Abstract
Nearly two decades after the last epidemic caused by a severe acute respiratory syndrome coronavirus (SARS-CoV), newly emerged SARS-CoV-2 quickly spread in 2020 and precipitated an ongoing global public health crisis. Both the continuous accumulation of point mutations, owed to the naturally imposed genomic plasticity of SARS-CoV-2 evolutionary processes, as well as viral spread over time, allow this RNA virus to gain new genetic identities, spawn novel variants and enhance its potential for immune evasion. Here, through an in-depth phylogenetic clustering analysis of upwards of 200,000 whole-genome sequences, we reveal the presence of previously unreported and hitherto unidentified mutations and recombination breakpoints in Variants of Concern (VOC) and Variants of Interest (VOI) from Brazil, India (Beta, Eta and Kappa) and the USA (Beta, Eta and Lambda). Additionally, we identify sites with shared mutations under directional evolution in the SARS-CoV-2 Spike-encoding protein of VOC and VOI, tracing a heretofore-undescribed correlation with viral spread in South America, India and the USA. Our evidence-based analysis provides well-supported evidence of similar pathways of evolution for such mutations in all SARS-CoV-2 variants and sub-lineages. This raises two pivotal points: (i) the co-circulation of variants and sub-lineages in close evolutionary environments, which sheds light onto their trajectories into convergent and directional evolution, and (ii) a linear perspective into the prospective vaccine efficacy against different SARS-CoV-2 strains.
Introduction
In the last two decades, human health has been threatened by the emergence of three important zoonotic and pathogenic betacoronaviruses, namely the severe acute respiratory syndrome coronavirus (SARS-CoV) [1], the Middle East respiratory syndrome coronavirus (MERS-CoV) [2] and, most recently, the causative agent of the Coronavirus Disease 2019 (COVID-19) pandemic, SARS-CoV-2 [3]. Likely originated from bats, pandemic SARS-CoV-2, like other endemic human alpha- (NL63 and 229E) and beta- (OC43 and HKU1) CoVs known for causing upper respiratory tract infections, overcame the interspecies barrier as a result of spillover and/or recombination events, and gained a pervasive ability to rapidly infect and spread around the globe [4–6].
The COVID-19 pandemic precipitated an intense genomic surveillance via data depositories and sequencing platforms and led to an unprecedented accumulation of public genomic data concerning a human pathogenic virus [5, 7]. The sheer amount of available sequencing data has the potential to facilitate higher-precision micro-evolutionary analyses mapping escape and point mutations in presumed positively selected sites and residues putatively associated to an increased virus fitness and pathogenesis and allows inferences concerning the dynamics of SARS-CoV-2 spread [8, 9].
Although the analysis of micro-evolutionary mechanisms is of paramount importance and may provide powerful information to promote the prediction of vaccination perspectives and the tracing of SARS-CoV-2 epidemiological chains, there is as yet a lack of data-based investigations examining the presence of eventual shared mutations and their evolutionary characteristics in classified SARS-CoV-2 Variants of Concern (VOC) and Variants of Interest (VOI) [10, 11].
Given the importance of monitoring mutations to track the emergence of novel variants, here we investigate the influence of directional selection and the dynamics of SARS-CoV-2 genomic plasticity in VOC and VOI by clustering partition high-scale phylogenetic and directional evolution (DEPS) approaches. Additionally, we show the presence of several mutations common for both VOI/VOC and convergently emerged sub-lineages, and provide a perspective of possible effects on the vaccination efficacy and the ongoing COVID-19 pandemic.
Methods
Sequence data and filtering strategy
High-coverage and complete HCoV-229E and HCoV-NL63 (alpha-CoVs), HCoV-OC43, HCoV-HKU1, MERS-CoV, SARS-CoV and SARS-CoV-2 VOC and VOI (beta-CoVs) genome sequences (≥ 29,000 bp), sampled from humans, were retrieved from the Global Initiative on Sharing Avian Influenza Data-EpiCoV (GISAID-EpiCoV) and GenBank databases at three different times, each date representing a snapshot of the COVID-19 spread at that time: February 12th (MERS-CoV, SARS-CoV and SARS-CoV-2), July 12th (HCoV-229E, HCoV-NL63, HCoV-OC43, HCoV-HKU1 and SARS-CoV-2) and updated in August 26th 2021 (SARS-CoV-2), totalling 238,990 sequences. With regards to SARS-CoV-2, we particularly focused on strains of countries from South America, China, India, and the United States of America (USA). At the time of analysis, India, the USA, and Brazil had reported the largest numbers of cumulative confirmed COVID-19 cases and deaths. This approach was used to compare putative mutual sites and residue changes under directional evolution over time [12].
Subsequently, sequences were filtered via Sequence Cleaner, a biopython-based program, utilising the following script: sequence_cleaner -q INPUT_DIRECTORY -o OUTPUT_DIRECTORY -ml 29,000 (MINIMUM_LENGTH) -mn 0 (PERCENTAGE_N)—remove_ambiguous. The outcome was a set of unambiguous sequences equal to and greater than 29,000 pb with zero percent of unknown nucleotides. Next, the datasets were aligned by adding coding-sequences related to references for HCoV-229E (NC_002645.1), HCoV-NL63 (NC_005831.2), HCoV-OC43 (NC_006213.1), HCoV-HKU1 (NC_006577.2), MERS-CoV (NC_038294.1), SARS-CoV (NC_004718.3), and SARS-CoV-2 (NC_045512.2), using default settings, with the rapid calculation of full-length multiple sequence alignment of closely-related viral genomes (MAFFT v.7 web-version program; https://mafft.cbrc.jp/alignment/software/closelyrelatedviralgenomes.html) and were edited by the UGENE v.38.1 [13].
