Abstract
Lactobacillus delbrueckii subsp. lactis ACA-DC 178, which was isolated from Greek Kasseri cheese, produces a cell-wall-bound proteinase. The proteinase was removed from the cell envelope by washing the cells with a Ca2+-free buffer. The crude proteinase extract shows its highest activity at pH 6.0 and 40°C. It is inhibited by phenylmethylsulfonyl fluoride, showing that the enzyme is a serine-type proteinase. Considering the substrate specificity, the enzyme is similar to the lactococcal PI-type proteinases, since it hydrolyzes β-casein mainly and α- and κ-caseins to a much lesser extent. The cell-wall-bound proteinase from L. delbrueckii subsp. lactis ACA-DC 178 liberates four main peptides from β-casein, which have been identified.
Lactic acid bacteria are fastidious organisms. For optimal growth, they are dependent on the presence of small peptides and free amino acids in the culture medium. Since the concentration of free amino acids and peptides present in milk is not sufficient for the growth of lactic acid bacteria, these bacteria must be able to degrade milk proteins; this is the basis of their utility in the dairy industry. Casein degradation and subsequent utilization of the degradation products requires a complex proteolytic system consisting of proteinases, peptidases, and amino acid and peptide carriers.
The proteolytic system of lactococci has been the subject of intensive biochemical and genetic research. Their cell-wall-bound proteinases have been divided into two main groups: the PI-type proteinases, which hydrolyze predominantly β-casein, and, the PIII-type proteinases, which degrade α- and κ-caseins in addition to β-casein. Proteinases showing a specificity pattern intermediate between the PI and PIII types have also been described. The intermediate-type proteinases cleave β-casein in a manner similar to that of the PI type but are also able to hydrolyze αs1-casein. In all lactococcal strains studied to date, proteinase genes are located on plasmids of different sizes. In close proximity to the proteinase gene prtP is another gene, named prtM. This gene encodes a membrane located lipoprotein, which is essential for activation of the proteinase (15, 25).
In contrast to the lactococcal proteolytic system, limited information is available on the proteolytic activity of lactobacilli. The most intensively studied proteinase system among the lactobacilli is that of Lactobacillus casei, which shows many parallels to that of lactococci (4, 6, 10, 12, 13, 24). The proteinase from Lactobacillus plantarum has similar properties to the L. casei enzyme, although there is conflicting evidence on its specificity (4, 12). A multiplicity of proteinase forms have been reported for Lactobacillus delbrueckii subsp. bulgaricus (5, 17). Finally, the cell-wall-associated serine proteinase of Lactobacillus helveticus has similar biochemical properties to the lactococcal proteinase PrtP (21, 30, 31).
Cell wall proteinases of lactic acid bacteria play an important role in cheese technology, since they contribute to the initial degradation of milk casein and to flavor defects due to the production of bitter peptides. Lactobacilli, both thermophilic and mesophilic, are widely involved in the production and ripening of many types of cheeses. However, to date, characterization of the peptides produced during casein degradation has been described only for L. helveticus (21, 30, 31) and to a lesser extent for L. casei (7).
In this paper, we describe the characterization of a cell-wall-associated proteinase from L. delbrueckii subsp. lactis ACA-DC 178 and provide information on the nature of peptides liberated from β-casein by this proteinase.
MATERIALS AND METHODS
Strains and growth conditions.
L. delbrueckii subsp. lactis ACA-DC 178 was isolated from Greek Kasseri cheese. It was subcultured twice in 10% (wt/vol) skim milk at 37°C; final growth was carried out at 37°C for 24 h on milk agar containing 8% (wt/vol) skim milk and 1.5% (wt/vol) agar. Lactococcus lactis MG 1363, containing plasmid pGKV552, was kindly provided by Jan Kok, University of Groningen, Groningen, The Netherlands. Plasmid pGKV552 harbored the Lactococcus lactis subsp. cremoris Wg2 prtP and prtM genes (9). Lactococcus lactis MG1363 was grown at 30°C in M17 supplemented with glucose (0.5%, wt/vol) and erythromycin (5 μg/ml).
Preparation of the cell wall extract.
