Abstract
The emergence of severe acute respiratory syndrome coronavirus 2 variants that have greater transmissibility and resistance to neutralizing antibodies has increased the incidence of breakthrough infections. We show that breakthrough infection increases neutralizing antibody titers to varying degrees depending on the nature of the breakthrough variant and the number of vaccine doses previously administered. Omicron breakthrough infection resulted in neutralizing antibody titers that were the highest across all groups, particularly against Omicron.
Keywords: breakthrough infection, neutralization, Omicron, SARS-CoV-2, vaccination
Vaccination and infection by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants are currently shaping immunity to SARS-CoV-2 in individuals and populations [1]. The exposure of individuals to SARS-CoV-2 antigens has become highly heterogeneous as breakthrough infections of vaccinated or previously infected individuals with variants that evade preexisting immunity have become common. In particular, Omicron (B.1.1.529) has a large number of amino acid changes compared to ancestral SARS-CoV-2 and is highly resistant to plasma neutralizing antibodies elicited by infection with prior variants and the 2-dose messenger RNA (mRNA) vaccination regimens. Nevertheless, vaccination after infection or administration of a third mRNA vaccine dose elicits high levels of antibodies that have broad activity and can neutralize Omicron to a degree [2–5]. To assess the impact of breakthrough infection neutralizing antibody potency and breadth, we compared neutralizing antibody titers against the ancestral and recently emergent SARS-CoV-2 variants in 54 individuals who had received 2 or 3 doses of mRNA vaccine and experienced breakthrough infection with the Delta (B.1.617) or Omicron SARS-CoV-2 variants.
METHODS
Participants were healthy adults who had been vaccinated with 2 or 3 doses of an mRNA vaccine and experienced breakthrough infections with Delta (n = 24; median age, 30 years [range, 21–63 years]; 67% male, 33% female) or Omicron (n = 30; median age, 33.5 years [range, 22–79 years]; 53% male, 47% female). Samples were obtained between 13 August 2021 and 28 January 2022 for an average of 27 days (range, 14–55 days) after the latest positive test for infection (for details, see Supplementary Table 1). The study was performed in compliance with all relevant ethical regulations, and the protocol (DRO-1006) for studies with human participants was approved by the institutional review board of The Rockefeller University. Uninfected vaccine recipient participants have been previously described [3] (Supplementary Table 1).
Blood Sample Processing and Storage
Heparinized plasma and serum samples were aliquoted and stored at −20 °C or below. Before experiments, aliquots of plasma samples were heat inactivated (56 °C for 1 hour) and then stored at 4 °C.
Pseudotyped Virus Neutralization Assays
293T cells were transfected with pNL4-3ΔEnv-nanoluc and pSARS-CoV-2-SΔ19 plasmids [6]. Forty-eight hours later, particles were harvested, filtered, and stored at −80 °C. The amino acid deletions and/or substitutions corresponding to SARS-CoV-2 variants were incorporated into a spike expression plasmid using synthetic gene fragments (Integrated DNA Technologies (IDT)) or overlap extension polymerase chain reaction–mediated mutagenesis and Gibson assembly. Specifically, the variant-specific deletions and substitutions introduced into the B1 sequence were verified by sequencing and included B.1.617 (Delta): T19R/Δ156-8/L452R/T478K/D614G/P681R/D950N; and B.1.1.529 (Omicron): A76V/Δ69-70/T95I/G214D/Δ143-145/Δ211/L212I/ins214EPE/G339D/S371L/S373P/S375F/K417N/N440K/G446S/S477N/T478K/E484A/Q493K/G498R/N501Y/Y505H/T547K/D614G/H655Y/N679K/P681H/N746K/D796Y/N856K/Q954H/N969K/L981F.
All spike proteins used in the pseudotype neutralization assays had a 19-amino acid C-terminal deletion and included the R683G substitution, which disrupts the furin cleavage site and increases particle infectivity without grossly affecting antibody sensitivity.
Five-fold serially diluted plasma samples from vaccinated individuals were incubated with SARS-CoV-2 pseudotyped virus for 1 hour at 37 °C. The mixture was subsequently added to an HT1080-based cell line engineered to express human angiotensin-converting enzyme 2 (HT1080.ACE2 cl14) [6]. The starting serum dilution on cells was 1:50. Nanoluc luciferase activity in lysates was measured 48 hours postinoculation using the Nano-Glo Luciferase Assay System (Promega) with a Glomax Navigator (Promega). Relative luminescence units were normalized to those derived from cells infected with SARS-CoV-2 pseudotyped virus in the absence of serum. The half-maximal neutralization titers for sera (NT50) were determined using 4-parameter nonlinear regression (least squares regression method without weighting; constraints: top = 1, bottom = 0) (GraphPad Prism), and median values were calculated for each sample from 2–3 independent experiments. Group comparisons were performed using Mann-Whitney test (GraphPad Prism).
