Abstract
Circoviridae is a family of circular single-stranded DNA viruses whose members infect a wide variety of hosts. While well characterized in avian and mammalian hosts, little is known about circoviruses associated with Antarctic animals. From 48 Weddell seal (Leptonychotes weddellii) fecal samples collected on the sea ice in McMurdo between Nov 2014 and Feb 2015, we identified and determined the genomes of novel viruses that fall within two genera of the family Circoviridae, i.e. Circovirus (n=7) and Cyclovirus (n=45). We named these viruses as werosea circovirus (WerCV) and werosea cyclovirus (WerCyV). The genomes of WerCV and WerCyV share ~63-64% genome-wide pairwise identity with classified circoviruses and cycloviruses, respectively. Based on the species demarcation threshold of 80% for members of the Circoviridae , the genomes of WerCV and WerCyV represent new species in their respective genera. Evidence indicated recombination in five of the 45 WerCyV genomes identified in this study. These are the first circoviruses found associated with Antarctic pinnipeds, adding to those recently identified associated with Adélie (Pygoscelis adeliae) and chinstrap penguins (P. antarcticus).
Keywords: Circoviridae, Weddell seals, pinnipeds, ssDNA virus
Introduction
Traditionally, the detection of viruses relied on their propagation in cell culture. Yet because many viruses cannot easily be propagated, they are difficult to identify in this way resulting in only a fraction of the global virome being determined. However, the emergence of metagenomic approaches is bridging the gap by allowing their identification in the absence of culture (Cantalupo and Pipas, 2019; Mokili et al., 2012; Shi et al., 2016; Sommers et al., 2021; Zhang et al., 2019; Zhang et al., 2018). This emerging field has begun to reveal a vast virome in the biosphere and to uncover the presence of viral types not previously known to exist in certain environments (Roux et al., 2016; Schulz et al., 2020; Shi et al., 2016; Tisza and Buck, 2021; Tisza et al., 2020). Viral metagenomics can also provide insight into virus-host interactions, evolution of viruses, and co-evolution between host and viral genomes (Roux et al., 2021).
Within the context of viral metagenomics, the use of rolling-circle amplification to preferentially amplify circular DNA, coupled with high-throughput sequencing, has led to rapid expansion in the identification and taxonomy of known and novel circular single-stranded DNA (ssDNA) viruses (Dayaram et al., 2016; de la Higuera et al., 2020; Fontenele et al., 2019; Kraberger et al., 2019; Krupovic et al., 2020; Tisza et al., 2020), including circoviruses.
Circoviruses (family Circoviridae) are small (1.7 – 2.1 kb) circular single-stranded DNA viruses (Breitbart et al., 2017) categorized into two genera, Circovirus and Cyclovirus, with a species demarcation threshold of 80% genome-wide pairwise identity (Rosario et al., 2017). The genomes of circoviruses encode two major open reading frames (ORFs) coding for the replication-associated protein (Rep) and capsid protein (CP). They are known to infect a wide variety of hosts, including fish, mammals, and invertebrates (Rosario et al., 2017). The most notable of the circoviruses are the disease-causing avian and swine viruses, such as beak and feather disease virus (BFDV) (Julian et al., 2013; Massaro et al., 2012) and porcine circovirus 2 (PCV2) (Segales, 2012; Segales et al., 2013). Infection in these animals is typically associated with potentially fatal illnesses including loss of condition and multisystem wasting, hemorrhagic enteritis, vasculitis, and necrosis of lymphatic tissues (Baekbo et al., 2012; Todd, 2000). Recently, circoviruses were identified in Adélie penguins (Pygoscelis adeliae) at Cape Crozier on Ross Island, East Antarctica (Morandini et al., 2019), and in Adélie and chinstrap penguins (P. antarcticus) on the Western Antarctic Peninsula (Levy et al., 2020). These were the first reports of circoviruses associated with Antarctic animals.
Weddell seals are the world’s southernmost-living mammal, residing year-round on the fast- and pack-ice around the coast of Antarctica (IUCN, 2014; LaRue et al., 2021; Rice, 1998). Adults typically reach ~350-550 kg in size, with a diet consisting primarily of Antarctic silverfish (Pleuragramma antarcticum) and other small notothenioid fish, and cephalopods (Goetz et al., 2017). They pup and molt on fast ice during Oct-Feb, with a diet consisting mostly of fish and cephalopods (Burns et al., 1998; Goetz et al., 2017; IUCN, 2014). There is also some regional variation in diet, with Weddell seals consuming more crustacean prey such as krill in the Western Antarctic Peninsula than in McMurdo Sound (Green and Burton, 1987). A health assessment of Weddell seals found them to be relatively naive in their exposure to known viral pathogens (Yochem et al., 2009), with early reports including herpesvirus (Harder et al., 1991; Stenvers et al., 1992), and a parapoxvirus (Tryland et al., 2005). Over the last decade, various small circular DNA viruses have been identified associated with Weddell seals in McMurdo Sound, East Antarctica. These include a polyomavirus (Varsani et al., 2017), numerous papillomaviruses (Smeele et al., 2018), and diverse anelloviruses (Fahsbender et al., 2017). Given that so little is known about viruses associated with Weddell seals, here we analyze fecal samples from 48 Weddell seals (Leptonychotes weddellii), collected on the fast ice of McMurdo Sound (Antarctic) during Nov 2014-Feb 2015 and identify circoviruses and cycloviruses.
Methods and materials
Sampling
Forty-eight fecal samples from Weddell seals were collected opportunistically on the fast ice of McMurdo Sound (Antarctic) between Nov 2014-Feb 2015 as part of a diet study in the Ross Sea. Despite tagged Weddell seals observed in close proximity of the sample collection sites, it was not possible to determine which individual animals deposited the feces.
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. Weddell seal samples were collected under National Marine Fisheries Service Marine Mammal permit #17411, Antarctic Conservation Act permit #2014-003, and University of Alaska Anchorage’s Institutional Animal Care and Use Committee approval #419971, with funding from the National Science Foundation grant ANT-1246463 to Jennifer M Burns (Varsani et al., 2017).
Viral DNA extraction and Illumina sequencing
Each of the 48 fecal samples was processed for viral analysis as described previously in Fahsbender et al. (2017). In brief, approximately 5 g of the fecal sample was homogenized in 20 ml of SM buffer (0.1 M NaCl, 50 mM Tris/HCl – pH 7.4, 10 mM MgSO4) and the suspension centrifuged at 10000 × g for 10 mins. The resulting supernatant was sequentially filtered through 0.45 µm and 0.2 µm syringe filters and viral particles precipitated with 15% w/v PEG, centrifuged at 10000 x g for 20 mins. The resulting pellet was resuspended in 2 ml of SM buffer. Of this, 200 µl was used to extract viral DNA using the High Pure Viral Nucleic Acid Kit (Roche Diagnostics, USA), and 5 µl of the viral DNA extract was used as template for rolling-circle amplification (RCA) using the TempliPhi™ kit (GE Healthcare, USA) to preferentially amplify circular nucleic acid.
