Abstract
Objectives
The coronavirus disease 2019 (COVID-19) is a resource-intensive global pandemic. It is important for healthcare systems to identify high-risk COVID-19-positive patients who need timely health care. This study was conducted to predict the hospitalization of older adults who have tested positive for COVID-19.
Methods
We screened all patients with COVID test records from 11 Mass General Brigham hospitals to identify the study population. A total of 1495 patients with age 65 and above from the outpatient setting were included in the final cohort, among which 459 patients were hospitalized. We conducted a clinician-guided, 3-stage feature selection, and phenotyping process using iterative combinations of literature review, clinician expert opinion, and electronic healthcare record data exploration. A list of 44 features, including temporal features, was generated from this process and used for model training. Four machine learning prediction models were developed, including regularized logistic regression, support vector machine, random forest, and neural network.
Results
All 4 models achieved area under the receiver operating characteristic curve (AUC) greater than 0.80. Random forest achieved the best predictive performance (AUC = 0.83). Albumin, an index for nutritional status, was found to have the strongest association with hospitalization among COVID positive older adults.
Conclusions
In this study, we developed 4 machine learning models for predicting general hospitalization among COVID positive older adults. We identified important clinical factors associated with hospitalization and observed temporal patterns in our study cohort. Our modeling pipeline and algorithm could potentially be used to facilitate more accurate and efficient decision support for triaging COVID positive patients.
Keywords: COVID-19, machine learning, electronic health record, temporal patterns, hospitalization
INTRODUCTION
The recent outbreak of coronavirus disease 2019 (COVID-19) was declared a public health emergency of international concern by the World Health Organization on January 30, 2020.1 As of December 2021, there are more than 275 million COVID-19 cases conformed worldwide, and over 5.35 million people have died.1 In the United States, around 97 000 people are currently in the hospital due to COVID-19.2 The high volume of patients during the pandemic has caused unprecedented pressure on healthcare systems. Many hospitals in the United States are over capacity due to limited clinical resources, including beds, intensive care units (ICUs), and ventilators, which are crucial for the treatment of COVID-19 patients with severe symptoms.3,4 Older patients are most susceptible to severe illness and have a higher mortality rate.5 It is critical for clinicians and hospitals to provide appropriate clinical care to patients in the right setting; for example, home for less severe COVID-19 cases while reserving hospital beds for the more severe cases requiring acute intervention. Given potentially large infected populations during future pandemic waves, it is important to develop accurate and efficient clinical decision support for triaging COVID positive patients before they are admitted to the hospital.6 There is an urgent need for an individual-level risk prediction model that could predict people in need of hospitalization to optimize this limited clinical resource.
With the wide adoption of electronic healthcare record (EHR) systems, machine learning predictive models have great potential to leverage the large volume of data and provide tools to support medical decision-making.7 An EHR-based predictive tool can improve COVID patient care by facilitating an informed, proactive decision-making process, which can be particularly useful in managing large populations.8 During the pandemic, many studies were conducted to develop machine learning-based models to predict COVID19 disease progression.9 However, most studies focused on predicting severe adverse outcomes (such as ICU admission, mechanical ventilation, and death) among hospitalized COVID19 patients.10–12 Few studies focused on predicting hospitalization among patients with confirmed COVID19, and the patient cohort used in those studies is now relatively old, spanning March 2020 to October 2020.6,13–19 Jehi et al16 developed a robust individualized prediction model among COVID positive patients using data from 2020 and identified important risk factors of hospitalization. They also provided strategies to integrate the model into clinical workflow and to link their informatics findings with clinical practice. Given the rapid progress of the pandemic, studies with newer data sets could be useful to reflect the current clinical status and further validate and expand previous studies. In this study, we developed and validated machine learning-based prediction models, using more recent EHR data (between March 2020 and May 2021) from the Mass General Brigham (MGB) Health system, to estimate hospitalization in confirmed cases. The outcome of the models is whether older COVID-19-positive patients are hospitalized within 14 days of the COVID-19 positive test date. Different from Jehi's study population, we focused on the patient population aged 65 and above particularly since older adults with multiple comorbidities have been shown to be at an increased risk. Our current goal is to predict hospitalization among COVID-19-positive patients using existing EHR information in an outpatient population. The cause of hospitalization is very likely to be COVID-19, as a hospital policy was in place at the time of study limiting elective services (eg, nonemergency procedures and surgeries were canceled or postponed) to ensure adequate beds were available for COVID patients. With further validation and optimization, our long-term goal is to provide a testable framework for subsequent validation and refinement towards a comprehensive prediction system to facilitate timely and appropriate care for COVID-19 patients.
