Abstract
The ntrC gene codes for a transcriptional activator protein that modulates gene expression in response to nitrogen. The cytochrome production pattern of a Rhizobium etli ntrC mutant (CFN2012) was studied. CO difference spectral analysis of membranes showed that CFN2012 produced a terminal oxidase similar to the symbiotic terminal oxidase of bacteroids in free-living cells under aerobic conditions, with a characteristic trough at 553 nm. CFN2012 produced two c-type cytochromes with molecular masses of 27 and 32 kDa, in contrast with the wild-type strain, which produced only a 32-kDa c-type cytochrome. The expression levels of the R. etli fixNOQP operon, which codes for terminal oxidase cbb3, were not affected by the ntrC mutation. However, the production levels of the two c-type cytochromes (27 and 32 kDa) were enhanced at least eightfold when the Bradyrhizobium japonicum fixNOQP operon was expressed in CFN2012 from the nptII promoter (pMSfixc), suggesting that these proteins are subunits FixO (27 kDa) and FixP (32 kDa) of cbb3 and that CFN2012/pMSfixc overproduced this terminal oxidase. CFN2012/pMSfixc showed a significant increase in its symbiotic performance as judged by the determination of nitrogenase activities of plants inoculated with this strain, suggesting that the overproduction of cbb3 terminal oxidase correlates with an enhancement in symbiotic nitrogen fixation.
In free-living diazotrophic bacteria combined nitrogen regulates the expression of the nitrogenase structural genes, inhibiting nitrogen fixation (10). The general nitrogen regulatory system (ntr system) activates the transcription of nitrogenase structural genes when combined nitrogen is not available (3). In contrast, in symbiotic nitrogen-fixing bacteria (genera Bradyrhizobium and Rhizobium) combined nitrogen has no effect on the expression of nitrogenase structural genes (8). Bacteria of these genera may establish a specific symbiotic relationship with their legume host plant. These bacteria elicit the formation of new organs, i.e., root nodules, in which bacteroids reduce atmospheric nitrogen to ammonia and supply the host plant with combined nitrogen. For the induction of root-nodules, members of Rhizobium express nod genes which code for enzymes involved in the formation of lipooligosaccharide factors (nod factors). However, in the presence of combined nitrogen, the formation of the root nodules is inhibited. It has been demonstrated that nitrogen negatively regulates bacterial nod gene expression (6, 7, 14, 27). In Rhizobium etli nitrogen repression of nod gene expression is mediated by the ntr system (14).
Symbiosis requires a respiratory chain that has a high affinity for O2 and is efficiently coupled to ATP production since nitrogen fixation is an energy-consuming process, requiring up to 20 ATP molecules to reduce just one molecule of N2. The genes of Bradyrhizobium japonicum which code for the bacteroid terminal oxidase have been identified as the fixNOQP operon (20). The sequence analysis of these genes, and biochemical characterization of the purified enzyme, showed that they code for a three-subunit terminal oxidase (cbb3) (11, 20, 21). FixN is a b-type heme- and copper-containing subunit, FixO is a single-heme-containing c-type cytochrome, and FixP is a diheme-containing c-type cytochrome (11, 20, 21). In R. etli, mutants that produce the cbb3 terminal oxidase under free-living conditions showed enhanced symbiotic nitrogen fixation (15).