For the purpose of high-throughput viral sequencing, a 5 µl aliquot of the RCA product from each of the fecal samples was pooled. This pooled nucleic acid was used to generate a 100 bp paired-end Illumina sequencing library and an Illumina 2500 (Illumina, USA) platform at Macrogen Inc. (South Korea). The raw reads were checked for quality and trimmed using Trimmomatic (Bolger et al., 2014) and then de novo assembled using metaSpades v3.12 (Bankevich et al., 2012) with k=33,55,77). Contigs of >750 nts were analyzed for viral-like sequences using BLASTx (Altschul et al., 1990) against a local viral sequence database.
Recovery and sequencing of circovirus genomes
Two contigs that were determined to be similar to circoviruses were identified, and based on these sequences, we designed two pairs of abutting primers (SP_circo_1210F: ACACCTGGAGCAAGCAATGGGAAG; SP_circo_1210R: GCCCGCGGGGATATTAATTCTTTC; SP_cyclo_16926F: GCATTATTGGCAAGGAACATTGTCCAACGAC; SP_cyclo_16926R: CGTATTTGCCATACTGGATGATGAAAGCCTTGATT) to screen and recover the complete circovirus genomes from each of the individual 48 fecal samples.
The RCA product for each fecal viral nucleic acid sample was used as a template for PCR amplification using Kapa HiFi Hotstart DNA polymerase (Kapa Biosystems, USA) with the following thermal cycling conditions: 95°C for 3 mins followed by 25 cycles at 98°C for 20 sec, 60°C for 15 sec, 72°C for 2 mins and then a final extension at 72°C for 2 min. The PCR amplicons were resolved on a 0.7% agarose gel, from which ~2 kb size fragments were excised, gel purified and cloned individually into the pJET1.2 plasmid vector (ThermoFisher, USA). The resulting recombinant plasmids were Sanger sequenced by primer walking at Macrogen Inc. (South Korea). The Sanger sequencing reads were assembled using Geneious Prime® 2021.1.1 (Biomatters Ltd, New Zealand).
Genome analysis and annotation
The genomes of the circoviruses and cycloviruses were characterized using ORFinder (https://www.ncbi.nlm.nih.gov/orffinder/) and annotated based on BLASTx (Altschul et al., 1990) based similarity to protein sequences. All genome-wide nucleotide, Rep and CP amino acid pairwise identities were determined using SDT v1.2 (Muhire et al., 2014).
Phylogenetic analysis
The genomes, Rep, and CP sequences from the circoviruses identified in this study were aligned with those downloaded from GenBank with MAFFT (Katoh and Standley, 2013). The aligned circovirus and cyclovirus genome sequence datasets were used to infer a maximum-likelihood phylogenetic tree with GTR+I+G used as the best substitution model determined using jModelTest (Darriba et al., 2012). The circovirus maximum-likelihood phylogenetic tree was rooted with reverse complementary sequences of representative cycloviruses and vice versa. Branches with less than 0.8 aLRT branch support were collapsed using TreeGraph2 (Stover and Muller, 2010).
In the case of the CP and Rep of the circovirus and cyclovirus datasets, the amino acid sequences were aligned using MAFFT (Katoh and Standley, 2013) and the resulting alignments used to infer maximum-likelihood phylogenetic trees with PhyML (Guindon et al., 2010). Best-fit models were determined using ProtTest (circovirus: RtREV+I+G+F for Rep and VT+I+G+F for CP; cyclovirus: LG+I+G for Rep and RtREV+I+G+F for CP) (Darriba et al., 2011). The circovirus Rep and CP maximum-likelihood phylogenetic trees were rooted with representative cycloviruses and vice versa. Branches with aLRT support of <0.8 were collapsed using TreeGraph2 (Stover and Muller, 2010).
Recombination analysis
RDP5 (Martin et al., 2021) was used to detect recombination in the aligned sequences of the circoviruses and cycloviruses identified in this study. RDP5 was used with default parameters and only recombination events detected by three or more methods coupled with a p-value <0.05 were considered credible.
Results and Discussion
Identification and recovery of circoviruses and cycloviruses
In the high throughput sequencing analysis of the pooled 48 fecal samples of Weddell seals, two contigs with high similarities to circoviruses were identified. Two sets of abutting primers were designed based on the de novo assembled sequence contigs to screen and recover complete genomes of these viruses from the samples (Table 1). Analyses revealed that they belong to the Circovirus and Cyclovirus genera (family Circoviridae). We named the viruses as werosea circovirus (WerCV) and werosea cyclovirus (WerCyV) with the name werosea derived from Weddell seal Ross Sea associated. WerCV was identified and recovered from seven fecal samples, whereas WerCyV in 45 samples. In six samples both WerCV and WerCyV were identified and only two samples (WSP25 and WSP37) did not have either of the viruses WerCV and WerCyV. (Table 1).
