Abstract
The genetic biodiversity of Clostridium botulinum type E strains was studied by pulsed-field gel electrophoresis (PFGE) with two macrorestriction enzymes (SmaI-XmaI and XhoI) and by randomly amplified polymorphic DNA (RAPD) analysis with two primers (OPJ 6 and OPJ 13) to characterize 67 Finnish isolates from fresh fish and fishery products, 15 German isolates from farmed fish, and 10 isolates of North American or North Atlantic origin derived mainly from different types of seafood. The effects of fish species, processing, and geographical origin on the epidemiology of the isolates were evaluated. Cluster analysis based on macrorestriction profiles was performed to study the genetic relationships of the isolates. PFGE and RAPD analyses were combined and resulted in the identification of 62 different subtypes among the 92 type E isolates analyzed. High genetic biodiversity among the isolates was observed regardless of their source. Finnish and North American or North Atlantic isolates did not form distinctly discernible clusters, in contrast with the genetically homogeneous group of German isolates. On the other hand, indistinguishable or closely related genetic profiles among epidemiologically unrelated samples were detected. It was concluded that the high genetic variation was probably a result of a lack of strong selection factors that would influence the evolution of type E. The wide genetic biodiversity observed among type E isolates indicates the value of DNA-based typing methods as a tool in contamination studies in the food industry and in investigations of botulism outbreaks.
A bacterial species is an assemblage of isolates which originated from a common ancestor population in which a steady generation of genetic divergence has resulted in clones (25). Clones are defined as genetically related isolates that are indistinguishable from each other by a variety of molecular typing methods (9). Genetic biodiversity arises from random nonlethal mutations that accumulate over time. If biodiversity within a bacterial species is wide enough, isolates can be characterized with DNA-based typing methods, and the results can be utilized for several applications. From a food microbiology perspective, these applications include the investigation of foodborne outbreaks and contamination routes of products and the establishment and maintenance of hazard analysis and critical control point (HACCP) systems at food manufacturing plants (25). From the standpoint of taxonomy, molecular subtyping of bacterial isolates may clarify the classification of bacterial species (33).
Very little is known about the genetic biodiversity of the foodborne pathogen Clostridium botulinum. The taxonomy of the species, based on botulinum neurotoxin (BoNT) production and phenotypic characteristics, is currently under reconsideration (8). The diagnostics of botulism outbreaks has traditionally concentrated on the detection of botulinum neurotoxin from clinical and food samples (12). Therefore, no effort has been made to develop methods that are able to characterize C. botulinum isolates to the subspecies level. Recently, pulsed-field gel electrophoresis (PFGE) (13, 22), randomly amplified polymorphic DNA analysis (RAPD) (18), repetitive element sequence-based PCR (18), and ribotyping (15) have been described as tools for genomic analysis of C. botulinum group I and II strains. Of these methods, PFGE and RAPD seem to be the most suitable for subtyping C. botulinum group II strains due to their high reproducibility and discriminatory power. The methods also seem to complement each other in terms of typeability, speed of performance, and ease of interpretation.
In recent surveys performed in Finland, high C. botulinum type E prevalences were detected in fish farm, freshwater, and Baltic Sea sediment samples (14, 16, 17). No other serotypes were found. The type E prevalence in raw fish varied from 10 to 40%, depending on the fish species studied (19). At the retail level, 5% of the vacuum-packaged and 3% of the air-packaged fishery products were positive for type E spores (19). The data clearly indicated that current fish processing practices are insufficient to eliminate botulinal spores from raw fish. A cluster of recent outbreaks in northern Europe (2, 3, 20, 27) has demonstrated the increased botulism risk associated with fishery products. Because they have a long shelf life, many products are distributed nationwide and internationally, enabling the spread of contaminating foodborne pathogens to a very large geographical area and thereby complicating the investigation of a potential foodborne botulism outbreak (23). More information about the epidemiology and biodiversity of type E is urgently needed to provide a basis for identification of critical control points and establishment of controlling practices in HACCP systems of fish manufacturing plants. Moreover, the diagnostics and investigation of human foodborne botulism outbreaks should also be updated to meet the requirements of modern epidemiological investigations with the ability to reliably confirm the link between a patient and vehicle.
The present study was performed to characterize C. botulinum type E strains isolated from fresh fish and from different types of fishery products and seafood by PFGE and RAPD analysis. The main objectives were to examine the biodiversity of type E strains and to evaluate the effects of fish species, harvest location, and processing on the epidemiology of the organism. We also performed a cluster analysis of type E isolates based on both SmaI-XmaI and XhoI macrorestriction profiles (MRPs) to study the genetic relationships of the isolates.
MATERIALS AND METHODS
C. botulinum type E strains.
Fifty-six type E strains were isolated from fresh fish caught or farmed at 21 different locations in Finland. Eleven isolates were derived from Finnish fishery product samples produced by six different manufacturers. Fifteen strains isolated from samples of German farmed fresh fish were included in the study. A detailed description of the origins of the strains studied is given in Table 1. The sampling and isolation of strains were performed during the period 1994 to 1996, as described previously (14, 19).
TABLE 1.