In an attempt to study if nitrogen regulation affects respiration in R. etli, we analyzed the cytochrome production pattern of an R. etli mutant with change affecting the ntr system. A mutation in ntrC, which codes for a transcriptional activator protein that modulates gene expression in response to nitrogen (16), was analyzed. This analysis showed that the ntrC mutant produces a terminal oxidase of the cbb3 type in free-living cultures. Analysis of the expression of the R. etli fixNOQP genes showed that the expression of these genes was not affected by the ntrC mutation. However, expression of the B. japonicum fixNOQP operon from a constitutive promoter in the R. etli ntrC mutant greatly enhanced the production level of the cbb3 terminal oxidase and also the symbiotic performance of this strain. These data indicate that symbiotic nitrogen fixation can be improved by the overproduction of the symbiotic terminal oxidase cbb3.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used are listed in Table 1. R. etli cells were cultured in minimal medium (MM), peptone yeast extract (PY) medium (17), or yeast extract succinate (YS) (15). To achieve microaerobic cultures (O2 pressure, 2 kPa) 2 ml of active culture was used to inoculate 40 ml of medium. These cultures had previously been evacuated and flushed with a 1,200-ml · min−1 sterile N2 stream for 10 min. Calculated volumes (9.8 ml) of sterilized, high-purity commercial air were injected by making use of disposable syringes, following extraction of the same volume of N2. Escherichia coli was grown in Luria broth medium. Antibiotics were used at the following concentrations: rifampin, 50 mg/liter; tetracycline, 5 mg/liter; kanamycin, 30 mg/liter; and streptomycin, 100 mg/liter.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Reference |
|---|---|---|
| Rhizobium etli | ||
| CE3 | Strr derivative of CFN42 | 16 |
| CFN2012 | ntrC::Tn5 mutant of CE3 | 15 |
| Plasmids | ||
| cGD101 | Cosmid clone from R etli symbiotic plasmid | 8 |
| pMSfixc | pTR101 containing the B. japonicum fixNOQP operon fused to the nptII promoter | 8 |
| pMP220 | Transcriptional lacZ fusion vector; Tcr | 24 |
| pOLfix10 | pMP220::1.1-kb PstI fragment; fixN-lacZ gene fusion | This work |
Str, streptomycin; Tc, tetracycline.
DNA manipulations.
Cloning, restriction mapping, transformation, plasmid isolation, and β-galactosidase measurements were done as described (12). SalI and PstI clones from cGD101 were subcloned into pBluescript SK(+) vector and sequenced (4,832 bp; GenBank accession no. U76906) at the automated DNA sequencing facility at the Molecular Genetics Core Facility in the Department of Microbiology and Molecular Genetics, University of Texas—Houston Medical School. Computer-assisted sequence analysis and comparisons with the GenBank sequence were done using the Gene Works 2.6 program from Intelligenetics, Inc.
Spectral and electrophoretic analysis of cytochromes.
Cells were grown overnight on PY medium. Cells were washed and diluted 50-fold on fresh YS medium and grown on a rotary shaker (200 rpm) at 30°C for 36 h. Cells were harvested by centrifugation, washed, and suspended to 30% (wt/vol) in 50 mM Tris hydrochloride (pH 7.4)–5 mM CaCl2–5 mM MgCl2. Cytochrome spectra of whole cells or membrane preparations in an SLM Aminco Midan II spectrophotometer were recorded. Samples were reduced with dithionite (a few grains) or oxidized with ammonium persulfate. Carbon monoxide difference spectra were obtained by bubbling with CO (2 min) to reduce a cell sample, and spectra were recorded against a reduced sample. Spectra were obtained at room temperature with 1.0-cm light path cuvettes. c-type proteins were analyzed after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of membrane samples, which were prepared mechanically as previously described (23). Protein blotting and heme peroxidase detection were done as reported (26), except that in Western blotting the detection reagents used for peroxidase detection were obtained from Pierce (SuperSignal Substrate; Rockford, Ill.); the use of these chemicals greatly enhanced the sensitivity of this technique. Protein was determined as described (13).
Nitrogen fixation determination.
For acetylene reduction measurements, Phaseolus vulgaris cv. negro jamapa was surface sterilized in hypochlorite and germinated on moist sterile filter paper. Three-day-old seedlings were transferred to plastic growth pots, inoculated with a bacterial suspension in PY medium, and grown with nitrogen-free salts in a greenhouse (23). Nitrogenase was determined by measuring the acetylene reduction of nodulated plant roots transferred to tubes with rubber seal stoppers by injecting acetylene to a final concentration of 10% of the gas phase. Samples were incubated 40 min at room temperature, and ethylene production was determined by gas chromatography in a Packard model 430 chromatograph (23). For each strain, the total nitrogen of 60-day-old nodulated plants was determined for three plants in four pots each (total, 12 plants) using an Antek 720 nitrogen detector as previously described (4).
Bacteroid preparation.
Nodules were harvested 30 days after inoculation. Bacteroids were isolated by layering a nodule extract on a sucrose gradient as previously reported (23).
RESULTS
Cytochrome production in an ntrC mutant of R. etli.
