Abstract
Cefiderocol (CFDC) is a novel chlorocatechol-substituted siderophore antibiotic approved to treat complicated urinary tract infections (cUTI) and hospital-acquired and ventilator-acquired pneumonia (HAP/VAP). Previous work determined that albumin-rich human fluids increase the minimum inhibitory concentration (MICs) of Acinetobacter baumannii against CFDC and reduce the expression of genes related to iron uptake systems. This latter effect may contribute to the need for higher concentrations of CFDC to inhibit growth. The presence of human urine (HU), which contains low albumin concentrations, did not modify MIC values of two carbapenem-resistant A. baumannii. Levels of resistance to CFDC were not modified by HU in strain AMA40 but were reduced in strain AB5075. Expanding the studies to other carbapenem-resistant A. baumannii isolates showed that the presence of HU resulted in unmodified or reduced MIC of CDFC values. The expression of piuA, pirA, bauA, and bfnH determined by qRT-PCR was enhanced in A. baumannii AMA40 and AB5075 by the presence of HU in the culture medium. All four tested genes code for functions related to recognition and transport of ferric-siderophore complexes. The effect of HU on expression of pbp1, pbp3, blaOXA-51-like, blaADC, and blaNDM-1, genes associated with resistance to β-lactams, as well as genes coding for efflux pumps and porins was variable, showing dependence with the strain analyzed. We conclude that the lack of significant concentrations of albumin and free iron in HU makes this fluid behave differently from others we tested. Unlike other albumin rich fluids, the presence of HU does not impact the antibacterial activity of CFDC when tested against A. baumannii.
Subject terms: Microbiology, Antimicrobials, Clinical microbiology
Introduction
Acinetobacter baumannii is a versatile pathogen responsible for hospital-acquired and ventilator-associated pneumonia (HAP/VAP), bacteremia, as well as complicated urinary tract (cUTI) and wound infections, especially in intensive care unit patients1–4. The World Health Organization (WHO) and the Centers for Disease Control and Prevention (CDC) identified A. baumannii as a “top priority” for the research and development of new antimicrobial therapies5, 6. Despite A. baumannii being the causative agent of common and catheter-associated cUTIs, and these infections accounting for about 20% of A. baumannii clinical isolates7, 8. The majority of research on this bacterium’s pathogenicity has focused on pneumonia and bloodstream infections to the detriment of the mechanisms that lead to infections in the human urinary tract.
Changes in gene expression when A. baumannii is exposed to human blood or serum has attracted significant interest. The presence of these fluids creates a hostile environment by limiting nutrients, such as iron, or actively damaging the bacterial cells through immunity mechanisms9, 10. For example, when A. baumannii is grown in human serum albumin (HSA), this pathogen responds by modifying the expression of critical genes10–16, such as those related to iron uptake systems and resistance to β-lactams17. Another effect that was observed in these conditions is an increase in the minimum inhibitory concentrations (MICs) to cefiderocol (CFDC), a novel antibiotic approved by Food and Drug Administration (https://www.accessdata.fda.gov/drugsatfda_docs/label/2019/209445s000lbl.pd). Since CFDC is a cephalosporin antibiotic conjugated to a siderophore, transport into the bacterial periplasm to inactivate penicillin binding proteins (PBPs) depends on receptors regulated by the iron concentration in the milieu. An attractive hypothesis to explain these results is that when A. baumannii grows in the presence of HSA-containing human fluids, there is down-regulation the expression of iron uptake system genes, reducing the ability of CDFC to reach the cell’s PBPs17. Since albumin is absent or present in trace concentrations in normal HU, we hypothesize that CFDC efficacy would not be reduced when A. baumannii is exposed to HU.
Previous studies showed that CFDC is safe and effective for the treatment of cUTI in patients at risk of multidrug-resistant (MDR) Gram-negative infections, including secondary bacteremia18, 19. In addition, CFDC did not demonstrate inferiority to imipenem-cilastatin19. However, patients with carbapenem-resistant infections were not included in this study. In this work, we evaluate the effect of HU on representative carbapenem-resistant A. baumannii (CRAB) strains with different genetic backgrounds focusing on changes in CFDC MICs and expression levels of iron uptake TonB-dependent receptors and β-lactam resistance genes.
Results and discussion
cUTIs and A. baumannii: a review of the clinical problem
In order to provide clinical context to this work, we focused our attention herein on A. baumannii infections in Argentina as a representative country to illustrate the magnitude of the medical problem.