Distribution of C. botulinum type E subtypes generated by PFGE with two macrorestriction enzymes (SmaI-XmaI and XhoI) and RAPD with two arbitrary primers (OPJ 6 and OPJ 13) among different fresh fish, fishery product, and seafood samples
| Source of sample | No. of isolates | Subtype(s) as determined bya:
|
|
|---|---|---|---|
| PFGE | RAPD | ||
| Finnish fresh fish | |||
| Rainbow trout intestines (Oncorhynchus mykiss) | 20 | IV, V, VI, VII, VIII (12), XVIII, XXIV, XLIV, XLV | I (2), II (8), III (4), XI, XII, XIII, XXI, XXV, XXVI |
| Rainbow trout surfaceb | 11 | I, II, XXIV, XXVI, XXVII, XXVIII (3), XXIX, XXX, XXXI | I (7), II, VI, VIII, IX |
| Burbot intestines (Lota lota) | 5 | X, XII, XV, XVI, XLIX | IV, XV, XVII, XVIII, XXX |
| Burbot surfaceb | 5 | XI, XIII, XIV, XVI, XVII | I, II, IV (2), XVI |
| Burbot roe | 1 | III | II |
| Whitefish intestines (Coregonus lavaretus) | 1 | XLIII | III |
| Whitefish surfaceb | 1 | III | X |
| Baltic herring (whole) (Clupea harengus membras) | 9 | XXXII, XXXIII (5), XXXIV, XXXV, XLVIII | I (7), IV, XXIX |
| Vendace (whole) (Coregonus albula) | 3 | XIX, XX, XXII | V (3) |
| Finnish fishery products | |||
| Cold-smoked rainbow trout | 2 | XXIV, UTc | IV, VI |
| Frozen rainbow trout roe | 2 | XXV, XLVI | I, XXVII |
| Frozen whitefish roe | 3 | IX, XXI (2) | IV, XIV, XIX |
| Hot-smoked whitefishd | 2 | XXIII, XLII | VII, XX |
| Hot-smoked salmond | 1 | XLVIII | XXVIII |
| Hot-smoked vendace | 1 | XXXVI | XXII |
| German fresh fish | |||
| Chub intestines (Leuciscus cephalus) | 2 | XXXVII, XXXVIII | III, XXIII |
| Chub surfaceb | 7 | XL (7) | III (7) |
| Bream intestines (Abramis brama) | 3 | XXXIX, XL (2) | III (3) |
| Bream surfaceb | 1 | XLI | XXIV |
| Brook trout surfaceb (Salvelinus fontinals) | 1 | XL | III |
| Brown trout surfaceb (Salmo trutta m. fario) | 1 | XL | III |
| North American or North Atlantic sources | |||
| Seafood | 7 | L (2), LI, LII, LIV (2), LV | XXXI (2), XXXII, XXXIII, XXXV, XXXVI, XXXVII |
| Dried mutton | 1 | LIV | XXXVI |
| Not known | 2 | LIII, LVI | XXXIV, XXXVIII |
Numbers of multiple isolates representing the same subtype are indicated within parentheses.
Surface samples consisted of skin, gills, fins, and peritoneum.
UT, untypeable.
Raw material of the product of Canadian origin.
Ten strains from our C. botulinum type E collection (31-2570, RS-1, Beluga, C-51, C-60, C-94, 250, 36208, KA-2, and 4062) were also included in the analysis. These strains were of North American and North Atlantic origin and were isolated mainly from different types of seafood over a period of 60 years. A more detailed description on the origin of each strain has been given previously (13).
Cultivation of strains.
Anaerobic egg yolk agar plates (1) were incubated for 3 days, and lipase-positive colonies were inoculated into tryptone-peptone-glucose-yeast (TPGY) extract (Difco, Detroit, Mich.) broth (10). All cultures were incubated at 26°C in an anaerobic cabinet with an internal atmosphere of 85% N2, 10% CO2, and 5% H2 (MK III; Don Whitley Scientific Ltd., Shipley, United Kingdom). The species and serotypes of C. botulinum type E cultures were ascertained by a BoNT-specific PCR assay (17). Dynazyme II DNA polymerase (Finnzymes, Espoo, Finland) and a 96-well DNA thermal cycler (MJ Research, Watertown, Mass.) were used for PCR amplifications. The size of the amplified PCR product was determined by agarose gel electrophoresis with comparison to standard DNA fragments (DNA molecular weight marker VI; Boehringer Mannheim GmbH, Mannheim, Germany).
DNA preparations.
Agarose-embedded DNA intended for PFGE analysis was isolated according to the method of Maslow et al. (26), with the modifications described by Hielm et al. (13). Briefly, overnight TPGY cultures in late log phase were chilled on ice and resuspended in PIV (10 mM Tris [pH 7.5], 1 M NaCl) containing 3.5 to 4.0% (vol/vol) formaldehyde solution and left on ice for 1 h. Cell suspensions were mixed with an equal amount of 2% (wt/vol) low-melting-temperature agarose (InCert agarose; FMC Bioproducts, Rockland, Maine) and cast in GelSyringe dispensers (New England Biolabs, Beverly, Mass.). The plugs were lysed for 2 h in lysis solution (6 mM Tris [pH 7.5], 1 M NaCl, 100 mM EDTA [pH 8.0], 0.5% Brij 58, 0.2% deoxycholate, 0.5% sodium lauroyl sarcosine, 20 μg of RNase/ml, 1 mg of lysozyme/ml, 10 U of mutanolysin/ml) with gentle shaking at 37°C. The isolation was completed with a 1-h wash in ESP (0.5 M EDTA [pH 8.0], 10% sodium lauroyl sarcosine, 100 μg of proteinase/ml) at 50°C, followed by phenylmethylsulfonyl fluoride inactivation of proteinase K.
