidobacterium species. Appl Microbiol Biotechnol. 1994;42:239–245. [Google Scholar]
- 7.Chung Y C, Kobayashi T, Kanai H, Akiba T, Kudo T. Purification and properties of extracellular amylase from the hyperthermophilic archaeon Thermococcus profundus DT5432. Appl Environ Microbiol. 1995;61:1502–1506. doi: 10.1128/aem.61.4.1502-1506.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong G, Vieille C, Savchenko A, Zeikus J G. Cloning, sequencing, and expression of the gene encoding extracellular α-amylase from Pyrococcus furiosus and biochemical characterization of the recombinant enzyme. Appl Environ Microbiol. 1997;63:3569–3576. doi: 10.1128/aem.63.9.3569-3576.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubois M, Gilles K A, Hamilton J K, Rebers P A, Smith F. Colorimetric method for determination of sugars related substances. Anal Chem. 1956;28:350–356. [Google Scholar]
- 10.Fujiwara S, Kakihara H, Sakaguchi K, Imanaka T. Analysis of mutations in cyclodextrin glucanotransferase from Bacillus stearothermophilus which affect cyclization characteristics and thermostability. J Bacteriol. 1992;174:7478–7481. doi: 10.1128/jb.174.22.7478-7481.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujiwara S, Kakihara H, Woo K B, Lejeune A, Kanemoto M, Sakaguchi K, Imanaka T. Cyclization characteristics of cyclodextrin glucanotransferase are conferred by the NH2-terminal region of the enzyme. Appl Environ Microbiol. 1992;58:4016–4025. doi: 10.1128/aem.58.12.4016-4025.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gantelet H, Duchiron F. Purification and properties of a thermoactive and thermostable pullulanase from Thermococcus hydrothermalis, a hyperthermophilic archaeon isolated from a deep-sea hydrothermal vent. Appl Microbiol Biotechnol. 1998;49:770–777. [Google Scholar]
- 13.Godfroy A, Meunier J-R, Guezennec J, Iesongeur F, Raguénès G, Rimbault A, Barbier G. Thermococcus fumicolans sp. nov., a new hyperthermophilic archaeon isolated from a deep-sea hydrothermal vent in the North Fiji Basin. Int J Syst Bacteriol. 1996;46:1113–1119. doi: 10.1099/00207713-46-4-1113. [DOI] [PubMed] [Google Scholar]
- 14.González J M, Kato C, Horikoshi K. Thermococcus peptonophilus sp. nov., a fast-growing, extremely thermophilic archaebacterium isolated from deep-sea hydrothermal vents. Arch Microbiol. 1995;164:159–164. [PubMed] [Google Scholar]
- 15.Hashimoto H The Amylase Research Society of Japan (ed.), editors . Handbook of amylase and related enzymes. Tokyo, Japan: Pergamon Press; 1988. Production and application of cyclodextrins; pp. 233–238. [Google Scholar]
- 16.Huber R, Stöhr J, Hohenhaus S, Rachel R, Burggraf S, Jannasch H W, Stetter K O. Thermococcus chitonophagus sp. nov., a novel, chitin-degrading, hyperthermophilic archaeum from a deep-sea hydrothermal vent environment. Arch Microbiol. 1995;164:255–264. [Google Scholar]
- 17.Jøgensen S, Vorgias C E, Antranikian G. Cloning, sequencing, characterization, and expression of an extracellular α-amylase from the hyperthermophilic archaeon Pyrococcus furiosus in Escherichia coli and Bacillus subtilis. J Biol Chem. 1997;272:16335–16342. doi: 10.1074/jbc.272.26.16335. [DOI] [PubMed] [Google Scholar]
- 18.Kaneko T, Kato T, Nakamura N, Horikoshi K. Spectrophotometric determination of cyclization activity of β-cyclodextrin-forming cyclomaltodextrin glucanotransferase. J Jpn Soc Starch Sci. 1987;34:45–48. [Google Scholar]
- 19.Kato T, Horikoshi K. Colorimetric determination of γ-cyclodextrin. Anal Chem. 1984;56:1738–1740. [Google Scholar]