A review of data analysis indicated that the incidence of CRAB in Argentina pre-COVID-19 was 8 in 10,000 patients, while after the arrival of COVID-19, the incidence in 2020 was 32 in 10,000. In addition, three out of the 32 infections were A. baumannii recovered from urinary sites while the pneumonia and bloodstream infections represent 55% and 25% respectively (Personal communication provided by the National Reference Laboratory and the National Commission for the Control of Antimicrobial Resistance, National Ministry of Health). On the other hand, during the period January 2021–2022, a total of 1758 strains isolated from urinary sites were recorded at the main adult hospital in the province of Tucumán (northern Argentina). Nineteen of these isolates (1.08%), corresponded to A. baumannii, of which 17 isolates (89.47%) were CRAB. While 86 A. baumannii strains (16.86%) were recovered from pneumonia infection out of a total of 510 isolates, of which 84 strains (97.67%) corresponded to CRAB. In bloodstream infections, out of a total of 502 isolates, 25 corresponded to A. baumannii (4.98%), of which 20 isolates (80%) were CRAB.
Modulation of the expression levels of genes associated with iron-uptake systems in the presence of HU
Iron uptake systems are expressed in early stages of A. baumannii infections under iron-restricted conditions20. The iron transporters bauA and bfnH, which are coupled to a TonB energy transduction system needed to translocate the siderophore-iron complex into the periplasm, were characterized in A. baumannii21. Jacobs et al.22 reported that the expression of bauA and bfnH were enhanced during an infection caused by A. baumannii. In addition, these genes were identified as vaccine candidates using reverse vaccinology23. Moreover, piuA and pirA encoded for two novel TonB-dependent receptors of A. baumannii, which are associated with iron uptake24.
To study A. baumannii response to HU (fluids with traces or undetectable level of HSA) and its possible effect on the expression level of iron transporters genes (piuA, pirA, bauA, and bfnH), we performed quantitative RT-PCR (qRT-PCR) assays. Total RNA extracted from two representative CRAB model strains (AB5075 and AMA40) cultured in iron-depleted cation adjusted Mueller Hinton broth (CAMHB), or CAMHB supplemented with 50% HU was used.
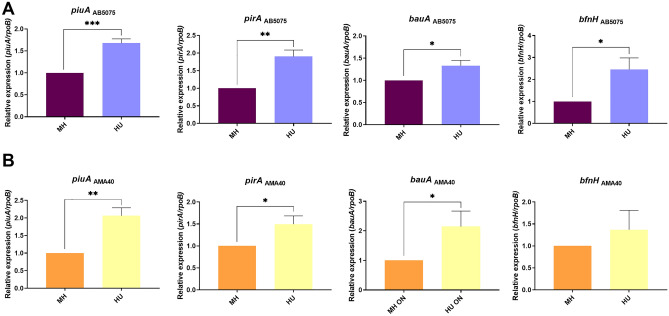
The expression of piuA, pirA, bauA, and bfnH was significantly enhanced in both strains when cultured in the presence of HU (Fig. 1A,B). Previous results showed that when the AB5075, AMA16 and AB0057 CRAB strains were exposed to pure 3.5% HSA, or 4% HPF or 100% HS, fluids containing HSA, the transcriptional expression levels of these genes were down regulated for most of the conditions and strains studied17. The total Fe concentration was determined in 3.5% HSA and HU, achieving 72 µM; no additional iron was detected, respectively (as determined by a colorimetric iron assay kit, Sigma-Aldrich, MO, USA). Moreover, the total Fe concentration in HPF is about 130 µM12. Therefore, we conclude that in HU, the expression levels of genes related to the acquisition of iron are upregulated (Fig. 1), in contrast to the effect observed with fluids containing high content of HSA.
Figure 1.
Genetic analysis of iron uptake genes of AB5075 (A) and AMA40 (B) A. baumannii strains. qRT-PCR of genes associated with iron uptake, pirA, piuA, bauA and bfnH expressed in cation adjusted Mueller Hinton or cation adjusted Mueller Hinton supplemented with 50% HU. Fold changes were calculated using double ΔCt analysis. At least three independent samples were used, and four technical replicates were performed from each sample. The cation adjusted Mueller Hinton was used as reference. Statistical significance (p < 0.05) was determined by ANOVA followed by Tukey’s multiple-comparison test, one asterisks: p < 0.05; two asterisks: p < 0.01 and three asterisks: p < 0.001.