METHODS
Database and cohort development
We used clinical databases within the MGB Healthcare system, which has a centralized clinical data warehouse for all types of clinical information from multiple Harvard-affiliated hospitals. Available data items include patient demographics, diagnoses, procedures, medications, laboratory tests, inpatient and outpatient encounter information, and provider data. For the current study, clinical data from 11 MGB hospitals were included.
Using the MGB database, we collected all patients with COVID test records (304 113 patient visits). We further identified 11 348 patients aged 65 and above and a positive COVID test result. We then removed inpatients and 6765 patients remained. After the removal of patients with high missing values and low data quality, 1495 patients remained in the final study cohort (Figure 1).
Figure 1.
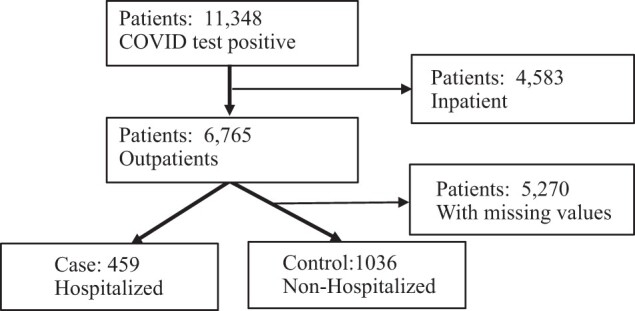
Process of data cleaning and study cohort development.
Clinician-guided phenotyping and feature selection
We identified risk factors for the severity of COVID-19 manifestation using an iterative combination of literature review, qualitative methods (interviews with clinical experts, physicians, who had experience in treating COVID-19 patients), and EHR data exploration (clinical data review and feature engineering). As a first step, we conducted a comprehensive literature review on previous COVID studies focusing on prediction models. Based on the literature, we developed a list of features used as predictors in COVID severity prediction models. Second, we validated the features using an online survey of clinicians treating COVID-19 patients from several hospitals within the MGB system. The goal of the survey was to rank predictors of poorer COVID-19 outcomes based on clinical experience. Third, we conducted a 1-hour interview with a group of physicians who were actively treating COVID patients to provide additional insight into top features based on clinical experience. The interview focused on 4 main areas (1) The clinical relevance of top features identified in the literature and validated in the survey. (2) Perceptions of factors indicative of COVID severity including early indicators. (3) Recommendations for feature collection time windows. (4) Perceived likelihood that these features are available for outpatients.
Time windows of outcome and input features
Our study design used the COVID test date as an objective proxy (index date) for the disease onset time (Figure 2). We used a 1-month time window (14 days before and 14 days after) surrounding the test date to extract both outcomes (hospitalization) and time-sensitive features (lab values), such that the model can represent the timely status (both outcome and input features) of patient conditions across the time window. The features of chronic conditions were extracted from the preceding 5 years of patient history. The definition of the model outcome is whether COVID positive patient (age 65+) will be hospitalized within 2 weeks before or 2 weeks after the COVID test date (Table 1).
Figure 2.
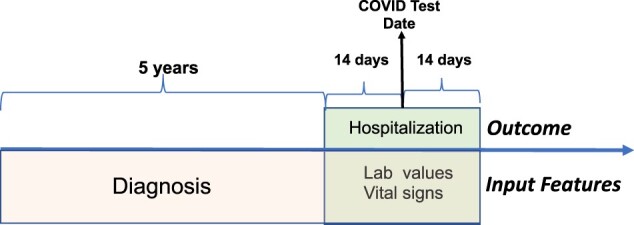
Time windows for features/outcomes.
Table 1.
Summary of input variables and outcome for model training
| Role | Definition | EHR measures |
|---|---|---|
| Outcome | Hospitalization | 14 days prior to COVID test and 14 days post-COVID test |
| Input variables | Demographic | Age, Gender |
| Vital signs/assessments | BMI, Smoking Status, SPO2, Temperature | |
| Diagnoses | Diabetes, Alzheimer Disease, Cancer, Cardiomyopathy, Cerebrovascular Disease, Chronic Kidney Disease, Chronic Respiratory Disease, Coronary Artery Disease, Cystic Fibrosis, Dementia, Dyslipidemia, Heart Failure, HIV/AIDS, Hypertensive Disease, Immunodeficiency, Liver Disease, Metastatic Solid Tumor, Sickle Cell Disease, Solid Organ Transplant | |
| Lab values | Albumin, White Blood Count, Blood Urea Nitrogen, Lymphocyte Count | |
| Temporal variable | 15 binary test date indicators for each month from March 2020 to May 2021 |
Model development and evaluation
Four hospitalization prediction models were developed using these EHR-derived features, including regularized logistic regression (LR), support vector machines (SVM), random forest (RF), and neural network (NN). These 4 models have varied model capacity in modeling complex relationships and are representative of the most popular machine learning models for various prediction tasks in clinical settings (summarized in Supplementary Table S1). To overcome the overfitting issue, we tuned models through cross-validation to select the best set of parameters and evaluated their performance on an independent test set.