Cells were collected and washed three times with 50 mM phosphate buffer (pH 7.0) containing 20 mM CaCl2. Washed cells were resuspended in 50 mM phosphate buffer (pH 7.0) (10 μl of buffer per μg [wet weight] of biomass) and incubated for 2 h at 30°C. The supernatant obtained after centrifugation (12,000 × g at 4°C for 5 min) was designated the cell wall extract. The release of lactate dehydrogenase (LDH) during incubation of cells was considered an indication of intracellular enzyme release. LDH was assayed by the method of Thomas (28).
DNA preparation and hybridization.
Plasmid DNA from L. delbrueckii subsp. lactis ACA-DC 178 was isolated by three different methods (1, 19, 26). Chromosomal DNA was isolated by the method of Leenhouts et al. (20). For Southern blotting experiments, DNA (3 μg) was digested with various restriction enzymes (BamHI, EcoRI, HindIII, BamHI-EcoRI, and HindIII-EcoRI), fractionated on a 0.8% (wt/vol) agarose gel, and transferred onto a Gene Screen Plus nylon membrane (NEN Research Products) by the protocol of Southern as modified by Chomczynski and Qasba (2). DNA was labeled with the digoxigenin DNA labeling and detection kit (Boehringer Mannheim). Probe labeling, hybridization conditions, and washing steps were performed as specified by the manufacturer.
Casein hydrolysis.
A whole-cell suspension (15 μl) or cell wall extract (15 μl) was incubated with 15 μl of casein solution (α-, β-, or κ-casein; 4 mg/ml) in 30 μl of 50 mM phosphate buffer (pH 6.0) at 40°C for 4, 8, or 24 h. For the whole-cell suspension, the reaction was stopped by centrifugation (12,000 × g for 5 min); the supernatant obtained was mixed in a 1:1 ratio with solubilization buffer (13), heated for 5 min at 100°C, and analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (12.5% acrylamide gels) by the method of Laemmli (16). For the cell wall extract, the reaction was stopped by addition of 60 μl of 12% trichloroacetic acid (TCA); after 10 min at room temperature, the sample was centrifuged (12,000 × g for 5 min). The sediment was dissolved in 120 μl of solubilization buffer and analyzed by SDS polyacrylamide gel electrophoresis as described above. The free amino acids and peptides liberated in the supernatant were determined by the o-phthaldialdehyde method (3).
Effect of pH on proteinase activity.
Cell wall extract (15 μl) was incubated with 15 μl of β-casein solution (4 mg/ml) for 8 h at 40°C in 30 μl of buffer of various pH values (pH 4 and 5 in 50 mM acetate buffer; pH 6 and 7 in 50 mM phosphate buffer; pH 8 and 9 in 50 mM borate buffer). The reaction was stopped by addition of 60 μl of 12% TCA; after 10 min at room temperature, the sample was centrifuged (12,000 × g for 5 min). The sediment was dissolved in 120 μl of solubilization buffer and analyzed by SDS-polyacrylamide gel electrophoresis as described above. The free amino acids and peptides liberated in the supernatant were determined as described above.
Effect of temperature on proteinase activity.
Cell wall extract (15 μl) was incubated with 15 μl of β-casein solution (4 mg/ml) for 8 h in 50 mM phosphate buffer (pH 6.0) at various temperatures (from 10 to 50°C). The reaction was stopped by addition of 60 μl of 12% TCA; after 10 min at room temperature, the sample was centrifuged (12,000 × g for 5 min). The sediment was dissolved in 120 μl of solubilization buffer and analyzed by SDS-polyacrylamide gel electrophoresis as described above. The free amino acids and peptides liberated in the supernatant were determined as described above.
Effect of inhibitors on proteinase activity.