Changes in the expression levels of β-lactam resistance genes in the presence of HU
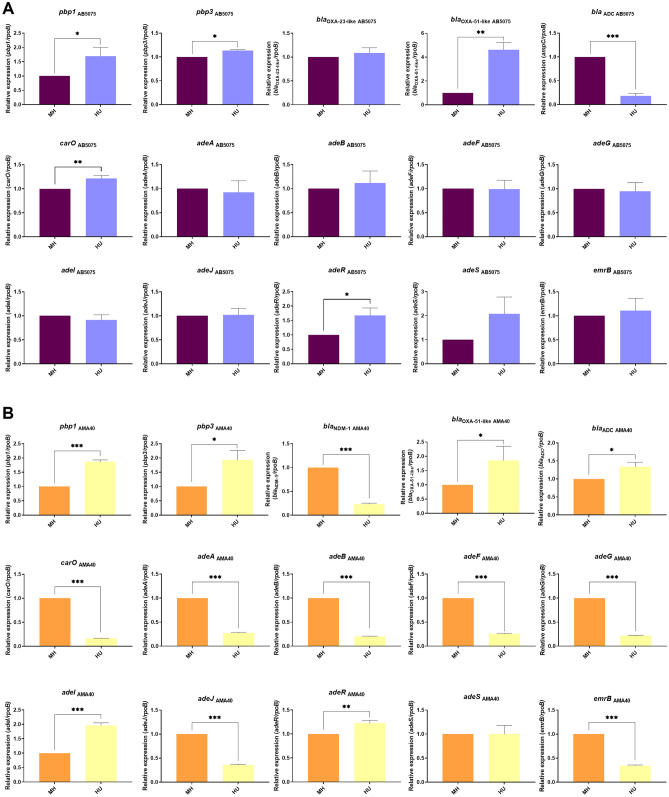
To further explore if changes in the expression of genes associated with β-lactam resistance are affected by HU in the selected CRAB strains, qRT-PCR was used to evaluate the expression of pbp1, pbp3, blaOXA-23, blaOXA-51-like, blaADC and blaNDM-1 in AB5075 and AMA40 exposed to HU (Fig. 2).
Figure 2.
Genetic analysis of β-lactamase and PBP genes of AB5075 (A) and AMA40 (B) A. baumannii strains. qRT-PCR of genes associated with β-lactams resistance, efflux pump, and porins expressed in cation adjusted Mueller Hinton, cation adjusted Mueller Hinton supplemented with 50% HU. Fold changes were calculated using double ΔCt analysis. At least three independent samples were used. LB was used as the reference condition. Statistical significance (p < 0.05) was determined by ANOVA followed by Tukey’s multiple-comparison test, one asterisks: p < 0.05; two asterisks: p < 0.01, and three asterisks: p < 0.001.
This transcriptional analysis revealed an up-regulation in the expression levels of pbp1, pbp3, and blaOXA-51-like in both strains analyzed. In contrast, a decrease in the level of expression of blaADC in AB5075, and blaNDM-1 in AMA40 clinical isolate was observed. Changes were not observed for blaOXA-23 in AB5075, while an increase in blaADC expression was seen for AMA40 (Fig. 2).
To further determine if changes at the transcriptional levels of genes codifying for efflux pumps such as AdeABC, AdeFGH, AdeIJK, and EmrAB, or the CarO porin could contribute to the altered CFDC resistance, qRT-PCR experiments were also carried out. The results showed an increase in the expression levels of carO in the AB5075 strain, while there was a decrease in the expression of this gene in the AMA40 strain (Fig. 2).
The analysis efflux pump associated genes varies according to the strain and gene analyzed (Fig. 2). The expression level of seven efflux pumps genes was significantly down-regulated in AMA40 under HU (Fig. 2), while in AB5075 most of the genes did not show a significant change in the expression when exposed to HU (Fig. 2). These results suggest that strains with different carbapenem resistance mechanisms could respond differently to modifications of external conditions.
RNA sequencing analysis of two carbapenem-resistant A. baumannii strains expose to HU
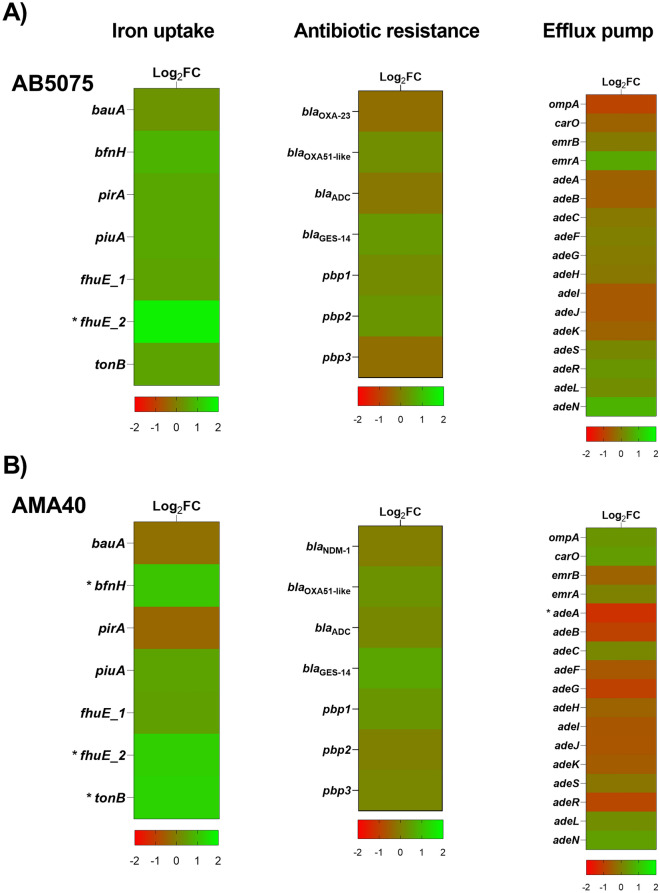
The RNA-seq analysis of A. baumannii strains AB5075 and AMA40 revealed that HU significantly affect the expression of 264 and 455 coding genes, respectively (FDR < 0.05 and log2 fold change > 1). In AB5075 strain, 148 genes were up-regulated and 116 were downregulated in the presence of HU. Furthermore, in AMA40 strain, 262 genes were up-regulated and 193 were downregulated in the presence of HU (Table S1).