Before training any of the models, we randomly split the data into 80% training and 20% test set. To ensure that our results would be generalizable, we repeated this random splitting process 30 times and reported the average model performance on the test set over the 30 splits. For a given split, we further divided the 80% training data into 5 equal-sized folds (stratified by class to ensure the minority class is present in equal proportion across all folds for hospitalization outcome). We trained the model on 4 folds and evaluated its performance on the fifth fold (validation set). We repeated this process 5 times while each time a different fold served as the validation set. Model performance was averaged across the 5 folds to determine the best hyperparameters.
To evaluate model performance, we used the area under the receiver operating characteristic curve (AUC), accuracy, sensitivity, specificity, precision, and F1 score. All metrics were calculated using the test set. The mean and standard deviation of each evaluation metric across the 30 test sets were reported.
LR, RF, and SVM were implemented using the scikit-learn library (0.21.3), and the NN was implemented using the Keras library (2.3.1) in Python (3.7.4). The hyperparameters we tuned for each model are given below. Parameters not mentioned were the same as the default set by the libraries.
LR: The regularization parameter C is tuned from to evenly on a log scale with base 10.
RF: The number of trees n_estimators is tuned from 100 to 800 with an increment of 100. The maximum number of levels in a tree max_depth is tune from 10 to 90 with an increment of 5. The minimum number of samples required to split a node min_samples_split is tuned with values in references 2,5,12,17,20. The minimum number of samples required min_samples_leaf is tuned with values in references 1,2,4,10.
SVM: The regularization parameter C is tuned from 0.1 to 100 evenly on a log scale with base 10. The kernel coefficient gamma is tuned from to 1 evenly on a log scale with base 10.
NN: The number of data points for each optimization cycle batch_size is tuned from 10 to 50 with an increment of 10. The number of times the entire training set is passed through epochs from 50 to 200 with an increment of 50. The number of nodes in the hidden layer neurons is tuned from 10 to 50 with an increment of 10. The dropout rate is 0.2. The activation function for the hidden layer is rectified linear unit. The activation function for the output layer is sigmoid. The loss is cross-entropy. The optimizer is “adam.”
This study was approved by the Institutional Review Boards (IRBs) at MGB (IRB Protocol# 2015P002472).
RESULTS
Cohort development and feature engineering/clinical phenotyping process
In the final study cohort, the case group (n = 459) had a record of hospitalization (with a hospital stay for more than 24 h after admission) during a 4-week window of the COVID-19 test date (14 days prior to and 14 days post COVID-19 test) and the control group (n = 1036) had no record of hospitalization (Figure 1). Similar age distributions were observed between case and control groups, although the case group was slightly older, more likely to be male, and slightly more likely to be a smoker (Supplementary Table S2).
We conducted a 3-stage clinician-guided feature engineering process (Table 2). First, we generated a list of 80 variables based on previous studies on COVID severity models. All these variables were used as predictors at least once. Second, we modified this list based on feedbacks from an online survey from 36 clinicians and generated a list with 45 variables. Third, through a clinician group interview and data quality assessment in the MGB database, we developed a final list with 29 variables, including demographics (age, gender), vital signs (such as SpO2 and temperature), lab tests (such as albumin), and chronic diseases (such as respiratory disease and heart failure) (Table 1 and Supplementary Table S3).
Table 2.
