Abstract
The major spliceosome mediates pre-mRNA splicing by recognizing the highly conserved sequences at the 5′ and 3′ splice sites and the branch point. More than 150 proteins participate in the splicing process and are organized in the spliceosomal A, B, and C complexes. FRA10AC1 is a peripheral protein of the spliceosomal C complex and its ortholog in the green alga facilitates recognition or interaction with splice sites. We identified biallelic pathogenic variants in FRA10AC1 in five individuals from three consanguineous families. The two unrelated Patients 1 and 2 with loss-of-function variants showed developmental delay, intellectual disability, and no speech, while three siblings with the c.494_496delAAG (p.Glu165del) variant had borderline to mild intellectual disability. All patients had microcephaly, hypoplasia or agenesis of the corpus callosum, growth retardation, and craniofacial dysmorphism. FRA10AC1 transcripts and proteins were drastically reduced or absent in fibroblasts of Patients 1 and 2. In a heterologous expression system, the p.Glu165del variant impacts intrinsic stability of FRA10AC1 but does not affect its nuclear localization. By co-immunoprecipitation, we found ectopically expressed HA-FRA10AC1 in complex with endogenous DGCR14, another component of the spliceosomal C complex, while the splice factors CHERP, NKAP, RED, and SF3B2 could not be co-immunoprecipitated. Using an in vitro splicing reporter assay, we did not obtain evidence for FRA10AC1 deficiency to suppress missplicing events caused by mutations in the highly conserved dinucleotides of 5′ and 3′ splice sites in an in vitro splicing assay in patient-derived fibroblasts. Our data highlight the importance of specific peripheral spliceosomal C complex proteins for neurodevelopment. It remains possible that FRA10AC1 may have other and/or additional cellular functions, such as coupling of transcription and splicing reactions.
Keywords: exome sequencing, homozygous, major spliceosome, splicing, intellectual disability
FRA10AC1 is a peripheral protein of the spliceosomal C complex. von Elsner et al. identify biallelic pathogenic FRA10AC1 variants in five individuals with intellectual disability, microcephaly, hypoplasia or agenesis of the corpus callosum, and growth retardation.
Introduction
The major spliceosome is required to remove introns from the vast majority of pre-mRNAs that results in the production of functional mRNAs. The spliceosomal complex is dynamic and composed of multiple uridine-rich (U1, U2, U4/U6, and U5) small nuclear RNAs (snRNAs) and associated ribonucleoproteins (snRNPs) as well as other proteins associated with these snRNPs.1-3 The major spliceosome recognizes the highly conserved sequences at the 5′ splice donor site (GT), the branch point, and the 3′ splice acceptor site (AG).3 Purification and characterization of spliceosomes indicated more than 150 proteins that are organized in distinct spliceosomal complexes.4,5 Each of the A, B, and C complexes contain up to 125 proteins and show a remarkable compositional exchange. The spliceosome composition during the splicing cycle follows three principles: First, several different groups of proteins are present in the spliceosome throughout the entire splicing process. Second, during the transition from the A to B complex and B to C complex, many additional spliceosomal proteins are recruited. Third, at each stage of the splicing cycle, many spliceosomal proteins are released or destabilized. Although the function of most of the spliceosomal proteins is known, several molecules have unclear functions. Less abundant and/or peripheral proteins may be involved in splicing regulation.1
The assembly of the spliceosome starts with the early spliceosome (E complex), composed of the U1 snRNP, splicing factor 1, and U2AF important for the initial recognition of the 5′ and 3′ splice sites and the branch point sequence of an intron. Displacement of splicing factor 1 by the U2 snRNP leads to the formation of the pre-spliceosome A complex. By recruiting the preassembled U4/U6.U5 tri-snRNP to the A complex, the pre-catalytic spliceosome (B complex) is formed. By releasing U1 and U4, the B complex is activated (Bact complex) and further remodeled to generate the catalytically activated spliceosome (B* complex), in which the step I splicing, including 5′ splice site-cleavage and the branching reaction, occurs. As a result, the spliceosome C complex is formed, which further rearranges to form step II catalytically activated spliceosome (C* complex) which carries out the step II splicing, including 3′ splice site-cleavage and the exon ligation. The resulting post-catalytic spliceosome (P complex) contains the spliced intron lariat and ligated exons. The P complex releases the intron lariat and disassembles into individual snRNPs and associated proteins that are recycled for additional splicing rounds.3,6
Mass spectrometry-based approaches for identification and quantification of proteins in pre-catalytic and catalytically active spliceosomes revealed FRA10AC1 (C10orf4) to be a specific component of the spliceosomal C complex.7,8 FRA10AC1 is an evolutionary conserved and mainly uncharacterized nuclear protein.9 Yeast two-hybrid studies revealed FRA10AC1 to interact with DGCR14 (DiGeorge-syndrome critical region 14; alternative names ES2 and ESS2), another peripheral spliceosomal protein of the C complex.7,8 Both DGCR14 and FRA10AC1 are present in the human spliceosome.10 Recent data suggest DGCR14 and FRA10AC1 ensure accurate pre-mRNA splicing when splice sites are compromised or contain non-canonical sequences.11–13
Here, we report biallelic FRA10AC1 variants to cause a disorder with developmental delay, borderline to profound intellectual disability, corpus callosum abnormalities, growth retardation, and craniofacial dysmorphism with long face, high forehead, broad and medial flaring of eyebrows, narrow palpebral fissures, long nose, smooth philtrum, and everted lower lip vermillion. The homozygous variants identified in two unrelated patients cause loss of FRA10AC1 protein, while the one-amino-acid deletion in the third family impacts FRA10AC1 stability without altering its localization in the nucleus. We confirm the peripheral spliceosomal C complex protein DGCR14 to be in complex with FRA10AC1, while other putative interaction partners implicated in splicing could not be co-immunoprecipitated.
Materials and methods
Patients
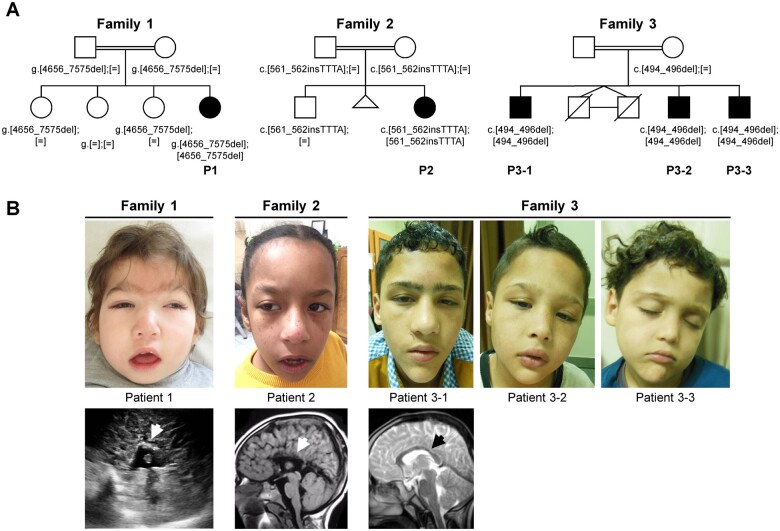
Informed consent for genetic analyses was obtained for all affected individuals, and genetic studies were performed as approved by the Institutional Review Boards of the relevant institutions. The parents of the affected individuals provided written informed consent for participation in the study, clinical data and specimen collection, genetic analysis, and publication of relevant findings. Permission to publish photographs and MRI scans was provided for individuals shown in Fig. 1.
Figure 1.
Pedigree, photographs, and MRI scans of patients with biallelic variants in FRA10AC1. (A) The identified FRA10AC1 variants in the five affected individuals of three families are given below each symbol. If tested, the genotype of healthy family members is also given. GenBank FRA10AC1 reference sequences: NG_016832.1 (Family 1); NM_145246.5 (Families 2 and 3). In Family 2, an abortion at 4 weeks of gestation is indicated. Monozygotic twins in Family 3 died at the age of 2 and 3 days of life because of respiratory distress. P = patient. (B) The five patients share several craniofacial features, such as high forehead, broad and medial flaring of eyebrows, narrow palpebral fissures, long nose, smooth philtrum, and everted lower lip vermillion. In the older Patients 2, 3–1, and 3–2, the shape of the face is triangular and all have a pointed chin. Photograph of Patient 1 at age 3 years and 2 months, of Patient 2 at 9 years, of Patient 3–1 at 15 years, of Patient 3–2 at 11 years, and of Patient 3–3 at 7 years. Brain ultrasound in Patient 1 at 4 months revealed corpus callosum agenesis (white arrow). MRI scans showed partial agenesis of the corpus callosum in Patient 2 at 8 years (white arrow) and thinning of the corpus callosum in Patient 3–1 at 14 years (black arrow).
Whole-exome sequencing, sequence data analysis, and variant validation
Patient 1
Genomic DNA was extracted from peripheral blood samples using standard procedures. Trio whole-exome sequencing (WES) with DNA samples of Patient 1 and healthy parents was performed as described before.14,15 Briefly, coding DNA fragments were enriched with the SureSelect Human All Exon V6 kit (Agilent), and captured libraries were then loaded and sequenced on the HiSeq platform (Illumina). Reads were aligned to the human reference genome (UCSC GRCh37/hg19) using the Burrows-Wheeler Aligner (BWA mem, v0.7.17-r1188).16 Genetic variants were detected with the Genome Analysis Toolkit (GATK, v3.8)17 and annotated using AnnoVar (v2018-04–16).18 Only private (absent in public database) and rare (with minor allele frequency <0.1% and not present in the homozygous state in public databases or the parents) exonic and intronic variants at exon-intron boundaries ranging from −10 to +10 were retained. Remaining variants were then prioritized by pathogenicity assessment using multiple in silico tools (CADD, REVEL, M-CAP, Human Splicing Finder 3.1, NetGene2-Server, and Berkeley Drosophila Genome Project-Database).
