Abstract
Bipolar disorder (BD) is a psychiatric condition characterized by depressive and manic episodes that affect 2% of the world population. The first-line long-term treatment for mood stabilization is lithium (Li). Induced pluripotent stem cell modeling of BD using hippocampal dentate gyrus-like neurons derived from Li-responsive (LR) and Li-non-responsive (NR) patients previously showed neuronal hyperexcitability. Li treatment reversed hyperexcitability only on the LR neurons. In this study we searched for specific targets of Li resistance in NR neurons and found that the activity of Wnt/β-catenin signaling pathway was severely affected, with a significant decrease in expression of LEF1. Li targets the Wnt/β-catenin signaling pathway by inhibiting GSK-3β and releasing β-catenin that forms a nuclear complex with TCF/LEF1, activating the Wnt/β-catenin transcription program. Therefore, we propose that downregulation of LEF1 may account for Li resistance in NR neurons. Our results show that valproic acid (VPA), a drug used to treat NR patients that also acts downstream of GSK-3β, upregulated LEF1 and Wnt/β-catenin gene targets, increased transcriptional activity of complex β-catenin/TCF/LEF1 and reduced excitability in NR neurons. Additionally, decreasing LEF1 expression in control neurons using shLEF1 caused hyperexcitability, confirming that the impact of VPA on excitability in NR neurons was connected to changes in LEF1 and in the Wnt/β-catenin pathway. Our results suggest that LEF1 may be a useful target for the discovery of new drugs for BD treatment.
Introduction
Bipolar disorder (BD) is a major psychiatric disorder characterized by mood cycles, with depressive and high-energy episodes (hypomanic or manic), and is associated with multiple psychiatric and medical comorbidities and a high risk of suicide [1–3]. BD is a spectrum of diagnosis based on the intensity, duration and frequency of mood episodes [4–6]. First symptoms appear during adolescence or in young adults but diagnosis may take up to 10 years after illness onset [7, 8]. In a World Health Organization (WHO) World Mental Health survey comparing 19 common disorders, BD was the second most disabling disorder, similar to neurological disorders and ranked three times higher than cancer [9].
Lithium (Li) has been used for 70 years and continues to be the first-line treatment to prevent manic and depressive episodes, and the only drug with an anti-suicidal effect [10, 11]. However, only about 30% of BD patients respond fully to Li monotherapy, which may need more than a year to reduce morbidity or to obtain full response; at least 20% of patients do not respond at all and the remaining 40–50% show a partial response [10, 12, 13]. Although efforts have been made to predict a patient’s response to Li, they have not been applicable in practice [14–16]. In addition, long-term treatment with Li is not easy to manage because of its multiple side effects, narrow therapeutic range, contraindications and extended time to response, frequently leading to patient non-adherence and treatment failure [14, 17, 18]. Second-generation antipsychotics have proven to be both effective and tolerable for treatment of acute mania [19] and are increasingly used as an alternative to mood stabilizers [18, 20]. Still, most international or national guidelines continue to propose Li as the first-line maintenance treatment for BD [21–23]. Frequently patients who do not respond to Li are treated with complex polypharmacy, including valproate (valproic acid or VPA) and other anticonvulsants, second-generation antipsychotics and antidepressants, leading to high medication burden [24]. Therefore, developing new treatments for BD that are more effective, tolerable and safe is urgently needed.
Development of new treatments would benefit from a better understanding of the underlying mechanism of action of current mood stabilizers. Li has multiple direct and indirect targets, namely in Wnt, phosphatidylinositol and calcium signaling pathways, in mitochondrial functions and in neuronal excitability [25–30]. The cellular effects of Li are extensive since many of the downstream targets are transcription factors, miRNAs, histone modifications and DNA methylation, all of which multiply the molecular complexity of the treatment [31, 32]. One of the most studied Li targets is glycogen synthase kinase 3 beta (GSK-3β), a component of the canonical or Wnt/β-catenin signaling pathway. In the absence of the Wnt signal, cytoplasmic β-catenin binds to the β-catenin destruction complex, is phosphorylated by casein kinase 1 (CK1) and GSK-3β and is targeted to degradation by the proteasome [33, 34]. When Wnt ligands bind to Frizzled (FZD) receptors and low-density lipoprotein receptor-related protein (LRP5/6) co-receptors, the β-catenin destruction complex disassembles and β-catenin translocates to the nucleus, where it binds to T-cell factor/lymphoid enhancer-binding (TCF/LEF) transcription factors [34]. Inhibition of GSK-3β by Li mimics the Wnt signal, resulting in accumulation of β-catenin in the nucleus and activation of β-catenin/TCF/LEF1 targets [34, 35]. Another molecule used to treat BD, the histone deacetylase inhibitor VPA, also increases Wnt/β-catenin signaling but possibly through an increase in β-catenin transcription [36, 37]. In agreement with Li and VPA targets, extensive evidence has shown defects in Wnt/β-catenin pathway in BD patients [38, 39].
Development of novel treatments for BD is limited by the lack of accurate animal models and relevant cell models. Recently, induced pluripotent stem cell (iPSC) technology has provided encouraging results for the study of neuropsychiatric disorders that result from a combination of pathological changes and compensatory mechanisms [40]. BD is an ideal case for examination in the context of iPSC technology, given its high heritability (up to 85%) [3, 41]. To study BD, neurons from different regions of the brain have been generated directly from fibroblasts by transdifferentiation or using intermediate iPSCs [42]. Interestingly, different iPSC-modeling studies using different techniques and cell types have all reported quantifiable responses to Li [43–46]. Mertens et al. showed that dentate gyrus (DG)-like neurons derived from iPSCs generated from BD patients displayed hyperexcitability that was normalized by Li treatment, but only in neurons derived from patients who had a clinical history of therapeutic Li response [47]. These results were reproduced using a different patient cohort, demonstrating that the hyperexcitability phenotype was robust and could be used to predict Li responsiveness [43]. More important than describing a new cellular phenotype, these studies strongly suggest that DG neuronal hyperexcitability correlates with patient clinical information and drug response, which can be used to find new drugs that could potentially be translated into new treatments.
In this study we searched for specific targets related to Li resistance. We first compared transcriptome profiles from DG neurons derived from BD patients who were clinically Li responsive (LR) and Li non-responsive (NR). Our results showed that the Wnt/β-catenin signaling pathway was affected in NR neurons, with a significant decrease in the expression of LEF1 transcription factor. VPA treatment increased expression of LEF1 targets and activity of the β-catenin/TCF/LEF1 complex, and it reduced excitability in NR neurons. Downregulation of LEF1 expression in control neurons caused hyperexcitability, similar to what was observed in BD neurons, suggesting that LEF1 expression is implicated in neuronal hyperexcitability. We propose that in NR patients the activity of β-catenin/TCF/LEF1 complex downstream of GSK-3β is a target for treatment.
Materials and Methods
Subjects
Patients with BD type I were participants in genetic studies at Dalhousie University and their clinical characteristics were described previously [43] (see also Table 1). All subjects (4 healthy controls, 3 BD patients responsive to Li and 3 BD patients non responsive to Li) were Caucasian males and provided written informed consent. The average age of the subjects was 47.8 (SD ± 15.90) for healthy individuals, 41.7 (SD ± 4.63) for LR patients and 49.7 (SD ± 5.24) for NR patients. The Nova Scotia Health Authority Research Ethics Board, University of California, San Diego (UCSD), the Salk Institute Institutional Review Board and the Embryonic Stem Cell Research Oversight Committee (IRB protocol #09–0003) approved all procedures.
Table 1.
Demographics of the subjects involved in the study
| Category | Age | Age at onset | Diagnosis | Sex | Ethnicity | Episodes off Li* | Episodes on Li | Years on Li at sampling | Li response score** |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Responder | 50 | 31 | Bipolar I | Male | Caucasian | 3M 4D | 0 | 5 | 9/10 |
| Responder | 41 | 34 | Bipolar I | Male | Caucasian | 5M 1D | 0 | 3 | 10/10 |
| Responder | 34 | 15 | Bipolar I | Male | Caucasian | 3M 1D | 0 | 4 | 9/10 |
| Non-responder | 51 | 35 | Bipolar I | Male | Caucasian | 1M 1D | 1M | 2 | 3/10 |
| Non-responder | 58 | 22 | Bipolar I | Male | Caucasian | 3M 5D | 2D 1RC | 6 | 1/10 |
| Non-responder | 40 | 24 | Bipolar I | Male | Caucasian | 4M 3D | 7M | 7 | 0/10 |
| Control | 62 | n/a | n/a | Male | Caucasian | n/a | n/a | n/a | n/a |
| Control | 25 | n/a | n/a | Male | Caucasian | n/a | n/a | n/a | n/a |
| Control | 51 | n/a | n/a | Male | Caucasian | n/a | n/a | n/a | n/a |
| Control | 53 | n/a | n/a | Male | Caucasian | n/a | n/a | n/a | n/a |
M mania, D depression, RC period of rapid cycling
on the treatment response scale; n/a, non applicable
We interviewed all participants with the Schedule for Affective Disorders and Schizophrenia (Lifetime version; SADS-L) [48] supplemented with queries about additional clinical details as appropriate. The interviews were conducted by pairs of experienced clinical researchers (psychiatrists and research nurses) who were blind to the diagnostic status of the subjects. All interview information and relevant medical records were then reviewed in a blind fashion by a panel of clinical researchers to obtain consensus diagnosis. Patients who received Li monotherapy for a minimum of 2 years were evaluated for treatment response using a previously validated and commonly used scale with documented good inter-rater reliability [49, 50]. The scale scores ranged from 0 to 10, with values of 7 to 10 indicating good response and scores of 0 to 6 indicating poor response [49].