Conventional DNA isolation for RAPD analysis was performed according to the method of Marmur (24) with the modifications previously described by Hyytiä et al. (18). Briefly, cells were resuspended in TE (10 mM Tris-HCl [pH 7.5], 1 mM EDTA [pH 8.0]) solution containing 8 mg of lysozyme per ml and 170 IU of mutanolysin per ml. The mixture was incubated at 37°C for 2 h with gentle shaking. Complete lysis was obtained by adding 50 μg of proteinase K per ml and 0.8% (vol/vol) sodium dodecyl sulfate and incubating the mixture with gentle shaking at 60°C for 1 h. RNA was removed by adding 165 μg of RNase. The purity and yield of the DNA were determined spectrophotometrically, and the DNA was diluted in TE buffer to a final concentration of 5 ng/μl.
The DNAs of all strains were isolated at least twice from separate colonies with both in situ and conventional isolation methods, and replicate runs by PFGE and RAPD were performed to filter out any variations.
Restriction enzyme digestions and PFGE.
Restriction endonuclease digestion of the agarose-embedded C. botulinum DNA was performed as described by the manufacturer by using three rare-cutting restriction enzymes (SmaI, XhoI, and XmaI [New England Biolabs]). All samples were electrophoresed on a Gene Navigator system (Pharmacia Biotech AB, Uppsala, Sweden) with a hexagonal electrode through a 1% (wt/vol) agarose gel (SeaKem Gold; FMC Bioproducts) in a 0.5× TBE buffer (Amresco, Solon, Ohio). Switch times were ramped from 1 to 24 s over 22 h at 14°C and 6 V/cm. The Low Range PFG marker (New England Biolabs) was used for fragment size determination. The gels were stained for 30 min in 1 liter of running buffer containing 0.5 mg of ethidium bromide and destained in running buffer until the appropriate contrast for either standard photography (28) or digital imaging with the Alpha Imager 2000 documentation system (Alpha Innotech, San Leandro, Calif.) was obtained.
RAPD analysis.
RAPD analysis was performed by using Ready-To-Go RAPD Analysis Beads (Pharmacia Biotech) as described by the manufacturer, with factors affecting reproducibility being carefully observed (32). The sample volume of 25 μl contained 10 ng of DNA and 25 pmol of a single oligonucleotide primer. All strains were analyzed by using the arbitrary primers OPJ 6 and OPJ 13 (Operon Inc., Alameda, Calif.). Amplifications were performed in a PTC-100 thermal cycler (MJ Research) for 45 cycles of 1 min at 95°C, 1 min at 36°C, and 2 min at 72°C, with a 5-min initial denaturation at 95°C and a 5-min final extension at 72°C. Amplification products were electrophoresed in 2.0% agarose gels (MetaPhor Agarose; FCM BioProducts) in 1× TAE buffer (Amresco) at 80 V for 5 h. The gels were stained for 20 min in 1.5 liters of distilled water containing 0.5 mg of ethidium bromide and destained for 40 min in distilled water before photography by standard methods (28). DNA molecular weight marker VI (Boehringer Mannheim GmbH) was used as a fragment size marker.
Fingerprint pattern analysis.
SmaI-XmaI and XhoI MRPs in the molecular size range of 50 to 350 kb were analyzed with GelCompar software (version 4.0; Applied Maths, Kortrijk, Belgium). The similarity between all MRPs was expressed as a Dice coefficient correlation and was determined with the equation SD = [2nAB/nA + nB] × 100, where nAB is the number of matched fragments and nA + nB is the total number of fragments in profiles A and B (4). The position tolerance for band matching was set at 1.4% of the total length of the pattern (300 kb), with no increase. The arrangement of SmaI-XmaI and XhoI MRPs into dendrograms was accomplished by the unweighted pair group method with arithmetic averages (UPGMA). The genotypes resulting from MRP analyses were clustered at a similarity level of 96% with SmaI-XmaI digests and 90% with XhoI digests, by referring to possible epidemiological relatedness according to the guidelines set out by Tenover et al. (31). These similarity levels corresponded roughly to a three-band difference.
The banding patterns generated by RAPD were interpreted visually. Patterns with two or more fragment size differences were classified as belonging to different clones.
RESULTS
Macrorestriction digests and cluster analysis.
There was a distinct difference in the capabilities of the restriction enzymes SmaI-XmaI and XhoI to digest C. botulinum type E DNA. Of the 30 isolates that were undigestible by SmaI, 13 were digested by XmaI, an isoschizomer of SmaI with the same restriction site but with a different cleaving point. Seventeen isolates (18%) were undigestible by both SmaI and XmaI, with 13 of these isolates being of German origin. Only one isolate (K-36, which was isolated from a fishery product) was undigestible by XhoI, and it was also not digested by SmaI-XmaI.