For copy number variation (CNV) detection, sequence reads were processed with FASTP (v0.20.1)19 to remove adapter and low quality sequences. Reads were aligned to the human reference assembly (GRCh38) using BWA mem (0.7.17).20 SAMtools (v.1.10)21 was employed for the removal of putative PCR duplicates, for sorting and format conversion. CNVs were called with ExomeDepth (v1.1.15)22 based on GENCODE v34 annotation using standard parameters. In this approach, read depth (coverage) of exons is compared to an optimized reference set of healthy control samples, thereby maximizing the power to detect CNVs (including homozygous and heterozygous deletions and duplications of one or two exons). Common CNVs and false-positive calls were excluded by filtering against CNV calls published by Conrad et al.23 and/or listed in the database of genomic variants (DGV),24 the dbVar database,25 and an in-house database.
To determine the breakpoints and exact size of the homozygous deletion on chromosome 10 in Patient 1’s DNA and for further segregation analysis, we PCR-amplified wild-type and junction fragments from leucocyte-derived DNA of Patient 1, her three healthy sisters and parents. To amplify a wild-type fragment of 1884 bp, we used forward primer FRA10AC1-In1-F located in FRA10AC1 intron 1 and reverse primer FRA10AC1-Ex3-R located in FRA10AC1 exon 3 (Supplementary Table 1). For generation of a junction fragment of unknown size, we combined forward primer FRA10AC1-5′-F located 5′ upstream of FRA10AC1 with reverse primer FRA10AC1-Ex3-R (Supplementary Table 1). Amplicons were directly sequenced using the ABI BigDye Terminator Sequencing kit (Applied Biosystems) and an automated capillary sequencer (ABI 3500, Applied Biosystems). Sequence electropherograms were analysed using Chromas Lite 2.1.1 (Technelysium Pty Ltd).
Patients 2, 3–2, and 3–3
Genomic DNA was extracted from peripheral blood samples of Patient 2, Patient 2’s healthy parents, and Patients 3–2 and 3–3 using standard protocols. WES libraries were generated using the SureSelect Human All Exome 50 Mb Kit (Agilent Technologies) or xGen exome research panel (Integrated DNA Technologies). Paired-end read sequences (100 or 150 bp) were obtained on a HiSeq4000 or NextSeq500 instrument (Illumina). Reads were aligned to the human reference genome (UCSC GRCh37/hg19) and variants were identified through the GATK pipeline or CLC Biomedical Genomics Workbench (Qiagen). Variant calling and filtering were performed following an established exome sequencing pipeline.26 Briefly, reads were aligned to the human reference genome (UCSC GRCh37/hg19) and variants were identified through the GATK pipeline or CLC Biomedical Genomics Workbench (Qiagen). Annotation was performed using in-house software, Annovar, Variant Effect Prediction software or CLC Biomedical Genomics Workbench to define population-specific allele frequencies from 1000 Genomes, the Greater Middle East Variome, dbSNP, and gnomAD, along with the transcript-specific predicted effect on the protein. Identified variants were filtered out if not consistent with autosomal recessive inheritance, if the minor allele frequency of gnomAD was >1:10 000, if the minor allele frequency of the local cohort was >1:1000, if not moderate or high impact, if the CADD PHRED score was ≤20, or if not predicted as damaging by either SIFT, PolyPhen, or MutationTaster.
Variant validation and segregation analysis were performed by Sanger sequencing with leucocyte- and/or fibroblast-derived DNA obtained from other members in Families 2 and 3. Their genotypes are annotated in the pedigrees in Fig. 1A and primer sequences are described in Supplementary Table 1.
Transcriptome sequencing and data analysis
Total RNA was extracted (Monarch Total RNA Miniprep Kit, New England Biolabs) from 42 primary skin fibroblast cultures of affected and healthy individuals including Patient 1. RNA concentration and purity of the samples were assessed by the EpochTM Microplate Spectrophotometer (BioTek).
RNA sequencing was done using polyA-enriched RNA sequencing (TruSeq Stranded mRNA Kit, Illumina). Paired-end sequencing (2 × 100 bp) was performed with an Illumina NovaSeq 6000 instrument at a sequencing depth of 60–100 million paired reads per sample and a minimum output of 12 Gb per sample. Output bcl files were converted to fastq files and demultiplexed using bcl2fastq (v2.20, Illumina). Adapter trimming was performed with Skewer (v0.2.2).27 Sequence reads were aligned to the human reference genome assembly (GRCh38.98) using STAR (v2.7.3a)28 in two-pass mode.
Detection of aberrantly expressed genes was performed with OUTRIDER (v1.5.0).29 All 42 samples were included in the analysis, but two of them were excluded from OUTRIDER‘s autoencoder and model fitting process to account for kinship. To increase detection power, additional samples of cultured fibroblasts were obtained from the Genotype-Tissue Expression (GTEx) portal on 08/28/2020. To account for possible technical biases, size factors were estimated for all samples using DESeq230 and only GTEx samples with size factors within the 15th to 85th percentile of those of the 42 sample cohort were included in subsequent analyses. 171 GTEx samples fit the criteria. Lowly expressed genes [fragments per kilobase per millions of reads (FPKM) ≤ 1 observed in at least 95% of all samples] were removed prior analysis. After filtering, 11 483 genes in combination with GTEx samples remained. OUTRIDER’s heuristic to automatically determine the autoencoder’s optimal encoding dimension suggested a value of 34 for our data. Accordingly, this value was applied. Significance was determined by adjusted P-value of 0.1.
For the detection and quantification of novel or known alternative splicing events, we used LeafCutter (v0.2.9).31 Transcriptome data derived from Patient 1 was compared to that of the 41 other samples. For intron clustering, the parameter for the maximum allowed length of introns was increased to 500 000 bp. For differential intron excision analysis the required minimum number of samples supporting an intron and the minimum number of samples per group was reduced to 1. Differentially spliced clusters with a false discovery rate (FDR) of 0.1 were visualized as Sashimi plots.
RNA isolation, qualitative and quantitative mRNA analyses
Total RNA was extracted (Monarch Total RNA Miniprep Kit, New England Biolabs) from cultured primary fibroblasts. RNA concentration and purity of the samples were assessed by the EpochTM Microplate Spectrophotometer (BioTek). Total RNA (1 µg) was reverse transcribed (LunaScript RT SuperMix Kit, New England Biolabs). Qualitative RT-PCR was performed on cDNA according to standard PCR protocols with the OneTaq Quick-Load 2× Master Mix (New England Biolabs). RT-PCR products were either directly Sanger-sequenced or cloned into the pCR2.1 TOPO TA Cloning Vector (Thermo Fisher Scientific) according to the manufacturer’s instructions. Escherichia coli clones were subjected to colony PCR, and PCR products from individual clones were Sanger-sequenced. Primer sequences are described in Supplementary Table 1.
For quantitative mRNA analysis, quantitative real-time PCR using a StepOnePlusTM system and predesigned TaqMan gene expression assays (Thermo Fisher Scientific, Supplementary Table 1) were performed as previously described.32 Expression of the housekeeping gene GAPDH was used as an internal control. Relative quantification was performed according to the comparative cycle threshold (ΔΔCT) method, and results were expressed in the linear form using the formula 2−ΔΔCT.
Antibodies
The following primary antibodies and dilutions were used: mouse monoclonal anti-β-actin (1:1000; Santa Cruz; #sc-47778; clone C4), rabbit polyclonal anti-CHERP (1:500; Abcam; #ab15951), rabbit polyclonal anti-DGCR14 (1:1000; Bethyl Laboratories Inc.; #A304-566A), mouse monoclonal anti-FLAG (1:5000; Sigma; #F1804; clone M2), mouse monoclonal anti-GFP (1:5000; BioLegend; #902601; clone B34), rabbit polyclonal anti-GFP (1:2000; Abcam; #ab290), rabbit polyclonal anti-FRA10AC1 (1:500; Bethyl Laboratories Inc.; #A304-738A), rabbit polyclonal anti-HA (1:200; Sigma-Aldrich; #H6908), rabbit polyclonal anti-nuclear factor κB-activating protein (NKAP; 1:500; Abcam; #ab229096), rabbit polyclonal anti-RED (1:1000; Bethyl Laboratories Inc.; #A301-708A), rabbit polyclonal anti-red fluorescent protein (RFP; 1:1000; Abcam; #ab62341), and rabbit polyclonal anti-SF3B2 (1:1000; Novus Biologicals; #NB100-79848).
The following secondary antibodies and dilutions were used: goat anti-rabbit IgG: StarBright Blue 700 antibody (1:10 000; Bio-Rad; #12004161), goat anti-mouse IgG: StarBright Blue 700 antibody (1:10 000; Bio-Rad; #12004158), hFAB Rhodamine anti-Tubulin IgG (1:10 000; Bio-Rad; #12004165), hFAB Rhodamine anti-GAPDH IgG (1:10 000; Bio-Rad; #12004167), goat anti-rabbit Alexa Fluor 488-conjugated secondary antibody (1:1000; Molecular Probes; #A-11008), anti-mouse-horseradish peroxidase (HRP) secondary antibody (1:5000; GE Healthcare; #NA931V), donkey anti-mouse IgG-HRP secondary antibody (1:5000; Jackson ImmunoResearch; 715–035-150), anti-rabbit-HRP secondary antibody (1:5000; GE Healthcare; #NA934V), donkey anti-rabbit IgG-HRP secondary antibody (1:5000; Jackson ImmunoResearch; 711–035-152), and anti-haemagglutinin (HA)-HRP secondary antibody (1:10 000; Sigma-Aldrich; #12013819001).
Cell culture and transfection
HEK293T cells and primary fibroblasts obtained from a skin biopsy of Patients 1 and 2, the mother of Patient 2 and two healthy individuals (controls) were cultured in Dulbecco’s modified Eagle medium (DMEM; Thermo Fisher Scientific) supplemented with 10% foetal bovine serum (GE Healthcare) and penicillin-streptomycin (100 U/ml and 100 µg/ml, respectively; Thermo Fisher Scientific) and incubated at 37°C in a humidified atmosphere with 5% CO2. Cells were tested for mycoplasma contamination by PCR and confirmed to be mycoplasma-free. Primer sequences are described in Supplementary Table 1.