In the present study, our analysis was focused on genes related to iron uptake, β-lactam resistance, and efflux pumps (Fig. 3 and Table S2). Results obtained confirmed the results determined by qRT-PCR. In addition, the RNA-seq study shows that the expression of fhuE_2 was significantly up-regulated when both strains were exposed to HU. The expression levels of bfnH and tonB were also increased under HU in AMA40. In addition, the expression of adeA was significantly down-regulated in AMA40 (Fig. 3 and Table S2).
Figure 3.
Genetic analysis of iron uptake, antibiotic resistance and efflux pump coding genes by RNA-seq. Heatmap outlining the differential expression of genes associated with iron uptake, antibiotic resistance and efflux pump in presence of HU in A. baumannii AB5075 (A) and AMA40 (B) strains. The asterisks represent the differentially expressed genes (DEGs) (adjusted p < 0.05 with log2fold change > 1).
CRAB strains susceptibility to CFDC in the presence of HU
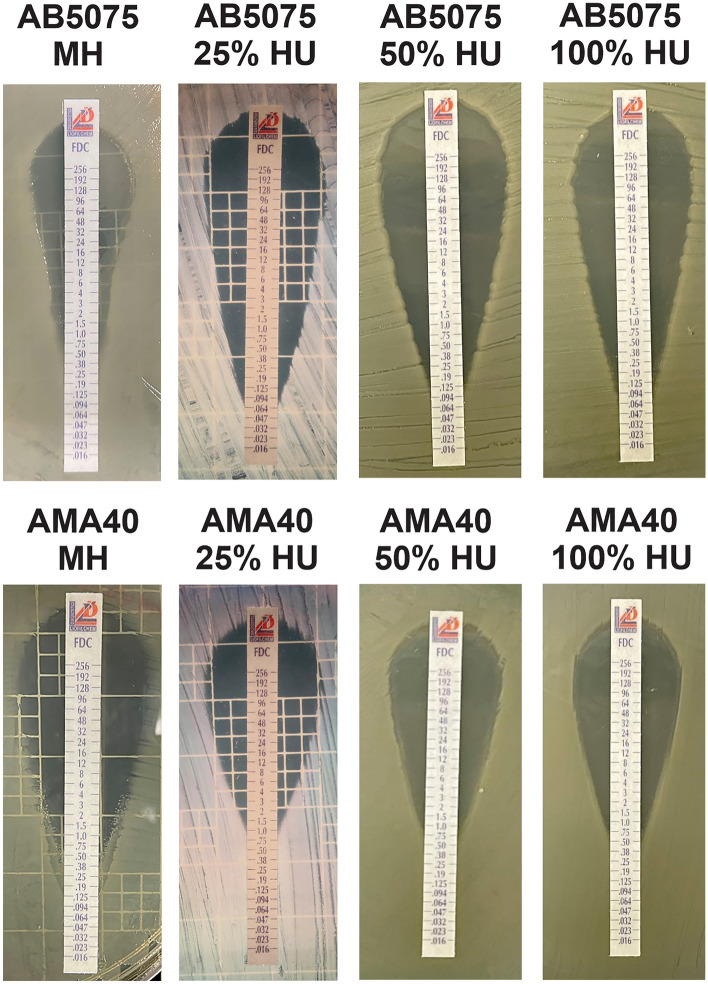
Based on these results, AB5075 and AMA40 cells grown in 100% HU or CAMHB that was supplemented with 0%, 25%, or 50% HU were used to determine CFDC MICs. Differences were not observed in CFDC MICs for AMA40 under the tested conditions. However, a twofold decrease in CFDC MIC was observed when AB5075 cells were grown in the presence of HU (Fig. 4 and Table 1). As the level of expression of blaNDM-1 was down-regulated when exposed to HU in the strain AMA40, MICs for imipenem (IMP) and meropenem (MEM) were performed. This analysis showed that MICs differences for MER was not seen; and a slight decrease (onefold decrease) was seen when testing susceptibility to IMI in presence of HU. We attribute this to the decrease in NDM-1 expression observed when AMA40 was grown in HU.
Figure 4.
Effect of HU on the antimicrobial susceptibility of A. baumannii strains AB5075 and AMA40 strains grew in MH broth, MH broth plus 25, 50 or 100% HU were used to performed cefiderocol (CFDC) susceptibility. Minimum inhibitory concentration (MIC) on cation adjusted Mueller Hinton agar was performed by MTS (Liofilchem S.r.l., Italy) following manufacturer’s recommendations.
