Abstract
We analyzed the duration of infectivity of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron variant by viral culture of respiratory samples collected daily from isolated patients with SARS-CoV-2 infection. The culture positivity rate of the Omicron variant was higher than that of the Delta variant within 8 days after symptom onset.
Keywords: viral shedding, SARS-CoV-2, Omicron variant, viral load, transmissibility
On December 2, 2021, the first outbreak of the Omicron (B.1.1.529) variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection was identified in South Korea among travelers returning from Nigeria. Eleven cases of Omicron variant infection were identified in the first cluster (Supplementary Figure 1). This report aims to describe the viral load kinetics by in vitro demonstration of the infectiousness of cell lines derived from serial respiratory samples obtained from patients hospitalized with SARS-CoV-2 Omicron variant infection.
All patients with SARS-CoV-2 infection, confirmed by positive real-time reverse transcriptase polymerase chain reaction testing, were included. Infection with the Omicron variant was confirmed by sequencing at the Korea Disease Control and Prevention Agency in December 2021. Patients with confirmed SARS-CoV-2 Omicron variant infection were promptly transferred to Incheon Medical Center (IMC), which was equipped with a negative-pressure isolation room. Early treatment was facilitated by rapid epidemiological investigation and early testing. As this was the first cluster of SARS-CoV-2 Omicron variant infection cases in South Korea, all patients were isolated in IMC and treated throughout the isolation period. Nasopharyngeal and oropharyngeal swab samples were collected from the patients daily until their discharge and transported in viral transport media (VTM). Eleven patients with coronavirus disease 2019 (COVID-19) were enrolled in this study. The median age of the patients was 38 years; 5 patients (45%) were male, and 6 (55%) were female. No patients were immunocompromised, and 2 patients (18%) received a second dose of the COVID-19 vaccine in October 2021. All patients had 1 or more COVID-19-related symptoms at diagnosis. Five patients (45%) had pneumonia, which was determined by the presence of signs of involvement of lung parenchyma on chest x-ray or high-resolution computed tomography. None of the patients received supplemental oxygen therapy, remdesivir, or anti-inflammatory drugs (steroids, interleukin-6 inhibitors, or Janus kinase inhibitors). Two patients (18%) received monoclonal antibody therapy. All patients had mild to moderate COVID-19 (Supplementary Table 1).
Viral Shedding Periods of the SARS-CoV-2 Omicron Variant
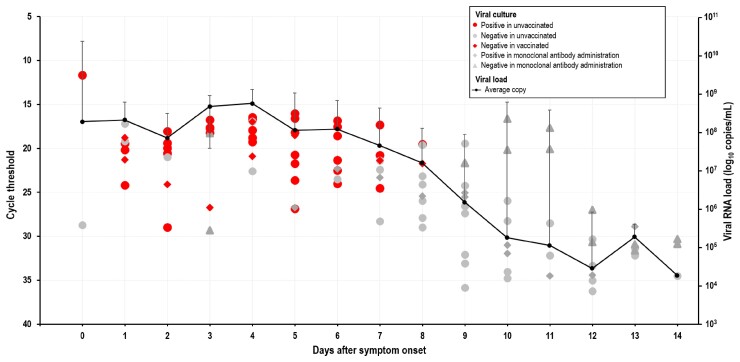
A total of 110 specimens from 11 patients with confirmed SARS-CoV-2 Omicron variant infection were cultured using the Vero E6 cell line within 14 days after symptom onset, and the viral culture positivity rate was evaluated (Figure 1). The Omicron variant was isolated in 48 specimens (43.6%) within 8 days after symptom onset in unvaccinated patients, while none were isolated from the vaccinated patients. Of 2 unvaccinated patients who received monoclonal antibody therapy, 1 had a negative culture, while the other had a positive culture within 8 days after symptom onset. The E gene cycle threshold (Ct) value for samples that produced culturable virus ranged from 11.66 to 28.96 (average, 19.86; n = 48), whereas the Ct values for samples that did not produce culturable virus ranged from 16.62 to 36.22 (average, 26.99; n = 62). The maximum Ct value for culturable virus was 28.96. The viral culture positivity rate of the SARS-CoV-2 Omicron variant ranged from 22% to 100% in unvaccinated patients within 0–8 days after symptom onset. In a comparison of the viral culture positivity rates of the Delta and Omicron variants, the Omicron variant had a shorter duration than the Delta variant (maximum, 8 days and 10 days, respectively) and a higher positivity rate (maximum, 100% and 74%, respectively) [1]. The average Ct within 14 days after symptom onset was converted to viral load to compare the quantitative differences based on viral load. Based on the results, patients with SARS-CoV-2 Omicron variant infection had a viral load ranging from 1.86 × 104 copies/mL to 5.69 × 108 copies/mL. The viral load in viral culture–negative samples was lower than 1.52 × 106 copies/mL.