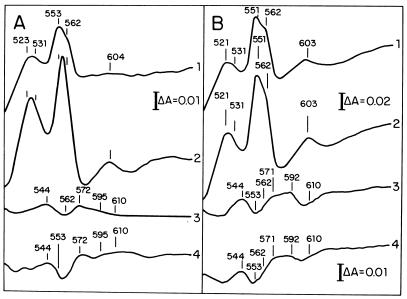
Cytochrome production was analyzed in cells cultured aerobically to the stationary phase of growth and in bacteroids of the wild-type (CE3) and the ntrC mutant (CFN2012) strains. Figure 1A shows difference spectra (spectra for reduced cells minus spectra for oxidized cells) of free-living cells, showing that CE3 produced c-type cytochromes (peak at 553 nm) and b-type cytochromes (shoulder at 562 nm) but no aa3 cytochromes (peak at 603 nm) under this culture condition. CFN2012 produced c-type, b-type, and aa3 cytochromes (Fig. 1A). Carbon monoxide (CO) difference spectra were obtained since CO reacts specifically with cytochrome terminal oxidases. CO difference spectra showed that the CE3 cells produced only cytochrome terminal oxidase o (peaks at 544 and 572 nm and trough at 562 nm) and no cytochrome aa3 (trough at 610 nm). In contrast, CFN2012 produced a different CO-reactive cytochrome with a spectrum signal showing a characteristic trough at 553 nm and no cytochrome aa3 (Fig. 1A). The lack of evidence for cytochrome aa3 in CO difference spectra of CFN2012 membranes suggests that the absorption peak at 603 nm found in difference spectra (spectra for reduced cells minus those for oxidized cells) of CFN2012 could be due to a different heme-containing protein that absorbs near 600 nm (e.g., catalase).
FIG. 1.
Difference spectra (spectra for reduced cells minus those for oxidized cells) (spectra 1 and 2 in panels A and B) and CO difference spectra (spectra 3 and 4 in panels A and B) of R. etli whole cells. (A) Spectra 1 and 3 are for CE3 cells cultured for 36 h (18.7 mg of protein ml−1), and spectra 2 and 4 are for CFN2012 cells cultured for 36 h (23.5 mg of protein ml−1). (B) Spectra of bacteroids of CE3 (17.7 mg of protein ml−1) (spectra 1 and 3) and CFN2012 (25.5 mg of protein ml−1) (spectra 2 and 4). Free-living cells were cultured aerobically in YS medium.
Bacteroids of the CE3 and CFN2012 strains had similar cytochrome production patterns, producing c-type (peak at 551 nm), b-type (shoulder at 562 nm), and aa3 (peak at 603 nm) cytochromes (Fig. 1B). Figure 1B also shows that bacteroids of these strains produced a CO-reactive cytochrome with a spectrum with a trough at 553 nm, very similar to the CO-reactive cytochrome produced by CFN2012 in free-living cultures, and also cytochrome aa3 (trough at 610 nm).
c-type cytochrome production by CFN2012.
