Abstract
Our study indicates sustained transmission (effective reproduction number, 1.3; serial interval, 4.2 days; regional doubling times, 3.3–11.4 days) of the severe acute respiratory syndrome coronavirus 2 Omicron (B.1.1.529) variant (N = 2351) in South Korea (25 November 2021–8 January 2022), implicating insufficient protection through vaccination and supporting nonpharmaceutical control measures.
Keywords: COVID-19, Korea, Omicron, reproduction number, SARS-CoV-2
Since its first identification on 24 November 2021, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron (B.1.1.529) variant has been detected in 149 countries [1]. In South Korea, the first 2 Omicron cases were identified on 25 November 2021 and on 7 December 2021; the first local outbreak was identified in Michuhol-gu, Incheon Metropolitan City (population 2.9 million) [2]. Six weeks after attempting a transition to “living with COVID-19,” coronavirus disease (COVID-19) infection-control measures were reinstated by the South Korean government on 18 December 2021, with stricter social distancing rules, such as banning gatherings of >4 fully vaccinated individuals [2]. On 13 December 2021, the Korea Disease Control and Prevention Agency reported 269 new infections (total n = 894), with a sharp increase in Omicron infections [2]. By 24 January 2022, Omicron accounted for >50% of all sequenced cases in South Korea, and replaced the Delta (B.1.617.2) variant as the dominant SARS-CoV-2 variant [2].
In late December 2021, South Korea, the United Kingdom, and Thailand shortened the standard 6-month COVID-19 booster-dose interval to 3 months to prepare for the impending Omicron wave [3]. Despite 85% of South Korea's 51 million population having been fully vaccinated, an unprecedented spike in COVID-19 cases was reported as of 1 February 2022 (>10 000 cases reported daily), and the epidemiology and transmission potential of Omicron is unclear. We investigated the transmission potential and epidemiological characteristics of Omicron in South Korea by analyzing the nationwide Omicron case data from 25 November 2021 to 8 January 2022.
METHODS
Modeling Methods
Individual-level data, including sex, age, diagnosis date, symptom-onset date, and autochthonous (local transmission)/imported case, of confirmed SARS-CoV-2 Omicron cases that were reported to the Korea Disease Control and Prevention Agency and Central Disease Control Headquarters, were analyzed. Symptom-onset dates were available for only 1116 of the 2351 Omicron cases. Therefore, we used empirical distribution of reporting delays from symptom onset to diagnosis to impute the missing dates of symptom onset for the remaining 1235 cases [4]. The epidemic curve is often described by the reporting dates, but dates of symptom onset are more suitable for analyzing the epidemic curve to reduce systematic or random error and artificial trends, such as a weekend drop, in surveillance data [5]. We reconstructed 300 epidemic curves by the symptom-onset dates and obtained the mean incidence curves of local and imported cases [4, 6]. The last 5 data points were excluded to adjust for reporting delays in the real-time analysis.
The frequency of doubling of the cumulative incidence was calculated, assuming
where and are the times at which the cumulative incidence doubled and the cumulative number of cases at the time , respectively. We assume and , and nd denotes the total number of times that the cumulative incidence doubles. Therefore, the sequence of doubling times is derived as
where j = 1, 2, 3,…, nd. Additionally, we incorporated parameter uncertainty into our analysis and obtained a 95% confidence interval (CI) using parametric bootstrapping with a Poisson error structure around the harmonic mean of the doubling times [7].
We estimated the effective reproduction number Rt using the incidence curves for imported and local cases. We initially calibrated the daily number of new local infections using a generalized growth model [8], which characterizes the ascending number of cases in an epidemic using the growth rate (r) and the deceleration of growth parameter (p) (Supplementary Methods) [8, 9], and thereby simulates the progression of incident cases at calendar time ti. We estimated the reproduction number using the following renewal equation:
assuming γ-distribution with a mean of 4.41 (standard deviation, 3.17) days for the generation interval (ρi) [7]. The local and imported incidence at calendar time ti are denoted by Ii and Ji, respectively, whereas the parameter α (0 ≤ α ≤ 1) is defined as the relative contribution of imported cases to secondary disease transmission. We estimated the Rt from 300 simulated curves by assuming a Poisson error structure, and derivegrd uncertainty bounds around the Rt curve [7].