Table 1.
Minimal Inhibitory Concentrations (MICs) of cefiderocol (CFDC) for AB5075 and AMA40 Carbapenem-resistant Acinetobacter baumanii representative strains performed using CFDC MTS strips (Liofilchem S.r.l., Italy) on Iron-depleted CAMHA (Cation Adjusted Mueller Hinton Agar).
| MICs E-test (mg/L) | ||||
|---|---|---|---|---|
| Strain | MH | MH 25% HU | MH 50% HU | 100% HU |
| AB5075 | 0.5 (S) | 0.094 (S) | 0.094 (S) | 0.125 (S) |
| AMA40 | 0.5 (S) | 1.5 (S) | 0.75 (S) | 0.5 (S) |
HU: Human Urine. A. baumannii cells were cultured in MH, MH supplemented with HU 25%, 50% or 100% HU.
To expand our observations to other CRAB strains with different genetic backgrounds and carbapenem resistance mechanisms, CFDC MICs of 11 CRAB selected strains grown in CAMHB or 50% HU were determined. The model strains AB0057 and AYE were included among the tested strains. A previously observed increase in the AB0057 MIC to CDFC when this isolate was cultured in presence of HPF was noted17. However, in this case, changes in MIC were not observed under HU conditions (Fig. S1 and Table S3).
Taken together we conclude that the presence of HU resulted in either an increase in susceptibility or no changes in the CFDC MIC in 4 and 6 out of the 11 tested strains, respectively. Lastly, a broader time span was employed to study the adaptive response of the A. baumannii to CFDC. To this end, MICs were performed with 48- and 72-h exposure to HU. Results show that CFDC MIC was twofold increase when AB5075 was expose to HU for 72 h, while was onefold increase in AMA40 (Table S4). These results suggest that there is not a clear adaptive response of A. baumannii to CFDC when exposed to HU in the condition tested.
Concluding remarks
UTIs are one of the most common bacterial infections caused by MDR Gram-negative bacteria. Among A. baumannii clinical isolates, one in five are from urinary sites according to a retrospective analysis of all Acinetobacter isolates identified in the BJC Healthcare System (BJC) from January 2007 through August 2017. Acinetobacter spp. pathogenicity has been extensively studied in pneumonia and bloodstream infection models. Past studies showed that the potency of CFDC is reduced when bacteria are exposed to human fluids that contain HSA, presumably due to iron bound to the protein.
Here we tested the effect of another human fluid, urine, which has none or negligible amounts of HSA or free iron. Interestingly, the MIC values of CFDC were not significantly modified by the presence of HU in the testing conditions used. An attractive hypothesis to explain the lack of effect in this bodily fluid as opposed to HSA and HPF is that the regulatory factor in the latter fluids is the iron attached HSA. The implications of these observations may define the more appropriate use of this and other antibiotics to treat MDR infections. Further studies are underway to explore this hypothesis.
Materials and methods
Bacterial strains
The model carbapenem-resistant A. baumannii strains AB5075 (blaOXA-23 and blaOXA-51)12, 25 and the carbapenem-resistant clinical AMA40 (blaNDM-1 and blaOXA-51) strain26, 27 were used in this work. In addition, a total of nine clinical isolates of carbapenem-resistant A. baumannii (CRAB) were included in this study. Among these strains, AB0057 isolated in the Walter Reed Army Medical Center28 and harboring blaTEM-1, blaOXA-23 and blaADC genes was included.
RNA extraction, quantitative reverse transcription polymerase chain reaction (qRT-PCR), sequencing, and transcriptomic analysis
Overnight cultures of A. baumannii strains were diluted 1:10 in fresh iron depleted cation adjusted Mueller Hinton or cation adjusted Mueller Hinton supplemented with 50% of human urine (HU) and incubated with agitation for 18 h at 37 °C. HU was obtained from a certified vendor (Innovative Research Inc, Novi, MI, USA). RNA was extracted from each strain using the Direct-zol RNA Kit (Zymo Research, Irvine, CA, USA) following manufacturer’s instructions. Total RNA extractions were performed in three biological replicates for each condition. The extracted and DNase treated RNA was used to synthesize cDNA using the manufacturer’s protocol provided within the iScriptTM Reverse Transcription Supermix for qPCR (Bio-Rad, Hercules, CA, USA). The cDNA concentrations were adjusted to 50 ng/µL and qPCR was conducted using the qPCRBIO SyGreen Blue Mix Lo-ROX following manufacturer’s protocol (PCR Biosystems,Wayne, PA, USA). At least three biological replicates of cDNA were used in triplets and were run using the CFX96 TouchTM Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). Transcriptional levels of each sample were normalized to the transcriptional level of rpoB. The relative quantification of gene expression was performed using the comparative threshold method 2−ΔΔCt. The ratios obtained after normalization were expressed as folds of change compared with cDNA samples isolated from bacteria cultures on LB. Asterisks indicate significant differences as determined by ANOVA followed by Tukey’s multiple comparison test (p < 0.05), using GraphPad Prism (GraphPad software, San Diego, CA, USA).