Primary fibroblasts were transiently transfected with JetOptimus (Polyplus Transfection) and HEK293T cells with TurboFect or Lipofectamine 2000 (Thermo Fisher Scientific) for 6 h according to the protocol provided and subsequently cultured in 10% DMEM overnight. Next day, whole-cell lysates were prepared.
Plasmid information and cloning procedures
The coding region of human FRA10AC1 (NM_145246.5) was amplified with specific primers using human fibroblast-derived cDNA as template and the PCR product was cloned into pENTR/D-TOPO vector (Thermo Fisher Scientific) according to the manufacturer’s protocol. The 3-bp deletion c.494_496delAAG was introduced into the FRA10AC1 coding region by using the Quik Change II Site Directed Mutagenesis Kit (Agilent) according to manufacturer’s instructions. The FRA10AC1 coding region of wild-type and mutant constructs was subsequently cloned into the GATEWAY compatible destination vectors pMT2SM-HA-DEST and pEGFP-C3-DEST by using the Gateway LR Clonase II Enzyme Mix (Invitrogen). To generate FRA10AC1-FLAG-IRES-GFP, wild-type and mutant (c.494_496delAAG) FRA10AC1 coding region with C-terminal FLAG tag were synthesized by Integrated DNA Technologies and subsequently inserted between NheI and XhoI restriction sites of pcDNA3.1(+)IRES GFP (Addgene, #51406) using NEBuilder HiFi DNA Assembly kit (New England Biolabs). A SMU1 cDNA open reading frame clone (human) in a pcDNA3.1+/C-(K)DYK vector was purchased from GenScript Biotech (#OHu13306D) and used to amplify the coding region of human SMU1 (NM_018225.3). Relevant restriction sites in primer sequences (NotI in the forward and XbaI in the reverse primer) allowed recombination of the PCR product in pMT2SM-HA vector by In-Fusion HD Cloning (TaKaRa) according to the manufacturer’s protocol. pGint, a reporter vector containing an enhanced GFP (EGFP) open reading frame divided into two exons by an artificial, constitutively spliced intron was a gift from Mariano Garcia-Blanco (Addgene plasmid # 24217; http://n2t.net/addgene:24217; RRID: Addgene_24217).33 To eliminate a predicted canonical splice donor site within the EGFP open reading frame, the triplet GTG encoding valine 2 was deleted by using the QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies) according to the manufacturer’s protocol. Mutations of the donor (+1G>A and +2T>G) and acceptor (−1G>C and −2A>G) splice sites within the artificial intron were introduced by site-directed mutagenesis. All constructs were regularly sequenced for integrity, and primer sequences for site-directed mutagenesis are described in Supplementary Table 1.
Immunoblot analysis
Whole-cell lysates from patient and control fibroblasts were prepared. HEK293T cells were transfected as indicated and lysed the next day. Immunoblotting was performed as previously described.34 Briefly, cells were harvested in ice-cold RIPA buffer, protein extracts were separated on SDS-PAGE and transferred to polyvinylidene fluoride membranes. Membranes were blocked followed by incubation with the indicated primary antibody overnight at 4°C and by HRP-linked or fluorescent dye-linked secondary antibodies and Rhodamine anti-Tubulin antibody at room temperature for 1 h. Chemiluminescent and fluorescent signals were digitally imaged using a ChemiDoc MP (Bio-Rad). Band intensities of fluorescence and chemiluminescence signals were quantified using the Image Lab v6.0 software (Bio-Rad). For quantification of the amount of ectopically expressed FRA10AC1 wild-type and mutant proteins in HEK293T cells, HA- or GFP-FRA10AC1 was normalized to tubulin and the respective ratio of wild-type was set to 1.
Immunocytochemistry
HEK293T cells were cultivated on glass coverslips coated with poly-L-lysine (0.1 mg/ml; Sigma Aldrich; #P9155) for 1 h at room temperature followed by five times washing with sterile water. Subsequently, 300 000 cells per well were seeded on coverslips in 6-well plates and co-transfected with the HA- or GFP-tagged FRA10AC1 constructs together with the RFP encoding vector pmRFP-N2. The next day, cells were rinsed with PBS, fixed with 4% paraformaldehyde (Morphisto) in PBS, and washed three times with PBS. Cells transfected with HA-FRA10AC1 constructs were treated with permeabilization/blocking solution (2% BSA, 3% goat serum, 0.5% Nonidet P40 in PBS) und incubated in antibody solution (3% goat serum and 0.1% Nonidet P40 in PBS) containing anti-HA antibody for 3 h. Cells were washed with PBS and incubated with the Alexa Fluor 488-coupled goat anti-rabbit secondary antibody in antibody solution. After extensive washing with PBS cells were embedded in ProLong Diamond Antifade Mountant with DAPI (Invitrogen, #P36962) on microscopic slides. Cells transfected with GFP-FRA10AC1 constructs were processed in parallel without adding an antibody during antibody incubation steps. Cells were imaged with a widefield Zeiss fluorescence microscope system (Zeiss Axiovert 200M) equipped with a 63x oil immersion objective lens and an Apotome unit. All images were taken with the same camera settings [RFP (red) 30 ms, Alexa Fluor 488 (green) 15 ms, DAPI (blue) 30 ms] and were not subjected to further processing. For HA-tagged constructs, 70 randomly chosen RFP-positive cells were imaged per experiment and condition and used for quantification (see image data analysis). For GFP-tagged constructs, images of representative cells were taken.
Image data analysis
Image analysis was performed using Icy (v2.1.4.0) open-source imaging software.35 RFP-positive (red) cell nuclei were manually selected as regions of interest and mean Alexa Fluor 488 signal intensity (green) was measured. A mean Alexa Fluor 488 signal intensity of >20 (arbitrary units) was defined as cut-off.
Co-immunoprecipitation
Co-immunoprecipitations of endogenous splice factors were performed with PierceTM Anti-HA Magnetic Beads (Thermo Fisher Scientific, #88836). One million HEK293T cells were plated on 100 mm culture dishes and two plates per plasmid were transiently transfected as indicated. Cells were lysed in 400 µl co-immunoprecipitation buffer (50 mM Tris-HCl pH 8, 120 mM NaCl, 0.5% Nonidet P40) or mRIPA buffer (50 mM HEPES pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Nonidet P40, 0.5% sodium deoxycholate, 0.05% sodium dodecyl sulfate, SDS), both supplemented with complete Mini Protease Inhibitors (Roche) and PhosphoStop (Roche), for 10 min at 4°C, pooled and cell debris was cleared by centrifugation for 10 min at 4°C. After removing an aliquot (whole-cell lysate), the remaining supernatant was incubated with the beads for 2 h at 4°C on a rotator. Subsequently, the beads were pelleted and washed four times with co-immunoprecipitation or mRIPA buffer. The bound target proteins were eluted by resuspending the beads in 30 µl 2× sample buffer (33% glycerol, 80 mM Tris-HCl pH 6.8, 0.3 M DTT, 6.7% SDS, 0.1% bromophenol blue; diluted in co-immunoprecipitation or mRIPA buffer) and subjected to immunoblot analysis.
In vitro splicing assay
Primary fibroblasts (150 000) were plated on 6-well plates. Next day, cells were co-transfected with pGint-Val2del vectors as indicated together with pmRFP-N2 vector as an internal control for monitoring transfection efficiency. Cells were lysed in 200 µl co-immunoprecipitation buffer, cleared by centrifugation, and supplemented with 4× sample buffer. Samples were analysed by SDS-PAGE and immunoblotting.
Data analysis and statistics
Quantitative data are presented by GraphPad Prism 8 software (Instat, GraphPad Software) as the mean ± standard deviation (SD). For quantification of FRA10AC1 mRNA and protein levels, statistical analysis via one-way ANOVA followed by a Bonferroni post hoc test for multiple comparisons was performed. For quantification of protein amounts of ectopically expressed FRA10AC1 wild-type and mutant proteins in HEK293T cells, statistical significance was determined by Mann-Whitney test. For quantification of differences in FRA10AC1 wild-type and mutant green fluorescence signal intensities in immunofluorescence images, a two-tailed unpaired t-test was used to determine statistical significance. For statistical analysis of the in vitro splicing assay, two-way ANOVA followed by a Tukey post hoc test was used. A P-value <0.05 was considered statistically significant (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001).
Data availability
The authors declare that all data supporting the findings of this study are available within the paper and its Supplementary material. Consent restrictions preclude sharing of full datasets and the consents do not cover the deposition of the WES data in a public database.
Results
FRA10AC1 biallelic variants in individuals with a neurodevelopmental disorder
Through GeneMatcher,36 we identified three consanguineous families with one or three children affected by developmental delay, mild or profound intellectual disability, microcephaly, and corpus callosum abnormalities. The five affected individuals, Patient 1 (Family 1), Patient 2 (Family 2), and Patients 3–1 to 3–3 (Family 3), carried biallelic pathogenic variants in the FRA10AC1 gene (Fig. 1A). Four of the five affected individuals had low birth weight (Supplementary Table 2). Neonatal complications comprised respiratory and/or feeding difficulties in three patients. At last examination, all five patients showed postnatal growth retardation, four had low weight and microcephaly, and in two of them growth hormone deficiency has been identified. Patients 1 and 2 had severe developmental delay, absent speech, and profound intellectual disability. Patient 1 had seizures that are well controlled on medication. The three male siblings of Family 3 were more mildly affected. They spoke first words between 30 and 40 months of age and had borderline to mild intellectual disability. All patients had hypotonia. Brain ultrasound in Patient 1 revealed corpus callosum agenesis. Partial agenesis of the corpus callosum in Patient 2 and thinning of the corpus callosum in Patients 3–1 to 3–3 were identified by brain MRI (Fig. 1B). Common craniofacial dysmorphism in the five patients comprised high forehead, broad and medial flaring of eyebrows, narrow palpebral fissures, long nose, smooth philtrum, and everted lower lip vermillion. As the affected individuals age, the shape of the face become triangular with pointed chin as seen in Patients 2, 3–1, and 3–2 at age 9, 15, and 11 years, respectively (Fig. 1B). Clinodactyly of 5th finger or 4th and 5th toes was observed in the five patients. While additional anomalies and malformations were absent in Patients 2, 3–1, 3–2, and 3–3, Patient 1 had nevus flammeus of the forehead, nose, and philtrum, complex heart defect, caudally located left kidney, and additional skeletal abnormalities, such as short sternum, left elbow flexion contracture, ulnar deviation of left hand, and proximal placement of thumbs (Table 1, Supplementary Table 2, and Supplementary material).
