Abstract
Objective
To determine whether a machine learning model can detect SARS-CoV-2 infection from physiological metrics collected from wearable devices.
Materials and Methods
Health care workers from 7 hospitals were enrolled and prospectively followed in a multicenter observational study. Subjects downloaded a custom smart phone app and wore Apple Watches for the duration of the study period. Daily surveys related to symptoms and the diagnosis of Coronavirus Disease 2019 were answered in the app.
Results
We enrolled 407 participants with 49 (12%) having a positive nasal SARS-CoV-2 polymerase chain reaction test during follow-up. We examined 5 machine-learning approaches and found that gradient-boosting machines (GBM) had the most favorable validation performance. Across all testing sets, our GBM model predicted SARS-CoV-2 infection with an average area under the receiver operating characteristic (auROC) = 86.4% (confidence interval [CI] 84–89%). The model was calibrated to value sensitivity over specificity, achieving an average sensitivity of 82% (CI ±∼4%) and specificity of 77% (CI ±∼1%). The most important predictors included parameters describing the circadian heart rate variability mean (MESOR) and peak-timing (acrophase), and age.
Discussion
We show that a tree-based ML algorithm applied to physiological metrics passively collected from a wearable device can identify and predict SARS-CoV-2 infection.
Conclusion
Applying machine learning models to the passively collected physiological metrics from wearable devices may improve SARS-CoV-2 screening methods and infection tracking.
Keywords: COVID-19, wearable device, machine learning, coronavirus, apple watch
INTRODUCTION
Infection prediction traditionally relies on the development of characteristic symptomatology, prompting confirmatory diagnostic testing. However, the SARS-CoV-2 infection poses a challenge to this traditional paradigm given its variable symptomatology, prolonged incubation period, high rate of asymptomatic infection, and variable access to testing.1,2 Ongoing case surges throughout the world, prompted by the delta variant, are characterized by greater infectivity and raise the possibility that SARS-CoV-2 may become endemic. While highly effective vaccines against SARS-CoV-2 have been developed, limited vaccine supplies, low vaccination rates in some communities and the evolution of variants, have prompted ongoing infectious spread.3 Novel means to identify and predict SARS-CoV-2 infection are needed.
Wearable devices are commonly used and can measure multi-modal continuous data throughout daily life.4 Increasingly, they have been applied to applications in health and disease.5 Researchers have previously demonstrated that the addition of wearable sensor data to symptom tracking apps can increase the ability to identify Coronavirus Disease 2019 (COVID-19) patients.6 Additionally, the combination of heart rate, activity, and sleep metrics measured from wearable devices was able to identify 63% of COVID-19 cases before symptoms, further demonstrating the promise of this approach.6,7
Our group launched the Warrior Watch Study, which employed a custom smartphone app to remotely monitor health care workers (HCWs) throughout the Mount Sinai Health System.8 This app delivered surveys to the subject’s iPhones and enabled passive collection of Apple Watch data. We previously demonstrated that significant changes in heart rate variability (HRV), the small differences in time between each heartbeat that reflect autonomic nervous system (ANS) function, collected from the Apple Watch, occurred up to 7 days before a COVID-19 diagnosis.8,9
OBJECTIVE
Building on these observations, our primary aim was to determine the feasibility to train and validate machine learning approaches combining HRV measurements with resting heart rate (RHR) metrics to predict COVID-19 before diagnosis via nasal polymerase chain reaction (PCR).
MATERIALS AND METHODS
Study design
We recruited HCWs for this prospective observational study from 7 hospitals in New York City (The Mount Sinai Hospital, Morningside Hospital, Mount Sinai West, Mount Sinai Beth Israel, Mount Sinai Queens, New York Eye and Ear Infirmary, Mount Sinai Brooklyn).8 Subjects were ≥18 years, employees at one of these hospitals, had at least an iPhone series 6, and were willing to wear an Apple Watch Series 4 or higher. Underlying autoimmune or inflammatory diseases, as well as medications known to interfere with ANS function, were exclusionary. The study was approved by the Mount Sinai Hospital Institutional Review Board, and all subjects provided informed consent prior to enrollment.