Table 1:
Summary of the genomes of werosea cycloviruses and werosea circoviruses identified in this study
| Lab sample ID | Field sample ID | Werosea circovirus | Werosea cyclovirus |
|---|---|---|---|
| WSP01 | SK3 | - | MN176096 |
| WSP02 | SK15 | - | MN176095 |
| WSP03 | SK20 | - | MN176094 |
| WSP04 | SK21 | - | MN176093 |
| WSP05 | SK22 | - | MN176092 |
| WSP06 | SK23 | - | MN176091 |
| WSP07 | SK24 | - | MN176090 |
| WSP08 | SK25 | - | MN176089 |
| WSP09 | SK26 | MN164712 | MN176088 |
| WSP10 | SK27 | - | MN176087 |
| WSP11 | SK28 | - | MN176086 |
| WSP12 | SK29 | MN164713 | MN176085 |
| WSP13 | SK30 | - | MN176084 |
| WSP14 | SK32 | MN164714 | MN176083 |
| WSP15 | SK34 | - | MN176082 |
| WSP16 | SK35 | - | MN176081 |
| WSP17 | SK36 | - | MN176080 |
| WSP18 | SK37 | - | MN176079 |
| WSP19 | SK38 | MN164715 | MN176078 |
| WSP20 | SK39 | - | MN176077 |
| WSP21 | BR1 | - | MN176076 |
| WSP22 | BR2 | - | MN176075 |
| WSP23 | BR3 | - | MN176074 |
| WSP24 | BR4 | - | MN176073 |
| WSP25 | HC1 | - | - |
| WSP26 | HC2 | - | MN176072 |
| WSP27 | HC3 | MN164716 | MN176071 |
| WSP28 | HC4 | - | MN176070 |
| WSP29 | HC5 | - | MN176069 |
| WSP30 | HC6 | - | MN176068 |
| WSP31 | HC7 | - | MN176067 |
| WSP32 | HC8 | - | MN176066 |
| WSP33 | HC9 | MN164717 | MN176065 |
| WSP34 | HC10 | - | MN176064 |
| WSP35 | HC11 | - | MN176063 |
| WSP36 | HC12 | - | MN176062 |
| WSP37 | HC13 | - | - |
| WSP38 | TR1 | - | MN176061 |
| WSP39 | TR2 | MN164718 | - |
| WSP40 | TR3 | - | MN176060 |
| WSP41 | TR4 | - | MN176059 |
| WSP42 | TR5 | - | MN176058 |
| WSP43 | RB5 | - | MN176057 |
| WSP44 | RB6 | - | MN176056 |
| WSP45 | WS14-01 | - | MN176055 |
| WSP46 | WS14-08 | - | MN176054 |
| WSP47 | WS14-11 | - | MN176053 |
| WSP48 | WS14-14 | - | MN176052 |
Sequence analysis of werosea circovirus
The werosea circovirus genomes have two intergenic regions (IR), one of which is upstream of the rep and cp ORFs start codons and contains a conserved origin of rolling circle replication (ori) ORFs (Rosario et al., 2017). The ori is characterized by a conserved nonanucleotide motif (TAGTATTAC) located at the apex of the potential stem loop structure. The genomes of the seven recovered WerCVs are 1,853 nucleotides in length and share > 99.6% genome-wide nucleotide sequence identity, >98.5% CP amino acid sequence identity, and >99.3% Rep amino acid sequence identity with each other. When compared with other circoviruses, they share between 54.7 and 64.2% genome-wide nucleotide sequence identity, 18-37.4% CP amino acid sequence identity, and 43-56.5% Rep amino acid sequence identity (Supplementary data 1).
These WerCVs, therefore, represent a new species of circovirus based on the species demarcation guidelines outlined in Rosario et al. (2017). The sequences are most closely related to that of a European catfish (Silurus glanis) circovirus (Lorincz et al., 2012), sharing a genome-wide identity of ~64%. The WerCVs phylogenetically cluster on their own but do form a clade with the fish circoviruses (Lorincz et al., 2011; Lorincz et al., 2012) and one human feces derived circovirus (Li et al., 2010) (Figure 1).
Figure 1:
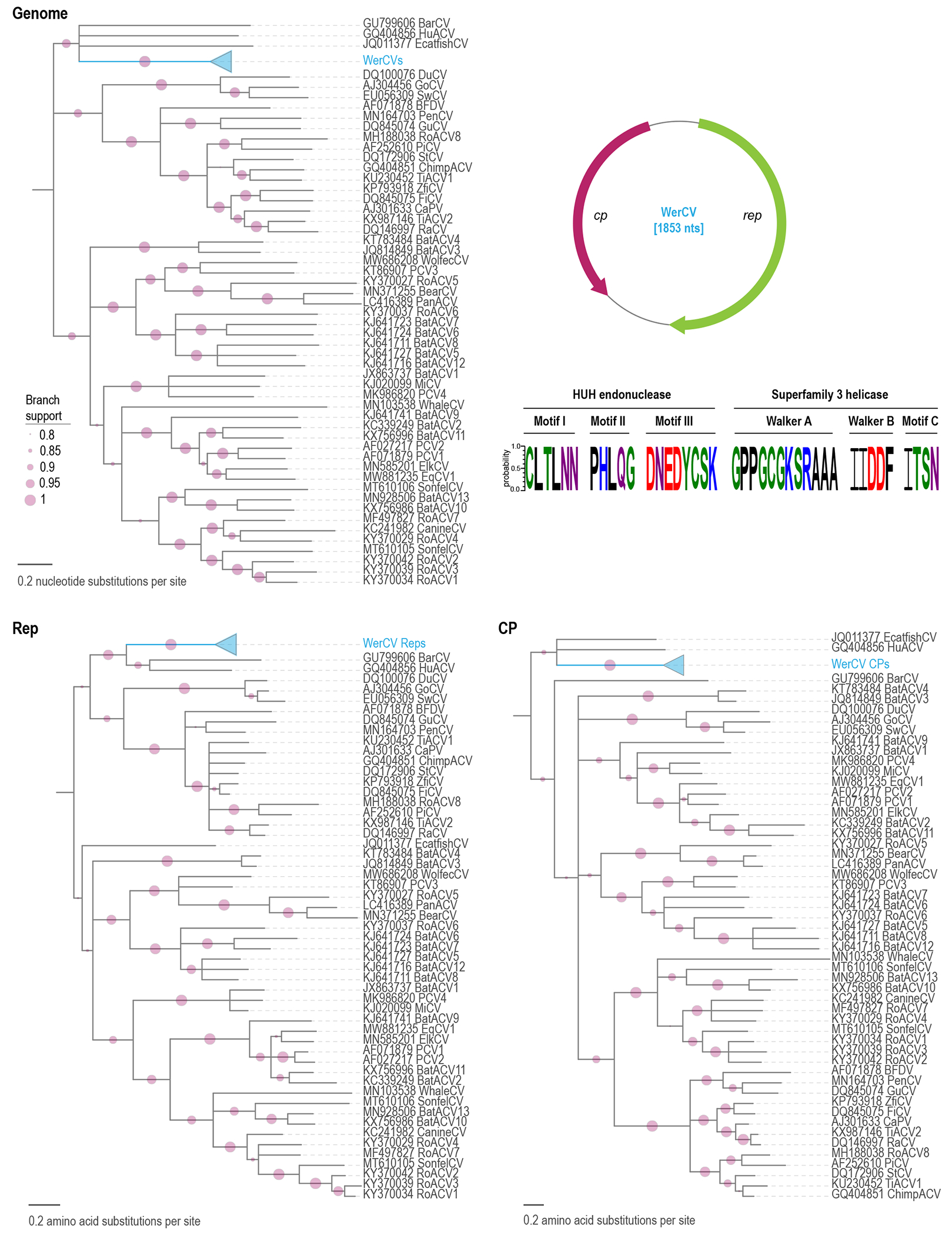
Maximum-likelihood phylogenetic tree of genome sequences of circoviruses, and the Rep and CP amino acid sequences. The seven WerCV sequences are shown as a collapsed clade. The phylogenetic trees have been rooted with representative sequences of cycloviruses and branches with <0.8 aLRT branch support have been collapsed with TreeGraph2 (Stover and Muller, 2010). On the top right the genome organization of WerCV is illustrated. The conserved motifs in the Rep of WerCV are shown with Weblogo (Crooks et al., 2004) and the residues are colored based on their chemical properties (polar, green; basic, blue; acidic, red; hydrophobic, black; neutral, purple).