RESULTS
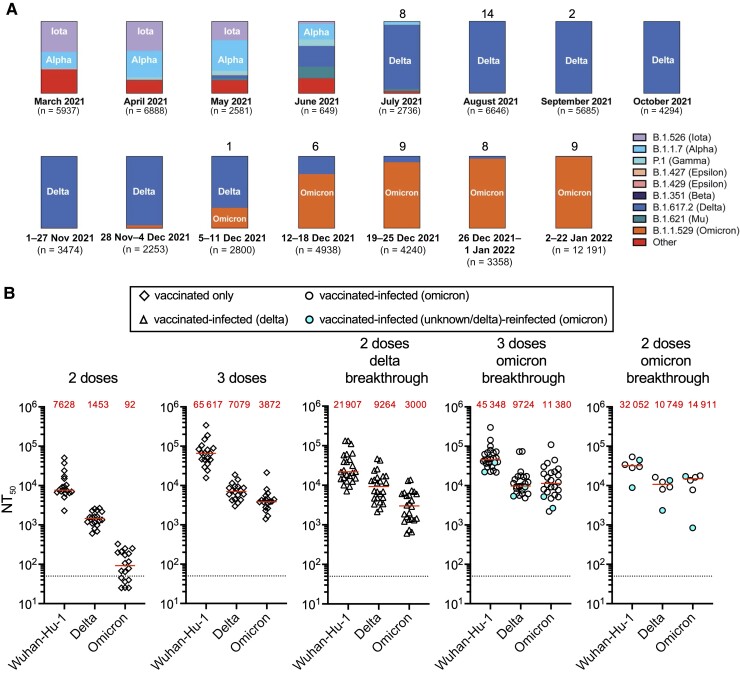
We collected samples from 54 individuals who had received 2 or 3 doses of mRNA-1273 (Moderna) or BNT162b2 (Pfizer) mRNA vaccines and were subsequently infected with SARS-CoV-2. Volunteer age was between 21 and 79 years (median age, 30 years); 32 (59%) were male and 22 (41%) were female (Supplementary Table 1). Breakthrough infection variants were deduced based on the prevalent variant circulating in New York City at the time of infection (Figure 1A; Supplementary Table 1). Specifically, Delta (B.1.617) was the predominant variant from July to early December 2021 but was rapidly displaced by Omicron by January 2022 (Figure 1A). As a consequence of evolving vaccination recommendations, 24 participants who experienced breakthrough infection with Delta had received 2 vaccine doses. Conversely, most Omicron breakthrough infections (24 of 30) were in participants who had received 3 vaccine doses.
Figure 1.
Variant prevalence and plasma neutralization titers. A, Frequency of variants over selected periods of weeks or months based on viral genome sequences obtained from samples collected in New York City and downloaded from the New York City Department of Health and Mental Hygiene (https://github.com/nychealth/coronavirus-data/blob/master/variants/variant-epi-data.csv) on 4 February 2022. Numbers on top of bar charts indicate number of polymerase chain reaction–positive samples identified over that period in our cohort. B, Half-maximal neutralization titer (NT50) values against Wuhan-Hu-1, Delta (B.1.617) variant, or Omicron variant (B.1.1.529). Plasma samples were collected from uninfected, vaccinated individuals 29–53 days after the second or third dose of a messenger RNA (mRNA) vaccine (BNT162b2 [Pfizer] or mRNA-1283 [Moderna]) or from individuals who had been vaccinated with 2 or 3 doses of an mRNA vaccine and experienced 1 or 2 breakthrough infections with samples collected between 14 and 55 days after the latest positive test for infection. Horizontal red lines and red values in each group indicate median NT50 values. Dotted black line indicates limit of detection of the assay.
In participants who did not experience breakthrough infection and received 2 vaccine doses, NT50 values against Delta and Omicron were 5.2- and 82.6-fold, respectively, lower than those against Wuhan-Hu-1, whereas in those who had received 3 vaccine doses, they were 9.2- and 16.9-fold lower (P < 0.0001 for all comparisons) (Figure 1B).