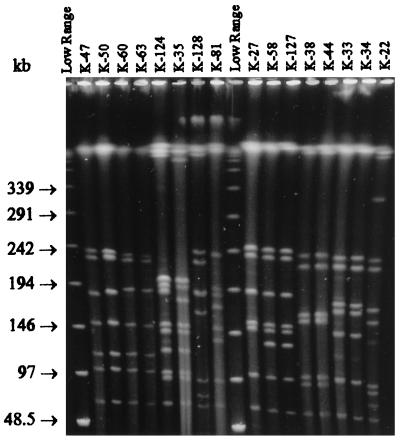
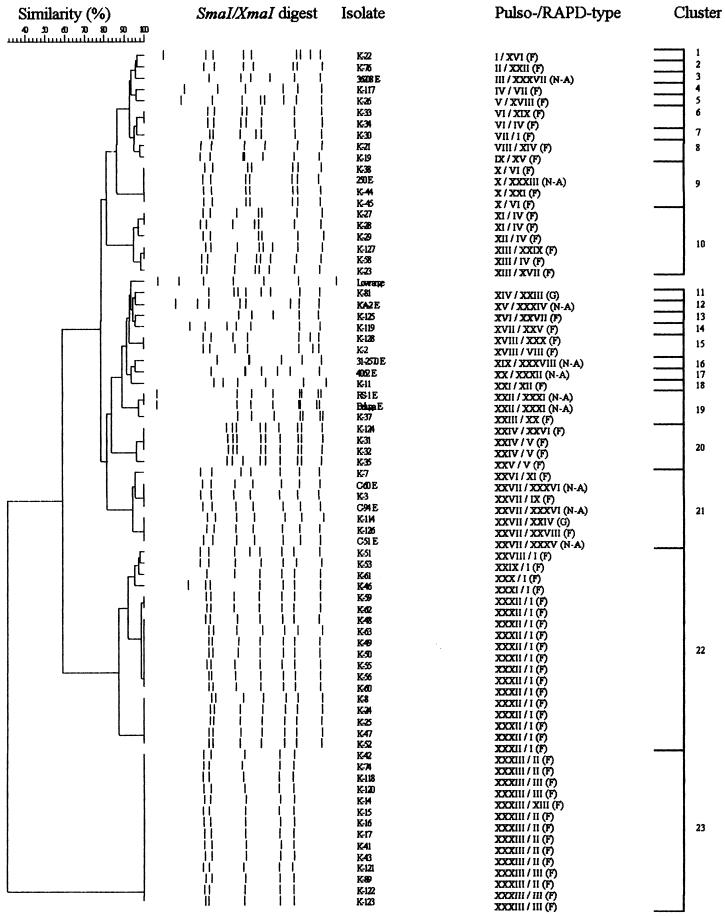
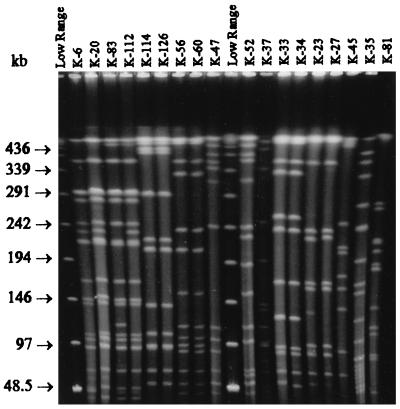
The SmaI-XmaI digests (Fig. 1) of the 75 typeable strains generated 33 different MRPs (I to XXXIII), forming 23 clusters at a similarity level of 96% (Fig. 2). The discriminatory power of XhoI was distinctly better: 51 different MRPs (I to LI) forming 37 clusters at a similarity level of 90% were detected when the XhoI digests (Fig. 3) of the 91 typeable strains were analyzed (Fig. 4). Combining the results of the SmaI-XmaI and XhoI digests increased the discriminatory power only slightly, yielding 56 different subtypes. The reproducibility of the banding patterns of different DNA lots was excellent with each enzyme used. Extensive genetic biodiversity between the strains isolated from different fish species as well as among isolates from one fish species was observed (Table 1). In some cases, strains isolated from intestinal and surface samples of the same fish (K-21 and K-22) (Fig. 2 and 4) had different MRPs. In both dendrograms, the 10 typeable Finnish fishery product isolates (K-19, K-33, K-34, K-37, K-38, K-46, K-76, K-117, K-125, and K-126) clustered together mainly with other epidemiologically unrelated isolates. When the results of both macrorestrictions were combined, three main PFGE types were observed: clone VIII, which included 12 Finnish rainbow trout isolates that were digested by XmaI but not by SmaI; clone XL, which was composed of 11 German isolates; and clone XXXIII, which was composed of 5 Finnish isolates from Baltic herring. Indistinguishable PFGE types were also found in several epidemiologically unrelated sample pairs, such as in two different fish species and in raw fish and prepared product.
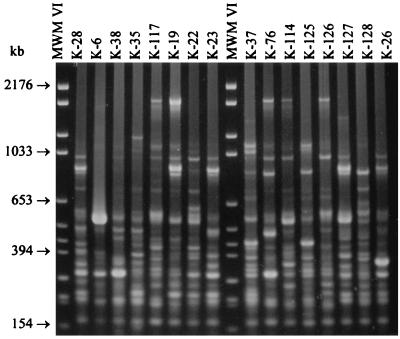
FIG. 1.