Clustering and sub-clustering analysis
A methodological approach to extract large-scale phylogenetic partitions was applied to identify transmission cluster chains on the large Maximum Likelihood (ML) phylogenetic trees of the SARS-CoV-2 variants circulating in South America, China, India and the USA on the basis of a depth-first search algorithm which unifies evaluation of node reliability, tree topology and patristic distance [14]. In this case, different datasets from each particular SARS-CoV-2 scenario were used in order to extract the clustering and sub-clustering data. Therefore, each ML tree was implemented in FastTree v.2.1.7 by using the standard implementation General Time Reversible (GTR) plus CAT with 20 gamma distribution parameters and a mix of Nearest-Neighbor Interchanges (NNI) and Sub-Tree-Prune-Regraft (SPR) [15]. Thereafter, in view of identifying SARS-CoV-2 cluster transmission events, for datasets comprising more than 100 sequences, we first selected sequences (one per cluster) from nodes/sub-trees with ≥ 2 distinct individuals and ≥ 90% reliability of statistical support (Shimodaira-Hasegawa test), where initially the patristic distance was adjusted to find a representative number of clusters (n = 100, which represent 100 sequences) from each large reconstructed ML tree. In addition to this strategy, a second approach included sub-clustering analysis as an indirect way to infer and investigate the possibility of co-circulating sub-lineages. For this, we selected sequences (two per cluster) with ≥ 95% node reliability of statistical support from a threshold of 0.05, thus corresponding to the 5th percentile when considering the whole-tree patristic distance distribution.
Recombination and directional evolution analyses
Before proceeding to directional evolution analysis, sequence datasets from the output provided by the clustering analyses were submitted to the Genetic Algorithm for Recombination Detection (GARD), a likelihood-based tool to pinpoint recombination breakpoints [16]. To check the outcome of the strategy described above, an additional test was conducted using the Pairwise Homoplasy Index (PHI; default settings) [17]. Evidence-based analysis through phylogenetic maximum-likelihood was then performed implementing the Datamonkey web-server and the program Hyphy v.2.5 to track directional selection in amino acid sequences (DEPS) [18]. The DEPS method identifies both the residue and sites evolving toward it with great accuracy and detects frequency-dependent selection-scenarios as well as selective sweeps and convergent evolution that can confound most existing tests [8]. Further, the DEPS method has shown better performance than (traditional) substitution rate-based analyses (dN/dS) in detecting transient and frequency-dependent selection and directionally evolving sites and residues. For the most part, a Beta-Gamma site-to-site rate variation was used to conduct the analysis. The best-fit protein substitution model was chosen according to the corrected Akaike Information Criterion (cAIC). Only target sites and residues with Empirical Bayes Factors for evidence in favour of a directional selection model equal to or greater than 100 were considered for further exploration. Certain datasets, randomly chosen, were run multiple times (more than eight) to confirm obtained results.
Statistical analysis
Data pertaining to SARS-CoV and MERS-CoV-related cases and deaths were extracted from the National Health Service (NHS, UK) (https://www.nhs.uk/conditions/sars/) and European Centre for Disease Prevention and Control (ECDC) (https://www.ecdc.europa.eu/en/publications-data/distribution-confirmed-cases-mers-cov-place-infection-and-month-onset-1), respectively. Information concerning SARS-CoV-2 was collected from the World Health Organization (WHO) (https://covid19.who.int/). Population demographic data were retrieved from the Our World in Data website (https://ourworldindata.org/grapher/world-population-by-world-regions-post 1820?tab = table&country = Oceania~~North+America~Europe~Africa~Asia).
Statistical analyses were performed using one-way analysis of variance (ANOVA) and nonparametric methods followed by post hoc Kruskal-Wallis and Friedman (both with Dunn’s Multiple Comparison), and Bartlett’s tests (Tukey’s, Newman-Keuls and Bonferroni’s multiple comparisons). Additionally, Mann Whitney and Wilcoxon matched-pairs signed-rank (T test) and Pearson/Spearman (Correlation), all one-tailed methods with 99% confidence interval (CI), were run. P-values equal to or less than 0.005 (p ≤ 0.005, SARS-CoV-2 from South America: DEPS [sites and/or residues] vs infections) and 0.05 (p ≤ 0.05, SARS-CoV-2 from Brazil, China, India and the USA: DEPS [residues] vs circulating variants and infections) were considered as statistically significant. Data analyses were carried out using GraphPad Prism v. 5.01 (GraphPad Software, San Diego, California, USA). Figures and data visualization were performed using the ggplot2 v.3.3.5 package in the R (RStudio v.1.4.1717) language environment. Final graphics were edited with the open-source software drawing tool Inkscape v.1.0.2.
Results and discussion
Recombination is known to be a crucial evolutionary process for many RNA viruses [19–21]; the process is frequently observed in the Coronaviridae family where recombination is likely facilitated by discontinuous transcription involving jumps of the replication-transcription complex during minus strand RNA synthesis. However, the consequences of recombination events occurring in the context of the current SARS-CoV-2 VOC/VOI evolutionary landscape are still speculative [22, 23]. Here, we address this knowledge gap, revealing the presence of recombination and shared mutations in the SARS-CoV-2 Spike-encoding protein, demonstrating them to be under directional and convergent evolution amongst SARS-CoV-2 VOC/VOI and sub-lineages, and tracing an interconnection with viral spread. First, endemic and epidemic human coronaviruses (HCoVs) were compared to identify similar evolutionary patterns that could help clarify the evolution of SARS-CoV-2. An initial recombination breakpoint analysis showed that four of six HCoVs analyzed presented such signals (Fig 1A). Endemic viruses OC43, NL63 and HKU1 also showed a similar pattern of residue accumulation and directional evolution, despite these viruses being subject to differing selective pressures [24].
