Abstract
We have developed a bioluminescent whole-cell biosensor that can be incorporated into biofilm ecosystems. RM4440 is a Pseudomonas aeruginosa FRD1 derivative that carries a plasmid-based recA-luxCDABE fusion. We immobilized RM4440 in an alginate matrix to simulate a biofilm, and we studied its response to UV radiation damage. The biofilm showed a protective property by physical shielding against UV C, UV B, and UV A. Absorption of UV light by the alginate matrix translated into a higher survival rate than observed with planktonic cells at similar input fluences. UV A was shown to be effectively blocked by the biofilm matrix and to have no detectable effects on cells contained in the biofilm. However, in the presence of photosensitizers (i.e., psoralen), UV A was effective in inducing light production and cell death. RM4440 has proved to be a useful tool to study microbial communities in a noninvasive manner.
Biofilms are complex structures in which bacterial populations are enclosed in a matrix. The cells form aggregates by adhering to each other or to the matrix (6). When nutrients are available in aquatic environments, biofilms are established and cells grow in communities. Studies of Pseudomonas aeruginosa have shown that cells in biofilms are phenotypically distinct from planktonic cells. Adhesion of cells triggers secretion of alginate in P. aeruginosa, allowing concentration of nutrients and adhesion of additional bacterial cells (15). Upon adhesion, synthesis of exopolysaccharide is coupled to cell division, which leads to the formation of microcolonies. Networks of water channels are formed in biofilms; these channels are responsible for the exchange of nutrients and metabolites with the bulk fluid (6).
Biosensors are devices that couple a biological component with a physical transducer. The biological element provides selectivity for the analyte being studied, while the transducer generates a measurable signal. Such biosensors are very useful in environmental settings because they can be integrated into a microbial community of interest. King et al. (17) used a Pseudomonas fluorescens biosensor for detection of naphthalene and its degradative intermediate, salicylate, in the environment. Vollmer et al. (25) developed an Escherichia coli biosensor for detection of DNA damage.
We have constructed a P. aeruginosa whole-cell biosensor for monitoring DNA-damaging agents for use in environmental settings (8) by fusing the promoter of the P. aeruginosa recA gene to the lux operon from Vibrio fischeri (8). The P. aeruginosa recA gene has been cloned previously (18) and shown to be inducible by DNA-damaging agents (20). The lux operon contains five structural genes, luxCDABE. Luciferase (encoded by luxAB) oxidizes reduced flavin mononucleotide and a long fatty acid aldehyde in the presence of oxygen to emit light with a 490-nm wavelength. The reductase complex (luxCDE) recycles the fatty acid, allowing autonomous bioluminescence (8).
Solar UV wavelengths of biological importance are mainly composed of UV A (320 to 400 nm) and UV B (290 to 320 nm). Besides its implication in damaging proteins and membranes, UV A indirectly damages DNA by creating reactive oxygen compounds (e.g., H2O2, O2−, etc.) by photooxidation of O2 which cause single-strand breaks in DNA (26). UV B is absorbed by DNA directly and alters nucleotides by creating cyclobutane pyrimidine dimers and photoproducts (9). UV C (100 to 290 nm) is the most energetic. Although completely blocked by the ozone layer, it has been studied extensively because of its germicidal effects (9).
Studies of ecosystems and the physiological states of microbial communities have long relied on using liquid bacterial cultures. These studies do not address the state of microorganisms in environments such as biofilms (21). The difficulty in analyzing biofilms lies in the lack of tools that allow noninvasive study. In this report, we study the response of a P. aeruginosa biofilm to stress by using a bioluminescent biosensor that responds to DNA damage.
MATERIALS AND METHODS
Bacterial strains and plasmids.
RM4440 is a P. aeruginosa FRD1 strain carrying the plasmid pMOE15. The plasmid contains a fusion of the recA promoter of P. aeruginosa to the luxCDABE operon of V. fischeri. The construction of the plasmid and the development of the strain have been described previously (8).
Maintenance media.
Luria broth (LB; Gibco-BRL, Gaithersburg, Md.) was used for viable counts (19). Pseudomonas minimal medium (PMM) was used as a nutrient source in the assays (19). RM4440 was grown in a 1-liter culture to midexponential growth phase in PMM at room temperature. The cells were collected by centrifugation, and the cell pellet was resuspended in a half-volume of saline and used to prepare the alginate matrix.
Alginate matrix.
One volume of resuspended cells was mixed with 2 volumes of sterile low-viscosity sodium alginate (3.5% [wt/wt] in 0.9% NaCl) and a 1/2 volume of sterile glycerol. The mixture was then aliquoted in 15-ml volumes and stored at −70°C.