SmaI digests of 16 C. botulinum type E isolates from fresh fish and fishery products. Lanes labeled “Low Range” contain a low-range PFG marker. The pulse time was ramped from 1 to 24 s for 22 h at 6 V/cm and 14°C.
FIG. 2.
Dendrogram of 75 C. botulinum type E isolates based on SmaI-XmaI MRPs. Schematic MRPs are shown, and a low-range PFG marker is included as an additional entry. Similarity analysis was performed by using the Dice coefficient, and clustering was done by UPGMA. RAPD types of each isolate are also included. Abbreviations (capital letters in parentheses): F, Finnish isolate; G, German isolate; N-A, North American or North Atlantic isolate.
FIG. 3.
XhoI digests of 18 C. botulinum type E isolates from fresh fish and fishery products. Lanes labeled “Low Range” contain a low-range PFG marker. The pulse time was ramped from 1 to 24 s for 22 h at 6 V/cm and 14°C.
FIG. 4.
Dendrogram of 91 C. botulinum type E isolates based on XhoI MRPs. Schematic MRPs are shown, and a low-range PFG marker is included as an additional entry. Similarity analysis was performed by using the Dice coefficient, and clustering was done by UPGMA. RAPD types of each isolate are also included. Abbreviations (capital letters in parentheses): F, Finnish isolate; G, German isolate; N-A, North American or North Atlantic isolate.
The Finnish type E isolates also exhibited a high level of local geographical biodiversity (Table 2) in macrorestriction analysis. Isolates originating in fish from lakes, fish farms, and manufacturing plants of interior Finland appeared to exhibit more extensive genetic variation than isolates from fish from the sea and from coastal Finland. Strains with differing genetic profiles could be isolated from fish originating in the same farm and from products of the same manufacturing plant. On the other hand, clonal MRPs were detected in isolates from diverse geographical locations in Finland (PFGE type VIII, Table 2). Additionally, some Finnish isolates (K-6, K-20, K-54, and K-126) also belonged to the same clusters (Fig. 4, clusters 1 and 6) as the German isolates, which were genetically very homogeneous. Similarly, the North American isolate 250 E clustered together with some Finnish isolates in both macrorestrictions (Fig. 2, cluster 9; Fig. 4, cluster 28).
TABLE 2.
Distribution of C. botulinum type E subtypes generated by PFGE with two macrorestriction enzymes (SmaI-XmaI and XhoI) and RAPD with two arbitrary primers (OPJ 6 and OPJ 13) according to the catching area or location of a fish farm or a manufacturing plant
| Catching area or farm location | No. of isolates | Subtypes as determined bya:
|
|
|---|---|---|---|
| PFGE | RAPD | ||
| Gulf of Finland | 40 | III, VII, VIII (7), IX, X, XI, XII, XIII, XIV, XV, XVI (2), XVII, XXI (2), XXIV (2), XXV, XXVI, XXVIII (2), XXIX, XXX, XXXI, XXXII, XXXIII (5), XXXIV, XXXV, XLVI, XLVIII | I (14), II (10), IV (5), VI, XIII, XIV, XV, XVI, XVII, XVIII, XIX, XXI, XXVII, XXIX |
| Gulf of Bothnia | 11 | III, VIII (5), XVIII, XXII, XXVII, XXVIII, XLV | I (3), II, III (4), V, X, XXVI |
| Interior of Finland | 16 | I, II, IV, V, VI, XIX, XX, XXIII, XXIV, XXXVI, XLII, XLIII, XLIV, XLVII, XLIX, UTb | I, III, IV, V (2), VI, VII, VIII, IX, XI, XII, XX, XXII, XXV, XXVIII, XXX |
| Germany | 15 | XXXVII, XXXVIII, XXXIX, XL (11), XLI | III (13), XXIII, XXIV |
| North America or North Atlantic | 10 | L (2), LI, LII, LIII, LIV (3), LV, LVI | XXXI (2), XXXII, XXXIII, XXXIV, XXXV, XXXVI (2), XXXVII, XXXVIII |
Numbers of multiple isolates representing the same subtype are indicated within the parentheses.
UT, untypeable.
RAPD analysis.