Table 1.
Genetic and clinical characteristics of patients with biallelic FRA10AC1 variants
| Family/Patient | 1/1 | 2/2 | 3/3–1 | 3/3–2 | 3/3–3 |
|---|---|---|---|---|---|
| General information | |||||
| Gender | Female | Female | Male | Male | Male |
| Consanguinity | First degree cousins | First degree cousins | Second degree cousins | Second degree cousins | Second degree cousins |
| Ethnicity | Arabic | Arabic | Arabic | Arabic | Arabic |
| FRA10CA1 variant | NG_016832.1: | NM_145246.5: | NM_145246.5: | NM_145246.5: | NM_145246.5: |
| g.4656_7575del | c.561_562insTTTA | c.494_496del | c.494_496del | c.494_496del | |
| p.? | p.(Ser188Phefs*6) | p.(Glu165del) | p.(Glu165del) | p.(Glu165del) | |
| Last examination | |||||
| Age | 3 years 1 month | 9 years | 15 years | 10 years | 7 years |
| Height in cm (z-score) | 85 (−2.8) | 109 (−4.3) | 134 (−4.8) | 113.5 (−4.2) | 109 (−2.9) |
| Weight in kg (z-score) | 9.4 (−3.4) | 15 (−5.3) | 32 (−3.9) | 20.5 (−3.6) | 19 (−1.8) |
| OFC in cm (z-score) | 45 (−4.1) | 47 (−4.8) | 50.3 (−3.8) | 49.4 (−3.1) | 50 (−2.0) |
| Development | |||||
| Motor delay | Severe, nonambulatory | Moderate, walking at 3 years | – | During infancy | – |
| Intellectual disability | Profound | Profound, IQ 35 | Mild, IQ 68 | Borderline, IQ 77 | Borderline, IQ 77 |
| Neurological and psychiatric features | |||||
| Muscular hypotonia | + (severe) | + | + | + | + |
| Seizures | + | – | – | – | – |
| Behavioural problems | – | + | – | – | – |
| Brain abnormalities (brain ultrasound, MRI) | Corpus callosum agenesis; mild hydrocephalus internus | Partial agenesis of corpus callosum; colpocephaly; unilateral retroorbital cyst | Thin stretched corpus callosum | Thin stretched corpus callosum | Thin stretched corpus callosum |
| Craniofacial dysmorphism | + | + | + | + | + |
| Other anomalies | |||||
| Abnormality of the skeletal system | Short sternum; contracture of left elbow; ulnar deviation of left hand; proximal placement of thumbs; bilateral 5th finger clinodactyly | Bilateral 5th finger clinodactyly | Clinodactyly of 4th and 5th toes | Clinodactyly of 4th and 5th toes | Clinodactyly of 4th and 5th toes |
| Other | Feeding problems; gastric feeding tube; recurrent airway infections | Feeding problems; recurrent airway infections in early childhood | Growth hormone deficiency; anaemia | Growth hormone deficiency | – |
+/– = feature present/absent; IQ = intelligence quotient; OFC = occipital frontal circumference.
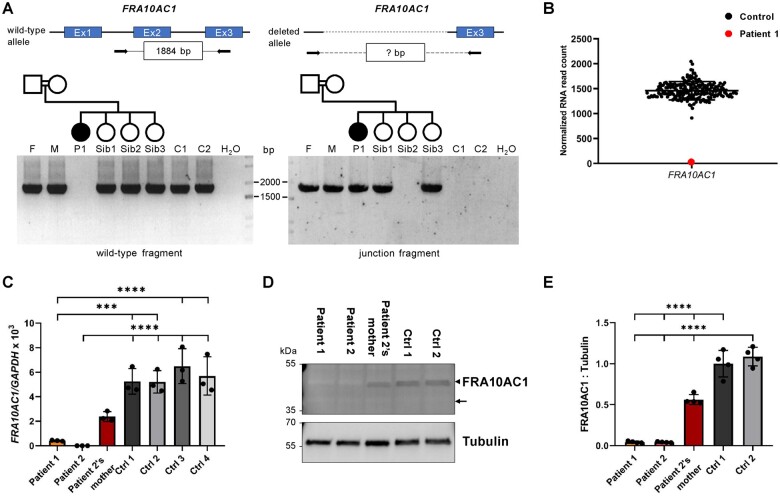
Using WES in Family 1 (index and healthy parents), we did not detect any heterozygous (de novo) or biallelic single nucleotide variant of pathogenic relevance in a disease gene or disease gene candidate in Patient 1 (Supplementary Table 3). CNV analysis of the trio exome data revealed a putative deletion of exons 1 and 2 in FRA10AC1 (NM_145246.5) in the homozygous state in Patient 1 and in the heterozygous state in parents. To confirm the presence of a partial homozygous FRA10AC1 deletion in Patient 1, we used a forward primer located in intron 1 (supposed to be deleted) and a reverse primer in exon 3 which did not produce an amplicon in Patient 1, while a wild-type fragment of 1884 bp was amplified from DNA of both parents, the three healthy sisters of Patient 1, and two controls (Fig. 2A). To generate a deletion-spanning fragment, we used a forward primer located upstream of FRA10AC1 exon 1 and the reverse primer in exon 3. A junction fragment of ∼1900 bp was obtained from Patient 1-derived DNA and DNA of both parents and two of the three siblings (Fig. 2A). Together, the data indicate homozygosity of the partial FRA10AC1 deletion in Patient 1 and heterozygosity in healthy parents and two siblings (Fig. 2A). We sequenced the junction fragment and identified a deletion of 2920 bp, NG_016832.1:g.4656_7575del [according to Human Genome Variation Society nomenclature; chr10:95,459,757–95,462,676 (hg19)], encompassing the non-coding exon 1, the coding exon 2, and the entire intron 1 of FRA10AC1, as well as 347 bp upstream of exon 1 and the first 30 bp of intron 2 (Supplementary Fig. 1). To analyse if the homozygous deletion leads to loss of FRA10AC1 expression, we performed transcriptome sequencing in Patient 1-derived fibroblasts and did not detect any sequence reads representing FRA10AC1 transcripts, while between 912–2045 normalized FRA10AC1 RNA read counts were identified in the control cohort (212 fibroblast cell lines; Fig. 2B).
Figure 2.
Biallelic FRA10AC1 loss-of-function variants in Patients 1 and 2. (A) FRA10AC1 junction PCR with DNA samples from Family 1. To validate the presence of the deletion of FRA10AC1 exons 1 and 2 in Family 1, we designed primers to amplify a FRA10AC1 wild-type fragment of 1884 bp (top left) and a deletion-spanning fragment of unknown size (question mark in the box in the top right panel). The wild-type fragment was not generated in Patient 1, but in her parents, the three healthy siblings, and two control individuals (bottom left). A junction fragment of ∼1900 bp amplified from Patient 1-derived DNA confirmed the FRA10AC1 deletion to be present in the homozygous state (bottom right). Amplification of both wild-type and junction fragment in the parents and siblings 1 and 3 indicate the FRA10AC1 deletion to be present in the heterozygous state (cf. bottom left and right panel). C1 and C2 = controls; F = father of Patient 1; M = mother of Patient 1; P1 = Patient 1; Sib1-Sib3 = healthy siblings of Patient 1. (B) Relative FRA10AC1 mRNA expression in fibroblasts of Patient 1. Transcriptome sequencing was performed with cDNA derived from 42 fibroblast cell lines including Patient 1 cells. The plot shows normalized FRA10AC1 RNA read count in the transcriptome data of the 42 fibroblast cell lines and included expression data of 171 fibroblast samples from the GTEx database. The red data-point represents FRA10AC1 read counts in Patient 1, while the black data-points represent those of the 212 other cell lines. The mean ± SD normalized read count of 213 samples is shown. (C) Quantification of the relative FRA10AC1 transcript levels in fibroblasts of Patients 1 and 2, the mother of Patient 2, and four controls by a TaqMan gene expression assay. GAPDH mRNA was used as an internal control. Relative quantification was performed according to the ΔCT method, and results were expressed in the linear form using the formula 2−ΔΔCT and multiplied by a factor of 1000. The mean ± SD of three independent experiments is shown. Statistical significance between controls and each patient was calculated by one-way ANOVA followed by Bonferroni post hoc test. ***P ≤ 0.001; ****P ≤ 0.0001. Datasets of three independent TaqMan gene expression assays with technical duplicates are provided in Supplementary Fig. 4. Ctrl = control. (D) Immunoblot of lysates obtained from fibroblasts of Patient 1, Patient 2, mother of Patient 2, and two controls. The amount of FRA10AC1 protein was monitored with an anti-FRA10AC1 antibody. An anti-Tubulin antibody was used to demonstrate equal loading. The predicted molecular mass of FRA10AC1 is ∼38 kDa (indicated by an arrow), although endogenous FRA10AC1 revealed a molecular mass of ∼45 kDa (indicated by an arrowhead). (E) Intensities of fluorescence signals were quantified using the ChemiDoc imaging system. The mean ± SD of four independent experiments is shown. One-way ANOVA with Bonferroni correction was used for statistical analysis: ****P ≤ 0.0001. Uncropped blots that were used for quantification are provided in Supplementary Fig. 5.