Neuronal differentiation and lentivirus
Epstein-Barr virus-immortalized B-lymphocytes from healthy controls and BD patients were reprogrammed to iPSCs using the Yamanaka Episomal vector set as previously described [43]. Quality control criteria for validation of the iPSC lines were absence of integration of episomal reprogramming plasmids, presence of normal karyotype, positive staining for pluripotency markers and authentication of cell lines using a comparison of a 16-loci short tandem repeat profile between the iPSC clones and the original lymphoblast line. iPSCs were cultured on matrigel-coated plates in mTeSR1 medium (STEMCELL Technologies) and differentiation into neural progenitor cells (NPCs) was performed as described [43, 51]. Embryoid bodies (EB) were generated from confluent iPSC cultures by mechanical dissociation with 1 mg/ml collagenase IV (Gibco) and then plated onto low-adherence plates in mTeSR1 medium containing 10 μM ROCK inhibitor (STEMCELL Technologies) and incubated overnight with shaking. The next day media was changed to DMEM/F12 Glutamax (Gibco) with N2 and B27 without vitamin A (both from Invitrogen; N2B27 media) with 0.5 μg/ml dickkopf Wnt signaling pathway inhibitor 1 (DKK1, PeproTech), 10 μM SB431542 (Thermo Fisher Scientific), 0.5 μg/ml Noggin (PeproTech) and 1 μM cyclopamine (LC Laboratories). Treatment continued for 20 days without shaking. EBs were plated onto 100 μg/ml poly-L-ornithine (poly-O, Sigma) and 5 μ/ml laminin (Invitrogen)-coated plates with N2B27 media and, after 1 week, rosettes were manually collected, dissociated with accutase (Chemicon) and plated with N2B27 media with 20 ng/ml fibroblast growth factor 2 (FGF2, Joint Protein Central) to obtain a monolayer of NPCs. For this study, DG-like neurons (expressing PROX1) were differentiated from frozen NPC stocks as described previously [43, 51]. Briefly, NPCs were plated onto poly-O/laminin-coated plates and differentiated in N2B27 media supplemented with 200 nM ascorbic acid (STEMCELL Technologies), 500 μg/ml cyclic AMP (Tocris Bioscience), 20 ng/ml brain-derived neurotrophic factor (BDNF, PeproTech), 20 ng/ml Wnt3a (R&D Systems) and 5 μg/ml laminin for 3 weeks. Media was changed 3 times a week and Wnt3a was removed after 3 weeks.
To generate a stable shLEF1 control line, NPCs were transduced with lentivirus expressing shRNAs and selected for puromycin resistance for 3 successive media changes using 0.5 μg/ml concentration. The shLEF1 (sc-35804-V) or shControl containing a universal scrambled sequence (sc-108080) virus was purchased from Santa Cruz. Fluorescent 7TGC lentivirus carrying a 7xTcf-eGFP reporter cassette and SV40-mCherry selection cassette was used to quantify activity of the Wnt signaling pathway [52]. 7TGC was a gift from Roel Nusse (Addgene plasmid #24304; http://n2t.net/addgene:24304; RRID:Addgene_24304) and recombinant lentivirus were purchased from Addgene.
Immunocytochemistry
Cells were fixed in 4% paraformaldehyde for 15 min. Antigen blocking and cell permeabilization were done using 10% horse serum and 0.1% Triton X-100 in PBS for 1 h at room temperature. Primary antibodies prepared in blocking solution were incubated overnight at 4°C, and the next day they were washed with PBS and incubated with secondary antibodies (1:250, Jackson Laboratories) for 1 h at room temperature. The primary antibodies used were chicken anti-MAP2 (1:1000, Abcam 5392) and goat anti-GFP (1:1000, Abcam 5450). Cells were counterstained with DAPI for cell nuclei visualization.
RNA extraction, library preparation and RNA sequencing
RNA-sequencing analysis was performed on bulk cultures of NPCs and neurons at 8, 16, 28 and 42 days post-differentiation. Cells (~5×106 cells) were collected in RNA-Bee solution (Tel-Test, Inc) and total RNA was extracted using the DNA-Free RNA Kit (Zymo Research) according to the manufacturer’s instructions. RNA-sequencing analysis was also performed on PROX1-positive and PSA-NCAM-positive sorted neurons at 16 days post-differentiation. Neuronal cultures were transduced at day 8 with Prox1-eGFP lentiviral vector [51]; 8 days later they were sorted by flow cytometry for cells positive for eGFP and APC fluorescence after staining with mouse anti-PSA-NCAM-APC (1:50, Miltenyi Biotec 130–120-437). Before sorting, cultures were treated with 1 mM LiCl, 1 mM VPA or vehicle (water) for 24 h. Total RNA was extracted from 5×105 sorted cells on TRIzol LS reagent (Invitrogen) using the DNA-Free RNA Kit (Zymo Research) as for bulk samples. RNA quality was assayed using Agilent Technologies 2200 TapeStation and libraries were prepared from samples with integrity superior to RNA Integrity Number (RIN) 7. Stranded mRNA-Seq libraries were prepared using the Illumina TruSeq Stranded mRNA Library Prep Kit according to the manufacturer’s instructions. All libraries were then quantified, pooled and sequenced at single-end 50 bp reads on Illumina HiSeq 2500 at the Salk Institute Next Generation Sequencing (NGS) Core at a depth of approximately 40 million reads per library. Sequenced reads were quality-tested using FASTQC [53]. Reads were trimmed with SolexaQA++ dynamic trim [54] and then aligned to GRCh38 with STAR [55]. Counts were normalized to transcripts per million (TPM) and then log + 1 transformed for all visualizations.
Statistical analyses
Differential expression was calculated using the software edgeR (v3.26.8) [56]. Raw counts were formatted into a DGE list and filtered for genes with counts > 1 in at least 3 samples. In order to capture all gene expression changes in response to VPA, all genes with counts > 0 in at least 1 sample were used. Counts were then normalized with the TMM method. Dispersion was estimated in 3 steps. First common dispersion was calculated with the conditional maximum likelihood method, followed by tagwise dispersion as a moving average, and then trended dispersion calculated as a spline. These dispersions were then used to calculate differential expression between the relevant 2 groups. P-values were then adjusted for multiple-testing using the p.adjust function in R (v3.6.1) with the fdr method. To identify the number of genes differentially expressed we used an FDR < 0.05 with no minimum logFC. Where differences in expression were calculated as a function of time, they were calculated using the edgeR GLM function with expression as a function of time + disease status. For bulk analyses, days 8–42 were combined and tested as a function of disease status using the described exact test functions. Results were reported as volcano plots with fold change transformed using base 2 and raw p-values transformed using base exp(1).
Functional enrichment analysis was performed using DAVID bioinformatics (v6.8) [57, 58]. All genes with FDR < 0.05 for a given differential expression test were uploaded as a gene list in official gene symbol format and corrected against the Homo sapiens background. A functional annotation chart was then calculated from the results.
Principal component analysis was performed on TPM values using the package pcaMethods (v1.76.0) with the svd algorithm.
LEF1-predicted downstream targets were identified using the Transfac database (gene id = 51176) [59].
Quantitative RT-PCR
Total RNA was extracted from neuronal cultures (~5×106 cells) using RNA-Bee solution (Tel-Test, Inc) and the DNA-Free RNA Kit (Zymo Research) according to the manufacturer’s instructions. cDNA was synthesized from 5 ng of RNA, using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer’s instructions. Quantitative PCR reactions were done using SYBR Green PCR Master Mix (Applied Biosystems) in 96-well plates. All reactions were performed in triplicate in each plate. qPCR results were analyzed using SDS Software v3.2 for 7900HT real-time PCR system. The amount of the LEF1 mRNA was normalized to the ACTB gene mRNA. Primer pairs used were: hLEF1_qFw ACAGATCACCCCACCTCTTG, hLEF1_qRv ATAGCTGGATGAGGGATGCC for the LEF1 gene; ACTB_Fw: CACCATTTGGCAATGAGCGGTTC, ACTB_Rv: AGGTCTTTGCGGATGTCCACGT for the β-actin gene.
Flow cytometry assay for Wnt/β-catenin activity
NPCs from 9 cell lines (3 controls, 3 LRs and 3 NRs) were seeded in 6-well coated plates for neuronal differentiation cultures and transduced with 7TGC lentivirus at day 8. To avoid interference with our assay, Wnt3a was removed from the differentiation media after 7 days. Drug treatments (1 mM LiCl or 1 mM valproic acid) were performed for 3 days before flow cytometry quantification using a BD LSRII cytometer (BD Biosciences) on 2.5-week-old neurons followed by analysis using FlowJo software (TreeStar). Negative gates were determined for each experiment using non-transduced cells. Experiments were performed with 3 experimental replicates and averaged for each cell line. Data in main figures show averages of 2 experiments for each cell line.
Multiwell microelectrode array (MEA) analysis
NPCs (15,000 cells) were plated in 96-well MEA plates coated with poly-O and laminin for neuronal differentiation as described before. Extracellular recordings were performed in a Maestro MEA system and AxIS software (Axion Biosystems) using a bandwidth with a filter for 200 Hz to 3 kHz cutoff frequencies. Spike detection was performed using an adaptive threshold set to 5.5 times the standard deviation of the estimated noise on each electrode. Each plate was acclimatized for 10 min in the Maestro Instrument and recorded for 10 min for quantification. Recordings were performed before media change. When drugs were added to the media, recordings were performed just before media change and drug treatment (1 mM LiCl or 1 mM VPA). Multi-electrode data analysis was performed using the Axion Biosystems Neural Metrics Tool. An electrode was considered active at a threshold of 5 spikes/min. Each experiment was repeated 3 times with 8 or 12 replicates per plate; replicates were averaged for each cell line.