- 20.Keller M, Braun F-J, Dirmeier R, Hafenbradl D, Burggraf S, Rachel R, Stetter K O. Thermococcus alcaliphilus sp. nov., a new hyperthermophilic archaeum growing on polysulfide at alkaline pH. Arch Microbiol. 1995;164:390–395. [PubMed] [Google Scholar]
- 21.Kitahata S The Amylase Research Society of Japan (ed.), editors . Handbook of amylase and related enzymes. Tokyo, Japan: Pergamon Press; 1988. Cyclomaltodextrin glucanotransferase; pp. 154–163. [Google Scholar]
- 22.Kitahata S The Amylase Research Society of Japan (ed.), editors . Enzyme chemistry and molecular biology of amylases and related enzymes. Tokyo, Japan: CRC Press; 1995. Cyclomaltodextrin glucanotransferase; pp. 6–17. [Google Scholar]
- 23.Kobayashi T, Kwak Y S, Akiba T, Kudo T, Horikoshi K. Thermococcus profundus sp. nov., a new hyperthermophilic archaeon isolated from a deep-sea hydrothermal vent. Syst Appl Microbiol. 1994;17:232–236. [Google Scholar]
- 24.Koch R, Spreinat A, Lemke K, Antranikian G. Purification and properties of a hyperthermoactive α-amylase from the archaeobacterium Pyrococcus woesei. Arch Microbiol. 1991;155:572–578. [Google Scholar]
- 25.Koch R, Zablowski P, Spreinat A, Antranikian G. Extremely thermostable amylolytic enzyme from the archaebacterium Pyrococcus furiosus. FEMS Microbiol Lett. 1990;71:21–26. [Google Scholar]
- 26.Kometani T, Terada Y, Nishimura T, Takii H, Okada S. Purification and characterization of cyclodextrin glucanotransferase from an alkalophilic Bacillus species and transglycosylation at alkaline pHs. Biosci Biotechnol Biochem. 1994;58:517–520. [Google Scholar]
- 27.Kubik S, Höller O, Steinert A, Tolksdorf M, Van der Leek Y, Wulff G. Molecular inclusion within polymeric carbohydrate matrices. In: van Bekkum H, Röber H, Voragen A G J, editors. Carbohydrates as organic raw materials. III. Weinheim, Germany: VCH; 1996. pp. 169–187. [Google Scholar]
- 28.Kuriki T, Imanaka T. Nucleotide sequence of the neopullulanase gene from Bacillus stearothermophilus. J Gen Microbiol. 1989;135:1521–1528. doi: 10.1099/00221287-135-6-1521. [DOI] [PubMed] [Google Scholar]
- 29.Lee J T, Kanai H, Kobayashi T, Akiba T, Kudo T. Cloning, nucleotide sequence, and hyperexpression of α-amylase gene from an archaeon, Thermococcus profundus. J Ferment Bioeng. 1996;82:432–438. [Google Scholar]
- 30.Lejeune A, Sakaguchi K, Imanaka T. A spectrophotometric assay for the cyclization activity of cyclomaltohexaose (α-cyclodextrin) glucanotransferase. Anal Biochem. 1989;181:6–11. doi: 10.1016/0003-2697(89)90385-0. [DOI] [PubMed] [Google Scholar]
- 31.Mesbah M, Premachandran U, Whitman W B. Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol. 1989;39:159–167. [Google Scholar]
- 32.Miroshnichenko M L, Bonch-Osmolovskaya E A, Neuner A, Kostrikina N A, Chernych N A, Alekseev V. Thermococcus stetteri sp. nov., a new extremely thermophilic marine sulfur-metabolizing archaebacterium. Syst Appl Microbiol. 1989;12:257–262. [Google Scholar]
- 33.Mori S, Hirose S, Oya T, Kitahata S. Purification and properties of cyclodextrin glucanotransferase from Brevibacterium sp. no. 9605. Biosci Biotechnol Biochem. 1994;58:1968–1972. [Google Scholar]
- 34.Nakajima R, Imanaka T, Aiba S. Comparison of amino acid sequences of eleven different α-amylases. Appl Microbiol Biotechnol. 1986;23:355–360. [Google Scholar]