Cell wall extract (15 μl) was incubated with 15 μl of β-casein solution (4 mg/ml) for 8 h at 40°C in 30 μl of 50 mM phosphate buffer (pH 6.0) containing various inhibitors at a final concentration of 10 mM. The inhibitors studied were EDTA, 1,10-phenanthroline, diisopropylofluorophosphate (DFP), phenylmethylsulfonyl fluoride (PMSF), iodoacetamide, and N-ethylmaleimide. PMSF and 1,10-phenanthroline (50 mM in isopropanol) were diluted to 10 mM in 50 mM phosphate buffer (pH 6.0) and then incubated with the enzyme as above. The effect of isopropanol on the enzyme was also investigated. The reaction was stopped by addition of 60 μl of 12% TCA; after 10 min at room temperature, the sample was centrifuged (12,000 × g for 5 min). The sediment was dissolved in 120 μl of solubilization buffer and analyzed by SDS-polyacrylamide gel electrophoresis as described above. The free amino acids and peptides liberated in the supernatant were determined as described above.
HPLC.
Cell wall extract (150 μl) was incubated with 150 μl of β-casein solution (20 mg/ml in 50 mM phosphate buffer [pH 6.0]) at 40°C for 8, 24, and 48 h. The reaction was stopped by addition of 12.5 μl of 25% trifluoroacetic acid (TFA) (1% final concentration). After 10 min at room temperature, the sample was centrifuged (12,000 × g for 5 min). The supernatant was filtered through a 0.22-μm-pore-size membrane filter (Millipore, Bedford, Mass.) and subjected to high-performance liquid chromatography (HPLC) analysis.
The 1% TFA-soluble fraction, which corresponded to noncasein fragments, was analyzed by reversed-phase HPLC on a Gilson instrument (Gilson Medical Electronics, Middleton, Wis.). The peptides were separated on a Nucleosil C18 column (4.6 mm [inner diameter] by 250 mm; Macherey-Nagel, Dueren, Germany) and detected by absorbance at 220 nm. The initial solvent A was 0.1% TFA in water. Peptides were eluted by a linear gradient from solvent A to solvent B: acetonitrile-water-TFA (600:399:1, vol/vol/vol) at 1 ml/min.
Peptide identification.
The amino acid sequences of the peptides were determined on a 476 Sequenator (Perkin-Elmer, Applied Biosystems Division) with on-line HPLC analysis of the phenylthiohydantoin derivatives. Mass spectrometry was performed with electrospray ionization either on a VG Bio-Q triple-quadrupole mass spectrometer (Micromass, Altrincham, United Kingdom) or on a hybrid-quadrupole time-of-flight Q-TOF instrument (Micromass, Wythenshawe, United Kingdom). Samples, 1:10 dilutions of the HPLC fractions in 0.1% formic acid–50% acetonitrile in water, were submitted by flow injection in a conventional electrospray source. Collision-induced fragmentation experiments were performed with argon as the collision gas.
RESULTS
The ability of L. delbrueckii subsp. lactis ACA-DC 178 to hydrolyze α-, β-, and κ-casein was tested after induction of the proteinase in milk and on milk-agar plates. Since LDH activity determined in the crude cell wall extract did not exceed 6% of the total cell LDH activity, it was concluded that the proteolytic activity detected was due to the action of a cell-wall-bound proteinase.
Plasmid DNA preparations obtained with three different protocols and separated on 0.8% agarose gel, contained only a faint band corresponding to chromosomal DNA. This suggested that the proteinase gene of L. delbrueckii subsp. lactis ACA-DC 178 was probably located in the chromosomal DNA. Nevertheless, both plasmid DNA preparations and chromosomal DNA were digested with restriction enzymes and used in hybridization experiments under both high- and low-stringency conditions. Two probes, the 4.2-kb BamHI-HindIII fragment of the prtP gene and the 0.884-kb ClaI-HindIII fragment of the prtM gene were used. Both chromosomal and plasmid DNA produced faint signals, which could be considered as background hybridization.
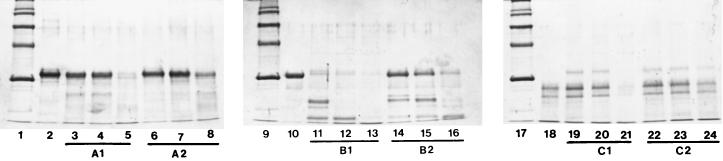
As shown in Fig. 1, the crude proteinase extract, obtained by washing the cells in a Ca2+-free buffer, hydrolyzed β-casein predominantly and α- and κ-casein at a much lower rate. The same results were obtained when instead of the cell wall extract, whole cells were acting on all three casein fractions. As expected, the proteolytic activity of the whole cells was much higher than that of the crude proteinase extract (Fig. 1). The results were also confirmed by photometric determination of the free amino groups in the respective noncasein fragments (data not shown).