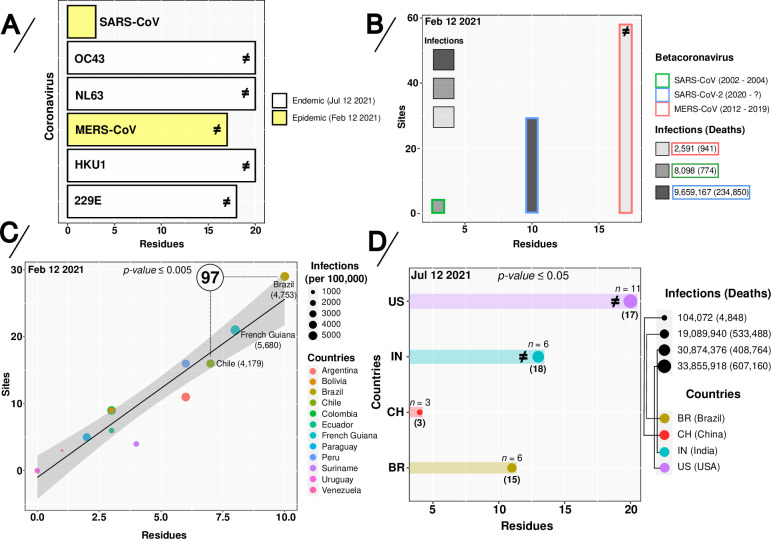
Fig 1. Directionally-evolving sites and residues in alpha- (229E and NL63) and beta- (OC43, HKU1, SARS-CoV, MERS-CoV and SARS-CoV-2) coronavirus sequences.
(A) Directed-evolving residues in six different endemic and epidemic human coronaviruses (HCoVs), (B) Sites and residues vs infections in epidemic and pandemic coronaviruses (CoVs). (C) Linear-regression curve and co-relationship on the absolute amount of sites and/or residues under directional-positive selection given SARS-CoV-2 infections per 100,000 people from South America countries and (D) the total number of SARS-CoV-2 infections in Brazil, China, India and the USA. In panels A, B and D, the symbol ≠ represents the presence of recombination breakpoints signals. In panel B, on the upper left, different intensities in grayscale are directly related to the number of infections. In panel C, the number inside the circle represents the amount of clusters found in Brazil and Chile. In panel D, n represents the amount of SARS-CoV-2 variants and the numbers in parentheses indicate sites under directional and convergent evolution in the Spike-encoding protein. P-values were obtained by considering the DEPS [sites and/or residues] vs infections per 100.000 inhabitants from one-way analysis of variance (ANOVA) and nonparametric methods, as described in the Statistical analysis section. Colors and symbols used in the panels are defined in the legend to the right of the figure.
A subsequent comparison of SARS-CoV-2 to the other two pathogenic HCoVs (SARS-CoV and MERS-CoV), highlighted differences in the number of directionally-evolving sites and residues (Fig 1B). These patterns, putatively reflecting the initial evolutionary paths of the individual viruses, may suggest that SARS-CoV was initially under lower positive evolutionary pressure than MERS-CoV and SARS-CoV-2. Nevertheless, deletions and mutations acquired by SARS-CoV have been shown to have had an impact on adaptation to human-to-human transmission, modifying both the capacity for viral proliferation and profiles of pathogenesis [25–27].
The evolution of SARS-CoV-2 was initially marked by genetic drift in a typical process of neutral evolution [28, 29]; the virus reached a large number of new and susceptible hosts and, although some mutations appeared along the genome, there was no significant shift [30]. However, as SARS-CoV-2 rapidly spread worldwide [31, 32], fitness changes resulting from mutations in the viral genome as well as the emergence of new variants were increasingly reported [33–35].
The first epidemic wave of SARS-CoV-2 severely affected most countries in South America as a probable result of multiple viral introductions [36]; rapid increases of case numbers were especially reported in Brazil, the largest and most populous country in Latin America [37, 38]. The uncontrolled viral spread created a favorable scenario for the emergence of new variants [39–42]. To identify the impact of directional-positive selection sites at the rate of infections under these particular conditions, we traced the evolutionary scenario of SARS-CoV-2 in South America (via analysis of a significant and representative amount of genome sequences).
Remarkably, our data showed that an increase of DEPS was correlated with viral spread dynamics, with Brazil exhibiting a lower proportion of COVID-19 cases (per 100,000 inhabitants) when compared to French Guiana and the same amount of SARS-CoV-2 clusters inferred in Chile (n = 97) (Fig 1C), probably due to a higher diversity of circulating viruses. Our results also highlighted a series of mutations; while certain mutations have previously been described, but have hitherto remained unidentified in SARS-CoV-2 VOC and VOI, multiple further mutations are identified for the first time in this study, to our knowledge (Table 1).
Table 1. Mutational landscape of SARS-CoV-2 Spike protein VOC and VOI based on the WHO label.