- 35.Nakamura A, Haga K, Yamane K. The transglycosylation reaction of cyclodextrin glucanotransferase is operated by a Ping-Pong mechanism. FEBS Lett. 1994;337:66–70. doi: 10.1016/0014-5793(94)80631-4. [DOI] [PubMed] [Google Scholar]
- 36.Nakamura A, Haga K, Yamane K. Four aromatic residues in the active center of cyclodextrin glucanotransferase from alkalophilic Bacillus sp. 1011: effects of replacements on substrate binding and cyclization characteristics. Biochemistry. 1994;33:9929–9936. doi: 10.1021/bi00199a015. [DOI] [PubMed] [Google Scholar]
- 37.Neuner A, Jannasch H W, Belkin S, Stetter K O. Thermococcus litoralis sp. nov.: a new species of extremely thermophilic marine archaebacteria. Arch Microbiol. 1990;153:205–207. [Google Scholar]
- 38.Norman B E, Jörgensen S T. Thermoanaerobacter sp. CGTase: its properties and application. Denpun Kagaku. 1992;39:101–108. [Google Scholar]
- 39.Reysenbach A-L, Pace N R. Reliable amplification of hyperthermophilic archaeal 16S rRNA genes by the polymerase chain reaction. In: Robb F T, Place A R, editors. Archaea, a laboratory manual. Thermophiles. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1995. pp. 101–105. [Google Scholar]
- 40.Ronimus R S, Reysenbach A-L, Musgrave D R, Morgan H W. The phylogenetic position of the Thermococcus isolate AN1 based on 16S rRNA gene sequence analysis: a proposal that AN1 represents a new species, Thermococcus zilligii sp. nov. Arch Microbiol. 1997;168:245–248. doi: 10.1007/s002030050495. [DOI] [PubMed] [Google Scholar]
- 41.Rüdiger A, Jorgensen P L, Antranikian G. Isolation and characterization of a heat-stable pullulanase from the hyperthermophilic archaeon Pyrococcus woesei after cloning and expression of its gene in Escherichia coli. Appl Environ Microbiol. 1995;61:567–575. doi: 10.1128/aem.61.2.567-575.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 43.Svensson B. Protein engineering in the α-amylase family: catalytic mechanism, substrate specificity, and stability. Plant Mol Biol. 1994;25:141–157. doi: 10.1007/BF00023233. [DOI] [PubMed] [Google Scholar]
- 44.Szejtli J. Helical and cyclic structures in starch chemistry. ACS Symp Ser. 1991;458:2–10. [Google Scholar]
- 45.Tachibana Y, Leclere M M, Fujiwara S, Takagi M, Imanaka T. Cloning and expression of the α-amylase gene from the hyperthermophilic archaeon Pyrococcus sp. KOD1, and characterization of the enzyme. J Ferment Bioeng. 1996;82:224–232. [Google Scholar]
- 46.Takata H, Kuriki T, Okada S, Takesada Y, Iizuka M, Minamiura N, Imanaka T. Action of neopullulanase: neopullulanase catalyzes both hydrolysis and transglycosylation at α-(1→4)- and α-(1→6)-glucosidic linkages. J Biol Chem. 1992;267:18447–18452. [PubMed] [Google Scholar]
- 47.Thompson J D, Higgins D S, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Williams W J. Handbook of anion determination. London, United Kingdom: Butterworths; 1979. pp. 570–572. [Google Scholar]
- 49.Wind R D, Liebl W, Buitelaar R M, Penninga D, Spreinat A, Dijkhuizen L, Bahl H. Cyclodextrin formation by the thermostable α-amylase of Thermoanaerobacterium thermosulfurigenes EM1 and reclassification of the enzyme as a cyclodextrin glycosyltransferase. Appl Environ Microbiol. 1995;61:1257–1265. doi: 10.1128/aem.61.4.1257-1265.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zillig W, Holz I, Janekovic D, Schäfer W, Reiter W D. The archaebacterium Thermococcus celer represents, a novel genus within the thermophilic branch of the archaebacteria. System Appl Microbiol. 1983;4:88–94. doi: 10.1016/S0723-2020(83)80036-8. [DOI] [PubMed] [Google Scholar]