Participants who received 2 vaccine doses and had Delta breakthrough infection had median (range) NT50 values of 21 907 (7089–133 843), 9264 (2131–46 555), and 3000 (605–13 749) against Wuhan-Hu-1, Delta, and Omicron, respectively (Figure 1B). These values were 2.8-, 4.9-, and 26.4-fold greater than uninfected 2-dose vaccine recipients, for whom the corresponding NT50 values were 7628 (2299–50 640), 1453 (606–2666), and 92.3 (25–327) against Wuhan-Hu-1, Delta, and Omicron, respectively (all comparisons P ≤ .0005). Participants who received 3 vaccine doses and had Omicron breakthrough infection had median (range) NT50 values of 45 348 (21 661–300 044), 9724 (4829–73 494), and 11 380 (2197–109 591) against Wuhan-Hu-1, Delta, and Omicron, respectively (Figure 1B). By comparison, uninfected 3-dose vaccine recipients had median (range) NT50 values against these 3 viruses of 65 617 (15 641–341 247), 7079 (2978–18 768), and 3872 (1411–21 301). Therefore, Omicron breakthrough infection increased titers by 2.9-fold (P < 0.0001) against Omicron and 1.4-fold (P = 0.0088) against Delta. For the few individuals with Omicron breakthrough infection after receiving only 2 vaccine doses, median (range) NT50 values were 32 052 (8880–53 494), 10 749 (2333–16 327), and 14 911 (845–17 756) against Wuhan-Hu-1, Delta, and Omicron, a respective increase in titer of 4.2-, 7.4-, and 161.5-fold (all P ≤ .0074) compared to uninfected 2-dose vaccine recipients (Figure 1B).
Finally, 4 individuals in the Omicron breakthrough infection group had been previously infected with other variants (Supplementary Table 1). We did not observe consistent differences in the NT50 values of these individuals compared to the rest of the group (Figure 1B), though the sample size was small.
DISCUSSION
The repeated emergence of new SARS-CoV-2 variants such as Delta and Omicron, with increasing levels of resistance to neutralizing antibodies, has led to an increase in incidence of breakthrough infections in vaccinated individuals [7, 8]. Erosion of the efficacy of vaccines based on the ancestral Wuhan-Hu-1 strain may impact future vaccination strategies. We determined the effect of breakthrough infection on neutralizing antibodies against original and variant SARS-CoV-2. Study limitations include the relatively small number and heterogeneity of participants that reflect the real-world circumstances at the time of recruitment. We note that correlation between neutralizing antibody levels and protection has been reported [9], even though additional correlates remain to be defined.
Consistent with prior reports from our group and others [3–5, 10–12], we show that plasma neutralizing antibodies in recipients of 2 mRNA vaccine doses are less potent against Delta and particularly against Omicron. However, administration of a third mRNA vaccine dose significantly increases neutralizing antibody titers against both variants (Figure 1B). Our data further demonstrate that in individuals who received 2 mRNA vaccine doses, breakthrough infection by either Delta or Omicron increases neutralizing antibody titers against both variants. While the magnitude of the increase against Delta is comparable in individuals who experienced either Delta or Omicron breakthrough infection, the increase in neutralization potency against Omicron is 161.5-fold in Omicron breakthrough infections compared to 26.4-fold in Delta breakthrough infections. Compared to individuals who received 3 mRNA vaccine doses, Omicron breakthrough infections also generated increased neutralizing antibody titers, but the magnitude of the effect was small.
Because Omicron breakthrough infection in 2-dose vaccine recipients resulted in broader plasma neutralization activity than Delta breakthrough infection, our data suggest that an Omicron-specific booster shot may have similar effects in those who received only 2 mRNA vaccine doses [12]. In individuals who have already received a third dose of the currently available mRNA vaccines, Omicron-specific boosters may modestly improve neutralizing responses against Omicron while not significantly impacting neutralization of other variants.
Supplementary Material
Acknowledgments
We are grateful to all participants who volunteered for this study. We thank M. Bergh, M. Okawa Frank, and R. B. Darnell for the occupational SARS-CoV-2 infection surveillance program and participant referrals.