Study procedures
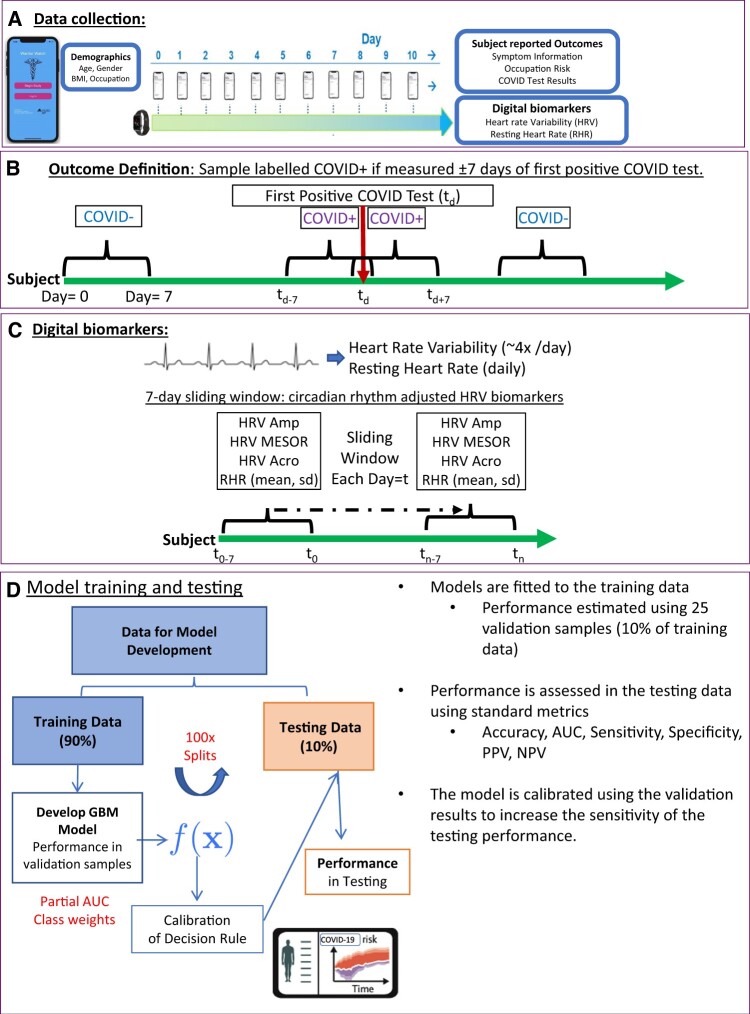
Subjects downloaded the Warrior Watch Study app, signed the electronic consent, and completed baseline demographic questionnaires. Prior COVID-19 diagnosis, medical history, and occupation classification within the hospital were collected via in-app assessments. Subjects completed daily surveys to report any COVID-19 related symptoms, symptom severity, the results for any SARS-CoV-2 nasal PCR tests, and SARS-CoV-2 antibody test results. A positive diagnosis was defined as a self-reported positive SARS-CoV-2 nasal PCR test. Each subject was asked to report the date he or she was diagnosed with a SARS-CoV-2 infection, which correlates with the date the nasal PCR took place. Subjects were asked to wear the Apple Watch for at least 8 hours per day (Figure 1A).
Figure 1.
General Strategy for training and testing statistical classifiers. Diagram illustrating the general strategy for developing the statistical classifier. (A) Subjects wore smartwatches that collect measurements of HRV and RHR. Subjects answer daily surveys to provide health outcomes including COVID test results. (B) Each day each subject is labeled as either; COVID+ if observation was made within ±7 days of the patients first positive COVID-19 test, otherwise the observation is labeled COVID−. (C) HRV measurements were too sparse to estimate HRV COSINOR parameters (MESOR, Amplitude, and Acrophase) for each day, thus, we estimated smoothed parameters using a 7-day sliding window. RHR (mean, standard deviation, minimum, and maximum) was also estimated over this window. (D) The data were split into 100 training and testing sets, models were fit to the training data and performance was estimated using 10-fold CV. 10-CV predictions were used define a decision rule that increases sensitivity, this decision rule was applied to the predictions in the testing data to get the final performance. COVID-19: Coronavirus Disease 2019; CV: cross-validation; HRV: heart rate variability; RHR: resting heart rate.
Wearable device
Subjects wore an Apple Watch Series 4 or higher, which are commercially available wearable devices that connect via Bluetooth to participants’ iPhones. The Apple Watch uses infrared and visible-light light-emitting diodes and photodiodes that act as a photoplethysmogram generating time series peaks from each heartbeat.10 There is a moving average window during which heart rate measurements are calculated while the device is worn. HRV is automatically calculated in ultra-short 60-second recording periods as the standard deviation (SD) of the inter-beat interval of normal sinus beats (SDNN), a time-domain index.9 SDNN reflects sympathetic and parasympathetic nervous system activity. The Warrior Watch Study app collects the generated SDNN and heart rate measurements at survey completion.
Data handling, model development, and statistical analysis
Our primary analysis consisted of measurements of HRV. HRV follows a circadian pattern that can be characterized by 3 parameters, namely the MESOR (M: the mean HRV during the day), amplitude (A: maximum HRV during the day), and the acrophase (Ψ: describing when the maximum occurs).8 We previously developed a mixed-effects COSINOR model to compare HRV circadian patterns at the group level and show that changes in those parameters were associated with infection.8 Given these findings, daily measurements of HRV were incorporated as potential diagnostic biomarkers for our machine-learning approach.