Summary of the 3-stage feature engineering process
| Number of features | Features selected |
|---|---|
| Stage 1. Initial feature selection based on previous studies | |
| 80 | Age, Gender (Higher in Males), BMI, High Maternal Age, Hypertension, CVD, Obesity, Diabetes Mellitus, Coronary Heart Disease, Cerebrovascular Disease, COPD, Kidney Disease, Malignancy, Respiratory Issue/Disease, Stroke, Dyslipidemia, Fever, Cough, Dyspnea, Myalgia, Shortness of Breath, Headache, Chest Pain, Sore Throat, Diarrhea, Rhinorrhea, Anosmia, Weakness, Arthralgias, Confusion, Hemoptysis, Nausea, Abdominal Pain, Vomiting, Loss of Appetite, Fatigue, O2 Saturation, Temperature, Anorexia, Malnutrition, Acute Kidney Injury, Blood Pressure, Olfactory Dysfunction, Smoking, Abnormal Liver, UP Cholesterol, UP WBC, UP Neutrophil, UP CRP, UP Ferritin, DOWN Eosinophil, DOWN Albumin, DOWN lymphocyte (CD4, CD8 T cell counts), DOWN CD3 CD19, UP IL-6, UP IL-10, UP Glucose, UP D-dimer, DOWN hemoglobin, UP BUN, UP Bilirubin, UP ALT, UP AST, Prolonged Prothrombin Time (PT), UP procalcitonin, DOWN Platelets, Change in Red Blood Cell Distribution, UP fibrinogen, UP erythrocyte sedimentation rate, UP LDH, UP Creatinine, UP Troponin, UP Serum Amyloid A (SAA), UP TNF-alpha, UP INF-gamma, UP NT-proBNP, DOWN Antithrombin, UP FDP, DOWN Thrombocytes, UP Anti-phopholipid Antibodies |
| Stage 2: Feature modification based on feedback of online clinician survey | |
| 45 | Age, Gender, BMI, Smoking Status, O2 status, Temperature, Diabetes, Alzheimer Disease, Cancer, Cardiomyopathy, Cerebrovascular Disease, Chronic Kidney Disease, Chronic Respiratory Disease, Coronary Artery Disease, Cystic Fibrosis, Dementia, Dyslipidemia, Heart Failure, HIV/AIDS, Hypertensive Disease, Immunodeficiency, Liver Disease, Metastatic Solid Tumor, Sickle Cell Disease, Solid Organ Transplant, Albumin, White Blood Count, Blood Urea Nitrogen, Lymphocyte Count, Obesity, Procalcitonin, Fibrinogen, Neutrophil Count, Creatinine, INF-gamma, C-Reactive Protein, Interleukin-6, Ferritin, Thrombocyte Count, Glucose, TNF-alpha, D-Dimer, Erythrocyte Sedimentation Rate, Lactate Dehydrogenase, Prothrombin Time |
| Stage 3: Feature finalization based on clinician interview and data quality assessment | |
| 29 | Age, Gender, BMI, Smoking Status, SPO2, Temperature, Diabetes, Alzheimer Disease, Cancer, Cardiomyopathy, Cerebrovascular Disease, Chronic Kidney Disease, Chronic Respiratory Disease, Coronary Artery Disease, Cystic Fibrosis, Dementia, Dyslipidemia, Heart Failure, HIV/AIDS, Hypertensive Disease, Immunodeficiency, Liver Disease, Metastatic Solid Tumor, Sickle Cell Disease, Solid Organ Transplant, Albumin, White Blood Count, Blood Urea Nitrogen, Lymphocyte Count |
We extracted the duration of hospital stay from the database and used 24-hour hospital stay as the definition of “hospitalization” based on clinicians' recommendation (Table 1).
Temporal pattern analysis and temporal variables
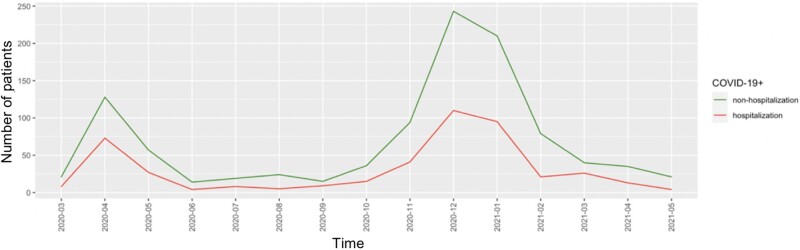
We investigated case and control patients' distributions by binning the cohort based on month (15 months in total) (Figure 3). We observed a similar distribution of case and control patients over time. There were 2 peaks over time at the months of April 2020 and December 2020, which could reflect the accumulated infection trend in our study cohort.
Figure 3.
Temporal patterns of COVID positive patients in our study cohort.
In addition, we created temporal variables to include in model training. Specifically, in accordance with the COVID test date, each month (15 months in our study duration) was encoded as a binary variable (0 for patients that not tested for that specific month and 1 for patients that tested for that specific month) for each patient from March 2020 to May 2021 (Table 1).
Model performance
We combined 29 clinical variables and 15 temporal variables (44 variables in total) as the model input. All predictive models have AUC > 0.80 (Table 3), indicating a good prediction was achieved with all 4 models, among which random forest, SVM had AUC = 0.83, while the other 3 models had AUC of 0.81 and 0.82. We did not observe a statistically significant difference in terms of AUC among the 4 models.