Results
NR neurons became transcriptionally distinct during differentiation
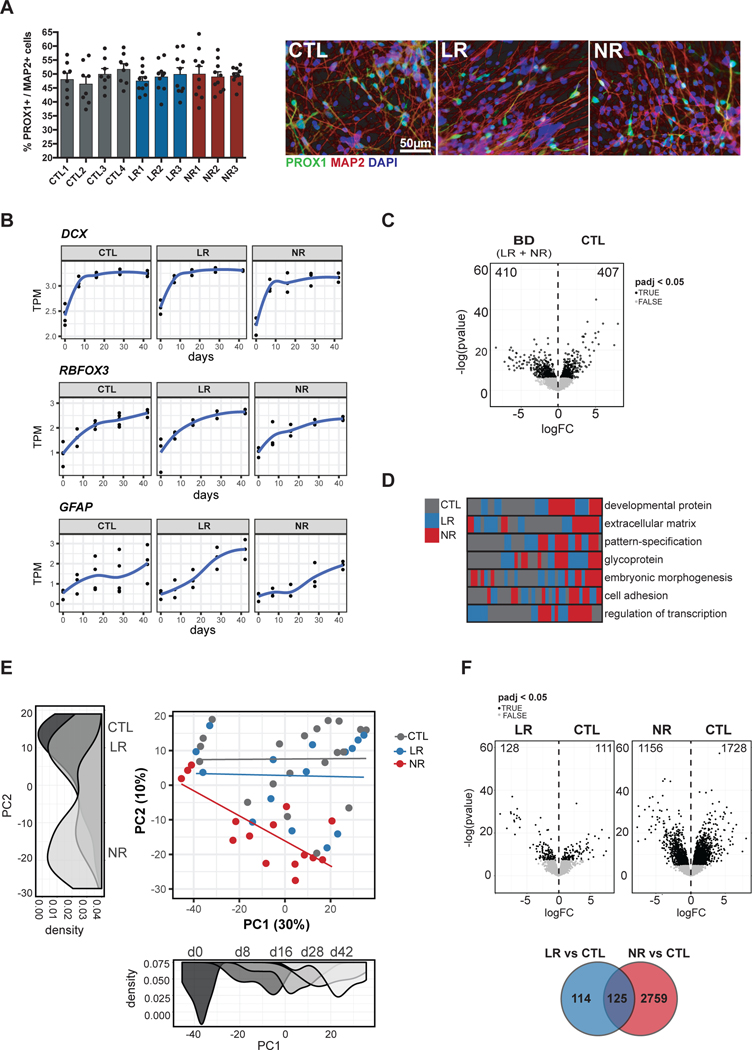
Our previous results showed that, although the hyperexcitability phenotype is characteristic of BD DG-like neurons, the electrophysiological properties and the response to Li are distinctive between LR and NR neurons [43, 44]. Therefore, we hypothesized that the comparison of gene expression in LR, NR and control (CTL) neurons could show Li response and/or Li resistance molecular signatures, in addition to a BD signature. We performed RNA sequencing from bulk cultures of NPCs (day 0) and neurons at 8, 16, 28 and 42 days post-differentiation (Fig. S1). As expected from previous observations [43], the neuronal and glial cell populations differentiated from NPCs were similar across the different cell lines from CTL, LR and NR groups. Immunofluorescence analysis of 28-day post-differentiation cell cultures showed similar proportions of neurons expressing prospero homeobox 1 (PROX1), a marker of DG neurons, and astrocytes expressing glial fibrillary acidic protein (GFAP) (Fig. 1a and S2). The transcriptional dynamics of neuronal progenitor and immature neurons (DCX “doublecortin”; padj < 4.1e-03), neurons (RBFOX3 “RNA binding Fox-1 homolog 3”, encoding NeuN; padj < 1.9e-07) and astrocytes (GFAP; padj < 5.17e-15) increased over time and were not significantly different between the 3 groups (Fig. 1b). In addition, the transcription dynamics of other gene markers of neuronal differentiation (TUBB3 “tubulin beta 3 class 3” or Tuj1, and MAP2 “microtubule associated protein 2”) or described specifically for DG-like neurons [60, 61] (PROX1, NEUROD1 “neuronal differentiation 1”, CALB1 “calbindin 1 ” and CALB2 “calbindin 2” or calretinin) were also similar between the 3 groups (Fig. S3).
Fig. 1.
NR neurons became transcriptionally distinct during differentiation. a Representative images of immunostainings and quantification of the fraction of neurons expressing the hippocampus DG cell marker PROX1 in the total population expressing the pan neuronal marker MAP2 at 28 days post-differentiation. PROX1 expression was detected using a lentivirus expressing eGFP under the control of PROX1 promoter. Data represent mean ± SEM for n=8–10 randomized images. b RNA-seq expression of neuronal markers (DCX and RBFOX3) and glial marker (GFAP) throughout differentiation starting from NPC (day 0) until day 42. c Differential expression results from all BD patients versus CTL days 8–42. d Sample clustering based on functional annotation. Each row is a functional term that is significantly dysregulated between CTL and BD. The rectangles represent the clustering of individual samples based on the gene expression profile for the respective functional term. e Unbiased PCA of all samples. Density plots are shown for each PC and colored by the top variable associated with the respective axis. PC1 = day, PC2 = disorder subtype. f-top Differential expression results from LR (left) or NR (right) versus CTL days 8–42. f-bottom Overlap between DEGs identified after comparing LR and CTL or NR and CTL.
We next studied genes that were differentially expressed between BD samples (LR and NR combined) and CTL samples in the neuronal state (8, 16, 28 and 42 days post-differentiation) and we identified 817 differentially expressed genes (DEGs) with an FDR threshold of 0.05 (Fig. 1c and Table S1). The top functional terms identified by DAVID bioinformatics [57, 58] that were enriched in this dataset were developmental protein (Benjamini p < 9.4e-19), extracellular matrix (Benjamini p < 4.5e-10), pattern-specification (Benjamini p < 1.0e-15), glycoprotein (Benjamini p < 5.9e-19), embryonic morphogenesis (Benjamini p < 3.8e-09), cell adhesion (Benjamini p < 5.1e-05), and regulation of transcription (Benjamini p < 1.2e-03), all of which are involved in neuronal differentiation. As expected, clustering all samples based on the significant genes within each functional category showed clustering of LR and NR samples distinctly from controls (Fig. 1d and Fig. S3), indicating that these gene sets represented pathways that were disrupted in both LR and NR bipolar patient-derived neurons.
Interestingly, unbiased principal component analysis (PCA) of all bulk samples revealed differences between NR and LR neurons. The first 2 components accounted for 30% and 10% of the variance for PC1 and PC2, respectively (Fig. 1e and Fig. S3). PC1 was most strongly associated with days post-differentiation (F-test: estimate = 1.39, p-value < 4.77e-10) as well as a slight effect of the NR status (F-test: estimate = −11.5, p-value < 2.0e-03) and no effect of LR status (F-test: estimate = 1.18, p-value = 0.73). PC2 was only associated with NR status (F-test: estimate = −20.6, p-value < 1.15e-06) and was not associated with either days post-differentiation (F-test: estimate = −0.15, p-value = 0.13) or LR status (F-test: estimate = −5.0, p-value = 0.18). The vector between PC1 and PC2 increasingly separated NR samples from CTLs and LRs after the NPC time point (day 0) as a function of developmental time (Fig. 1e). Given that NR neurons separated from both LR and CTL neurons in PC analysis, we next asked whether additional gene sets could uniquely characterize LR or NR neurons. We performed differential expression for all bulk samples by combining all neuronal samples (days 8–42) and testing as a function of disease status; either LR and CTL or NR and CTL and identified 239 and 2,884 DEGs, respectively (FDR threshold = 0.05) (Fig. 1f and Table S1). The striking 12-fold increase in DEGs in the NR samples underscored the separation observed in PC analysis, indicating that NRs represented a transcriptionally distinct subset of neurons when compared to CTLs and LRs.
To further examine the distinction based on drug responsiveness, we performed a direct comparison between LR and NR bulk neurons and identified 1,511 and 1,237 DEGs up-regulated in LR and NR, respectively (Fig. S3). We compared the DEGs identified between LR and NR with those identified between each group of patients and controls. As expected, the DEGs identified between LR and NR were highly overlapping with the DEGs identified between CTL and NR (Chi-square p < 2.2 e-302; Fig. S3).
In summary, these results show that NR neurons are distinct from CTLs and LR neurons and that these differences increase throughout differentiation.
Extensive dysregulation of Wnt/β-catenin signaling in NR neurons
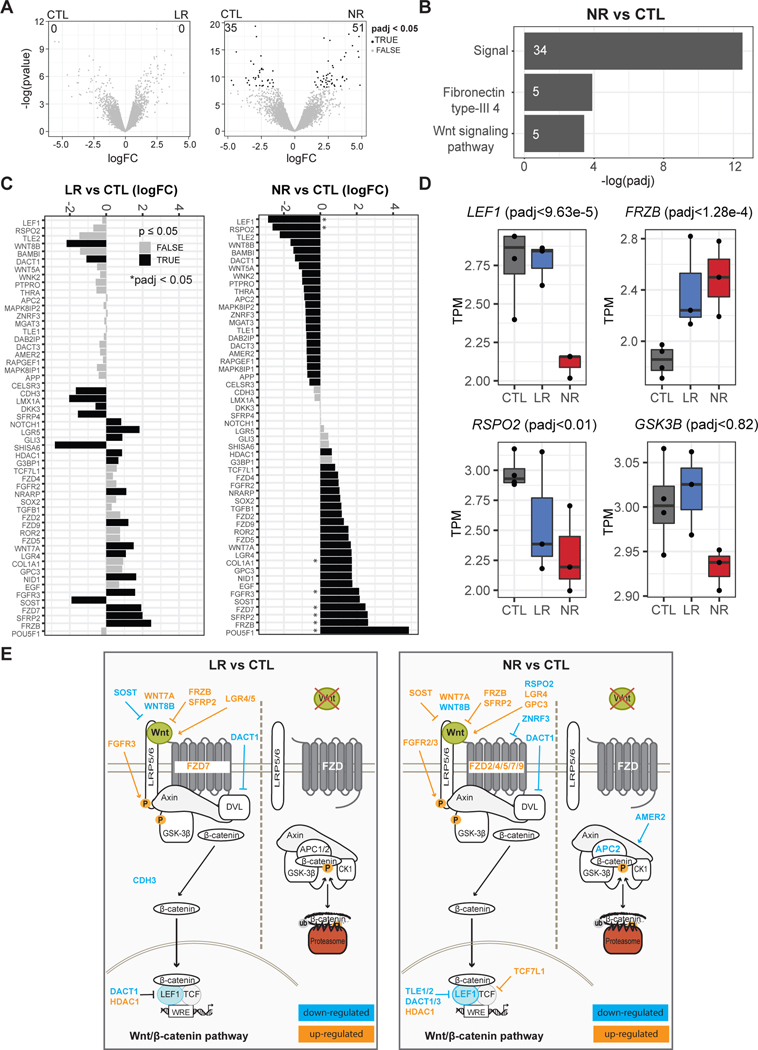
To specifically assess transcription in a homogeneous population of DG-like neurons, differentiating cultures from NPCs were sorted by flow cytometry for PROX1-positive and polysialylated neural cell adhesion molecule (PSA-NCAM)-positive cells at day 16 post-differentiation (Fig. S4). This time point was chosen because it was in the earliest time window for detection of spontaneous firing in bipolar neurons compared to controls [43]. Differential expression analysis between LR and CTL neurons returned no significant genes after multiple-testing correction (Fig. 2a). In contrast, NR neurons exhibited a unique transcriptional signature of 86 genes (padj < 0.05), primarily represented by genes involved in signaling (Fig. 2b and Table S1). The Wnt signaling genes LEF1 (padj < 9.6e-05) and FRZB (frizzled related protein) (padj < 1.3e-04) were some of the top DEGs between NR and CTL neurons. Upon further inspection of the full list of canonical (GO:0060070, 290 genes) and non-canonical Wnt pathway genes (GO:0035567, 57 genes, 13 unique to non-canonical), we identified 57 DEGs with a raw p-value ≤ 0.05 (8 DEGs padj ≤ 0.05) that were differentially regulated between either LR and CTL neurons (25 genes raw p-value, 0 genes padj) or NR and CTL neurons (45 genes raw p-value, 8 genes padj) (Fig. 2c and Table S2). In general, the genes commonly dysregulated in both LR and NR exhibited fold-change differences from CTL neurons in the same direction, with a larger change in NR than LR (Fig. 2c, d). Common genes were mostly components of the canonical or Wnt/β-catenin pathway; WNT7A/8B (wnt family member 7A/8B), FZD7 (frizzled class receptor 7), FRZB, SFRP2 (secreted frizzled related protein 2), SOST (sclerostin) and HDAC1 (histone deacetylase 1) (Fig. 2c–e; Table S2). The FGFR3 (fibroblast growth receptor 3) gene was also upregulated in LR and NR neurons. Other than the known role of FGF signaling in brain development, the mechanism of FGF receptor gene expression in neurons is unknown. It is possible, however, that FGFR3, as well as FGFR2 and EGFR induced only in NR neurons, activates Wnt/β-catenin signaling by LRP6 and β-catenin phosphorylation, as observed in RCS and HEK293 cells [62].