HRV measurements for each day were sparse and were not taken at regular intervals. Thus, daily estimates of HRV COSINOR parameters M, A, and Ψ could not be calculated. Due to this limitation, we estimated the daily HRV parameters for each subject and day (tn) using HRV data from a 7-day sliding window (tn–tn–6), thereby creating daily smoothed estimates reflecting changes in the last 7 days (Figure 1B). To aid the optimization procedures, each subject’s initial estimates are obtained using the first 2 weeks of data from each subject fitted to a mixed-effect COSINOR model with A, M, and Ψ as random effects.8 From this model, the subject-specific COVID-negative baseline A, M, and Ψ is derived and used to initialize the iterative 7-day smoothed estimates within each subject. If the number of days in the 7-day window was <3, the window was expanded to 14 days (tn–14). In rare cases, no data were available over 14-days, and parameters were imputed using the last observation carried forward imputation method. During each window, we also measured the maximum, minimum, mean, and SD of the RHR. For each day and subject, there were a total of 8 digital biomarkers used to develop our predictive models: HRV-amplitude, HRV-MESOR, HRV-acrophase, daily RHR, RHR-max, RHR-min, RHR-sd, RHR-mean, and 3 demographic variables known to impact HRV-body mass index (BMI), age, and gender.11 This smoothed approach ensures that small and transient changes in HRV profile will not dramatically effect daily HRV metrics, rather, our feature engineering approach detects large and sustained changes from the subjects COVID-negative baseline.
Data were split into independent training and testing sets, ensuring that observations with proximity in time (±4 days), for the same subject, were in the same set. The rational being those measurements taken on chronologically similar days (eg, day 6 and day 7), would have similar HRV metrics, and thus would create time-dependency bias if they appeared in different sets (eg, day 6 in training, day 7 in testing). This procedure created 100 training and testing sets, containing 90% and 10% of the data, respectively. Care was also taken to ensure that the prevalence of COVID-19 positive (COVID+) diagnoses in each set was similar to the prevalence of the full data set.
Machine learning model training and evaluation were performed using caret and pROC packages, with tuning parameters estimated using 25 validation sets, selected using the same sampling procedure as the testing data. To safeguard against biases induced by the low prevalence of COVID+ samples, we considered several sampling methods to balance the data during model training, ultimately using class weights to give more weighting to the minority class. Models were trained on each of the 100 training sets, and their performance (area under the receiver operating characteristic [auROC], partial-auROC, area under the precision recall curve [auPRC], accuracy, precision, sensitivity/recall, specificity, and balanced accuracy) was assessed on the corresponding testing set and presented as mean with 95% confidence interval (CI). The sensitivity of the diagnostic algorithm was prioritized since the application of wearable devices as a noninvasive screening modality would be to prompt a confirmatory PCR test. Our models were trained to maximize partial-auROC (sensitivity boundary of >75%), with tuning parameters estimated using the 25-validation sample. When exploring the training data, validation performance for several different machine-learning algorithms was assessed (gradient-boosting machines [GBM], elastic-net, partial least squares, support vector machines, and random forests). However, a GBM model was selected as the best performing and was used to develop our statistical classifier.
When calibrating the model, the validation predictions were used to optimize the probability threshold such that the sensitivity was above >78%. The average value of this probability threshold, over all 100 iterations, was then used to define the final decision rule where cases with a predicted probability above this threshold were considered COVID+. We used a previously described method to estimate each feature’s relative influence/importance in the model, over all 100 training sets.12 All analyses were performed by R, version 4.0.2, including the caret and pROC packages.13,14
RESULTS
Study population
Four hundred and seven HCWs were enrolled between April 29, 2020 and March 2, 2021 (Table 1). The mean age of participants at enrollment was 38 years (SD 9.8), and 34.2% were men. A positive SARS-CoV-2 nasal PCR was reported by 12.0% (49/407) of participants during follow-up (Figure 1C). The median follow-up time was 73 days (range, 3–253 days) for a total of 28 528 days of observations. A median of 4 HRV samples were collected at varying times per participant per day, and daily measures of RHR. Subjects who were diagnosed with COVID-19 were less likely to report a baseline negative SARS-CoV-2 nasal PCR test (73.5% vs 96.6%, respectively; P < .001).
Table 1.
Baseline characteristics of study participants
| Total cohort (n = 407) | Never COVID-19 positive (n = 358) | COVID-19 positive (n = 49) | P value | |
|---|---|---|---|---|
| Age, mean (SD) | 37.9 (9.82) | 37.9 (9.73) | 37.3 (10.55) | .65 |
| Body Mass Index, mean (SD) | 25.7 (5.47) | 25.7 (5.50) | 25.8 (5.31) | .91 |
| Male gender (%) | 139 (34.2) | 128 (35.8) | 11 (22.4) | .09 |
| Race (%) | .07 | |||
| Asian | 111 (27.3) | 104 (29.1) | 7 (14.3) | |
| Black | 43 (10.6) | 40 (11.2) | 3 (6.1) | |
| Hispanic/Latino | 71 (17.4) | 58 (16.2) | 13 (26.5) | |
| Other | 23 (5.7) | 21 (5.9) | 2 (4.1) | |
| White | 159 (39.1) | 135 (37.7) | 24 (49.0) | |
| Baseline negative SARS-CoV-2 Nasal PCR (%) | 382 (93.9) | 346 (96.6) | 36 (73.5) | <.001 |
| Baseline negative SARS-CoV-2 serum antibody (%) | 367 (90.2) | 325 (90.8) | 42 (85.7) | .39 |
| Baseline smoking status (%) | .61 | |||
| Never/rarely smoker | 343 (84.3) | 300 (83.8) | 43 (87.8) | |
| Current/past smoker | 64 (15.7) | 58 (16.2) | 6 (12.2) |
COVID-19, coronavirus disease 2019; PCR, polymerase chain reaction; SD, standard deviation.