Sample preparation and induction of recA.
Fifteen milliliters of the alginate cell suspension was passed through a 20-gauge needle (Becton Dickinson, Franklin Lakes, N.J.) and dripped into a 0.1 M strontium chloride (J. T. Baxter, Phillipsburg, N.J.) solution to form small beads of 4 mm in diameter. The mixture was stirred for 15 min. Alginate cross-links upon mixing with strontium chloride, immobilizing the cells within the matrix. The beads were then taken out of the cross-linking solution and placed in an electrophoresis tray. An electrophoresis apparatus was used as an incubation chamber. Incubation was started by passing PMM through a monolayer of beads at room temperature at a flow rate of 3.5 ml per min. In different experiments, the alginate beads were exposed to various doses of UV radiation. UV C was delivered by a germicidal bulb (General Electric Corporation, Atlanta, Ga.) with a maximum emission at 256 nm. UV A and UV B were delivered by UV A and UV B lamps (Spectronics Corporation, Westbury, N.Y.). UV doses were determined with a UVX radiometer (Ultra Violet Products, San Gabriel, Calif.) fitted with sensors for UV C or UV B.
Viable cell counts.
Viable cell counts were taken before and immediately after UV exposure of alginate bead biofilms. Before exposure to UV, five beads were removed and transferred to 1 ml of (NaPO3)n. The beads were mixed until completely dissolved. Serial dilutions were made in saline, and 50 μl was plated on LB. After exposure to UV, five beads were removed and the procedure was repeated. Plates were incubated at 37°C and counted. Post-UV C and -UV B incubations were conducted in the dark to prevent light repair.
Light measurements.
A flexible liquid light guide (Oriel Instruments, Stratford, Colo.) was used to transmit light from the sample into a photomultiplier (PMT) tube, model 77340 (Oriel Instruments), which produces an induced current. The PMT was connected to a PMT readout, model 7070 (Oriel Instruments), with a digital display to read the current in amperes. The PMT readout was sensitive to 1 nA of induced current. The PMT readout was connected to a chart pen recorder to allow continuous light reading.
Electron microscopy.
Scanning electron microscopy was done on beads containing entrapped RM4440 cells before and after exposure to 10 J of UV C per m2. The electron microscope used was a JEOL 35 U. The beads were placed in a fixative solution (3% glutaraldehyde and 3% sucrose) for 3 h. They were then washed with 0.2 M sodium cacodylate buffer (pH 7.4). Following refrigeration overnight, the beads were dehydrated in a graded concentration series of ethanol (50, 70, 90, 95, and 100%). They were dried to the critical point required for electron microscopy. An acceleration voltage of 25 kV was used.
RESULTS
Growth of RM4440 and FRD1.
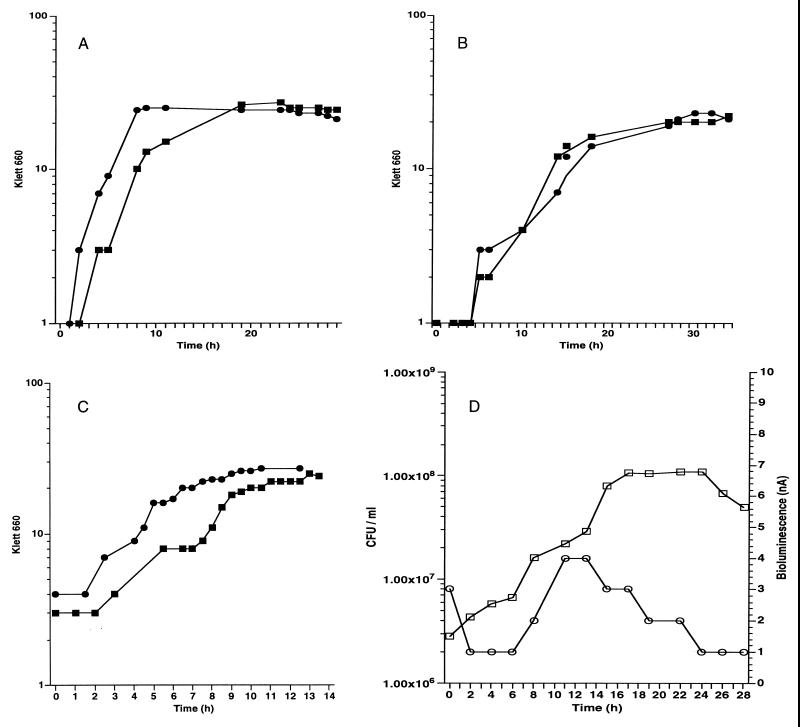
In order to study the growth characteristics of the biosensor, growth curves were done on the host strain (FRD1) without the plasmid (Fig. 1A) and RM4440 containing pMOE15 (Fig. 1B). Two 50-ml PMM batch cultures were inoculated from an overnight culture. The flasks were incubated at room temperature with vigorous shaking. Growth was measured with a Klett-Summerson colorimeter (660-nm filter). Under these conditions, FRD1 had an average doubling time of G = 135 min. RM4440 had a slightly longer doubling time of G = 150 min. The growth rates for FRD1 and RM4440 were μ = 0.44 h−1 and μ = 0.4 h−1, respectively. The lower growth rate of RM4440 is probably due to the presence of the plasmid, which adds more of a burden on the cell to replicate the extra DNA.