Figure 1.
Viral load, culture positivity rate, and duration of symptoms as indicators of infectious virus shedding. Viral RNA loads (log10 RNA copies/mL) in the respiratory samples vs duration of symptoms (days). Data are the cycle threshold values for the E gene of SARS-CoV-2 tested using a real-time reverse transcription polymerase chain reaction assay. Red circles: virus culture positive in unvaccinated patients; gray circles: virus culture negative in unvaccinated patients; red rhombus: virus culture negative in fully vaccinated patients; gray rhombus: virus culture positive in patients who received monoclonal antibody treatment; gray triangles: virus culture negative in patients who received monoclonal antibody treatment. Abbreviation: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Clinical Course and Culture Positivity
Patient 4 had a fever for 11 days after symptom onset. However, he had a positive culture for 5 days after symptom onset. He had respiratory symptoms but did not experience a reduction in the saturation of percutaneous oxygen saturation. High-resolution computed tomography, performed 5 and 11 days after symptom onset, showed increasing pneumonic infiltration (Supplementary Figure 2). Patient 6 had a fever for 4 days after symptom onset, and the virus culture was positive until 8 days after symptom onset. Patient 8 had a fever for 3 days after symptom onset, and the virus culture was positive until 6 days after symptom onset.
DISCUSSION
This study analyzed the duration of the infectious stage of individuals infected with the SARS-CoV-2 Omicron variant using viral culture of respiratory samples collected daily from isolated patients with confirmed Omicron variant infection. The viral cultures were negative for all samples taken 9 days after symptom onset. This study suggests that the Omicron variant may have a shorter infectious stage than the Delta variant. However, the culture-positive rate of the Omicron variant was higher than that of the Delta variant. Using culture positivity as a surrogate measure for infectivity [2], the results of our study help to explain the recent worldwide surge of COVID-19 cases caused by the Omicron variant.
The receptor-binding domain (RBD) of the SARS-CoV-2 spike (S) protein interacts with the host cellular receptor angiotensin-converting enzyme 2 (ACE 2) [3, 4]. An RBD mutation directly affects the interaction between the virus and ACE 2. SARS-CoV-2, harboring the D614G mutation, where glutamic acid (D) at 614 is substituted with glycine (G) on the S protein, has a higher transmissibility [5]. Furthermore, a mutation in the furin cleavage domain at 681–687 prevents proper S1/S2 unit formation, thereby altering viral infectivity and pathogenicity [6]. The Omicron variant harbors a mutation at 614G in the S protein RBD, as well as G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, and 681H mutations at the furin cleavage site of the S protein [7, 8]. The increased transmissibility of the Omicron variant has been attributed to the 614G and S protein RBD multiple mutations, while its altered viral infection and pathogenicity have been attributed to the 681H mutation [8]. The results of our study infer the potential for increased transmissibility and infectivity of the Omicron variant. The Omicron variant exhibited a higher culture positivity rate than the Delta variant within 8 days after symptom onset. However, the viral load of the Omicron variant was slightly lower than that of the Delta variant. In addition, the RBD mutations with K417N, N501Y, and Q498R of the Omicron variant S protein increased with host cell ACE 2 interaction, resulting in increased transmissibility [9, 10].
Worldwide, many guidelines on release from isolation take the resolution of fever into account. However, our study results show that the duration of fever is not well correlated with the time to viral culture conversion. Improvement of symptoms, including fever, is an important indicator of a decrease in infectivity, but there is a lack of evidence that simply emphasizing improvement in fever provides a scientific basis for release from isolation, and further studies are needed to determine whether improvement in fever has a significant association with decreased infectivity.
Our study has some limitations. First, since the index cases were travelers and given the strict hospital-based isolation procedures for this cohort, the validity of their symptom onset dates and their willingness to admit, for example, that they traveled internationally while symptomatic must be taken into consideration. Second, it is important to note that the sample size of this study was small, and all the patients had mild to moderate illness. Thus, these results should be verified in a larger study involving different patient groups.