To estimate the serial interval, we identified 202 infector–infectee pairs for whom the date of symptom onset of both the infector and infectee was available. The transmission pair was defined as including an infector who had a laboratory-confirmed infection and an infectee who reported a single epidemiological link with that infector. Using the onset dates of transmission pairs, the Epicontact R library was utilized to analyze pairwise contact between individuals and to estimate the serial interval [10]. The maximum-likelihood estimation of the normal distribution was performed to estimate the distribution of the serial interval. Furthermore, we applied the correction of the estimates of serial interval to address possible sampling bias during the early exponential growth phase of the epidemic (Supplementary Methods) [11].
This study was conducted in accordance with the Declaration of Helsinki and was exempt from ethical approval due to a waiver that was granted by the Institutional Review Board of Soongsil University (SSU-202202-HR-381-1).
RESULTS
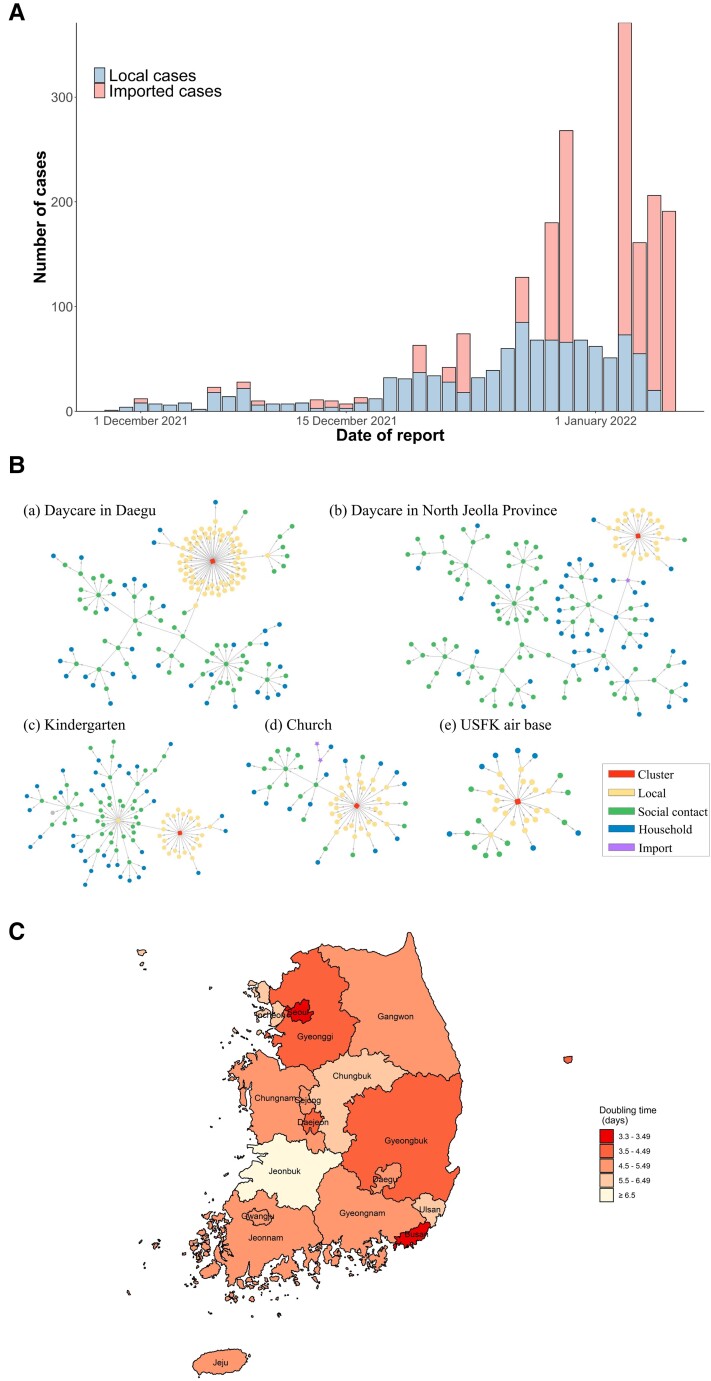
During the study period (25 November 2021–8 January 2022), 1276 imported and 1075 local Omicron cases were detected in South Korea (Figure 1A). Among the local cases, 30 (3%) were epidemiologically linked with imported cases and 845 (79%) with local cases; 200 (18%) cases were not epidemiologically linked.