| Sequences collection date | Inferred substitutions (Spike location) | SARS-CoV-2 carrying this mutation (from WHO) | Additional SARS-CoV-2 variants carrying this mutation (from this study) | Empirical Bayes Factors | Time interval (months) |
|---|---|---|---|---|---|
| Obs.: new mutations are underlined | |||||
| Convergent evolution: ● | |||||
| L18F ● (NTD) | Beta and Gamma | Alpha | >105 | ||
| T20N ● (NTD) | Gamma | Alpha | >105 | ||
| P26S/P26L* ● (NTD) | Gamma | Alpha and Epsilon/Zeta* | 2129.2 | ||
| D138H/D138Y* ● (NTD) | Gamma | Alpha and Epsilon/Delta* | >105 | ||
| Feb 12th 2021 | R190S ● (NTD) | Gamma | Delta | 9869.0 | |
| K417T ● (RBD) | Gamma | - | 1966.6 | ||
| E484K/E484Q ● (RBD) | Beta, Gamma, Eta, Iota, Mu, Theta and Zeta | - | >105 | ||
| N501Y ● (RBD) | Alpha, Beta, Gamma, Omicron, Mu and Theta | Eta, Kappa and Lambda | >105 | ||
| T1027I ● (CH) | Gamma | - | >105 | 5 | |
| S13I ● (SP-NTD) | Epsilon | Alpha | 166.9 | ||
| R21I/R21T* ● (NTD) | - | Gamma/Epsilon* | 168.0 | ||
| R34L/R34P* (NTD) | - | Unsigned/Eta* | 262.6 | ||
| S50L (NTD) | - | unsigned | 124.0 | ||
| L54F ● (NTD) | - | Gamma | >105 | ||
| W152L*/W152C (NTD) | Epsilon | Gamma* | 226.2 | ||
| Jul 12th 2021 Obs.: without Delta variant | S255F ● (NTD) | - | Gamma, Delta and Kappa | 160.0 | |
| N501Y ● (RBD) | Alpha, Beta, Gamma, Omicron, Mu and Theta | Eta, Kappa and Lambda | 428.8 | ||
| A570D ● (CT1) | Alpha | Eta, Kappa and Lambda | >105 | ||
| P681H* (CT2) | Alpha, Omicron, Mu and Theta | Gamma* and Lambda* | >105 | ||
| A688V ● (S1/S2) | - | Alpha, Gamma and Zeta | 113.4 | ||
| T716I ● (S1/S2) | Alpha | Epsilon | 2054.1 | ||
| D1118H/D1118Y* (CD1) | Alpha | Lambda/Zeta* | >105 | ||
| C1235F ● (CTail) | - | unsigned | 317.0 | 0 | |
| S13I ● (SP-NTD) | Epsilon | Alpha | 137.1 | ||
| T19R/T19I* ● (NTD) | Delta | Eta* | >105 | ||
| R21I/R21T* ● (NTD) | - | Gamma/Epsilon* | 138.2 | ||
| R34L (NTD) | - | unsigned | 241.4 | ||
| L54F ● (NTD) | - | Gamma | 327.6 | ||
| G142D/G142S* ● (NTD) | Delta and Kappa | Zeta* | >105 | ||
| W152L (NTD) | - | Gamma | 201.2 | ||
| R237M ● (NTD) | - | unsigned | 418.7 | ||
| Jul 12th 2021 Obs.: with Delta variant | L452R/L452Q ● (RBD) | Delta, Epsilon, Iota, Lambda and Kappa | - | >105 | |
| T478K ● (RBD) | Delta and Omicron | - | >105 | ||
| E484K (RBD) | Beta, Gamma, Eta, Iota, Mu, Theta and Zeta | - | >105 | ||
| N501Y ● (RBD) | Alpha, Beta, Gamma, Omicron, Mu and Theta | Eta, Kappa and Lambda | >105 | ||
| A570D ● (CT1) | Alpha | Eta, Kappa and Lambda | >105 | ||
| D574Y ● (CT1) | - | unsigned | 236.3 | ||
| P681R/P681H* ● (CT2) | Alpha, Delta, Omicron, Kappa, Mu and Theta | Gamma* and Lambda* | >105 | ||
| T716I ● (S1/S2) | Alpha | Epsilon | 1013.4 | ||
| D936Y● (HR1) | - | Gamma and Kappa | 236.6 | ||
| S982A ● (HR1) | Alpha | - | 9852.5 | ||
| D1118H ● (CD1) | Alpha | Lambda | >105 | ||
| D1163G ● (HR2) | - | Gamma | 237.4 |
Obs.: R158- and G142- deletions were also found in the Delta and Theta SARS-CoV-2 variants, respectively. NTD, N-terminal domain; RBD, receptor binding domain; CD1, connector domain 1; CH, center helix; CT1, C-terminal domain 1; CT2, C-terminal domain 2; CTail, cytoplasmic tail; HR1, heptad repeat 1; HR2, heptad repeat 2; S1/S2, cleavage site and SP-NTD, Signal peptide-N-terminal domain. The asterisks (*) represent mutations also found in such particular variant(s). Sources: CDC (https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html), ECDC (https://www.ecdc.europa.eu/en/covid-19/variants-concern) and Stanford Coronavirus Antiviral & Resistance Database (CoVDB) (https://covdb.stanford.edu/page/mutation-viewer/).
Spike mutations such as E484K, N501Y, L452R, S13I and W152C, seem to be fundamentally important in the process of adaptation of SARS-CoV-2 to human hosts, this by enhancing the affinity to the human ACE2 receptor and mediating immune system evasion [43–45]. Our analyzes allowed us to follow SARS-CoV-2 spread dynamics over time in Brazil, showing an increasing number of sites under DEPS, primarily in the Spike-encoding protein. Nine sites are highlighted prior to February 2021, followed by fourteen sites until July 2021 (SARS-CoV-2 Delta variant not included). With the introduction of the Delta variant, both the presence of recombination signals as well as an increase of sites under DEPS were detected (Table 1), allowing for inferences concerning a SARS-CoV-2 reproductive number increase. An increase in virus circulation augments the chance of viral coinfection, which in turn (and as a prerequisite for recombination), can heighten the risk of emergence of new variants [46, 47].
The Delta variant, first identified in late 2020 in India as B.1.617.2 [48], harbors a constellation of non-synonymous mutations in the Spike protein [49] and had become the leading VOC worldwide by the end of July 2021, this VOC accounted for 90% of all sequenced samples [50, 51]. Brazil, India and the USA, the countries most severely affected by the pandemic, are now once again threatened by this highly contagious variant. Analysis of the molecular evolution of SARS-CoV-2 taking into account the influence of local demography in these specific scenarios has the potential to generate important insights into the spread and infection dynamics of this pathogen.
Using SARS-CoV-2 sequences from China (the most populated country in the world and also a country where control measures against COVID-19 infections have been deployed very effectively) as reference, we analyzed all datasets from Brazil, India, and the USA via a large-scale phylogenetic partitions analysis. Increases in SARS-CoV-2 infections were observed to be proportional to locally circulating variants and were not (in the scenarios analyzed), correlated with any particular demography (Fig 1D); this indirectly reinforces the importance of measures implemented to avoid viral propagation. Analysis of phylogenetic partition clusters along the length of the circa 30 kb CoV genome evidenced several directionally-evolving sites under convergent evolution. Thus, a possible association between the number of infections from locally circulating SARS-CoV-2 variants carrying distinct residue profiles as well as sites in the Spike-encoding protein under DEPS may be established (Fig 1D). Interestingly, this supports a hypothesis of convergent evolution due to repeated and multiple site-specific substitutions in distinct SARS-CoV-2 VOC and VOI (see Table 1 and Fig 2).