Fig. 2.
Dysregulation of Wnt/β-catenin signaling in NR neurons. a Differential expression results from sorted CTL and LR neurons (left) or sorted CTL and NR neurons (right). Numbers denote DEGs after padj < 0.05. b Top functional terms with Benjamini p < 0.05 from genes identified from NR vs CTL. Numbers indicate the gene count of differentially expressed genes in the category. c Barplots of logFC between LR and CTL or NR and CTL for all Wnt genes (canonical-GO:0060070, and non-canonical-GO:0035567) that were significant with a p raw < 0.05 from at least one comparison. * = padj < 0.05. d Boxplots for key Wnt-signaling related genes. e Graphical representation of the Wnt/β-catenin pathway and the genes showed in c.
A total of 27 genes were uniquely dysregulated in NR neurons compared to CTLs (Fig. 2c–e; Fig. S5; Table S2). Strong downregulation of LEF1 and RSPO2 (R-spondin 2) genes in addition to GSK3B and upregulation of repressor encoding TCF7L1 suggested inhibition of Wnt/β-catenin signaling in NR neurons (Fig. 2c–e). In support of this hypothesis, APC2 (APC regulator of Wnt signaling pathway 2), which encodes a scaffold protein of the β-catenin destruction complex, and AMER2 (APC membrane recruitment protein 2) were downregulated. Upregulation of TGFB1 (transforming growth factor beta 1) and downregulation of BAMBI (bone morphogenic protein and activin membrane-bound inhibitor) genes suggested that TGF-β signaling was increased in NR neurons compared to CTL neurons; however, this pathway might have conflicting effects in Wnt/β-catenin signaling [63, 64]. POU5F1 (POU class 5 homeobox 1) (formerly Oct4) and SOX2 (SRY-box transcription factor 2), which play a role in regulating Wnt signaling [65, 66], were also upregulated in NR neurons compared to CTL neurons (Fig. 2c and Table S2). The non-canonical pathway was less impacted but WNT5A was downregulated (Fig. S5 and Table S2). WNT5A is particularly important for adult neurogenesis in the DG [67, 68]. In summary, these results show that Wnt signaling, and in particular the Wnt/β-catenin pathway, is dysregulated in NR neurons compared to CTL neurons.
LEF1 is downregulated and Wnt/β-catenin signaling is impaired in NR neurons
Downregulation of LEF1 gene expression was specific to NR neurons. We confirmed that this was also the case in a second cohort of BD patients. As with the previous cohort, we compared bulk samples from NR and CTL neurons and identified 1,370 DEGs (Table S1). This cohort replicated the differential expression of both LEF1 (padj < 7.8e-04) and RSPO2 (padj < 2.3e-12) (Fig. S6).
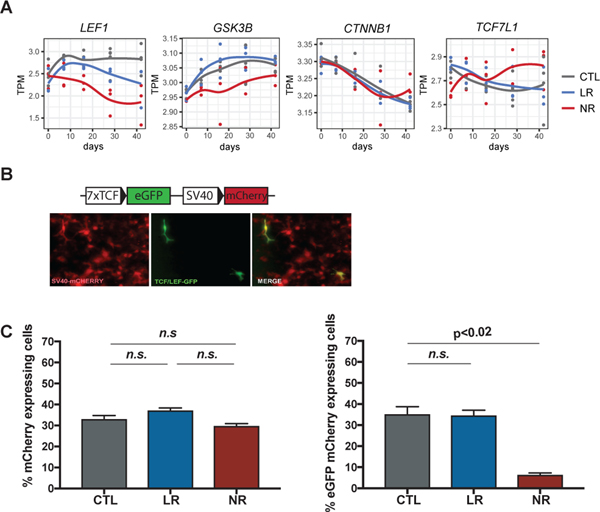
We next addressed the question of when LEF1 transcription was reduced in NR neurons. Analysis of time course expression of LEF1 showed that expression was not significantly different in NPCs but decreased sharply after 8 days post-differentiation (Fig. 3a). Closer inspection of LEF1 partners in Wnt/β-catenin signaling showed varying levels of differential expression. Expression of CTNN1B, TCF7 (also known as TCF1) and TCF7L2 (also known as TCF4) did not change, but GSK3B expression was reduced at all times although not significantly (Fig. 2d and 3a). However, expression of the canonical Wnt repressor TCF7L1 (also known as TCF3) was higher in NR compared to CTL neurons (Fig. 3a and Table S1), suggesting that as NR neurons differentiate, Wnt/β-catenin signaling decreases progressively, which may contribute to the separation over time of NR transcriptome profile from CTLs and LRs.
Fig. 3.
Downregulation of LEF1 gene and Wnt/β-catenin signaling impairment in NR neurons. a Expression of key Wnt-signaling related genes throughout differentiation starting from NPC (day 0) until day 42. b Vector carrying 7xTcf-eGFP in which eGFP fluorescence is a measure of β-catenin/TCF/LEF1-dependent transcription activity and mCherry fluorescence is a control for the transduced cells. c Barplots show percentage of mCherry-positive cells or percentage of mCherry-positive eGFP-positive cells. Data represent mean ± SEM for n=3 cell lines. Statistical analysis: Mixed linear model. Data distribution for 2 experiments for each cell line is shown in Fig. S7.
We postulated that downregulation of LEF1 transcription might be implicated in Li resistance in NR neurons. Considering that Li targets GSK-3β phosphorylation and β-catenin translocation into the nucleus, if LEF1 expression was low, formation of β-catenin/TCF/LEF1 complex and activation of transcription would be compromised (Fig. 2e and Fig. S5). Therefore, we investigated if β-catenin/TCF/LEF1 transcription activity was reduced in NR neurons. Neuronal differentiation cultures were transduced at day 8 with 7TGC lentivirus carrying 7xTcf-eGFP reporter and SV40-mCherry selection cassettes [52] and fluorescence was analyzed at day 18 using flow cytometry (Fig. 3b and Fig. S7). This reporter virus allows quantification of the β-catenin/TCF/LEF1-dependent transcription by measuring eGFP fluorescence, for which the synthesis is dependent on 7 TCF-binding sequences in the promoter, and simultaneous selection for the transduced cells by measuring mCherry fluorescence. The transduction efficiency of CTL, LR and NR neuronal cultures was over 30% and similar in all cell lines (Fig. 3c and Fig. S7). Using a mixed model [eGFP ~ disease + (1|patient)] controlling for patient, we observed that expression of eGFP was comparable between CTL and LR neurons but reduced by 4.5-fold (p-value < 0.02) in NR neurons (Fig. 3c and Fig. S7). These results confirmed that β-catenin/TCF/LEF1-dependent transcription activity was strongly reduced in NR neurons.
β-catenin-dependent signaling increases with VPA treatment in NR neurons
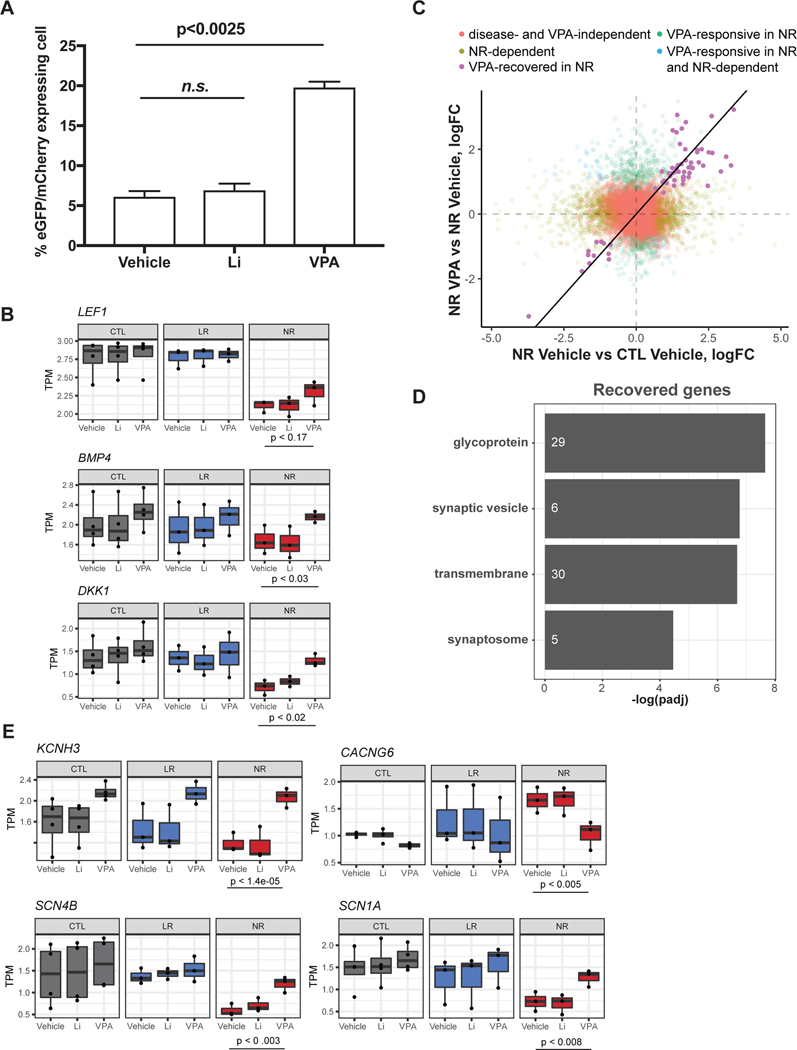
VPA is frequently used to treat BD patients who do not respond to Li. This drug also increases Wnt/β-catenin signaling but its mode of action is less well known than Li [36, 37]. Therefore, we investigated whether treatment with VPA or Li altered activity of the Wnt/β-catenin pathway using the 7xTcf-eGFP reporter in NR neurons. VPA raised activity by 2.65-fold (p-value < 2.5 e-03) the percentage of cells expressing mCherry and eGFP, indicating activation of β-catenin/TCF/LEF1-dependent transcription (Fig. 4a). Importantly, Li treatment did not change Wnt/β-catenin signaling, supporting the hypothesis that inhibition of the pathway in NR neurons is predominantly downstream of GSK-3β.