FIG. 1.
Growth of FRD1 and RM4440. (A) FRD1 in liquid PMM. ■, experiment 1; ●, experiment 2. (B) RM4440 in liquid PMM. ■, experiment 1; ●, experiment 2. (C) RM4440 in liquid PMM supplemented with 1% alginate. ■, experiment 1; ●, experiment 2. (D) RM4440 immobilized in the alginate matrix. Bioluminescence was continuously monitored. □, CFU per milliliter, ○, bioluminescence (nanoamperes). All experiments were done twice.
RM4440 uses alginate as a nutrient.
To investigate whether RM4440 is able to use unlinked alginate as a carbon source, another batch culture was monitored for growth. A 1% alginate solution was added to PMM, and the culture was incubated at room temperature with vigorous shaking. Klett-Summerson measurements were taken periodically. The experiment was done twice (Fig. 1C). The average doubling time of RM4440 was G = 60 min, and the growth rate was μ = 1.0 h−1. The growth rate was twofold higher in the presence of alginate than in its absence, which indicates that RM4440 uses alginate as a carbon source under batch culture conditions.
Growth of RM4440 in the alginate matrix.
When cells are immobilized in an alginate matrix, the composition of the medium is different from batch cultures because the alginate is covalently cross-linked into a network.
Growth of RM4440 was analyzed under immobilization conditions. RM4440 was immobilized into 4-mm alginate beads. A monolayer of beads was fed continuously with PMM at a flow rate of 3.5 ml/min. Samples of viable counts were taken every 2 h by dissolving five beads and plating serial dilutions on LB plates. Growth in the matrix was monitored for a period of 30 h. Under these conditions, RM4440 showed a growth pattern similar to a batch culture (Fig. 1D). The exponential growth phase showed a doubling time for RM4440 of G = 168 min, and the growth rate was μ = 0.36 h−1. A phase with a constant number of cells followed the exponential phase. The growth rate in the alginate matrix was higher than that of the batch culture with alginate because the immobilized cells were fed continuously with PMM while the batch culture had an exhaustible source of nutrients.
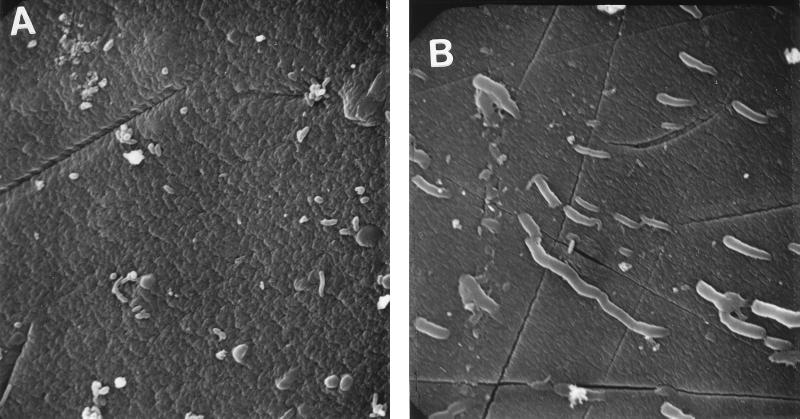
Physical appearance of P. aeruginosa entrapped in alginate beads.
RM4440 cells were physically entrapped in the beads of the strontium alginate matrix. The beads were 4 mm in diameter and transmitted a maximum of 13% of UV C, 31% of UV B, and 33% of UV A radiation to which the biofilm was exposed. Scanning electron microscopy was done on the beads to observe the distribution and the effects of UV C stress (10 J/m2) on the cells in the matrix (Fig. 2). The cells on the surface of untreated beads were distributed individually and showed no obvious clumping. Most cells were partially buried in the matrix, although the harsh treatment and washing of the beads prior to microscopy may have washed some cells out. The cells exposed to UV C were filamentous.