RNA sequencing was outsourced to Novegene (Novogene Corporation, Sacramento,CA, USA), where the RNA-seq library preparation (Illumina, San Diego, CA, USA) and HiSeq 2500 paired-end 150 bp sequencing of three independent biological replicates in the presence or absence of HU was performed for AB5075 and AMA40 strains. Trimming of low-quality bases at the ends of the reads to a minimum length of 100 bp and removal of Illumina adaptor sequences was performed using Trimmomatic29, yielding an average of 7.6 million paired reads per sample. FastQC http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) (January 2019) was used to assess the quality of the reads before and after trimming. Burrows–Wheeler Alignment software (BWA) was used to align the RNA-seq reads to the genome of Acinetobacter baumannii30. The alignments were visualized using the Integrated Genome Viewer software31. FeatureCounts (REF) was used to calculate the read counts per gene32, and differential expression analysis was performed using DEseq33. Features exhibiting FDR < 0.05 and log2fold change > 1 were considered statistically significant. Both RNA-seq data were deposited in the Gene Expression Omnibus (GEO) database under the accession number GSE201259.
Antimicrobial susceptibility testing
Antibiotic susceptibility assays were performed following the procedures recommended by the Clinical and Laboratory Standards Institute (CLSI)34. After OD adjustment, 100 µL of cells grown in fresh cation adjusted Mueller Hinton broth (CAMHB) or CAMHB supplemented with 25, 50 or 100% HU were inoculated on CAMHB agar plates as previously described12. Antimicrobial commercial E-strips (Liofilchem S.r.l., Roseto degli Abruzzi, Italy) for cefiderocol (CFDC) was used. Mueller–Hinton agar plates were incubated at 37 °C for 18 h. CLSI breakpoints were used for interpretation34. E. coli ATCC 25922 was used for quality control purposes.
Determination of total iron concentration
Total iron concentration of HU and LB supplemented with 3.5% HSA was determined using the Iron Assay Kit (Sigma-Aldrich) following the manufacturer’s recommendations. In addition, the iron content was also measured in LB supplemented with 4% HPF confirming previous published results12.
Supplementary Information
Acknowledgements
The authors are grateful to Mariel Agustina Cáceres from Hospital “Ángel C. Padilla”, Province of Tucumán for statistical data provided.
Author contributions
B.N., J.E., M.R.T., T.S., C.P., N.G., V.M., F.P., C.R., R.A.B., M.E.T. and M.S.R. conceived the study and designed the experiments. B.N., J.E., M.R.T., T.S., C.P., N.G., V.M., F.P., C.R., R.S. and M.S.R. performed the experiments and genomics and bioinformatics analyses. M.R.T., T.S., F.P., R.A.B., M.E.T. and M.S.R. analyzed the data and interpreted the results. R.A.B., M.E.T. and M.S.R. contributed reagents/materials/analysis tools. M.R.T., T.S., F.P., R.A.B., M.E.T. and M.S.R. wrote and revised the manuscript. All authors read and approved the final manuscript.
Funding
The authors’ work was supported by NIH SC3GM125556 to MSR, R01AI100560, R01AI063517, R01AI072219 to RAB, and 2R15 AI047115 to MET. This study was supported in part by funds and/or facilities provided by the Cleveland Department of Veterans Affairs, Award Number 1I01BX001974 to RAB from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development and the Geriatric Research Education and Clinical Center VISN 10 to RAB. JE and CP were supported by grant MHRT 2T37MD001368 from the National Institute on Minority Health and Health Disparities, National Institute of Health. SA and AC were supported by Project RAISE, U.S. Department of Education HSI-STEM award number P031C160152. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Department of Veterans Affairs. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Department of Veterans Affairs. MRT and TS are recipient of a postdoctoral fellowship from CONICET. C.R. and R.S. are a staff member from CONICET.
Data availability
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request. The datasets generated and analyzed during the current study are available in the Gene Expression Omnibus (GEO) database repository under the accession number GSE201259.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Brent Nishimura and Jenny Escalante.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-12829-7.
References
- 1.Peleg AY, de Breij A, Adams MD, Cerqueira GM, Mocali S, Galardini M, Nibbering PH, Earl AM, Ward DV, Paterson DL, Seifert H, Dijkshoorn L. The success of acinetobacter species; genetic, metabolic and virulence attributes. PLoS ONE. 2012;7:e46984. doi: 10.1371/journal.pone.0046984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: Emergence of a successful pathogen. Clin. Microbiol. Rev. 2008;21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spellberg B, Bonomo RA. The deadly impact of extreme drug resistance in Acinetobacter baumannii. Crit. Care Med. 2014;42:1289–1291. doi: 10.1097/CCM.0000000000000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harding CM, Hennon SW, Feldman MF. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat. Rev. Microbiol. 2018;16:91–102. doi: 10.1038/nrmicro.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CDC. Antibiotic Resistance Threats in the United States (US Department of Health and Human Services, CDC, 2019).