Performance in training and 10 cross-validation, and model calibration
Given the low prevalence of COVID+ observations (<1% of all daily observations were COVID+), and to avoid biased performance metrics resulting from a single split, the data were split into 100 training (including ∼90% of the data) and testing (∼10%) sets, using a strategy that guarantees independence between testing and training sets. This procedure produced robust estimates of the model performance in the testing set as well as 95% CI (Figure 1D). The validation performance of several different machine-learning methods was explored, but ultimately, GBM had the most favorable performance, particularly compared to linear methods such as elastic net regularization (Table 2), suggesting a non-linear relationship between HRV and SARS-CoV-2 infection.
Table 2.
Validation performance of GBM and Elastic Net machine learning methods
| Machine learning method | Area under receiver operating characteristic | Area under partial receiver operating characteristic (sensitivity > 0.75) | Area under precision recall curve |
|---|---|---|---|
| Gradient boosting machines | 0.85 | 0.79 | 0.19 |
| Elastic net regularization | 0.60 | 0.60 | 0.03 |
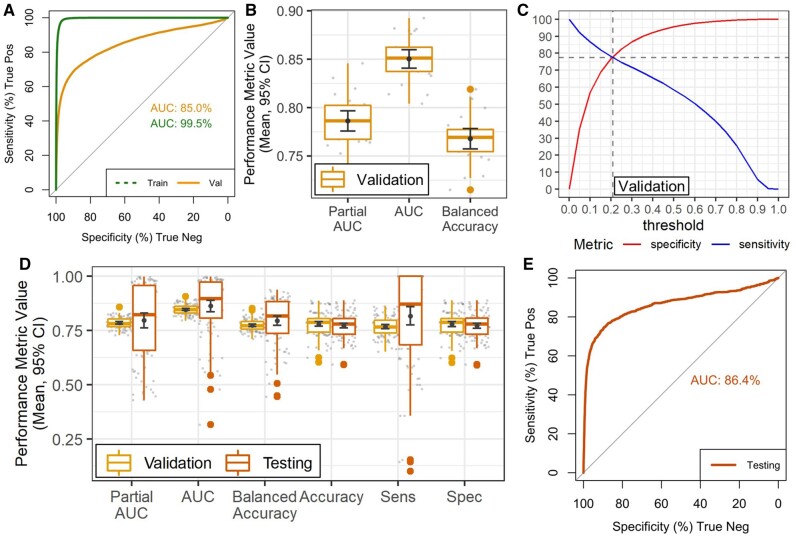
As would be expected, ROC curves calculated for GBM using all training samples show a high AUC (>99%) (Figure 2A and B), whilst performance in validation sets achieved AUC= 85%. The validation sets were selected to minimize time-correlation between training and validation, and to provide less biased performance estimates. We also calculated the auPRC, a metric that is more informative for imbalanced data, which achieved 19%, much higher than the prevalence of positive outcomes.15 It is important to note that, since this wearable device-based algorithm would be used as a screening test, we optimized the model to value sensitivity/recall metrics rather than metrics based on precision.
Figure 2.
Model performance in training and testing data. (A) ROC curve and AUC over all training and validation samples. (B) Boxplots show distribution of validation performance metrics in over all 100 training sets. (C) Plot shows specificity (red, upward sloping line) and sensitivity (blue, downward sloping line) at different response thresholds for all validation samples, a threshold ∼0.21 achieved a sensitivity of 77% and a specificity of 78%. (D) Boxplots show distribution of performance metrics over all 100 training and test sets using the 0.21 threshold decision rule. (E) ROC curve and AUC over all testing samples. AUC: area under the curve; ROC: receiver operating characteristic.
We calibrated the final decision rule to guarantee high sensitivity, as a wearable device-based algorithm would be utilized as a screening test (Table 3). This calibrated decision rule increases the true positive rate by allowing for a larger rate of false positive results. To keep the testing performance unbiased, we used the validation data to optimize the decision rule to guarantee a sensitivity >78% (Figure 2C). This optimal decision rule was 0.21 (Figure 2C) and produced an average validation Accuracy (Figure 2D) of 78% (CI ±∼1%), with 77% sensitivity and 78% specificity, thus indicating a specificity loss of 18%, for a 19% gain in sensitivity compared to the standard 0.5 decision threshold. When the calibrated diagnostic rule was applied to testing data, an AUC >85% (Figure 2D and E) was achieved. Accuracy was 77%, specificity was 77% (CI ±∼1%) (Figure 2D). The mean sensitivity was 82% (CI ±∼4%).
Table 3.