FIG. 1.
The action of whole cells (A1) and cell wall crude proteinase (A2) on α-casein (lane 2) after 4 h (lanes 3 and 6), 8 h (lanes 4 and 7), and 24 h (lanes 5 and 8), the action of whole cells (B1) and cell wall crude proteinase (B2) on β-casein (lane 10) after 4 h (lanes 11 and 14), 8 h (lanes 12 and 15), and 24 h (lanes 13 and 16), and the action of whole cells (C1) and cell wall crude proteinase (C2) on κ-casein (lane 18) after 4 h (lanes 19 and 22), 8 h (lanes 20 and 23), and 24 h (lanes 21 and 24) are shown. Molecular mass markers are 116, 97, 66, 45, and 29 kDa (from top to the bottom) (lanes 1, 9, and 17). The reaction mixtures were incubated at 40°C and pH 6.0. Electrophoretic conditions were 12.5% acrylamide gels in 0.025 M Tris HCl–0.19 M glycine buffer (pH 8.3).
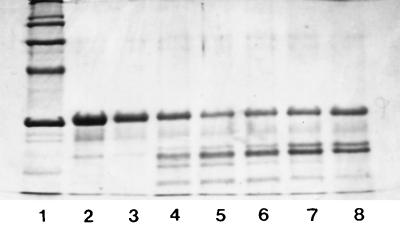
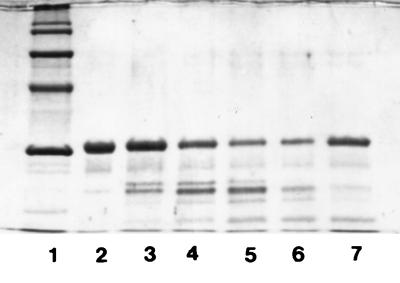
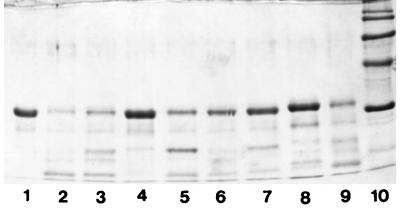
The action of the crude proteinase on β-casein was tested under various assay conditions. According to the results obtained, the crude proteinase showed maximum activity at pH 6.0 (Fig. 2) and at 40°C (Fig. 3). The crude proteinase was strongly inhibited by PMSF; however, the effect of DFP on the enzyme was weaker. N-Ethylmaleimide had no effect on enzyme activity; in contrast, inhibition was observed when iodoacetamide was used. The crude proteinase was not significantly influenced by EDTA or 1,10-phenanthroline (Fig. 4). The results were also confirmed by photometric determination of the free amino groups in the respective noncasein fragments (data not shown).
FIG. 2.
Action of the cell wall crude proteinase on β-casein at various pH values. Lanes: 1, Molecular weight markers; 2, β-casein; 3, pH 4; 4, pH 5; 5, pH 6; 6, pH 7; 7, pH 8; 8, pH 9. Reaction mixtures were incubated at 40°C for 8 h. Electrophoretic conditions were as in Fig. 1.
FIG. 3.
Action of the cell wall crude proteinase on β-casein at various temperatures. Lanes: 1, molecular weight markers; 2, β-casein; 3, 10°C; 4, 20°C; 5, 30°C; 6, 40°C; 7, 50°C. Reaction mixtures were incubated at pH 6.0 for 8 h. Electrophoretic conditions were as in Fig. 1.
FIG. 4.
Action of the cell wall proteinase on β-casein in the presence of various inhibitors (10 mM). Lanes: 1, β-casein; 2, proteolysis assay in the absence of inhibitor; 3, proteolysis assay in the absence of inhibitor but in the presence of 3 μl of isopropanol; 4, PMSF; 5, DFP; 6, EDTA; 7, 1,10-phenanthroline; 8, iodoacetamide; 9, N-ethylmaleimide; 10, molecular mass markers (116, 97, 66, 45, and 29 kDa from top to bottom). Reaction mixtures were incubated at 40°C and pH 6.0 for 8 h. Electrophoretic conditions were as in Fig. 1.