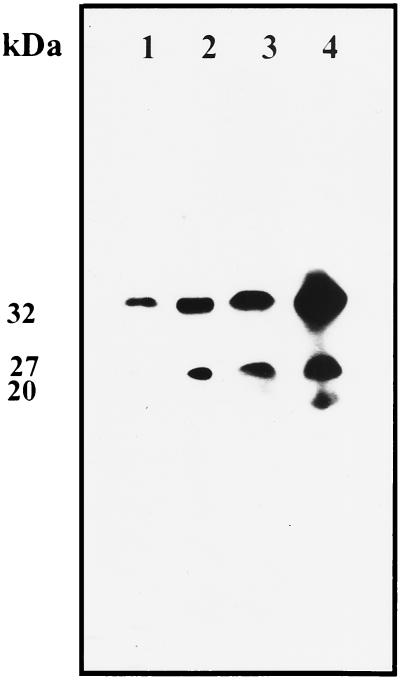
The c-type cytochromes produced by CE3 and CFN2012 strains cultured aerobically in YS medium to the stationary growth phase were analyzed. CE3 and CFN2012 cells harboring plasmid pMSfixc, which contains the B. japonicum fixNOQP operon fused to the nptII promoter (24), were included as controls for the production of FixO (27 kDa) and FixP (32 kDa) c-type cytochrome subunits of terminal oxidase cbb3. Figure 2 shows that CE3 cell membranes contained detectable levels of a 32-kDa c-type cytochrome, as was previously reported (23); in contrast, CFN2012 produced higher levels of the 32-kDa protein and produced an additional c-type cytochrome of 27 kDa (Fig. 2). CE3/pMSfixc produced both the 32- and 27-kDa proteins, while CFN2012/pMSfixc showed very high production levels of both c-type cytochromes and also detectable levels of a 20-kDa c-type cytochrome (Fig. 2). The production of the two c-type cytochromes by the ntrC mutant, as well as the results of the CO difference spectral analysis (Fig. 1A), suggests that in this strain a terminal oxidase similar to a cbb3 terminal oxidase was produced in aerobic cultures of free-living stationary-phase cells. We quantified the production levels of cbb3 terminal oxidase by measuring the 32- and 27-kDa c-type proteins produced when fixNOQP was transcribed from the nptII promoter. This analysis showed that CFN2012 produced eightfold-higher levels of this oxidase than CE3.
FIG. 2.
Proteins containing c-type heme from membrane particles. Each lane contains 50 μg of protein from membrane particles from CE3 cells (lane 1), CFN2012 cells (lane 2), CE3/pMSfixc cells (lane 3), or CFN2012/pMSfixc cells (lane 4) grown aerobically in YS medium for 36 h.
Expression of R. etli fixNOQP.
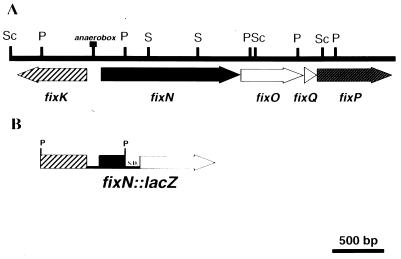
In order to further analyze the free-living cell production of the cbb3 terminal oxidase in CFN2012, we decided to study the expression of the R. etli fixNOQP operon. A cosmid clone (cGD101) of the R. etli symbiotic plasmid pd (9), which hybridized against heterologous gene probes from fixK (from Sinorhizobium meliloti) (1) and fixN (from B. japonicum) (20), was identified (data not shown). In order to localize the fixNOQP operon, 4,832 bp were sequenced (GenBank accession no. U76906).
Five open reading frames were identified as the R. etli fixK and fixNOQP operon (Fig. 3). The predicted FixK protein is comprised of 239 residues. FixK is a transcriptional activator involved in fixNOQP induction in S. meliloti (1). This protein has 63% identity with FixK of Rhizobium leguminosarum bv. viciae (19) (GenBank accession no. Z70305) and lacks the cysteine-amino-terminal domain present in Fnr. The predicted FixN protein is comprised of 540 residues. R. etli FixN has significant homology with FixN from different organisms, i.e., 78% identity with FixN of S. meliloti (GenBank accession no. Z21854) and 92% identity with both copies of fixN gene products in R. leguminosarum (GenBank accession no. Z80339 and Z80340). FixN contains the six conserved histidine residues proposed to bind the low-spin b-type heme (H117 and H406), CuB (H266, H136, and H317), and the high-spin b-type heme (H404). FixO, comprised of 244 residues, has 86% identity with FixO of R. leguminosarum and 75% identity with FixO of S. meliloti. FixO contains two cysteine residues, at positions 69 and 72, which are likely to be involved in the attachment of c-type heme. FixQ, comprised of 50 residues, has 88% identity with FixQ from R. leguminosarum and 66% identity with FixQ from S. meliloti. Finally, FixP, comprised of 287 residues, has identities of 81 and 61% with FixP of R. leguminosarum and of S. meliloti, respectively, and contains the four conserved cysteine residues involved in the attachment of two c-type hemes (C121, C124, C216, and C219).
FIG. 3.