The circovirus Reps are distinguished by three rolling circle replication (RCR) motifs {Rosario, 2017 #18} at the N-terminus: motif I [CLTLNN], motif II [PHLQG], and motif III [DNEDYCSK]. They also contain three superfamily 3 (SF3) helicase motifs (Rosario et al., 2017), including walker-A [GPPGCGKSRAA], walker-B [IIDDF], and motif C [ITSN] (Figure 1).
Since the WerCVs phylogenetically cluster with fish-derived circoviruses, they are most likely derived from infected fish consumed by Weddell seals. Circovirus-like endogenous sequence elements have been identified in Weddell seals (Dennis et al., 2019) but the WerCVs appear to be distantly related to these, sharing 50-56% amino acid identity to the Rep-like translated genomic sequences in genomic scaffolds KB715046 (751891→752082; 814789→814670 nt position), KB715143 (1026075→1025894; 1026300→1026227; 1026060→1025833 nt position) and KB718192 (10471→10277; 10716→10588 nt position).
Sequence analysis of werosea cyclovirus
The 45 WerCyV genomes range between 2,005 to 2,065 nts in size (Figure 2). They share >79.5% genome-wide nucleotide sequence identity, >79.5% Rep amino acid and >89.7 CP amino acid sequence identity with each other. When compared to other cycloviruses, they share 54.7-63% genome-wide nucleotide sequence identity, 37.6-57.1% Rep amino and 15.6-30.3% CP amino acid sequence identity (Supplementary data 2).
Figure 2:
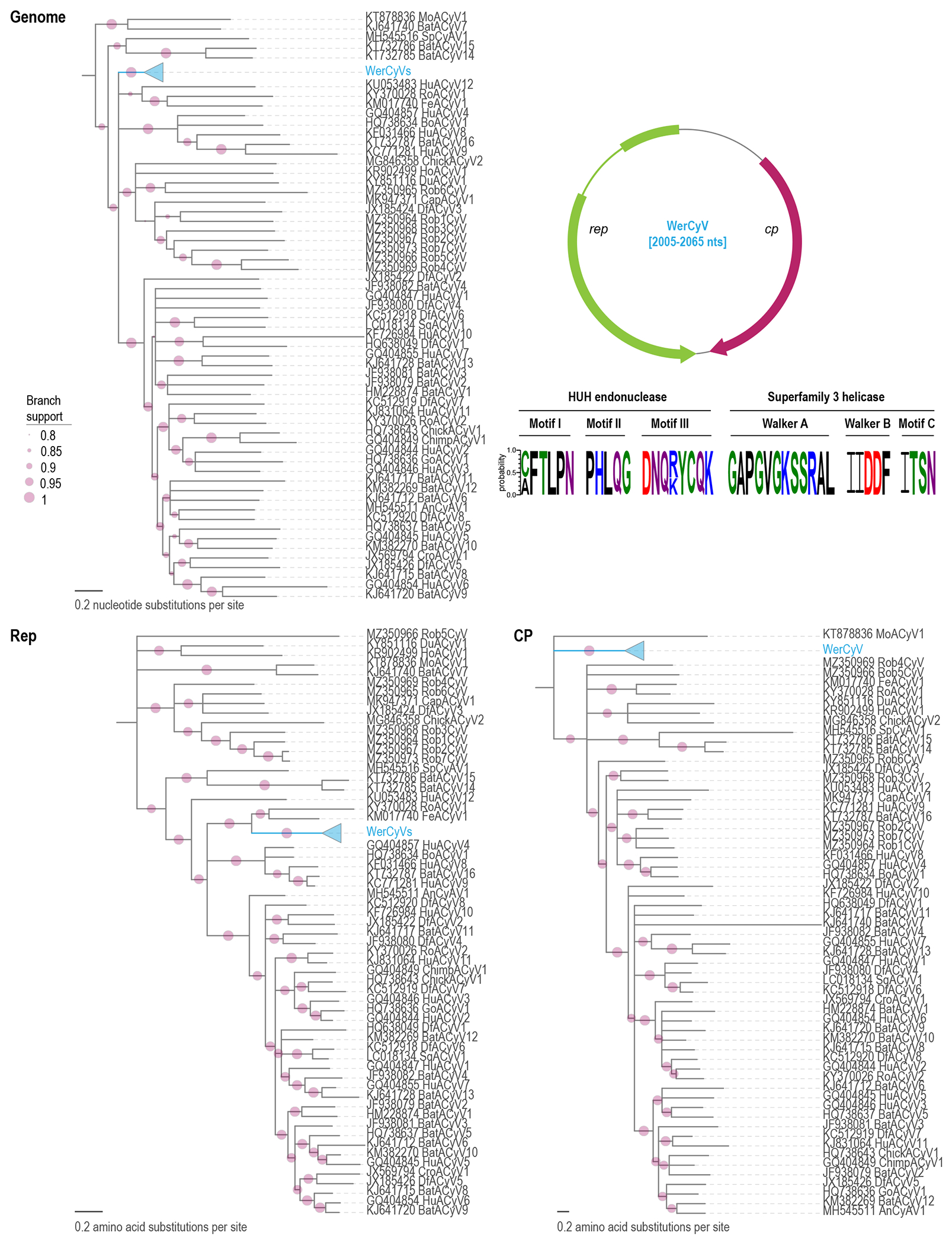
Maximum-likelihood phylogenetic tree of genome sequences of cycloviruses, and the Rep and CP amino acid sequences. The 45 WerCyV sequences are shown as a collapsed clade. An expansion of this clade is shown in Figure 3. The phylogenetic trees have been rooted with representative sequences of circoviruses and branches with <0.8 aLRT branch support have been collapsed with TreeGraph2 (Stover and Muller, 2010). On the top right the genome organization of WerCyV is illustrated. The conserved motifs in the Rep of WerCyV are shown with Weblogo (Crooks et al., 2004) and the residues are colored based on their chemical properties (acidic, red; basic, blue; hydrophobic, black; neutral, purple; polar, green).