Performance summary of the gradient boosting machine learning model in validation and testing sets before and after calibration
| 10-fold cross-validation before calibration (n = 100) | 10-fold cross-validation after calibration (n = 100) | Testing sets (n = 100) | |
|---|---|---|---|
| AUC | 84.7% (CI ±∼0.1%) | 84.7% (CI ±∼0.1%) | 86.4% (CI ±∼3%) |
| AUC partial | 78.5% (CI ±∼1%) | 78.5% (CI ±∼1%) | 79.6% (CI ±∼3%) |
| Accuracy | 95.4% (CI ±∼0.1%) | 78% (CI ±∼1%) | 77.2% (CI ±∼1%) |
| Sensitivity | 57.4% (CI ±∼1%) | 76.8% (CI ±∼1%) | 81.7% (CI ±∼4%) |
| Specificity | 95.7% (CI ±∼0.1%) | 78.0% (CI ±∼1%) | 77.2% (CI ±∼1%) |
| Balanced accuracy | 76.5% (CI ±∼0.1%) | 77.4% (CI ±∼1%) | 79.5% (CI ±∼2%) |
| auPRC | 19.3% (CI ±∼1%) | 19.3% (CI ±∼0.1%) | 18.0% (CI ±∼3%) |
AUC, area under the curve; auPRC, area under the precision recall curve; CI: confidence interval.
Feature importance and interpretation
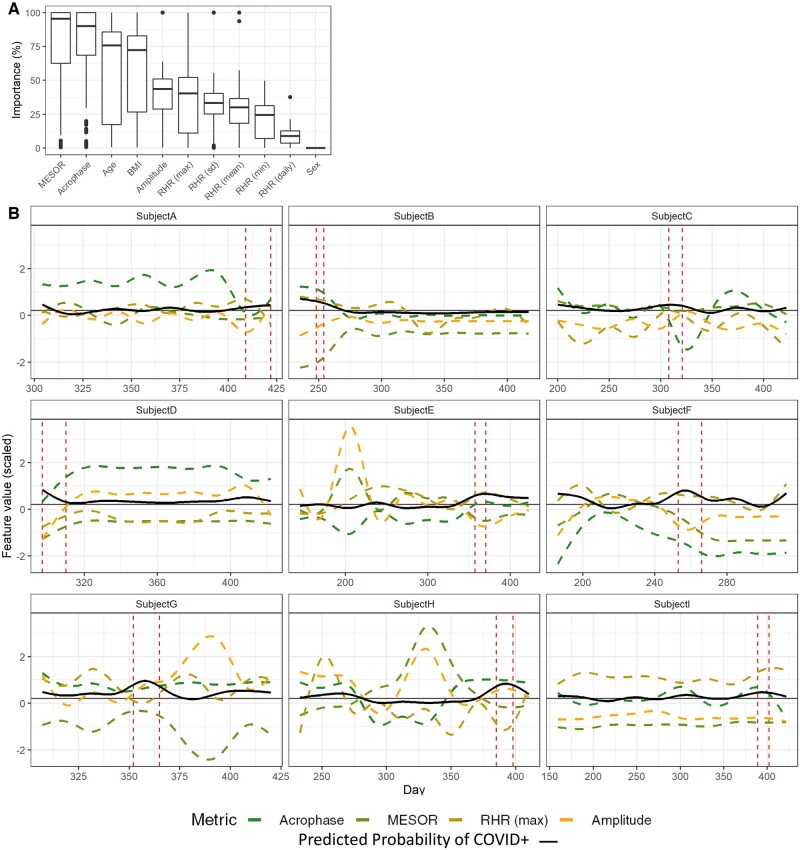
The 4 most important/influential predictors were HRV acrophase, HRV MESOR, age, and BMI (Figure 3A), with median importance >70%. RHR metrics (maximum, minimum, SD, mean) as well as HRV amplitude, were less influential (median importance 25–50%). Sex had importance equal to 0 in most models. To visualize the relationship between feature values and model prediction, we selected the 9 patients for which the model was best able to predict COVID-19 (AUC > 79% validation), and plotted the acrophase, amplitude, MESOR and max RHR, as well as the predicted probability, for each day (Figure 3). This analysis revealed a complex relationship between HRV parameters and SARS-CoV-2 infection. It was notable that, for some subjects, the predicted probability increased when HRV amplitude decreased, which is consistent with our previously published analysis.8
Figure 3.
Changes in HRV parameters and model predictions over time. (A) Box plots show the importance of each variable selected by the GBM models over all 100 training sets. (B) Line plots show daily measurements of HRV parameters (Acrophase, MESOR, and Amplitude), and Maximum resting heart rate, as well as the probability of infection (black, solid line) predicted by the model. Feature values are centered, scaled and smoothed to facilitate comparison. Daily measurements for 9 subjects are shown, predictions for each of these 9 subjects all had AUC > 65% in validation. Vertical red-dashed lines indicate the infection window for each patient, horizontal gray solid line indicates the .18 probability threshold used for the decision rule. AUC: area under the curve; GBM: gradient-boosting machines; HRV: heart rate variability.
DISCUSSION
Our results demonstrate that a machine learning approach applied to the physiological metrics measured by a wearable device identifies and predicts SARS-CoV-2 infections, in a manner suitable for a screening test. This highlights the potential utility of assessing individual changes in passively collected physiological data from wearable devices to facilitate the management of the COVID-19 pandemic.