FIG. 2.
Scanning electron micrographs of the alginate beads containing RM4440 cells. (A) Unirradiated control. (B) Exposure to 10 J of UV C per m2. Stressed cells show filamentation due to inhibition of cell division. Magnification, ca. ×2,000.
RM4440 response to UV C stress.
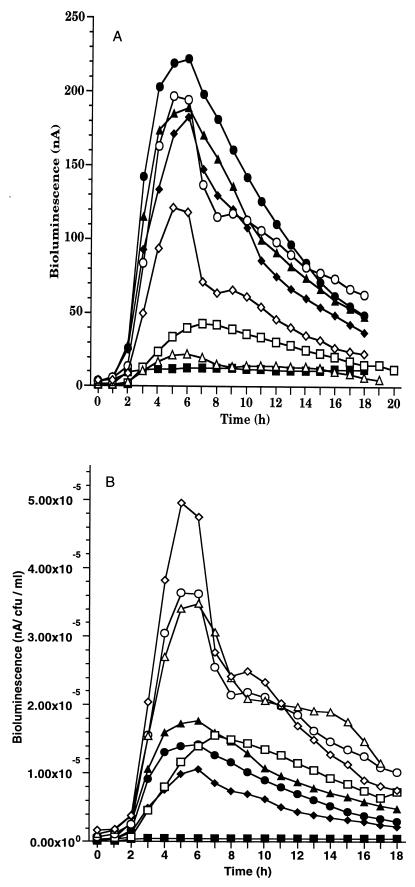
RM4440 cells were trapped in a cross-linked alginate matrix and exposed to increasing doses of UV C light. The dose of UV C radiation was delivered as a single pulse, and visible (490-nm) lux-dependent light production was monitored continuously following exposure (Fig. 3A). Viable counts were taken before and after irradiation to account for cell death. If RM4440 was not exposed to UV radiation, only a basal level of light was produced.
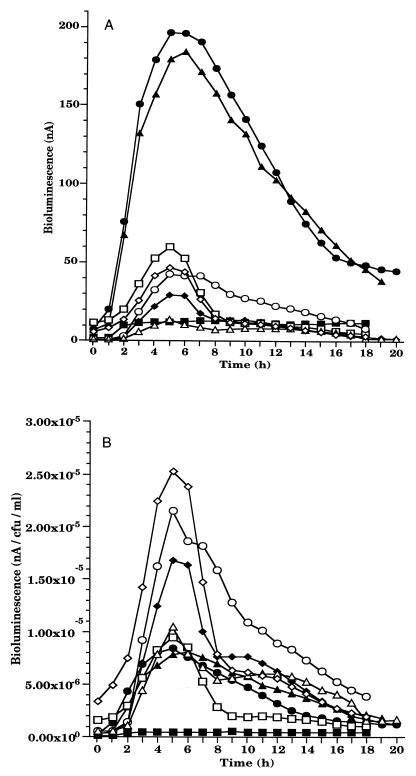
FIG. 3.
Bioluminescence response of RM4440 to increasing doses of UV C in biofilm. (A) Total bioluminescence (nanoamperes). (B) Normalized bioluminescence (nanoamperes per CFU per milliliter). UV C doses (J/m2): ■, 0; ●, 2.5; ▴, 5; ⧫, 10; □, 12.5; ○, 15; ▵, 17.5; ◊, 20. Each point represents the mean of three independent experiments.