Fig. 4.
VPA increases Wnt/β-catenin signaling and induces LEF1 expression. a Barplot shows percentage of mCherry-positive cells or percentage of mCherry-positive eGFP-positive cells. Data represent mean ± SEM for n=3 cell lines. Statistical analysis: Mann-Whitney U test. Data distribution for 2 experiments for each cell line is shown in Fig. S7. b Scatterplot of logFC from the comparison between NR in vehicle and CTL in vehicle versus NR in VPA and CTL in VPA. Each dot represents a gene. Line indicates a 1:1 relationship. c Expression pattern of key Wnt-signaling genes. P-value from comparison between NR with vehicle and NR treated with VPA. d Top functionally enriched terms from genes identified to be recovered in NR after the addition of VPA. e Channel-related genes that are recovered in NR after treatment with VPA.
Next, to examine the molecular changes induced by VPA, we performed RNA sequencing on sorted PROX1-positive PSA-NCAM-positive neurons treated with VPA or vehicle from all cell lines (Table S1 and Fig. S4). The top associated functional terms identified by DAVID bioinformatics were Pleckstrin homology-type (Benjamini p < 1.9e-03), membrane fraction (Benjamini p < 2.1e-02), and chromatin regulator (Benjamini p < 1.6e-02). Importantly, DEGs in NR neurons identified between VPA and vehicle (FDR = 0.05, gene count = 855) were significantly enriched for predicted targets of LEF1 based on the TRANSFAC database (gene count = 224 overlapping out of 5,480 predicted targets; hypergeometric p < 2.13e-19; Table S1). This overlap further supported the role of increased LEF1-dependent transcriptional regulation in response to VPA.
To gain further insight into the molecular changes induced by VPA in NR, we directly compared transcription in NR neurons after treatment with Li, VPA, or vehicle to CTL. As expected, Li treatment did not significantly change gene expression compared to vehicle in NR neurons, but VPA treatment showed 68 DEGs with a p-adj threshold of 0.05 (Table S1). Enriched functional terms identified by DAVID bioinformatics were ion homeostasis (Benjamini p < 1.9e-3), regulation of transcription, DNA-dependent (Benjamini p < 8.5e-4), and glycoprotein (Benjamini p < 1.4e-5), suggesting changes in transcription and in membrane receptors and transporters (Table S1). Examination of the full list of canonical and non-canonical Wnt pathway genes revealed more DEGs in NR neurons treated with VPA than with vehicle (Fig. S8). Collectively, the observed changes in expression were in agreement with the increase in β-catenin/TCF/LEF1 activity in VPA-treated neuronal cultures of NRs (Fig. 4a). For instance, there was an upregulation of CAV1 (caveolin 1), FZD4/7/9, LGR4, and GPC3 and downregulation of APC2 and DKK2 (Fig. S8). Expression of LEF1 was only slightly increased but expression of known β-catenin/TCF/LEF1 gene targets was upregulated, suggesting increased β-catenin/TCF/LEF1 activity. For example, DKK1, SP5 transcription factor and BMP4 (bone morphogenic protein 4), which is also a ligand of TGF-β signaling and a regulator of LEF1 expression, were upregulated [69] (Fig. 4b and Table S1). Our hypothesis is that LEF1 downregulation was a major blocker of Wnt/β-catenin signaling in NR neurons. RNA-seq data confirmed increased transcriptional activity of β-catenin/TCF/LEF1 and upregulation of LEF1. In addition, alterations in other targets likely contributed to the VPA effect in the Wnt/β-catenin pathway in NR neurons.
The search for genes differentially expressed between NR and CTL neurons that were recovered in NR neurons in response to VPA treatment returned 63 genes that were significantly enriched for transmembrane proteins (Benjamini p < 1.9e-05), glycoproteins (Benjamini p < 3.6e-06), synaptic vesicles (Benjamini p < 1.0e-05), synaptosome (Benjamini p < 3.1e-04) and calcium ion binding (Benjamini p < 5.5e-04) (Fig. 4c and d; Fig. S8). This finding suggests that functions that change to normal levels are related to synaptic activity. We reported previously that DG NR neurons were hyperexcitable, and indeed, 4 DEGs encoded voltage-gated channels: SCN1A (sodium voltage-gated channel alpha subunit 1), SCN4B (sodium voltage-gated channel beta subunit 4), KCNH3 (potassium voltage-gated channel subfamily H member 3) and CACNG6 (calcium voltage-gated channel auxiliary subunit gamma 6) (Fig. 4e and Fig. S8). The DG NR neurons showed altered potassium and sodium currents that could be responsible for hyperexcitability [43, 45]. KCNH3 encodes the potassium channel subunit Kv12.2, whose activity reduces hyperexcitability in hippocampal pyramidal neurons [70]. Therefore, upregulation of SCN1A, SCN4B and KCNH3 suggests that excitability may be reduced by VPA treatment in NR neurons. In accordance with these results, gene MITF (melanocyte inducing transcription factor) was upregulated by 2.5-fold in NR neurons treated with VPA compared to CTL neurons (padj <0.0076). MITF was recently described as a regulator of neuronal activity by transcriptional activation of Kv4.3 potassium channel in olfactory bulb projection neurons [71].
In summary, we show that VPA increased Wnt/β-catenin signaling, indicating that a pharmacological increase in LEF1 expression and its gene targets is possible with the clinically used drug VPA but not Li in NR neurons.
Hyperexcitability is modulated by VPA and LEF1 expression
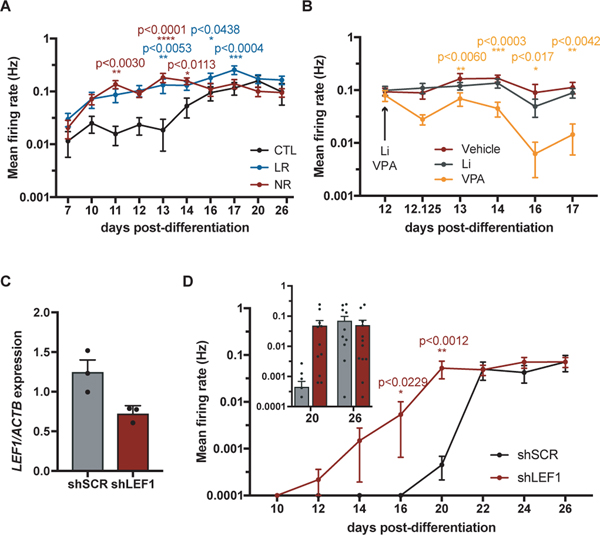
Given that RNA-seq results suggested that hyperexcitability in NR neurons may be reduced by treatment with VPA, we tested this possibility using MEA technology, in which extracellular electrodes embedded in cell culture plates allow recording of spontaneous firing over time. Control and BD cell lines were differentiated from NPCs simultaneously and recorded for a month. We first confirmed that the hyperexcitability phenotype was detected using the MEA platform. This was the case; mean firing rate was significantly higher in LR and NR neurons compared to control neurons in the first 2.5 weeks of differentiation (Fig. 5a and Fig. S9). After recording at day 12, when spontaneous firing was maximal in NR neurons, the culture media was changed for media containing Li, VPA or vehicle and recordings were performed after 3 h until day 17. Although Li had no effect on neuronal firing, VPA significantly decreased the mean firing rate following treatment (Fig. 5b and Fig. S9). This result was confirmed by electrophysiology experiments using the whole cell patch clamp technique similar to that previously described [43]. VPA treatment of NR neurons caused a significant reduction in both spontaneous and evoked firing as well as a decrease in sodium and potassium currents (Fig. S10).
Fig. 5.
Hyperexcitability is modulated by VPA and LEF1 expression. a, b, d Extracellular spontaneous firing detected in MEA platform. a Data represent mean ± SEM for n=4 CTL, n=3 LR or n=3 NR cell lines. Statistical analysis: Dunett’s multiple comparisons test LR and NR compared to CTL. Data distribution for 8 wells for each cell line from a representative experiment is shown in Fig. S9. b Data represent mean ± SEM for n=3 NR cell lines. Statistical analysis: Dunett’s multiple comparisons test Li and VPA compared to vehicle. Cell line distribution shown in Fig. S9. c Quantitative PCR showing the mRNA levels of LEF1 gene normalized to ACTB. Data represent mean ± SEM for 3 independent experiments. d Data represent mean ± SEM for n=12 wells for 1 control cell line expressing shLEF1 or shSCR (scrambled control). Statistical analysis: unpaired t-test. e Model of action of VPA and LEF1 on excitability.
Considering that VPA decreased excitability and increased Wnt/β-catenin signaling in NR neurons, we next tested if downregulation of LEF1 expression was sufficient to change excitability in a control cell line. NPCs were transduced with lentivirus-expressing shLEF1 or a scrambled control and were selected for stable populations using the puromycin antibiotic. The LEF1 mRNA level detected by quantitative PCR was reduced by approximately half in shLEF1-expressing NPC populations (Fig. 5c). Spontaneous firing was measured in the MEA platform in differentiating neuronal cultures from NPCs and a significant increase in mean firing rate was observed in neurons downregulating LEF1 (Fig. 5d). This result shows that LEF1 expression regulates hyperexcitability in the DG neuronal model.