The cyclovirus genomes are similar to those of the circoviruses, with a few key differences. They contain the RCR motif I [(C/A)FTLPN], motif II [PHLQG], and motif III [DNQ(R/K)YCQK], as well at the SF3 helicase walker-A [GAPGVGKSSRAL], walker-B [IIDDF], and motif C [ITSN]. The conserved nonanucleotide motif (TAGTATTAC) was identified on the cp-encoding strand. While there is an IR that is similarly sized to those seen in the circoviruses between the 5’ ends of the rep and cp ORFs, the IR between the 3’ ends of the cp and rep ORFs is much smaller (Rosario et al., 2017). All of the cyclovirus sequences displayed evidence of the Rep protein being expressed from a spliced transcript (Figure 2).
The 45 cyclovirus genomes represent a new species in the genus Cyclovirus and phylogenetically group into three well supported clades (Figure 3). We identified one recombinant region in WerCyV sequence MN176089 in clade III with the recombinant region (derived from clade II sequences) of 480 nts spanning the larger intergenic region and the N-termini of the rep and cp ORFs (Figure 3). All four of the sequences in clade II have a ~480nt recombinant region in the cp ORF that is derived from a clade I sequence. A summary of the recombination events detected in the WerCyVs is provided in Figure 3.
Figure 3:
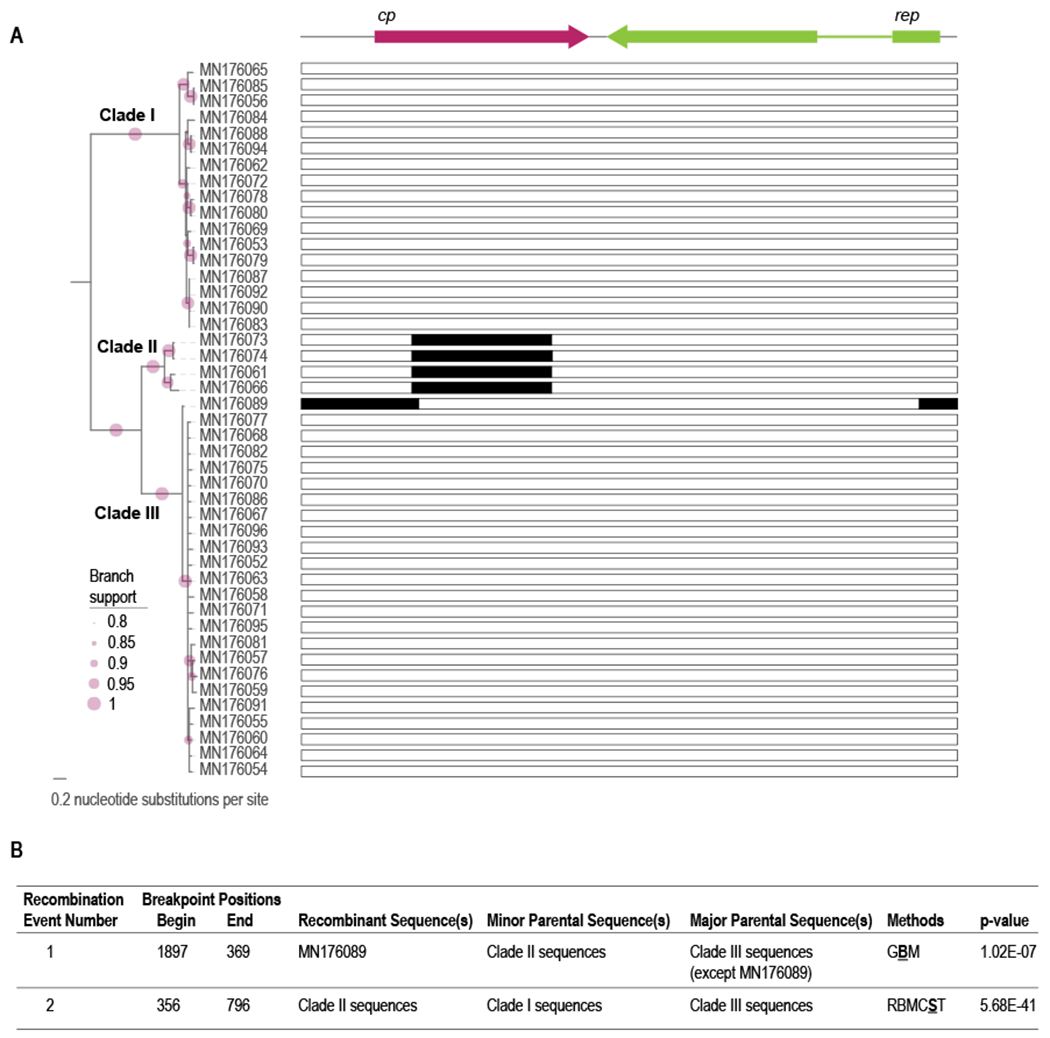
A. Maximum-likelihood phylogenetic tree of genome sequences of the 45 cycloviruses and an illustration of the recombinant (black boxes) regions is provided to the right. B. Summary of the recombination events detected using RDP5 (Martin et al., 2021). The methods used to detect recombination are RDP (R), GENCONV (G), BOOTSCAN (B), MAXCHI (M), CHIMERA (C), SISCAN (S) and 3SEQ (T). The method with the highest p-value for each recombination event is shown in bold font and underlined. Clade I, II and III are the three well supported clades and when they are referred to in the table they include all the sequences in the clade unless stated otherwise.
A BLASTn based analysis of the WerCyV sequences revealed sequences with 67-80% nucleotide pairwise identity from the genome assemblies of the parasitic tapeworm, Spirometra erinaceieuropaei (family Diphyllobothriidae, class Cestoda). We downloaded and analyzed the S.erinaceieuropaei genome sequences and identified that they align to the rep ORF of WerCyV (Figure 4). In four of the five aligned sequences of S. erinaceieuropaei to the rep of WerCyV, we identified nonsense mutations. In one of the sequences we also identified an indel that results in a frameshift. In sequence LN016718, there are two elements of WerCyV-like sequences, and in LN032853 there are three elements and one is in the reverse complementary orientation (Figure 4). Following translation of these rep-like sequences in the S. erinaceieuropaei genome, we noted that they share 68-87% amino acid identity with the WerCyV Reps. Based on the WerCyV rep-like sequence with nonsense mutations, indels, as well as sequence elements in different segments of the S. erinaceieuropaei genome sequence, these appear to be relics of endogenized rep-like sequences that are non-functional. Endogenized Rep-like sequences have been found in the genomes of various animals (Dennis et al., 2019; Liu et al., 2011). It is therefore likely, based on similarity of the endogenized rep-like sequences to the WerCyV Reps, that WerCyVs infect related cestodes that are associated with Weddell seals. Weddell seals are generally heavily infested with various cestodes, with one study estimating a mean intensity at 1,300,000 tapeworms per seal (Iurakhno and Mal’tsev, 1997; McFarlane et al., 2009; Scholz et al., 2019). Since viruses are shed via fecal matter from the gut and epithelial cells, it is likely that WerCyVs detected in the feces are from tapeworm parasites in the gut that originated in the ingested fish diet (Rocka, 2017).