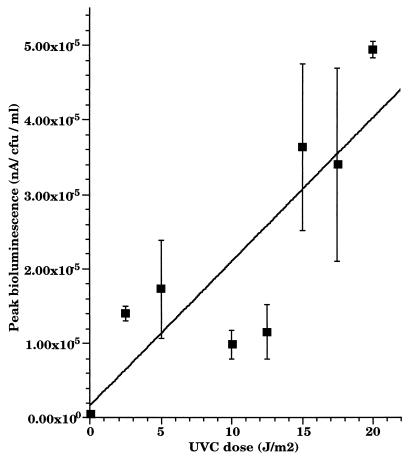
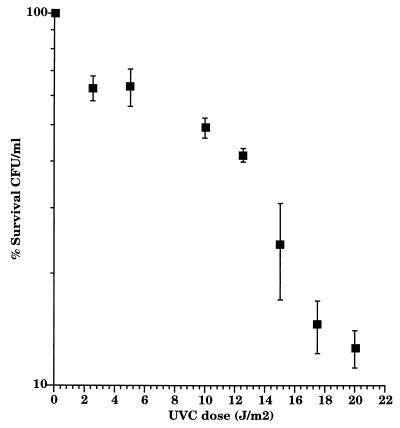
The bioluminescent response was normalized by dividing the amount of light by the number of CFU after each UV C exposure. RM4440 showed a rapid and reproducible increase in bioluminescence in response to UV C stress. The response profile was similar for all doses: a lag period of 30 min followed the pulse of UV C before visible light production began to increase steadily (Fig. 3B). Peak bioluminescence was observed 4.5 h after irradiation for all doses except for the 12.5-J/m2 dose, for which it was 6.5 h. The response of RM4440 showed an overall dose-response profile consistent with results obtained previously in batch culture experiments (10) (Fig. 4). The survival rate of RM4440 in the matrix decreased as the dose of UV C increased (Fig. 5); it was, however, higher than the survival rate observed in rich liquid medium (23). Although UV C is the UV most poorly transmitted by the matrix (13% maximum transmittance), it has shown the most bioluminescent response.
FIG. 4.
Peak bioluminescence after exposure of RM4440 biofilm to increasing doses of UV C. The peak response was 5 h postirradiation for most doses. The data represent the means of three independent experiments.
FIG. 5.
Survival rate of RM4440 under increasing doses of UV C. Data represent the means of three independent experiments.
RM4440 response to UV B stress.
RM4440 was exposed to increasing doses of UV B radiation in a fashion similar to the procedure used for UV C irradiation. In order for the study to be environmentally relevant, UV B doses were chosen based on average daily doses of UV B reaching the earth (3). Similar to the results obtained following UV C irradiation, the response profile of the irradiated biofilms was marked by a lag period of 30 min and peak responses were observed 4.5 h postirradiation (Fig. 6A).
FIG. 6.
Bioluminescence response of RM4440 to increasing doses of UV B in biofilm. (A) Total bioluminescence (nanoamperes). (B) Normalized bioluminescence (nanoamperes per CFU per milliliter). UV B doses (J/m2): ■, 0; ●, 25; ▴, 50; ⧫, 75; □, 100; ○, 110; ▵, 120; ◊, 125. Each point represents the mean of three independent experiments.
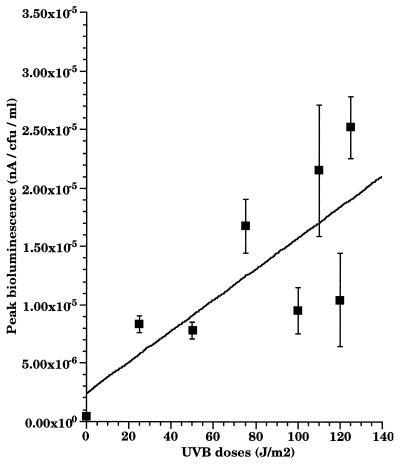
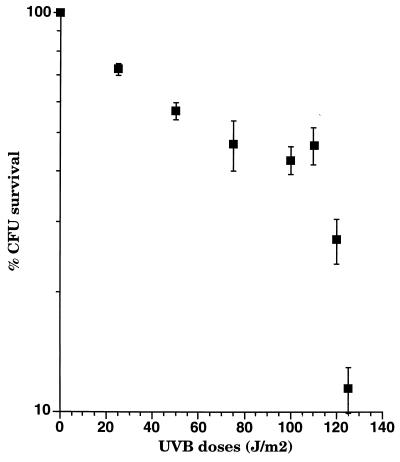
When the response was normalized, dose dependence was not as apparent with UV B as it was with UV C (Fig. 6B). However, when peak responses (5 h for most doses) were plotted as a function of the UV B dose administered, there was a tendency for bioluminescence to increase as dose increased (Fig. 7). The survival rate of cells under various doses was monitored (Fig. 8). Survival rate decreased with higher doses of UV B. The survival rate of cells in the biofilm was greater than that of free cells (16) owing to the physical shielding of the alginate matrix. This is important in improving our understanding of the role of biofilms. In nature, it appears that sessile microbial communities are likely to surpass planktonic cells in coping with UV stress in environmental settings.
FIG. 7.
Peak bioluminescence after exposure of RM4440 to increasing doses of UV B in biofilm. The peak response was 5 h postirradiation for most doses. The data represent the means of three independent experiments.
FIG. 8.
Survival rate of RM4440 under increasing doses of UV B. Data represent the means of three independent experiments.