Figure 1.
Epidemiological characteristics of the severe acute respiratory syndrome coronavirus 2 Omicron variant in South Korea. A, Epidemic curve of the daily Omicron cases based on the reporting date (N = 2351). B, The Omicron variant's transmission network in South Korea: transmission networks associated with the daycare in Daegu (n = 152; a), the daycare in North Jeolla province (n = 152; b), the kindergarten in North Jeolla province (n = 101; c), the church in Incheon (n = 61; d), and the United States Forces Korea (USFK) air base in North Jeolla province (n = 35; e). C, Harmonic mean of the arithmetic means of the estimated doubling time for the Omicron variant in South Korea, between 25 November 2021 and 8 January 2022, for each administrative division.
The 5 major transmission networks of Omicron infection clusters in South Korea are shown in Figure 1B; the characteristics of each cluster, as of 8 January 2022, are summarized in Table 1. The first Omicron infection cluster was associated with a church in Michuhol-gu, Incheon, South Korea (n = 61), and coincided with the country's first confirmed Omicron cases comprising a couple who returned from Nigeria and were diagnosed on 24 November 2021 [2]. Another cluster outbreak, associated with the United States Forces Korea Air Base in Gunsan, North Jeolla province (n = 35), that led to community transmission was subsequently identified. Three other Omicron infection clusters are shown in Figure 1B.
Table 1.
Demographic Characteristics of the 2351 Omicron Cases and the Major Outbreak Clusters in South Korea
| Characteristic | Total | Major Cluster Outbreaks | ||||
|---|---|---|---|---|---|---|
| Daycare in Daegu | Daycare in North Jeolla Province | Kindergarten | Church | USFK Air Base | ||
| Overall, No. | 2351 | 152 | 152 | 101 | 61 | 35 |
| Imported | 1276 (54.3) | 0 (0) | 1 (0.7) | 0 (0) | 2 (3.3) | 0 (0) |
| Pandemic period | 25 Nov 2021– 8 Jan 2022 | 24 Dec 2021– 5 Jan 2022 | 5 Dec 2021– 4 Jan 2022 | 16–30 Dec 2021 | 25 Nov– 16 Dec 2021 | 24 Dec 2021– 3 Jan 2022 |
| Sex | ||||||
| Male | 1165 (49.6) | 61 (40.1) | 71 (46.7) | 46 (45.5) | 33 (54.1) | 20 (57.1) |
| Female | 1186 (50.4) | 91 (59.9) | 81 (53.3) | 55 (54.5) | 28 (45.9) | 15 (42.9) |
| Age group, y | ||||||
| ≤7 | 181 (7.7) | 46 (30.3) | 22 (14.5) | 48 (47.5) | 10 (16.4) | 0 (0) |
| 8–19 | 281 (12.0) | 23 (15.1) | 4 (2.6) | 5 (5.0) | 6 (9.8) | 6 (17.1) |
| 20–39 | 1129 (48.0) | 40 (26.3) | 50 (32.9) | 22 (21.8) | 24 (39.3) | 16 (45.7) |
| 40–59 | 565 (24.0) | 42 (27.6) | 42 (27.6) | 17 (16.8) | 15 (24.6) | 11 (31.4) |
| 60–74 | 170 (7.2) | 1 (0.7) | 24 (15.8) | 9 (8.9) | 6 (9.8) | 2 (5.7) |
| ≥75 | 25 (1.1) | 0 (0) | 10 (6.6) | 0 (0) | 0 (0) | 0 (0) |
| Clinical features, asymptomatic | 459 (19.5) | 11 (7.2) | 25 (16.4) | 29 (28.7) | 9 (14.8) | 6 (17.1) |
| COVID-19 vaccination status | ||||||
| 1 dose | 72 (3.1) | … | … | … | … | … |
| 2 doses | 1249 (53.1) | … | … | … | … | … |
| 3 doses | 121 (3.5) | … | … | … | … | … |
Data are presented as No. (%) unless otherwise indicated.
Abbreviations: COVID-19, coronavirus disease 2019; USFK, United States Forces Korea.