Figure 4:
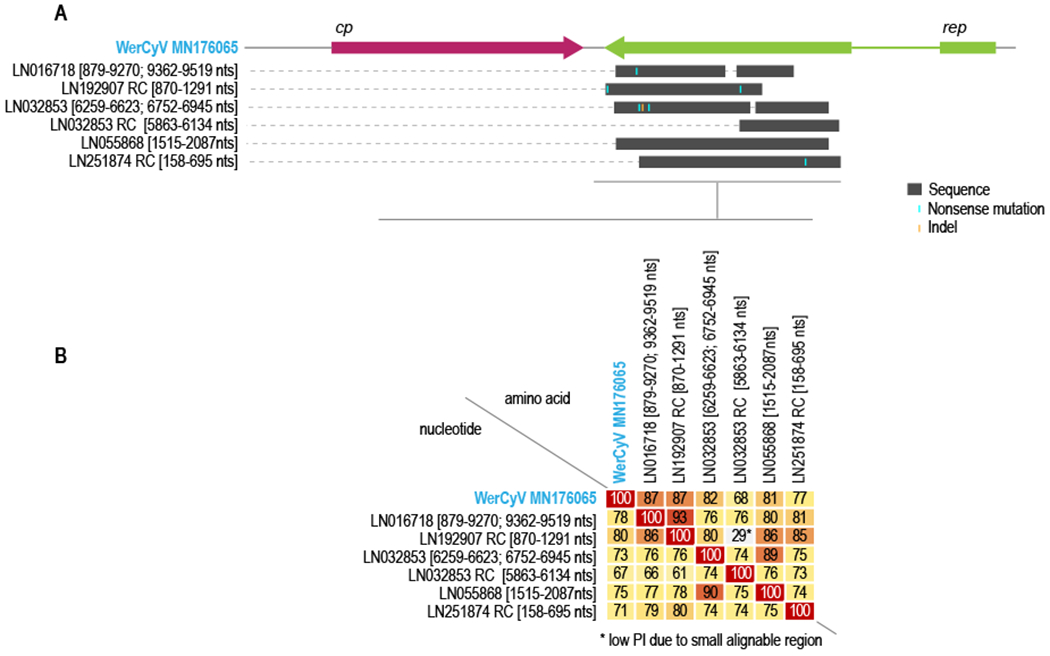
A. Positions in the alignment of rep-like sequences in Spirometra erinaceieuropaei (family Diphyllobothriidae, class Cestoda) genome sequence scaffolds with WerCyV genome. The scaffold accession number and position on the scaffold is provided. Nonsense mutations and indels are shown. RC - reverse complementary sequences. B. A pairwise identity matrix of the nucleotide and amino acid sequences of the rep-like elements with the rep nucleotide and Rep amino acid sequences. * marks a low pairwise identity and this is attributed to a small alignable sequence region.
Concluding remarks
In this study we identified seven WerCV genomes and 45 WerCyV genomes in Weddell seal fecal samples. Given they were isolated from fecal matter, we were unable to definitively identify the true hosts of WerCV and WerCyV. The WerCVs are most closely related to a catfish circoviruses (Lorincz et al., 2011; Lorincz et al., 2012) and one human feces derived circovirus (Li et al., 2010). Thus, their identification in the fecal samples could indicate that they may be derived from the fish eaten by the seals see (Goetz et al., 2017). However, it is important to note that this is based on the phylogeny of the known circoviruses and, given the fact that there remain large gaps in our knowledge of circovirus diversity within marine ecosystems, it is plausible that these viruses have non-fish hosts. It is interesting that the WerCyV sequences share similarity to Rep-like sequences found in the genome assemblies of the diphyllobothriid cestode, S. erinaceieuropaei. Based on the high degrees of similarity between the WerCyV rep and the Rep-like sequences of S. erinaceieuropaei, it is likely that WerCyV infects cestodes that are associated with Weddell seals. Further research is, however, needed to better characterize the host ranges of these viruses.
The identification of the viruses in this study adds to the growing knowledge of viruses found associated with the marine fauna of the high latitude Southern Ocean, but also opens up new questions about their hosts. Although advancements within viral metagenomics have enabled an expansion of research into the virosphere, as with almost all other biomes on the planet much remains to be determined about the complex webs of host and virus associations that underpin the Antarctic virome.