In summary, VPA treatment or LEF1 upregulation activates Wnt/β-catenin signaling, which reduces neuronal excitability in NR neurons.
Discussion
In previous studies we found that hyperexcitability of hippocampal DG-like BD neurons is a robust phenotype that is reproducible across patient cohorts and can be used to predict Li responsiveness [43, 44]. Here, we searched for specific targets related to Li resistance in NR neurons and observed that the Wnt/β-catenin signaling pathway is profoundly affected, with a significant decrease in expression of LEF1. Downregulation of LEF1 may account for Li resistance in NR neurons since Li acts upstream by inhibiting GSK-3β and releasing β-catenin. As a proof of concept that existing compounds used as BD medication can modulate LEF1 expression and excitability, we tested the effect of VPA, which is consistently used to treat NR patients. The mode of action of VPA is not well known but it is supposed to act upon multiple targets, including downstream of GSK-3β [36, 37, 72]. We found that VPA increased expression of LEF1 and, most importantly, Wnt canonical gene targets were also upregulated. In addition, VPA reduced excitability in NR neurons. Decreasing LEF1 expression in control neurons using shLEF1 caused hyperexcitability, suggesting that the impact of VPA on excitability in NR neurons was connected to changes in LEF1 and the Wnt canonical pathway (Fig. 6). We propose that β-catenin/TCF/LEF1 transcription activity and hyperexcitability are useful phenotypes for drug development.
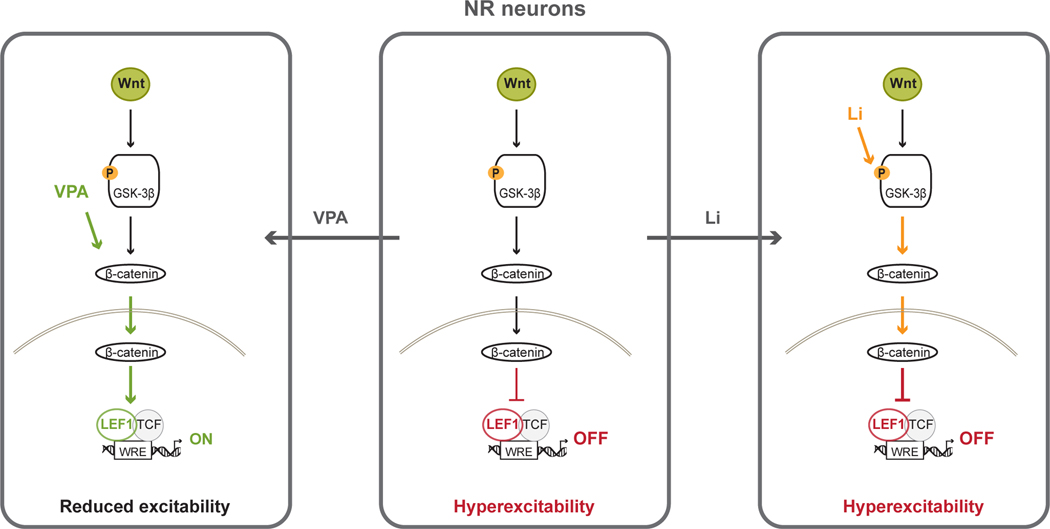
Fig. 6.
Model of action of Li and VPA on LEF1 expression and hyperexcitability in NR neurons. NR neurons show downregulation of LEF1, decreased activity of Wnt/β-catenin signaling pathway and hyperexcitability. Li acts by inhibiting GSK-3β and releasing β-catenin and does not change LEF1 expression, Wnt/β-catenin signaling or excitability. VPA upregulates LEF1 and increases transcription activity of complex β-catenin/TCF/LEF1 and reduces excitability in NR neurons.
Our study focused on iPSC-derived neurons that have the cellular markers of granule cells that are generated in the DG subgranular zone from embryonic development to adulthood [73]. Magnetic resonance imaging studies in patients with BD showed consistently smaller volumes of the hippocampus and DG subfields that are directly associated with illness progression and number of manic episodes [74–80]. Long-term Li treatment compensates for the loss of hippocampal volume [76, 77, 79]. The Wnt canonical pathway is essential for hippocampus embryonic development and adult neurogenesis [81, 82]. Studies of embryos from WNT3A, LEF1 and LRP6 null mice revealed that WNT3A is necessary for hippocampus formation and that LEF1 and LRP6 null embryos have a smaller DG and lack granule cells [82–84]. PROX1, which is a target of TCF/LEF1, is also required for maturation of granule cells during DG embryonic development and maintenance of intermediate progenitors during adult neurogenesis [61, 85]. Interestingly, Benes et al. detected LEF1 downregulation in hippocampus from post-mortem tissue [86]. Therefore, it is possible that LEF1 is downregulated in the hippocampus of BD patients, taking part in the reduction of hippocampal volume.
Increasing evidence suggests a neurodevelopmental component in the etiology of BD [87, 88]. The “immature dentate gyrus” endophenotype of BD was proposed by Hagihara et al. [89], based on the observation in several mouse models of psychiatric disorders that granule cells were arrested in development at the stage of immature neurons. This endophenotype was also described in BD patients [90]. The pattern of LEF1 expression in NR neurons compared to LR and CTL neurons follows the global transcriptome curve; it is similar in progenitors but is progressively downregulated over time during differentiation. A switch from canonical to non-canonical signaling by WNT5A occurs during adult neurogenesis in vivo and in vitro [67, 68]. However, WNT5A expression decreases sharply in the first two weeks of differentiation in NR neurons, suggesting a negative impact of canonical and non-canonical signaling in differentiation of NR neurons. Expression of genes associated with neuronal differentiation, such as MAP2, and with DG granule cell maturation, such as PROX1, is similar in NR, LR and CTL neurons, implying that the neuronal differentiation process follows a similar temporal trajectory in this model.
One hypothesis to explain hyperexcitability is the dysregulation of Wnt signaling. This hypothesis is supported by the decrease in excitability by Li and VPA treatments in LR and NR neurons, respectively, and by the induction of hyperexcitability in control neurons by downregulation of LEF1. The relationship between Wnt signaling, neuronal excitability and synapse formation in the hippocampus has been observed previously [35, 91–96]. For example, WNT5A and WNT7A stimulate excitatory synapse formation in hippocampal neurons. WNT7A-DVL1-deficient mice show defects in spine morphogenesis and mossy fiber-CA3 synaptic transmission [92]. WNT5A depolarizes hippocampal CA1 neurons and increases neuronal excitability dependent on receptor tyrosine kinase-like orphan receptor 2 (ROR2) and Wnt/calcium signaling pathway, leading to increased surface expression of N-methyl-d-aspartate receptor (NMDAR) at the synapse [91]. In our study, we found that treatment of NR neurons with VPA caused a change in transcription of the genes involved in synaptic activity and voltage-gated channels. Interestingly two of these genes, MITF and KCNH3, encode regulators of excitability [70, 71]. This study and our previous studies [43, 44] showed that hyperexcitability of BD DG-like neurons was a reproducible phenotype that correlated with clinical response of patients to medication. In future studies, it will be interesting to extend this research to understand whether cortical neurons generated from iPSCs from these patients exhibit similar phenotypes.
This study has some limitations that should be considered. Although our results are robust and reproduced in 2 independent patient cohorts, the sample size is small and represented only by males. Studies using iPSC disease modeling are limited to the number of cell lines that can be analyzed at the same time in the same conditions. However, we have shown that excitability and Wnt signaling phenotypes can be replicated in 2 different cohorts, and expanding the study to a third cohort with female patients will be necessary. We report a second target in the Wnt canonical pathway. Our results suggest that, in patients who do not respond to Li, the levels of LEF1 transcription factor also need to be increased by the treatment. Currently, this can only be accomplished by multitarget compounds. The difficulty will be to find a compound that acts on both GSK-3β/β-catenin and LEF1, nevertheless we show here that VPA, which is used as a medication for NR patients, acts on these 2 targets and increases Wnt signaling and decreases excitability.
In conclusion, our results show that iPSC-derived neurons from NR and LR bipolar patients exhibit markedly different molecular changes that are associated with hyperexcitability, and that modulation of LEF1 may be key in ameliorating disease pathology in NR patients.
Supplementary Material
Acknowledgments
We thank the Salk Institute Core Facilities: Next Generation Sequencing (NGS) Core Facility with funding from NIH-NCI CCSG: P30 014195, the Chapman Foundation and the Helmsley Charitable Trust; The Razavi Newman Integrative Genomics and Bioinformatics Core Facility with funding from NIH-NCI CCSG: P30 014195; and Flow Cytometry Core Facility with funding from NIH-NCI CCSG: P30 014195. We would also like to thank M.L. Gage for editorial comments. This work was supported by the National Cancer Institute (Grant No. P30 CA014195) and the National Institutes of Health (Grant No. R01AG05651) and by the National Cooperative Reprogrammed Cell Research Groups (NCRCRG; Grant No. U19 MH106434). The Gage laboratory is supported in part by the JPB Foundation, Annette C. Merle-Smith, and the Robert and Mary Jane Engman Foundation. S.S. acknowledges Zuckerman STEM leadership program funding.
Footnotes
Conflict of interest
No competing interests to be reported.
Data and materials availability
GEO Accession number is GSE159487.