Physical-genetic map of the fixK-fixNOQP sequenced region. (A) Physical-genetic map of the R. etli fixK-fixNOQP genetic region. The restriction sites used in subcloning for sequencing are shown. Sc, SacI; P, PstI; S, SalI. The position of the anaerobox sequence is shown. (B) Diagram of fixN-lacZ transcriptional gene fusion construction. S.D., Shine-Dalgarno sequence. Arrows indicate the directions of transcription.
Similar to R. leguminosarum bv. viciae (19), in R. etli fixK and fixNOQP are transcribed in opposite directions with a spanning DNA region of 230 bp (Fig. 3). These data suggest that the promoters of both genes are contained in this DNA region. The “anaerobox” sequence (TTGATGTAGATCAA) is located 88 bp in front of the beginning of fixN (Fig. 3).
In order to generate transcriptional lacZ gene fusions of fixN (plasmid pOLfix10) promoter, a 1.1-kb PstI fragment containing the amino-terminal part of FixN and FixK was cloned in plasmid pMP220, which carries the E. coli lacZ gene (25) (Fig. 3B).
The expression of fixNOQP in CE3 and CFN2012 strains cultured aerobically or microaerobically was studied. The expression of fixN was induced 15-fold in cells cultured microaerobically, as has been shown in several other Rhizobium species, showing that in R. etli as well, oxygen is the most important metabolic signal triggering fixNOQP expression. The level of expression of fixN was slightly lower in the wild-type than in the CFN2012 strain under stationary-phase aerobic (60%) and microaerobic (70%) conditions. This analysis revealed that the production of cbb3 terminal oxidase by CFN2012 was not due to the enhanced expression of fixNOQP, suggesting that NtrC modulates cbb3 production at a level different from fixNOQP transcription. When cells were grown on YS medium under atmospheric O2 tension for 8 h, under atmospheric O2 tension for 36 h, and under 2 kPa O2 tension for 8 h, the CE3 strain carrying the fixN-lacZ(pOLfix10) reporter gene fusion showed β-galactosidase activities of 474, 342, and 6,199 U/mg of protein, respectively, and the CFN2012 strain carrying the same fusion showed activities of 526, 557, and 8,719 U/mg of protein, respectively (values are expressed after subtraction of activities of strain without any plasmid [range, 50 to 90 U/mg of protein]).
Symbiotic nitrogen fixation of plants inoculated with different R. etli strains.
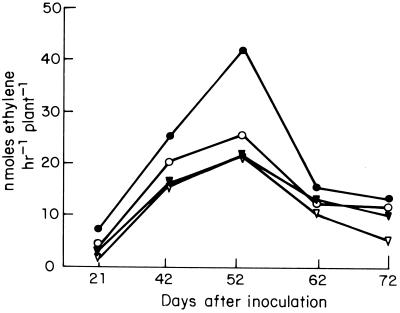
P. vulgaris cv. negro jamapa plants were inoculated with CE3 and CFN2012 strains and the same strains harboring pMSfixc, and nitrogenase activity was determined at different days after inoculation. Figure 4 shows that for plants inoculated with CE3 the highest nitrogenase activity (23 nmol of ethylene produced h−1 per plant) was reached at 52 days after inoculation and decreased gradually afterwards. No effect of plasmid pMSfixc was found in this strain. Plants inoculated with CFN2012 demonstrated nitrogenase activity more rapidly than those inoculated with CE3, but similar nitrogenase activities were observed. CFN2012 strain harboring pMSfixc induced nitrogenase activity more rapidly and achieved a twofold-higher nitrogenase activity than the CE3 strain (Fig. 4). In addition, nitrogen fixation was estimated by the total nitrogen content of plants 60 days after inoculation with the different strains. CE3/pMSfixc-inoculated plants contained 90% (3.51 ± 0.092 mg of nitrogen [100 mg dry weight]−1; n = 4) of the nitrogen found in CE3-inoculated plants (3.90 ± 0.031 mg of nitrogen [100 mg dry weight]−1; n = 4), whereas CFN2012- and CFN2012/pMSfixc-inoculated plants contained 8% (4.25 ± 0.102 mg of nitrogen [100 mg dry weight]−1; n = 4) and 18% (4.57 ± 0.065 mg of nitrogen [100 mg dry weight]−1; n = 4) more nitrogen, respectively, than CE3-inoculated plants.
FIG. 4.