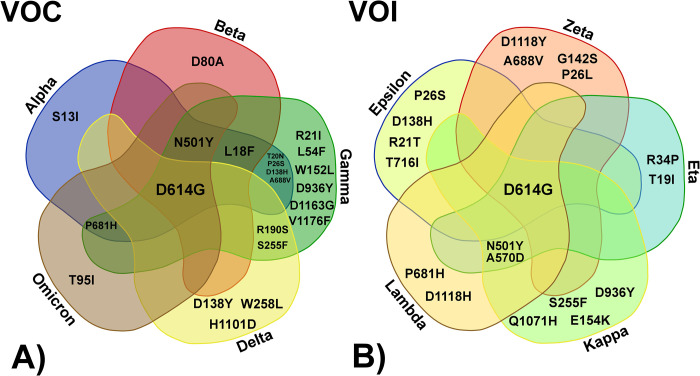
Fig 2.
Compiled data Table 1 and Fig 1D Venn diagram showing the shared mutations in distinct SARS-CoV-2 Variants of Concern (VOC) (A) as well as the additional Variants of Interest (VOI) (B) carrying such substitutions. The diagram was created through VIB web tool https://bioinformatics.psb.ugent.be/webtools/Venn/.
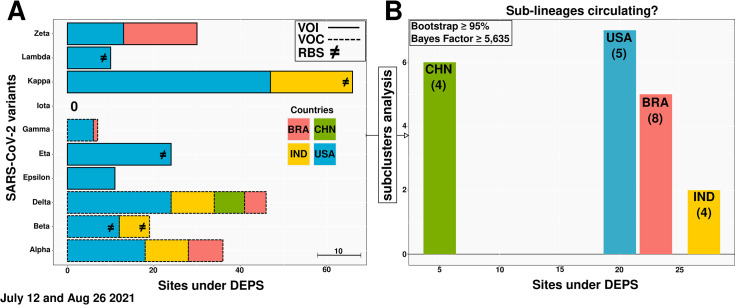
Additionally, we also inferred the possible appearance of SARS-CoV-2 sub-lineages and traced the influence of an environment favoring directional evolution acting on SARS-CoV-2 variants. We showed different patterns among sites in the VOC and VOI, with a particular emphasis on the Kappa VOI currently circulating in the USA. We further demonstrated recombination among SARS-CoV-2 VOC and VOI from India (Beta, Eta and Kappa) and the USA (Beta, Eta and Lambda) (Fig 3A).
Fig 3. Transmission clustering and sub-clustering analyses on SARS-CoV-2 Variants of Concern (VOC) and Variants of Interest (VOI) sequences.
(A) Directionally evolving sites in the VOC (dashed line) and VOI (unbroken line) sampled from the model-based phylogenetic Maximum Likelihood (ML) method and (B) the number of sub-clusters inferred in strains circulating in Brazil, China, India and the USA. Each color represents a particular country. RBS stands for recombination breakpoints signal (≠) and the scale bar shows the proportion of ten sites under DEPS (A). The numbers in parentheses indicate Spike-encoding protein sites under convergent evolution (B). Obs.: (1) In panel A, the majority of these sites, distributed across the genome, are also under convergent evolution and (2) the data extracted from the India’s SARS-CoV-2 Eta dataset did not show sites under DEPS, but did exhibit signals of recombination (see Data Availability).
As one of the first countries in the world to develop efficient immunizations and implement a vaccination policy, the USA vaccinated more than 30% of its population by April 2021 [52, 53]. By September 2021, 60% of the booster-immunized population possessed neutralizing antibodies against several viral variants [54]. Similar outcomes were observed following widespread vaccination with various SARS-CoV-2 vaccines (different technologies leveraged for vaccine production) in many other regions, including South America and India [55, 56]. We hypothesize that the site-specific mutations found under convergent evolution at strategic positions in the SARS-CoV-2 VOC/VOI Spike protein targeted by vaccine-induced neutralizing antibodies, as shown by Andreata-Santos et al. (2022) [57], may also help to explain these findings (Fig 4). Nonetheless, viral circulation in the face of incomplete immunization has been described as one of the probable causes of the emergence of new variants [42] (Sabino et al., 2021). Accordingly, our own analysis identified SARS-CoV-2 VOC and VOI sub-clusters (Fig 3B), thus indicating co-circulation of variants and sub-lineages carrying new mutations under convergent evolution. Surprisingly, the same evolutionary pattern was also observed for other endemic and epidemic CoVs studied (see Data availability).
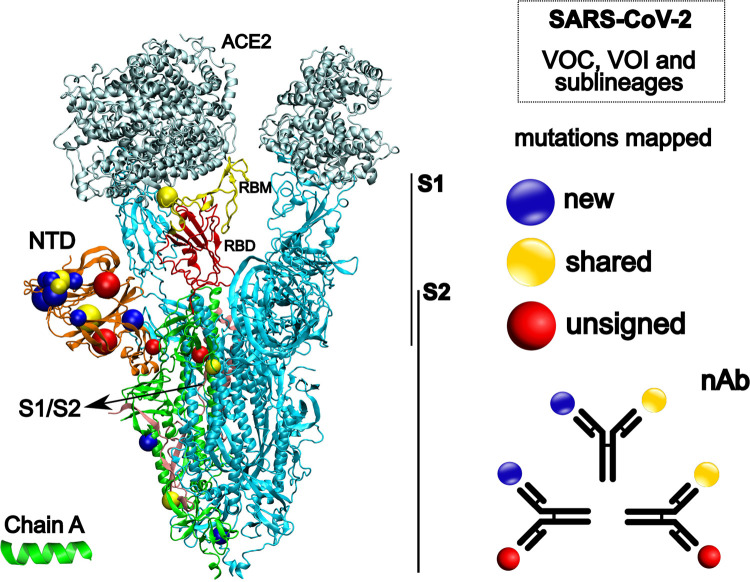
Fig 4. Structural representation of the SARS-CoV-2 spike glycoprotein (PDB 7A98).