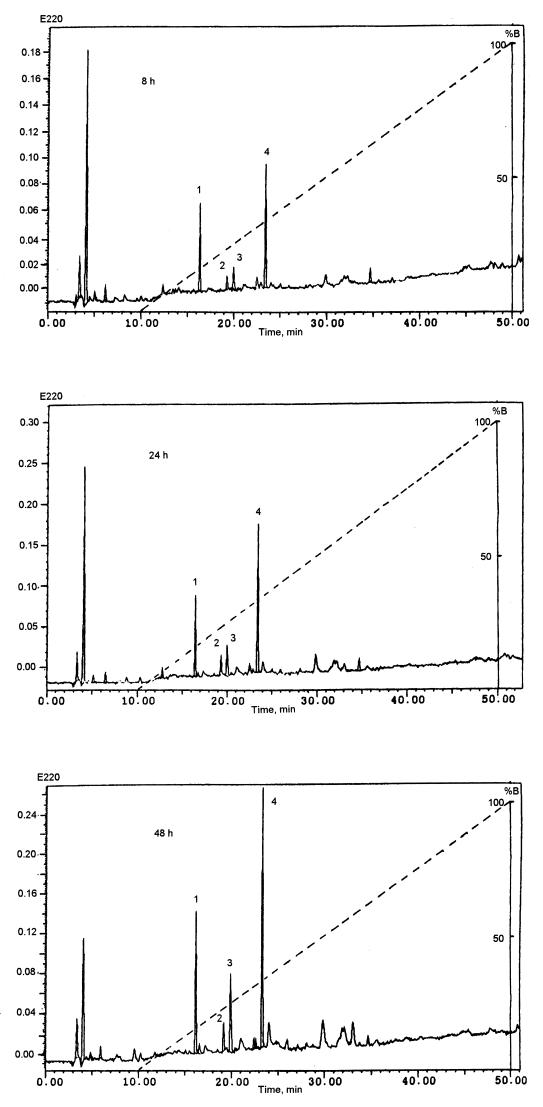
When β-casein hydrolysate was studied by HPLC, four main peptides (peptides 1, 2, 3, and 4) appeared after 8 h of reaction; they were present on the chromatograms in increasing concentrations after 24 h and 48 h of incubation (Fig. 5). The liberation rate of peptide 2 remained constant, while peptides 1, 3, and 4 were more rapidly produced after 24 h of incubation; the liberation rates of peptides 1 and 4 were similar to each other. Subsequently, the peptides were identified by sequence analysis and mass spectrometry. Their sizes varied from 5 to 10 residues. All four peptides were located in the C-terminal part of β-casein (Fig. 6); peptide 1 at Pro186 to Tyr193, peptide 2 at Phe157 to Ser161, peptide 3 at His145 to Thr154, and peptide 4 at Ser166 to Gln175. The peptide masses were determined as follows: peptide 1, 964.04 Da (theoretical, 964.17 Da); peptide 2, 573.80 Da (theoretical, 573.28 Da); peptide 3, 1,150.24 Da (theoretical, 1,150.59 Da); and peptide 4, 1,081.39 Da (theoretical, 1,081.60 Da).
FIG. 5.
Separation by reversed-phase HPLC of the peptides from the 1% TFA soluble fraction obtained after 8, 24, and 48 h of hydrolysis of β-casein by the cell wall proteinase of L. delbrueckii subsp. lactis ACA-DC 178.
FIG. 6.
Localization of the cell wall proteinase-generated peptides 1 to 4 in the amino acid sequence of β-casein.
DISCUSSION
The importance of the cell-wall-bound proteinases of lactic acid bacteria has been discussed in a number of papers over the last several years (15, 25). Most experimental data were obtained with lactococcal enzymes. Literature data on the biochemical properties, genetics, and specificity patterns of extracellular proteolytic enzymes in lactobacilli are, however, limited. This paper describes the characterization of a cell-wall-associated proteinase from L. delbrueckii subsp. lactis ACA-DC 178 and provides information about the nature of peptides liberated from β-casein by this proteinase.