Acetylene reduction activity in plants inoculated with strains CE3 (▿), CE3/pMSfixc (▾), CFN2012 (○), and CFN2012/pMSfixc (●). Acetylene reduction activity for four plants in each of two pots was determined on the specified days. Data are means for two pots (eight plants); variations were 30% or less.
DISCUSSION
Symbiotic nitrogen fixation is an energy-consuming process requiring up to 20 ATP molecules to reduce one molecule of N2. Also, symbiotic nitrogen fixation occurs at very low oxygen tensions (2). Therefore, a high-affinity oxidase, of the cbb3 type, efficiently coupled to ATP production is produced during symbiosis. During free life, Rhizobium species do not produce the cbb3 terminal oxidase, mainly due to a low level of expression of the fixNOQP operon in the presence of O2 (2).
In this work we present data showing that, in R. etli, in addition to oxygen regulation, NtrC represses the free-living cell production of the cbb3 symbiotic terminal oxidase. The analysis of cytochrome production revealed that in aerobic cultures an ntrC mutant produced a terminal oxidase very similar to that produced in bacteroids, in contrast with the wild-type strain. This suggests that nitrogen availability could modulate symbiotic cytochrome production in R. etli. The molecular mechanism by which NtrC represses cbb3 production is still unclear. However, two results indicate that this regulation is exerted after transcription of the structural fixNOQP genes: (i) no difference was found in fixNOQP expression between CFN2012 and CE3; and (ii) under conditions where fixNOQP was expressed under a strong promoter, CFN2012 produced at least eightfold-higher levels of cbb3 than CE3, suggesting that a posttranscriptional step involved in cbb3 biogenesis was negatively regulated by NtrC. In several Rhizobium species, including R. etli, several genes that participate in the correct assembly of the cbb3 terminal oxidase have been identified. These include genes necessary for the covalent attachment of heme to c-type apoproteins (5) and genes (fixGHIS) that are probably involved in the transport of copper ions, which are essential for oxygen reduction by cbb3 (5, 22).
The ntrC mutation in CFN2012 had a positive effect on nitrogenase activity only when the fixNOQP genes were expressed from a strong promoter; also, CFN2012/pMSfixc produced an eightfold-higher level of cbb3 terminal oxidase than CE3/pMSfixc in culture. These data suggest that increased cbb3 terminal production correlates with enhanced nitrogen fixation. However, because ntrC is known to be global in its regulatory role, it cannot be ruled out that other effects of ntrC mutation, in addition to the elevated levels of the terminal oxidase, could also participate in the enhanced nitrogen fixation capacity of this strain. In fact plants inoculated with CFN2012 accumulated more nitrogen than CE3-inoculated plants. It is important to study the respiratory physiology of ntrC mutant-derived bacteroids.
Total nitrogen determinations of plants inoculated with the different strains correlated roughly with nitrogenase activities. However, plasmid pMSfixc had a negative effect on nitrogen accumulation for plants inoculated with the CE3 strain even though plants inoculated with these strains showed very similar nitrogenase activities. Therefore, we cannot rule out the possibility that in CFN2012/pMSfixc-inoculated plants nitrogen accumulation was also affected by the presence of this plasmid. At this moment we have no explanation for the negative effect of this construct, although the process of replication of the plasmid or enhanced transcription could compete for ATP with nitrogen fixation. It is important to introduce the nptII-fixNOQP construct into the genome to determine the reason for the negative effect of plasmid pMSfixc on plant nitrogen accumulation.
Two possible reasons for the enhanced symbiotic nitrogen fixation observed for CFN2012/pMSfixc can be proposed: one is a higher nitrogenase activity due to enhanced supply of ATP to nitrogenase, and the other is a more-efficient performance during infection and bacteroid development. R. etli bacteroids repress ntrC expression, and NtrC protein could not be detected (18). This explains the similar cytochrome production patterns found in bacteroids of CE3 and CFN2012 strains. However, it was shown that bacteria inside the infection thread and young bacteroids could express ntrC (18). Since NtrC is not produced in well-developed bacteroids, it seems possible that the difference in nitrogenase activity between CE3/pMSfixc and CFN2012/pMSfixc could be due to a better performance of CFN2012 during infection. This could suggest that the repression of cbb3 terminal oxidase production by NtrC could be relevant during the infection process whereas another terminal oxidase could support growth. Alternatively, the negative regulation of the production of the symbiotic terminal oxidase by NtrC could be an additional control point for the repression of symbiosis when combined nitrogen is available.