On the left side, different colors represent ACE2, angiotensin-converting enzyme 2 (silver); NTD, N-terminal domain (orange); RBM, receptor binding motif (yellow); RBD, receptor binding domain (red) and S1/S2, cleavage site to S2 (pink). The structures in blue represent the chain B and C, respectively. Colored spheres highlight the mutations mapped in the study. On the right side, our hypotheses about a linear perspective into the prospective vaccine efficacy against different SARS-CoV-2 strains. nAb, neutralizing antibody. Spike protein image was created with the Visual Molecular Dynamics (VMD) v.1.9.3 [58].
This study demonstrates the influence of positive directional evolution on SARS-CoV-2 circulating in South America and in those countries most severely affected by the COVID-19 pandemic. Furthermore, our methodology allowed the identification of distinct transmission sub-clusters and recombination breakpoints in many SARS-CoV-2 variants, to our knowledge, not previously shown so far. We were able to indirectly infer transmission of a viral epidemiological chain and the generation of new variants. Through sequence data collected in February, July and August 2021, we also further identified and classified several convergently emerged shared mutations in different SARS-CoV-2 VOC and VOI. Lastly, we hypothesize that the co-circulation of SARS-CoV-2 variants and their possible sub-lineages takes place within a very close evolutionary environment, which can be translated to a setting of strong convergent and directional evolution. The latter may have affected the number of different lineages within each country, with highly infected countries being impacted the most (as can be seen from comparison against China). This agrees with the general virological perspective that the larger the number of viruses circulating, as well as the number of lineages within a region, the more likely it becomes that sub-lineages will originate and spread. Our results confirm the importance of critical assessment, monitoring, and control of SARS-CoV-2 lineages and sub-lineages throughout the pandemics, even more so in countries with large populations where many opportunities arise for positive selection, due to both recurrent substitutions and recombination events.
Acknowledgments
We gratefully acknowledge the authors and both the originating and submitting laboratories for the sequence data in GISAID EpiCoV and GenBank on which this work is based. The authors also thank the Rede Corona-Ômica/MCTI/FINEP, the National Laboratory for Scientific Computing (LNCC/MCTI, Brazil) for providing HPC resources of the Santos Dumont supercomputer (ID #45691), and Prof. Luiz Mário Ramos Janini for fruitful discussion.
Data Availability
The data underlying the results presented in the study are available from https://github.com/rduraescarvalho/SARS-CoV-2_DEPS.
Funding Statement
This work was supported by the Fundação de Amparo à Pesquisa doEstado384 de São Paulo (FAPESP), Brazil, grants 2019/01255-9 and 2021/03684-4(Young385 Investigator Program) (RD-C), and by the Conselho Nacional de Desenvolvimento386 Científico e Tecnológico (CNPq), Brazil, grant 405691/2018-1 (C.T.B). DRNis387 recipient of an institutional scholarship from the Coordenação de Aperfeiçoamento388 de Pessoal de Nível Superior (CAPES), Brazil, grant 88887.506234/2020-00.
References
- 1.Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, Cheung CL, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302: 276–8. doi: 10.1126/science.1087139 [DOI] [PubMed] [Google Scholar]
- 2.Zaki AM, Boheemen S van, Bestebroer TM, Osterhaus ADME, Fouchier RAM. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367: 1814–20. doi: 10.1056/NEJMoa1211721 [DOI] [PubMed] [Google Scholar]
- 3.Costa VG da Moreli ML, Saivish MV. The emergence of SARS, MERS and novel SARS-2 coronaviruses in the 21st century. Arch Virol. 2020;165: 1517–26. doi: 10.1007/s00705-020-04628-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corman VM, Muth D, Niemeyer D, Drosten C. Hosts and Sources of Endemic Human Coronaviruses. Adv Virus Res. 2018;100: 163–88. doi: 10.1016/bs.aivir.2018.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boni MF, Lemey P, Jiang X, Lam TT-Y, Perry BW, Castoe TA, et al. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat Microbiol. 2020;5: 1408–17. doi: 10.1038/s41564-020-0771-4 [DOI] [PubMed] [Google Scholar]
- 6.V’kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021:19: 155–70. doi: 10.1038/s41579-020-00468-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munnink BBO, Worp N, Nieuwenhuijse DF, Sikkema RS, Haagmans B, Fouchier RAM, et al. The next phase of SARS-CoV-2 surveillance: real-time molecular epidemiology. Nat Med. 2021;27: 1518–24. doi: 10.1038/s41591-021-01472-w [DOI] [PubMed] [Google Scholar]
- 8.Pond SLK, Poon AFY, Brown AJL, Frost SDW. A maximum likelihood method for detecting directional evolution in protein sequences and its application to influenza A virus. Mol Biol Evol. 2008;25: 1809–24. doi: 10.1093/molbev/msn123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alteri C, Cento V, Piralla A, Costabile V, Tallarita M, Colagrossi L, et al. Genomic epidemiology of SARS-CoV-2 reveals multiple lineages and early spread of SARS-CoV-2 infections in Lombardy, Italy. Nat Commun. 2021;12: 434. doi: 10.1038/s41467-020-20688-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention (CDC). SARS-CoV-2 Variant Classifications and Definitions. 01 December 2021. Available at: https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html.