The Omicron variant is thought to spread more easily than the original SARS-CoV-2 and Delta variants [11]. The findings of this study support the high transmissibility of the Omicron variant. Improvement of symptoms, including fever, is an important indicator of a decrease in infectivity, but our study results show that duration of fever might not correlate well with time to viral culture conversion.
Supplementary Material
Acknowledgments
Financial support. Not applicable.
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Patient consent. This study was conducted in accordance with the guidelines of the Declaration of Helsinki. This study, which involved human participants, was reviewed and approved by the Institutional Review Board of the Korea Disease Control and Prevention Agency (number: 2020-03-01-P-A). The study was designated a service to public health during the outbreak; therefore, the board waived the requirement for written informed consent from the participants’ legal guardian/next of kin to participate in this study, in accordance with the national legislation and the institutional requirements.
Contributor Information
Young R Jang, Division of Infectious Diseases, Department of Internal Medicine, Incheon Medical Center, Incheon, South Korea.
Jeong-Min Kim, Division of Emerging Infectious Diseases, Bureau of Infectious Disease Diagnosis Control, Korea Disease Control and Prevention Agency, Cheongju, South Korea.
Jee E Rhee, Division of Emerging Infectious Diseases, Bureau of Infectious Disease Diagnosis Control, Korea Disease Control and Prevention Agency, Cheongju, South Korea.
Dongju Kim, Division of Emerging Infectious Diseases, Bureau of Infectious Disease Diagnosis Control, Korea Disease Control and Prevention Agency, Cheongju, South Korea.
Nam-Joo Lee, Division of Emerging Infectious Diseases, Bureau of Infectious Disease Diagnosis Control, Korea Disease Control and Prevention Agency, Cheongju, South Korea.
Hyeokjin Lee, Division of Emerging Infectious Diseases, Bureau of Infectious Disease Diagnosis Control, Korea Disease Control and Prevention Agency, Cheongju, South Korea.
Jong-Hun Kim, Department of Social and Preventive Medicine, Sungkyunkwan University School of Medicine, Suwon, South Korea.
Eun-Jin Kim, Division of Emerging Infectious Diseases, Bureau of Infectious Disease Diagnosis Control, Korea Disease Control and Prevention Agency, Cheongju, South Korea.
Jin Y Kim, Division of Infectious Diseases, Department of Internal Medicine, Incheon Medical Center, Incheon, South Korea.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
References
- 1. Kim JM, Rhee JE, Yoo M, et al. Increase in viral load in patients with SARS-CoV-2 Delta variant infection in the Republic of Korea. Front Microbiol 2022; 13:819745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jefferson T, Spencer EA, Brassey J, Heneghan C. Viral cultures for coronavirus disease 2019 infectivity assessment: a systematic review. Clin Infect Dis 2021; 73:e3884–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bahrami A, Ferns GA. Genetic and pathogenic characterization of SARS-CoV-2: a review. Future Virol 2020; 15:533–49. [Google Scholar]
- 4. Deshpande A, Harris BD, Martinez-Sobrido L, Kobie JJ, Walter MR. Epitope classification and RBD binding properties of neutralizing antibodies against SARS-CoV-2 variants of concern. Front Immunol 2021; 12:691715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Volz E, Hill V, McCrone JT, et al. Evaluation of the effects of the SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell 2021; 184:64–75.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Johnson BA, Xie X, Bailey AL, et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature 2021; 591:293–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Scripps Research . Variant of concern reports. Available at: https://outbreak.info/situation-reports. Accessed 26 March 2022.
- 8. Lubinski B, Fernandes MHV, Frazier L, et al. Functional evaluation of the P681H mutation on the proteolytic activation of the SARS-CoV-2 variant B.1.1.7 (Alpha) spike. iScience 2022; 25:103589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. US Centers for Disease Control and Prevention . Science brief: Omicron (B.1.1.529) variant. Published 2 December 2021. Available at: https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/scientific-brief-omicron-variant.html. Accessed 26 March 2022.
- 10. Fratev F. N501Y and K417N mutations in the spike protein of SARS-CoV-2 alter the interactions with both hACE2 and human-derived antibody: a free energy of perturbation retrospective study. J Chem Inf Model 2021; 61:6079–84. [DOI] [PubMed] [Google Scholar]
- 11. UK Health Security Agency . SARS-CoV-2 variants of concern and variants under investigation - technical briefing 33. Published December 2021. Available at: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1043807/technical-briefing-33.pdf. Accessed 26 March 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.