WES in Patient 2 and healthy parents revealed the homozygous 4-bp insertion c.561_562insTTTA in exon 9 of FRA10AC1 (NM_145246.5; total of 14 exons) predicting a frameshift and introduction of a premature stop codon [p.(Ser188Phefs*6)] in the affected girl. Any other homozygous variant that could underlie her phenotype was not detected (Supplementary Table 4). The FRA10AC1 variant is absent in public databases including gnomAD and predicted to be damaging by CADD (score: 32).37 The insertion was validated in Patient 2’s DNA. Her parents and healthy brother were heterozygous carriers of the variant (Fig. 1A and Supplementary Fig. 2A).
WES in two of the affected boys in Family 3 (Patients 3–2 and 3–3) revealed the shared homozygous 3-bp deletion c.494_496delAAG in FRA10AC1, predicting the in-frame deletion p.(Glu165del). No other shared homozygous variant was detected. The variant is absent in gnomAD, predicted to be damaging by CADD (score: 23.8),37 and Glu165 is evolutionarily highly conserved (Supplementary Fig. 3) and intolerant to variation as predicted by MetaDome.38 Sanger sequencing confirmed the variant to be present in all affected boys in the homozygous state and in their mother in the heterozygous state (father’s DNA was not available; Fig. 1A and Supplementary Fig. 2B). FRA10AC1 is slightly intolerant to functional genetic variation, as only 89% of the expected missense (observed/expected score: 0.89) and 78% of the expected loss-of-function variants (observed/expected score: 0.78) were found in gnomAD.39 Together, based on the absence of the identified FRA10AC1 variants in population databases, two homozygous loss-of-function variants, deletion of an evolutionary highly conserved amino acid residue with a possible damaging effect on protein function, as well as overlapping clinical features in the five affected individuals, biallelic FRA10AC1 variants likely underlie the neurodevelopmental disorder in all of them.
Loss of FRA10AC1 mRNA and protein in fibroblasts of Patients 1 and 2
To characterize the effect of biallelic FRA10AC1 variants on mRNA level, we generated fibroblast-derived cDNA and performed qualitative RT-PCR analysis followed by Sanger sequencing. FRA10AC1 transcripts with the c.561_562insTTTA variant were revealed in Patient 2, while only wild-type transcripts could be detected in her mother, suggesting nonsense-mediated decay of transcripts expressed from the mutant allele (Supplementary Fig. 2A). We next determined relative FRA10AC1 mRNA levels. FRA10AC1 mRNA amount was reduced to 6–8% and 0% in fibroblasts of Patients 1 and 2, respectively, compared with control cells (Fig. 2C and Supplementary Fig. 4). Detection of a minimal amount of FRA10AC1 mRNAs in Patient 1 by the TaqMan gene expression assay was likely due to non-specific amplification. Fibroblasts of Patient 2’s mother had 37–47% of FRA10AC1 mRNA levels compared to control cells (Fig. 2C and Supplementary Fig. 4). These data are in line with the lack of FRA10AC1 mRNAs in Patient 1’s transcriptome and underscore effective nonsense-mediated decay of FRA10AC1 mRNAs harboring loss-of-function variants.
We next checked FRA10AC1 protein in whole-cell lysates from cultured primary fibroblasts using immunoblotting. The predicted molecular mass of FRA10AC1 is 38 kDa. However, endogenous FRA10AC1 had a molecular mass of ∼45 kDa in control cells (Fig. 2D and Supplementary Fig. 5), suggesting potential posttranslational modification(s) of the mature protein. Qualitatively, no FRA10AC1 was detected in fibroblasts of Patients 1 and 2 compared to control cells (Fig. 2D and Supplementary Fig. 5). Quantification of immunoblot data, however, suggested ∼4% of FRA10AC1 in both Patients (Fig. 2E) although this was likely an artifactual signal due to a slightly low signal-to-noise ratio of the applied anti-FRA10AC1 antibody. In cells of Patient 2’s mother, 52–56% of FRA10AC1 was detected compared with control cells (Fig. 2D and E and Supplementary Fig. 5). The data suggests that Patients 1 and 2 carry biallelic FRA10AC1 complete loss-of-function alleles, while the impact of the FRA10AC1 c.494_496delAAG, p.(Glu165del) variant on the endogenous protein could not be determined due to lack of patient-derived cells.
The p.Glu165del variant impacts FRA10AC1 stability but not its subcellular localization
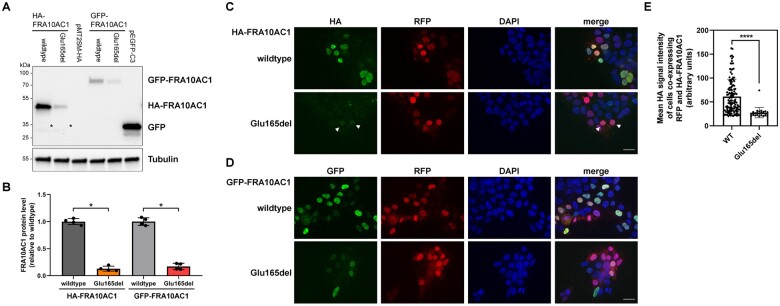
To characterize the effect of the one-amino-acid deletion p.Glu165del on FRA10AC1 protein level, we generated HA- and GFP-tagged FRA10AC1 wild-type and Glu165del mutant constructs, transiently transfected HEK293T cells, and detected FRA10AC1 protein in whole-cell lysates using anti-HA and anti-GFP antibodies. Ectopically expressed HA-FRA10AC1 wild-type and Glu165del mutant had a molecular mass of ∼45 kDa (Fig. 3A and Supplementary Fig. 6), similar to the endogenous protein (Fig. 2D and Supplementary Fig. 5), and the GFP-tagged proteins run at ∼70 kDa (Fig. 3A and Supplementary Fig. 6). Levels of FRA10AC1-Glu165del were drastically reduced compared with wild-type: ∼13% of HA-tagged and ∼17% of GFP-tagged FRA10AC1-Glu165del were detected compared with HEK293T cells expressing the respective wild-type protein (Fig. 3B). By using pcDNA3.1(+) plasmids with FRA10AC1-FLAG-IRES-GFP, enabling simultaneous translation of FLAG-tagged FRA10AC1 and GFP from a single transcript, the amount of C-terminally FLAG-tagged FRA10AC1-Glu165del in HEK293T cells was only ∼19% compared to wild-type level, while the GFP amount was similar in cells expressing wild-type and mutant FRA10AC1 (Supplementary Fig. 7). These data show the same transfection efficiency for FRA10AC1 expression constructs and indicate intrinsic instability of FRA10AC1 lacking glutamate 165.
Figure 3.
Ectopic expression of FRA10AC1 wild-type and the Glu165del mutant in HEK293T cells. (A) HEK293T cells were transfected with HA-tagged or GFP-tagged FRA10AC1 wild-type and FRA10AC1-Glu165del mutant constructs as indicated. pMT2SM-HA and pEGFP-C3 vectors were used as negative controls. FRA10AC1 proteins were detected in whole-cell lysates by immunoblotting using an anti-GFP or anti-HA antibody. An anti-Tubulin antibody was used to demonstrate equal loading. A non-specific band at ∼30 kDa was detected with the anti-HA antibody (marked with an asterisk). Uncropped blots that were used for quantification are shown in Supplementary Fig. 6. (B) Band intensities of fluorescence and chemiluminescence signals were quantified using the ChemiDoc imaging system. The mean ± SD of four independent experiments is shown, and individual data-points from each experiment are shown. Mann-Whitney test was used for statistical analysis: *P ≤ 0.05. (C and D) HEK293T cells were cultured on poly-L-lysine coated coverslips and co-transfected with HA-FRA10AC1 expression constructs as indicated together with RFP expressing plasmid. The next day, cells were fixed and HA-FRA10AC1 was stained by rabbit anti-HA antibody followed by anti-rabbit Alexa Fluor 488-conjugated antibody (green). Nuclear DNA was labelled by DAPI (blue). Cells were imaged by widefield fluorescence microscopy and representative images are shown. Arrowheads point to cells expressing RFP and a low level of the HA-FRA10AC1-Glu165del mutant (C). Scale bar = 20 µm. (E) Quantification of HA-FRA10AC1 wild-type and Glu165del mutant expressing cells. Seventy RFP-positive cells per condition and experiment (n = 3) were imaged and the mean Alexa Fluor 488 signal intensity (green) was measured. A mean signal intensity >20 arbitrary units were defined as HA-positive cells and are depicted as data-points. A two-tailed unpaired t-test was used for statistical analysis: ****P ≤ 0.0001. WT = wild-type.
EGFP-tagged FRA10AC1 has been reported to localize to the nucleus and most prominently in the nucleoplasm.9 To examine if the p.Glu165del variant influences subcellular localization of FRA10AC1, we transiently expressed HA- or GFP-tagged FRA10AC1 wild-type and the Glu165del mutant in HEK293T cells followed by immunofluorescence microscopy. For quantification reasons, we co-transfected cells with an RFP-expressing plasmid. FRA10AC1 wild-type and Glu165del mutant proteins consistently localized to the nucleus (Fig. 3C and D). To quantify fluorescence intensity of HA-tagged FRA10AC1 wild-type versus Glu165del mutant transfected cells, we imaged RFP-positive cell nuclei randomly and measured the mean HA signal intensity (green) within the nucleus. A total of 210 RFP-positive cells were imaged for signal intensity of HA-FRA10AC1 wild-type and HA-FRA10AC1-Glu165del each (from three independent experiments). Cell nuclei with a HA mean signal intensity of >20 arbitrary units were defined as HA-positive cells (Fig. 3E). Out of the 210 RFP-positive cells, 122 HA-FRA10AC1 wild-type and 24 HA-FRA10AC1-Glu165del mutant expressing cells passed this filter. The mean signal intensity was reduced by ∼50% in HA-FRA10AC1-Glu165del mutant expressing cells compared to HA-FRA10AC1 wild-type expressing cells [mean signal intensity of 61.38 (arbitrary units) for wild-type compared to 27.59 (arbitrary units) for the Glu165del mutant] (Fig. 3E). Taking the lower number of cells expressing both RFP and the HA-FRA10AC1-Glu165del mutant into account, the overall measured signal intensity of HA-FRA10AC1-Glu165del mutant expressing cells was ∼9% of that of HA-FR10AC1 wild-type expressing cells, reflecting the results from immunoblot experiments (Fig. 3B). Together, these data indicate that the Glu165del variant has an impact on FRA10AC1 stability, while it does not affect FRA10AC1’s localization in the cell.