L. delbrueckii subsp. lactis ACA-DC 178 produced a cell-wall-bound proteinase, which hydrolyzed β-casein predominantly and α- and κ-caseins at a much lower rate. This suggested that the enzyme resembled that of the lactococcal PI-type proteinases (8, 18, 27). The crude proteinase showed maximum activity at pH 6.0 and at 40°C. In this aspect, the L. delbrueckii subsp. lactis ACA-DC 178 proteinase was similar to the lactococcal enzymes (18) but also to enzymes described for other lactobacilli (5, 7, 17, 30).
In contrast to the lactococcal proteinase genes, which are all plasmid-borne (14), the L. delbrueckii subsp. lactis ACA-DC 178 proteinase gene seemed to be located in the chromosomal DNA. Furthermore, the absence of a positive hybridization signal even under low-stringency conditions when using the 4.2-kb BamHI-HindIII fragment of the prtP gene and the 0.884-kb ClaI-HindIII fragment of the prtM gene as probes was an indication that at the nucleotide level and for the regions tested, the proteinase of the L. delbrueckii subsp. lactis ACA-DC 178 had low homology to the Lactococcus lactis subsp. cremoris Wg2 proteinase.
Although enzyme inhibition by DFP was weaker than that by PMSF, the strong inhibition by the latter indicated that the enzyme belonged to the serine group of proteinases. This means that the enzyme resembled the lactococcal proteinases (15, 25). Among the two sulfhydryl blockers tested, N-ethylmaleimide had no effect on the enzyme activity. In contrast, strong inhibition, although less strong than with PMSF, was observed when iodoacetamide was used. Although the specificity of iodoacetamide at high concentrations (10 mM in this study) decreases, the observed inactivation might be due to the presence of sulfhydryl groups close to the active center of the proteinase. The involvement of sulfhydryl groups in the enzyme mechanism has been reported for the proteinase of L. delbrueckii subsp. bulgaricus (17). Low inhibition of the L. helveticus L89 proteinase by E-64, a thiol proteinase inhibitor, was reported by Martin-Hernandez et al. (21). Limited inhibition was observed in the presence of both EDTA and 1,10-phenanthroline, suggesting that the proteinase was not a metalloenzyme. It has been reported that EDTA partially inhibits the activity of lactobacillus proteinases when these enzymes are released from the cells without using it (21, 23, 30).
Four main peptides were produced by the action of the proteinase on β-casein. They were all located in the C-terminal part of the molecule, which is in agreement with the literature data about lactococcal but also lactobacillus proteinases (11, 22, 29–31). Three bonds cleaved by the proteinase of L. delbrueckii subsp. lactis ACA-DC 178, namely, Leu165-Ser166, Gln175-Lys176, and Tyr193-Gln194, belong to the most highly recognized β-casein bonds by other proteinases. The other five cleavage sites detected, Met144-His145, Thr154-Val155, Met156-Phe157, Ser161-Val162, and Met185-Pro186, were less expected to be cleaved as these sites are not frequently observed to be cleaved by other proteinases studied to date (15).
Identification of the peptides produced during β-casein hydrolysis by lactobacilli has been only described for two L. helveticus strains (30, 31). The major degradation products of the low-molecular-weight peptides were virtually identical to each other and similar to those produced by the lactococcal proteinases. In the present study, one of the eight cleavage sites described, namely, the Gln175-Lys176 bond, has also been reported as such for the proteinase of L. helveticus. In contrast, the L. delbrueckii subsp. lactis ACA-DC 178 proteinase hydrolyzed the Met144-His145 bond, which was not attacked by the L. helveticus enzyme. All the other cleavage sites described in this study were slightly shifted in comparison to those reported for L. helveticus. Finally, in contrast to the L. helveticus proteinase, no cleavage site could be detected in the part of the β-casein molecule upstream of residue 144.