ACKNOWLEDGMENTS
We thank Alejandra Bravo for revising the manuscript, Guadalupe Espín for providing us the CFN2012 strain, Yolanda Mora for total nitrogen determinations, and Jose Luis Zitlalpopoca for technical assistance.
This work was partially supported by the European Communities through the International Scientific Co-operation Program, contract CI1*-CT94-0042, by CONACyT contract no. 3372-N9309, and by DGAPA contract no. IN204697.
REFERENCES
- 1.Batut J, Daveran-Mingot M-L, David M, Jacobs J, Garnerone A M, Kahn D. fixK, a gene homologous with fnr and crp from Escherichia coli, regulates nitrogen fixation genes positively and negatively in Rhizobium meliloti. EMBO J. 1989;8:1279–1286. doi: 10.1002/j.1460-2075.1989.tb03502.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batut J, Boistard P. Oxygen control in Rhizobium. Antonie Leeuwenhoek. 1994;66:129–150. doi: 10.1007/BF00871636. [DOI] [PubMed] [Google Scholar]
- 3.Buck M. Transcriptional activation of nitrogen fixation genes in Klebsiella pneumoniae. In: Gresshohoff P M, Roth L E, Stacey G, Newton W E, editors. Nitrogen fixation: achievements and objectives. New York, N.Y: Chapman and Hall; 1990. pp. 451–457. [Google Scholar]
- 4.Cevallos M A, Encarnación S, Leija A, Mora Y, Mora J. Genetic and physiological characterization of a Rhizobium etli mutant strain unable to synthesize poly-β-hydroxybutyrate. J Bacteriol. 1996;178:1646–1654. doi: 10.1128/jb.178.6.1646-1654.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delgado M J, Bedmar E J, Downie A. Genes involved in the formation and assembly of rhizobial cytochromes and their role in symbiotic nitrogen fixation. Adv Microb Physiol. 1998;40:191–231. doi: 10.1016/s0065-2911(08)60132-0. [DOI] [PubMed] [Google Scholar]
- 6.Dusha I, Bakos A, Kondorosi A, de Bruijn F J, Shell J. The Rhizobium meliloti early nodulation genes (nodABC) are nitrogen-regulated: isolation of a mutant strain with efficient nodulation capacity of alfalfa in the presence of ammonium. Mol Gen Genet. 1989;219:89–96. [Google Scholar]
- 7.Dusha I, Kondorosi A. Genes at different regulatory levels are required for the ammonia control of nodulation in Rhizobium meliloti. Mol Gen Genet. 1993;240:435–444. doi: 10.1007/BF00280398. [DOI] [PubMed] [Google Scholar]
- 8.Gallon J R. Reconciling the incompatible: N2 fixation and O2. New Phytol. 1992;122:571–609. [Google Scholar]
- 9.Girard M D L, Flores M, Brom S, Romero D, Palacios R, Dávila G. Structural complexity of the symbiotic plasmid of Rhizobium leguminosarum bv. phaseoli. J Bacteriol. 1991;173:2411–2419. doi: 10.1128/jb.173.8.2411-2419.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hom S S M, Hennecke H, Shanmungan K T. Regulation of nitrogenase biosynthesis in Klebsiella pneumoniae: effect of nitrate. J Gen Microbiol. 1980;117:169–179. doi: 10.1099/00221287-117-1-169. [DOI] [PubMed] [Google Scholar]
- 11.Mandon K, Kaminski P A, Elmerich C. Functional analysis of the fixNOQP region of Azorhizobium caulinodans. J Bacteriol. 1994;176:2560–2568. doi: 10.1128/jb.176.9.2560-2568.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. 1st ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 13.Markwell M A K, Haas S M, Vieber Z Z, Tolbert N E. A modification of the Lowry procedure to simplify protein determinations in membrane and lipoprotein samples. Anal Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- 14.Mendoza A, Leija A, Martinez-Romero E, Hernandez G, Mora J. The enhancement of ammonium assimilation in Rhizobium etli prevents nodulation of Phaseolus vulgaris. Mol Plant-Microbe Interact. 1995;8:584–592. doi: 10.1094/mpmi-8-0584. [DOI] [PubMed] [Google Scholar]