Table 3.
Prediction model performance
| AUC (sd) | Sensitivity (sd) | Specificity (sd) | Accuracy (sd) | Precision (sd) | F1 score (sd) | |
|---|---|---|---|---|---|---|
| Logistic regression | 0.82 (0.02) | 0.77 (0.08) | 0.76 (0.06) | 0.76 (0.03) | 0.59 (0.06) | 0.66 (0.04) |
| Random forest | 0.83 (0.02) | 0.78 (0.06) | 0.75 (0.06) | 0.76 (0.03) | 0.59 (0.05) | 0.67 (0.04) |
| Support vector machine | 0.82 (0.03) | 0.76 (0.08) | 0.76 (0.08) | 0.76 (0.04) | 0.60 (0.08) | 0.66 (0.04) |
| Neural network | 0.81 (0.02) | 0.75 (0.08) | 0.76 (0.08) | 0.75 (0.04) | 0.59 (0.07) | 0.65 (0.03) |
Most important factors associated with hospitalization
Among the statistically significant factors associated with hospitalization from the logistic regression model (Table 4), albumin, a plasma protein which is an important index for nutritional status, was found to have the most impact on the outcome. Multiple EHR variables, including vital signs, lab values, and chronic diseases, were also important for the prediction. Two temporal variables, August 2020 and May 2021 were also identified as important associated factors.
Table 4.
Top features from logistic regression
| Top 12 significant factors associated with hospitalization (based on logistic regression) |
|---|
| Albumin (Standardized coefficient = −1.01); SPO2 (−0.41); Temperature (−0.22); Cancer (−0.17); Cystic Fibrosis (0.15); Nitrogen (0.14); HIV/AIDS (0.08); Diabetes (0.06); 2020–2008 (−0.05); Metastatic Solid Tumor (−0.05); 2021–2005 (−0.05); Solid Organ Transplant (0.05) |
DISCUSSION
In this study, we developed a clinician-guided machine learning predictive algorithm to identify high-risk COVID positive patients by using EHR-defined “hospitalization” as the outcome and HER-derived variables (input). We used a multihospital study cohort and an iterative feature engineering process for model development. Our final input variable list was based on considerations of previous studies, expert opinions, and the quality of the EHR-based clinical data set. Feedback from clinicians played an important role in optimizing the input variables for model training. For example, suggestions like “fever (especially prolonged fever) may be an early indicator” and “measure for frailty/vulnerable baseline state (such as albumin) would be useful” helped us to narrow down and finalize the feature list. More importantly, during the survey and interview with clinicians, valuable knowledge of the practical advantages/disadvantages of each input variable was carefully reviewed. Since we are focusing on the outpatient setting, there is less information available and data quality is more unstable compared with inpatient settings. Selecting useful and more clinically available features was an important task during our feature engineering process and reflected in the final feature list. In general, guidance from the clinical team enabled our model to represent real clinical settings and significantly improved our models' performance and practical value.
One important goal of our study is to identify risk factors for severe COVID patients. Our results suggest that albumin could be a strong factor associated with hospitalization (protector) of hospitalization risk, which is consistent with previous studies.8,20,21 Our study further validated that albumin is an important factor associated with COVID-related hospitalization, along with SPO2 and temperature. Since albumin tests are commonly conducted among patients and can be easily obtained from EHR data set,22 it can potentially serve as a useful marker for severe COVID patients. Because all of our input variables are routinely available patient features, we expect that the algorithm can be adjusted and applied in other health care systems in the current and potential future pandemic. Moreover, we expect the proposed algorithm could help health care providers to identify those at high risk who need timely in-patient services among COVID-19-positive patients in the community to optimize the use of the limited clinical resources.
Gaining a deeper understanding of “Long COVID” generally used to describe the long-term effects of COVID infection is an increasingly important topic.23,24 “Hospitalization,” as an objective and comprehensive indication of patient status, could be a very useful phenotype for this kind of study. Also, how to utilize fast-growing large-scale clinical data sets to develop practical COVID tools is a challenge for the informatics field. We believe that the integrated model leveraging informatics/clinical components presented in this study provide a useful framework for other researchers and future studies. For example, the database with national-level information (eg, National COVID Cohort Collaborative (N3C))25 will be a good target to further test and improve the algorithm in the future.