Thermophilic lactobacilli, including L. delbrueckii subsp. lactis, are involved in the production of various types of cheeses (e.g., Swiss-type cheeses). The biochemical and genetic characterization of their proteolytic system will help elucidate their contribution in cheese ripening. This work provides information about the cell-wall-bound PI-type proteinase of L. delbrueckii subsp. lactis ACA-DC 178. The enzyme exhibits similar biochemical properties to those reported previously for lactococcal and lactobacillus enzymes. Still, further genetic information is necessary to improve our knowledge of its functional properties.
REFERENCES
- 1.Casey M G, Jimeno J. Lactobacillus delbrueckii subsp. lactis plasmids. Neth Milk Dairy J. 1989;43:279–286. [Google Scholar]
- 2.Chomczynski P, Qasba P K. Alkaline transfer of DNA to plastic membrane. Biochem Biophys Res Commun. 1984;122:340–344. doi: 10.1016/0006-291x(84)90480-7. [DOI] [PubMed] [Google Scholar]
- 3.Church F C, Swaisgood H E, Porter D H, Catignani G L. Spectrophotometric assay using o-phthaldialdehyde for determination of proteolysis in milk and isolated milk proteins. J Dairy Sci. 1983;66:1219–1227. [Google Scholar]
- 4.El Soda M, Desmazeaud M J, Le Bars D, Zevaco C. Cell-wall associated proteinases in Lactobacillus casei and Lactobacillus plantarum. J Food Prot. 1986;49:361–365. doi: 10.4315/0362-028X-49.5.361. [DOI] [PubMed] [Google Scholar]
- 5.Ezzat N, Zevaco C, El Soda M, Gripon J C. Partial purification and characterization of a cell wall associated proteinase from Lactobacillus bulgaricus. Milchwissenschaft. 1987;42:95–97. [Google Scholar]
- 6.Ezzat N, El Soda M, El Shafei H. The cell bound proteinase system of Lactobacillus casei—purification and characterization. Int J Food Microbiol. 1988;6:327–332. doi: 10.1016/0168-1605(88)90026-8. [DOI] [PubMed] [Google Scholar]
- 7.Fernandez de Palencia P, Pelaez C, Romero C, Martin-Hernandez M C. Purification and characterization of the cell wall proteinase of Lactobacillus casei subsp. casei IFPL 731 isolated from raw goat’s milk cheese. J Agric Food Chem. 1997;45:3401–3405. [Google Scholar]
- 8.Haandrikman A J. Ph.D. thesis. Groningen, The Netherlands: University of Groningen; 1990. [Google Scholar]
- 9.Haandrikman A J, Meesters R, Laan H, Konings W L, Kok J, Venema G. Processing of the lactococcal extracellular serine proteinase. Appl Environ Microbiol. 1991;57:1899–1904. doi: 10.1128/aem.57.7.1899-1904.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holck A, Naes H. Cloning, sequencing and expression of the gene encoding the cell-envelope associated proteinase from Lactobacillus paracasei subsp. paracasei NCDO151. J Gen Microbiol. 1992;138:1353–1364. doi: 10.1099/00221287-138-7-1353. [DOI] [PubMed] [Google Scholar]
- 11.Juillard V, Laan H, Kunji E R S, Jeronimus-Stratingh C M, Bruins A P, Konings W N. The extracellular PI type proteinase of Lactococcus lactis hydrolyzes β-casein into more than one hundred different oligopeptides. J Bacteriol. 1995;177:3472–3478. doi: 10.1128/jb.177.12.3472-3478.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khalid N M, Marth E H. Proteolytic activity by strains of Lactobacillus plantarum and Lactobacillus casei. J Dairy Sci. 1990;73:3068–3076. [Google Scholar]
- 13.Kojic M, Fira D, Banina A, Topisirovic L. Characterization of the cell wall bound proteinase of Lactobacillus casei HN14. Appl Environ Microbiol. 1991;57:1753–1757. doi: 10.1128/aem.57.6.1753-1757.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kok J. Genetics of the proteolytic system of lactic acid bacteria. FEMS Microbiol Rev. 1990;87:15–42. doi: 10.1111/j.1574-6968.1990.tb04877.x. [DOI] [PubMed] [Google Scholar]