- 11.Peacock TP, Penrice-Randal R, Hiscox JA, Barclay WS. SARS-CoV-2 one year on: evidence for ongoing viral adaptation. J Gen Virol. 2021;102: 001584. doi: 10.1099/jgv.0.001584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durães-Carvalho R, Ludwig-Begall LF, Salemi M, Lins RD, Marques ETA. Influence of directional positive Darwinian selection-driven evolution on arboviruses Dengue and Zika virulence and pathogenesis. Mol Phylogenet Evol. 2019;140: 106607. doi: 10.1016/j.ympev.2019.106607 [DOI] [PubMed] [Google Scholar]
- 13.Okonechnikov K, Golosova O, Fursov M. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 2012;28: 1166–7. doi: 10.1093/bioinformatics/bts091 [DOI] [PubMed] [Google Scholar]
- 14.Prosperi MCF, Ciccozzi M, Fanti I, Saladini F, Pecorari M, Borghi V, et al. A novel methodology for large-scale phylogeny partition. Nature Commun. 2011;2: 321. doi: 10.1038/ncomms1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5: e9490. doi: 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pond SLK, Posada D, Gravenor MB, Woelk CH, Frost SDW. GARD: a genetic algorithm for recombination detection. Bioinformatics. 2006;22: 3096–8. doi: 10.1093/bioinformatics/btl474 [DOI] [PubMed] [Google Scholar]
- 17.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2005;23: 254–67. doi: 10.1093/molbev/msj030 [DOI] [PubMed] [Google Scholar]
- 18.Pond SLK, Poon AFY, Velazquez R, Weaver S, Hepler NL, Murrell B, et al. HyPhy 2.5-A Customizable Platform for Evolutionary Hypothesis Testing Using Phylogenies. Mol Biol Evol. 2020;37: 295–99. doi: 10.1093/molbev/msz197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai MMC. Genetic Recombination in RNA Viruses. Curr Top Microbiol Immunol. 1992;176: 21–32. doi: 10.1007/978-3-642-77011-1_2 [DOI] [PubMed] [Google Scholar]
- 20.Lemey P, Salemi M, Vandamme A-M. The Phylogenetic Handbook: A Practical Approach to Phylogenetic Analysis and Hypothesis Testing. 2nd ed. Cambridge University Press; 2009. [Google Scholar]
- 21.Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, et al. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol. 2016;24: 490–502. doi: 10.1016/j.tim.2016.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pollett S, Conte MA, Sanborn M, Jarman RG, Lidl GM, Modjarrad K, et al. A comparative recombination analysis of human coronaviruses and implications for the SARS-CoV-2 pandemic. Sci Rep. 2021;11: 17365. doi: 10.1038/s41598-021-96626-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He Y, Ma W, Dang S, Chen L, Zhang R, Mei S, et al. Possible recombination between two variants of concern in a COVID-19 patient. Emerg Microbes Infect. 2022;11: 552–55. doi: 10.1080/22221751.2022.2032375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forni D, Cagliani R, Arrigoni F, Benvenuti M, Mozzi A, Pozzoli U, et al. Adaptation of the endemic coronaviruses HCoV-OC43 and HCoV-229E to the human host. Virus Evol. 2021;7: veab061. doi: 10.1093/ve/veab061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muth D, Corman VM, Roth H, Binger T, Dijkman R, Gottula LT, et al. Attenuation of replication by a 29 nucleotide deletion in SARS-coronavirus acquired during the early stages of human-to-human transmission. Sci Rep. 2018;8: 15177. doi: 10.1038/s41598-018-33487-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pereira F. Evolutionary dynamics of the SARS-CoV-2 ORF8 accessory gene. Infect Genet Evol. 2020;85: 104525. doi: 10.1016/j.meegid.2020.104525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pereira F. SARS-CoV-2 variants lacking a functional ORF8 may reduce accuracy of serological testing. J Immunol Methods. 2021;488: 112906. doi: 10.1016/j.jim.2020.112906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dearlove B, Lewitus E, Bai H, Li Y, Reeves DB, Joyce MG, et al. A SARS-CoV-2 vaccine candidate would likely match all currently circulating variants. Proc Natl Acad Sci USA. 2020;117: 23652–62. doi: 10.1073/pnas.2008281117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacLean OA, Lytras S, Weaver S, Singer JB, Boni MF, Lemey P, et al. Natural selection in the evolution of SARS-CoV-2 in bats created a generalist virus and highly capable human pathogen. PLoS Biol. 2021;19: e3001115. doi: 10.1371/journal.pbio.3001115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin DP, Weaver S, Tegally H, San JE, Shank SD, Wilkinson E, et al. The emergence and ongoing convergent evolution of the N501Y lineages coincides with a major global shift in the SARS-CoV-2 selective landscape. Cell. 2021;5189–5200.e7. doi: 10.1016/j.cell.2021.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yadav PD, Potdar VA, Choudhary ML, Nyayanit DA, Agrawal M, Jadhav SM, et al. Full-genome sequences of the first two SARS-CoV-2 viruses from India. Indian J Med Res. 2020;151: 200–09. doi: 10.4103/ijmr.IJMR_663_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hodcroft EB, Zuber M, Nadeau S, Vaughan TG, Crawford KHD, Althaus CL, et al. Spread of a SARS-CoV-2 variant through Europe in the summer of 2020. Nature. 2021;595: 707–12. doi: 10.1038/s41586-021-03677-y [DOI] [PubMed] [Google Scholar]