Analysis of the 202 infector–infectee transmission pairs indicated that the mean serial interval fitted to a normal distribution was 4.2 (95% CI, 3.8–4.5) days (Supplementary Table 1). The mean effective reproductive number as of 8 January 2022 was approximately 1.3 (95% CI, 1.2–1.4), with a growth rate (r) of 0.3 (95% CI, .2–.4) and deceleration of growth parameter (p) of 0.9 (95% CI, .8–1.0); this indicates the subexponential growth dynamics of the Omicron variant in South Korea (Supplementary Figures 1 and 2).
During the study period, the harmonic mean of the arithmetic means of doubling times, which were estimated from the cumulative incidence, ranged from 3.3 (95% CI, 2.7–4.4) days in Busan to 11.4 (95% CI, 7.5–15.9) days in North Jeolla province, and was 4.0 (95% CI, 3.9–4.1) days for South Korea. The harmonic means of the doubling times in Busan, Seoul, Gyeonggi province, and North Gyeongsang province were <4.0 days (Figure 1C).
DISCUSSION
In February 2022, a surge of SARS-CoV-2 Omicron cases was reported worldwide, even in countries with relatively high COVID-19 vaccine coverage; the basic reproduction number in the absence of control measures was >8 [12]. Our study quantifies the transmission potential of Omicron in South Korea and explains the recent unprecedented national surge in COVID-19 cases. Using reconstructed onset dates for the first reported cases, we estimated an effective reproduction number of 1.3, which is lower than previous estimates in other settings (1.9−3.2) [13, 14], but clearly indicates sustained Omicron transmission in South Korea. The serial interval of Omicron infections was estimated as 4.2 days (Supplementary Figure 3), which is longer than that of previous estimates (2.8–2.9 days) in South Korea [15, 16] but shorter than that in Spain (4.8 days) [17].
Furthermore, 1370 of the 2351 (58%) Omicron infections occurred in individuals who had received at least 2 vaccine doses, and this finding concurs with reports of Omicron's immune escape contributing to the lower efficacy of COVID-19 vaccines as well as a greater replication rate compared with the Delta variant [18]. Insufficient prevention of symptomatic Omicron infections among otherwise healthy individuals who had received 3 doses of COVID-19 messenger RNA vaccines has been demonstrated, and emphasizes the need for nonpharmaceutical control measures [18].
Imported cases accounted for more than half of the reported cases (1276/2351 [54%]), and thus contributed significantly to infection transmission in the early phase of the Omicron wave. These findings validate the international travel–related interventions that were implemented by the Korean government to control the outbreak.
This study had some limitations. First, we could not account for unreported cases of asymptomatic or mild Omicron infections. Second, our results were based on a statistical reconstruction of epidemic curves using symptom-onset dates [11]. Third, given the relatively high COVID-19 vaccine coverage and low prevalence of COVID-19 in the pre-Omicron era in South Korea, our estimate of transmissibility is nongeneralizable to resource-limited countries. Nonetheless, the findings highlight the importance of maintaining nonpharmaceutical control measures in South Korea to mitigate the impact of the Omicron variant.
Supplementary Material
Acknowledgments
We are grateful to the Korea Disease Control and Prevention Agency and the Central Disease Control Headquarters for providing access to nationwide COVID-19 case data for research purposes.
Author contributions . E. S.: conceptualization, methodology, validation, formal analysis, investigation, writing–original draft, supervision, project administration, and funding acquisition. D. K. and T. K.: data curation. W. C. and Y. S.: data visualization. All authors: writing–review and editing.
Patient consent . This study did not include factors necessitating patient consent.
Financial support . This work was supported by the National Research Foundation of Korea funded by the Korean government (MSIT) (grant number 2018R1C1B6001723 to E. S.); and the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education (grant number 2021R1A6A1A10044154 to E. S., Y. S., and W. C.).
Potential conflicts of interest . All authors: No reported conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Contributor Information
Eunha Shim, Department of Mathematics, Soongsil University, Seoul, Republic of Korea.
Wongyeong Choi, Department of Mathematics, Soongsil University, Seoul, Republic of Korea.
Donghyok Kwon, Division of Public Health Emergency Response Research, Korea Disease Control and Prevention Agency, Osong, Republic of Korea.
Taeyoung Kim, Division of Public Health Emergency Response Research, Korea Disease Control and Prevention Agency, Osong, Republic of Korea.