- 15.Kunji E R S, Mierau I, Hagting A, Poolman B, Konings W N. The proteolytic system of lactic acid bacteria. Antonie Leeuwenhoek. 1996;70:187–221. doi: 10.1007/BF00395933. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Laloi P, Atlan D, Gilbert C, Portalier R. Cell wall associated proteinase of Lactobacillus delbrueckii subsp. bulgaricus CNRZ 397: differential extraction, purification and properties of the enzyme. Appl Microbiol Biotechnol. 1991;36:196–204. doi: 10.1007/BF00164419. [DOI] [PubMed] [Google Scholar]
- 18.Law J, Haandrikman A. Proteolytic enzymes of lactic acid bacteria. Int Dairy J. 1997;7:1–11. [Google Scholar]
- 19.Leenhouts K J, Kok J, Venema G. Campel-like intergration of heterologous plasmid DNA into the chromosome of Lactococcus lactis subsp. lactis. Appl Environ Microbiol. 1989;55:394–400. doi: 10.1128/aem.55.2.394-400.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leenhouts K J, Kok J, Venema G. Stability of integrated plasmids in the chromosome of Lactococcus lactis. Appl Environ Microbiol. 1990;56:2726–2735. doi: 10.1128/aem.56.9.2726-2735.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin-Hernandez M C, Alting A C, Exterkate F A. Purification and characterization of the mature, membrane-associated cell-envelope proteinase of Lactobacillus helveticus L89. Appl Microbiol Biotechnol. 1994;40:828–834. [Google Scholar]
- 22.Monnet V, Le Bars D, Gripon J C. Specificity of a cell wall proteinase from Streptococcus lactis NCDO763 towards bovine β-casein. FEMS Microbiol Lett. 1986;36:127–131. [Google Scholar]
- 23.Naes H, Chrzanowska J, Blom H. Partial purification and characterization of a cell wall bound proteinase from Lactobacillus casei. Food Chem. 1991;42:65–79. [Google Scholar]
- 24.Naes H, Nissen-Meyer J. Purification and N-terminal amino acid sequence determination of the cell wall bound proteinase from Lactobacillus paracasei subsp. paracasei. J Gen Microbiol. 1992;138:313–318. doi: 10.1099/00221287-138-2-313. [DOI] [PubMed] [Google Scholar]
- 25.Pritchard G G, Coolbear T. The physiology and biochemistry of the proteolytic system in lactic acid bacteria. FEMS Microbiol Rev. 1993;12:179–206. doi: 10.1111/j.1574-6976.1993.tb00018.x. [DOI] [PubMed] [Google Scholar]
- 26.Somkuti G A, Steinberg D H. General method for plasmid DNA isolation from thermophilic lactic acid bacteria. J Biotechnol. 1986;3:323–332. [Google Scholar]
- 27.Tan P S T, Poolman B, Konings W N. Proteolytic enzymes of Lactococcus lactis. J Dairy Res. 1993;60:269–286. doi: 10.1017/s0022029900027606. [DOI] [PubMed] [Google Scholar]
- 28.Thomas T D. Tagatose-1,6-diphosphate activation of lactate dehydrogenase from Streptococcus cremoris. Biochem Biophys Res Commun. 1975;63:1035–1042. doi: 10.1016/0006-291x(75)90673-7. [DOI] [PubMed] [Google Scholar]
- 29.Visser S, Robben A J P M, Slangen C J. Specificity of a cell-envelope-located proteinase (PIII type) from Lactococcus lactis subsp. lactis AM1 in its action on bovine β-casein. Appl Microbiol Biotechnol. 1991;35:477–483. doi: 10.1007/BF00169753. [DOI] [PubMed] [Google Scholar]
- 30.Yamamoto N, Akino A, Takano T. Purification and specificity of a cell wall associated proteinase from Lactobacillus helveticus CP790. J Biochem. 1993;114:740–745. doi: 10.1093/oxfordjournals.jbchem.a124247. [DOI] [PubMed] [Google Scholar]
- 31.Zevaco C, Gripon J C. Properties and specificity of a cell wall proteinase from Lactobacillus helveticus. Lait. 1988;68:393–408. [Google Scholar]