In addition, a better understanding of the temporality of COVID is a very important topic. Different COVID strains may have played a role in pandemic progression and the mechanisms of the disease could have dynamic changes. We did not include COVID strain information in this study due to the lack of data, but this is an important topic for future research. We did however, explore the temporal patterns of the disease from 2 angles (1) we visualized the trend of hospitalization and identified 2 peak time during the study period consistent with the trend of accumulated infection events in MGB system; (2) we included time components in the prediction model and estimated their strength in predicting hospitalization. For example, 2 particular months were important associated factors, and both months corresponded with the downward duration of the COVID infection trend. This could be informative for future studies to provide a better understanding of the relationship between time and COVID-related outcomes. A good example is that in the potential future waves, 3–6 months after onset would be an appropriate time point for developing severity models.
The current study has several limitations. First, our study focused on patients age 65 and above. A large number of COVID positive patient ages 64 and below were removed during the cohort development stage. Also, we are focusing on the outpatient setting. Many features in the initial feature list (Table 1) have high missing rates for outpatients, for example, Medication records (eg, hospitalized patients had more complete records). Therefore, we did not include medications and other data types with high missingness in our model. We also removed significant number of patients with high missing values to develop the final study cohort, which allows us to have high-quality data set for model training, but also could create a certain level of bias for our study population. Second, only structured data were used for the current study, unstructured data (clinical notes) may provide additional predictive power. Third, we did not observe a statistically significant difference in terms of AUC among the 4 models. This observation that more advanced machine learning models did not perform better compared to regularized logistic regression could be due to the relatively small sample size and presence of a few strongly predictive features (eg, albumin, SpO2, and temperature), under which the power of machine learning models in handling large and complex data could not be fully leveraged. Fourth, our study cohort mainly comes from a prevaccine stage (March 2020 and May 2021), so we did not include vaccine status in our model. Due to this characteristic of the study population, this model will be more closely applicable to a nonvaccinated population. In addition, studies have shown that different COVID variants can have different responses to current vaccines. Omicron is about 2.7–3.7 times more infectious than Delta in vaccinated and boosted people.26 This will be an important topic for our future studies as more vaccination data is becoming available. Fifth, we extracted both model outcome (hospitalization) and time-sensitive input features (lab values) from the 4-week time window surrounding the COVID test date, leading to the possibility the outcome might precede the inputs. This could bias the relationship between input and outcome in our model and limit its prediction ability. Our current model design is based on known limitations of outpatient EHR data sets and the undetermined relationship between COVID test date (known) and COVID actual onset time (unknown). Several data challenges existed over the course of the pandemic including limited understanding of COVID progression and incubation period, limited testing capacity and delayed results reporting, limited medical resources, and the inconsistent workflows and EHR documentation patterns in outpatient settings. These limitations are not unique to our project but rather limitations of the availability of testing and the speed at which tests were processed at different periods during the COVID pandemic. Based on available data, we used the test date to approximate the onset time and the difference between these 2 time points can vary depending on different COVID incubation periods and testing systems. During our study period (especially in the early days of the pandemic), COVID tests were difficult to get and it took relatively longer for patients to get results after COVID testing. Therefore, some patients were sick with COVID but were not tested until they were hospitalized. Today COVID tests are widely available and patients can get the results within 24 hours. But with increasing home-based testing, this may continue to be a problem with data sets since the results of home tests are not consistently reported. Therefore, a COVID positive patient (home test) may not have an EHR-documented COVID test result until hospitalization. We used a 14-day time window surrounding the COVID test date (before and after) to capture the potential COVID incubation period. This is based on clinical expert opinion and published standards (WHO: 0–14 days and ECDC: 2–12 days).27 We used a fixed time window to standardize the model pipeline. As part of our future studies, we are working on improving the model by refining the input and outcome time windows and using newer data sets, to make the algorithm more accurate. But this also requires a deeper understanding of the basic science of COVID, which is slowly evolving. This knowledge will lead to a better definition of the COVID onset time point. Lastly, the current study is only using data from the MGB site, which could limit the generalizability and utility of the final algorithms. We are currently conducting the second stage of this project by getting comparable data from an independent site, which would allow testing of the generalizability of this algorithm with an extended data set and further validation of the key features identified here.
CONCLUSION
In the current study, we developed prediction models for general hospitalization among older COVID positive patients. Our input variables are routinely available patient features, and the model development was guided by a group of clinicians with direct experience on the front lines of COVID treatment. Our modeling pipeline and algorithm can be used to facilitate an accurate and efficient decision-making system for triaging COVID positive patients before they are admitted to the hospital.