- 33.Velazquez-Salinas L, Zarate S, Eberl S, Gladue DP, Novella I, Borca MV. Positive Selection of ORF1ab, ORF3a, and ORF8 Genes Drives the Early Evolutionary Trends of SARS-CoV-2 During the 2020 COVID-19 Pandemic. Front Microbiol. 2020;11: 550674. doi: 10.3389/fmicb.2020.550674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang L, Jackson CB, Mou H, Ojha A, Peng H, Quinlan BD, et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat Commun. 2020;11: 6013. doi: 10.1038/s41467-020-19808-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plante JA, Liu Y, Liu J, Xia H, Johnson BA, Lokugamage KG, et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592: 116–21. doi: 10.1038/s41586-020-2895-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Candido DS, Claro IM, de Jesus JG, Souza WM, Moreira FRR, Dellicour S, et al. Evolution and epidemic spread of SARS-CoV-2 in Brazil. Science. 2020;369: 1255–60. doi: 10.1126/science.abd2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paiva MHS, Guedes DRD, Docena C, Bezerra MF, Dezordi FZ, Machado LC, et al. Multiple Introductions Followed by Ongoing Community Spread of SARS-CoV-2 at One of the Largest Metropolitan Areas of Northeast Brazil. Viruses. 2020;12: 1414. doi: 10.3390/v12121414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stefanelli P, Faggioni G, Presti AL, Fiore S, Marchi A, Benedetti E, et al. Whole genome and phylogenetic analysis of two SARS-CoV-2 strains isolated in Italy in January and February 2020: additional clues on multiple introductions and further circulation in Europe. Eurosurveill. 2020;25: 2000305. doi: 10.2807/1560-7917.ES.2020.25.13.2000305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Voloch CM, da Silva Francisco R Jr, Almeida LGP de, Cardoso CC, Brustolini OJ, Gerber AL, et al. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. J Virol. 2021;95: e00119–21. doi: 10.1128/JVI.00119-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Faria NR, Mellan TA, Whittaker C, Claro IM, Candido D da S, Mishra S, et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021;372: 815–21. doi: 10.1126/science.abh2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Resende PC, Gräf T, Paixão ACD, Appolinario L, Lopes RS, Mendonça AC da F, et al. A Potential SARS-CoV-2 Variant of Interest (VOI) Harboring Mutation E484K in the Spike Protein Was Identified within Lineage B.1.1.33 Circulating in Brazil. Viruses. 2021;13: 724. doi: 10.3390/v13050724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sabino EC, Buss LF, Carvalho MPS, Prete CA, Crispim MAE, Fraiji NA, et al. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet. 2021;397: 452–5. doi: 10.1016/S0140-6736(21)00183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCallum M, Bassi J, Marco AD, Chen A, Walls AC, Iulio JD, et al. SARS-CoV-2 immune evasion by the B.1.427/B.1.429 variant of concern. Science. 2021;373: 648–54. doi: 10.1126/science.abi7994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greaney AJ, Loes AN, Crawford KHD, Starr TN, Malone KD, Chu HY, et al. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29: 463–76. doi: 10.1016/j.chom.2021.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19: 409–24. doi: 10.1038/s41579-021-00573-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haddad D, John SE, Mohammad A, Hammad MM, Hebbar P, Channanath A, et al. SARS-CoV-2: Possible recombination and emergence of potentially more virulent strains. PLoS One. 2021;16: e0251368. doi: 10.1371/journal.pone.0251368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Centers for Disease Control and Prevention (CDC). What You Need to Know About Variants. 02 February 2022. Available at: https://www.cdc.gov/coronavirus/2019-ncov/variants/delta-variant.html.
- 48.Kirola L. Genetic emergence of B.1.617.2 in COVID-19. New Microbes New Infect. 2021;43: 100929. doi: 10.1016/j.nmni.2021.100929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCallum M, Walls AC, Sprouse KR, Bowen JE, Rosen L, Dang HV, et al. Molecular basis of immune evasion by the delta and kappa SARS-CoV-2 variants. Science. 2021;374: 1621–26 doi: 10.1126/science.abl8506 [DOI] [PubMed] [Google Scholar]
- 50.Lamarca AP, Almeida LGP de, da Silva Francisco R Jr, Cavalcante L, Machado DT, Brustolini O, et al. Genomic surveillance tracks the first communitary outbreak of Delta (B.1.617.2) variant in Brazil. J Virol. 2022;96: e0122821.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pan American Health Organization (PAHO). Epidemiological Update: Increase of the Delta variant and its potential impact in the Region of the Americas. 8 August 2021. Available at: https://www.paho.org/en/documents/epidemiological-update-increase-delta-variant-and-its-potential-impact-region-americas-8. [Google Scholar]
- 52.Centers for Disease Control and Prevention (CDC). COVID-19 Incidence and Death Rates Among Unvaccinated and Fully Vaccinated Adults with and Without Booster Doses During Periods of Delta and Omicron Variant Emergence—25 U.S. Jurisdictions, April 4–December 25, 2021. 28 January 2022. Available at: https://www.cdc.gov/mmwr/volumes/71/wr/mm7104e2.htm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.U.S. Food & Drug Administration (FDA). Spikevax and Moderna COVID-19 Vaccine. 31 January 2022. Available at https://www.fda.gov/emergency-preparedness-and-response/coronavirus-disease-2019-covid-19/moderna-covid-19-vaccine. [Google Scholar]
- 54.Pegu A, O’Connell S, Schmidt SD, O’Dell S, Talana CA, Lai L, et al. Durability of mRNA-1273 vaccine-induced antibodies against SARS-CoV-2 variants. Science. 2021;373: 1372–77. doi: 10.1126/science.abj4176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li X-N, Huang Y, Wang W, Jing Q-L, Zhang C-H, Qin P-Z, et al. Effectiveness of inactivated SARS-CoV-2 vaccines against the Delta variant infection in Guangzhou: a test-negative case-control real-world study. Emerg Microbes Infect. 2021;10: 1751–59. doi: 10.1080/22221751.2021.1969291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bernal JL, Andrews N, Gower C, Gallagher E, Simmons R, Thelwall S, et al. Effectiveness of Covid-19 Vaccines against the B.1.617.2 (Delta) Variant. New Engl J Med. 2021;385: 585–94. doi: 10.1056/NEJMoa2108891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Andreata-Santos R, Janini LMR, Durães-Carvalho R. From Alpha to Omicron SARS-CoV-2 variants: What their evolutionary signatures can tell us? J Med Virol. 2022. Online ahead of print. doi: 10.1002/jmv.27555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Humphrey W, Dalke A, Schulten K. VMD—Visual Molecular Dynamics. J Mol Graph. 1996;14: 33–8. doi: 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying the results presented in the study are available from https://github.com/rduraescarvalho/SARS-CoV-2_DEPS.