- 6.WHO. Antimicrobial Resistance Global Report on surveillance 2014. (World Health Organization, WHO).
- 7.Di Venanzio G, Flores-Mireles AL, Calix JJ, Haurat MF, Scott NE, Palmer LD, Potter RF, Hibbing ME, Friedman L, Wang B, Dantas G, Skaar EP, Hultgren SJ, Feldman MF. Urinary tract colonization is enhanced by a plasmid that regulates uropathogenic Acinetobacter baumannii chromosomal genes. Nat. Commun. 2019;10:2763. doi: 10.1038/s41467-019-10706-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding R, Li X, Zhang X, Zhang Z, Ma X. The epidemiology of symptomatic catheter-associated urinary tract infections in the intensive care unit: A 4-year single center retrospective study. Urol. J. 2019;16:312–317. doi: 10.22037/uj.v0i0.4256. [DOI] [PubMed] [Google Scholar]
- 9.Murray GL, Tsyganov K, Kostoulias XP, Bulach DM, Powell D, Creek DJ, Boyce JD, Paulsen IT, Peleg AY. Global gene expression profile of Acinetobacter baumannii during bacteremia. J Infect Dis. 2017;215:S52–S57. doi: 10.1093/infdis/jiw529. [DOI] [PubMed] [Google Scholar]
- 10.Quinn B, Rodman N, Jara E, Fernandez JS, Martinez J, Traglia GM, Montana S, Cantera V, Place K, Bonomo RA, Iriarte A, Ramirez MS. Human serum albumin alters specific genes that can play a role in survival and persistence in Acinetobacter baumannii. Sci. Rep. 2018;8:14741. doi: 10.1038/s41598-018-33072-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez J, Razo-Gutierrez C, Le C, Courville R, Pimentel C, Liu C, Fung SE, Tuttobene MR, Phan K, Vila AJ, Shahrestani P, Jimenez V, Tolmasky ME, Becka SA, Papp-Wallace KM, Bonomo RA, Soler-Bistue A, Sieira R, Ramirez MS. Cerebrospinal fluid (CSF) augments metabolism and virulence expression factors in Acinetobacter baumannii. Sci. Rep. 2021;11:4737. doi: 10.1038/s41598-021-81714-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pimentel C, Le C, Tuttobene MR, Subils T, Martinez J, Sieira R, Papp-Wallace KM, Keppetipola N, Bonomo RA, Actis LA, Tolmasky ME, Ramirez MS. Human pleural fluid and human serum albumin modulate the behavior of a hypervirulent and multidrug-resistant (MDR) Acinetobacter baumannii representative strain. Pathogens. 2021;10:471. doi: 10.3390/pathogens10040471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodman Nyah MJ, Sammie F, Jun N, Myers AL, Harris CM, Emily D, Fernandez JS, Christine L, Mendoza AM, Veronica J, Nikolas N, Brennan CA, Bonomo RA, Rodrigo S, Soledad RM. Human pleural fluid elicits pyruvate and phenylalanine metabolism in Acinetobacter baumannii to enhance cytotoxicity and immune evasion. Front. Microbiol. 2019;10:1581. doi: 10.3389/fmicb.2019.01581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez J, Fernandez JS, Liu C, Hoard A, Mendoza A, Nakanouchi J, Rodman N, Courville R, Tuttobene MR, Lopez C, Gonzalez LJ, Shahrestani P, Papp-Wallace KM, Vila AJ, Tolmasky ME, Bonomo RA, Sieira R, Ramirez MS. Human pleural fluid triggers global changes in the transcriptional landscape of Acinetobacter baumannii as an adaptive response to stress. Sci. Rep. 2019;9:17251. doi: 10.1038/s41598-019-53847-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le C, Pimentel C, Tuttobene MR, Subils T, Nishimura B, Traglia GM, Perez F, Papp-Wallace KM, Bonomo RA, Tolmasky ME, Ramirez MS. Interplay between meropenem and human serum albumin on expression of carbapenem resistance genes and natural competence in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2021;65:e01019-21. doi: 10.1128/AAC.01019-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pimentel C, Le C, Tuttobene MR, Subils T, Papp-Wallace KM, Bonomo RA, Tolmasky ME, Ramirez MS. Interaction of Acinetobacter baumannii with human serum albumin: Does the host determine the outcome? Antibiotics. 2021;10:833. doi: 10.3390/antibiotics10070833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le C, Pimentel C, Pasteran F, Tuttobene MR, Subils T, Escalante J, Nishimura B, Arriaga S, Carranza A, Mezcord V, Vila AJ, Corso A, Actis LA, Tolmasky ME, Bonomo RA, Ramírez MS. Human serum proteins and susceptibility of Acinetobacter baumannii to cefiderocol: Role of iron transport. Biomedicines. 2022;10:600. doi: 10.3390/biomedicines10030600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Portsmouth S, Echols R, Den Nagata T. Cefiderocol for treatment of complicated urinary tract infections. Lancet Infect. Dis. 2019;19:23–24. doi: 10.1016/S1473-3099(18)30721-7. [DOI] [PubMed] [Google Scholar]