Financial support . This work was supported by grants from the National Institutes of Health (grant numbers R01AI501111 to P. D. B.; R01AI78788 to T. H.; P01-AI138398-S1 and 2U19AI111825 to M. C. N.; and P01AI65075 to P. D. B., T. H., and M. C. N.). C. G.’s work is supported by the Robert S. Wennett Post-Doctoral Fellowship, National Center for Advancing Translational Sciences (National Institutes of Health Clinical and Translational Science Award program, grant number UL1 TR001866) and the Shapiro–Silverberg Fund for the Advancement of Translational Research. P. D. B. and M. C. N. are Howard Hughes Medical Institute (HHMI) investigators. This article is subject to HHMI’s Open Access to Publications policy. HHMI laboratory heads have previously granted a nonexclusive CC BY 4.0 license to the public and a sublicensable license to HHMI in their research articles.
Potential conflicts of interest . P. D. B. has received compensation from Pfizer for consultation in the area of SARS-CoV-2 vaccines. All other authors report no potential conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Contributor Information
Christian Gaebler, Laboratory of Molecular Immunology, The Rockefeller University, New York, New York, USA.
Justin DaSilva, Laboratory of Retrovirology, The Rockefeller University, New York, New York, USA.
Eva Bednarski, Laboratory of Retrovirology, The Rockefeller University, New York, New York, USA.
Frauke Muecksch, Laboratory of Retrovirology, The Rockefeller University, New York, New York, USA.
Fabian Schmidt, Laboratory of Retrovirology, The Rockefeller University, New York, New York, USA.
Yiska Weisblum, Laboratory of Retrovirology, The Rockefeller University, New York, New York, USA.
Katrina G Millard, Laboratory of Molecular Immunology, The Rockefeller University, New York, New York, USA.
Martina Turroja, Laboratory of Molecular Immunology, The Rockefeller University, New York, New York, USA.
Alice Cho, Laboratory of Molecular Immunology, The Rockefeller University, New York, New York, USA.
Zijun Wang, Laboratory of Molecular Immunology, The Rockefeller University, New York, New York, USA.
Marina Caskey, Laboratory of Molecular Immunology, The Rockefeller University, New York, New York, USA.
Michel C Nussenzweig, Laboratory of Molecular Immunology, The Rockefeller University, New York, New York, USA; Howard Hughes Medical Institute, Rockefeller University, New York, New York, USA.
Paul D Bieniasz, Laboratory of Retrovirology, The Rockefeller University, New York, New York, USA; Howard Hughes Medical Institute, Rockefeller University, New York, New York, USA.
Theodora Hatziioannou, Laboratory of Retrovirology, The Rockefeller University, New York, New York, USA.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
References
- 1. Wang Z, Muecksch F, Schaefer-Babajew D, et al. Naturally enhanced neutralizing breadth against SARS-CoV-2 one year after infection. Nature 2021; 595:426–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schmidt F, Weisblum Y, Rutkowska M, et al. High genetic barrier to SARS-CoV-2 polyclonal neutralizing antibody escape. Nature 2021; 600:512–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schmidt F, Muecksch F, Weisblum Y, et al. Plasma neutralization of the SARS-CoV-2 Omicron variant. N Engl J Med 2022; 386:599–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cele S, Jackson L, Khoury DS, et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature 2021; 602:654–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Planas D, Saunders N, Maes P, et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 2021; 602:671–5. [DOI] [PubMed] [Google Scholar]
- 6. Schmidt F, Weisblum Y, Muecksch F, et al. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J Exp Med 2020; 217:e20201181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kuhlmann C, Mayer CK, Claassen M, et al. Breakthrough infections with SARS-CoV-2 Omicron despite mRNA vaccine booster dose. Lancet 2022; 399:625–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nemet I, Kliker L, Lustig Y, et al. Third BNT162b2 vaccination neutralization of SARS-CoV-2 Omicron infection. N Engl J Med 2022; 386:492–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Khoury DS, Cromer D, Reynaldi A, et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med 2021; 27:1205–11. [DOI] [PubMed] [Google Scholar]
- 10. Gruell H, Vanshylla K, Tober-Lau P, et al. mRNA booster immunization elicits potent neutralizing serum activity against the SARS-CoV-2 Omicron variant. Nat Med 2022; 28:477–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu L, Iketani S, Guo Y, et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature 2021; 602:676–81. [DOI] [PubMed] [Google Scholar]
- 12. Wratil PR, Stern M, Priller A, et al. Three exposures to the spike protein of SARS-CoV-2 by either infection or vaccination elicit superior neutralizing immunity to all variants of concern. Nat Med 2022; 28:496–503. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.