- 15.Miranda J, Membrillo-Hernandez J, Tabche M L, Soberón M. Rhizobium etli cytochrome mutants with derepressed expression of cytochrome terminal oxidases and enhanced nitrogen accumulation. Appl Microbiol Biotechnol. 1996;45:182–188. [Google Scholar]
- 16.Moreno S, Patriarca E J, Chiaruzzi M, Meza R, Defez R, Lamberti A, Riccio A, Iaccarino M, Espín G. Phenotype of a Rhizobium leguminosarum ntrC mutant. Res Microbiol. 1992;143:161–171. doi: 10.1016/0923-2508(92)90005-9. [DOI] [PubMed] [Google Scholar]
- 17.Noel K D, Sanchez A, Fernandez L, Leemans J, Cevallos M A. Rhizobium phaseoli symbiotic mutants with transposon Tn5 insertions. J Bacteriol. 1984;158:148–155. doi: 10.1128/jb.158.1.148-155.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patriarca E J, Taté R, Fedorova E, Riccio A, Defez R, Maurizio I. Down regulation of the Rhizobium ntr system in determinate nodule of Phaseolus vulgaris identifies a specific developmental zone. Mol Plant-Microbe Interact. 1996;9:243–251. [Google Scholar]
- 19.Patschowski T, Schluter A, Priefer U B. Rhizobium leguminosarum bv viciae contains a second fnr/fixK-like gene and an unusual fixL homologue. Mol Microbiol. 1996;21:267–280. doi: 10.1046/j.1365-2958.1996.6321348.x. [DOI] [PubMed] [Google Scholar]
- 20.Preisig O, Anthamatten D, Hennecke H. Genes for a microaerobically induced oxidase complex in Bradyrhizobium japonicum are essential for a nitrogen-fixing endosymbiosis. Proc Natl Acad Sci USA. 1993;90:3309–3313. doi: 10.1073/pnas.90.8.3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Preisig O, Zufferey R, Thöny-Meyer L, Appleby C A, Hennecke H. A high-affinity cbb3-type cytochrome oxidase terminates the symbiosis-specific respiratory chain of Bradyrhizobium japonicum. J Bacteriol. 1996;178:1532–1538. doi: 10.1128/jb.178.6.1532-1538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Preisig O, Zufferey R, Hennecke H. The Bradyrhizobium japonicum fixGHIS genes are required for the formation of the high-affinity cbb3-type cytochrome oxidase. Arch Microbiol. 1996;165:297–305. doi: 10.1007/s002030050330. [DOI] [PubMed] [Google Scholar]
- 23.Soberón M, Williams H D, Poole R K, Escamilla E. Isolation of a Rhizobium phaseoli cytochrome mutant with enhanced respiration and symbiotic nitrogen fixation. J Bacteriol. 1989;171:465–472. doi: 10.1128/jb.171.1.465-472.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soberón M, Lopez O, Miranda J, Tabche M L, Morera C. Genetic evidence for 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) as a negative effector of cytochrome terminal oxidase cbb3 production in Rhizobium etli. Mol Gen Genet. 1997;254:665–673. doi: 10.1007/s004380050464. [DOI] [PubMed] [Google Scholar]
- 25.Spaink H P, Okker R J H, Wijffelman C A, Pees E, Lugtenberg B J J. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol Biol. 1987;9:27–39. doi: 10.1007/BF00017984. [DOI] [PubMed] [Google Scholar]
- 26.Vargas C, McEwan A G, Downie J A. Detection of c-type cytochromes using enhanced chemiluminescence. Anal Biochem. 1993;209:323–326. doi: 10.1006/abio.1993.1127. [DOI] [PubMed] [Google Scholar]
- 27.Wang S P, Stacey G. Ammonia regulation of nod genes in Bradyrhizobium japonicum. Mol Gen Genet. 1990;223:329–331. doi: 10.1007/BF00265071. [DOI] [PubMed] [Google Scholar]