Supplementary Material
Acknowledgements
David G Ainley is supported by the US National Science Foundation (NSF; ANT-0944411). Arvind is supported by the US National Science Foundation (NSF; ANT-1935870, ANT-1947040) All the samples were collected with logistics supplied from the US Antarctic Program. Stacy Kim was supported by NSF ANT-0944747. Jennifer Burns and Michelle Shero were supported by the US National Science Foundation (NSF; ANT-1246463). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation. Amy Kirkham and Roxanne Beltran were supported by Institutional Development Awards (IDeA) Networks of Biomedical Research Excellence Assistantships (grant number P20GM103395) from the National Institute of General Medical Sciences of the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily reflect the official views of the NIH. All the wet bench molecular work and sequencing was supported by philanthropic funds from Arvind Varsani.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ, 1990. Basic local alignment search tool. J Mol Biol 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Baekbo P, Kristensen CS, Larsen LE, 2012. Porcine circovirus diseases: a review of PMWS. Transbound Emerg Dis 59 Suppl 1, 60–67. [DOI] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA, 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19, 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B, 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M, Delwart E, Rosario K, Segales J, Varsani A, Ictv Report C, 2017. ICTV Virus Taxonomy Profile: Circoviridae. J Gen Virol 98, 1997–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns JM, Trumble SJ, Castellini MA, Testa JW, 1998. The diet of Weddell seals in McMurdo Sound, Antarctica as determined from scat collections and stable isotope analysis. Polar Biology 19, 272–282. [Google Scholar]
- Cantalupo PG, Pipas JM, 2019. Detecting viral sequences in NGS data. Curr Opin Virol 39, 41–48. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE, 2004. WebLogo: a sequence logo generator. Genome Res 14, 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D, 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D, 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9, 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayaram A, Galatowitsch ML, Arguello-Astorga GR, van Bysterveldt K, Kraberger S, Stainton D, Harding JS, Roumagnac P, Martin DP, Lefeuvre P, Varsani A, 2016. Diverse circular replication-associated protein encoding viruses circulating in invertebrates within a lake ecosystem. Infect Genet Evol 39, 304–316. [DOI] [PubMed] [Google Scholar]
- de la Higuera I, Kasun GW, Torrance EL, Pratt AA, Maluenda A, Colombet J, Bisseux M, Ravet V, Dayaram A, Stainton D, Kraberger S, Zawar-Reza P, Goldstien S, Briskie JV, White R, Taylor H, Gomez C, Ainley DG, Harding JS, Fontenele RS, Schreck J, Ribeiro SG, Oswald SA, Arnold JM, Enault F, Varsani A, Stedman KM, 2020. Unveiling Crucivirus Diversity by Mining Metagenomic Data. mBio 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis TPW, de Souza WM, Marsile-Medun S, Singer JB, Wilson SJ, Gifford RJ, 2019. The evolution, distribution and diversity of endogenous circoviral elements in vertebrate genomes. Virus Res 262, 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahsbender E, Burns JM, Kim S, Kraberger S, Frankfurter G, Eilers AA, Shero MR, Beltran R, Kirkham A, McCorkell R, Berngartt RK, Male MF, Ballard G, Ainley DG, Breitbart M, Varsani A, 2017. Diverse and highly recombinant anelloviruses associated with Weddell seals in Antarctica. Virus Evol 3, vex017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenele RS, Lacorte C, Lamas NS, Schmidlin K, Varsani A, Ribeiro SG, 2019. Single Stranded DNA Viruses Associated with Capybara Faeces Sampled in Brazil. Viruses 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz KT, Burns JM, Hückstӓdt LA, Shero MR, Costa DP, 2017. Temporal variation in isotopic composition and diet of Weddell seals in the western Ross Sea. Deep Sea Research Part II: Topical Studies in Oceanography 140, 36–44. [Google Scholar]
- Green K, Burton H, 1987. Seasonal and Geographical Variation in the Food of Weddell Seals, Leptonychotes-Weddelii, in Antarctica. Wildlife Research 14, 475–489. [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O, 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59, 307–321. [DOI] [PubMed] [Google Scholar]
- Harder TC, Plötz J, Liess B, 1991. Antibodies against european phocine herpesvirus isolates detected in sera of Antarctic seals. Polar Biology 11. [Google Scholar]
- IUCN, 2014. Leptonychotes weddellii: Hückstädt L. IUCN Red List of Threatened Species. [Google Scholar]
- Iurakhno MV, Mal’tsev VN, 1997. [Cestode infection of Antarctic seals]. Parazitologiia 31, 81–89. [PubMed] [Google Scholar]
- Julian L, Piasecki T, Chrzastek K, Walters M, Muhire B, Harkins GW, Martin DP, Varsani A, 2013. Extensive recombination detected among beak and feather disease virus isolates from breeding facilities in Poland. J Gen Virol 94, 1086–1095. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM, 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraberger S, Schmidlin K, Fontenele RS, Walters M, Varsani A, 2019. Unravelling the Single-Stranded DNA Virome of the New Zealand Blackfly. Viruses 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M, Varsani A, Kazlauskas D, Breitbart M, Delwart E, Rosario K, Yutin N, Wolf YI, Harrach B, Zerbini FM, Dolja VV, Kuhn JH, Koonin EV, 2020. Cressdnaviricota: a Virus Phylum Unifying Seven Families of Rep-Encoding Viruses with Single-Stranded, Circular DNA Genomes. J Virol 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRue M, Salas L, Nur N, Ainley D, Stammerjohn S, Pennycook J, Dozier M, Saints J, Stamatiou K, Barrington L, Rotella J, 2021. Insights from the first global population estimate of Weddell seals in Antarctica. Science Advances. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy H, Fiddaman SR, Djurhuus A, Black CE, Kraberger S, Smith AL, Hart T, Varsani A, 2020. Identification of Circovirus Genome in a Chinstrap Penguin (Pygoscelis antarcticus) and Adelie Penguin (Pygoscelis adeliae) on the Antarctic Peninsula. Viruses 12, 858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Kapoor A, Slikas B, Bamidele OS, Wang C, Shaukat S, Masroor MA, Wilson ML, Ndjango JB, Peeters M, Gross-Camp ND, Muller MN, Hahn BH, Wolfe ND, Triki H, Bartkus J, Zaidi SZ, Delwart E, 2010. Multiple diverse circoviruses infect farm animals and are commonly found in human and chimpanzee feces. J Virol 84, 1674–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Fu Y, Li B, Yu X, Xie J, Cheng J, Ghabrial SA, Li G, Yi X, Jiang D, 2011. Widespread horizontal gene transfer from circular single-stranded DNA viruses to eukaryotic genomes. BMC Evol Biol 11, 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorincz M, Csagola A, Farkas SL, Szekely C, Tuboly T, 2011. First detection and analysis of a fish circovirus. J Gen Virol 92, 1817–1821. [DOI] [PubMed] [Google Scholar]
- Lorincz M, Dan A, Lang M, Csaba G, Toth AG, Szekely C, Csagola A, Tuboly T, 2012. Novel circovirus in European catfish (Silurus glanis). Arch Virol 157, 1173–1176. [DOI] [PubMed] [Google Scholar]
- Martin DP, Varsani A, Roumagnac P, Botha G, Maslamoney S, Schwab T, Kelz Z, Kumar V, Murrell B, 2021. RDP5: a computer program for analyzing recombination in, and removing signals of recombination from, nucleotide sequence datasets. Virus Evol 7, veaa087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massaro M, Ortiz-Catedral L, Julian L, Galbraith JA, Kurenbach B, Kearvell J, Kemp J, van Hal J, Elkington S, Taylor G, Greene T, van de Wetering J, van de Wetering M, Pryde M, Dilks P, Heber S, Steeves TE, Walters M, Shaw S, Potter J, Farrant M, Brunton DH, Hauber M, Jackson B, Bell P, Moorhouse R, McInnes K, Varsani A, 2012. Molecular characterisation of beak and feather disease virus (BFDV) in New Zealand and its implications for managing an infectious disease. Arch Virol 157, 1651–1663. [DOI] [PubMed] [Google Scholar]
- McFarlane RA, de B. Norman RJ, Jones HI, 2009. Diseases and Parasites of Antarctic and Sub-Antarctic Seals, in: Kerry KR, Riddle M. (Eds.), Health of Antarctic Wildlife: A Challenge for Science and Policy. Springer Berlin Heidelberg, Berlin, Heidelberg, pp. 57–93. [Google Scholar]
- Mokili JL, Rohwer F, Dutilh BE, 2012. Metagenomics and future perspectives in virus discovery. Curr Opin Virol 2, 63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morandini V, Dugger KM, Ballard G, Elrod M, Schmidt A, Ruoppolo V, Lescroel A, Jongsomjit D, Massaro M, Pennycook J, Kooyman GL, Schmidlin K, Kraberger S, Ainley DG, Varsani A, 2019. Identification of a Novel Adelie Penguin Circovirus at Cape Crozier (Ross Island, Antarctica). Viruses 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhire BM, Varsani A, Martin DP, 2014. SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS One 9, e108277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice DW, 1998. Marine Mammals of the World: Systematics and Distribution.