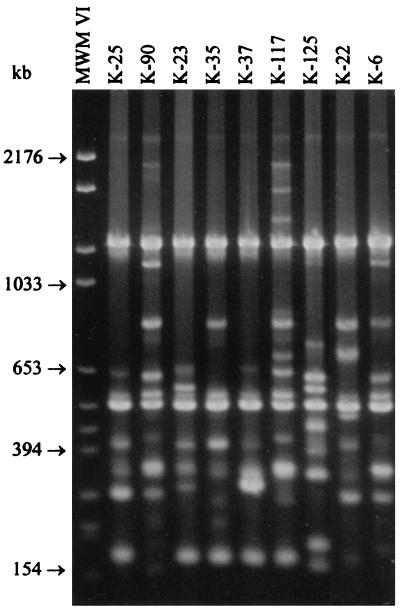
All 92 strains were typeable by RAPD with both primers used. Interpretation of RAPD banding patterns was difficult due to a large number of small fragments and frequent occurrence of faint bands (Fig. 5 and 6). Therefore, RAPD fingerprints were not used for the computed cluster analysis. Primers OPJ 6 and OPJ 13 generated 27 and 19 different banding patterns, respectively. Despite the occurrence of faint bands, the reproducibility of the banding patterns between different DNA lots was good. When the results obtained with both primers were combined, 38 different RAPD types (I to XXXVIII) were observed (Table 1). Fifty-six isolates (61%) belonged to the five most prevalent RAPD types (I to V), which were distributed throughout different types of samples. In five cases, the discriminatory power of RAPD was superior to that of PFGE. For example, strains K-33 and K-34 were isolated from the same package of frozen salted whitefish roe and appeared to be clonal according to the SmaI and XhoI MRPs (Fig. 2 and 4). However, a two-band difference was reproducibly observed in fingerprints generated by primer OPJ 13. When the results of PFGE with two restriction enzymes and RAPD analysis with two primers were combined, 62 different genetic profiles were detected among the 92 type E isolates analyzed.
FIG. 5.
RAPD banding patterns of 16 C. botulinum type E isolates generated by primer OPJ 6. Lanes labeled MWM VI contain molecular weight marker VI.
FIG. 6.
RAPD banding patterns of nine C. botulinum type E isolates generated by primer OPJ 13. The first lane contains molecular weight marker VI (MWM VI).
DISCUSSION
The 92 C. botulinum type E strains characterized in the present study each belonged to one of three main groups: Finnish isolates, German isolates, and North American or North Atlantic isolates. In general, high genetic biodiversity among the isolates was found regardless of the isolation source or geographical origin, with the exception of the genetically homogeneous group of German isolates. North American or North Atlantic isolates mainly grouped in the middle of both dendrograms. These ten strains, most of which were isolated several decades ago, belonged to seven different clusters in the XhoI dendrogram. Some of these clusters either contained Finnish isolates or showed close relatedness with clusters containing Finnish and German isolates. Characterization of the Finnish strains suggested that processing of fishery products did not seem to favor the survival of any particular genotype, while all 11 isolates had differing genetic profiles, despite the fact that some of the isolates originated in the same manufacturing plant or same product package. Moreover, isolates originating in narrow epidemiological fresh fish sources, such as rainbow trout from one farm or burbots caught from small catching areas, had high levels of genetic divergence. On the other hand, isolates that were clonal by all typing methods could be isolated from catching areas or farms that were distant from each other. These results raise intriguing questions about the evolution of type E. As an environmental organism, type E is in general not exposed to strong selection factors that would influence its genetic evolution and favor the survival of certain genotypes. Additionally, high mutational capacity might facilitate the adjustment of strains into several different ecological niches that exist in the aquatic environment. As a consequence, a high level of genetic biodiversity has evolved. More characterization of isolates and the creation of an international data bank for C. botulinum fingerprints are needed before any accurate estimations about the worldwide prevalence of different type E genotypes can be made.
In contrast to the wide genetic divergence observed among Finnish and North American or North Atlantic strains, the German isolates were found to be genetically homogeneous. All these isolates originated in the same fish farm among four different fish species. There are no surveys available about the prevalence of type E in German freshwater sediments and in wild fish. However, in a small-scale study performed in the early 1970s, Bach et al. (5) were able to identify C. botulinum type E in mud and fish samples originating from a German fish farm. Fish farming has been shown to maintain a reserve of botulinal spores, despite the low natural contamination levels in the surrounding environment (7). The few strains that are introduced into farms with fish derived from outside the farm or with fish feed become dominant, resulting in low genetic variation. Additionally, at this particular farm, the practice of recycling water from one fish pond to another probably enhanced the spreading of this dominant genotype.
The large number of isolates undigestible by SmaI was a problem in this study. Hielm et al. (14) suspected CG methylation as a cause of nondigestion and addressed the problem by changing from SmaI to its isoschizomer XmaI. However, only 13 of 30 isolates undigestible by SmaI in the present study were digested by XmaI. These isolates were clonal both by XmaI (Fig. 2, cluster 23) and XhoI (Fig. 4, cluster 3) macrorestriction. The 13 German isolates undigestible by SmaI-XmaI were all closely related by XhoI macrorestriction (Fig. 4, cluster 1) and were clonal by RAPD analysis (Fig. 4, RAPD type III). Interestingly, three of the four Finnish isolates untypeable by SmaI-XmaI (K-6, K-20, and K-54) belonged to the same XhoI cluster as the German strains. Additionally, this cluster was related at a similarity level of 82% to XhoI cluster 3, which contained the XmaI-digested isolates. The close genetic relatedness of these epidemiologically unrelated isolates suggests that there is a specific genetic basis for nondigestion by SmaI and to some extent XmaI. Samore et al. (30) described a similar genetic relatedness between C. difficile isolates that were untypeable by SmaI but typeable by restriction enzyme analysis and RAPD. They suggested that DNA degradation by endonucleases was the cause for nondigestion. DNase activity has indeed been recognized in some clostridial species (6, 21). However, in this study it appeared that only one strain was untypeable due to active DNases, because it was not digested by either SmaI-XmaI or XhoI. The rest of the isolates untypeable by SmaI were still digested by XhoI, which proved that the DNA was not severely degraded. Instead, it appears that the strains represented by these particular genotypes possess a specific DNA modification system, possibly methylation, that rendered the DNA undigestible by SmaI. Since the worldwide prevalence of this genotype is unknown, it is not advisable to use SmaI as the only restriction enzyme in the characterization of type E isolates.