DGCR14, a component of the spliceosomal C complex, co-immunoprecipitates with HA-tagged FRA10AC1
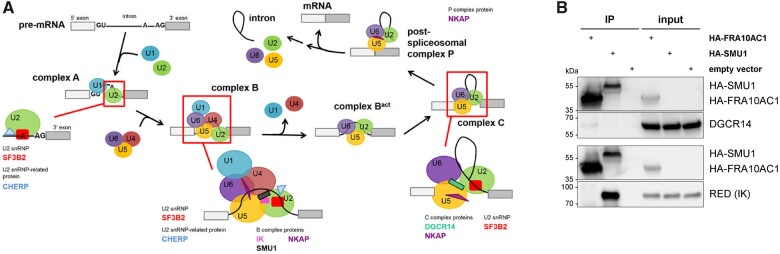
The function of the spliceosomal FRA10AC1 protein as a C complex-specific protein is mainly unknown.7,8 By searching the protein pattern and domain databases Pfam, PROSITE, and SMART, we did not find any significant homology of FRA10AC1 to known protein motifs or domains. Similarly, by using Phyre2 and SWISS-MODEL no potential structural homology of FRA10AC1 to known protein structures could be predicted.40 The BioGrid database lists 40 putative FRA10AC1 interacting proteins.41 The function of the 40 proteins and/or associated pathways can be categorized into 13 groups; 15 proteins play a role in the pre-mRNA splicing process (Supplementary Table 5). To study complex formation between FRA10AC1 and some of the proposed binding partners, we selected SF3B2, CHERP, IK, NKAP, and DGCR14 for this experiment because (i) they have a well-known role in splicing (see below); (ii) they represent components of the spliceosomal A, B, and C complexes (Fig. 4A); and (iii) established antibodies are available for these proteins. SF3B2 (splicing factor 3B subunit 2) is one of seven subunits of the splicing factor 3B (SF3B), an essential component of the U2 snRNP that interacts with the pre-mRNA at the branch site sequence and is present in all spliceosomal complexes during pre-mRNA splicing.7,42–46 Calcium homeostasis endoplasmic reticulum protein (CHERP) is a 17S U2-related protein that is only associated with the U2 snRNP during the formation of the spliceosomal A and B complexes.7,45,47 During B complex assembly, IK (also called RED protein) interacts with the spliceosome and is released after B to Bact transition. RED binds to SMU1, another B complex-specific protein.7,44,48–50 Nuclear factor κB-activating protein is part of the B, C, and P complexes promoting pre-mRNA processing and exon ligation.7,51–53 DGCR14 is a non-core component of the spliceosome C complex and proposed to ensure accurate splicing (Fig. 4A).7,10–13,51
Figure 4.
FRA10AC1 is in complex with DGCR14, a component of the spliceosomal C complex. (A) Schematics of the pre-mRNA splicing process. The illustration was adapted from Wahl et al.1 The stepwise interaction of the spliceosomal snRNPs (coloured circles U1, U2, U4–U6) in the assembly and disassembly cycle of the major spliceosome is shown. Putative FRA10AC1 interaction partners listed in the BioGrid database and examined in co-immunoprecipitation experiments are indicated (protein names and polygons in the scheme are given in different colours). SMU1 is a component of the spliceosomal B complex and was used as a positive control in co-immunoprecipitation experiments. Only spliceosomal complexes implicated in mammalian splicing are depicted; non-snRNP proteins (except putative FRA10AC1 interaction partners and SMU1) are not displayed for simplicity. (B) HEK293T cells were transfected with the HA-FRA10AC1 wild-type construct as indicated. HA-SMU1 was used as a positive control to co-immunoprecipitate endogenous RED (IK). An empty vector (pMT2SM-HA) was used as negative control. Overexpressed HA-FRA10AC1 was immunoprecipitated from whole-cell lysates using HA-specific magnetic beads. Co-precipitated endogenous splice factors were detected by immunoblotting using anti-DGCR14 and anti-RED antibodies. HA-FRA10AC1 and HA-SMU1 were monitored with an anti-HA antibody. Representative blots from two separate experiments of four independent experiments each are shown. HA-SMU1 showed a very weak expression and cannot be visualized in whole-cell lysates (input) under the applied conditions, but was detectable after enrichment in immunoprecipitate (IP) samples. Uncropped blots are provided in Supplementary Fig. 9.
We performed co-immunoprecipitation assays to study complex formation between ectopically expressed HA-FRA10AC1 and the endogenous splicing factors CHERP, DGCR14, NKAP, RED, and SF3B2 by using specific antibodies. As a positive control, we ectopically expressed HA-tagged SMU1, a known interaction partner of RED (see above). We successfully and specifically detected endogenous CHERP, NKAP, RED, and SF3B2 in total cell lysates of HEK293T cells ectopically expressing HA-FRA10AC1 or HA-SMU1 (Fig. 4B and Supplementary Figs. 8 and 9). We could demonstrate efficient co-immunoprecipitation of endogenous RED with HA-SMU1 (Fig. 4B and Supplementary Fig. 9). While we did not observe any co-precipitation of CHERP, NKAP, RED, and SF3B2 with HA-tagged FRA10AC1 (Fig. 4B and Supplementary Figs 8 and 9), endogenous DGCR14 was found to co-immunoprecipitate with HA-FRA10AC1 (Fig. 4B and Supplementary Fig. 9). The data suggest that FRA10AC1 is in complex with DGCR14 and both form part of the spliceosomal C complex, while none of the other investigated splice factors co-immunoprecipitated with ectopically expressed FRA10AC1 in HEK293T cells.
FRA10AC1 deficiency did not suppress missplicing events in an in vitro splicing assay in fibroblasts
Cumulative data obtained in the plant Arabidopsis thaliana, the green alga Chlamydomonas reinhardtii, and the worm Caenorhabditis elegans revealed DGCR14 to discriminate between canonical and non-canonical splice sites and to promote accurate pre-mRNA splicing when splice sites are compromised.11–13 A similar function in splicing was identified for Fra10ac1: a frameshift mutation in C. reinhardtii fra10 acts as a suppressor and leads to restoration of wild-type IFT121 pre-mRNA splicing despite the presence of a 5′ splice site mutation in the IFT121 gene.11 To investigate the effect of FRA10AC1 deficiency on recognition of compromised (mutated) canonical splice sites, we performed an in vitro splicing assay in primary fibroblasts. For this purpose, we used the slightly modified reporter vector pGint-Val2del (see methods), which encodes EGFP with the open reading frame split into two exons by an artificial intron containing strong canonical splice sites, branch point, and polypyrimidine tract sequences.33 We mutated each of the dinucleotides of the donor and acceptor splices sites of the artificial intron in pGint-Val2del (Supplementary Fig. 10 and Supplementary Table 1). Transient transfection of the wild-type pGint-Val2del plasmid resulted in comparable EGFP protein amounts in cells of Patient 1, Patient 2’s mother and controls (Supplementary Fig. 10A). EGFP protein levels in Patient 2’s fibroblasts were increased by 1.6- and 1.4-fold compared to cells of control 1 and 2, respectively (Supplementary Fig. 10B). However, we did not expect this to be a general effect of FRA10AC1 deficiency as EGFP levels in fibroblasts of Patient 1 were not significantly increased compared to control cells. Mutation in +2 intronic position of the splice donor [pGint-Val2del(+2T>G)] caused a reduction in EGFP levels by 80–90% compared to EGFP levels in fibroblasts transfected with pGint-Val2del, however, EGFP levels in patient-derived fibroblasts did not differ from control cells, including Patient 2’s mother cells (Supplementary Fig. 10). Mutation in +1 [pGint-Val2del(+1G>A)] and −1 [pGint-Val2del(−1G>C)] intronic positions resulted in no detectable EGFP protein in all cell lines (Supplementary Fig. 10A), while mutation in −2 intronic position [pGint-Val2del(−2A>G)] caused expression of very low and not quantifiable EGFP levels in the five fibroblast cells lines (Supplementary Fig. 10A). Taken together, FRA10AC1 deficiency did not suppress aberrant splicing of the artificial EGFP intron harboring a mutation in each of the invariant dinucleotides of the donor and acceptor sites in an in vitro splicing assay in human fibroblast cells.
Alternative splicing is not significantly disturbed in Patient 1 fibroblasts
We finally evaluated transcriptome sequencing data of Patient 1’s fibroblasts for different forms of alternative splicing events, such as skipping of exons, mutually exclusive exons, alternative 5′ and 3′ splice sites selection, and retained introns, and identified three significant alternative splicing events for the genes SRGAPC2, TMEM63A, and TBC1D31 (Supplementary Fig. 11). Partial skipping of SRGAPC2 exon 3 (190 bp) (NM_001329984.2) and of TMEM63A exon 11 (81 bp) (NM_014698.3), as well as complete skipping of the TBC1D31 exon 19 (195 bp) (NM_145647.4) were observed in Patient 1 cells (Supplementary Fig. 11A–C). We confirmed these alternative splicing events by performing RT-PCR using fibroblast-derived cDNA of Patient 1 and three controls (Supplementary Fig. 11D). To analyse these alternative splicing events in Patient 2, we included fibroblast-derived cDNA of Patient 2 and Patient 2’s mother in the RT-PCRs. We did not observe any preferential exon skipping event for SRGAPC2 exon 3, TMEM63A exon 11, and TBC1D31 exon 19 in Patient 2 fibroblasts, while cells of Patient 2’s mother showed partial skipping of TMEM63A exon 11, similar to Patient 1 (Supplementary Fig. 11D). We conclude that the alternative splicing events for SRGAPC2, TMEM63A, and TBC1D31 in Patient 1 cells were transcript variant switching events whose biological significance is unclear. As these alternative splicing events were not detected in Patient 2 cells, alternative splicing integrity is not significantly disturbed in fibroblasts of individuals with biallelic pathogenic FRA10AC1 variants.