- 19.Portsmouth S, van Veenhuyzen D, Echols R, Machida M, Ferreira JCA, Ariyasu M, Tenke P, Nagata TD. Cefiderocol versus imipenem-cilastatin for the treatment of complicated urinary tract infections caused by Gram-negative uropathogens: A phase 2, randomised, double-blind, non-inferiority trial. Lancet Infect. Dis. 2018;18:1319–1328. doi: 10.1016/S1473-3099(18)30554-1. [DOI] [PubMed] [Google Scholar]
- 20.Aghajani Z, Rasooli I, Mousavi Gargari SL. Exploitation of two siderophore receptors, BauA and BfnH, for protection against Acinetobacter baumannii infection. APMIS. 2019;127:753–763. doi: 10.1111/apm.12992. [DOI] [PubMed] [Google Scholar]
- 21.Sheldon JR, Skaar EP. Acinetobacter baumannii can use multiple siderophores for iron acquisition, but only acinetobactin is required for virulence. PLoS Pathog. 2020;16:e1008995. doi: 10.1371/journal.ppat.1008995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobs AC, Sayood K, Olmsted SB, Blanchard CE, Hinrichs S, Russell D, Dunman PM. Characterization of the Acinetobacter baumannii growth phase-dependent and serum responsive transcriptomes. FEMS Immunol. Med. Microbiol. 2012;64:403–412. doi: 10.1111/j.1574-695X.2011.00926.x. [DOI] [PubMed] [Google Scholar]
- 23.Ni Z, Chen Y, Ong E, He Y. Antibiotic resistance determinant-focused Acinetobacter baumannii vaccine designed using reverse vaccinology. Int. J. Mol. Sci. 2017;18:458. doi: 10.3390/ijms18020458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moynie L, Luscher A, Rolo D, Pletzer D, Tortajada A, Weingart H, Braun Y, Page MG, Naismith JH, Kohler T. Structure and function of the PiuA and PirA siderophore-drug receptors from Pseudomonas aeruginosa and Acinetobacter baumannii. Antimicrob. Agents Chemother. 2017;61:e02531-16. doi: 10.1128/AAC.02531-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobs AC, Thompson MG, Black CC, Kessler JL, Clark LP, McQueary CN, Gancz HY, Corey BW, Moon JK, Si Y, Owen MT, Hallock JD, Kwak YI, Summers A, Li CZ, Rasko DA, Penwell WF, Honnold CL, Wise MC, Waterman PE, Lesho EP, Stewart RL, Actis LA, Palys TJ, Craft DW, Zurawski DV. AB5075, a highly virulent isolate of Acinetobacter baumannii, as a model strain for the evaluation of pathogenesis and antimicrobial treatments. MBio. 2014;5:e01076-14. doi: 10.1128/mBio.01076-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodgers D, Pasteran F, Calderon M, Jaber S, Traglia GM, Albornoz E, Corso A, Vila AJ, Bonomo RA, Adams MD, Ramirez MS. Characterisation of ST25 NDM-1-producing Acinetobacter spp. strains leading the increase in NDM-1 emergence in Argentina. J. Glob. Antimicrob. Resist. 2020;23:108–110. doi: 10.1016/j.jgar.2020.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adams MD, Pasteran F, Traglia GM, Martinez J, Huang F, Liu C, Fernandez JS, Lopez C, Gonzalez LJ, Albornoz E, Corso A, Vila AJ, Bonomo RA, Ramirez MS. Distinct mechanisms of dissemination of NDM-1 metallo-beta-lactamase in Acinetobacter spp. in Argentina. Antimicrob. Agents Chemother. 2020;64:e00324-20. doi: 10.1128/AAC.00324-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hujer KM, Hujer AM, Hulten EA, Bajaksouzian S, Adams JM, Donskey CJ, Ecker DJ, Massire C, Eshoo MW, Sampath R, Thomson JM, Rather PN, Craft DW, Fishbain JT, Ewell AJ, Jacobs MR, Paterson DL, Bonomo RA. Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob. Agents Chemother. 2006;50:4114–4123. doi: 10.1128/AAC.00778-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liao Y, Smyth GK, Shi W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 33.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.(CLSI) CLSI. Performance standards for antimicrobial susceptibility testing: Thirty Edition informational supplement. CLSI Document M100-S30:2020 (Clinical Lab Standards Institute, 2020).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request. The datasets generated and analyzed during the current study are available in the Gene Expression Omnibus (GEO) database repository under the accession number GSE201259.