Infections alter physiological metrics differentiating infected and uninfected states. Changes in vital signs in the setting of infection, including increased heart rate, elevated respiratory rate, and altered body temperature, have been well described.16,17 In addition to these traditional physiological metrics, ANS function, measured by HRV, is altered during illness. Several small studies have shown that changes in HRV can identify and predict infections.18,19 Building on these observations and the growing capabilities of wearable technology, wearable devices have been increasingly explored in the setting of infection. They provide a unique means to measure physiological parameters and offer an advantage over periodic assessments in the clinical setting by collecting real-time continuous measurements.20 This approach can identify trends in individual physiological outputs. On a population level, retrospective analysis of physical activity and heart rate data collected from Fitbits was shown to improve influenza-like illness predictions.21 This approach applied to an individual level was explored during the COVID-19 pandemic.
SARS-CoV-2 alters physiological metrics commonly measured by wearable devices.22 Quer et al6 collected symptom data and physiological metrics from smartwatches. They found that while RHR could not discriminate SARS-CoV-2 infections from negative cases (AUC of 0.52), when combined with sleep, activity, and symptom-based data, the AUC increased to 0.80. They demonstrated that the addition of wearable-based data significantly improved the ability of symptoms alone to discriminate between those positive or negative for COVID-19. Similarly, Mishra et al7 demonstrated that heart rate, physical activity, and sleep time collected from wearable devices could detect COVID-19. They found that 26 of the 32 COVID-19 positive subjects in their cohort had significant alterations of these metrics before diagnosis or symptom development and that 63% of cases could be detected before symptom onset.
HRV has been evaluated in SARS-CoV-2 infections. A small study of 17 subjects with SARS-CoV-2 found that rises in inflammation markers were preceded by low HRV, while another study on 14 subjects with SARS-CoV-2 in the intensive care unit demonstrated that high frequency HRV was higher and SDNN was lower in patients who later passed away.23,24 These findings were followed in a larger study of 271 subjects hospitalized with SARS-CoV-2 infections, which calculated HRV from 10 seconds of electrocardiogram recordings at admission. SDNN was predictive of survival (hazard ratio= 0.53) in subjects over 70 years of age.25 These studies demonstrate that changes in HRV are useful in the context of COVID-19. While they demonstrate a relationship in a cross-sectional fashion, we sought to leverage the longitudinal nature of HRV collection using wearable devices to expand upon these observations. Our group previously demonstrated that changes in the circadian pattern of HRV were associated with a COVID-19 diagnosis.8 We demonstrated that significant changes, particularly in the amplitude of SDNN, were observed over the 7 days before diagnosis in both symptomatic and asymptomatic individuals. Based on this observation, we built a machine learning algorithm that incorporated HRV circadian rhythm, RHR parameters, and demographic characteristics that can easily be collected from wearable device users. We trained a predictive model and then demonstrated the ability to accurately predict COVID-19 status in new data with relatively high sensitivity (82%) and specificity (77%), compared to the current gold standard of SARS-CoV-2 nasal PCR testing. This model’s high sensitivity and the minimal demographic data required lend itself to easy deployment. Our model has an advantage over prior publications evaluating the relationship between wearable-based data and a COVID-19 diagnosis, in that we trained a predictive model and then demonstrated its accuracy in predicting COVID-19 status in a test set not used in training or validation.6–8
Our findings highlight how changes in circadian features of HRV can be used to identify inflammatory events, such as SARS-CoV-2 infections. Traditionally, HRV analyses rely on assessing relative sympathetic and parasympathetic ANS tone. However, by evaluating subtle alterations in HRV architecture, nuanced changes in the ANS can be identified to perhaps enhance identification of physiological changes. In our model, alteration of HRV features were more influential predictors of infection compared to heart rate metrics. This observation warrants further exploration in other disease states as well and may identify a physiological feature that can improve predictive wearable-based algorithms in other diseases. It is important to recognize that while wearable device derived physiological metrics offer the ability to identify SARS-CoV-2 infections, these changes are likely not specific to this condition. Other infections, such as influenza, or exacerbations of chronic inflammatory conditions, can result in physiological deviations in HRV and other metrics.21,26 Chronic diseases were excluded in our study, however, recognition of this limitation, in all wearable-based algorithms, is important especially when applications to real-world data are considered. Operationalization of such algorithms therefore requires a minimum prevalence of the condition to be predicted which will improve its positive predictive value. While our study was able to control for prior infection with SARS-CoV-2 in the analysis, our prior work demonstrated that its impact on HRV circadian pattern was short lived with statistically significant alterations for 7 days from the date of COVID-19 diagnosis, mitigating the long-term impact of prior infection on machine learning models incorporating this data.