FUNDING
This study was funded by the National Institute on Aging (NIA), 3P30AG031679-10S3, supplement Assisted Living Communities: Transforming Predictive Data into Proactive Care for COVID-19.
AUTHOR CONTRIBUTIONS
PD and RJ initialed the study and developed the study cohort. LL and MK conducted literature review and MS extracted data set. WS and LZ designed and conducted data analysis. PD, NP, MK, RJ, and SL interpreted the results. NP and AM provided important clinical opinions. PD and RJ were involved in study supervision and provided critical revision of the manuscript. All authors are participated in manuscript development and are accountable for integrity of this work.
ETHICS APPROVAL
This project was reviewed and approved by the Mass General Brigham Human Subjects Committee.
Supplementary material
Supplementary material is available at Journal of the American Medical Informatics Association online.
CONFLICT OF INTEREST STATEMENT
None declared.
Supplementary Material
Contributor Information
Wenyu Song, Department of Medicine, Brigham & Women’s Hospital, Boston, Massachusetts, USA; Department of Medicine, Harvard Medical School, Boston, Massachusetts, USA.
Linying Zhang, Department of Biomedical Informatics, Columbia University, New York, New York, USA.
Luwei Liu, Department of Medicine, Brigham & Women’s Hospital, Boston, Massachusetts, USA.
Michael Sainlaire, Department of Medicine, Brigham & Women’s Hospital, Boston, Massachusetts, USA.
Mehran Karvar, Department of Medicine, Harvard Medical School, Boston, Massachusetts, USA; Department of Surgery, Brigham & Women’s Hospital, Boston, Massachusetts, USA.
Min-Jeoung Kang, Department of Nursing, College of Nursing, The Catholic University of Korea, Seoul, South Korea.
Avery Pullman, Department of Medicine, Brigham & Women’s Hospital, Boston, Massachusetts, USA.
Stuart Lipsitz, Department of Medicine, Brigham & Women’s Hospital, Boston, Massachusetts, USA; Department of Medicine, Harvard Medical School, Boston, Massachusetts, USA.
Anthony Massaro, Department of Medicine, Brigham & Women’s Hospital, Boston, Massachusetts, USA; Department of Medicine, Harvard Medical School, Boston, Massachusetts, USA.
Namrata Patil, Department of Medicine, Harvard Medical School, Boston, Massachusetts, USA; Department of Surgery, Brigham & Women’s Hospital, Boston, Massachusetts, USA.
Ravi Jasuja, Department of Medicine, Brigham & Women’s Hospital, Boston, Massachusetts, USA; Department of Medicine, Harvard Medical School, Boston, Massachusetts, USA.
Patricia C Dykes, Department of Medicine, Brigham & Women’s Hospital, Boston, Massachusetts, USA; Department of Medicine, Harvard Medical School, Boston, Massachusetts, USA.
DATA AVAILABILITY STATEMENT
The clinical data set generated and/or analyzed during the current study are not publicly available due to patient privacy and IRB regulation.
REFERENCES
- 1. World Health Organization, Coronavirus disease (COVID-2019) situation reports. Secondary World Health Organization, Coronavirus disease (COVID-2019) situation reports. 2021. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/. Accessed May 17, 2022.
- 2. Ritchie H, Mathieu E, Rodés-Guirao L, et al. “Coronavirus Pandemic (COVID-19)”. Published online at OurWorldInData.org. Secondary “Coronavirus Pandemic (COVID-19)”. Published online at OurWorldInData.org. 2020; https://ourworldindata.org/coronavirus. Accessed May 17, 2022.