RM4440 response to UV A stress.
UV A doses of up to 20,000 J/m2 were used to test the response of biofilms containing RM4440 to near-UV light. Bioluminescence stayed at basal levels, indicating little or no induction of recA expression. Doses used in this study were within the range of the average dose of UV A light that reaches the earth daily.
Effects of psoralen treatment on RM4440.
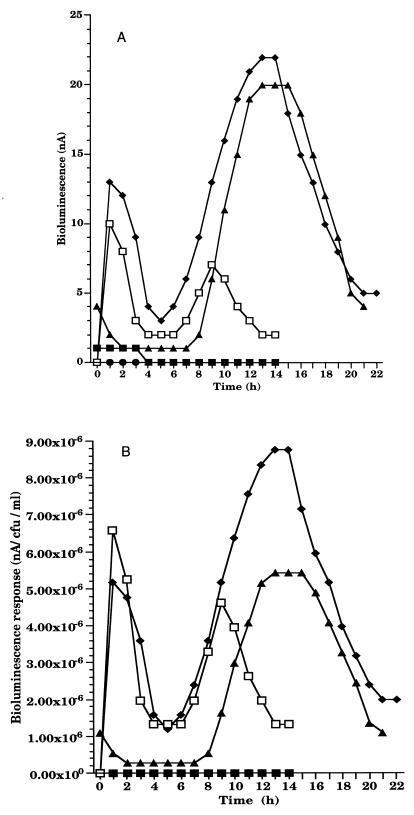
In order to investigate the effects of UV A in the presence of a photosensitizer on microbial communities in biofilms, beads were stirred in 100 ml of PMM containing psoralen (final concentration of 10 mg/liter) for 30 min and subsequently exposed to an hour of ambient light or a pulse of UV A (8,000 and 16,000 J/m2). Psoralen was also added to the biofilm in the dark as a control. Treatment of RM4440 with psoralen and UV A resulted in increased bioluminescence (Fig. 9). A loss in viability was also noted.
FIG. 9.
Bioluminescence response of RM4440 after exposure to UV A and psoralen. (A) Total bioluminescence (nanoamperes). (B) Normalized bioluminescence (nanoamperes per CFU per milliliter). The biofilm beads were stirred in PMM containing psoralen. ■, UV A only; ●, psoralen only; ▴, ambient light and psoralen; ⧫, 8,000 J of UV A per m2 and psoralen; □, 16,000 J of UV A per m2 and psoralen.
DISCUSSION
We characterized the response of a P. aeruginosa recA promoter-luxCDABE fusion under conditions that more closely resemble the natural setting of microorganisms found in freshwater habitats. Minimal medium was used to maintain a low-nutritional-content environment. We immobilized cells in an alginate matrix to simulate the way that mucoid P. aeruginosa strains engulf themselves in secreted exopolymers (primarily alginate) when establishing a biofilm in nature. Biofilms have been shown to be the preferred mode of growth of P. aeruginosa in aquatic environments (5). The alginate excreted by cells in their native habitat is believed to be used by cells for establishment of biofilms upon adhesion of cells to solid surfaces in nature (5, 7, 10). Alginate biofilm matrices have been shown to be important in complications of cystic fibrosis (11) and other diseases such as urinary tract infections (22). In addition, alginate endows P. aeruginosa with protection against antibiotics (14). Bacteria entrapped in alginate have also been shown to play a major role in fouling of human-made materials (4). In this investigation, we observed that the alginate biofilm transmits only a small amount of UV radiation (13% of UV C, 31% of UV B, and 33% of UV A), thus protecting the cells from exposure and suggesting that the exopolymer may be a natural defense mechanism used to attenuate UV light exposure in nature. This attenuation was evident from the higher rates of survival of alginate-entrapped cells than were observed with liquid cultures after exposure to various doses of UV (8, 16).
The alginate-entrapped cells that were exposed to UV C were filamentous, indicating that cell division was inhibited in a manner similar to that of the SOS-associated filamentation observed for E. coli (13). This phenomenon ensures the proper partitioning of genetic materials to daughter cells. In E. coli, two pathways coordinate DNA replication and cell division, an sfi-dependent and an sfi-independent pathway. In the sfi-dependent pathway, the gene sfiA is induced as part of the SOS response, and the protein SfiA binds FtsZ, an essential protein involved in septal biogenesis. This binding prevents septation and results in the formation of filaments (12). In sfi-independent filamentation, SfiA is not required; however, DNA damage is necessary, as well as the derepression of the SOS regulon (13).