There are several limitations to our study. First, HRV was collected sporadically by the Apple Watch. We employed statistical modeling to account for this. However, a denser data set using continuous data would likely further improve our predictions. Second, the model we employed used a 7-day smoothing approach. This approach observed infection-induced changes in HRV later than if HRV was estimated using a single-day method. Thus, the approach we employed is conservative. It is important to mention that our approach relies on first establishing a COVID-negative baseline HRV profile for each patient and attempts to learn when changes from this baseline are associated with being COVID-19 positive. Thus, to mimic the clinical implementation of this approach, we used a data splitting approach that allowed samples from the same patient to be in training and test, albeit at different time points. This approach is not beyond critique since the testing and training sets are not fully independent and could lead to an overestimate of performance. Although we argue that our approach appropriately emulates the real-world application of this algorithm, we acknowledge we would need to externally validate our machine learning algorithm in another cohort to get a more accurate estimate of performance.
An additional limitation is that the Apple Watch provides HRV measurements only in the SDDN time domain. This limits assessments between other types of HRV measurements and COVID-19 outcomes. Additionally, other factors might impact HRV, which we were not able to capture and control for in the analysis. Furthermore, we were not routinely checking for SARS-CoV-2 infections and relied on subjects reporting a COVID-19 diagnosis. Therefore, infections could have occurred that are not accounted for, or the date of a COVID-19 diagnosis could vary from the true date, due to subject reporting errors. Another limitation is it is not known whether different SARS-CoV-2 variants, and their slightly different physiological effects, can be identified using the same machine learning algorithm. While our recruitment included through the period when the Delta variant was circulating in the United States, we did not recruit HCWs while the omicron variant was present.
CONCLUSION
We demonstrate that a machine learning algorithm combining circadian features of HRV with features of RHR derived from the Apple Watch achieves high sensitivity and specificity in predicting the development of COVID-19. While further validation is necessary, this non-invasive and passive modality may be helpful to monitor large numbers of people for possible infection with SARS-CoV-2 and help direct testing toward high-risk individuals.
FUNDING
Support for this study was provided by the Ehrenkranz Lab For Human Resilience, the BioMedical Engineering and Imaging Institute, The Hasso Plattner Institute for Digital Health at Mount Sinai, The Mount Sinai Clinical Intelligence Center, The Dr. Henry D. Janowitz Division of Gastroenterology and by K23DK129835 (to RPH).
AUTHOR CONTRIBUTIONS
RPH, GN, MD, and ZAF developed the study concept. RPH and LT assisted with the drafting of the manuscript. RPH, LT, MD, EG, MZ, AK, DH, AB, RP, DC, EPB, LK, GNN, MSF, and ZAF critically revised the article for important intellectual content. RPH, LT, MD, EG, MZ, AK, DH, AB, RP, DC, EPB, LK, GNN, MSF, and ZAF provided final approval of the version of the article to be published and agree to be accountable for all aspects of the work. All authors approve the authorship list. All authors had full access to all the data in the article and had final responsibility for the decision to submit for publication. RPH, ZAF, MSF, MD, and LT have verified the underlying data.
CONFLICT OF INTEREST STATEMENT
None declared.
DATA AVAILABILITY
The database that supports this study’s findings includes information about health care workers. Due to privacy issues researchers interested in gaining access to the data can contact the corresponding author.
Contributor Information
Robert P Hirten, Department of Medicine, The Dr. Henry D. Janowitz Division of Gastroenterology, Icahn School of Medicine at Mount Sinai, New York, New York, USA; The Hasso Plattner Institute for Digital Health at the Mount Sinai, New York, New York, USA.
Lewis Tomalin, Department of Population Health Science and Policy, Center for Biostatistics, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Matteo Danieletto, The Hasso Plattner Institute for Digital Health at the Mount Sinai, New York, New York, USA; Department of Genetics and Genomic Sciences, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Eddye Golden, The Hasso Plattner Institute for Digital Health at the Mount Sinai, New York, New York, USA; Department of Genetics and Genomic Sciences, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Micol Zweig, The Hasso Plattner Institute for Digital Health at the Mount Sinai, New York, New York, USA; Department of Genetics and Genomic Sciences, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Sparshdeep Kaur, The Hasso Plattner Institute for Digital Health at the Mount Sinai, New York, New York, USA.
Drew Helmus, Department of Medicine, The Dr. Henry D. Janowitz Division of Gastroenterology, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Anthony Biello, Department of Medicine, The Dr. Henry D. Janowitz Division of Gastroenterology, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Renata Pyzik, The BioMedical Engineering and Imaging Institute, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Erwin P Bottinger, The Hasso Plattner Institute for Digital Health at the Mount Sinai, New York, New York, USA.