Youngji Song, Department of Mathematics, Soongsil University, Seoul, Republic of Korea.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
References
- 1. World Health Organization . Enhancing response to Omicron SARS-CoV-2 variant. Geneva, Switzerland: WHO; 2022. [PubMed] [Google Scholar]
- 2. Korea Disease Control and Prevention Agency . The updates of COVID-19 in Republic of Korea. 2022. https://www.kdca.go.kr/board/board.es?mid=a20501010000&bid=0015. Accessed 8 January 2022.
- 3. Beasley D. As Omicron threatens a global surge, some countries shorten COVID-19 booster timelines. 2021. https://www.reuters.com/business/healthcare-pharmaceuticals/omicron-threatens-global-surge-some-countries-shorten-covid-19-booster-timelines-2021-12-20/. Accessed 14 May 2022.
- 4. Chowell G, Blumberg S, Simonsen L, Miller MA, Viboud C. Synthesizing data and models for the spread of MERS-CoV, 2013: key role of index cases and hospital transmission. Epidemics 2014; 9:40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hennessee I, Clennon JA, Waller LA, Kitron U, Bryan JM. Considerations for improving reporting and analysis of date-based COVID-19 surveillance data by public health agencies. Am J Public Health 2021; 111:2127–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shim E, Tariq A, Choi W, Lee Y, Chowell G. Transmission potential and severity of COVID-19 in South Korea. Int J Infect Dis 2020; 93:339–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shim E, Tariq A, Chowell G. Spatial variability in reproduction number and doubling time across two waves of the COVID-19 pandemic in South Korea, February to July, 2020. Int J Infect Dis 2021; 102:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Viboud C, Simonsen L, Chowell G. A generalized-growth model to characterize the early ascending phase of infectious disease outbreaks. Epidemics 2016; 15:27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chowell G, Viboud C. Is it growing exponentially fast? Impact of assuming exponential growth for characterizing and forecasting epidemics with initial near-exponential growth dynamics. Infect Dis Model 2016; 1:71–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nagraj VP, Jombart T, Randhawa N, Sudre B, Campbell F, Crellen T. Epicontacts: handling, visualisation and analysis of epidemiological contacts. 2017. https://cran.r-project.org/web/packages/epicontacts/index.html. Accessed 30 May 2022. [DOI] [PMC free article] [PubMed]
- 11. Xin H, Wong JY, Murphy C, et al. The incubation period distribution of coronavirus disease 2019: a systematic review and meta-analysis. Clin Infect Dis 2021; 73:2344–52. [DOI] [PubMed] [Google Scholar]
- 12. Liu Y, Rocklov J. The effective reproduction number for the Omicron SARS-CoV-2 variant of concern is several times higher than Delta [manuscript published online ahead of print 9 March 2022]. J Travel Med 2022. doi:10.1093/jtm/taac037. [Google Scholar]
- 13. Ito K, Piantham C, Nishiura H. Relative instantaneous reproduction number of Omicron SARS-CoV-2 variant with respect to the Delta variant in Denmark. J Med Virol 2021; 94:2265–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim D, Ali AT, Kim S, et al. Estimation of serial interval and reproduction number to quantify the transmissibility of SARS-CoV-2 Omicron variant in South Korea. Viruses 2022; 14:533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee JJ, Choe YJ, Jeong H, et al. Importation and transmission of SARS-CoV-2 B.1.1.529 (Omicron) variant of concern in Korea, November 2021. J Korean Med Sci 2021; 36:e346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Song JS, Lee J, Kim M, et al. Serial intervals and household transmission of SARS-CoV-2 Omicron variant, South Korea, 2021. Emerg Infect Dis 2022; 28:756–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Del Águila-Mejía J, Wallmann R, Calvo-Montes J, Rodríguez-Lozano J, Valle-Madrazo T, Aginagalde-Llorente A. Secondary attack rate, transmission and incubation periods, and serial interval of SARS-CoV-2 Omicron variant, Spain [manuscript published online ahead of print 7 April 2022]. Emerg Infect Dis 2022. doi: 10.3201/eid2806.220158. [DOI] [PMC free article] [PubMed]
- 18. Kuhlmann C, Mayer CK, Claassen M, et al. Breakthrough infections with SARS-CoV-2 Omicron despite mRNA vaccine booster dose. Lancet 2022; 399:P625–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.