The ability of RM4440 to monitor the effects of UV C and UV B on microorganisms in biofilms was demonstrated. The response profiles for UV C and UV B were similar, supporting the observation that they cause similar types of damage to cells (9). UV C is more efficient in damaging DNA (9), and low doses caused significant induction of recA expression. No bioluminescence response was observed when alginate-immobilized RM4440 cells were exposed to UV A radiation. This is most likely due to the absorption of UV A by the alginate matrix. However, in the presence of a diffusible, exogenous chromophore, exposure to similar doses of UV A radiation caused indirect damage to RM4440 in the alginate biofilm. UV A has been shown to induce recA expression in marine isolates (3) and in P. aeruginosa when exposed in a liquid medium (16). This suggests an increased UV sensitivity of planktonic cells in the environment. In the study reported here, P. aeruginosa cells were sheltered by the alginate matrix, allowing an apparent increased UV resistance. The presence of photosensitizers such as psoralen in the environment has the potential to render UV A an important stress even in biofilm communities. recA expression was induced in response to covalent binding of psoralen to DNA. The psoralen-dark control did not show a bioluminescent response; it did, however, cause cell death. The profile of the response to psoralen and UV A was different from that of the responses to UV B and UV C. Bioluminescence showed two peaks, an early one that is reminiscent of the rapid response to UV B or UV C exposure and a delayed response that appeared several hours later. This profile may be due to an early phase of damage that is caused by DNA-bound psoralen which causes immediate injury. The SOS system is induced in response, and some damage is repaired. The second phase of damage may be an effect of residual psoralen that is trapped in the matrix or of secondary damage to other macromolecules (i.e., proteins) (1). These results are consistent with previous studies showing the involvement of the SOS system in repair of psoralen damage (2, 24). When psoralen was added to the RM4440 biofilm in the dark, a genotoxic effect was observed. However there was no noticeable recA expression. Quinto et al. (22a) showed that, in the dark, psoralen causes frameshift mutagenesis in bacterial cells, through noncovalent interaction with DNA. This type of mutagenesis may explain the genotoxic effects observed with RM4440 after the dark-psoralen treatment.
RM4440 has the potential to be used as a monitor for biological stress in various environments. As a biosensor, it can be incorporated into sessile, as well as planktonic, bacterial communities.
ACKNOWLEDGMENTS
We thank Tricia Reid and Steve Hutchens for their invaluable help with the experiments.
This study was supported by a grant from the Oklahoma Center for the Advancement of Science and Technology, no. H97-051.
REFERENCES
- 1.Averbeck D. Recent advances in psoralen phototoxicity mechanism. Photochem Photobiol. 1989;50:859–882. doi: 10.1111/j.1751-1097.1989.tb02917.x. [DOI] [PubMed] [Google Scholar]
- 2.Bauluz C, Paramio J M, de Vidania R. Further studies on the lethal and mutagenic effects of 8-methoxypsoralen-induced lesions on plasmid DNA. Cell Mol Biol. 1991;37:481–500. [PubMed] [Google Scholar]
- 3.Booth M G. Solar ultraviolet radiation and the role of RecA in marine bacteria. Ph.D. dissertation. Stillwater: Oklahoma State University; 1997. [Google Scholar]
- 4.Christensen B E, Characklis W G. Physical and chemical properties of biofilms. In: Characklis W G, Marshall K C, editors. Biofilms. New York, N.Y: John Wiley & Sons, Inc.; 1990. pp. 93–130. [Google Scholar]
- 5.Costerton J W, Cheng K J, Geesey G G, Ladd T I, Nickel J C, Dasgupta M, Marrie T J. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- 6.Costerton J W, Lewandowski Z, Caldwell D E, Korber D R, Lappin-Scott H M. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 7.Davies D G, Geesey G G. Regulation of the alginate biosynthesis gene algC in Pseudomonas aeruginosa during biofilm development in continuous culture. Appl Environ Microbiol. 1995;61:860–867. doi: 10.1128/aem.61.3.860-867.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elasri M O, Miller R V. A Pseudomonas aeruginosa biosensor responds to exposure to ultraviolet radiation. Appl Microbiol Biotechnol. 1998;50:455–458. doi: 10.1007/s002530051320. [DOI] [PubMed] [Google Scholar]