Of the individual typing protocols used in this study, PFGE with XhoI macrorestriction showed the highest discriminating power. Slightly better discrimination was achieved when the results of XhoI MRPs and RAPD patterns generated by primers OPJ 6 and OPJ 13 were combined, with 60 different subtypes being observed among 92 isolates. The use of SmaI-XmaI did not increase the discrimination. The distinct advantages of RAPD analysis were 100% typeability and rapid performance. The incongruity in the results of the two typing methods for some sets of isolates reflects the fact that the molecular bases of PFGE and RAPD are very different. As a consequence, the discriminating power of the methods can vary considerably for particular sets of isolates (29). Therefore, for the characterization of type E strains, we recommend an approach which combines XhoI MRPs and RAPD analysis performed with two primers. The complicated interpretation of RAPD patterns due to a substantial variation in the intensity of individual fragments was also described by Samore et al. (30), when they characterized C. difficile strains by using RAPD. To overcome this problem, we strongly suggest that all isolates be analyzed twice from separate colonies and that only bands which are detected reproducibly be included in the fingerprint.
The wide genetic biodiversity observed among C. botulinum type E isolates and the use of molecular typing methods introduce new strategies into the investigation of epidemiological problems caused by type E. Subtyping of isolates facilitates contamination studies both at fish farms and in the food industry. Critical control points can be recognized, and thereafter appropriate measures can be taken to control the high-risk production phases to ensure the safety of products with respect to type E. Molecular subtyping of isolates is also the key to more accurate and reliable investigation of botulism outbreaks. RAPD analysis can be used to rapidly characterize the isolates from patient samples and suspected foods with confirmation by the better discriminating, albeit more time-consuming, PFGE. It is advisable to analyze multiple isolates from one food sample, because a single sample may harbor strains with different genetic profiles. Genotyping all type E isolates associated with botulism outbreaks would facilitate the creation of an international database for type E fingerprints and thereby help in tracking international outbreaks (11).
ACKNOWLEDGMENTS
This work was supported by grants from the Academy of Finland, Technology Development Centre, the Walter Ehrström Foundation, and the Finnish Veterinary Foundation.
We are grateful to Kirsi Ristkari, Maria Stark, and Sirkku Ekström for their invaluable technical assistance.
REFERENCES
- 1.American Public Health Association. Compendium of methods for the microbiological examination of foods. 3rd ed. Washington, D.C: American Public Health Association; 1992. [Google Scholar]
- 2.Anonymous. Botulism. Epid-aktuellt (Stockholm, Sweden) 1991;14:9. [Google Scholar]
- 3.Anonymous. Fallbericht: Botulismus nach dem Verzehr von geräucherten Lachsforellen. Epidemiol Bull (Münster, Germany) 1998;4:20. [Google Scholar]
- 4.Applied Maths. GelCompar version 4.0 manual. Kortrijk, Belgium: Applied Maths; 1996. [Google Scholar]
- 5.Bach R, Wenzel S, Müller-Prasuhn G, Gläsker H. Teichforellen als Träger von Clostridium botulinum and Ursache von Botulismus. Arch Lebensmittelhyg. 1971;22:107–112. [Google Scholar]
- 6.Blaschek H P, Klacik M A. Role of DNase in recovery of plasmid DNA from Clostridium perfringens. Appl Environ Microbiol. 1984;48:178–181. doi: 10.1128/aem.48.1.178-181.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cann D C, Taylor L Y, Hobbs G. The incidence of Clostridium botulinum in farmed trout raised in Great Britain. J Appl Bacteriol. 1975;39:331–336. doi: 10.1111/j.1365-2672.1975.tb00579.x. [DOI] [PubMed] [Google Scholar]
- 8.Collins M D, East A K. Phylogeny and taxonomy of the food-borne pathogen Clostridium botulinum and its neurotoxins. J Appl Microbiol. 1998;84:5–17. doi: 10.1046/j.1365-2672.1997.00313.x. [DOI] [PubMed] [Google Scholar]
- 9.Farber J M. An introduction to the hows and whys of molecular typing. J Food Prot. 1996;59:1091–1101. doi: 10.4315/0362-028X-59.10.1091. [DOI] [PubMed] [Google Scholar]
- 10.Food and Agricultural Organization. Manual of food quality control. Vol. 12. Rome, Italy: United Nations Food and Agricultural Organization; 1991. [Google Scholar]
- 11.Griffin P M. Abstracts of the 98th General Meeting of the American Society for Microbiology 1998. Washington, D.C: American Society for Microbiology; 1998. The role of subtyping by pulsed-field gel electrophoresis (PFGE) in outbreak identification and investigation, abstr. Y-164; p. 27. [Google Scholar]