References
- 1.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition. American Psychiatric Press, Inc.: Washington, DC, 2013. [Google Scholar]
- 2.WHO. The ICD-10 classification of mental and behavioural disorders: diagnostic criteria for research Geneva: World Health Organization,1993. [Google Scholar]
- 3.Vieta E, Berk M, Schulze TG, Carvalho AF, Suppes T, Calabrese JR, et al. Bipolar disorders. Nat Rev Dis Primers 2018; 4: 18008. [DOI] [PubMed] [Google Scholar]
- 4.Angst J and Gamma A. A new bipolar spectrum concept: a brief review. Bipolar Disord 2002; 4 Suppl 1: 11–14. [DOI] [PubMed] [Google Scholar]
- 5.Merikangas KR, Jin R, He JP, Kessler RC, Lee S, Sampson NA, et al. Prevalence and correlates of bipolar spectrum disorder in the World Mental Health Survey Initiative Arch Gen Psychiatry 2011; 68: 241–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hede V, Favre S, Aubry JM, Richard-Lepouriel H. Bipolar spectrum disorder: what evidence for pharmacological treatment? A systematic review. Psychiatry Res 2019; 282: 112627. [DOI] [PubMed] [Google Scholar]
- 7.Phillips ML and Kupfer DJ. Bipolar disorder diagnosis: challenges and future directions. Lancet 2013; 381: 1663–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fritz K, Russell AMT, Allwang C, Kuiper S, Lampe L, Malhi GS. Is a delay in the diagnosis of bipolar disorder inevitable? Bipolar Disord 2017; 19: 396–400. [DOI] [PubMed] [Google Scholar]
- 9.Alonso J, Petukhova M, Vilagut G, Chatterji S, Heeringa S, Üstün TB, et al. Days out of role due to common physical and mental conditions: results from the WHO World Mental Health Surveys. Mol Psychiatry 2011; 16: 1234–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alda M. Lithium in the treatment of bipolar disorder: pharmacology and pharmacogenetics. Mol Psychiatry 2015; 20: 661–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller JN and Black DW. Bipolar disorder and suicide: a review. Curr Psychiatry Rep 2020; 22: 6. [DOI] [PubMed] [Google Scholar]
- 12.Harrison PJ, Cipriani A, Harmer CJ, Nobre AC, Saunders K, Goodwin GM, et al. Innovative approaches to bipolar disorder and its treatment. Ann N Y Acad Sci 2016; 1366: 76–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahrens B, Müller-Oerlinghausen B, Grof P. Length of lithium treatment needed to eliminate the high mortality of affective disorders. Br J Psychiatry Suppl 1993; 21: 27–29. [PubMed] [Google Scholar]
- 14.Scott J, Hidalgo-Mazzei D, Strawbridge R, Young A, Resche-Rigon M, Etain B, et al. Prospective cohort study of early biosignatures of response to lithium in bipolar-I-disorders: overview of the H2020-funded R-LiNK initiative. Int J Bipolar Disord 2019; 7: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hui TP, Kandola A, Shen L, Lewis G, Osborn DPJ, Geddes JR, et al. A Systematic review and meta-analysis of clinical predictors of lithium response in bipolar disorder. Acta Psychiat Scand 2019; 140: 94–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunsberger JG, Chibane FL, Elkahloun AG, Henderson R, Singh R, Lawson J, et al. Novel integrative genomic tool for interrogating lithium response in bipolar disorder. Transl Psychiatry 2015; 5: e504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tondo L, Alda M, Bauer M, Bergink V, Grof P, Hajek T, et al. Clinical use of lithium salts: guide for users and prescribers. Int J Bipolar Disord 2019; 7: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tournier M, Neumann A, Pambrun E, Weil A, Chaffiol JP, Alla F, et al. Conventional mood stabilizers and/or second-generation antipsychotic drugs in bipolar disorders: a population-based comparison of risk of treatment failure. J Affect Disord 2019; 257: 412–420. [DOI] [PubMed] [Google Scholar]
- 19.Cipriani A, Barbui C, Salanti G, Rendell J, Brown R, Stockton S,t et al. Comparative efficacy and acceptability of antimanic drugs in acute mania: a multiple-treatments meta-analysis. Lancet 2011; 378: 1306–1315. [DOI] [PubMed] [Google Scholar]
- 20.Tibrewal P, Ng T, Bastiampillai T, Dhillon R, Koh DH, Kulkarni S. Why is lithium use declining? Asian J Psychiatr 2019; 43: 219–220. [DOI] [PubMed] [Google Scholar]
- 21.National Collaborating Centre for Mental Health (UK). Bipolar Disorder: The NICE Guideline on the Assessment and Management of Bipolar Disorder in Adults, Children and Young People in Primary and Secondary Care. London: The British Psychological Society and The Royal College of Psychiatrists; 2014. [PubMed] [Google Scholar]
- 22.Goodwin GM, Haddad PM, Ferrier IN, Aronson JK, Barnes T, Cipriani A, et al. Evidence-based guidelines for treating bipolar disorder: revised third edition recommendations from the British Association for Psychopharmacology. J Psychopharmacol 2016; 30: 495–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Won E and Kim YK. An oldie but goodie: lithium in the treatment of bipolar disorder through neuroprotective and neurotrophic mechanisms. Int J Mol Sci 2017; 18: 2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinstock LM, Gaudiano BA, Epstein-Lubow G, Tezanos K, Celis-deHoyos CE, Miller IW. Medication burden in bipolar disorder: a chart review of patients at psychiatric hospital admission. Psychiatry Res 2014; 216: 24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jope RS. Lithium and GSK-3: one inhibitor, two inhibitory actions, multiple outcomes. Trends Pharmacol Sci 2003; 24: 441–443. [DOI] [PubMed] [Google Scholar]
- 26.Butler-Munro C, Coddington EJ, Shirley CH, Heyward PM. Lithium modulates cortical excitability in vitro. Brain Res 2010; 1352: 50–60. [DOI] [PubMed] [Google Scholar]
- 27.Tselnicker IF, Tsemakhovich V, Rishal I, Kahanovitch U, Dessauer CW, Dascal N. Dual regulation of G proteins and the G-protein-activated K+ channels by lithium. Proc Natl Acad Sci U S A 2014; 111: 5018–5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bachmann RF, Wang Y, Yuan P, Zhou R, Li X, Alesci S, et al. Common effects of lithium and valproate on mitochondrial functions: protection against methamphetamine-induced mitochondrial damage. Int J Neuropsychopharmacol 2009; 12: 805–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roux M and Dosseto A. From direct to indirect lithium targets: a comprehensive review of omics data. Metallomics 2017; 9: 1326–1351. [DOI] [PubMed] [Google Scholar]
- 30.Maurer IC, Schippel P, Volz H-P. Lithium-induced enhancement of mitochondrial oxidative phosphorylation in human brain tissue. Bipolar Disord 2009; 11: 515–522. [DOI] [PubMed] [Google Scholar]
- 31.Bellivier F and Marie-Claire C. Molecular signatures of lithium treatment: current knowledge. Pharmacopsychiatry 2018; 51: 212–219. [DOI] [PubMed] [Google Scholar]
- 32.Kidnapillai S, Wade B, Bortolasci CC, Panizzutti B, Spolding B, Connor T, et al. Drugs used to treat bipolar disorder act via microRNAs to regulate expression of genes involved in neurite outgrowth. J Psychopharmacol 2020; 34: 370–379. [DOI] [PubMed] [Google Scholar]
- 33.Stamos JL and Weis WI. The β-catenin destruction complex. Cold Spring Harb Perspect Biol 2013; 5: a007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cadigan KM and Waterman ML. TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb Perspect Biol 2012; 4: a007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pérez-Palma E, Andrade V, Caracci MO, Bustos BI, Villaman C, Medina MA, et al. Early transcriptional changes induced by Wnt/β-catenin signaling in hippocampal neurons. Neural Plast 2016; 2016: 4672841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gupta A, Schulze TG, Nagarajan V, Akula N, Corona W, Jiang XY, et al. Interaction networks of lithium and valproate molecular targets reveal a striking enrichment of apoptosis functional clusters and neurotrophin signaling. Pharmacogenomics J 2012; 12: 328–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phiel CJ, Zhang F, Huang EY, Guenther MG, Lazar MA, Klein PS. Histone deacetylase is a direct target of valproic acid, a potent anticonvulsant, mood stabilizer, and teratogen. J Biol Chem 2001; 276: 36734–36741. [DOI] [PubMed] [Google Scholar]
- 38.Hoseth EZ, Krull F, Dieset I, Morch RH, Hope S Gardsjord ES, et al. Exploring the Wnt signaling pathway in schizophrenia and bipolar disorder. Transl Psychiatry 2018; 8: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pandey GN, Rizavi HS, Tripathi M, Ren X. Region-specific dysregulation of glycogen synthase kinase-3β and β-catenin in the postmortem brains of subjects with bipolar disorder and schizophrenia. Bipolar Disord 2015; 17: 160–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang M, Zhang L, Gage FH. Modeling neuropsychiatric disorders using human induced pluripotent stem cells. Protein Cell 2020; 11: 45–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Craddock N and Sklar P. Genetics of bipolar disorder. Lancet 2013; 381: 1654–1662. [DOI] [PubMed] [Google Scholar]
- 42.Miller ND and Kelsoe JR. Unraveling the biology of bipolar disorder using induced pluripotent stem-derived neurons. Bipolar Disord 2017; 19: 544–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stern S, Santos R, Marchetto MC, Mendes APD, Rouleau GA, Biesmans S, et al. Neurons derived from patients with bipolar disorder divide into intrinsically different sub-populations of neurons, predicting the patients’ responsiveness to lithium. Mol Psychiatry 2018; 23: 1453–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mertens J, Wang QW, Kim Y, Yu DX, Pham S, Yang B, et al. Differential responses to lithium in hyperexcitable neurons from patients with bipolar disorder. Nature 2015; 527: 95–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stern S, Sarkar A, Stern T, Mei A, Mendes APD, Stern Y, et al. Mechanisms underlying the hyperexcitability of CA3 and dentate gyrus hippocampal neurons derived from patients with bipolar disorder. Biol Psychiatry 2020; 8: 139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang JL, Shamah SM, Sun AX, Waldman ID, Haggarty SJ, Perlis RH. Label-free, live optical imaging of reprogrammed bipolar disorder patient-derived cells reveals a functional correlate of lithium responsiveness. Transl Psychiatry 2014; 4: e428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bardy C, van den Hurk M, Eames T, Marchand C, Hernandez RV, Kellogg M, et al. Neuronal medium that supports basic synaptic functions and activity of human neurons in vitro. Proc Natl Acad Sci U S A 2015; 112: E2725–E2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Endicott J and Spitzer RL. A diagnostic interview: the schedule for affective disorders and schizophrenia. Arch Gen Psychiatry 1978; 35: 837–844. [DOI] [PubMed] [Google Scholar]