- 9.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C: ASM Press; 1995. [Google Scholar]
- 10.Gacesa P. Bacterial alginate biosynthesis—recent progress and future prospects. Microbiology. 1998;144:1133–1143. doi: 10.1099/00221287-144-5-1133. [DOI] [PubMed] [Google Scholar]
- 11.Govan J R, Harris G S. Pseudomonas aeruginosa and cystic fibrosis: unusual bacterial adaptation and pathogenesis. Microbiol Sci. 1986;3:302–308. [PubMed] [Google Scholar]
- 12.Higashitani A, Higashitani N, Horiuchi K. A cell division inhibitor SulA of Escherichia coli directly interacts with FtsZ through GTP hydrolysis. Biochem Biophys Res Commun. 1995;209:198–204. doi: 10.1006/bbrc.1995.1489. [DOI] [PubMed] [Google Scholar]
- 13.Hill T M, Sharma B, Valjavec-Gratian M, Smith J. sfi-independent filamentation in Escherichia coli is lexA dependent and requires DNA damage for induction. J Bacteriol. 1997;179:1931–1939. doi: 10.1128/jb.179.6.1931-1939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hodges N A, Gordon C A. Protection of Pseudomonas aeruginosa against ciprofloxacin and beta-lactams by homologous alginate. Antimicrob Agents Chemother. 1991;35:2450–2452. doi: 10.1128/aac.35.11.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoyle B D, Williams L J, Costerton J W. Production of mucoid exopolysaccharide during development of Pseudomonas aeruginosa biofilms. Infect Immun. 1993;61:777–780. doi: 10.1128/iai.61.2.777-780.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kidambi S P, Booth M G, Kokjohn T A, Miller R V. recA-dependence of the response of Pseudomonas aeruginosa to UVA and UVB irradiation. Microbiology. 1996;142:1033–1040. doi: 10.1099/00221287-142-4-1033. [DOI] [PubMed] [Google Scholar]
- 17.King J M H, Digarzia P M, Applegate B, Burlage R, Sanseverino J, Dunbar P, Larimer F, Sayler G S. Rapid, sensitive bioluminescence reporter technology for naphthalene exposure and biodegradation. Science. 1990;249:778–781. doi: 10.1126/science.249.4970.778. [DOI] [PubMed] [Google Scholar]
- 18.Kokjohn T A, Miller R V. Molecular cloning and characterization of the recA gene of Pseudomonas aeruginosa PAO. J Bacteriol. 1985;163:568–572. doi: 10.1128/jb.163.2.568-572.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller R V, Ku C-M C. Characterization of Pseudomonas aeruginosa mutants deficient in the establishment of lysogeny. J Bacteriol. 1978;134:875–883. doi: 10.1128/jb.134.3.875-883.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller R V, Kokjohn T A. Expression of the recA gene of Pseudomonas aeruginosa PAO is inducible by DNA-damaging agents. J Bacteriol. 1988;170:2385–2387. doi: 10.1128/jb.170.5.2385-2387.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller R V, Poindexter J S. Strategies and mechanisms for field research in environmental bioremediation. A call for a national environmental bioremediation field research program. Washington, D.C: American Academy of Microbiology; 1994. [Google Scholar]
- 22.Nickel J C, Ruseska I, Costerton J W. Tobramycin resistance of cells of Pseudomonas aeruginosa growing as a biofilm on urinary catheter material. Antimicrob Agents Chemother. 1985;27:619–624. doi: 10.1128/aac.27.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a.Quinto I, Averbeck D, Moustacchi E, Hrisoho Z, Moron J. Frameshift mutagenicity in Salmonella typhimurium of furocoumarins in the dark. Mutat Res. 1984;136:49–54. doi: 10.1016/0165-1218(84)90133-2. [DOI] [PubMed] [Google Scholar]
- 23.Simonson C S, Kokjohn T A, Miller R V. Inducible UV repair potential of Pseudomonas aeruginosa PAO. J Gen Microbiol. 1990;136:1241–1249. doi: 10.1099/00221287-136-7-1241. [DOI] [PubMed] [Google Scholar]
- 24.Sladek F M, Melian A, Howard-Flanders P. Incision by UvrABC excinuclease is a step in the path to mutagenesis by psoralen crosslinks in Escherichia coli. Proc Natl Acad Sci USA. 1989;86:3982–3986. doi: 10.1073/pnas.86.11.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vollmer A C, Belkin S, Smulski D R, Van Dyk T K, LaRossa R A. Detection of DNA damage by use of Escherichia coli carrying recA′::lux, uvrA′::lux, or alkA′::lux reporter plasmids. Appl Environ Microbiol. 1997;63:2566–2571. doi: 10.1128/aem.63.7.2566-2571.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.World Health Organization. Ultraviolet radiation. Geneva, Switzerland: World Health Organization; 1994. [Google Scholar]