- 3. Moghadas SM, Shoukat A, Fitzpatrick MC, et al. Projecting hospital utilization during the COVID-19 outbreaks in the United States. Proc Natl Acad Sci USA 2020; 117 (16): 9122–6. [32245814][Mismatch [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Soria A, Galimberti S, Lapadula G, et al. The high volume of patients admitted during the SARS-CoV-2 pandemic has an independent harmful impact on in-hospital mortality from COVID-19. PLoS One 2021; 16 (1): e0246170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Perrotta F, Corbi G, Mazzeo G, et al. COVID-19 and the elderly: insights into pathogenesis and clinical decision-making. Aging Clin Exp Res 2020; 32 (8): 1599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wollenstein-Betech S, Cassandras CG, Paschalidis IC.. Personalized predictive models for symptomatic COVID-19 patients using basic preconditions: hospitalizations, mortality, and the need for an ICU or ventilator. Int J Med Inform 2020; 142: 104258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Obermeyer Z, Emanuel EJ.. Predicting the future - big data, machine learning, and clinical medicine. N Engl J Med 2016; 375 (13): 1216–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Abdeen Y, Kaako A, Ahmad Amin Z, et al. The prognostic effect of serum albumin level on outcomes of hospitalized COVID-19 patients. Crit Care Res Pract 2021; 2021: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Syeda HB, Syed M, Sexton KW, et al. Role of machine learning techniques to tackle the COVID-19 crisis: systematic review. JMIR Med Inform 2021; 9 (1): e23811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang JM, Liu W, Chen X, McRae MP, McDevitt JT, Fenyö D.. Predictive modeling of morbidity and mortality in patients hospitalized with COVID-19 and its clinical implications: algorithm development and interpretation. J Med Internet Res 2021; 23 (7): e29514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vaid A, Jaladanki SK, Xu J, et al. Federated learning of electronic health records improves mortality prediction in patients hospitalized with COVID-19. JMIR Med Inform2021; 9 (1): e24207. doi: 10.2196/24207. [DOI] [PMC free article] [PubMed]
- 12. Heldt FS, Vizcaychipi MP, Peacock S, et al. Early risk assessment for COVID-19 patients from emergency department data using machine learning. Sci Rep 2021; 11 (1): 4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen Z, Russo NW, Miller MM, Murphy RX, Burmeister DB.. An observational study to develop a scoring system and model to detect risk of hospital admission due to COVID-19. J Am Coll Emerg Physicians Open 2021; 2 (2): e12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shao Y, Ahmed A, Liappis AP, Faselis C, Nelson SJ, Zeng-Treitler Q.. Understanding demographic risk factors for adverse outcomes in COVID-19 patients: explanation of a deep learning model. J Healthc Inform Res 2021; 5 (2): 181–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Estiri H, Strasser ZH, Murphy SN.. Individualized prediction of COVID-19 adverse outcomes with MLHO. Sci Rep 2021; 11 (1): 5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jehi L, Ji X, Milinovich A, et al. Development and validation of a model for individualized prediction of hospitalization risk in 4,536 patients with COVID-19. PLoS One 2020; 15 (8): e0237419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Patricio A, Costa RS, Henriques R.. Predictability of COVID-19 hospitalizations, intensive care unit admissions, and respiratory assistance in Portugal: longitudinal cohort study. J Med Internet Res 2021; 23 (4): e26075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hao B, Sotudian S, Wang T, et al. Early prediction of level-of-care requirements in patients with COVID-19. Elife 2020; 9: e60519. doi: 10.7554/eLife.60519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Murtas R, Morici N, Cogliati C, et al. Algorithm for individual prediction of COVID-19-related hospitalization based on symptoms: development and implementation study. JMIR Public Health Surveill 2021; 7 (11): e29504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Violi F, Cangemi R, Romiti GF, et al. Is albumin predictor of mortality in COVID-19? Antioxid Redox Signal 2021; 35 (2): 139–42. [DOI] [PubMed] [Google Scholar]
- 21. Levine DM, Lipsitz SR, Co Z, Song W, Dykes PC, Samal L.. Derivation of a clinical risk score to predict 14-day occurrence of hypoxia, ICU admission, and death among patients with coronavirus disease 2019. J Gen Intern Med 2021; 36 (3): 730–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moman RN, Gupta N, Varacallo M.. Physiology, Albumin. StatPearls. Treasure Island (FL: ): StatPearls; 2021. [PubMed] [Google Scholar]
- 23. Raveendran AV, Jayadevan R, Sashidharan S.. Long COVID: an overview. Diabetes Metab Syndr 2021; 15 (3): 869–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lopez-Leon S, Wegman-Ostrosky T, Perelman C, et al. More than 50 long-term effects of COVID-19: a systematic review and meta-analysis. Sci Rep2021; 11 (1): 16144. doi: 10.1038/s41598-021-95565-8. [DOI] [PMC free article] [PubMed]
- 25. Haendel MA, Chute CG, Bennett TD, et al. ; N3C Consortium. The National COVID Cohort Collaborative (N3C): rationale, design, infrastructure, and deployment. J Am Med Inform Assoc 2021; 28 (3): 427–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Khan K, Karim F, Cele S, et al. Omicron infection of vaccinated individuals enhances neutralizing immunity against the Delta variant. medRxiv 2022; doi: 10.1101/2021.12.27.21268439.
- 27. Zaki N, Mohamed EA.. The estimations of the COVID-19 incubation period: a scoping reviews of the literature. J Infect Public Health 2021; 14 (5): 638–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The clinical data set generated and/or analyzed during the current study are not publicly available due to patient privacy and IRB regulation.