Laurie Keefer, Department of Medicine, The Dr. Henry D. Janowitz Division of Gastroenterology, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Dennis Charney, Office of the Dean, Icahn School of Medicine at Mount Sinai, New York, New York, USA; Nash Family Department of Neuroscience, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Girish N Nadkarni, The Hasso Plattner Institute for Digital Health at the Mount Sinai, New York, New York, USA; The Department of Medicine, Icahn School of Medicine at Mount Sinai, New York, New York, USA; The Charles Bronfman Institute for Personalized Medicine, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Mayte Suarez-Farinas, Department of Population Health Science and Policy, Center for Biostatistics, Icahn School of Medicine at Mount Sinai, New York, New York, USA; Department of Genetics and Genomic Sciences, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Zahi A Fayad, The BioMedical Engineering and Imaging Institute, Icahn School of Medicine at Mount Sinai, New York, New York, USA; Department of Diagnostic, Molecular and Interventional Radiology, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
REFERENCES
- 1. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 02 2020; 395 (10223): 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. He X, Lau EHY, Wu P, et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med 2020; 26 (5): 672–5. [DOI] [PubMed] [Google Scholar]
- 3. Anand NM, Liya DH, Pradhan AK, et al. A comprehensive SARS-CoV-2 genomic analysis identifies potential targets for drug repurposing. PLoS One 2021; 16 (3): e0248553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hirten RP, Stanley S, Danieletto M, et al. Wearable devices are well accepted by patients in the study and management of inflammatory Bowel disease: a survey study. Dig Dis Sci 2020; 66 (6): 1836–44. [DOI] [PubMed] [Google Scholar]
- 5. Li X, Dunn J, Salins D, et al. Digital health: tracking physiomes and activity using wearable biosensors reveals useful health-related information. PLoS Biol 2017; 15 (1): e2001402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Quer G, Radin JM, Gadaleta M, et al. Wearable sensor data and self-reported symptoms for COVID-19 detection. Nat Med 2020; 27 (1): 73–7. [DOI] [PubMed] [Google Scholar]
- 7. Mishra T, Wang M, Metwally AA, et al. Pre-symptomatic detection of COVID-19 from smartwatch data. Nat Biomed Eng 2020; 4 (12): 1208–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hirten RP, Danieletto M, Tomalin L, et al. Use of physiological data from a wearable device to identify SARS-CoV-2 infection and symptoms and predict COVID-19 diagnosis: observational study. J Med Internet Res 2021; 23 (2): e26107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shaffer F, Ginsberg JP.. An overview of heart rate variability metrics and norms. Front Public Health 2017; 5: 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Monitor your heart rate with Apple Watch. https://support.apple.com/en-us/HT204666. Accessed October 22, 2020.
- 11. Heart rate variability. Standards of measurement, physiological interpretation, and clinical use. Task Force of the European Society of Cardiology and the North American Society of Pacing and Electrophysiology. Eur Heart J. Mar 1996; 17 (3): 354–81. [PubMed] [Google Scholar]
- 12. Friedman JH. Greedy function approximation: a gradient boosting machine. Ann Statist 2001; 29 (5): 1189–232. [Google Scholar]
- 13. Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 2011; 12: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kuhn M. Building predictive models in R using the caret package. J Stat Soft 2008; 28 (5): 1– 26. [Google Scholar]
- 15. Saito T, Rehmsmeier M.. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS One 2015; 10 (3): e0118432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Karjalainen J, Viitasalo M.. Fever and cardiac rhythm. Arch Intern Med 1986; 146 (6): 1169–71. [PubMed] [Google Scholar]
- 17. Nicolò A, Massaroni C, Schena E, Sacchetti M.. The importance of respiratory rate monitoring: from healthcare to sport and exercise. Sensors (Basel) 2020; 20 (21): 6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kovatchev BP, Farhy LS, Cao H, Griffin MP, Lake DE, Moorman JR.. Sample asymmetry analysis of heart rate characteristics with application to neonatal sepsis and systemic inflammatory response syndrome. Pediatr Res 2003; 54 (6): 892–8. [DOI] [PubMed] [Google Scholar]
- 19. Ahmad S, Ramsay T, Huebsch L, et al. Continuous multi-parameter heart rate variability analysis heralds onset of sepsis in adults. PLoS One 2009; 4 (8): e6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ates HC, Yetisen AK, Güder F, Dincer C.. Wearable devices for the detection of COVID-19. Nat Electron 2021; 4 (1): 13–4. [Google Scholar]
- 21. Radin J, Wineinger NE, Topol EJ, Steinhubl SR.. Harnessing wearable device data to improve state-level real-time surveillance of influenza-like illness in the USA: a population-based study. Lancet Digital Health 2020; 2 (2): e85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Richardson S, Hirsch JS, Narasimhan M, et al. ; the Northwell COVID-19 Research Consortium. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City Area. JAMA 2020; 323 (20): 2052–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hasty F, García G, Dávila CH, Wittels SH, Hendricks S, Chong S.. Heart rate variability as a possible predictive marker for acute inflammatory response in COVID-19 patients. Mil Med 2020; 186: e34–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Aragón-Benedí C, Oliver-Forniés P, Galluccio F, et al. Is the heart rate variability monitoring using the analgesia nociception index a predictor of illness severity and mortality in critically ill patients with COVID-19? A pilot study. PLoS One 2021; 16 (3): e0249128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mol MBA, Strous MTA, van Osch FHM, et al. Heart-rate-variability (HRV), predicts outcomes in COVID-19. PLoS One 2021; 16 (10): e0258841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hirten RP, Danieletto M, Scheel R, et al. Longitudinal autonomic nervous system measures correlate with stress and ulcerative colitis disease activity and predict flare. Inflamm Bowel Dis 2021; 27 (10): 1576–84. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The database that supports this study’s findings includes information about health care workers. Due to privacy issues researchers interested in gaining access to the data can contact the corresponding author.