- 12.Hatheway C L. Botulism: the present status of the disease. Curr Top Microbiol Immunol. 1995;195:55–75. doi: 10.1007/978-3-642-85173-5_3. [DOI] [PubMed] [Google Scholar]
- 13.Hielm S, Björkroth J, Hyytiä E, Korkeala H. Genomic analysis of Clostridium botulinum group II by pulsed-field gel electrophoresis. Appl Environ Microbiol. 1998;64:703–708. doi: 10.1128/aem.64.2.703-708.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hielm S, Björkroth J, Hyytiä E, Korkeala H. Prevalence of Clostridium botulinum in Finnish trout farms: PFGE typing reveals extensive genetic diversity between type E isolates. Appl Environ Microbiol. 1998;64:4161–4167. doi: 10.1128/aem.64.11.4161-4167.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hielm, S., J. Björkroth, E. Hyytiä, and H. Korkeala. Ribotyping as an identification tool for Clostridium botulinum species causing human botulism. Int. J. Food Microbiol., in press. [DOI] [PubMed]
- 16.Hielm S, Hyytiä E, Andersin A-B, Korkeala H. A high prevalence of Clostridium botulinum type E in Finnish freshwater and Baltic Sea sediment samples. J Appl Microbiol. 1998;84:133–137. doi: 10.1046/j.1365-2672.1997.00331.x. [DOI] [PubMed] [Google Scholar]
- 17.Hielm S, Hyytiä E, Ridell J, Korkeala H. Detection of Clostridium botulinum in fish and environmental samples using polymerase chain reaction. Int J Food Microbiol. 1996;31:357–365. doi: 10.1016/0168-1605(96)00984-1. [DOI] [PubMed] [Google Scholar]
- 18.Hyytiä, E., J. Björkroth, S. Hielm, and H. Korkeala. Characterization of Clostridium botulinum groups I and II by randomly amplified polymorphic DNA analysis and repetitive element sequence-based PCR. Submitted for publication. [DOI] [PubMed]
- 19.Hyytiä E, Hielm S, Korkeala H. Prevalence of Clostridium botulinum type E in Finnish fish and fishery products. Epidemiol Infect. 1998;120:245–250. doi: 10.1017/s0950268898008693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korkeala H, Stengel G, Hyytiä E, Vogelsang B, Bohl H, Wihlman H, Pakkala P, Hielm S. Type E botulism associated with vacuum-packaged hot-smoked whitefish. Int J Food Microbiol. 1998;43:1–5. doi: 10.1016/s0168-1605(98)00080-4. [DOI] [PubMed] [Google Scholar]
- 21.Kristjánsson M, Samore M H, Gerding D N, DeGirolami P C, Bettin K M, Karchmer A W, Arbeit R D. Comparison of restriction endonuclease analysis, ribotyping, and pulsed-field gel electrophoresis for molecular differentiation of Clostridium difficile strains. J Clin Microbiol. 1994;32:1963–1969. doi: 10.1128/jcm.32.8.1963-1969.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin W-J, Johnson E A. Genome analysis of Clostridium botulinum type A by pulsed-field gel electrophoresis. Appl Environ Microbiol. 1995;61:4441–4447. doi: 10.1128/aem.61.12.4441-4447.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Majkowski J. Strategies for rapid response to emerging foodborne microbial hazards. Emerg Infect Dis. 1997;3:551–554. doi: 10.3201/eid0304.970420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marmur J. Procedures for the isolation of deoxyribonucleic acid from micro-organisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 25.Maslow J N, Mulligan M E, Arbeit R D. Molecular epidemiology: application of contemporary techniques to the typing of microorganisms. Clin Infect Dis. 1993;17:153–164. doi: 10.1093/clinids/17.2.153. [DOI] [PubMed] [Google Scholar]
- 26.Maslow J N, Slutsky A M, Arbeit R D. Application of pulsed-field gel electrophoresis to molecular epidemiology. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C: ASM Press; 1993. pp. 563–572. [Google Scholar]
- 27.Öberg, S. 1994. Botulism. Epid-aktuellt (Stockholm, Sweden) 17:5.
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Samore M, Killgore G, Johnson S, Goodman R, Shim J, Venkataraman L, Sambol S, DeGirolami P, Tenover F, Arbeit R, Gerding D. Multicenter typing comparison of sporadic and outbreak Clostridium difficile isolates from geographically diverse hospitals. J Infect Dis. 1997;176:1233–1238. doi: 10.1086/514117. [DOI] [PubMed] [Google Scholar]
- 30.Samore M H, Kristjansson M, Venkataraman L, DeGirolami P C, Arbeit R D. Comparison of arbitrarily-primed polymerase chain reaction, restriction enzyme analysis and pulsed-field gel electrophoresis for typing Clostridium difficile. J Microbiol Methods. 1996;25:215–224. [Google Scholar]
- 31.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tyler K D, Wang G, Tyler S D, Johnson W M. Factors affecting reliability and reproducibility of amplification-based DNA fingerprinting of representative bacterial pathogens. J Clin Microbiol. 1997;35:339–346. doi: 10.1128/jcm.35.2.339-346.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vandamme P, Pot B, Gillis M, De Vos P, Kersters K, Swings J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol Rev. 1996;60:407–438. doi: 10.1128/mr.60.2.407-438.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]