- 49.Grof P, Duffy A, Cavazzoni P, Grof E, Garnham J, MacDougall M, et al. Is response to prophylactic lithium a familial trait? J Clin Psychiatry 2002; 63: 942–947. [DOI] [PubMed] [Google Scholar]
- 50.Manchia M, Adli M, Akula N, Ardau R, Aubry JM, Backlund L, et al. Assessment of response to lithium maintenance treatment in bipolar disorder: a Consortium on Lithium Genetics (ConLiGen) report. PLoS One 2013; 8: e65636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu DX, Di Giorgio FP, Yao J, Marchetto MC, Brennand K, Wright R, et al. Modeling hippocampal neurogenesis using human pluripotent stem cells. Stem Cell Reports 2014; 2: 295–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fuerer C and Nusse R. Lentiviral vectors to probe and manipulate the Wnt signaling pathway. PLoS One 2010; 5: e9370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Andrews S, FastQC: a quality control tool for high throughput sequence data. in Reference Source (2010). [Google Scholar]
- 54.Cox MP, Peterson DA, Biggs PJ. SolexaQA: at-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics 2010; 11: 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013; 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010; 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang DW, Sherman BT, Lempicki, RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 58.Huang DW, Sherman BT, Lempicki, RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009; 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res 2006; 34 (Database issue): D108–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Iwano T, Masuda A, Kiyonari H, Enomoto H, Matsuzu F. Prox1 postmitotically defines dentate gyrus cells by specifying granule cell identity over CA3 pyramidal cell fate in the hippocampus. Development 2012; 139: 3051–3062. [DOI] [PubMed] [Google Scholar]
- 61.Lavado A, Lagutin OV, Chow LM, Baker SJ, Oliver G. Prox1 is required for granule cell maturation and intermediate progenitor maintenance during brain neurogenesis. PLoS Biol 2010; 8: e1000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krejci P, Aklian A, Kaucka M, Sevcikova E, Prochazkova J, Masek JK, et al. Receptor tyrosine kinases activate canonical WNT/β-catenin signaling via MAP kinase/LRP6 pathway and direct β-catenin phosphorylation. PLoS One 2012; 7: e35826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu Y, Chen W, Wu L, Jiang L, Liang N, Tan L, et al. TGF-β1 restores hippocampal synaptic plasticity and memory in Alzheimer model via the PI3K/Akt/Wnt/β-Catenin signaling pathway. J Mol Neurosci 2019; 67: 142–149. [DOI] [PubMed] [Google Scholar]
- 64.Falk S, Wurdak H, Ittner LM, Ille F, Sumara G, Schmid MT, et al. Brain area-specific effect of TGF-β signaling on Wnt-dependent neural stem cell expansion. Cell Stem Cell 2008; 2: 472–483. [DOI] [PubMed] [Google Scholar]
- 65.Ying L, Mills JA, French DL, Gadue P. OCT4 coordinates with WNT signaling to pre-pattern chromatin at the SOX17 locus during human ES cell differentiation into definitive endoderm. Stem Cell Reports 2015; 5: 490–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Agathocleous M, Iordanova I, Willardsen MI, Xue XY, Vetter ML, Harris WA, et al. A directional Wnt/β-catenin-Sox2-proneural pathway regulates the transition from proliferation to differentiation in the Xenopus retina. Development 2009; 136: 3289–3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arredondo SB, Guerrero FG, Herrera-Soto A, Jensen-Flores J, Bustamante DB, Oñate-Ponce A, et al. Wnt5a promotes differentiation and development of adult-born neurons in the hippocampus by noncanonical Wnt signaling. Stem Cells 2020; 38: 422–436. [DOI] [PubMed] [Google Scholar]
- 68.Schafer ST, Han J, Pena M, von Bohlen Und Halbach O, Peters J, Gage FH. The Wnt adaptor protein ATP6AP2 regulates multiple stages of adult hippocampal neurogenesis. J Neurosci 2015; 35: 4983–4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Armenteros T, Andreu Z, Hortigüela R, Lie, DC, Mira H. BMP and WNT signalling cooperate through LEF1 in the neuronal specification of adult hippocampal neural stem and progenitor cells. Sci Rep 2018; 8: 9241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang X, Bertaso F, Yoo JW, Baumgärtel K, Clancy SM, Lee V, et al. Deletion of the potassium channel Kv12.2 causes hippocampal hyperexcitability and epilepsy. Nat Neurosci 2010; 13: 1056–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Atacho DAM, Reynisson H, Petursdottir AT, Eysteinsson T, Steingrimsson E, Petersen, PH. Mitf links neuronal activity and long-term homeostatic intrinsic plasticity. eNeuro 2020; 7: ENEURO.0412–0419.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chiu CT, Wang Z, Hunsberger JG, Chuang DM. Therapeutic potential of mood stabilizers lithium and valproic Acid: beyond bipolar disorder. Pharmacol Rev 2012; 65: 105–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Berg DA, Su Y, Jimenez-Cyrus D, Patel A, Huang N, Morizet D, et al. A common embryonic origin of stem cells drives developmental and adult neurogenesis. Cell 2019; 177: 654–668.e615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rimol LM, Hartberg CB, Nesvåg R, Fennema-Notestine C, Hagler DJ Jr, Pung CJ, et al. Cortical thickness and subcortical volumes in schizophrenia and bipolar disorder. Biol Psychiatry 2010; 68: 41–50. [DOI] [PubMed] [Google Scholar]
- 75.Haukvik UK, Westlye LT, Mørch-Johnsen L, Jørgensen KN, Lange EH, Dale AM. et al. In vivo hippocampal subfield volumes in schizophrenia and bipolar disorder. Biol Psychiatry 2015; 77: 581–588. [DOI] [PubMed] [Google Scholar]
- 76.Hajek T, Bauer M, Simhandl C, Rybakowski J, O’Donovan C, Pfennig A, et al. Neuroprotective effect of lithium on hippocampal volumes in bipolar disorder independent of long-term treatment response. Psychol Med 2014; 44: 507–517. [DOI] [PubMed] [Google Scholar]
- 77.Otten M and Meeter M. Hippocampal structure and function in Individuals with bipolar disorder: a systematic review J Affect Disord 2015; 174: 113–125. [DOI] [PubMed] [Google Scholar]
- 78.Han KM, Kim A, Kang W, Kang Y, Kang J, Won E, et al. Hippocampal subfield volumes in major depressive disorder and bipolar disorder. Eur Psychiatry 2019; 57: 70–77. [DOI] [PubMed] [Google Scholar]
- 79.Simonetti A, Sani G, Dacquino C, Piras F, De Rossi P, Caltagirone C, et al. Hippocampal subfield volumes in short- and long-term lithium-treated patients with bipolar I disorder. Bipolar Disord 2016; 18: 352–362. [DOI] [PubMed] [Google Scholar]
- 80.Cao B, Passos IC, Mwangi B, Amaral-Silva H, Tannous J, Wu MJ, et al. Hippocampal subfield volumes in mood disorders. Mol Psychiatry 2017; 22: 1352–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lie DC, Colamarino SA, Song HJ, Désiré L, Mira H, Consiglio A, et al. Wnt signalling regulates adult hippocampal neurogenesis. Nature 2005; 437: 1370–1375. [DOI] [PubMed] [Google Scholar]
- 82.Lee SM, Tole S, Grove E, McMahon AP. A Local Wnt-3a signal is required for development of the mammalian hippocampus. Development 2000; 127: 457–467. [DOI] [PubMed] [Google Scholar]
- 83.Galceran J, Miyashita-Lin EM, Devaney E, Rubenstein JL, Grosschedl R. Hippocampus development and generation of dentate gyrus granule cells is regulated by LEF1. Development 2000; 127: 469–482. [DOI] [PubMed] [Google Scholar]
- 84.Zhou CJ, Zhao C, Pleasure SJ. Wnt signaling mutants have decreased dentate granule cell production and radial glial scaffolding abnormalities. J Neurosci 2004; 24: 121–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Karalay Ö, Doberauer K, Vadodaria KC, Knobloch M, Berti L, Miquelajauregui A, et al. Prospero-related homeobox 1 gene (Prox1) is regulated by canonical Wnt signaling and has a stage-specific role in adult hippocampal neurogenesis. Proc Natl Acad Sci U S A 2011; 108: 5807–5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Benes FM, Lim B, Matzilevich D, Walsh JP, Subburaju S, Minns M. Regulation of the GABA cell phenotype in hippocampus of schizophrenics and bipolars. Proc Natl Acad Sci U S A 2007; 104: 10164–10169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kloiber S, Rosenblat JD, Husain MI, Ortiz A, Berk M, Quevedo J, et al. Neurodevelopmental pathways in bipolar disorder. Neurosci Biobehav Rev 2020; 112: 213–226. [DOI] [PubMed] [Google Scholar]
- 88.O’Shea KS and McInnis MG. Neurodevelopmental origins of bipolar disorder: iPSC models. Mol Cell Neurosci 2016; 73: 63–83. [DOI] [PubMed] [Google Scholar]
- 89.Hagihara H, Takao K, Walton NM, Matsumoto M, Miyakawa T. Immature dentate gyrus: an endophenotype of neuropsychiatric disorders. Neural Plast 2013; 2013: 318596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Walton NM, Zhou Y, Kogan JH, Shin R, Webster M, Gross AK, et al. Detection of an immature dentate gyrus feature in human schizophrenia/bipolar patients. Transl Psychiatry 2012; 2: e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.McQuate A, Latorre-Esteves E, Barria A. A Wnt/calcium signaling cascade regulates neuronal excitability and trafficking of NMDARs. Cell Rep 2017; 21: 60–69. [DOI] [PubMed] [Google Scholar]
- 92.Ciani L, Boyle KA, Dickins E, Sahores M, Anane D, Lopes DM, et al. Wnt7a signaling promotes dendritic spine growth and synaptic strength through Ca2+/Calmodulin-dependent protein kinase II. Proc Natl Acad Sci U S A 2011; 108: 10732–10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sahores M, Gibb A, Salinas PC. Frizzled-5, a receptor for the synaptic organizer Wnt7a, regulates activity-mediated synaptogenesis. Development 2010; 137: 2215–2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Davis EK, Zou Y, Ghosh A. Wnts acting through canonical and noncanonical signaling pathways exert opposite effects on hippocampal synapse formation. Neural Dev 2008; 3: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Parodi J, Montecinos-Oliva C, Varas R, Alfaro IE, Serrano FG, Varas-Godoy M, et al. Wnt5a inhibits K+ currents in hippocampal synapses through nitric oxide production. Mol Cell Neurosci 2015; 68: 314–322. [DOI] [PubMed] [Google Scholar]
- 96.Oliva CA and Inestrosa NC. A novel function for Wnt signaling modulating neuronal firing activity and the temporal structure of spontaneous oscillation in the entorhinal-hippocampal circuit. Exp Neurol 2015; 269: 43–55. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GEO Accession number is GSE159487.