Discussion
Here we demonstrate biallelic pathogenic variants in FRA10AC1 cause a neurodevelopmental disorder, characterized by global developmental delay, mild to profound intellectual disability, microcephaly, absent or hypoplastic corpus callosum, hypotonia, and growth retardation. The five affected individuals had a recognizable facial gestalt with high forehead, broad and medial flaring of eyebrows, narrow palpebral fissures, long nose, smooth philtrum, and everted lower lip vermillion. With age, the shape of the face became long and triangular and the chin pointed. While four of the five affected individuals showed predominant neurological impairment, Patient 1 was more severely affected and had additional clinical features. She had seizures, a complex heart defect, left displaced kidney, and several skeletal abnormalities. Patients 1 and 2 carried homozygous FRA10AC1 null alleles that are possibly associated with a more severe clinical picture, such as profound intellectual disability in both individuals and organ anomalies in Patient 1. The three siblings with the homozygous FRA10AC1 p.Glu165del variant had speech delay, mild intellectual disability, and a thin corpus callosum suggesting a less severe impact of the variant on FRA10AC1 function. Indeed, the FRA10AC1-Glu165del mutant protein was detected in the cell nucleus under overexpression, albeit to a much lower amount than wild-type. Similarly, clinical severity of the neurodegenerative phenotype in individuals with biallelic variants in PPIL1 and PRP17, encoding core spliceosomal proteins, has been correlated with the type of variant having different impacts on protein stability or function.26 The findings suggest a possible genotype-phenotype correlation in patients with biallelic pathogenic FRA10AC1 variants and a broad clinical spectrum that may range from severe neurological abnormalities with various organ malformations to mild intellectual disability as main clinical feature. Six male patients with a pathogenic missense variant in the X-linked FAM50A gene, encoding a protein of the spliceosomal C complex similar to FRA10AC1, also show a syndromic intellectual disability phenotype with postnatal growth retardation, ocular findings, heart defects in three, and genitourinary anomalies including horseshoe kidney and unilateral renal agenesis in two.54 It is well known that individuals affected by a spliceosomopathy can show syndromic phenotypes. Patients with craniofacial spliceosomopathies and pathogenic variants in genes encoding core components of the spliceosome (SNRPB, RNU4ATAC, SF3B4, PUF60, EFTUD2, TXNL4, EIF4A3, and CWC27) exhibit intellectual disability, microcephaly, micrognathia, malar hypoplasia, external ear anomalies, eye anomalies, limb and heart defects. Abnormalities of the kidney have rarely been reported.55 Nevertheless, additional genetic and environmental factors may also modulate the phenotype of spliceosomopathies. For example, exome data in Patient 1 revealed the homozygous missense variant c.207G>C (p.Glu69Asp) in XIRP2 (absent in gnomAD) (Supplementary Table 3). Ablation of Xirp2 in the mouse caused abnormal heart shape, ventricular septal defects, severe growth retardation, and postnatal lethality,56 and further studies revealed an important role of the encoded protein in cardiac development and function.57,58 As no CNV associated with heart anomalies has been detected in Patient 1 (F. L. Harms and K. Kutsche, personal communication), the XIRP2 variant might have contributed to her congenital heart defects.
We showed that endogenous DGCR14 co-precipitates with ectopically expressed FRA10AC1, while other spliceosomal proteins, such as endogenous CHERP, NKAP, RED, and SF3B2, failed to co-immunoprecipitate with HA-tagged FRA10AC1. Although part of the spliceosomal C complex, DGCR14 and FRA10AC1 are not core spliceosomal proteins.10 In line with this, we only observed minor changes in alternative pre-mRNA splicing of the genes SRGAPC2, TMEM63A, and TBC1D31 in the transcriptome sequencing data of Patient 1-derived fibroblasts. These transcript variant switching events could not be recapitulated in fibroblast-derived RNA (cDNA) of Patient 2 by RT-PCR analysis indicating that FRA10AC1 deficiency did not result in disruption of gene-specific or global alternative splicing integrity. Transcriptome sequencing in C. reinhardtii dgr14 and fra10 single mutants and the double mutant revealed only few changes in pre-mRNA splicing and RNA abundance,11 that is in accordance with the absence of any splicing defect in the C. elegans Dgcr14 mutant.12 The single C. reinhardtii mutants dgr14 and fra10 did not show any defects in viability, mating efficiency, and mobility,11 that is consistent with a normal phenotype of yeast, C. elegans, and A. thaliana mutated in the respective Dgcr14 homologue.12,13,59 Of note, fission yeast bis1 (Dgcr14 orthologue) mutant cells show a reduced cell survival in the stationary phase.59
In C. reinhardtii Fra10ac1 facilitates splicing as a mutant alga with a Fra10ac1 1-bp deletion tolerates splice site mutations and shows wild-type splicing of the mutated gene.11 Using an in vitro splicing assay in fibroblasts from controls and individuals with heterozygous or biallelic FRA10AC1 variants, we were not able to recapitulate the published data in human cells. Mutations disrupting each of the highly conserved dinucleotides of the splice donor and acceptor in the intron of the splicing reporter plasmid did not lead to enhanced wild-type splicing in human fibroblasts deficient of FRA10AC1. These data together with no obvious severe defects in pre-mRNA splicing in Patient 1’s fibroblasts point toward a cell type-specific function of FRA10AC1 in the spliceosomal C complex. FRA10AC1 is an ubiquitously expressed gene (https://gtexportal.org/home/gene/FRA10AC1). However, during early human development FRA10AC1 shows expression in the cerebrum and cerebellum (https://www.ebi.ac.uk/gxa/home)60 that might be important for normal neuronal development.
Distribution of DGCR14 and FRA10AC1 across multiple eukaryotic kingdoms suggests absence of FRA10AC1 in organisms with low intron density and complexity.11 Thus, Lin et al.11 proposed the requirement of additional spliceosomal proteins for recognition of exon-intron boundaries and enhancement of splicing. The importance of pre-mRNA splicing integrity for normal brain development has recently been demonstrated by the identification of hemizygous FAM50A variants in male individuals with Armfield X-linked intellectual disability syndrome and biallelic pathogenic variants in genes encoding the spliceosome components PPIL1 and PRP17 in individuals with a neurodegenerative form of pontocerebellar hypoplasia with microcephaly (PCHM).26,54 A higher susceptibility of neurons to alterations in pre-mRNA splicing and/or perturbation of neural-specific splicing has been suggested to underlie neurodegenerative and other neurological phenotypes in humans.26,61 In addition to FRA10AC1’s likely role in pre-mRNA splicing, it may have additional functions as suggested by the finding of many putative interactors not involved in splicing (Supplementary Table 5). For example, DGCR14 that is a component of the spliceosomal C complex together with FRA10AC1 associates with the retinoid-related orphan nuclear receptor gamma (RORγ). RORγ is a transcription factor and DGCR14 acts as a transcriptional coactivator for RORγ.62,63 The DGCR14 yeast orthologue Bis1 interacts with Ish1, a protein of the nuclear envelope, and both Bis1 and Ish1 play a role in maintaining yeast viability in the stationary phase.59 These data may suggest other cellular functions of DGCR14 and also FRA10AC1, such as coupling of transcription and splicing reactions as proposed for DGCR14.63
In conclusion, our data show the importance of specific peripheral spliceosomal C complex proteins for neurodevelopment. In the absence of experimental evidence for FRA10AC1 to be involved in pre-mRNA splicing in patient-derived cells, this poorly characterized nuclear protein may have to-be-discovered cellular functions.
Supplementary Material
Acknowledgements
We thank all probands and families for their participation in this study, Inka Jantke and Dennis Zorndt for skillful technical assistance, and the UKE Microscopy Imaging Facility (UMIF) for their technical support.
Funding
This work was supported by grants from the Deutsche Forschungsgemeinschaft (KU 1240/6–2 and KU 1240/10–1 to K.K.), NIH grants (R01NS048453 and R01NS09800), Rady Children’s Institute for Genomic Medicine (to J.G.G.), and Egypt Science and Technology Development fund (STDF) project 26040.