- Rocka A, 2017. Cestodes and Nematodes of Antarctic Fishes and Birds, in: Klimpel S, Kuhn T, Mehlhorn H (Eds.), Biodiversity and Evolution of Parasitic Life in the Southern Ocean. Springer International Publishing, Cham, pp. 77–107. [Google Scholar]
- Rosario K, Breitbart M, Harrach B, Segales J, Delwart E, Biagini P, Varsani A, 2017. Revisiting the taxonomy of the family Circoviridae: establishment of the genus Cyclovirus and removal of the genus Gyrovirus. Arch Virol 162, 1447–1463. [DOI] [PubMed] [Google Scholar]
- Rosario K, Duffy S, Breitbart M, 2012. A field guide to eukaryotic circular single-stranded DNA viruses: insights gained from metagenomics. Arch Virol 157, 1851–1871. [DOI] [PubMed] [Google Scholar]
- Roux S, Brum JR, Dutilh BE, Sunagawa S, Duhaime MB, Loy A, Poulos BT, Solonenko N, Lara E, Poulain J, Pesant S, Kandels-Lewis S, Dimier C, Picheral M, Searson S, Cruaud C, Alberti A, Duarte CM, Gasol JM, Vaque D, Tara Oceans C, Bork P, Acinas SG, Wincker P, Sullivan MB, 2016. Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature 537, 689–693. [DOI] [PubMed] [Google Scholar]
- Roux S, Matthijnssens J, Dutilh BE, 2021. Metagenomics in Virology. Encyclopedia of Virology, 133–140. [Google Scholar]
- Scholz T, Kuchta R, Brabec J, 2019. Broad tapeworms (Diphyllobothriidae), parasites of wildlife and humans: Recent progress and future challenges. Int J Parasitol Parasites Wildl 9, 359–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz F, Roux S, Paez-Espino D, Jungbluth S, Walsh DA, Denef VJ, McMahon KD, Konstantinidis KT, Eloe-Fadrosh EA, Kyrpides NC, Woyke T, 2020. Giant virus diversity and host interactions through global metagenomics. Nature 578, 432–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segales J, 2012. Porcine circovirus type 2 (PCV2) infections: clinical signs, pathology and laboratory diagnosis. Virus Res 164, 10–19. [DOI] [PubMed] [Google Scholar]
- Segales J, Kekarainen T, Cortey M, 2013. The natural history of porcine circovirus type 2: from an inoffensive virus to a devastating swine disease? Vet Microbiol 165, 13–20. [DOI] [PubMed] [Google Scholar]
- Shi M, Lin XD, Tian JH, Chen LJ, Chen X, Li CX, Qin XC, Li J, Cao JP, Eden JS, Buchmann J, Wang W, Xu J, Holmes EC, Zhang YZ, 2016. Redefining the invertebrate RNA virosphere. Nature 540, 539–543. [DOI] [PubMed] [Google Scholar]
- Smeele ZE, Burns JM, Van Doorsaler K, Fontenele RS, Waits K, Stainton D, Shero MR, Beltran RS, Kirkham AL, Berngartt R, Kraberger S, Varsani A, 2018. Diverse papillomaviruses identified in Weddell seals. J Gen Virol 99, 549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommers P, Chatterjee A, Varsani A, Trubl G, 2021. Integrating Viral Metagenomics into an Ecological Framework. Annu Rev Virol. [DOI] [PubMed] [Google Scholar]
- Stenvers O, Plotz J, Ludwig H, 1992. Antarctic seals carry antibodies against seal herpesvirus. Arch Virol 123, 421–424. [DOI] [PubMed] [Google Scholar]
- Stover BC, Muller KF, 2010. TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics 11, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisza MJ, Buck CB, 2021. A catalog of tens of thousands of viruses from human metagenomes reveals hidden associations with chronic diseases. Proc Natl Acad Sci U S A 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisza MJ, Pastrana DV, Welch NL, Stewart B, Peretti A, Starrett GJ, Pang YS, Krishnamurthy SR, Pesavento PA, McDermott DH, Murphy PM, Whited JL, Miller B, Brenchley J, Rosshart SP, Rehermann B, Doorbar J, Ta’ala BA, Pletnikova O, Troncoso JC, Resnick SM, Bolduc B, Sullivan MB, Varsani A, Segall AM, Buck CB, 2020. Discovery of several thousand highly diverse circular DNA viruses. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd D, 2000. Circoviruses: Immunosuppressive threats to avian species: A review. Avian Pathology 29, 373–394. [DOI] [PubMed] [Google Scholar]
- Tryland M, Klein J, Nordoy ES, Blix AS, 2005. Isolation and partial characterization of a parapoxvirus isolated from a skin lesion of a Weddell seal. Virus Res 108, 83–87. [DOI] [PubMed] [Google Scholar]
- Varsani A, Frankfurter G, Stainton D, Male MF, Kraberger S, Burns JM, 2017. Identification of a polyomavirus in Weddell seal (Leptonychotes weddellii) from the Ross Sea (Antarctica). Arch Virol 162, 1403–1407. [DOI] [PubMed] [Google Scholar]
- Yochem PK, Stewart BS, Gelatt TS, Siniff DB, 2009. Health Assessment of Weddell Seals, Leptonychotes weddellii, in McMurdo Sound, Antarctica. Health of Antarctic Wildlife, 123–138. [Google Scholar]
- Zhang YZ, Chen YM, Wang W, Qin XC, Holmes EC, 2019. Expanding the RNA Virosphere by Unbiased Metagenomics. Annu Rev Virol 6, 119–139. [DOI] [PubMed] [Google Scholar]
- Zhang YZ, Shi M, Holmes EC, 2018. Using Metagenomics to Characterize an Expanding Virosphere. Cell 172, 1168–1172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