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
Abbreviations
- CNV
copy number variation
- snRNPs
small nuclear ribonucleoproteins
References
- 1. Wahl MC, Will CL, Luhrmann R.. The spliceosome: Design principles of a dynamic RNP machine. Cell. 2009;136(4):701–718. [DOI] [PubMed] [Google Scholar]
- 2. Will CL, Luhrmann R.. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011;3(7):a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shi Y. Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat Rev Mol Cell Biol. 2017;18(11):655–670. [DOI] [PubMed] [Google Scholar]
- 4. Rappsilber J, Ryder U, Lamond AI, Mann M.. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12(8):1231–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhou Z, Licklider LJ, Gygi SP, Reed R.. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419(6903):182–185. [DOI] [PubMed] [Google Scholar]
- 6. Matera AG, Wang Z.. A day in the life of the spliceosome. Nat Rev Mol Cell Biol. 2014;15(2):108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bessonov S, Anokhina M, Will CL, Urlaub H, Lührmann R.. Isolation of an active step I spliceosome and composition of its RNP core. Nature. 2008;452(7189):846–850. [DOI] [PubMed] [Google Scholar]
- 8. Schmidt C, Gronborg M, Deckert J, et al. Mass spectrometry-based relative quantification of proteins in precatalytic and catalytically active spliceosomes by metabolic labeling (SILAC), chemical labeling (iTRAQ), and label-free spectral count. RNA. 2014;20(3):406–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sarafidou T, Kahl C, Martinez-Garay I, et al. ; European Collaborative Consortium for the Study of ADLTE . Folate-sensitive fragile site FRA10A is due to an expansion of a CGG repeat in a novel gene, FRA10AC1, encoding a nuclear protein. Genomics. 2004;84(1):69–81. [DOI] [PubMed] [Google Scholar]
- 10. Hegele A, Kamburov A, Grossmann A, et al. Dynamic protein-protein interaction wiring of the human spliceosome. Mol Cell. 2012;45(4):567–580. [DOI] [PubMed] [Google Scholar]
- 11. Lin H, Zhang Z, Iomini C, Dutcher SK.. Identifying RNA splicing factors using IFT genes in Chlamydomonas reinhardtii. Open Biol. 2018;8(3):170211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Noma K, Goncharov A, Jin Y.. Systematic analyses of rpm-1 suppressors reveal roles for ESS-2 in mRNA splicing in Caenorhabditis elegans. Genetics. 2014;198(3):1101–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kanno T, Venhuizen P, Wu MT, et al. A collection of Pre-mRNA splicing mutants in Arabidopsis thaliana. G3 (Bethesda). 2020;10(6):1983–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schneeberger PE, Nayak SS, Fuchs S, Kutsche K, Girisha KM.. Roberts syndrome in an Indian patient with humeroradial synostosis, congenital elbow contractures and a novel homozygous splice variant in ESCO2. Am J Med Genet A. 2020;182(11):2793–2796. [DOI] [PubMed] [Google Scholar]
- 15. Schneeberger PE, von Elsner L, Barker EL, et al. Bi-allelic pathogenic variants in HS2ST1 cause a syndrome characterized by developmental delay and corpus callosum, skeletal, and renal abnormalities. Am J Hum Genet. 2020;107(6):1044–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li H, Durbin R.. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang K, Li M, Hakonarson H.. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen S, Zhou Y, Chen Y, Gu J.. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics (Oxford, England). 2018;34(17):i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li H, Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li H, Handsaker B, Wysoker A, et al. ; 1000 Genome Project Data Processing Subgroup . The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Plagnol V, Curtis J, Epstein M, et al. A robust model for read count data in exome sequencing experiments and implications for copy number variant calling. Bioinformatics. 2012;28(21):2747–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Conrad DF, Bird C, Blackburne B, et al. Mutation spectrum revealed by breakpoint sequencing of human germline CNVs. Nat Genet. 2010;42(5):385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. MacDonald JR, Ziman R, Yuen RK, Feuk L, Scherer SW.. The Database of Genomic Variants: A curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42(Database issue):D986–D992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lappalainen I, Lopez J, Skipper L, et al. DbVar and DGVa: public archives for genomic structural variation. Nucleic Acids Res. 2013;41(Database issue):D936–D941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chai G, Webb A, Li C, et al. Mutations in spliceosomal genes PPIL1 and PRP17 cause neurodegenerative pontocerebellar hypoplasia with microcephaly. Neuron. 2021;109(2):241–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jiang H, Lei R, Ding SW, Zhu S.. Skewer: A fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics. 2014;15:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dobin A, Davis CA, Schlesinger F, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brechtmann F, Mertes C, Matusevičiūtė A, et al. OUTRIDER: A statistical method for detecting aberrantly expressed genes in RNA sequencing data. Am J Hum Genet. 2018;103(6):907–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li YI, Knowles DA, Humphrey J, et al. Annotation-free quantification of RNA splicing using LeafCutter. Nat Genet. 2018;50(1):151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Di Lorenzo G, Velho RV, Winter D, et al. Lysosomal proteome and secretome analysis identifies missorted enzymes and their nondegraded substrates in mucolipidosis III mouse cells. Mol Cell Proteomics. 2018;17(8):1612–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bonano VI, Oltean S, Garcia-Blanco MA.. A protocol for imaging alternative splicing regulation in vivo using fluorescence reporters in transgenic mice. Nat Protoc. 2007;2(9):2166–2181. [DOI] [PubMed] [Google Scholar]
- 34. Schneeberger PE, Kortüm F, Korenke GC, et al. ; Undiagnosed Diseases Network . Biallelic MADD variants cause a phenotypic spectrum ranging from developmental delay to a multisystem disorder. Brain. 2020;143(8):2437–2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. de Chaumont F, Dallongeville S, Chenouard N, et al. Icy: An open bioimage informatics platform for extended reproducible research. Nat Methods. 2012;9(7):690–696. [DOI] [PubMed] [Google Scholar]
- 36. Sobreira N, Schiettecatte F, Boehm C, Valle D, Hamosh A.. New tools for Mendelian disease gene identification: PhenoDB variant analysis module; and GeneMatcher, a web-based tool for linking investigators with an interest in the same gene. Hum Mutat. 2015;36(4):425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J.. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wiel L, Baakman C, Gilissen D, Veltman JA, Vriend G, Gilissen C.. MetaDome: Pathogenicity analysis of genetic variants through aggregation of homologous human protein domains. Hum Mutat. 2019;40(8):1030–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Karczewski KJ, Francioli LC, Tiao G, et al. ; Genome Aggregation Database Consortium . The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Waterhouse A, Bertoni M, Bienert S, et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Oughtred R, Stark C, Breitkreutz BJ, et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019;47(D1):D529–D541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Will CL, Schneider C, Reed R, Lührmann R.. Identification of both shared and distinct proteins in the major and minor spliceosomes. Science. 1999;284(5422):2003–2005. [DOI] [PubMed] [Google Scholar]
- 43. Will CL, Urlaub H, Achsel T, Gentzel M, Wilm M, Lührmann R.. Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. Embo J. 2002;21(18):4978–4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Deckert J, Hartmuth K, Boehringer D, et al. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol Cell Biol. 2006;26(14):5528–5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Agafonov DE, Deckert J, Wolf E, et al. Semiquantitative proteomic analysis of the human spliceosome via a novel two-dimensional gel electrophoresis method. Mol Cell Biol. 2011;31(13):2667–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sun C. The SF3b complex: Splicing and beyond. Cell Mol Life Sci. 2020;77(18):3583–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lin-Moshier Y, Sebastian PJ, Higgins L, Sampson ND, Hewitt JE, Marchant JS.. Re-evaluation of the role of calcium homeostasis endoplasmic reticulum protein (CHERP) in cellular calcium signaling. J Biol Chem. 2013;288(1):355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Spartz AK, Herman RK, Shaw JE.. SMU-2 and SMU-1, Caenorhabditis elegans homologs of mammalian spliceosome-associated proteins RED and fSAP57, work together to affect splice site choice. Mol Cell Biol. 2004;24(15):6811–6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chung T, Wang D, Kim CS, Yadegari R, Larkins BA.. Plant SMU-1 and SMU-2 homologues regulate pre-mRNA splicing and multiple aspects of development. Plant Physiol. 2009;151(3):1498–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Keiper S, Papasaikas P, Will CL, Valcárcel J, Girard C, Lührmann R.. Smu1 and RED are required for activation of spliceosomal B complexes assembled on short introns. Nat Commun. 2019;10(1):3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bessonov S, Anokhina M, Krasauskas A, et al. Characterization of purified human Bact spliceosomal complexes reveals compositional and morphological changes during spliceosome activation and first step catalysis. RNA. 2010;16(12):2384–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Burgute BD, Peche VS, Steckelberg AL, et al. NKAP is a novel RS-related protein that interacts with RNA and RNA binding proteins. Nucleic Acids Res. 2014;42(5):3177–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fica SM, Oubridge C, Wilkinson ME, Newman AJ, Nagai K.. A human postcatalytic spliceosome structure reveals essential roles of metazoan factors for exon ligation. Science. 2019;363(6428):710–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lee YR, Khan K, Armfield-Uhas K, et al. Mutations in FAM50A suggest that Armfield XLID syndrome is a spliceosomopathy. Nat Commun. 2020;11(1):3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Beauchamp MC, Alam SS, Kumar S, Jerome-Majewska LA.. Spliceosomopathies and neurocristopathies: Two sides of the same coin? Dev Dyn. 2020;249(8):924–945. [DOI] [PubMed] [Google Scholar]
- 56. Wang Q, Lin JL, Reinking BE, et al. Essential roles of an intercalated disc protein, mXinbeta, in postnatal heart growth and survival. Circ Res. 2010;106(9):1468–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang Q, Lin JL, Erives AJ, Lin CI, Lin JJ.. New insights into the roles of Xin repeat-containing proteins in cardiac development, function, and disease. Int Rev Cell Mol Biol. 2014;310:89–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Guo H, Lu YW, Lin Z, et al. Intercalated disc protein Xinbeta is required for Hippo-YAP signaling in the heart. Nat Commun. 2020;11(1):4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Taricani L, Tejada ML, Young PG.. The fission yeast ES2 homologue, Bis1, interacts with the Ish1 stress-responsive nuclear envelope protein. J Biol Chem. 2002;277(12):10562–10572. [DOI] [PubMed] [Google Scholar]
- 60. Cardoso-Moreira M, Halbert J, Valloton D, et al. Gene expression across mammalian organ development. Nature. 2019;571(7766):505–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lin L, Zhang M, Stoilov P, Chen L, Zheng S.. Developmental attenuation of neuronal apoptosis by neural-specific splicing of Bak1 microexon. Neuron. 2020;107(6):1180–1196.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Takada I. DGCR14 induces Il17a gene expression through the RORgamma/BAZ1B/RSKS2 complex. Mol Cell Biol. 2015;35(2):344–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Takada I, Tsuchiya M, Yanaka K, et al. Ess2 bridges transcriptional regulators and spliceosomal complexes via distinct interacting domains. Biochem Biophys Res Commun. 2018;497(2):597–604. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the paper and its Supplementary material. Consent restrictions preclude sharing of full datasets and the consents do not cover the deposition of the WES data in a public database.